Abstract
Thyroid hormone signaling has long been implicated in mammalian testicular function, affecting steroidogenesis in testicular Leydig cells. However, its molecular mechanism is not well understood. Here, we investigated the molecular action of thyroid hormone receptor-α (TRα) on mouse testicular steroidogenesis. TRα/thyroid hormone (T3) signaling differentially affected the expression of steroidogenic enzyme genes, mainly regulating their promoter activity. TRα directly regulated the promoter activity of the cytochrome P450 17α-hydroxylase/C17–20 lyase gene, elevating its expression in the presence of T3. TRα also indirectly regulated the expression of steroidogenic enzyme genes, such as steroidogenic acute regulatory protein and 3β-hydroxysteroid dehydrogenase, by modulating the transactivation of Nur77 on steroidogenic enzyme gene promoters through protein-protein interaction. TRα enhanced Nur77 transactivation by excluding histone deacetylases from Nur77 in the absence of T3, whereas liganded TRα inhibited Nur77 transactivation, likely due to interfering with the recruitment of coactivator such as the steroid receptor coactivator-1 to Nur77. Together, these findings suggest a role of TRα/T3 in testicular steroidogenesis and may provide molecular mechanisms for the differential regulation of steroidogenic enzyme genes by thyroid hormone.
Thyroid hormone plays a role in the development, growth, and function of many organs and tissues including the testis. In the testis, levels of thyroid hormone are changed during early development, affecting testicular maturation and function (1). Thyroid hormone regulates the proliferation and differentiation of testicular Sertoli and Leydig cells. For example, neonatal transient hypothyroidism induced by treatment with 6-propyl-2-thiouracil increases the number of testicular cells including Sertoli and germ cells, and consequently increases the production of sperm (2). In addition, neonatal-prepubertal transient hypothyroidism causes a delay in the differentiation of precursors into Leydig cells, whereas hyperthyroidism accelerates the differentiation of precursor cells and the proliferation of adult-type Leydig cells (3).
The action of thyroid hormone in target tissues is mediated by thyroid hormone receptors (TRs). TRs belong to the family of nuclear receptors (NRs) and are ligand-dependent transcription factors. There are 2 TR isoforms, TRα and TRβ, which are encoded by separate genes, THRA and THRB, respectively (4). TRα and TRβ have very similar structures, consisting of an N-terminal transactivation domain, a central DNA-binding domain, and a C-terminal ligand-binding domain. TRs regulate gene expression by binding to thyroid response elements (TREs) in the promoter region of target genes as TR homodimer or heterodimers with retinoic X receptors (5). The binding of ligands to TRs changes the conformation of NRs, reverses silencing, and induces transcriptional activation that releases corepressors and recruits coactivators. This positive regulation of gene expression by thyroid hormones is well known. Thyroid hormones also negatively regulate target genes. However, in contrast to positive regulation, the mechanism for negative gene regulation by thyroid hormones is less well understood.
TRs have been identified in many testicular cells such as sperm, developing germ, Sertoli, Leydig, and peritubular cells (6). TRα1 isoform is expressed in human and rat testis at different levels throughout development, and TRβ1 is also detected in the testis, although its levels are very low (6, 7). The level of TRα1 expression in rat testis is higher during late fetal and perinatal life than in adult life (1). It has been also reported that mesenchymal and immature Leydig cells contain higher levels of TRα mRNA than adult Leydig cells (8). Recent studies with TRα and TRβ knockout (KO) mice revealed that TRα KO mice had or trended toward an increase in Sertoli cell number, daily sperm production, and testicular weight, whereas TRβ KO mice had similar values for these variables as wild-type mice (9). In addition, the effects of neonatal hyperthyroidism on testicular development in wild-type mice were diminished or reduced in TRα KO mice, but not in TRβ KO mice, suggesting a major role of the TRα isoform in testicular development.
Steroidogenesis is the process of testosterone biosynthesis, which is controlled by pituitary gonadotropin LH in testicular Leydig cells. Steroid hormone synthesis requires several essential enzymes: steroidogenic acute regulatory protein (StAR), cholesterol side chain cleavage cytochrome P450 (P450scc), cytochrome P450 17α-hydroxylase/C17–20 lyase (P450c17), and 3β-hydroxysteroid dehydrogenase/isomerase (3β-HSD). StAR translocates cholesterol from the outer to the inner mitochondrial membrane in Leydig cells. In the inner mitochondrial membrane, cholesterol is converted to pregnenolone by P450scc. Pregnenolone is then transported to the smooth endoplasmic reticulum and becomes testosterone by a series of enzymes, 3β-HSD, and P450c17 (10).
The expression of steroidogenic enzyme genes is generally regulated at the transcription level, and the orphan nuclear receptor Nur77 is one of the major transcription factors involved in the regulation of steroidogenic enzyme genes in Leydig cells (11, 12). Nur77 is a transcription factor belonging to the superfamily of NRs and recognizes a specific nucleotide sequence, the monomeric NGF1-B response element (NBRE) or dimeric Nur response element (NurRE) (13, 14). Nur77 recruits coactivators for its function similarly as other NRs. Steroid receptor coactivator-1 (SRC-1), SRC-2, p300, and P300/CBP-associated factor (PCAF) coactivators have been shown to directly interact with the activation function 1 region of Nur77 (15). LH, the regulator of testicular steroidogenesis, induces Nur77 gene expression through the second messenger cAMP-signaling pathway in Leydig cells (16), and Nur77 has been shown to regulate the expression of steroidogenic enzyme genes including steroid 21-hydroxylase, 20α-hydroxysteroid dehydrogenase, and P450c17 (11, 17, 18). In addition, Nur77-binding regions within the promoter of rat P450c17 and mouse StAR and human 3β-HSD type 2 (3β-HSD2) genes have been defined (11, 19, 20).
Several studies have shown that thyroid hormone regulates the expression of some steroidogenic enzyme genes and the biosynthesis of testosterone. Acute thyroid hormone (T3) treatment of mouse Leydig cells increased the levels of StAR mRNA and protein, as well as steroid production, whereas chronic stimulation with T3 (beyond 8 hours) resulted in an attenuation of StAR expression. T3 stimulated the expression of StAR through the mediation of steroidogenic factor-1 (SF-1) in mLTC-1 Leydig cells and thus the inhibition of SF-1 expression by DAX-1 abolished the T3-mediated response (21). It has also been observed that chronic T3 treatment in Leydig cells increased the mRNA levels of P450scc, whereas it decreased those of 3β-HSD (21). In addition, 2,2′,4,4′-tetrahydroxybenzophenone, which acts as endocrine disruptors, affected the expression of steroidogenic enzyme genes such as StAR, P450c17, and P450scc in a manner opposite to that of T3 signaling, disrupting steroid hormone synthesis as a thyroid disruptor (22). These findings suggest that thyroid hormone and the expression of steroidogenic enzyme genes coordinately regulate steroid hormone biosynthesis in Leydig cells. However, molecular mechanisms whereby thyroid hormone affects the expression of steroidogenic enzyme genes in Leydig cell are still unclear.
In the present study, we have demonstrated that TR/thyroid hormone affects the expression of steroidogenic enzyme genes via direct and indirect actions in Leydig cells. TRα/T3 directly acts on the P450c17 promoter, which contains putative thyroid response elements (TREs), and indirectly acts on the promoter of steroidogenic enzyme genes such as StAR and 3β-HSD by modulating Nur77 transactivation on these promoters. TRα-mediated modulation of Nur77 transactivation occurs via their direct physical interaction. These findings may provide a mechanistic explanation for the previous observation that thyroid hormone signaling affects testicular steroidogenesis through the alteration of steroidogenic enzyme gene expression.
