This study indicates that MCAK contributes to the mechanosensing-mediated regulation of MT dynamics through a myosin-II–dependent mechanism that becomes uncoupled in response to 3D ECM engagement specifically within EC branches.
Abstract
Compliance and dimensionality mechanosensing, the processes by which cells sense the physical attributes of the extracellular matrix (ECM), are known to drive cell branching and shape change largely through a myosin-II–mediated reorganization of the actin and microtubule (MT) cytoskeletons. Subcellular regulation of MT dynamics is spatially controlled through a Rac1–Aurora-A kinase pathway that locally inhibits the MT depolymerizing activity of mitotic centromere–associated kinesin (MCAK), thereby promoting leading-edge MT growth and cell polarization. These results suggest that the regulation of MT growth dynamics is intimately linked to physical engagement of the cell with the ECM. Here, we tested the hypothesis that MCAK contributes to compliance and dimensionality mechanosensing-mediated regulation of MT growth dynamics through a myosin-II–dependent signaling pathway. We cultured endothelial cells (ECs) on collagen-coupled stiff or compliant polyacrylamide ECMs to examine the effects of MCAK expression on MT growth dynamics and EC branching morphology. Our results identify that MCAK promotes fast MT growth speeds in ECs cultured on compliant 2D ECMs but promotes slow MT growth speeds in ECs cultured on compliant 3D ECMs, and these effects are myosin-II dependent. Furthermore, we find that 3D ECM engagement uncouples MCAK-mediated regulation of MT growth persistence from myosin-II–mediated regulation of growth persistence specifically within EC branched protrusions.
INTRODUCTION
Cell shape, morphology, and migration behaviors are known to respond to the physical and mechanical attributes of the extracellular environment (Pelham and Wang, 1997; Wang, 1998; Hu et al., 2002; Beningo et al., 2004; Fischer et al., 2009; Kniazeva and Putnam, 2009; Bailey et al., 2011; Ghibaudo et al., 2011; Myers et al., 2011; Barthelemi et al., 2012; Mousavi et al., 2013; Aung et al., 2014; Bae et al., 2014; Edgar et al., 2014a; Underwood et al., 2014; Carey et al., 2015; Case and Waterman, 2015; Doyle et al., 2015). On compliant extracellular matrices (ECMs), cells become less rigid, extend numerous branched protrusions, and typically migrate quickly but with reduced directional persistence (Pelham and Wang, 1997; Lo et al., 2000; Myers et al., 2011). In comparison, cells cultured on less compliant ECMs generate fewer branched protrusions and display slower, more directionally persistent migration patterns (Fischer et al., 2009; Doyle and Yamada, 2015; Lee et al., 2016). In addition to matrix compliance, three-dimensional (3D) ECM engagement also contributes to significant morphological adaptations of various cell types compared with cell culture in two-dimensional (2D) ECMs (Doyle and Yamada, 2015; Kutys and Yamada, 2015; Riching and Keely, 2015). Experimental outcomes of 3D ECM engagement include enhanced cell branching and reduced migration velocities (Beningo et al., 2004; Fischer et al., 2009); these effects result from cell engagement of the ECM on both the dorsal and ventral surfaces (Fischer et al., 2009; Doyle and Yamada, 2015). Thus, experimental evidence suggests that cells cultured in a 3D ECM display an increased propensity for ECM engagement that enhances physical/mechanical cues promoting branched morphologies that are specific to a 3D environment and involve increased cell–ECM contacts (Fischer et al., 2009; Edgar et al., 2014a,b; Doyle et al., 2015; Doyle and Yamada, 2015; Thievessen et al., 2015). In turn, increased numbers of cell–ECM contacts may regulate productive, directional cell migration and contribute to the development of a polarized morphology (Doyle et al., 2009). Additionally, the combination of ECM compliance and dimensionality mechanosensing further complicates cellular navigation through a complex extracellular environment, although the effects of compliance and dimensionality contribute synergistically to branching morphology and migration behaviors (Fischer et al., 2009).
