Abstract
The secretory and autophagy pathways can be thought of as the biosynthetic (i.e., anabolic) and degradative (i.e., catabolic) branches of the endomembrane system. In analogy to anabolic and catabolic pathways in metabolism, there is mounting evidence that the secretory and autophagy pathways are intimately linked and that certain regulatory elements are shared between them. Here we highlight the parallels and points of intersection between these two evolutionarily highly conserved and fundamental endomembrane systems. The intersection of these pathways may play an important role in remodeling membranes during cellular stress.
INTRODUCTION
The term autophagy (“auto”: self; “phagy”: eating) was coined by Christian De Duve, who noted that cells are able to “eat” their own cytoplasmic contents by delivering them to lysosomes (De Duve and Wattiaux, 1966). Autophagy is an evolutionarily conserved process that is responsible for the degradation of superfluous or damaged proteins and organelles. Macroautophagy (hereafter referred to as autophagy) is induced by starvation. Superfluous cellular contents are sequestered in de novo–formed double-membrane–bound vesicles called autophagosomes, which mature through fusion with endomembrane vesicles and lysosomes. Ultimately, the cargo within autophagolysosomes is degraded, and metabolic intermediates are recycled. Because autophagy fulfills an important homeostatic function, it is not surprising that it plays an important role during development and that disruption of this process contributes to the pathogenesis of immune disorders, cancer, and neurodegeneration.
The basic machinery includes >30 autophagy-related (ATG) genes that govern the biogenesis and maturation of autophagosomes. Although these have been best characterized through elegant genetic, biochemical, and cell biological studies in yeast, our understanding of autophagy regulation in mammalian cells is rapidly emerging (Mehrpour et al., 2010). Various stimuli, including nutrients, energy, and/or growth factor deprivation, activate the autophagy-inducing kinase, called Atg1 in yeast and ULK1/ULK2 in mammals, at the site of autophagosome formation (Mehrpour et al., 2010). Although there are examples of ULK1/2-independent autophagy (Cheong et al., 2011; Lin and Hurley, 2016), ULK/Atg1 forms a conserved complex with ATG13 and FIP200/Atg17 and regulates multiple steps in the canonical autophagy pathway. ULK1/2 phosphorylates multiple components of the autophagy-inducing class III phosphoinositide 3 kinase (PI3K)-VPS34 complex, leading to the generation of phosphatidylinositol 3-phosphate (PI3P), the recruitment of PI3P-binding proteins, and autophagosome membrane nucleation (Russell et al., 2013; Park et al., 2016). ULK/Atg1 also promotes membrane recycling from various sources, including the trans-Golgi and recycling endosomes, via the multipass transmembrane protein Atg9 (Young et al., 2006; Yamamoto et al., 2012). Subsequently additional autophagy-related proteins, including components of the ubiquitin-like conjugation pathways that lead to lipidation of Atg8, regulate the elongation and closure of autophagosomes.
The process of transporting cargo-containing vesicles is not unique to autophagy and is also an important feature of the secretory pathway, which is responsible for the biogenesis and synthesis of one-third of the cellular proteome. After synthesis and quality control in the endoplasmic reticulum (ER), secretory proteins are exported via coat protein complex II (COPII)–coated carriers that form on ribosome-free regions of the rough ER called ER exit sites (ERES). The COPII coat consists of the Sec23-Sec24 dimer and a Sec13-Sec31 tertrameric complex (Zanetti et al., 2011). In yeast, COPII vesicles fuse directly with the Golgi, but in mammalian cells, COPII vesicles transport cargo to the Golgi via the ER–Golgi intermediate compartment (ERGIC). The bidirectional cycle of transport in the mammalian ER-Golgi system is completed by another class of coated vesicles, COPI, that transport proteins from the ERGIC to the ER.