Materials and Methods
Plasmids
The mammalian expression vectors for Nur77, pCDNA3-HA-Nur77 and pCDNA3-Flag-Nur77, and reporter plasmids of NurRE-Luc and NBRE-Luc were previously described (16). The mammalian expression vectors of human (h)TRα and glutathione S-transferase (GST)-TRα were kindly gifted by Dr. Y-C. Lee (Chonnam National University, South Korea). Hemagglutinin (HA)-TRα expression plasmid was constructed by cloning the TRα into pCDNA3-HA vector. GST-Nur77, pCDNA3-Flag-SF-1, mouse P450c17 (−1040)-Luc, rat wild-type (WT) (−447/−399)P450c17-Luc, mutant (Mut) (−447/−339Δ2)P450c17-Luc, mouse StAR(−2200/+3)-Luc, mouse 3β-HSD(−4700/+40)-Luc, and sft4-Luc were previously described (11, 23, 24), as were histone deacetylase (HDAC)1–6 and SRC-1 (25).
Cell culture and purification of primary Leydig cells
MA-10 cells were maintained in RPMI 1640 medium supplemented with 15% horse serum and antibiotics (26). Cos-7 and human embryonic kidney 293T cells were maintained in DMEM supplemented with 10% fetal bovine serum and antibiotics. All cells were cultured at 37°C under an atmosphere of 5% CO2. Purification of mouse primary Leydig cells was carried out as previously described (26). Briefly, testicular cells were dispersed by treating the decapsulated testes with collagenase type I (0.25 mg/mL, Sigma-Aldrich) in M199 medium (Invitrogen) for 20 minutes. Following incubation, the dispersed tissues were filtered by a 40-mm cell strainer (BD Biosciences). The interstitial cells were precipitated by centrifugation of the filtrate, washed with M199, and then seeded in RPMI 1640 medium supplemented with 15% horse serum and antibiotics. Enrichment for Leydig cells was estimated using 3β-HSD immunocytochemistry. The population of Leydig cells was 60%–70% of the total purified cells.
Transient transfection assay
Transfections were performed using Superfect (QIAGEN) transfection reagent for human embryonic kidney 293T cells and Lipofectamine 2000 (Invitrogen) transfection reagent for MA-10 cells according to the manufacturer's instructions. For hormone treatments, MA-10 cells were plated in medium containing 5% charcoal-stripped serum, and 8-bromo-cAMP (8Br-cAMP, Sigma-Aldrich) and T3 (Sigma-Aldrich) were added to the medium 24 hours later. Cells were transfected with expression vectors, a reporter gene, and the control lacZ expression plasmid pCMVβ (Clontech). The total amount of DNA was kept constant by adding the pcDNA3 empty vector. After 24 hours of transfection, cells were treated with T3 for 24 hours. Cells were lysed with lysis buffer containing 0.1% Triton X-100 and 0.2 M Tris-HCl (pH 8.0). Luciferase and β-galactosidase activities were assayed as described previously (23). The levels of luciferase activity were normalized by that of lacZ expression.
Immunohistochemistry
Adult mouse testes were fixed in 4% formaldehyde solution. Immunohistochemistry for TRs was performed using the Histostain-Plus kit (Zymed Laboratories, Inc), according to the manufacturer's instructions. Briefly, deparaffinized and rehydrated slides from mouse testes were first blocked with CAS-Block (Invitrogen) and then incubated with anti-TRα1 antibody (PA1–211A, lot no. NL177632; Thermo Scientific) diluted 1:50 in blocking solution for overnight at 4°C. For a negative control, adjacent sections were incubated with normal rabbit IgG (sc-2027, lot no. K1412; Santa Cruz Biotechnology). After incubating with the secondary antibody, slides were developed using the AEC substrate solution until signal was seen. Slides were mounted with GVA mounting solution, and observed under light microscope with bright-field illumination.
Northern blot analysis
Total RNA was prepared from MA-10 cells and rat testes using Tri reagent (Molecular Research Center, Inc). Total RNA (20 μg) was separated on a 1.2% denaturing agarose gel, transferred to a Zeta probe nylon membrane (Bio-Rad Laboratories), and immobilized by UV cross-linking. The membrane was hybridized with random-primed α-32P-labeled StAR, P450c17, P450scc, 3β-HSD, and Nur77 cDNA probes as described previously (23). The membrane was reprobed for glyceraldehyde-3-phosphate dehydrogenase as a loading control.
Quantitative real-time PCR analysis
Total RNA (2 μg) isolated from cells, which were treated 8Br-cAMP with or without T3 for 8 hours, were used for reverse transcription (RT) with M-MLVRT (Promega Corp). Quantitative real-time PCR was performed using a Rotor-Gene 6000 real-time amplification operator (Corbett Research). The 25-μL reaction mixture contained 10 nM primers, 1 μL cDNA, and 12.5 μL SYBR Green Master Mix kit (QIAGEN). A260/A280 and A260/A230 ratios of RNA samples, which were isolated using Tri reagent (Molecular Research Center, Inc), were more than 1.7 and 2.0, respectively. The primer sequences for mouse genes were P450c17-F (forward): 5′-CCAGGACCCAAGTGTGTTCT-3′; P450c17-R (reverse): 5′-CCTGATACGAAGCACTTCTCG-3′; StAR-F: 5′-TGTCAAGGAGATCAAGGTCCTG-3′; StAR-R: 5′-CGATAGGACCTGGTTGATGAT-3′; 3β-HSD-F: 5′-ATGGTCTGCCTGGGAATGAC-3′; 3β-HSD-R: 5′-ACTGCAGGAGGTCAGAGCT-3′; P450scc-F: 5′-CTGCCTCCAGACTTGTTTCG-3′; P450scc-R: 5′-TTCTTGAAGGGCAGCTTGTT-3′ (27); Nur77-F: 5′-CATCTTCTGCTCAGGCCTGGT-3′; Nur77-R: 5′-AGACGTGACAGGCAGCTGGC-3′; TRα-F: 5′-GGCTGTGCTGCTAATGTCAA-3′; TRα-R: 5′-CGGAGGTCAGTCACCTTCAT-3′. All the primers were ordered from GenoTech Corp. PCR was performed using the following PCR condition: denaturing at 94°C for 20 seconds, annealing at 57°C (StAR, 3β-HSD, P450scc, and TRα) or 62°C (P450c17 and Nur77) for 20 seconds, and elongation at 72°C for 30 seconds. mRNA levels were normalized by β-actin (23). The 2-ΔΔCt method was used to calculate relative mRNA expression levels.
Western blot analysis
Cells were harvested with radioimmune precipitation assay cell lysis buffer (50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 1% nonyl phenoxypolyethoxylethanol [NP-40], and 5 mM EDTA at pH 8.0). Total protein was separated by SDS-PAGE and transferred to a nitrocellulose membrane (Sigma-Aldrich). Western blot analyses were performed with anti-TRα/β (sc-772, lot no. F1113; 1:500), anti-Nur77 (sc-5569, lot no. K2309; 1:500), anti-P450c17 (sc-46081, lot no. F1807; 1:500), anti-StAR (sc-25806, lot no. G0706; 1:1000), anti-3β-HSD (sc-30820, lot no. D2108; 1:300), anti-LaminB (sc-6216, lot no J2909; 1:1000) antibodies (Santa Cruz Biotechnology) and anti-P450scc (AB1244, lot no. LV1569825, 1:500) antibodies (Millipore Corp). The signals were then detected with an ECL kit (Amersham Pharmacia).
Coimmunoprecipitation
Cos-7 cells were transfected with 4 μg of flag-Nur77 and 6 μg of HA-TRα for 48 hours. Cells were lysed with NP-40 buffer (10 mM sodium phosphate at pH 7.2, 150 mM NaCl, and 1% NP-40) containing protease inhibitors (20 μg/mL leupeptin, 10 μg/mL pepstatin A, 2 μg/mL aprotinin, and 1 mM phenylmethylsulfonyl fluoride). Whole-cell lysate (500 μg) was incubated with 2 μg of anti-HA antibody (sc-7392X, lot no. K0912) overnight at 4°C and further incubated for another 2 hours after adding 20 μL of Protein A/G PLUS-agarose beads (Santa Cruz Biotechnology). Agarose beads were washed with NP-40 buffer containing protease inhibitors at 4°C. Bound proteins were analyzed by 8% SDS-PAGE and subsequent immunoblotting using anti-Nur77 and anti-TRα/β antibodies.