ECM compliance and dimensionality mechanosensing drive cell branching largely through reorganization of the actin–myosin and microtubule (MT) cytoskeletons (Pelham and Wang, 1997; Myers et al., 2011). Cell culture on stiff 2D ECMs promotes slow, long-lived MT growth and reduces cell branching, whereas cell culture on soft 2D ECMs promotes fast, short-lived MT growth and increases cell branching. In addition, the cellular response to matrix rigidity and density modulates force transmission and transduction via a biphasic force relationship on substrates of intermediate compliance (∼7–10 kPa), further highlighting the importance of mechanophysical signaling cues as mediators of cytoskeletal dynamics (Elosegui-Artola et al., 2016). The effects of substrate stiffness on MT growth dynamics occur through a myosin-II–dependent pathway. Of interest, when cells are cultured on 3D ECMs, MT growth persistence becomes uniformly long and is no longer sensitive to ECM stiffness-mediated down-regulation of myosin-II (Myers et al., 2011). These data suggest that engagement of a 3D ECM induces differential control of the dynamic instability of a subset of the total MT array. Further, these data suggest that distinct signaling pathways regulate MT growth speed and lifetime, depending on whether cells are cultured in 2D or 3D ECMs.
MT dynamics are regulated by families of MT-associated proteins (MAPs) capable of enhancing MT growth or promoting MT stability via inhibition of MT disassembly. The kinesin-13 family of MAPs consists of protein members that function as MT catastrophe factors or depolymerases (Howard and Hyman, 2007). The best-studied member of the kinesin-13 MAPs is mitotic centromere–associated kinesin (MCAK). MCAK functions to enzymatically catalyze MT disassembly, a process that is critical to proper spindle formation and is regulated during spindle disassembly in order to drive the correct separation of sister chromatids during mitosis (Gorbsky, 2004; Helenius et al., 2006; Knowlton et al., 2009). During interphase, MCAK activity is spatially regulated at the leading and trailing edges of endothelial cells (ECs) through a Rac1–Aurora-A kinase pathway. Within this pathway, enhanced Rac1 activation in leading-edge lamellipodia of polarized ECs results in localized activation of Aurora-A kinase, which directly targets and phosphorylates MCAK to inhibit MCAK’s MT depolymerase activity. The outcome of Aurora-A–mediated phosphoinhibition of MCAK is enhanced MT growth within the leading edge of the cell, which further promotes leading-edge polarization and directional migration of ECs during wound healing (Braun et al., 2014).
The finding that MCAK is locally inhibited within the leading edge in order to promote trailing-edge MT disassembly, coupled with the finding that 3D ECM engagement induces significant increases in MT growth lifetimes that no longer respond to ECM compliance mechanosensing, suggests that MCAK activity may be spatially controlled by ECM compliance and dimensionality-regulated signaling pathways. Here we tested the hypothesis that MCAK contributes to compliance and dimensionality mechanosensing-mediated regulation of MT growth dynamics through a myosin-II–dependent signaling pathway. Human umbilical vein endothelial cells (HUVECs) were cultured on stiff versus compliant (55 vs. 0.7 kPa) collagen-coupled polyacrylamide (PA) ECMs to examine the effects of MCAK expression on MT growth dynamics and HUVEC branching morphology. Our results show that MCAK promotes fast MT growth speeds in ECs cultured on compliant 2D ECMs but promotes reduced MT growth speeds in ECs cultured on compliant 3D ECMs. In 2D ECMs, MCAK-mediated regulation of MT growth speed and growth persistence is myosin-II dependent. Comparison of EC branches and nonbranched regions of the cell reveals that MT growth is long-lived within branches and that 3D ECM engagement uncouples MCAK-mediated regulation of MT growth persistence from myosin-II–mediated regulation of growth persistence specifically within EC branched protrusions.
