ABSTRACT
Necrosis culminates in spilling cellular content through the permeabilized plasma membrane, thereby releasing potentially immunostimulatory molecules in the pericellular space of dead cells. Accordingly, molecules involved in necroptotic signaling, such as receptor-interacting serine/threonine-protein kinase 3 (RIPK3) and mixed lineage kinase-like (MLKL) have been found to stimulate anticancer immune responses in mouse models of chemotherapy. mRNAs encoding prominent pro-necrotic gene products (RIPK1, RIPK3, MLKL, PGAM5 and DFNA5) were correlated with immune-related metagenes in several cancer types (breast, colorectal, lung, ovary, melanoma), revealing the strongest associations in breast cancer. In two independent breast cancer cohorts, the expression of MLKL and DFNA5 was decreased at the mRNA levels in tumor as compared with normal tissues. Moreover, MLKL expression exhibited a strong positive correlation with genes reflecting the presence of B, NK and T lymphocytes in the tumor bed, in multiple distinct breast cancer subtypes. In contrast, the positive correlation between RIPK3 and lymphoid cells was restricted to HER2+ and triple negative/basal-like breast cancer. Moreover, the expression of DFNA5, which mediates post-apoptotic secondary necrosis, mostly correlated with the monocytic lineage and macrophages in ER+/luminal A breast cancers. MLKL (and to some extent RIPK1 and RIPK3) was strongly associated with the local expression of genes involved in interferon-α and interferon-γ responses. Altogether, these results support the idea that pro-necrotic signaling facilitates intratumoral immune responses in human breast cancer.
KEYWORDS: Apoptosis, immunogenic cell death, immunosurveillance, necrosis
Introduction
Although it had been initially thought that necrosis, a cell death modality culminating in plasma membrane permeabilization, would occur in a non-regulated fashion,1 it has become clear that, at least in some cases, necrosis can occur in a highly regulated fashion following the activation of a series of pro-necrotic signaling molecules.2 In the so-called necroptotic pathway, the central signaling module involves phosphorylation of the pseudokinase mixed lineage kinase-like (MLKL) by receptor-interacting serine/threonine-protein kinase 3 (RIPK3).2,3 RIPK3 is often activated by another kinase of the same family, RIPK14 and can associate with yet another putative pro-necrotic protein, namely, the mitochondrial protein phosphatase PGAM5.5 MLKL associates with the plasma membrane and causes its permeabilization as a result of uncontrolled ion fluxes.2,3,6 A homolog of MLKL, non-syndromic hearing impairment protein 5 (also known as deafness associated tumor suppressor, DFNA5), is activated in secondary necrosis (i.e. necrosis after apoptosis) as a result of its partial proteolysis by the pro-apoptotic caspase-3.7
Although it had been widely thought that apoptosis would be a non-immunogenic cell death modality, while necrosis would be immunogenic,8 it turned out that the activation of caspases, which is a hallmark of apoptosis, may be required for immunogenic signaling when cell death is stimulated by chemotherapy or radiotherapy.9-12 In addition, ER stress, autophagy and type-1 interferon signaling contribute to elicit immune responses to dead-cell antigens.13-16 Recent research revealed that pro-necrotic molecules, in particular RIPK3 and MLKL, can contribute to immunogenic cell death (ICD) signaling.17-19 Of note, cancer cells often lose the expression of either RIPK3 or MLKL,20,21 which might increase their cell-intrinsic resistance to lethal signals and facilitate their escape from immunosurveillance. Altogether, these observations underscore the probable clinical relevance of ICD in determining the control of cancers by the immune system.
Breast cancer is under strong immunosurveillance,22-25 and there is ample evidence that the suppression of ICD-relevant molecules and pathways has a negative prognostic impact in breast cancer patients, correlating with a poor ratio of CD8+ cytotoxic T lymphocytes over FOXP3+ regulatory T cells as a sign of poor local immunosurveillance.14,26-30 However, it has not yet been investigated whether the expression of pro-necrotic gene products may affect immunosurveillance in patient samples. Here, we show that low expression of several pro-necrotic proteins, in particular RIP3, MLKL and DFNA5, may negatively affect the density of particular immune cell subtypes infiltrating breast cancers.