COMPONENTS OF THE SECRETORY PATHWAY AS A MEMBRANE SOURCE FOR AUTOPHAGY
In yeast, autophagosome assembly is restricted to a discrete site that is located in close proximity to the vacuole (pre-autophagosomal structure [PAS]), whereas in mammals, autophagosome formation is initiated at multiple PI3P-enriched cup-like subdomains of the ER (Axe et al., 2008). ERES reside adjacent to the growing edge of the autophagosomal membrane (Graef et al., 2013; Suzuki et al. 2013). In addition, morphological studies revealed contact sites between nascent autophagosomes and ERES, mitochondria, the Golgi, plasma membrane, recycling endosomes, and late endosomes or lysosomes (Lamb et al., 2013; Biazik et al., 2015). The functional significance of many of these contact sites, however, has not been explored, and it is not clear whether these sites are transferring membrane or other components. In support of the importance of membrane exchange is the growing evidence that shuttling of Atg9 between the Golgi and other reservoirs to autophagosomes plays an important role in recycling membranes from these various sources upon autophagy induction (Webber et al., 2007; Yamamoto et al., 2012). There may also be differences in the role of these contacts among different species or indirect consequences of disrupting contact sites. For example, although ER–mitochondria contact sites in mammalian cells were reported to play a role in an early stage of autophagosome formation (Hamasaki et al., 2013), these findings have not been corroborated in yeast (Graef et al., 2013). It is not clear whether this represents a fundamental difference between autophagosome biogenesis in yeast and mammals or the proximity between ERES and ER–mitochondria contact sites in higher eukaryotes has added confusion to the interpretation of the data (Tan et al., 2013).
Nevertheless, the ER is a primary site for autophagosome biogenesis in both yeast and mammals (Graef et al., 2013; Lamb et al., 2013; Suzuki et al. 2013; Sanchez-Wandelmer et al., 2015). Proteomics has identified interactions between constituents of ERES and the Atg machinery in yeast (Graef et al., 2013). In mammals, superresolution microscopy has revealed that ULK1/ATG13-positive structures emerge from regions where ATG9-positive vesicles align with the ERGIC (Karanasios et al., 2016). The formation of these pre-autophagosomal structures requires COPII and COPI function (Karanasios et al., 2016). There is also evidence that ERGIC membranes support LC3/Atg8 lipidation, a key step in autophagosome formation, in vitro (Ge et al., 2013). Thus, whereas certain secretory compartments (e.g., Golgi, endosomes) may contribute membrane to autophagosomes via ATG9, early compartments of the secretory pathway (i.e., ERES and ERGIC) have a more direct role in the biogenesis of nascent autophagosomes.
MULTITASKING PROTEINS IN SECRETION AND AUTOPHAGY
Role of COPII components in the secretion–autophagy cross-talk
ERES may be a preferred site for autophagosome biogenesis because of the curved membranes that are available at ERES. Alternatively, it is possible that autophagy and the secretory pathway share common machinery that resides at ERES. The first indication that core components of the ER export machinery regulate autophagy came from the observation that yeast strains with genetic defects in certain COPII components or their regulators are unable to generate autophagosomes (Ishihara et al., 2001). More recently, several proteins, including Sar1, Ypt1/Rab1, and the CK1 kinase Hrr25, which phosphorylates the COPII coat, have been implicated in autophagy (Zoppino et al., 2010; Wang et al., 2015; Davis et al., 2016). Inhibition of the GTPase Sar1, which interacts with its membrane-bound exchange factor Sec 12 and initiates COPII vesicle transport, impairs autophagy in yeast and mammalian cells (Zoppino et al., 2010; Graef et al., 2013), implying an evolutionarily conserved dependence of autophagy on ER export and/or COPII components. In support of the former, treating mammalian cells with the ER-export inhibitor FLI-06 (Krämer et al., 2013) reduces the number of autophagosomes under starvation conditions (Karanasios et al., 2016). Moreover, a recent study in yeast demonstrated that ER export of the ER-localized Qa/t-SNARE Ufe1 is induced in response to starvation and that Ufe1 is targeted to sites containing Atg8 and Atg9, where it contributes to autophagosome formation (Lemus et al., 2016).