RESULTS
In 2D ECMs, MCAK increases MT growth speed and eliminates the sensitivity of MT growth persistence to ECM compliance
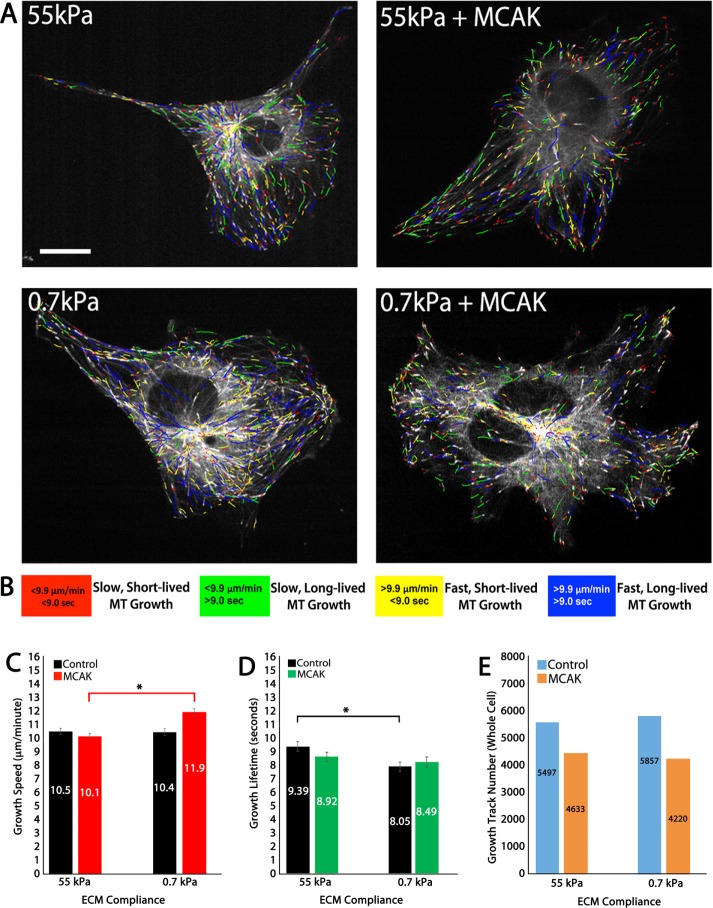
Previous studies have demonstrated that increasing the compliance (also known as softness) of the ECM results in increased MT growth speeds and reduced MT growth lifetimes (also known as persistence; Myers et al., 2011), suggesting that ECM compliance mechanosensing may function by signaling to proteins that regulate MT growth dynamics. To test the hypothesis that MCAK-mediated regulation of MT dynamics responds to ECM compliance mechanosensing, we measured MT dynamics in control HUVECs and compared them to results in HUVECs overexpressing MCAK that were cultured on either stiff (55 kPa) or soft (0.7 kPa) type 1 collagen–coupled PA ECMs (Figure 1 and Supplemental Figure S1). MT dynamics were visualized by tracking mApple EB3, a marker of MT plus-end growth, and MT growth speeds and MT growth lifetimes were analyzed using plusTipTracker software (Matov et al., 2010; Applegate et al., 2011). MT plusTipTracker software detects and tracks fluorescently labeled MT plus ends to calculate MT growth speeds and lifetimes (Figure 1, C and D). plusTipTracker also generates image overlays of MT plus-end motion tracks, with the magnitudes of MT growth speed and growth lifetime color coded to allow qualitative visualization of differences in these values throughout the cell (Figure 1, A and B). Thresholds for classifying EB3 tracks as “slow” versus “fast” or “short-lived” versus “long-lived” were based on the mean value for each parameter from the entire population of cells analyzed over all experimental conditions (Figure 1B). MT dynamics for individual cells were averaged within experimental groups, and then the means were statistically analyzed between groups (Supplemental Figure S2).
FIGURE 1:
In 2D ECMs, MCAK increases MT growth speed and eliminates the sensitivity of MT growth persistence to ECM compliance. (A) Color-coded MT growth track subpopulation overlays from 2-min time-lapse movies of mApple-EB3 (frame rate, 2 s) on representative cells for comparison between a cell cultured on 2D stiff (55 kPa) or soft (0.7 kPa) polyacrylamide gels under conditions of endogenous MCAK expression (left) or in MCAK-overexpressing HUVECs (+MCAK; right). (B) Color scheme for the four subpopulations of MT growth tracks derived by plusTipTracker software and depicted in A. (C, D) Whole-cell comparisons of mean MT growth speeds (C) and mean MT growth lifetimes (D) in cells under the conditions in A. (E) Quantification of total MT growth excursion number in cells under the conditions in A. Red and black horizontal lines designate experimental conditions meeting statistical significance. Error bars indicate SEM. For 55 kPa control, n = 9; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 10; 0.7 kPa MCAK, n = 7. *p < 0.05. Scale bar, 20 μm.