Results and discussion
Impact of pro-necrotic molecules on immunosurveillance in different cancer types
Previous work by our group and that of others indicate that cell death occurring within established cancer can stimulate local immune responses9,11,13,14,31-35 and that pro-necrotic signaling can contribute to this immunogenicity.17-19 In a first step of this bioinformatics analysis, we took advantage from publicly available data, focusing on five frequent malignancies that are known to be under immunosurveillance (breast cancer, colorectal cancer, non-small cell lung cancer (NSCLC), melanoma and ovarian cancer). We determined the correlation of the mRNAs coding for prominent proteins involved in pro-necrotic signaling (RIPK1, RIPK3, MLKL, PGAM5) and secondary necrosis (DFNA5) with that of genes indicating the local presence of different stromal cell populations, based on the microenvironment cell populations-counter (MCP-counter) method, which facilitates estimating the absolute abundance of eight immune and two stromal cell populations.36 As indicated by volcano plots in which the Spearman correlation values (ρ) were plotted against the p values, MLKL correlated positively with 6 out of 8 among the immune cell subtypes (T cells, CD8 T cells, CTL, NK cells, B cells, monocytic lineage and myeloid dendritic cells) in breast cancer with a rather low p value (p < 10−30) (Fig. 1). This portion of extremely high positive correlations (p < 10−30) dropped to 2 out of 8 (cytotoxic T lymphocytes and NK cells) for MLKL in the case of lung cancer and was not attained by any of the other cancer cell types (Fig. 1). Of note, MLKL and immune-related metagenes were not more abundant in breast cancer than in other cancer types (Fig. S1), a finding that excludes the breast-cancer-specific correlation between MLKL and immune subtypes is due to their high expression level. As a result, we decided to concentrate our subsequent analysis on breast cancer.
Figure 1.
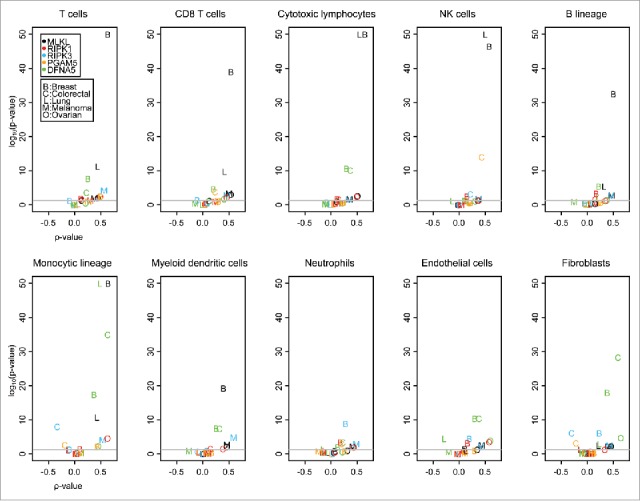
Volcano plots of Spearman correlation tests (“ρ value” versus p value). The Spearman tests were applied on correlations between selected genes (MLKL, RIPK1, RIPK3, PGAM5 and DFNA5) and between activities of different immune cells, measured within MCP-counter. Datasets of different cancers (primary tumors) are considered: breast, colorectal, lung, melanoma, ovarian. p values are adjusted for each immune cell type following the Benjamini Hochberg method. The lowest p values are indicated as 10e-50.
Reduced expression of pro-necrotic molecules in breast cancer
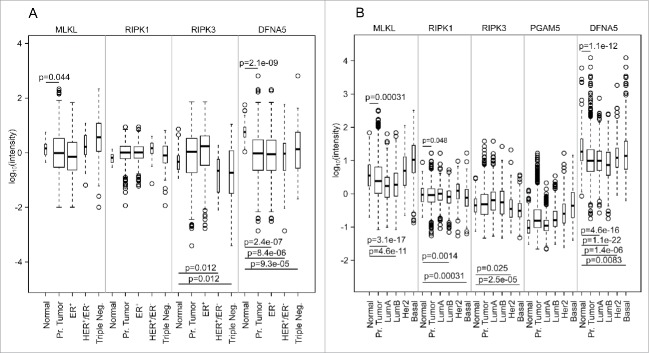
We determined the mRNA expression levels of RIPK1, RIPK3, MLKL, PGAM5 and DFNA5 in two major breast cancer-relevant databases, namely TCGA and molecular taxonomy of breast cancer international consortium (METABRIC37). We expected that such genes would be downregulated in malignancies due to their implication in cell-autonomous cell death pathways, as well as possibly in immunostimulatory signaling. When comparing the expression level of these genes in the total population of breast cancer patients, DFNA5 was found to be significantly (p < 10−8, one-sided Student t-test) downregulated (both in TCGA and METABRIC) and this downregulation was again significant (p < 0.01) for all subgroups of breast cancers (Fig. 2). A global tendency for downregulation was also found for MLKL (p < 0.05 in both TCGA and METABRIC) and RIPK1 (p < 0.05 only in METABRIC). Subgroup analyses indicated that RIPK3 was only downregulated in HER2+/ER− and triple-negative breast cancers (in TCGA) and HER2+ and basal-like breast cancers (METABRIC). Hence, analyses of two independent cohorts (Fig. 2) indicate that breast cancers tend to reduce the expression level of major pro-necrotic mediators.
Figure 2.
Expression of MLKL, RIPK1, RIPK3, PGAM5 and DFNA5 in breast cancer (normal tissue, primary [Pr.] tumors and tumor subgroups), from the TCGA data set (A) and the METABRIC data set (B). p values are indicated when the expression is significantly lower compared with normal tissue (one-sided t-test). TCGA breast cancer subgroups are slightly different than the usual ones because of missing information about tumor tissues. PGAM5 expression is not available for TCGA data set because no probeset is available for this gene.
