A recently identified frameshift mutation in the C-terminus of caveolin-1 is characterized. Human fibroblasts carrying this mutation proliferated twice as rapidly due to SMAD1/5/8 hyperphosphorylation. These results shed new light on the importance of the final 20 amino acids of caveolin-1.
Abstract
A heterozygous caveolin-1 c.474delA mutation has been identified in a family with heritable pulmonary arterial hypertension (PAH). This frameshift mutation leads to a caveolin-1 protein that contains all known functional domains but has a change in only the final 20 amino acids of the C-terminus. Here we studied how this mutation alters caveolin-1 function, using patient-derived fibroblasts. Transmission electron microscopy showed that fibroblasts carrying the c.474delA mutation form typical caveolae. Expression of mutated caveolin-1 in caveolin-1–null mouse fibroblasts failed to induce formation of caveolae due to retention of the mutated protein in the endoplasmic reticulum. However, coexpression of wild-type caveolin-1 with mutated caveolin-1 restored the ability to form caveolae. Importantly, fibroblasts carrying the mutation showed twofold increase in proliferation rate associated with hyperphosphorylation of Smad1/5/8. This mutation impaired the antiproliferative function of caveolin-1. Inhibition of type I TGFβ receptors ALK1/2/3/6 responsible for phosphorylation of Smad1/5/8 reduced the hyperproliferation seen in c.474delA fibroblasts. These results demonstrate the critical role of the final 20 amino acids of caveolin-1 in modulating fibroblast proliferation by dampening Smad signaling and suggest that augmented Smad signaling and fibroblast hyperproliferation are contributing factors in the pathogenesis of PAH in patients with caveolin-1 c.474delA mutation.
INTRODUCTION
Caveolae are bulb-shaped, 50- to 100-nm plasma membrane invaginations found in many cell types, including adipocytes, endothelial cells, and fibroblasts. They serve as signaling hubs and are involved in endocytosis and tension sensing (Parton and del Pozo, 2013). Caveolae can flatten in response to stretching of the plasma membrane and reduce membrane tension (Sinha et al., 2011), and caveolae in part also regulate both rapid and long-term dilation of blood vessels in response to increased shear stress (Yu et al., 2006). Caveolin and cavin families of proteins are essential for the characteristic shape of caveolae. Deletion of caveolin-1 (Cav1) leads to loss of caveolae (Drab et al., 2001), except in cardiac and skeletal muscle, in which caveolin-3 compensates for the loss (Galbiati et al., 2001). Another member of this family, caveolin-2, interacts with Cav1 but is not required to generate caveolae (Nassar and Parat, 2015). The cavin family of proteins consists of four members (cavins 1–4), which form homo- and hetero-oligomers in the cytoplasm and are recruited to caveolae, where they stabilize caveolin oligomers (Kovtun et al., 2015; Nassar and Parat, 2015). Cavin-1 appears to be especially important because it interacts with caveolin-1, and knockout of cavin-1 leads to loss of caveolae (Liu et al., 2008). The caveolar coat is believed to contain ∼150–200 caveolin subunits and 50 cavins, which function cooperatively to establish the caveolae shape (Gambin et al., 2014).
Cav1 contains an N-terminal cytoplasmic end responsible for oligomerization (amino acids 61–101; Sargiacomo et al., 1995) and a scaffolding domain (amino acids 82–101) responsible for interaction with a number of proteins, including endothelial nitric oxide synthase and transforming growth factor β (TGFβ) receptors (Li et al., 1995; Boscher and Nabi, 2012). The adjacent stretch of hydrophobic amino acids (102–134) inserts into the plasma membrane (Monier et al., 1995). The importance of the cytoplasmic C-terminal domain (amino acids 135–178) is less clear. This region appears to be required for interactions of Cav1 oligomers and binding of Cav1 to trans-Golgi membranes during posttranslational modification of the protein before export to the plasma membrane (Schlegel and Lisanti, 2000). The C-terminus also contains three palmitoylation sites (cysteines 133, 143, and 156) believed to stabilize Cav1 oligomers (Monier et al., 1996) and regulate their membrane association (Bakhshi et al., 2013).
Several germline and somatic mutations in Cav1 have been identified. A germline homozygous nonsense mutation at position 38 of Cav1 (p.Glu38X) was identified in a patient with Berardinelli–Seip congenital lipodystrophy, which is characterized by the near absence of adipose tissue (Kim et al., 2008). Hypertriglyceridemia and partial lipodystrophy affecting the upper part of the body were associated with a heterozygous nonsense mutation at position 134 of Cav1 (p.I134fsdelA-X137; Cao et al., 2008). A heterozygous c.474delA frameshift mutation, which alters the final 20 amino acids (p.P158PfsX22), was identified in multiple members of a family with heritable pulmonary arterial hypertension (PAH; Austin et al., 2012). It is unclear how the frameshift c.474delA mutation at the C-terminal of Cav1, which alters only the final 20 amino acids but leaves other domains intact, affects the function of caveolae. Because the molecular basis for the disease has not been studied, here we address the signaling abnormalities associated with this mutation, using dermal fibroblasts from the family carrying the c.474delA mutation (Austin et al., 2012). Studying this mutation might provide clues about the pathogenesis of PAH seen in these patients.
RESULTS
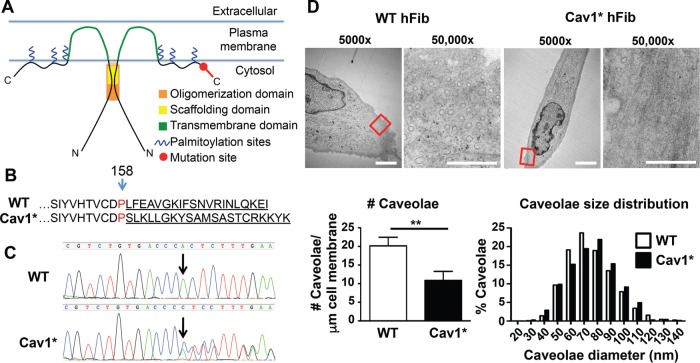
The model in Figure 1A describes specific Cav1 domains and the site of the c.474delA mutation. The resulting frameshift alters the final C-terminal 20 amino acids of Cav1 (Cav1*, Figure 1B). Genomic sequencing confirmed the presence of Cav1* c.474delA mutation in primary patient fibroblasts from these patients (Figure 1C). All patients were heterozygous for the c.474delA mutation, because the juxtaposition of one wild-type (WT) and one mutated sequence led to a double sequencing signal after the deletion. Transmission electron microscopy showed that Cav1* fibroblasts formed caveolae having a normal size distribution (mean diameter 75.3 ± 0.5 nm in control fibroblasts vs. 76.0 ± 0.7 nm in Cav1* fibroblasts); however, their number in Cav1* fibroblasts was half that in controls (Figure 1D and Supplemental Figure S1).