GST pull-down assay
GST, GST-Nur77, and GST-hTRα fusion proteins were expressed in Escherichia coli BL21 cells and isolated with glutathione-Sepharose-4B beads (Amersham Bioscience AB). The immobilized GST fusion proteins were then incubated with [35S] methionine-labeled TRα, Nur77, HDAC3, or SRC-1 proteins produced by in vitro translation using the TNT-coupled transcription-translation system (Promega Corp). [35S]methionine-labeled TRα was preincubated with or without T3 for 20 minutes. The binding reactions were performed in 400 μL of GST binding buffer (20 mM Tris-HCl, pH 7.9; 100 mM NaCl; 10% glycerol; 0.05% NP-40; 5 mM MgCl2; 0.5 mM EDTA; 1 mM dithiothreitol; and 1.5% BSA) overnight at 4°C. For in vitro competition assays, [35S]methionine-labeled TRα and HDAC3 proteins or [35S] methionine-labeled TRα and SRC-1 proteins produced by in vitro translation were added to the binding reaction containing GST-Nur77 protein. The beads were washed 4 times with 1 mL of GST binding buffer. Bound proteins were eluted by adding 20 μL of SDS-PAGE sample buffer, separated by SDS-PAGE, and visualized using a Phosphorimager (BAS-7000, Fuji) (28). The GST-fused proteins used in each reaction were analyzed by SDS-PAGE and confirmed for the relative quantity by Coomassie blue staining. Ten percent of the in vitro translated proteins used in each reaction were loaded as inputs.
EMSA
The GST fusion proteins (GST-Nur77 and GST-TRα) were expressed in E. coli BL21 cells and purified with glutathione-Sepharose-4B beads (Pharmacia, Biotech AB) for EMSA. The NurRE oligonucleotide (5′-GGGGTGATATTTACCTCCAAATGCCA-3′) was annealed to its complementary oligonucleotide to form a double-strand sequence, labeled with [α-32P] dCTP, and purified using Sephadex G50 spin columns. EMSA was performed as described previously, using gel mobility shift assay solution containing 1 mM Tris (pH 7.6), 50 mM KCl, 4% glycerol, 1 mM EDTA, and 2 μg of polydeoxyinosinic deoxycytidylic acid (29). For competition experiments, proteins were preincubated for 10 minutes at 4°C with unlabeled NurRE or unrelated oligonucleotide (ARE, 50-fold excess) at 30 minutes prior to the incubation with the labeled NurRE probe. For supershift of Nur77, GST-Nur77 proteins prepared as described above were preincubated with the anti-Nur77 antibody for 10 minutes prior to the addition of NurRE probe. DNA-protein complexes were analyzed on 5% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. Gels were dried and analyzed by autoradiography.
Chromatin immunoprecipitation (ChIP) assay
MA-10 cells were cotreated with 0.5 mM 8Br-cAMP and 100 nM T3 for 6 hours. In total, 7 × 106 cells were cross-linked with 1% formaldehyde and processed for ChIP assays as previously described (29). In brief, cell lysates in sodium dodecyl sulfate-lysis buffer were sonicated to shear DNA about 500 bp, immunoprecipitated with anti-HDAC3 (sc-11417X, lot no. D2611), anti-TRα/β, anti-Nur77, and anti-SRC1 (sc-8995, lot no. C282; Santa Cruz Biotechnology) antibodies overnight at 4°C and further incubated for another 4 hours after adding 20 μL of Protein A/G PLUS-agarose beads. Protein A/G bead-antibody-chromatin complexes were washed sequentially with low-salt immune complex, high-salt immune complex, and LiCl immune complex wash buffer. After reverse cross-linking, purified immunoprecipitated DNA and input-sheared DNA were subjected to PCR using a primer pair for the mouse StAR promoter (forward, 5′-TGATGCACCTCAGTTACTGG-3′; and reverse, 5′-GCTGTGCATCATCACTTGAG-3′), which amplifies the proximal region (−299 to −41 bp) of the StAR promoter (19). A primer pair for mouse β-actin promoter was included as a negative control (23).
RIA
Culture media of MA-10 cells and primary Leydig cells, which were treated with 0.5 mM 8Br-cAMP and 100 nM T3 for 48 hours, were collected. Testosterone concentrations were measured by RIA, as described previously (26).
Statistical analysis
To identify significant differences, data were analyzed by GraphPad prism version 5.0. Single comparisons between 2 experimental groups were done using an unpaired Student's t test. The data are presented as the mean ± SEM of at least 3 independent experiments. For all statistical analyses, P < .05 was considered significant.
Results
Differential effects of thyroid hormone on cAMP-induced expression of steroidogenic enzyme genes in Leydig cells
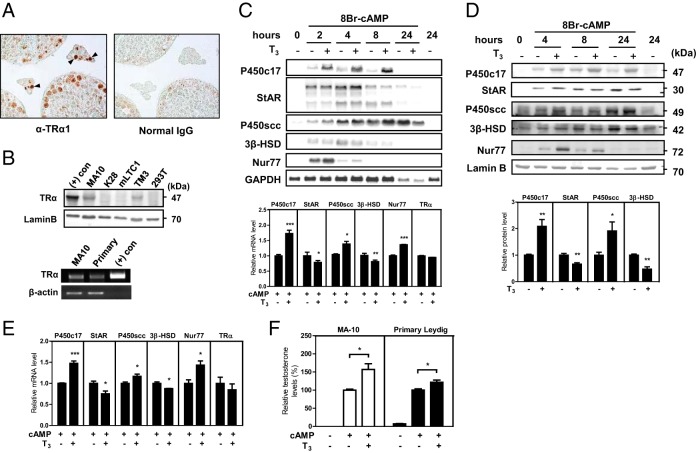
TRs have been reported to be expressed in various testicular cells, including Leydig, Sertoli, and germ cells in rats (6), which we confirmed in mice by immunohistochemistry. In adult mouse testis, the expression of TRα1, which is the dominant isoform in rat testis (6), was detected in interstitial Leydig cells as well as germ cells within seminiferous tubules (Figure 1A). The expression of TRα was also detected in several Leydig cell lines, including MA-10 and TM3, and primary Leydig cells (Figure 1B).
Figure 1.
Differential effects of thyroid hormone on the expression of steroidogenic enzyme genes in Leydig cells. A, Expression of TRα1 in the testis. TRα1 expression was determined by immunohistochemistry using anti-TRα1 antibody in adult mouse testes. Positive signals were detected in Leydig cells (arrows) and germ cells. The section for negative control was incubated with normal IgG prior to incubation with the secondary antibody (magnification, ×400). B, Expression of TRα in Leydig cell lines and mouse primary Leydig cells. Extracts of various Leydig cell lines were subjected to Western blot analysis. Extracts of MA-10 cells overexpressing TRα and 293T human kidney cells were used as a positive and negative control, respectively (top). TRα expression in mouse primary Leydig cells was analyzed by RT-PCR. TRα expression plasmid was included as a positive control (bottom). LaminB and β-actin expression was used as a loading control. C–E, Effect of thyroid hormone on cAMP-induced expression of steroidogenic enzyme genes. C and D, The mouse Leydig cell line MA-10 was cultured overnight in medium containing 5% charcoal-stripped serum. Cells were cotreated with 8Br-cAMP (0.5 mM) and T3 (100 nM) and were harvested at different time points. C, Total RNAs (20 μg) were analyzed by Northern blot analyses with steroidogenic enzyme (P450C17, StAR, 3β-HSD, and P450scc) and Nur77 cDNA probes (top). The mRNA levels of steroidogenic enzyme genes, Nur77 and TRα, were analyzed by quantitative real-time PCR with cells treated with 8Br-cAMP and T3 for 8 hours (bottom). D, Total cell extracts (120 μg of protein) were subjected to Western blot analysis for the protein levels of steroidogenic enzymes and Nur77 (top). The protein levels of steroidogenic enzyme genes in cells, which were treated with 8Br-cAMP and T3 for 24 hours, were quantified by Image J software (bottom). E, Primary Leydig cells were cultured and cotreated with 8Br-cAMP and T3 for 8 hours as in panel C. mRNA levels of steroidogenic enzyme genes, Nur77 and TRα, were analyzed by quantitative real-time PCR. F, Effect of thyroid hormone on cAMP-induced testosterone production. Culture media of MA-10 and primary Leydig cells, which were treated with 8Br-cAMP and T3 for 48 hours, were analyzed for testosterone levels by RIA. In panels A, B, and C (top) and D (top), data are representative of 3 independent experiments. In panels C (bottom), D (bottom), E, and F, values represent means ± SEM of 3 independent experiments. *, P < .05; **, P < .01; ***, P < .001.