To understand the mechanisms through which the expression of pro-necrotic genes is diminished in breast cancer compared with normal tissue, we determined promoter methylation (according to Koo et al.21 methylation of RIPK3 represses necrosis in cancer), mutations and copy number variations. Of note, mutations affecting these genes were infrequent, while a reduction in copy numbers was observed in <2% of breast cancers (Fig. S2, from cBioPortal, http://www.cbioportal.org,38,39). DNA hypermethylation did not affect any of the pro-necrotic genes investigated here (Fig. S3, from Wanderer, http://maplab.imppc.org/wanderer/,40). As a result, the mechanisms that account for the relative downregulation of MLKL, RIPK3 and DFNA5 in breast cancer tissue remain elusive. Nevertheless, we did not find any significant effect of the expression of these genes on overall survival.
Positive correlation between the expression of pro-necrotic mediators and the immune infiltrate
MLKL stood out for positive correlations with many different stromal cell types, in particular T cells, CD8+ T cells, cytotoxic T lymphocytes, NK cells, B cells, monocytic cell and myeloid dendritic cells (but not neutrophils, endothelial cells and fibroblasts), both in TCGA (Fig. 3) and in METABRIC (Fig. 4), irrespective of whether all breast cancers were analyzed together or whether they were analyzed as subgroups. Such positive (though more moderate) correlations were also found for RIPK3, but mostly in the subgroups of triple-negative breast cancers (TCGA, Fig. 3) and basal-like tumors (METABRIC). DFNA5 exhibited positive correlations with several lymphoid and myeloid subpopulations across the entire breast cancer cohort (Fig. 3, 4), as well as in Her2+ (TCGA, Fig. 3) and luminal A breast cancers (METABRIC, Fig. 4). Of note, in normal tissue, no significant correlations were found between mRNAs coding for pro-necrotic molecules and those indicating the presence of immune cells in the TCGA (Fig. 3), while such associations were found in several incidences (and in particular for MLKL) in the METABRIC cohort (Fig. 4), perhaps reflecting the fact that in TCGA “normal” tissue is from patients without cancer, while in METABRIC “normal” tissues has been retrieved from cancer patients, adjacent to the malignant tumor. Irrespective of this discrepancy, it appears that the expression levels MLKL, RIPK3 and DFNA5 often correlate with distinct immune cell subtypes. These positive correlations suggest that the abundance of immunologically relevant cell types is globally reduced in cancer tissues as compared with normal ones, as this applies to the expression of necrosis-associated genes. Indeed, immune cell type activities are lower in tumor tissues as compared with normal breast tissues in the METABRIC data set. However, this tendency was not found for the TCGA data set (Fig. S4). The reason for this discrepancy is not clear, yet may be linked to the disparate definition of “normal” tissue for the 2 data sets.
Figure 3.
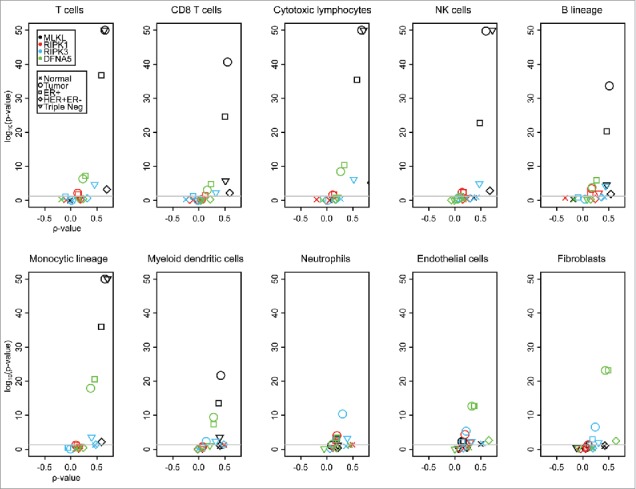
Volcano plots of Spearman correlation test (“ρ value” and associated p value) in TCGA breast cancer (normal tissue, primary [Pr.] tumor and the indicated tumor subgroups). The Spearman tests were applied on correlation between selected genes (MLKL, RIPK1, RIPK3, PGAM5 and DFNA5) and between the activities of different immune cells, measured within MCP-counter, in TCGA breast cancer data set. Subgroups are slightly different than the usual ones because of missing information. PGAM5 expression is not available for the TCGA data set because there is no associated probeset. p values were adjusted for each immune cell type (Benjamini-Hochberg method). The lowest p values are indicated as 10e-50.
Figure 4.