FIGURE 1:
Characterization of fibroblasts carrying c.474delA mutation in caveolin-1. (A) Domains of Cav1 and location of the frameshift mutation. The oligomerization domain (residues 61–101), scaffolding domain (82–101), transmembrane domain (residues 102–134), and palmitoylation sites (133, 143, 156). (B) Amino acid sequence of Cav1*. The one-base deletion of adenine 474 causes a frameshift and changes the final 20 amino acids. (C) Sequencing of Cav1 in WT and Cav1* patient fibroblasts. Arrow indicates loss of adenine 474 due to deletion. The sequence chromatogram confirmed heterozygosity of patient cells. The one-base deletion in Cav1* sequence caused a shift that resulted in juxtaposition of both sequences after the deletion site. (D) Electron microscopy of WT and Cav1* patient fibroblasts showed lower overall number of caveolae in patient fibroblasts. Size distribution of caveolae was similar in both groups. Images were obtained from 17 cells from two different WT cell lines and 18 Cav1* cells from two different patient cell lines. For size distribution, 1140 WT and 689 Cav1* caveolae were analyzed. Scale bar, 5 μm (5000×), 0.5 μm (50,000×).
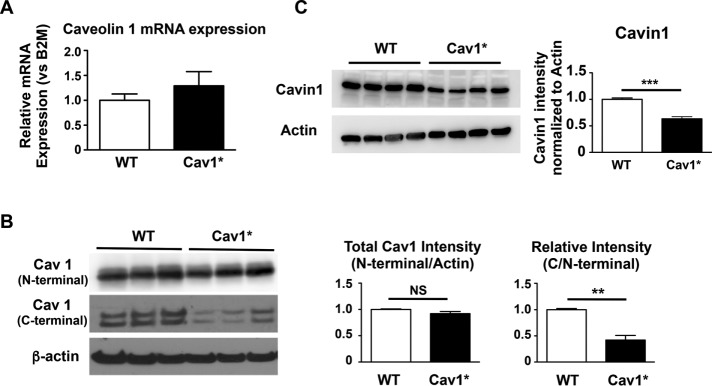
Cav1 mRNA expression was not significantly different between fibroblasts from healthy control subjects and Cav1* patients (Figure 2A). Total Cav1 protein level was investigated using an antibody against the N-terminal in WT Cav1 and Cav1*. We observed no significant change in total Cav1 protein expression in Cav1* fibroblasts (Figure 2B, top). An antibody against the C-terminal, which is changed due to the frameshift mutation in Cav1*, detected only nonmutated Cav1. In line with the sequencing data, we observed that the patient fibroblasts were heterozygous for c.474delA mutation, as they contained ∼50% nonmutated Cav1 expression compared with WT fibroblasts (Figure 2B, middle). Moreover, the expression of cavin-1 was reduced (Figure 2C). Cavin-1 expression correlates with the number of caveolae (Voldstedlund et al., 2003; Hill et al., 2008); therefore these results are in accordance with reduced caveolae numbers found by electron microscopy.
FIGURE 2:
Gene and protein expression of Cav-1 and cavin-1 in human fibroblasts. (A) Cav1 gene expression was similar among three fibroblast lines derived from control subjects and three lines derived from Cav1* patients. (B) Total Cav1 protein levels were investigated using an antibody against the N-terminal in WT Cav1 and Cav1*. There was no significant change in total Cav1 protein level in Cav1* fibroblasts. An antibody against the C-terminal, which is changed due to the frameshift mutation in Cav1*, was used to detect nonmutated/WT Cav1. Results indicated that all three patient cells were heterozygous for c.474delA mutation; that is, cells contained ∼50% nonmutated Cav1 expression compared with WT fibroblasts. (C) Cavin1 levels were significantly decreased in Cav1* human fibroblasts.
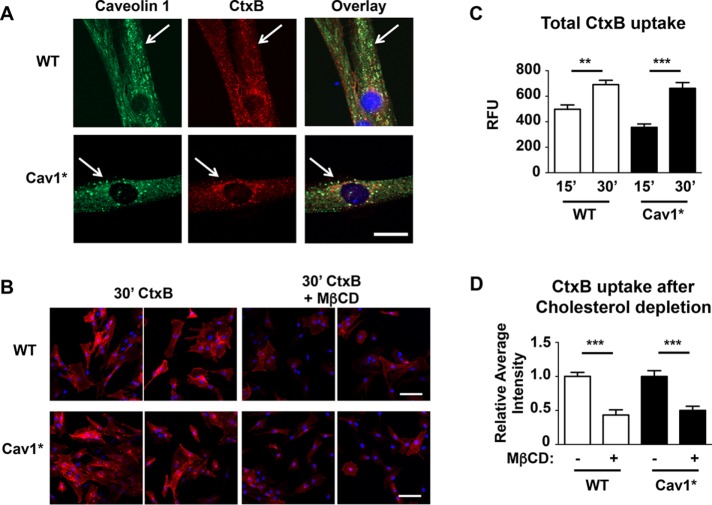
We next studied endocytosis of fluorescently labeled cholera toxin subunit B (CtxB). After an acid wash to remove extracellular CtxB, all cells showed colocalization of Cav1 and CtxB (Figure 3A), with similar uptake of CtxB in WT and Cav1* fibroblasts (Figure 3, B and C). Furthermore, extraction of cholesterol using methyl-β-cyclodextrin (MβCD) reduced the uptake of CtxB by 50% in both control and Cav1* fibroblasts (Figure 3, B and D). Caveolae fuse with early endosomes, and CtxB is subsequently transported to the Golgi apparatus (Pelkmans et al., 2004). To discriminate from clathrin-mediated endocytosis, we also looked at the uptake of CtxB in transferrin receptor (CD71)–positive endosomes. We found a similar uptake of CtxB in the Golgi apparatus of both groups (colocalization with the Golgi marker GM130; Supplemental Figure S2, A and B), with almost the entire Golgi network stained by CtxB (Supplemental Figure S2C). In contrast, low levels of endosomal labeling were seen in both groups (Supplemental Figure S2, D and E).
FIGURE 3:
Endocytosis of CtxB. (A) Human fibroblasts were incubated for 30 min with 7.5 μg/ml Alexa 594–labeled CtxB at 37°C in the presence of 0.1 mg/ml BSA, fixed, and stained for Cav1 (green). Similar colocalization and uptake were evident in both control and Cav1* fibroblasts. Scale bar, 10 μm. (B–D) Uptake of CtxB over time is similar in WT and Cav1* human fibroblasts, and cholesterol depletion by 5 mM MβCD similarly reduced CtxB uptake in WT and Cav1* human fibroblasts. For the time course, images from five independent coverslips were obtained for each group, and fluorescence intensities were analyzed by tracing the cell contour (>100 cells/group). For CtxB uptake analysis, images from eight independent coverslips were analyzed for each group (>200 cells/group). Scale bar, 100 μm.