Leydig cells in the testis express steroidogenic enzyme genes and produce testosterone in response to LH/cAMP signaling from the pituitary. To mimic physiological conditions, we treated cultured Leydig cells with cAMP and explored the effects of thyroid hormone on steroidogenesis. The effects of thyroid hormone on cAMP-induced expression of steroidogenic enzyme genes were first assessed at their mRNA and protein levels by incubating MA-10 Leydig cells with or without T3 (100 nM) in the presence of cAMP for 0–24 hours. Northern blot analysis revealed that T3 treatment resulted in an increase of the P450c17 mRNA levels, whereas 3β-HSD mRNA levels were decreased by T3. On the other hand, mRNA levels of StAR were increased until 4 hours and mRNA levels of P450scc were increased until 8 hours, both of which were decreased thereafter by T3 treatment (Figure 1C, top). Interestingly, the mRNA levels of Nur77, one of the major transcription factors that regulate the expression of steroidogenic enzyme genes in Leydig cells, were increased by T3 treatment. As expected, quantitative real-time PCR analysis of mRNA levels at 8 hours treatment of T3 in the presence of cAMP revealed that T3 treatment significantly increased mRNA levels of P450c17, P450scc, and Nur77, whereas it decreased mRNA levels of StAR and 3β-HSD (Figure 1C, bottom). Western blot analyses showed a similar T3 effect on the protein levels of steroidogenic enzymes, although the effects were less prominent and time delayed (Figure 1D, top). At 24 hours treatment of T3 in the presence of cAMP, protein levels of P450c17 and P450scc were significantly increased, whereas protein levels of StAR and 3β-HSD were decreased (Figure 1D, bottom).
Consistent with the effects of T3 in MA-10 cells, T3 treatment of primary Leydig cells for 8 hours in the presence of cAMP increased P450c17 and P450scc mRNA levels, whereas it decreased StAR and 3β-HSD mRNA levels (Figure 1E). Remarkably, cAMP-induced testosterone production in both MA-10 cells and primary Leydig cells was increased by T3 treatment (Figure 1F). These data suggest that TRα is expressed in mouse Leydig cells and T3 affects the cAMP-induced expression of steroidogenic enzyme genes, resulting in an increase of testosterone production.
Differential effect of thyroid hormone and TRα on the promoter activity of steroidogenic enzyme genes
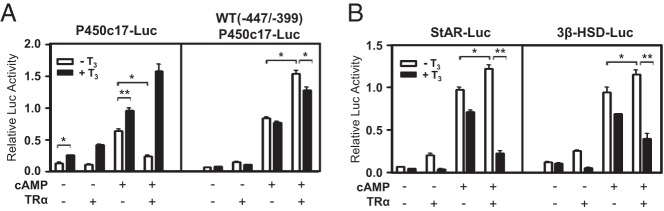
Thyroid hormones and TR have been implicated in the regulation of steroidogenic enzyme gene expression (21). Because TR is a ligand-inducible transcription factor and T3 affects the expression of steroidogenic enzyme genes (Figure 1,C–E), we first analyzed the promoter sequences of steroidogenic enzyme genes (P450c17, StAR, and 3β-HSD) for TREs. Sequence analysis of approximately 1.0 kb upstream of the mouse P450c17 promoter, but not StAR and 3β-HSD promoter, revealed one putative TRE which consists of an everted repeat (5′-TGACCT nnnnnn AGGTCA-3′) (5) at −452 to −435 region of the promoter (data not shown). Therefore, it is possible that TRα/T3 directly acts on the promoter of at least the P450c17 gene.
We then investigated the effect of TR/T3 on the promoter activity of steroidogenic enzyme genes using TRα expression vector and steroidogenic enzyme gene promoter-reporters in MA-10 cells. T3 treatment increased the transcriptional activity of P450c17 promoter containing the putative TRE (Figure 2A, left). As reported with functional TREs (4), TRα overexpression in the absence of T3 decreased the transcriptional activity of P450c17 promoter, especially with cAMP induction (Figure 2A, left). These effects of T3 treatment and TRα overexpression were diminished with the rat WT (−447/−399)P450c17 promoter (23), which contains only the NBRE and no TRE (Figure 2A, right), suggesting the presence of a functional TRE in mouse P450c17 promoter. Rather, very interestingly, the cAMP-dependent promoter activity of WT (−447/−399)P450c17 containing the NBRE was increased by TRα overexpression and was then decreased by T3 treatment (Figure 2A, right). Similar effects were observed with the StAR and 3β-HSD promoters, in which NBREs were previously identified (19, 20), but no putative TRE was found (Figure 2B). Taken together, these data show that thyroid hormone T3 and TRα differently regulate the promoter activity of steroidogenic enzyme genes.
Figure 2.
Differential effect of TRα/T3 signaling on the promoter activity of steroidogenic enzyme genes. A and B, Effects of TRα and/or T3 on steroidogenic enzyme gene promoters were tested using promoter-luciferase reporter constructs. MA-10 cells were transfected with 100 ng of TRα expression plasmids and 150 ng of the indicated luciferase reporter gene. Cells were treated with 8Br-cAMP (0.5 mM) and T3 (100 nM) and assayed for luciferase activity after 24 hours' incubation. In all panels, values are the means ± SEM of 3 independent experiments performed in duplicate. *, P < .05; **, P < .01.
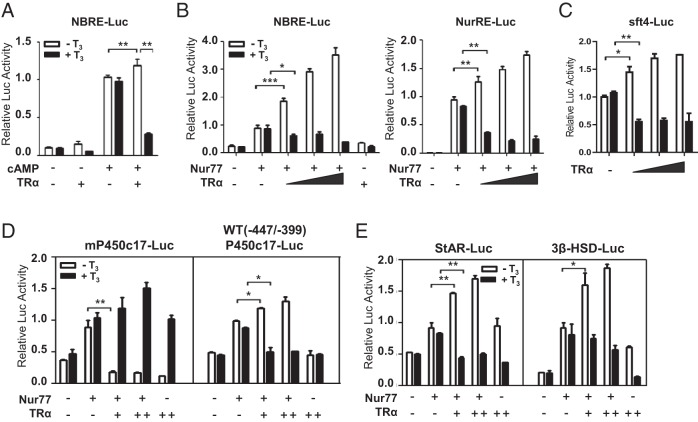
Effect of TRα/T3 on Nur77 transactivation in the promoter of steroidogenic enzyme genes
The orphan nuclear receptor Nur77, which is induced by LH-cAMP signaling (16, 23), is one of the major transcription factors that regulate the expression of steroidogenic enzyme genes in testicular Leydig cells. Interestingly, the activity of the WT (−447/−399) P450c17 promoter containing only the NBRE was affected by TRα/T3 (Figure 2A). In addition, as WT (−447/−399) P450c17-Luc, the promoter activity of NBRE-Luc was increased by TRα overexpression in the absence of T3, and T3 treatment decreased the promoter activity (Figure 3A). Therefore, we investigated the effect of TRα/T3 on Nur77-activated promoter activity using Nur77 and TRα expression vectors and reporter constructs in MA-10 cells. TRα enhanced the transactivation of Nur77 with both NBRE-Luc and NurRE-Luc in a concentration-dependent manner, and T3 diminished the TRα effect, inhibiting further Nur77 transactivation (Figure 3B). These effects of TRα/T3 on the Nur77-activated promoter activity showed a similar pattern to that on the cAMP-induced promoter activity of NBRE-Luc (Figure 3, panel A vs panel B, left).