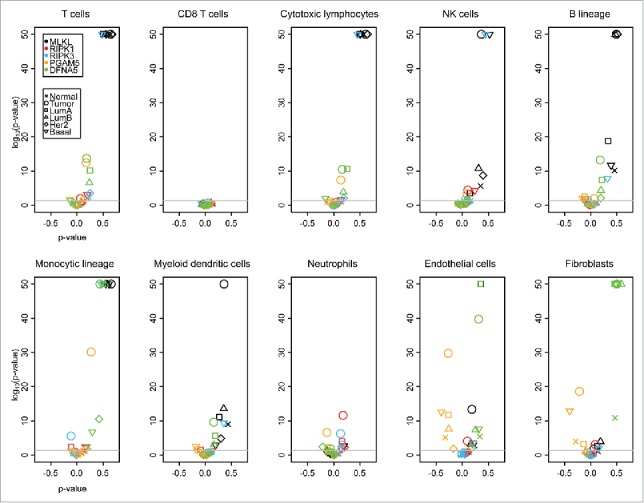
Volcano plots of Spearman correlation test (“ρ value” and associated p value) in METABRIC breast cancer (normal tissue, primary [Pr.] tumor and the indicated tumor subgroups). The Spearman tests were applied on correlation between selected genes (MLKL, RIPK1, RIPK3 and DFNA5) and between activities of different immune cells, measured within MCP-counter, in the METABRIC breast cancer data set. p values were adjusted for each immune cell type (Benjamini-Hochberg method). The lowest p values are indicated as 10e-50. Note that CD8+ T cells are uniquely defined by the CD8A gene and that the probe detecting this gene may be inappropriate in the METABRIC data set.
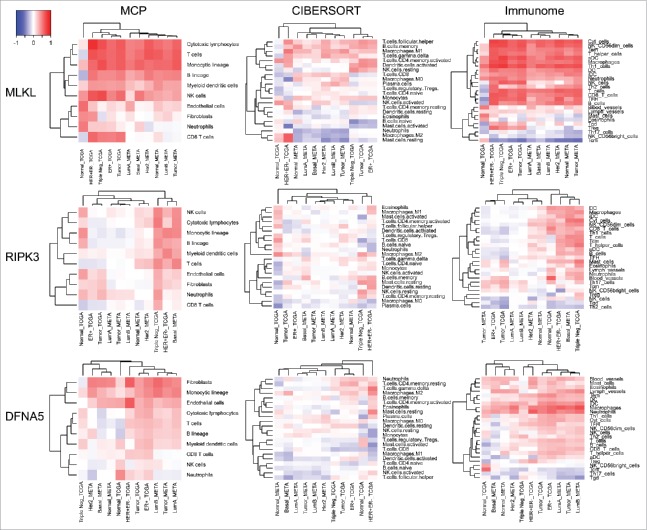
At a next step, we determined whether the positive correlations obtained by means of the MCP counter36 (Fig. 3, 4) could be reproduced using other methods for extracting information on the abundance of immune cell subsets from microarray data, namely CIBERSORT (which informs on the relative rather than the absolute abundance of immune subsets in a tissue)41 and IMMUNOME (which yields data on the absolute abundance of immune subsets, implemented in Ref.42 based on Ref.43). MLKL expression strongly correlated with multiple different immune cell subtypes, both according to the MCP and the immunome methods. The correlation between MLKL expression and particular immune subsets was less impressive when the CIBERSORT method was used, yet appeared clear for follicular helper T cells, memory B cells, M1 macrophages and γ/δ T cells (Fig. 5). Analyses of the levels of MLKL expression found in distinct immunological cell subtypes (from “The Human Protein Atlas”, http://www.proteinatlas.org/,44-47 and from the “Immunological Genome Project”, https://www.immgen.org/,48) suggest a particular strong expression in the spleen and in stem cell subpopulations (Fig. S5), yet revealed no clear overlap with the correlations found in breast cancer (Fig. 5). The correlation between RIPK3 and metagenes corresponding to different immune subtypes was only detectable by MCP and IMMUNOME but not CIBSERSORT analyses. With respect to DFNA5, the positive correlations across distinct breast cancer types were only concordant for one particular immune subset, namely monocytic cells (MCP) and macrophages (IMMUNOME) (Fig. 5). However, the fact that DFNA5 is particular abundant in granulocytes (Fig. S5) might explain its association with neutrophil infiltration in breast cancers (Fig. 5). Altogether, these results suggest that a few particular associations may reflect functional interactions between pro-necrotic molecules and immune cell subsets in human breast cancer. The CIBERSORT analysis differs from the MCP and IMMUNOME analyses, probably because CIBERSORT estimates the relative ratio of different immune cell types, while MCP and IMMUNOME methods estimate the absolute abundance of immune effectors.
Figure 5.
Heatmap representation of Spearman correlation coefficients. Correlations between three genes (MLKL, RIPK3 and DFNA5) and between activities of different immune cells are indicated. Immune cell densities were estimated by three different methods: MCP-counter, CIBERSORT and IMMUNOME. Datasets are from TCGA and METABRIC breast cancer microarrays (normal tissue, all primary tumors and tumor subgroups).
Positive correlation between the expression of pro-necrotic mediators and molecular processes
In a final step, we sought to correlate the expression levels of pro-necrotic molecules with annotated molecular processes rather than with specific immune subpopulations in breast cancer transcriptomes (Fig. 6). The strongest enrichment that was fully reproducible (between the TCGA and the METABRIC data sets) affected the interferon-α and interferon-γ responses, both of which exhibited a strong correlation with MLKL expression and, to a lower degree, with that of RIPK1 and RIPK3 (Fig. 6). Both interferon responses are well-known to play a major role in general immunosurveillance49 and in particular in the immune control of breast cancer,15,16 strongly supporting the idea that pro-necrotic molecules (and in particular MLKL) favor local antitumor immune responses.