The COPII machinery may have a role beyond regulating the trafficking of autophagy regulators and instead may be redirected to autophagy in response to stress. For example, a recent study indicated that phosphorylation of three amino acids on the membrane-distal surface of the COPII cargo adapter Sec24 enhances the interaction of Sec24 with Atg9 at the PAS (Davis et al., 2016; Figure 1). Although phosphorylation of the distal sites on Sec24 is needed to increase autophagosome number, which facilitates cellular homeostasis during starvation, it is not required for ER-to-Golgi transport (Davis et al., 2016). The Sec24 phosphorylation sites are conserved in mammalian SEC24A, and the highly conserved kinase Hrr25 is one of the kinases that phosphorylate this site. The redirection of COPII vesicle machinery for autophagy has also been documented in mammals. Studies demonstrate that the induction of autophagy results in class III PI3 kinase–dependent relocation of ERES components to the ERGIC (Ge et al., 2014).
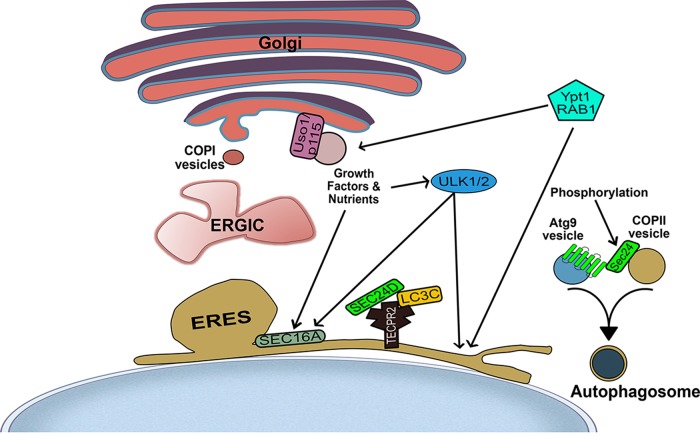
FIGURE 1:
Schematic representations of selected modes of cross-talk between autophagy and the secretory pathway. Right, the small GTPase Ypt1/Rab1 regulates the tethering of ER-derived, COPII-coated vesicles to the Golgi in rich medium. During starvation, however, Sec24 is phosphorylated, which promotes the fusion of COPII vesicles with Atg9 vesicles to regulate autophagosome abundance. Middle, growth factors and nutrients signal to the ERES regulator SEC16A. The autophagy-initiating kinases ULK1/2 are also activated by nutrient signaling and phosphorylate SEC16A. Finally (middle, bottom), the autophagy-regulating protein TECPR2 regulates COPII carrier biogenesis in a manner that depends on LC3C, thereby bridging secretion and autophagy.
TECPR2 as a bridge between the autophagy and secretory machinery
Another example of a multitasking regulator is tectonin β-propeller–containing protein 2 (TECPR2), a protein implicated in neurodegenerative diseases (Oz-Levi et al., 2012). TECPR2 binds to ATG8 (LC3 in mammals) and positively regulates autophagy (Behrends et al., 2010; Oz-Levi et al., 2012; Figure 1). ATG8 is a small, ubiquitin-like molecule that is conjugated to phosphatidylethanolamine in the growing autophagosome membrane through a cascade involving other autophagy proteins (Suzuki et al., 2001; Ichimura et al., 2004). Recently TECPR2 was found to interact with the COPII component SEC24D and to regulate the number of ERES in an LC3C-dependent manner (Stadel et al., 2015). The mechanism by which TECPR2 switches from being an ERES regulator to an autophagy regulator remains to be determined.
Rab1 and its dual role in autophagy and secretion
Rab GTPases regulate all membrane-bound intracellular trafficking pathways (Zhen and Stenmark, 2015). Therefore it is not surprising that certain Rabs (Ypt in yeast) regulate both autophagy and secretion. For example, Ypt1 activation on COPII vesicles leads to the recruitment of Uso1, which tethers the vesicles to the Golgi (Cao et al., 1998). Active Ypt1 is also required for starvation-induced autophagy and provides a means of targeting autophagy (i.e., ULK/Atg1) components to the PAS (Wang et al., 2013; Figure 1). Given the degree of evolutionary conservation in this class of proteins, it is perhaps not surprising that mammalian Ypt1 (Rab1) and Trs85, a component of the multimeric guanine nucleotide exchange factor (TRAPPIII) that activates Ypt1/Rab1, have been implicated in the regulation of autophagy (Behrends et al., 2010; Lynch-Day et al., 2010; Zoppino et al., 2010; Wang et al., 2013).