As a control, we repeated the foregoing experiments in Cav1−/− fibroblasts obtained from Cav1−/− mice and found that MβCD treatment of these cells had no significant effect on CtxB uptake (Supplemental Figure S3A). Endosomal uptake was increased in Cav1−/− fibroblasts (Supplemental Figure S3B) perhaps as a compensation for the loss of Cav1. These results show that patient Cav1* fibroblasts did not phenocopy Cav1−/− fibroblasts.
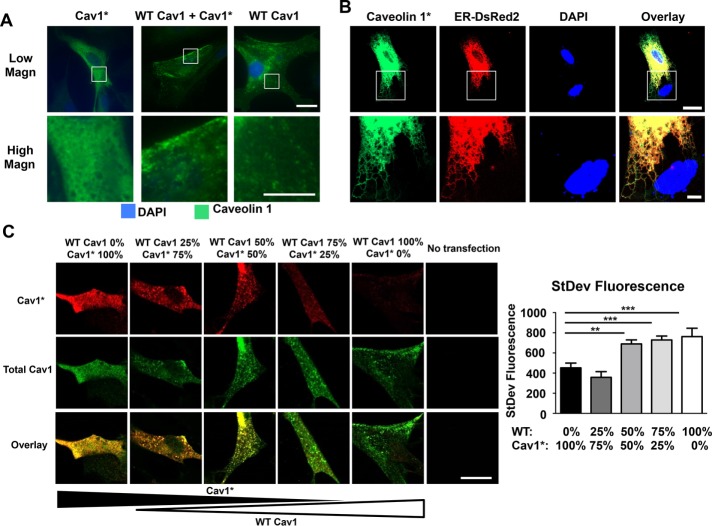
Next, to assess the functional significance of the mutant protein in Cav1* fibroblasts, we used fibroblasts from Cav1−/− mice (Zhao et al., 2009) and expressed control human WT Cav1, human Cav1*, or a combination of both to mimic heterozygous patient fibroblasts. Sequencing of constructs after site-specific mutagenesis confirmed the deletion of adenine 474 (Supplemental Figure S4A). As shown in Figure 4A, staining for Cav1 in transfected cells demonstrated that Cav1−/− fibroblasts transfected with Cav1* had a diffuse cytoplasmic staining pattern. In contrast, fibroblasts transfected with WT Cav1 or fibroblasts having 1:1 WT Cav1 and Cav1* showed a punctate staining pattern (Figure 4A) resembling the staining pattern observed in human Cav1* fibroblasts (Figure 3A).
FIGURE 4:
Dependence of caveolae formation on varying WT Cav1 and mutated Cav1* ratios. (A) Fibroblasts derived from Cav1−/− mice were transfected with either human WT Cav1 or human mutated Cav1*, followed by immunofluorescence analysis. Punctate staining was seen in fibroblasts expressing WT Cav1, whereas the pattern was diffuse after expression of Cav1*. On carrying out double transfection with both plasmids, fibroblasts showed relatively normal punctate staining pattern. Scale bar, 20 μm (top), 10 μm (bottom). (B) High-magnification view of Cav1−/− fibroblasts cotransfected with Cav1* and p-DsRed2-ER, showing localization of Cav1* in ER. Scale bar, 20 μm (top), 5 μm (bottom). (C) Cav1−/− fibroblasts were transfected with different ratios of WT Cav1 and Myc-tagged Cav1* (total amount of plasmid DNA was kept constant at 1 μg of plasmid DNA/150,000 cells), and the cells were stained with an antibody against Myc (red) and total Cav1 (green). Puncta formation was assessed by measuring the SD of the green fluorescent signal. Diffuse staining was seen in cells expressing only Cav1*, whereas higher expression of WT Cav1 led to a more heterogeneous and punctate staining. We also observed mutant Cav1* in puncta, as shown by yellow staining in cells transfected with a 1:1 ratio of Cav1 and mutant Cav1*. A higher-magnification view is shown in Supplemental Figure S4C. Scale bar, 20 μm.
To assess further the intracellular localization of Cav1* cells, we cotransfected Cav1* and endoplasmic reticulum (ER)–targeted DsRed and found that cells expressing only Cav1* showed accumulation of the protein in the ER (Figure 4B), which we did not observe in cells expressing WT Cav1 or WT Cav1 in combination with Cav1* (Supplemental Figure S4B). We also performed experiments using different ratios of WT and mutant Cav1*. We observed a diffuse staining pattern at low levels of WT Cav1 (0–25%). However, as the proportion of WT Cav1 increased relative to Cav1*, we observed distinctly more punctate staining (Figure 4C). Moreover, using a Myc-tagged Cav1* construct, we found that Cav1* has a punctate staining pattern in the presence of WT Cav1, suggesting that WT Cav1 rescues the defective diffuse staining phenotype of mutant Cav1* (Figure 4C; see Supplemental Figure S4C for higher-magnification images).
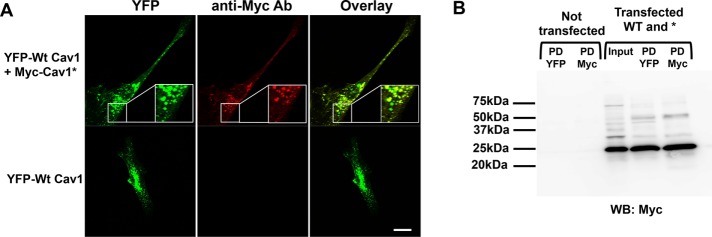
Given the apparent colocalization of WT Cav1 and mutant Cav1*, we next addressed whether there was any direct interaction between the two. For this, we generated two plasmids: WT Cav1 labeled with yellow fluorescent protein (YFP; Zimnicka et al., 2016) and Cav1* labeled with a Myc tag. As shown in Figure 5A, the YFP and Myc tag significantly overlapped, suggesting colocalization of WT and mutant Cav1*. Specificity of the Myc staining was evident by the absence of staining in cells that were only transfected with WT Cav1 (Figure 5A, bottom). Immunoprecipitation of WT Cav1 pulled down a significant amount of Myc-labeled mutant Cav1*, demonstrating that WT and mutant Cav1 indeed interacted (Figure 5B). Cav1 molecules normally form oligomers consisting of 14–16 individual proteins (Sargiacomo et al., 1995), and we found that oligomerization was not impaired in Cav1* fibroblasts, as the ratio of oligomer/monomer was identical between WT and Cav1* human fibroblasts (Supplemental Figure S5A).
FIGURE 5:
Interaction between WT and mutant Cav1. (A) Cotransfection of WT Cav1 tagged with YFP and Cav1* tagged with Myc in Cav1−/− fibroblasts confirmed that mutant Cav1* colocalized with WT Cav1. Cells transfected with YFP served as a negative control, demonstrating the specificity of Myc staining. Scale bar, 20 μm. (B) Immunoprecipitation with antibody against YFP was able to pull down Myc-labeled Cav1*, demonstrating interaction of WT and mutant Cav1*. Nontransfected cells were used as controls to exclude nonspecific bands due to light or heavy chain contamination. PD YFP, pull down of wild type Cav1; PD Myc, pull down of Cav1*.