Based on this approach, comparison of control HUVECs on stiff (55 kPa) 2D type 1 collagen ECMs revealed that MCAK expression alone had no effect on MT growth speeds. Comparison of MT growth lifetimes revealed a significant reduction in control HUVECs cultured on 0.7 kPa (9.39 vs. 8.05 s; Figure 1D), a result consistent with previous investigations (Myers et al., 2011). Comparison of stiff and compliant ECMs (55 and 0.7 kPa) revealed that MCAK expression promoted fast MT growth (Figure 1, A, yellow and blue tracks, and C) and eliminated the effects of compliance mechanosensing on MT growth lifetime (Figure 1D). The number of MT growth tracks did not differ substantially in response to ECM compliance, but the number of growth tracks was reduced by MCAK expression (Figure 1E), suggesting that compliance mechanosensing alone does not alter the ability of the cell to grow MTs. Together these results suggest that MCAK increases the speed of growing MTs on soft ECMs while eliminating the sensitivity of MT growth lifetimes to ECM compliance, in addition to reducing the total number of MT growth tracks within the cell.
In 2D ECMs, MCAK-mediated effects on MT growth speed and growth persistence are myosin-II dependent
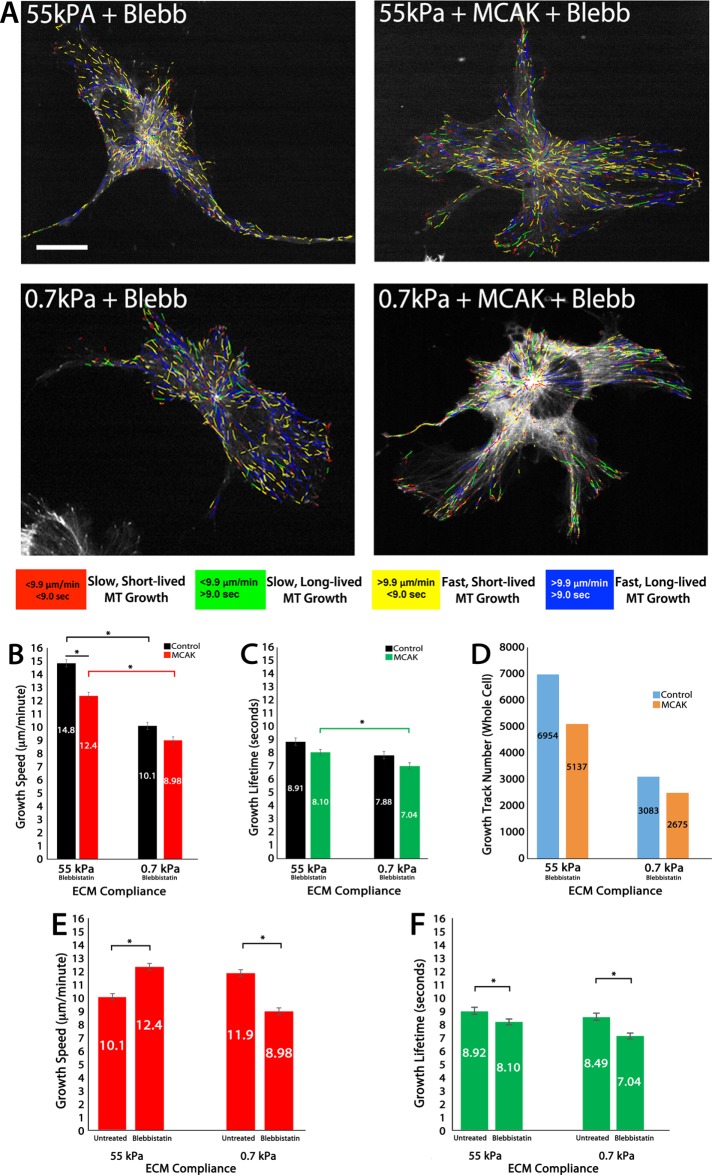
Cell engagement of compliant ECMs is known to cause the down-regulation of myosin-II contractility (Discher et al., 2005; Fischer et al., 2009), and this results in increased MT growth speeds and reduced MT growth lifetimes (Myers et al., 2011). Therefore we sought to determine the role of myosin-II contractility in MCAK-mediated regulation of MT growth dynamics in response to ECM compliance mechanosensing. To do this, we treated HUVECs with blebbistatin (20 µM) 60 min before imaging and maintained them in blebbistatin-containing medium throughout the course of the experiment. On stiff (55 kPa) ECMs, inhibition of myosin-II contractility in control HUVECs resulted in fast, short-lived MT growth (growth speed, 10.5 vs. 14.8 µm/min; growth lifetime, 9.39 vs. 8.91 s; compare Figures 1, C and D, and 2, B and C). The combination of blebbistatin treatment and MCAK expression reduced MT growth speeds in HUVECs cultured on stiff ECMs and had no significant effect on growth speeds in HUVECs cultured on soft ECMs (Figure 2B). This effect was opposite of that measured for control cells (Figure 1C), where MCAK promoted increased MT growth speeds in response to compliance mechanosensing. These data support the finding that MCAK functions to increase MT growth speeds and further suggest that myosin-II contractility is required for MCAK-mediated increases in MT growth speed.