ULK1/2 are multitasking kinases that regulate autophagy and secretion
ULK1/2, known for their role in the induction and regulation of autophagy (Chan and Tooze, 2009), were recently shown to play a role in ER–Golgi traffic during normal growth (Joo et al., 2016). ULK1/2 localize to ERES and phosphorylate SEC16A (Figure 1), a scaffold protein that facilitates the biogenesis and maintenance of ERES (Sprangers and Rabouille, 2015). Of interest, the phosphorylation of SEC16A by ULK1/2 is required for the trafficking of specific cargo from the ER to the Golgi (Joo et al., 2016). Because SEC16A is a target of several kinases, its posttranslational modification may provide a means of fine-tuning the regulation of COPII transport with the availability of growth factors and nutrients (Farhan et al., 2010; Zacharogianni et al., 2011; Tillmann et al., 2015). Although inhibition of COPII transport impairs starvation-induced autophagy (Ishihara et al., 2001; Zoppino et al., 2010; Graef et al., 2013), additional studies are required to determine whether SEC16A phosphorylation by ULK1/2 is required for autophagy and to precisely elucidate how the multitasking function of SEC16A is orchestrated in the context of secretion and autophagy.
UVRAG and its dual role in autophagy and retrograde COPI transport
In the absence of cellular stress, the PI3P-binding protein UVRAG localizes to the ER, where it assembles into a RINT-1–containing tethering complex that regulates the arrival of COPI vesicles involved in Golgi-to-ER retrograde trafficking (He et al., 2013). On the induction of autophagy, UVRAG dissociates from the RINT-1 complex and associates with the class III PI3K complex to mediate the mobilization of ATG9 from the Golgi (He et al., 2013; Figure 1). UVRAG also regulates autophagosome maturation by binding to the class C Vps complex (Liang et al., 2008). Thus UVRAG is a multitasking protein whose function is altered in response to cellular stress.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
Connections between secretory traffic and autophagy are rapidly emerging, and, as discussed, we have already identified several “multitasking” proteins that participate in both pathways. Yet, the extent of cross-talk between these pathways remains to be determined, and there are many unanswered questions. For example, how do autophagy-inducing stimuli affect the secretory pathway with respect to structure, function, and molecular composition? What are the signals and molecular events that allow proteins to switch tasks in response to changes in metabolism? Do basal autophagy and starvation-induced autophagy have different dependences on ER export?
Although a few systematic screens have been performed on either the autophagy or the secretory pathway, we are far from a systematic understanding of the cross-talk between these two pathways (Farhan, 2015). A combination of RNA interference or clustered regularly interspaced short palindromic repeats (CRISPR) screening with chemical biology will be required to obtain an integrative view of the coregulation of secretory trafficking and autophagy. This is very important because alterations of the secretory pathway or autophagy have been implicated in several human diseases. For instance, defects in trafficking to and from the Golgi apparatus and alterations in Golgi structure are common in neurodegenerative disorders and may be direct or indirect consequences of certain disease-causing mutations (Joshi et al., 2015). Similarly, alterations in the autophagy-lysosomal pathway have been implicated in pathogenesis of neurodegenerative diseases (Nixon, 2013). The role of the cross-talk between autophagy and secretion in diseases is an exciting topic for future studies.
Abbreviations used:
- ATG
autophagy-related gene
- ER
endoplasmic reticulum
- ERES
ER exit sites
- ERGIC
ER-Golgi intermediate compartment
- PAS
pre-autophagosomal structure.