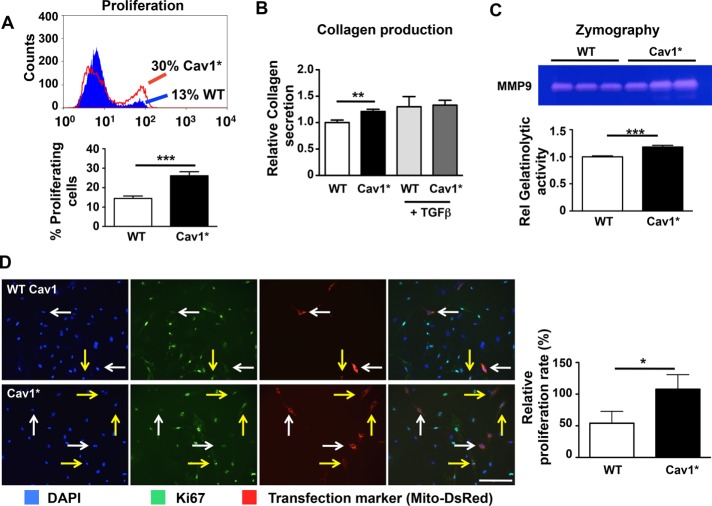
We observed a number of functional abnormalities in Cav1* human fibroblasts, with increased proliferation being the most striking. Cav1* fibroblasts proliferated at twofold greater rate than control fibroblasts (Figure 6A). In addition, Cav1* human fibroblasts had a modest increase in collagen3A1 expression (Supplemental Figure S5B) and collagen secretion (Figure 6B). Synthesis of matrix metalloproteinase 9 (MMP9) was also increased (Figure 6C), suggesting that Cav1* fibroblasts were activated. Because Cav1 is known to suppress fibroblast proliferation (Razani et al., 2001a), we next addressed whether Cav1* could also inhibit proliferation. Messenger mRNA levels revealed a similar transfection efficiency of both control Cav1 and Cav1* in Cav1−/− fibroblasts (Supplemental Figure S5C). We observed, however, that transfection of WT Cav1 decreased fibroblast proliferation by 50%, whereas transfection with Cav1* had no effect (Figure 6D).
FIGURE 6:
Functional characterization of Cav1* fibroblasts. (A) Flow cytometry was used to measure the proliferation rate of human fibroblasts. Cells were incubated with 10 μM thymidine analogue EdU for 1 h, followed by fluorescence staining of cells that incorporated EdU. Cav1* fibroblasts proliferated at a twofold higher rate than control fibroblasts. (B) Collagen secretion was increased by 20% in Cav1* human fibroblasts compared with control fibroblasts. Treatment of cells with TGFβ increased collagen secretion by 30% in control fibroblasts but had minimal effect on Cav1* fibroblasts. (C) Zymography using gelatin showed that Cav1* fibroblasts secreted greater matrix metalloproteinase 9 (MMP9 or gelatinase B) than controls, based on increased gelatinolytic band at 92 kDa in Cav1* fibroblasts vs. controls. (D) Reintroduction of human WT Cav1 in Cav1−/− fibroblasts reduced proliferation by 50%, whereas introduction of Cav1* in Cav1−/− fibroblasts did not alter proliferation. To identify transfected cells, we cotransfected cells with Mito-DsRed plasmid (red fluorescence labeling of mitochondria). Proliferation was measured using Ki67. White arrows point to transfected cells that are not proliferating; yellow arrows point to transfected proliferating cells. Scale bar, 200 μm.
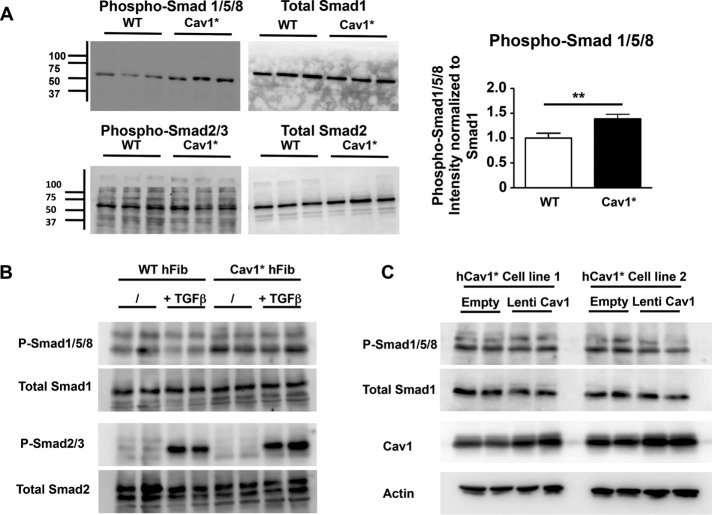
Because proliferation was markedly increased in fibroblasts from patients carrying the Cav1* mutation (Figure 6A), we addressed possible mechanisms. Members of the TGFβ family of receptors interact with Cav1 (Razani et al., 2001b). In most cases, signaling through these receptors leads to phosphorylation of mothers against decapentaplegic protein (Smad) 2/3 in response to TGFβ and activins, whereas Smad1/5/8 is phosphorylated in response to bone morphogenetic protein (BMP; Horbelt et al., 2012). At baseline, we observed marked increases in Smad1/5/8 phosphorylation in Cav1* fibroblasts, whereas Smad2/3 phosphorylation was not altered compared with controls (Figure 7A), consistent with activation of the BMP pathway. Stimulation with TGFβ increased Smad2/3 phosphorylation to a similar extent in both cell types (Figure 7B), confirming that signaling downstream of TGFβ is normal both at baseline and after stimulation. Furthermore, increasing WT Cav1 levels in human Cav1* fibroblasts did not reduce Smad1/5/8 phosphorylation (Figure 7C), arguing against Cav1 haploinsufficiency as an explanation for increased Smad1/5/8 phosphorylation.
FIGURE 7:
Increased Smad1/5/8 signaling in Cav1* fibroblasts. (A) Immunoblot analysis in control and Cav1* fibroblasts showing Smad1/5/8 hyperphosphorylation and normal Smad2/3 phosphorylation in Cav1* fibroblasts. Proteins were isolated from cells grown in DMEM plus 10%FBS. (B) Stimulation with 10 ng/ml TGFβ for 1 h similarly increased Smad2/3 phosphorylation in both cell types, confirming that signaling downstream of TGFβ is normal both at baseline and after stimulation. (C) To rescue WT Cav1 expression in human Cav1* fibroblasts, we transduced two different human Cav1* fibroblast cell lines at MOI 5, leading to doubling of Cav1 levels. We did not observe a change in phosphorylation of Smad1/5/8 in response to WT Cav1 up-regulation, arguing against haploinsufficiency as an explanation for increased phosphorylation of Cav1* patient fibroblasts.