Figure 3.
Effect of TRα/T3 on Nur77 transactivation in the promoter of steroidogenic enzyme genes. A, MA-10 cells were transfected with TRα expression plasmids and NBRE-Luc reporter construct in medium containing 5% charcoal-stripped serum. Cells were treated with 8Br-cAMP (0.5 mM) and T3 (100 nM) after 24 hours of transfection and harvested for measurement of luciferase activity after an additional 24 hours. B–E, MA-10 cells were transfected with Nur77 expression plasmid (50 ng) and increasing amounts of TRα expression plasmid (20, 50, and 100 ng) (panel B), with increasing amounts of only TRα expression plasmid (20, 50, and 100 ng) (panel C) or with Nur77 expression plasmid (100 ng) and TRα expression plasmid (+: 100 ng, ++: 200 ng) (panels D and E), and the indicated reporter construct. Cells were treated with T3 after 24 hours of transfection and harvested for measurement of luciferase activity after an additional 24 hours. In all panels, values are the means ± SEM of 3 independent experiments performed in duplicate. *, P < .05; **, P < .01; ***, P < .001.
TRα overexpression also activated and inhibited SF-1 transactivation in the absence and presence of T3, respectively, but the extent of TRα/T3 effects was less than that with Nur77 (Figure 3C). SF-1 is another transcription factor involved in the regulation of steroidogenic enzyme genes in Leydig cells and expressed abundantly in MA-10 cells (19). Therefore, without cAMP induction, TRα/T3 effects, if any, on the promoter of steroidogenic enzyme genes such as StAR and 3β-HSD might be attributable to the effects of TRα/T3 on SF-1 (Figure 2B).
We next explored the effect of TRα/T3 on the Nur77-activated promoter activity of steroidogenic enzyme genes, such as P450c17, StAR, and 3β-HSD by transient transfection assays. When Nur77 and TRα were coexpressed, TRα repressed Nur77 transactivation with the P450c17 promoter whereas treatment with T3 enhanced Nur77 transactivation (Figure 3D, left). However, with the WT (−447/−399) P450c17 promoter, TRα enhanced Nur77 transactivation in the absence of T3, whereas it repressed Nur77 transactivation in the presence of T3 (Figure 3D, right). The Mut (−447/−339Δ2) P450c17 promoter containing the mutated Nur77-binding site did not show any significant effect with Nur77 and TRαexpression in the presence or absence of /T3 (data not shown). We observed similar response patterns of StAR and 3β-HSD promoters to TRα/T3 signaling with the WT (−447/−399) P450c17 promoter (Figure 3E). These results with the Nur77-activated promoters of steroidogenic enzyme genes are reminiscent of the results with the cAMP-induced promoters of steroidogenic enzyme genes (Figure 2). Together, these data suggest that TRα/T3 signaling in Leydig cells regulates the expression of steroidogenic enzyme genes by modulating Nur77 transactivation on steroidogenic enzyme gene promoters, as well as directly targeting promoters containing TRE such as the P450c17 promoter.
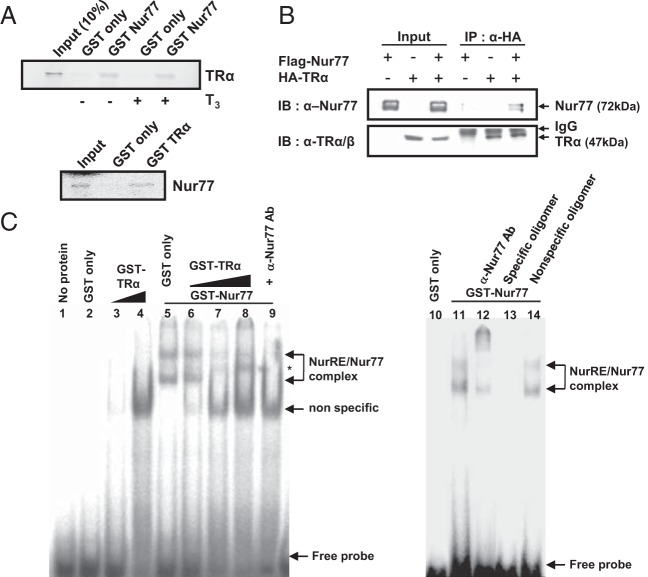
Physical association of Nur77 with TRα
Because TRα functionally interacted with Nur77 (Figure 3), we assessed their physical interaction by GST pull-down assays using [35S]methionine-labeled TRα and Nur77. TRα protein was retained by GST-Nur77 irrespective of T3, and Nur77 protein was retained by GST-TRα, which suggests that Nur77 directly interacts with TRα in vitro (Figure 4A). To confirm the physical interaction between Nur77 and TRα, we performed coimmunoprecipitation assays with Cos-7 cells, which were transfected with Flag-Nur77 and HA-TRα expression vectors. When the TRα protein complex was precipitated with an anti-HA antibody, Nur77 was detected with TRα (Figure 4B). Taken together, these results indicate that Nur77 interacts with TRα.
Figure 4.
Physical association of Nur77 with TRα. A, Nur77 directly interacts with TRα in vitro. [35S]methionine-labeled TRα and Nur77 produced by in vitro translation were allowed to interact with the GST-fusion protein of Nur77 and GST-fusion protein of TRα, respectively. B, Nur77 associates with TRα in cells. Cos-7 cells were cotransfected with flag-Nur77 and HA-TRα expression plasmids and coimmunoprecipitation was performed with anti-HA antibody. Western blot analyses were conducted with anti-Nur77 or anti-TRα/β antibody. C, The physical interaction of TRα with Nur77 on DNA was accessed by EMSA. The GST-Nur77 fusion protein was incubated with α-32P-labeled NurRE oligonucleotide, along with purified GST only (lane 5) or increasing amounts of GST-TRα (lanes 6, 7, and 8) proteins. A 50-fold excess of cold NurRE oligomer (lane 13) or nonspecific oligomer (lane 14) was added. Positions of the specific protein-DNA complex, nonspecific complex, and free probe were indicated. *, A new complex formed by coincubation with TRα protein. In all panels, data are representative of 3 independent experiments. IB, immunoblotting; IP, immunoprecipitation.
The physical interaction of TRα with Nur77 on DNA was then accessed by EMSA using α-32P-labeled NurRE oligonucleotide (Figure 4C). The α-32P-labeled NurRE oligonucleotide formed 2 complexes with GST-Nur77 protein, monomeric and dimeric Nur77 complexes (lanes 5 and 11), the formation of which was diminished by 50-fold excess of cold NurRE oligomer but not with 50-fold excess of nonspecific oligomer (lanes 13 and 14), indicating the formation of specific complexes. Supershift of the complex by anti-Nur77 antibody further verified the formation of specific complexes (lane 12). Coincubation of GST-TRα with GST-Nur77 protein, along with α-32P-labeled NurRE oligonucleotide, produced a new complex between the monomeric Nur77 and dimeric Nur77 complexes, diminishing the levels of NurRE-GST-Nur77 complexes in a dose-dependent manner (lanes 6, 7, and 8). The new complex as well as the monomeric and dimeric Nur77 complexes disappeared by coincubating the reaction mixture with anti-Nur77 antibody (lane 9), suggesting that the complex contains Nur77 protein, likely as a heterodimer with TRα. These results suggest that TRα physically interacts with Nur77 bound to DNA, modulating Nur77 transactivation.