Footnotes
REFERENCES
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biazik J, Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy. 2015;11:439–451. doi: 10.1080/15548627.2015.1017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Tooze SA. Evolution of Atg1 function and regulation. Autophagy. 2009;5:758–765. doi: 10.4161/auto.8709. [DOI] [PubMed] [Google Scholar]
- Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Wang J, Zhu M, Stahmer K, Lakshminarayan R, Ghassemian M, Jiang Y, Miller EA, Ferro-Novick S. Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. Elife. 2016;5:e21167. doi: 10.7554/eLife.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Farhan H. Systems biology of the secretory pathway: what have we learned so far. Biol Cell. 2015;107:205–217. doi: 10.1111/boc.201400065. [DOI] [PubMed] [Google Scholar]
- Farhan H, Wendeler MW, Mitrovic S, Fava E, Silberberg Y, Sharan R, Zerial M, Hauri HP. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J Cell Biol. 2010;189:997–1011. doi: 10.1083/jcb.200912082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I, Pirooz SD, Zhao Z, Bharatham N, Li B, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Wang B, Frankel E, Ge L, Xu L, Iyengar R, Li-Harms X, Wright C, Shaw TI, Lindsten T, et al. The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol Cell. 2016;62:491–506. doi: 10.1016/j.molcel.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Bekier ME, 2nd, Wang Y. Golgi fragmentation in Alzheimer’s disease. Front Neurosci. 2015;9:340. doi: 10.3389/fnins.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. doi: 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Mentrup T, Kleizen B, Rivera-Milla E, Reichenbach D, Enzensperger C, Nohl R, Täuscher E, Görls H, Ploubidou A, et al. Small molecules intercept Notch signaling and the early secretory pathway. Nat Chem Biol. 2013;9:731–738. doi: 10.1038/nchembio.1356. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Lemus L, Ribas JL, Sikorska N, Goder V. An ER-localized SNARE protein is exported in specific COPII vesicles for autophagosome biogenesis. Cell Rep. 2016;14:1710–1722. doi: 10.1016/j.celrep.2016.01.047. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol. 2016;39:61–68. doi: 10.1016/j.ceb.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- Nixon RA ( The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Oz-Levi D, Ben-Zeev B, Ruzzo EK, Hitomi Y, Gelman A, Pelak K, Anikster Y, Reznik-Wolf H, Bar-Joseph I, Olender T, et al. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am J Hum Genet. 2012;91:1065–1072. doi: 10.1016/j.ajhg.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, Moriarity B, Kim YM, Starker C, Nho RS, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12:547–564. doi: 10.1080/15548627.2016.1140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Wandelmer J, Ktistakis NT, Reggiori F. ERES: sites for autophagosome biogenesis and maturation? J Cell Sci. 2015;128:185–192. doi: 10.1242/jcs.158758. [DOI] [PubMed] [Google Scholar]
- Sprangers J, Rabouille C. SEC16 in COPII coat dynamics at ER exit sites. Biochem Soc Trans. 2015;43:97–103. doi: 10.1042/BST20140283. [DOI] [PubMed] [Google Scholar]
- Stadel D, Millarte V, Tillmann KD, Huber J, Tamin-Yecheskel BC, Akutsu M, Demishtein A, Ben-Zeev B, Anikster Y, Perez F, et al. TECPR2 Cooperates with LC3C to regulate COPII-dependent ER export. Mol Cell. 2015;60:89–104. doi: 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Cai Y, Wang J, Zhang J, Menon S, Chou HT, Ferro-Novick S, Reinisch KM, Walz T. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci USA. 2013;110:19432–19437. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann KD, Reiterer V, Baschieri F, Hoffmann J, Millarte V, Hauser MA, Mazza A, Atias N, Legler DF, Sharan R, et al. Regulation of Sec16 levels and dynamics links proliferation and secretion. J Cell Sci. 2015;128:670–682. doi: 10.1242/jcs.157115. [DOI] [PubMed] [Google Scholar]
- Wang J, Davis S, Menon S, Zhang J, Ding J, Cervantes S, Miller E, Jiang Y, Ferro-Novick S. Ypt1/Rab1 regulates Hrr25/CK1δ kinase activity in ER-Golgi traffic and macroautophagy. J Cell Biol. 2015;210:273–285. doi: 10.1083/jcb.201408075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Menon S, Yamasaki A, Chou HT, Walz T, Jiang Y, Ferro-Novick S. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci USA. 2013;110:9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Young AR, Tooze SA. Atg9 trafficking in mammalian cells. Autophagy. 2007;3:54–56. doi: 10.4161/auto.3419. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zacharogianni M, Kondylis V, Tang Y, Farhan H, Xanthakis D, Fuchs F, Boutros M, Rabouille C. ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 2011;30:3684–3700. doi: 10.1038/emboj.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128:3171–3176. doi: 10.1242/jcs.166074. [DOI] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]