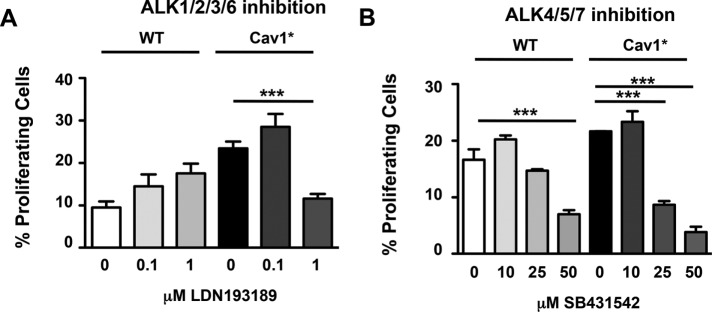
Because phosphorylation of Smad1/5/8 occurs through activin receptor–like kinase (ALK) 1/2/3/6 (Goumans et al., 2002; Schmierer and Hill, 2007), we next determined whether ALK1/2/3/6 activity was responsible for the hyperproliferation seen in Cav1* fibroblasts. We used LDN193189, an inhibitor of these ALK receptors that blocks Smad1/5/8 activation (Cuny et al., 2008). As shown in Figure 8A, LDN193189 significantly reduced proliferation of Cav1* fibroblasts but not WT cells. In contrast, SB431542, an inhibitor of ALK4/5/7 upstream of Smad2/3, equally reduced proliferation rates in both WT and Cav1* fibroblasts (Figure 8B).
FIGURE 8:
Inhibition of ALK1/2/3/6 specifically reduces proliferation of Cav1* patient fibroblasts. (A) Fibroblasts were treated with LDN193189, an inhibitor of ALK1/2/3/6, for 24 h, and the proliferation rate of fibroblasts was determined. LDN193189 (1 μM) significantly reduced proliferation of Cav1* fibroblasts to the level of WT cells at baseline. In contrast, WT fibroblast proliferation was not altered by LDN193189. (B) Experiments were also made using SB431542, an inhibitor of ALK4/5/7, which induced similar reductions in WT and Cav1* fibroblast proliferation.
DISCUSSION
Our results provide insights into the phenotype of the Cav1 c.474delA mutation of Cav1 at the C-terminus and underlying basis of PAH in these patients. The p.P158PfsX22 frameshift mutation in Cav1 occurred at the final 20 amino acids of the C-terminus, leaving the rest of the protein unaffected, specifically the Cav1 oligomerization domain (residues 61–101), scaffolding domain (82–101), and transmembrane domain (residues 102–134; Li et al., 1995). In the ER, 14–16 Cav1 proteins oligomerize through their oligomerization domain (residues 61–101; Sargiacomo et al., 1995). Subsequently, in the Golgi apparatus, several oligomers interact and combine with cholesterol to form detergent-resistant complexes, after which fully formed caveolae are transported to the plasmalemma (Hayer et al., 2010). Oligomerization in the ER is a prerequisite for successful trafficking of Cav1 to the plasma membrane (Ren et al., 2004).
We observed that the Cav1* patient fibroblasts formed normal-appearing and -sized caveolae, although their overall number was reduced by half. We found, using Cav1−/− fibroblasts, that expression of Cav1* alone resulted in the failure of fibroblasts to generate caveolae, and Cav1* was trapped in the endoplasmic reticulum. Supplying WT Cav1 to these cells expressing Cav1* rescued the trafficking defect of Cav1*. We also observed that Cav1* colocalized and interacted with WT Cav1, consistent with the finding that oligomerization appeared normal in human Cav1* fibroblasts. This is likely due to the intact oligomerization domain of Cav1*, which enabled mutant Cav1* to interact normally with WT Cav1. The reason for the reduction in number of caveolae in Cav1* fibroblasts to half of normal values is not clear. One possibility is that the frameshift mutation occurred at the final 20 amino acids of the C-terminus in proximity to palmitoylation sites interfered with formation and stability of caveolae (Schlegel and Lisanti, 2000).
We used CtxB to study endocytosis. CtxB binds to ganglioside GM1 on the cell membrane (Pang et al., 2004) and is internalized by endocytosis. Several pathways for endocytosis, including clathrin-dependent and -independent pathways, have been described. Among the clathrin-independent pathways are endocytosis mediated by caveolae and the recently described pathway clathrin-independent carriers (CLIC)/glycosylphosphatidyl inositol-anchored proteins–enriched early endosomal compartment (GEEC) pathway. Depending on the cell type and number of caveolae, different routes are used for endocytosis. For example, in fibroblasts, the majority of CtxB (∼80%) was internalized via a caveolae-dependent pathway; however, cell types with a low level of Cav1 mostly internalized CtxB through the clathrin-dependent pathway (Singh et al., 2003). Here we discriminated between clathrin-dependent and -independent endocytosis by staining for transferrin receptor CD71 (clathrin-mediated endocytosis; Supplemental Figure S2). We found that human WT and Cav1* fibroblasts primarily used a clathrin-independent pathway to internalize CtxB. A further complicating factor is that the CLIC/GEEC pathway plays an important role in the uptake of CtxB in fibroblasts (Howes et al., 2010) and this pathway is up-regulated in the absence of caveolae (Chaudhary et al., 2014). Therefore it is possible that the observed normal CtxB uptake in human Cav1* fibroblasts, despite the reduction in number of caveolae, may be related to compensatory upregulation of the CLIC/GEEC pathway.
Another important difference between Cav1* fibroblasts and WT fibroblasts was the twofold greater proliferation rate of Cav1* fibroblasts. Cav1* mutation was responsible for fibroblast hyperproliferation, because transfection of WT human Cav1 but not human Cav1* into mouse Cav1−/− fibroblasts reduced fibroblast proliferation. We also investigated the activation of signaling pathways mediating the response. We observed a marked difference between human WT and Cav1* fibroblasts in Smad signaling, as evidenced by increased phosphorylation of Smad1/5/8 in Cav1* fibroblasts. Ligands from the TGFβ family (TGFβ, activins, BMPs) induce the association of two type I receptors (ALK1 to ALK7) with two type II receptors (Schmierer and Hill, 2007) and downstream phosphorylation of Smads 1/2/3/5/8 (Schmierer and Hill, 2007). The phosphorylated Smads interact with Smad4 to activate gene transcription (Moustakas et al., 2001). Distinct type I receptors phosphorylate different Smads: ALK1/2/3/6 phosphorylate Smads1/5/8, whereas ALK 4/5/7 phosphorylate Smads2/3 (Goumans et al., 2002; Schmierer and Hill, 2007). We used LDN193189 to inhibit ALK1/2/3/6 (Cuny et al., 2008) and SB431542 to inhibit ALK4/5/7 (Inman et al., 2002). Inhibition of ALK1/2/3/6 significantly reduced proliferation of Cav1* fibroblasts compared with control fibroblasts, suggesting that ALK1/2/3/6-mediated Smad1/5/8 phosphorylation was critical in mediating Cav1* fibroblast hyperproliferation. Fibroblast hyperproliferation has been linked to the pathogenesis of PAH (Das et al., 2000), and, as our findings suggest, it may also be involved in the pathogenesis of PAH associated with c.474delA mutation of Cav1.