Competition of TRα with HDACs for the regulation of Nur77 transactivation in the absence of T3
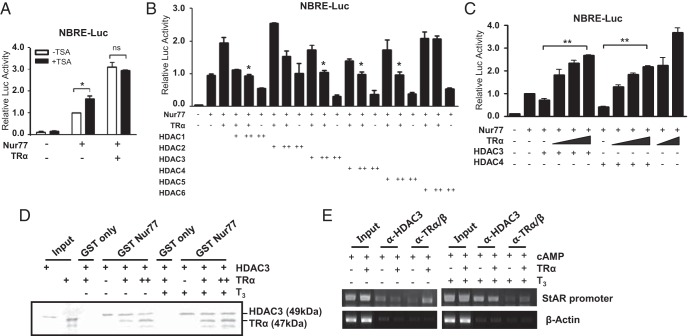
Nur77 transactivation is endogenously repressed by histone deacetylase (HDAC) activities (30). To explore the mechanism by which TRα enhances Nur77 transactivation in the absence of T3, we investigated the involvement of HDACs using the HDAC inhibitor trichostatin (TSA) (Figure 5A). The endogenous repression of Nur77 transactivation by HDACs was recovered with TSA treatment. However, such a relief effect was completely diminished by TRα overexpression, although increased expression of TRα highly enhanced Nur77 transactivation above the TSA-relieved level. These results suggest that TRα enhances Nur77 transactivation, at least in part, by excluding HDACs from Nur77.
Figure 5.
Competition of TRα with HDACs for the modulation of Nur77 transactivation in the absence of T3. A, HDAC activity is involved in TRα-mediated enhancement of Nur77 transactivation in the absence of T3. MA-10 cells were transfected with Nur77 (50 ng) and TRα (25 ng) expression plasmids along with the NurRE-Luc reporter. After 24 hours of transfection, cells were treated with 100 nM of trichostatin A (TSA) and harvested for the measurement of luciferase activity after an additional 24 hours. B, TRα-mediated enhancement of Nur77 transactivation in the absence of T3 is inhibited by HDACs. Nur77 (50 ng), TRα (25 ng), and HDAC expression plasmids (+, 100 ng; ++, 200 ng) were transfected into MA-10 cells. Statistical significance compared with the control with Nur77 and TRα coexpression only. C, HDAC-suppression of Nur77 transactivation is relieved by coexpression. Nur77, increasing amounts of TRα (25, 50, and 100 ng; 25 and 100 ng), and the indicated HDAC expression plasmids (200 ng) were transfected into MA-10 cells. D, TRα competes with HDAC3 for binding to Nur77. The increasing amounts of in vitro-translated 35S-radiolabeled TRα were added to GST-Nur77 bound to glutathione-Sepharose beads and the constant amounts of in vitro-translated 35S-radiolabeled HDAC3. After washing, the bound proteins were eluted and separated by SDS-PAGE. E, TRα inhibits the recruitment of HDAC3 to the Nur77-binding site in the StAR promoter in the absence of T3. ChIP assays were performed using anti-HDAC3 and anti-TRα/β antibodies. In panels, A, B, and C, values are the means ± SEM of 3 independent experiments performed in duplicate. *, P < .05; **, P < .01; ns, not significant. In panels D and E, data are representative of 3 independent experiments.
We then examined different classes of HDACs for their effects on the TRα-mediated enhancement of Nur77 transactivation in the absence of T3 by transient transfection assays (Figure 5B). Interestingly, several HDACs including HDAC3 and -4, which are highly expressed in testicular Leydig cells (data not shown), efficiently repressed the TRα-enhanced Nur77 transactivation. As expected, increased expression of TRα recovered the Nur77 transactivation repressed by HDAC3 or HDAC4 in a concentration-dependent manner (Figure 5C). These results indicate that HDAC-mediated suppression of Nur77 transactivation is partially relieved by TRα in the absence of T3, suggesting functional competition between HDACs and TRα for the modulation of Nur77 transactivation.
To determine whether TRα physically competes with HDACs to bind to Nur77 in the absence of T3, we first performed GST pull-down competition assays in vitro using 35S-labeled HDAC3 and 35S-labeled TRα. HDAC3 interacted with the GST-Nur77 fusion protein irrespective of T3, and the interaction of HDAC3 and GST-Nur77 was inhibited by binding of TRα to GST-Nur77 in the absence of T3, but not in the presence of T3 (Figure 5D). The competition between HDAC3 and TRα to Nur77 binding was further confirmed in the StAR promoter, in which Nur77 is recruited to DNA by cAMP induction in MA-10 Leydig cells (19). ChIP assays with HDAC3 and TRα/β antibodies showed that TRα overexpression in the absence of T3 resulted in a decrease of HDAC3 recruitment to the Nur77 binding region in the StAR promoter, while increasing TRα recruitment (Figure 5E). The TRα overexpression-induced dissociation of HDAC3 from the promoter was not observed in the presence of T3. These results suggest that TRα enhances the transactivation of Nur77, at least in part, by interfering with the binding of HDACs to Nur77 in the absence of T3.
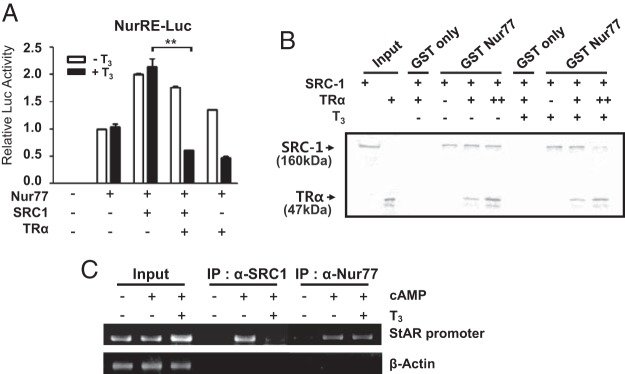
Inhibition of SRC-1 recruitment to Nur77 by liganded TRα
SRC-1 has been identified as a coactivator for Nur77 (31). As expected, overexpression of SRC-1 enhanced Nur77 transcriptional activity, but coexpression of TRα strongly blocked SRC-1 enhancement of Nur77 transactivation in the presence of T3 (Figure 6A). To determine whether TRα physically blocks the binding of SRC-1 to Nur77 in the presence of T3, we performed competitive GST pull-down assays using 35S-labeled SRC-1 and 35S-labeled TRα. As shown in Figure 6B, SRC-1 recruitment to GST-Nur77 was decreased by increased TRα binding to GST-Nur77 in the presence of T3. In the absence of T3, TRα hardly blocked SRC-1 recruitment to Nur77. The competitive binding between SRC-1 and liganded TRα to Nur77 was further confirmed in the StAR promoter by ChIP assays using SRC-1 and Nur77 antibodies. In MA-10 cells, SRC-1 was recruited to the Nur77 binding region in the StAR promoter in response to cAMP similarly as Nur77, but this association was decreased by T3 treatment (Figure 6C). These results suggest that TRα reduces the transactivation of Nur77 by interfering with the recruitment of SRC-1 to Nur77 in the presence of T3.
Figure 6.
Inhibition of SRC-1 recruitment to Nur77 by liganded TRα. A, SRC-1-induced Nur77 transactivation is repressed by TRα in the presence of T3. 293T cells were transfected with Nur77, TRα, and SRC-1 expression plasmids along with NurRE-Luc reporter and then treated with T3 for 24 hours. Values are the means ± SEM of 3 independent experiments performed in duplicate. **, P < .01. B, TRα and SRC-1 bind competitively to Nur77. 35S-labeled in vitro-translated TRα and SRC-1 were incubated with GST or GST-Nur77 in the absence or presence of T3 in GST pull-down assays. Data are representative of 3 independent experiments. C, TRα disturbs the binding of SRC-1 to Nur77 on DNA in the presence of T3. ChIP assays were performed using anti-SRC-1 and anti-Nur77 antibodies with the StAR promoter containing the Nur77-binding site. Data are representative of 2 independent experiments. IP, immunprecipitation.