Figure 6.
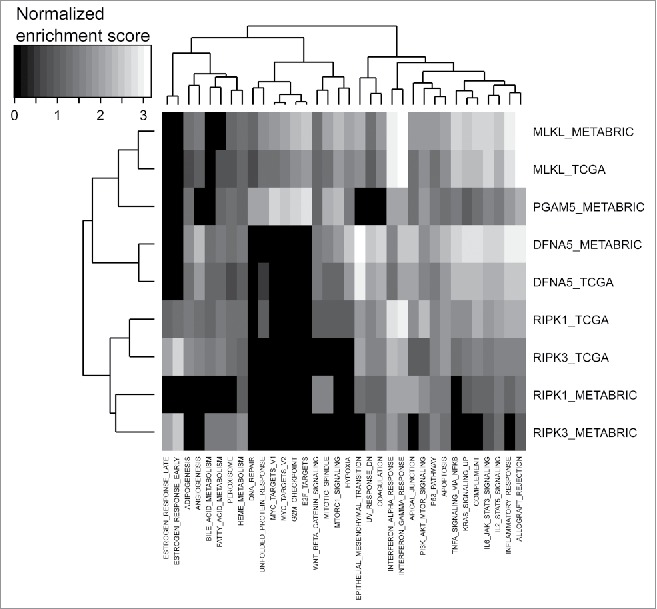
Heatmap representation of GSEA hallmark enrichment scores. GSEA analysis was based on Spearman correlation coefficients, between genome-wide expression and between selected genes (MLKL, RIPK1, RIPK3, PGAM5, DFNA5), in the two breast tumor data sets TCGA and METABRIC. PGAM5 expression is not available for the TCGA data set because there is no associated probeset.
Concluding remarks
The bioinformatics analyses that we performed on two independent breast cancer cohorts revealed a marked tendency to reduce the expression of the two central mediators of necroptosis (RIPK3 and MLKL) as well as that of the central mediator of secondary necrosis (DFNA5) in breast cancers compared with adjacent tissues. The molecular mechanisms of this relative downregulation remain elusive because the frequency of promoter hypermethylation, mutation or allelic loss of the corresponding genes/loci was too small to have a statistical impact on gene expression. Importantly, it appeared that high expression of MLKL (and to a less extent that of RIPK3) correlated with an enhanced density of multiple effectors of the cellular immune system including myeloid and lymphoid elements, perhaps reflecting the fact that these types of cells often are expressed in an “organized” (highly correlated) fashion in breast cancer.42,50,51 In contrast, DFNA5 expression was particularly well-correlated with that of myeloid cells, in particular macrophages. These correlations were partially dependent on the breast cancer subtype. For instance, RIPK3 correlated with the lymphoid infiltrate in particular in HER2+ and triple negative/basal-like breast cancer, perhaps reflecting the fact that these classes of mammary carcinomas is under particularly strong immunosurveillance.52
The precise mechanisms accounting for poor immune infiltration in mammary tumors with reduced expression of pro-necrotic gene products remain elusive. Preclinical experiments performed on RIPK3- or MLKL-deficient cancers revealed that such tumors were relatively unable to recruit immune cells post-chemotherapy, likely due to the absence of cellular release of ATP (which acts on purinergic receptors to attract myeloid cells into the tumor bed) and HMGB1 (which acts on toll-like receptor-4 to induce the activation/maturation of dendritic cell precursors).18 Furthermore, RIPK3- or MLKL-deficient cancer cells showed a reduced activation of interferon response genes upon in vitro exposure to anthracyclines,18 and similarly reduced expression of MLKL (and to some extent RIPK1 and RIPK3) in breast cancer correlated with a diminished expression of genes involved in interferon-α and interferon-γ responses. Future preclinical work as well as clinical studies must determine whether measures to compensate for such defects (e.g. ATPase inhibitors to increase extracellular ATP levels, synthetic toll-like receptor-4 agonists, stimulators of the interferon response) may overcome the obstacle to immunosurveillance that results from deficient expression of pro-necrotic molecules.
Materials and methods
Datasets
The two biggest data sets of breast cancer microarrays, which are publicly available, were used are as follows: TCGA Breast cancer and METABRIC.37 For the latter, when a gene has several probesets, we used the one with the biggest variance. For other cancers, we used the biggest microarrays data set that has at least one probeset for MLKL and one for DFNA5: a multi-cancers data set (http://www.intgen.org/, “Bittner”) for colorectal cancer, a lung cancer data set,53,54 a skin cancer datataset55 for melanoma and an ovarian cancer data set.56 The multi-cancer data set was also used for producing Fig. S1.