In summary, our findings provide the first molecular understanding of the c.474delA Cav1 mutation associated with PAH in patients. This mutation in the C-terminus of Cav1 induced Smad1/5/8 hyperphosphorylation and increased fibroblast proliferation. The results highlight an underappreciated role of the final 20 amino acids in the C-terminus of Cav1 in mediating Smad signaling and fibroblast hyperproliferation.
MATERIAL AND METHODS
Fibroblast culture
Primary skin fibroblasts from healthy subjects (n = 3) and members of the family carrying the c.474delA mutation in Cav1 (n = 3) were obtained from a punch biopsy as described previously (Austin et al., 2012). Written informed consent for all studies was obtained according to protocols approved by the institutional review board at Vanderbilt University. Fibroblasts were maintained in culture in DMEM supplemented with 10% fetal bovine serum (FBS), l-glutamine, nonessential amino acids, and penicillin/streptomycin (Life Technologies, Grand Island, NY). For studies using mouse fibroblasts, cells were isolated from tail-tip biopsies of newborn Cav1−/− mice (Zhao et al., 2009). After trypsinization for 20 min, fibroblasts were cultured in DMEM with 10% FBS, L-glutamine, nonessential amino acids, and penicillin/streptomycin.
Fibroblast proliferation assay
Proliferation was quantified by determining the incorporation of the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) with the Click-It EdU kit according to manufacturer’s instructions (Invitrogen, Carlsbad, CA). Cells were incubated with 10 μM EdU for 1 h. Specific ALK-pathway inhibitors LDN193189 (targeting the ALK1/ALK2/ALK3/ALK6 family; Cuny et al., 2008) and SB431542 (targeting the ALK4/ALK5/ALK7 family; Inman et al., 2002) were obtained from Stemcell Technologies (Vancouver, Canada). These agents were incubated with cells for 24 h before EdU administration.
Collagen production and zymography
Collagen production was measured using the QuickZyme BioSciences soluble collagen assay kit (Leiden, Netherlands). Fibroblasts (50,000) were grown for 4 d in six-well plates and washed with phosphate-buffered saline (PBS), and collagen was solubilized by adding 0.5 M acetic acid overnight. Measurements were normalized for total protein content. As positive control, cells were exposed for 4 d to 5 ng/ml recombinant human TGFβ1 (R&D Systems, Minneapolis, MN). For zymography, conditioned medium was loaded on a 10% polyacrylamide gel containing 1% bovine skin gelatin (Sigma Aldrich, St. Louis, MO). Gels were incubated for 48 h and then stained with 1% bromophenol blue; subsequently gels were destained with 1% acetic acid.
Gene expression
Total RNA was extracted using the PureLink Micro-to-midi total RNA isolation kit (Invitrogen). Reverse transcription was performed on 500 ng of RNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA). Quantification was performed on a 7900HT fast real-time PCR system (Applied Biosystems) using FastStart Universal Sybr Green master mix (Roche Applied Science, Indianapolis, IN). Primer sequences used are shown in Supplemental Table S1. Results were normalized to β2-microglobulin (B2M) and compared with control samples (2-ΔΔCt method).
Site-specific mutagenesis and sequencing
Human Cav1 was cloned into the pcDNA3 vector, and a c.474delA mutation was introduced using the QuikChange Lightning Site-Directed mutagenesis kit (Agilent, Santa Clara, CA) with specific primers (Supplemental Table S1). To confirm the mutation, Sanger sequencing was performed using 3730xl DNA Analyzer (Figure 3; Life Technologies).
Immunoblotting and immunofluorescence
Immunoblotting was performed on proteins isolated using RIPA buffer. Protein concentration was determined using the DC protein assay kit (Bio-Rad, Hercules, CA). Primary antibodies used were against C-terminal Cav1 (Ab32577), N-terminal Cav1 (Ab2910), and Ki67 (Ab16667), all purchased from Abcam (Cambridge, MA), and cavin-1 (18892-1-AP; Rosemont, IL). To confirm equal protein loading, blots were stripped in Restore Western Blot stripping buffer (ThermoFisher Scientific, Rockford, IL) for 10 min and reprobed with anti–β-actin (Sigma-Aldrich). Band intensities were quantified using ImageJ. After background subtraction, ratios were calculated (intensity of caveolin-1 band divided by intensity of corresponding actin band) and normalized to average ratio in the WT group. To assess the ratio of oligomerized Cav1 to free monomer, samples were collected in 2% n-octyl-β-d-glucopyranoside buffer and lysed using sonication before loading on SDS–PAGE gel as previously described (Bakhshi et al., 2013). For immunofluorescence stainings, cells were fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100 in PBS. After blocking with 2% bovine serum albumin (BSA) in PBS plus 0.05% Tween 20, primary antibodies were applied overnight at 4°C. Matching fluorescently labeled secondary antibodies were applied for 1 h and coverslips were mounted using ProLong Gold antifade mountant containing 4′,6-diamidino-2-phenylindole (Invitrogen). Images were obtained on a LSM710 confocal microscope (Carl Zeiss, Göttingen, Germany) using a 63×/1.4 Oil Plan-Apochromat objective and a 25-mW argon laser.
Immunoprecipitation
We cloned human Cav1 cDNA into a pcDNA3 vector (Invitrogen) containing an N-terminal Myc-His tag and introduced the c.474delA mutation using site-specific mutagenesis as described earlier. Wild-type, C-terminal, YFP-tagged caveolin-1 was generated as previously described (Chen et al., 2012). HEK293T cells were transfected with both plasmids, and after 3 d, proteins were isolated according to the Crosslink IP kit protocol (Pierce, Rockford, IL). A total of 1000 μg of proteins was immunoprecipitated using 2 μg of antibodies against either Myc tag (MA1–21316; ThermoFisher Scientific) or GFP/YPF (Ab6556; Abcam). After overnight incubation, proteins were eluted, and an immunoblot against Myc tag was performed.
Transfections
Fibroblasts were transfected with FuGENE HD transfection reagent (Promega, Madison, WI) at a ratio of 2 μg of DNA/8 μl of FuGENE according manufacturer’s recommendations. Plasmids used were either pcDNA3vectors containing human Cav1 or p-DsRed2-ER plasmid to fluorescently label endoplasmic reticulum (632409; Clontech, Mountain View, CA).