Discussion
Thyroid gland disorders have been shown to associate with alterations in the hypothalamus-pituitary-testicular axis. In hyperthyroid patients, levels of plasma testosterone were increased (32, 33), whereas levels of plasma testosterone in hypothyroid patients were decreased compared with normal men (34, 35). Hypothyroidism in rats also caused a significant decrease in serum testosterone levels (36). These observations are quite consistent with our data that T3 enhances cAMP-induced testosterone production in Leydig cells (Figure 1F). In addition, current analysis of published data indicates that active TR isoforms are also found in interstitial and germ cells, not only during neonatal development but also in the adult testis (6, 37). These data together suggest that TRs and thyroid hormone play important roles in the development and functions of Leydig cells. However, the effects and action mechanisms of TRs and thyroid hormone on Leydig cell functions are still unclear. In this study, we therefore investigated the molecular action of TRα/T3 signaling on testicular steroidogenesis, the major function of Leydig cells.
Leydig cells are constantly exposed to LH/cAMP signaling from the brain once the hypothalamus-pituitary-testicular axis is established. To mimic the physiological condition, we carried out experiments with cAMP-treated Leydig cells, investigating T3 effects on the expression of steroidogenic enzyme genes. Interestingly, we observed that acute (4 hours) treatment with T3 increased cAMP-induced StAR and P450scc expression, and chronic (24 hours) T3 treatment decreased their expression (Figure 1). The positive acute response of steroidogenic enzyme gene expression to T3 might be due to the early (2 hours) strong stimulation of Nur77 expression by T3 with the high level of Nur77 expression induced by cAMP (Figure 1, C and 1D). In such conditions, the negative indirect effect of TRα/T3 signaling on Nur77 transactivation is probably much less than that in the condition of low Nur77 expression at the later time point (24 hours). A similar acute and chronic dual effect of T3 was previously reported with basal StAR expression in the absence of cAMP stimulation in mLTC-1 mouse Leydig cell line (21). Further studies are necessary to elucidate the mechanisms responsible for the dual effect of T3 on the expression of steroidogenic enzyme genes.
In our study, T3 treatment decreased the promoter activity of StAR and 3β-HSD gene in Leydig cells. In addition to well-known positive regulation of gene expression by thyroid hormones, negative regulation that ligand binding to TRs represses gene expression has also been reported. However, the mechanism of negative regulation by thyroid hormones is not well understood. TREs are usually composed of 2 or more receptor-binding “half sites” arranged as direct repeats, inverted repeats, or everted repeats (5). In one model for negative regulation, TR binds as a monomer to a negative TRE, which exists as TRE half-sites widely spaced in target gene promoters, as shown in the TSH and TRH promoter (38–40). StAR and 3β-HSD promoters have several half-sites of TRE sequence when analyzed using the Transcriptional Regulatory Element Database. Therefore, it is possible that expression of StAR and 3β-HSD is negatively regulated by thyroid hormone through the putative negative TREs. However, such an regulation is not sufficient to explain the similar pattern of negative regulation between Nur77-only binding promoters [WT (−447/−399) P450c17, NurRE, and NBRE] and steroidogenic enzyme gene promoters (StAR and 3β-HSD) by TRα/T3 signaling (Figure 3).
Previous studies have shown that the orphan nuclear receptor Nur77 regulates the transcription of steroidogenic enzyme genes in Leydig cells (16). Another orphan nuclear receptor, steroidogenic factor-1 (SF-1) is also known to regulate the expression of several steroidogenic enzyme genes as well as genes involved in sex determination and endocrine function (41). Nur77 and SF-1 bind to a similar regulatory element (14), which suggests that these 2 orphan nuclear receptors may regulate a common set of genes. Indeed, several steroidogenic enzyme genes are regulated by both SF-1 and Nur77, which include the human HSD3B2 (20), rat P450c17 (11), and mouse Hsd3b1 (23). However, Nur77 is induced by LH/cAMP signaling at the puberty stage (16), whereas SF-1 is constitutively expressed in Leydig cells from the embryonic stage (42). We have observed that TRα activated and inhibited SF-1 transactivation in the absence and presence of T3, respectively, as observed with Nur77, although the extent of TRα/T3 effects was less than that with Nur77 (Figure 3). These results suggest that TRα/T3 signaling also regulates SF-1 transactivation and may play an important role during early development of the testis and Leydig cells.
In the present study, we investigated the effect of TRα/T3 signaling on testicular steroidogenesis by examining the altered promoter activity of steroidogenic enzyme genes. We verified that Nur77 transactivation in the promoters was enhanced by TRα-mediated dissociation of HDACs in the absence of T3 and was inhibited by TRα/T3-mediated blockage of SRC-1 binding in the presence of T3. TRα-mediated regulation of Nur77 transactivation appears to occur through direct protein-protein interaction between Nur77 and TRα. These findings may contribute to our understanding of the effect of thyroid hormones and thyroid disruptors on testicular steroidogenesis.
Acknowledgments
We thank Drs M. Ascoli (University of Iowa, Iowa) and Y-C. Lee (Chonnam National University, Republic of Korea) for kindly providing us with MA-10 mouse Leydig cell line and expression vectors of hTRα and GST-TRα, respectively.
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2012R1A2A2A01008388) and by a grant from the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (A100304). E.P. and Y.K. were supported by the Brain Korea 21 Research Fellowship.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2012R1A2A2A01008388) and by a grant from the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (A100304). E.P. and Y.K. were supported by the Brain Korea 21 Research Fellowship.
Footnotes
- 8Br-cAMP
- 8-bromo-cAMP
- ChIP
- chromatin immunoprecipitation
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- HDAC
- histone deacetylase
- 3β-HSD
- 3β-hydroxysteroid dehydrogenase
- KO
- knockout
- Mut
- mutant
- NBRE
- NGF1-B response element
- NP-40
- nonyl phenoxypolyethoxylethanol
- NR
- nuclear receptor
- NurRE
- Nur response element
- P450c17
- cytochrome P450 17α-hydroxylase/C17–20 lyase
- P450scc
- cytochrome P450
- SF-1
- steroidogenic factor-1
- SRC-1
- steroid receptor coactivator-1
- StAR
- steroidogenic acute regulatory protein
- TRα
- thyroid hormone receptor α
- TRE
- thyroid response element
- TSA
- trichostatin
- WT
- wild type.