Immune infiltrate estimation
We used the R-package associated with MCP-counter36 to estimate the density of infiltration by distinct immune cell types from microarray data. CIBERSORT41 and IMMUNOME (metagenes associated with immune cells defined in Ref.42 based in selected genes/probeset from Ref.43) were also used.
Gene set enrichment of correlation
We ranked the genes/probesets according to the Spearman correlation coefficient (with MLKL, RIPK1, RIPK3, PGAM5, DFNA5 in various cases) and applied GSEA57 to these lists (“GseaPreranked”). Moreover, we used the “h.all.v5.2.symbols.gmt [hallmarks]” gene set database.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
GK is supported by the Ligue contre le Cancer (équipe labelisée); Agence National de la Recherche (ANR) – Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Institut National du Cancer (INCa); Institut Universitaire de France; Fondation pour la Recherche Médicale (FRM); the European Commission (ArtForce); the European Research Council (ERC); the LeDucq Foundation; the LabEx Immuno-Oncology; the RHU Torino Lumière, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); the Paris Alliance of Cancer Research Institutes (PACRI) and the TCGA Research Network. YM is supported by National Natural Science Foundation of China (Grant No 81671630), Natural Science Foundation of Jiangsu Province (Grant No SBK2016040379), Chinese Academy of Medical Sciences research grant (2015RC310003) and a Start-up package from the National Thousand Young Talent Program in China.
References
- 1.Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev 2010; 24:2592-602; PMID:21123646; http://dx.doi.org/27959630 10.1101/gad.1984410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L, Kepp O, Chan FK, Kroemer G. Necroptosis: mechanisms and relevance to disease. Annu Rev Pathol 2016; 12:103-30; PMID:27959630; http://dx.doi.org/ 10.1146/annurev-pathol-052016-100247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X et al.. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148:213-27; PMID:22265413; http://dx.doi.org/ 10.1016/j.cell.2011.11.031 [DOI] [PubMed] [Google Scholar]
- 4.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008; 135:1311-23; PMID:19109899; http://dx.doi.org/ 10.1016/j.cell.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012; 148:228-43; PMID:22265414; http://dx.doi.org/24418759 10.1016/j.cell.2011.11.030 [DOI] [PubMed] [Google Scholar]
- 6.Galluzzi L, Kepp O, Kroemer G. MLKL regulates necrotic plasma membrane permeabilization. Cell Res 2014; 24:139-40; PMID:24418759; http://dx.doi.org/ 10.1038/cr.2014.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun 2017; 8:14128; PMID:28045099; http://dx.doi.org/ 10.1038/ncomms14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol 2009; 9:353-63; PMID:19365408; http://dx.doi.org/ 10.1038/nri2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casares N, Pequignot MO, Tesniere A, Ghiringhelli F, Roux S, Chaput N, Schmitt E, Hamai A, Hervas-Stubbs S, Obeid M et al.. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J Exp Med 2005; 202:1691-701; PMID:16365148; http://dx.doi.org/ 10.1084/jem.20050915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obeid M, Panaretakis T, Joza N, Tufi R, Tesniere A, van Endert P, Zitvogel L, Kroemer G. Calreticulin exposure is required for the immunogenicity of gamma-irradiation and UVC light-induced apoptosis. Cell Death Differ 2007; 14:1848-50; PMID:17657249; http://dx.doi.org/ 10.1038/sj.cdd.4402201 [DOI] [PubMed] [Google Scholar]
- 11.Obeid M, Tesniere A, Panaretakis T, Tufi R, Joza N, van Endert P, Ghiringhelli F, Apetoh L, Chaput N, Flament C et al.. Ecto-calreticulin in immunogenic chemotherapy. Immunol Rev 2007; 220:22-34; PMID:17979837; http://dx.doi.org/19165151 10.1111/j.1600-065X.2007.00567.x [DOI] [PubMed] [Google Scholar]
- 12.Panaretakis T, Kepp O, Brockmeier U, Tesniere A, Bjorklund AC, Chapman DC, Durchschlag M, Joza N, Pierron G, van Endert P et al.. Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death. EMBO J 2009; 28:578-90; PMID:19165151; http://dx.doi.org/ 10.1038/emboj.2009.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G et al.. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011; 334:1573-7; PMID:22174255; http://dx.doi.org/ 10.1126/science.1208347 [DOI] [PubMed] [Google Scholar]
- 14.Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M et al.. An immunosurveillance mechanism controls cancer cell ploidy. Science 2012; 337:1678-84; PMID:23019653; http://dx.doi.org/ 10.1126/science.1224922 [DOI] [PubMed] [Google Scholar]
- 15.Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remédios C et al.. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med 2014; 20:1301-9; PMID:25344738; http://dx.doi.org/ 10.1038/nm.3708 [DOI] [PubMed] [Google Scholar]
- 16.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol 2015; 15:405-14; PMID:26027717; http://dx.doi.org/27050509 10.1038/nri3845 [DOI] [PubMed] [Google Scholar]