Lentiviral generation
Human WT caveolin-1 cDNA was cloned into the pLJM1 vector (19319; Addgene) using Nhe1 and EcoR1 restriction enzymes (NEB, Ipswich, MA) using the primers listed in the Supplemental Methods. These restriction enzymes also removed the GFP sequence of pLJM1. Plasmids were transfected together with pMD2.G and psPAX2 plasmids (12259 and 12260; Addgene) into HEK 293T cells to allow for lentiviral production. After 2 and 3 d, supernatant was collected, filtered through a 0.45-μm filter, and concentrated using PEG-it solution (System Biosciences, Palo Alto, CA). For transduction, 4 μg/ml Polybrene was added to the medium (Santa Cruz Biotechnology, Dallas, TX). Viral titers were determined on genomic DNA of fibroblasts 3 d after transduction using primers against the viral RRE element and the UCR111 region of human genomic DNA (Supplemental Table S1). The absolute number of infectious units was calculated using a standard provided in the Global UltraRapid Lentiviral Titer Kit (System Biosciences). Fibroblasts were infected at a multiplicity of infection (MOI) 5.
CtxB endocytosis assay
Endocytosis was assessed using CtxB. Cells were labeled with 7.5 μg/ml Alexa 594–labeled CtxB for 15 or 30 min at 37°C in the presence of 0.1 mg/ml BSA. For cholesterol depletion, cells were pretreated with 5 mM MβCD (Sigma-Aldrich) for 30 min before adding CtxB. Excess CtxB at cell surface was removed by acid stripping (30 s with 0.2 M acetic acid, 0.5 M NaCl, pH 2.5). Cells were then fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, and stained for Cav1. For quantification of CtxB uptake in the Golgi or endosomes, cells were stained with GM130 or CD71 (both from ThermoFisher Scientific), and images were obtained using a Zeiss LSM880 confocal microscope using a Plan-Apochromat 63×/1.40 oil objective. Using ImageJ, the Otsu threshold tool was used to generate a binary image that was used as a mask to select the same region in the CtxB channel. Mean pixel intensity and area were measured for the total cell and the masked region, and percentage uptake in the Golgi was subsequently calculated after subtracting background intensities. Imaging parameters and settings for the generation of binary images were identical for all images. For colocalization analysis, we used Zeiss Zen 2.1 software.
Electron microscopy
Cells were grown on 35-mm dishes, rinsed with prewarmed, serum-free DMEM, and fixed with prewarmed 2.5% glutaraldehyde in DMEM for 30 min. Samples were then washed with PBS, postfixed with 1% osmium tetroxide (in 0.1 M sodium phosphate buffer, pH 7.2–7.4) for 1 h, rinsed with the same buffer, and then dehydrated through an ascending series of ethanols (30–100%). Cell layers were infiltrated for 30 min in a 50/50 mixture of 100% ethanol/epoxy resin, followed by 100% epoxy resin (LX-112; Ladd Research, Williston, VT), after which they were placed in a 60°C oven for 2 d. Blocks with monolayer cells were thin-sectioned (70–80 nm) using an UCT ultramicrotome (Leica, Buffalo Grove, IL). Resulting sections were stained with uranyl acetate and lead citrate. Digital images were acquired using a JEOL JEM-1220 transmission electron microscope (JEOL, Peabody, MA), equipped with an Erlangshen ES1000W 11 MP charge-coupled device camera (Gatan, Pleasanton, CA). Caveolae number and size were measured using ImageJ.
Statistics
All values are given as mean ± SEM. Intergroup differences were assessed by unpaired Student’s t test or analysis of variance with post hoc analysis using Tukey’s test when multiple groups were analyzed. Wherever applicable, normality was confirmed with the Kolmogorov–Smirnov test. p < 0.05 was considered statistically significant. Significance is indicated as *** for p < 0.001, ** for p < 0.01, and * for p < 0.05.
ACKNOWLEDGMENTS
We thank the Electron Microscopy Service in the Research Resource Center of the University of Illinois at Chicago and in particular Figen Seiler for preparation and imaging of transmission electron microscopy images. We also thank Kraig Theriault for technical assistance with cell culture experiments. This work was supported in part by a Parker B. Francis Fellowship and American Heart Scientist Development Grant 15SDG23250002 (G.M.), National Institutes of Health Grant R01-GM094220 (J.R.), National Institutes of Health/National Heart, Lung, and Blood Institute Grant R01-HL111259 (E.D.A.), and National Institutes of Health Grants R01-HL045638, R01-HL090152, R01-HL118068, P01-HL060678, and P01-HL077806 (A.B.M.).
Abbreviations used:
- ALK
activin receptor–like kinase
- Cav1
caveolin 1
- CtxB
cholera toxin subunit B
- Smad
mothers against decapentaplegic protein
- TGFβ
transforming growth factor β.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.1177) on March 15, 2017.