References
- 1. Jannini EA, Dolci S, Ulisse S, Nikodem VM. Developmental regulation of the thyroid hormone receptor α 1 mRNA expression in the rat testis. Mol Endocrinol. 1994;8:89–96. [DOI] [PubMed] [Google Scholar]
- 2. Hess RA, Cooke PS, Bunick D, Kirby JD. Adult testicular enlargement induced by neonatal hypothyroidism is accompanied by increased Sertoli and germ cell numbers. Endocrinology. 1993;132:2607–2613. [DOI] [PubMed] [Google Scholar]
- 3. Teerds KJ, de Rooij DG, de Jong FH, van Haaster LH. Development of the adult-type Leydig cell population in the rat is affected by neonatal thyroid hormone levels. Biol Reprod. 1998;59:344–350. [DOI] [PubMed] [Google Scholar]
- 4. Bassett JH, Harvey CB, Williams GR. Mechanisms of thyroid hormone receptor-specific nuclear and extra nuclear actions. Mol Cell Endocrinol. 2003;213:1–11. [DOI] [PubMed] [Google Scholar]
- 5. Flamant F, Gauthier K, Samarut J. Thyroid hormones signaling is getting more complex: STORMs are coming. Mol Endocrinol. 2007;21:321–333. [DOI] [PubMed] [Google Scholar]
- 6. Buzzard JJ, Morrison JR, O'Bryan MK, Song Q, Wreford NG. Developmental expression of thyroid hormone receptors in the rat testis. Biol Reprod. 2000;62:664–669. [DOI] [PubMed] [Google Scholar]
- 7. Canale D, Agostini M, Giorgilli G, et al. . Thyroid hormone receptors in neonatal, prepubertal, and adult rat testis. J Androl. 2001;22:284–288. [PubMed] [Google Scholar]
- 8. Hardy MP, Sharma RS, Arambepola NK, et al. . Increased proliferation of Leydig cells induced by neonatal hypothyroidism in the rat. J Androl. 1996;17:231–238. [PubMed] [Google Scholar]
- 9. Holsberger DR, Cooke PS. Understanding the role of thyroid hormone in Sertoli cell development: a mechanistic hypothesis. Cell Tissue Res. 2005;322:133–140. [DOI] [PubMed] [Google Scholar]
- 10. Payne AH, Youngblood GL. Regulation of expression of steroidogenic enzymes in Leydig cells. Biol Reprod. 1995;52:217–225. [DOI] [PubMed] [Google Scholar]
- 11. Zhang P, Mellon SH. Multiple orphan nuclear receptors converge to regulate rat P450c17 gene transcription: novel mechanisms for orphan nuclear receptor action. Mol Endocrinol. 1997;11:891–904. [DOI] [PubMed] [Google Scholar]
- 12. Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Philips A, Lesage S, Gingras R, et al. . Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol. 1997;17:5946–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson TE, Fahrner TJ, Milbrandt J. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol Cell Biol. 1993;13:5794–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wansa KD, Harris JM, Muscat GE. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J Biol Chem. 2002;277:33001–33011. [DOI] [PubMed] [Google Scholar]
- 16. Song KH, Park JI, Lee MO, Soh J, Lee K, Choi HS. LH induces orphan nuclear receptor Nur77 gene expression in testicular Leydig cells. Endocrinology. 2001;142:5116–5123. [DOI] [PubMed] [Google Scholar]
- 17. Stocco CO, Lau LF, Gibori G. A calcium/calmodulin-dependent activation of ERK1/2 mediates JunD phosphorylation and induction of nur77 and 20α-hsd genes by prostaglandin F2α in ovarian cells. J Biol Chem. 2002;277:3293–3302. [DOI] [PubMed] [Google Scholar]
- 18. Wilson TE, Mouw AR, Weaver CA, Milbrandt J, Parker KL. The orphan nuclear receptor NGFI-B regulates expression of the gene encoding steroid 21-hydroxylase. Mol Cell Biol. 1993;13:861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin LJ, Boucher N, Brousseau C, Tremblay JJ. The orphan nuclear receptor NUR77 regulates hormone-induced StAR transcription in Leydig cells through cooperation with Ca2+/calmodulin-dependent protein kinase I. Mol Endocrinol. 2008;22:2021–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin LJ, Tremblay JJ. The human 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase type 2 promoter is a novel target for the immediate early orphan nuclear receptor Nur77 in steroidogenic cells. Endocrinology. 2005;146:861–869. [DOI] [PubMed] [Google Scholar]
- 21. Manna PR, Kero J, Tena-Sempere M, Pakarinen P, Stocco DM, Huhtaniemi IT. Assessment of mechanisms of thyroid hormone action in mouse Leydig cells: regulation of the steroidogenic acute regulatory protein, steroidogenesis, and luteinizing hormone receptor function. Endocrinology. 2001;142:319–331. [DOI] [PubMed] [Google Scholar]
- 22. Kim Y, Ryu JC, Choi HS, Lee K. Effect of 2,2′,4,4′-tetrahydroxybenzophenone (BP2) on steroidogenesis in testicular Leydig cells. Toxicology. 2011;288:18–26. [DOI] [PubMed] [Google Scholar]
- 23. Hong CY, Park JH, Ahn RS, et al. . Molecular mechanism of suppression of testicular steroidogenesis by proinflammatory cytokine tumor necrosis factor α. Mol Cell Biol. 2004;24:2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teixeira J, Fynn-Thompson E, Payne AH, Donahoe PK. Müllerian-inhibiting substance regulates androgen synthesis at the transcriptional level. Endocrinology. 1999;140:4732–4738. [DOI] [PubMed] [Google Scholar]
- 25. Jeong BC, Hong CY, Chattopadhyay S, et al. . Androgen receptor corepressor-19 kDa (ARR19), a leucine-rich protein that represses the transcriptional activity of androgen receptor through recruitment of histone deacetylase. Mol Endocrinol. 2004;18:13–25. [DOI] [PubMed] [Google Scholar]
- 26. Lee SY, Park E, Kim SC, Ahn RS, Ko C, Lee K. ERα/E2 signaling suppresses the expression of steroidogenic enzyme genes via cross-talk with orphan nuclear receptor Nur77 in the testes. Mol Cell Endocrinol. 2012;362:91–103. [DOI] [PubMed] [Google Scholar]
- 27. Volle DH, Decourteix M, Garo E, et al. . The orphan nuclear receptor small heterodimer partner mediates male infertility induced by diethylstilbestrol in mice. J Clin Invest. 2009;119:3752–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qamar I, Gong EY, Kim Y, et al. . Anti-steroidogenic factor ARR19 inhibits testicular steroidogenesis through the suppression of Nur77 transactivation. J Biol Chem. 2010;285:22360–22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee SY, Gong EY, Hong CY, et al. . ROS inhibit the expression of testicular steroidogenic enzyme genes via the suppression of Nur77 transactivation. Free Radic Biol Med. 2009;47:1591–1600. [DOI] [PubMed] [Google Scholar]
- 30. Kang SA, Na H, Kang HJ, Kim SH, Lee MH, Lee MO. Regulation of Nur77 protein turnover through acetylation and deacetylation induced by p300 and HDAC1. Biochem Pharmacol. 2010;80:867–873. [DOI] [PubMed] [Google Scholar]
- 31. Kelly SN, McKenna TJ, Young LS. Coregulatory protein-orphan nuclear receptor interactions in the human adrenal cortex. J Endocrinol. 2005;186:33–42. [DOI] [PubMed] [Google Scholar]
- 32. Vermeulen A, Verdonck L. Some studies on the biological significance of free testosterone. J Steroid Biochem. 1972;3:421–426. [DOI] [PubMed] [Google Scholar]
- 33. Hudson RW, Edwards AL. Testicular function in hyperthyroidism. J Androl. 1992;13:117–124. [PubMed] [Google Scholar]
- 34. Wortsman J, Rosner W, Dufau ML. Abnormal testicular function in men with primary hypothyroidism. Am J Med. 1987;82:207–212. [DOI] [PubMed] [Google Scholar]
- 35. Cavaliere H, Abelin N, Medeiros-Neto G. Serum levels of total testosterone and sex hormone binding globulin in hypothyroid patients and normal subjects treated with incremental doses of L-T4 or L-T3. J Androl. 1988;9:215–219. [DOI] [PubMed] [Google Scholar]
- 36. Chiao YC, Lee HY, Wang SW, et al. . Regulation of thyroid hormones on the production of testosterone in rats. J Cell Biochem. 1999;73:554–562. [PubMed] [Google Scholar]
- 37. Rao JN, Liang JY, Chakraborti P, Feng P. Effect of thyroid hormone on the development and gene expression of hormone receptors in rat testes in vivo. J Endocrinol Invest. 2003;26:435–443. [DOI] [PubMed] [Google Scholar]
- 38. Hollenberg AN, Monden T, Flynn TR, Boers ME, Cohen O, Wondisford FE. The human thyrotropin-releasing hormone gene is regulated by thyroid hormone through two distinct classes of negative thyroid hormone response elements. Mol Endocrinol. 1995;9:540–550. [DOI] [PubMed] [Google Scholar]
- 39. Breen JJ, Hickok NJ, Gurr JA. The rat TSHβ gene contains distinct response elements for regulation by retinoids and thyroid hormone. Mol Cell Endocrinol. 1997;131:137–146. [DOI] [PubMed] [Google Scholar]
- 40. Shibusawa N, Hollenberg AN, Wondisford FE. Thyroid hormone receptor DNA binding is required for both positive and negative gene regulation. J Biol Chem. 2003;278:732–738. [DOI] [PubMed] [Google Scholar]
- 41. Sadovsky Y, Dorn C. Function of steroidogenic factor 1 during development and differentiation of the reproductive system. Rev Reprod. 2000;5:136–142. [DOI] [PubMed] [Google Scholar]
- 42. Parker KL, Rice DA, Lala DS, et al. . Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res. 2002;57:19–36. [DOI] [PubMed] [Google Scholar]