- 17.Aaes TL, Kaczmarek A, Delvaeye T, De Craene B, De Koker S, Heyndrickx L, Delrue I, Taminau J, Wiernicki B, De Groote P et al.. Vaccination with necroptotic cancer cells induces efficient anti-tumor immunity. Cell Rep 2016; 15:274-87; PMID:27050509; http://dx.doi.org/ 10.1016/j.celrep.2016.03.037 [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Ma Y, Chen G, Zhou H, Yamazaki T, Klein C, Pietrocola F, Vacchelli E, Souquere S, Sauvat A et al.. Contribution of RIP3 and MLKL to immunogenic cell death signaling in cancer chemotherapy. Oncoimmunology 2016; 5:e1149673; PMID:27471616; http://dx.doi.org/ 10.1080/2162402X.2016.1149673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatim N, Jusforgues-Saklani H, Orozco S, Schulz O, Barreira da Silva R, Reis e Sousa C, Green DR, Oberst A, Albert ML. RIPK1 and NF-kappaB signaling in dying cells determines cross-priming of CD8(+) T cells. Science 2015; 350:328-34; PMID:26405229; http://dx.doi.org/ 10.1126/science.aad0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geserick P, Wang J, Schilling R, Horn S, Harris PA, Bertin J, Gough PJ, Feoktistova M, Leverkus M. Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell Death Dis 2015; 6:e1884; PMID:26355347; http://dx.doi.org/25952668 10.1038/cddis.2015.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo GB, Morgan MJ, Lee DG, Kim WJ, Yoon JH, Koo JS, Kim SI, Kim SJ, Son MK, Hong SS et al.. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res 2015; 25:707-25; PMID:25952668; http://dx.doi.org/ 10.1038/cr.2015.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroemer G, Senovilla L, Galluzzi L, Andre F, Zitvogel L. Natural and therapy-induced immunosurveillance in breast cancer. Nat Med 2015; 21:1128-38; PMID:26444637; http://dx.doi.org/ 10.1038/nm.3944 [DOI] [PubMed] [Google Scholar]
- 23.Pol J, Vacchelli E, Aranda F, Castoldi F, Eggermont A, Cremer I, Sautès-Fridman C, Fucikova J, Galon J, Spisek R et al.. Trial Watch: Immunogenic cell death inducers for anticancer chemotherapy. Oncoimmunology 2015; 4:e1008866; PMID:26137404; http://dx.doi.org/ 10.1080/2162402X.2015.1008866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoll G, Zitvogel L, Kroemer G. Differences in the composition of the immune infiltrate in breast cancer, colorectal carcinoma, melanoma and non-small cell lung cancer: a microarray-based meta-analysis. Oncoimmunology 2016; 5:e1067746; PMID:27057431; http://dx.doi.org/ 10.1080/2162402X.2015.1067746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vacchelli E, Sistigu A, Yamazaki T, Vitale I, Zitvogel L, Kroemer G. Autocrine signaling of type 1 interferons in successful anticancer chemotherapy. Oncoimmunology 2015; 4:e988042; PMID:26405588; http://dx.doi.org/ 10.4161/2162402X.2014.985940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg AD, De Ruysscher D, Agostinis P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: a large-scale meta-analysis. Oncoimmunology 2016; 5:e1069938; PMID:27057433; http://dx.doi.org/ 10.1080/2162402X.2015.1069938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladoire S, Enot D, Andre F, Zitvogel L, Kroemer G. Immunogenic cell death-related biomarkers: impact on the survival of breast cancer patients after adjuvant chemotherapy. Oncoimmunology 2016; 5:e1082706; PMID:27057465; http://dx.doi.org/ 10.1080/2162402X.2015.1082706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladoire S, Enot D, Senovilla L, Ghiringhelli F, Poirier-Colame V, Chaba K, Semeraro M, Chaix M, Penault-Llorca F, Arnould L et al.. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy 2016; 12(5):864-75; PMID:26979828; http://dx.doi.org/ 10.1080/15548627.2016.1154244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladoire S, Penault-Llorca F, Senovilla L, Dalban C, Enot D, Locher C, Prada N, Poirier-Colame V, Chaba K, Arnould L et al.. Combined evaluation of LC3B puncta and HMGB1 expression predicts residual risk of relapse after adjuvant chemotherapy in breast cancer. Autophagy 2015; 11:1878-90; PMID:26506894; http://dx.doi.org/ 10.1080/15548627.2015.1082022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semeraro M, Adam J, Stoll G, Louvet E, Chaba K, Poirier-Colame V, Sauvat A, Senovilla L, Vacchelli E, Bloy N et al.. The ratio of CD8+/FOXP3 T lymphocytes infiltrating breast tissues predicts the relapse of ductal carcinoma in situ. Oncoimmunology 2016; 5:e1218106. PMID:27853639; http://dx.doi.org/ 10.1080/2162402X.2016.1218106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garg AD, Krysko DV, Verfaillie T, Kaczmarek A, Ferreira GB, Marysael T, Rubio N, Firczuk M, Mathieu C, Roebroek AJ et al.. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J 2012; 31:1062-79; PMID:22252128; http://dx.doi.org/ 10.1038/emboj.2011.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013; 31:51-72; PMID:23157435; http://dx.doi.org/ 10.1146/annurev-immunol-032712-100008 [DOI] [PubMed] [Google Scholar]
- 33.Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer 2012; 12:860-75; PMID:23151605; http://dx.doi.org/ 10.1038/nrc3380 [DOI] [PubMed] [Google Scholar]