REFERENCES
- Austin ED, Ma L, LeDuc C, Berman Rosenzweig E, Borczuk A, Phillips JA, 3rd, Palomero T, Sumazin P, Kim HR, Talati MH, et al. Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ Cardiovasc Genet. 2012;5:336–343. doi: 10.1161/CIRCGENETICS.111.961888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshi FR, Mao M, Shajahan AN, Piegeler T, Chen Z, Chernaya O, Sharma T, Elliott WM, Szulcek R, Bogaard HJ, et al. Nitrosation-dependent caveolin 1 phosphorylation, ubiquitination, and degradation and its association with idiopathic pulmonary arterial hypertension. Pulm Circ. 2013;3:816–830. doi: 10.1086/674753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscher C, Nabi IR. Caveolin-1: role in cell signaling. Adv Exp Med Biol. 2012;729:29–50. doi: 10.1007/978-1-4614-1222-9_3. [DOI] [PubMed] [Google Scholar]
- Cao H, Alston L, Ruschman J, Hegele RA. Heterozygous CAV1 frameshift mutations (MIM 601047) in patients with atypical partial lipodystrophy and hypertriglyceridemia. Lipids Health Dis. 2008;7:3. doi: 10.1186/1476-511X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary N, Gomez GA, Howes MT, Lo HP, McMahon KA, Rae JA, Schieber NL, Hill MM, Gaus K, Yap AS, Parton RG. Endocytic crosstalk: cavins, caveolins, and caveolae regulate clathrin-independent endocytosis. PLoS Biol. 2014;12:e1001832. doi: 10.1371/journal.pbio.1001832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Bakhshi FR, Shajahan AN, Sharma T, Mao M, Trane A, Bernatchez P, van Nieuw Amerongen GP, Bonini MG, Skidgel RA, et al. Nitric oxide-dependent Src activation and resultant caveolin-1 phosphorylation promote eNOS/caveolin-1 binding and eNOS inhibition. Mol Biol Cell. 2012;23:1388–1398. doi: 10.1091/mbc.E11-09-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18:4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Dempsey EC, Bouchey D, Reyland ME, Stenmark KR. Chronic hypoxia induces exaggerated growth responses in pulmonary artery adventitial fibroblasts: potential contribution of specific protein kinase c isozymes. Am J Respir Cell Mol Biol. 2000;22:15–25. doi: 10.1165/ajrcmb.22.1.3536. [DOI] [PubMed] [Google Scholar]
- Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- Galbiati F, Engelman JA, Volonte D, Zhang XL, Minetti C, Li M, Hou H, Jr, Kneitz B, Edelmann W, Lisanti MP. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J Biol Chem. 2001;276:21425–21433. doi: 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- Gambin Y, Ariotti N, McMahon KA, Bastiani M, Sierecki E, Kovtun O, Polinkovsky ME, Magenau A, Jung W, Okano S, et al. Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. Elife. 2014;3:e01434. doi: 10.7554/eLife.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. EMBO J. 2002;21:1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer A, Stoeber M, Bissig C, Helenius A. Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic. 2010;11:361–382. doi: 10.1111/j.1600-0854.2009.01023.x. [DOI] [PubMed] [Google Scholar]
- Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbelt D, Denkis A, Knaus P. A portrait of Transforming Growth Factor beta superfamily signalling: background matters. Int J Biochem Cell Biol. 2012;44:469–474. doi: 10.1016/j.biocel.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Howes MT, Kirkham M, Riches J, Cortese K, Walser PJ, Simpson F, Hill MM, Jones A, Lundmark R, Lindsay MR, et al. Clathrin-independent carriers form a high capacity endocytic sorting system at the leading edge of migrating cells. J Cell Biol. 2010;190:675–691. doi: 10.1083/jcb.201002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Kim CA, Delepine M, Boutet E, El Mourabit H, Le Lay S, Meier M, Nemani M, Bridel E, Leite CC, Bertola DR, et al. Association of a homozygous nonsense caveolin-1 mutation with Berardinelli-Seip congenital lipodystrophy. J Clin Endocrinol Metab. 2008;93:1129–1134. doi: 10.1210/jc.2007-1328. [DOI] [PubMed] [Google Scholar]
- Kovtun O, Tillu VA, Ariotti N, Parton RG, Collins BM. Cavin family proteins and the assembly of caveolae. J Cell Sci. 2015;128:1269–1278. doi: 10.1242/jcs.167866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Okamoto T, Chun M, Sargiacomo M, Casanova JE, Hansen SH, Nishimoto I, Lisanti MP. Evidence for a regulated interaction between heterotrimeric G proteins and caveolin. J Biol Chem. 1995;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- Liu L, Brown D, McKee M, Lebrasseur NK, Yang D, Albrecht KH, Ravid K, Pilch PF. Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 2008;8:310–317. doi: 10.1016/j.cmet.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S, Dietzen DJ, Hastings WR, Lublin DM, Kurzchalia TV. Oligomerization of VIP21-caveolin in vitro is stabilized by long chain fatty acylation or cholesterol. FEBS Lett. 1996;388:143–149. doi: 10.1016/0014-5793(96)00519-4. [DOI] [PubMed] [Google Scholar]
- Monier S, Parton RG, Vogel F, Behlke J, Henske A, Kurzchalia TV. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- Nassar ZD, Parat MO. Cavin family: new players in the biology of caveolae. Int Rev Cell Mol Biol. 2015;320:235–305. doi: 10.1016/bs.ircmb.2015.07.009. [DOI] [PubMed] [Google Scholar]
- Pang H, Le PU, Nabi IR. Ganglioside GM1 levels are a determinant of the extent of caveolae/raft-dependent endocytosis of cholera toxin to the Golgi apparatus. J Cell Sci. 2004;117:1421–1430. doi: 10.1242/jcs.01009. [DOI] [PubMed] [Google Scholar]
- Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Burli T, Zerial M, Helenius A. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell. 2004;118:767–780. doi: 10.1016/j.cell.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem. 2001a;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- Razani B, Zhang XL, Bitzer M, von Gersdorff G, Bottinger EP, Lisanti MP. Caveolin-1 regulates transforming growth factor (TGF)-beta/SMAD signaling through an interaction with the TGF-beta type I receptor. J Biol Chem. 2001b;276:6727–6738. doi: 10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- Ren X, Ostermeyer AG, Ramcharan LT, Zeng Y, Lublin DM, Brown DA. Conformational defects slow Golgi exit, block oligomerization, and reduce raft affinity of caveolin-1 mutant proteins. Mol Biol Cell. 2004;15:4556–4567. doi: 10.1091/mbc.E04-06-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargiacomo M, Scherer PE, Tang Z, Kubler E, Song KS, Sanders MC, Lisanti MP. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, Lisanti MP. A molecular dissection of caveolin-1 membrane attachment and oligomerization. Two separate regions of the caveolin-1 C-terminal domain mediate membrane binding and oligomer/oligomer interactions in vivo. J Biol Chem. 2000;275:21605–21617. doi: 10.1074/jbc.M002558200. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- Singh RD, Puri V, Valiyaveettil JT, Marks DL, Bittman R, Pagano RE. Selective caveolin-1-dependent endocytosis of glycosphingolipids. Mol Biol Cell. 2003;14:3254–3265. doi: 10.1091/mbc.E02-12-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, et al. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144:402–413. doi: 10.1016/j.cell.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voldstedlund M, Thuneberg L, Tranum-Jensen J, Vinten J, Christensen EI. Caveolae, caveolin and cav-p60 in smooth muscle and renin-producing cells in the rat kidney. Acta Physiol Scand. 2003;179:179–188. doi: 10.1046/j.1365-201X.2003.01183.x. [DOI] [PubMed] [Google Scholar]
- Yu J, Bergaya S, Murata T, Alp IF, Bauer MP, Lin MI, Drab M, Kurzchalia TV, Stan RV, Sessa WC. Direct evidence for the role of caveolin-1 and caveolae in mechanotransduction and remodeling of blood vessels. J Clin Invest. 2006;116:1284–1291. doi: 10.1172/JCI27100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YY, Zhao YD, Mirza MK, Huang JH, Potula HH, Vogel SM, Brovkovych V, Yuan JX, Wharton J, Malik AB. Persistent eNOS activation secondary to caveolin-1 deficiency induces pulmonary hypertension in mice and humans through PKG nitration. J Clin Invest. 2009;119:2009–2018. doi: 10.1172/JCI33338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimnicka AM, Husain YS, Shajahan AN, Sverdlov M, Chaga O, Chen Z, Toth PT, Klomp J, Karginov AV, Tiruppathi C, et al. Src-dependent phosphorylation of caveolin-1 Tyr-14 promotes swelling and release of caveolae. Mol Biol Cell. 2016;27:2090–2106. doi: 10.1091/mbc.E15-11-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]