- 34.Vacchelli E, Ma Y, Baracco EE, Sistigu A, Enot DP, Pietrocola F, Yang H, Adjemian S, Chaba K, Semeraro M et al.. Chemotherapy-induced antitumor immunity requires formyl peptide receptor 1. Science 2015; 350:972-8; PMID:26516201; http://dx.doi.org/15246252 10.1126/science.aad0779 [DOI] [PubMed] [Google Scholar]
- 35.Zitvogel L, Casares N, Pequignot MO, Chaput N, Albert ML, Kroemer G. Immune response against dying tumor cells. Adv Immunol 2004; 84:131-79; PMID:15246252; http://dx.doi.org/ 10.1016/S0065-2776(04)84004-5 [DOI] [PubMed] [Google Scholar]
- 36.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH et al.. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016; 17:218; PMID:27765066; http://dx.doi.org/ 10.1186/s13059-016-1070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y et al.. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486:346-52; PMID:22522925; http://dx.doi.org/ 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E et al.. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-4; PMID:22588877; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6:pl1; PMID:23550210; http://dx.doi.org/ 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 2015; 8:22; PMID:26113876; http://dx.doi.org/25822800 10.1186/s13072-015-0014-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Met 2015; 12:453-7; PMID:25822800; http://dx.doi.org/ 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoll G, Bindea G, Mlecnik B, Galon J, Zitvogel L, Kroemer G. Meta-analysis of organ-specific differences in the structure of the immune infiltrate in major malignancies. Oncotarget 2015; 6:11894-909; PMID:26059437; http://dx.doi.org/ 10.18632/oncotarget.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A et al.. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013; 39:782-95; PMID:24138885; http://dx.doi.org/ 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 44.Ponten F, Jirstrom K, Uhlen M. The human protein atlas–a tool for pathology. J pathol 2008; 216:387-93; PMID:18853439; http://dx.doi.org/ 10.1002/path.2440 [DOI] [PubMed] [Google Scholar]
- 45.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C et al.. A human protein atlas for normal and cancer tissues based on antibody proteomics. MCP 2005; 4:1920-32; PMID:16127175; http://dx.doi.org/25613900 10.1074/mcp.M500279-MCP200 [DOI] [PubMed] [Google Scholar]
- 46.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C et al.. Proteomics. Tissue-based map of the human proteome. Science 2015; 347:1260419; PMID:25613900; http://dx.doi.org/ 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 47.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S et al.. Towards a knowledge-based human protein atlas. Nat Biotechnol 2010; 28:1248-50; PMID:21139605; http://dx.doi.org/ 10.1038/nbt1210-1248 [DOI] [PubMed] [Google Scholar]
- 48.Heng TS, Painter MW. The immunological genome project: networks of gene expression in immune cells. Nat Immunol 2008; 9:1091-4; PMID:18800157; http://dx.doi.org/ 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- 49.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol 2011; 29:235-71; PMID:21219185; http://dx.doi.org/ 10.1146/annurev-immunol-031210-101324 [DOI] [PubMed] [Google Scholar]
- 50.Stoll G, Enot D, Mlecnik B, Galon J, Zitvogel L, Kroemer G. Immune-related gene signatures predict the outcome of neoadjuvant chemotherapy. Oncoimmunology 2014; 3:e27884; PMID:24790795; http://dx.doi.org/ 10.4161/onci.27884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoll G, Iribarren K, Michels J, Leary A, Zitvogel L, Cremer I, Kroemer G. Calreticulin expression: interaction with the immune infiltrate and impact on survival in patients with ovarian and non-small cell lung cancer. Oncoimmunology 2016; 5:e1177692; PMID:27622029; http://dx.doi.org/ 10.1080/2162402X.2016.1177692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R et al.. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 2015; 33:983-91; PMID:25534375; http://dx.doi.org/ 10.1200/JCO.2014.58.1967 [DOI] [PubMed] [Google Scholar]
- 53.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S et al.. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 2012; 72:100-11; PMID:22080568; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-1403 [DOI] [PubMed] [Google Scholar]
- 54.Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y et al.. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS One 2012; 7:e43923; PMID:23028479; http://dx.doi.org/ 10.1371/journal.pone.0043923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS et al.. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics 2008; 1:13; PMID:18442402; http://dx.doi.org/ 10.1186/1755-8794-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshihara K, Tajima A, Yahata T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H et al.. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One 2010; 5:e9615; PMID:20300634; http://dx.doi.org/ 10.1371/journal.pone.0009615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545-50; PMID:16199517; http://dx.doi.org/ 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.