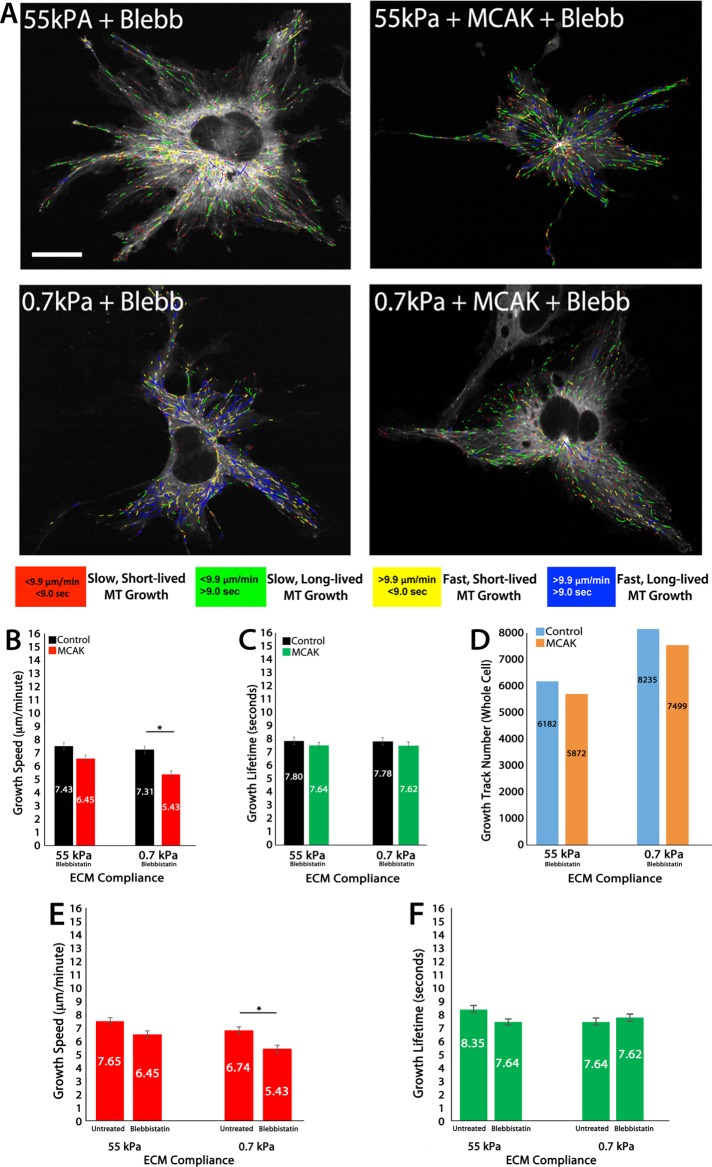
FIGURE 2:
In 2D ECMs, MCAK-mediated effects on MT growth speed and growth persistence are myosin-II dependent. (A) Color-coded MT growth track subpopulation overlays from 2-min time-lapse movies of mApple-EB3 (frame rate, 2 s) on representative cells for comparison between a cell cultured on 2D stiff (55 kPa) or soft (0.7 kPa) polyacrylamide gels under conditions of blebbistatin treatment (20 μM) with endogenous MCAK expression (+Blebb; left) or in MCAK-overexpressing HUVECs (+MCAK +Blebb; right). Color code for classification of MT growth excursion subpopulations is shown below. (B, C) Whole-cell comparisons of mean MT growth speeds (B) and mean MT growth lifetimes (C) in cells under the conditions in A. (D) Quantification of total MT growth excursion number in cells under the conditions in A. (E, F) Comparison of MT growth speeds in MCAK-expressing HUVECs cultured on stiff vs. compliant ECMs in the absence (untreated) or presence of 20 μM blebbistatin. Red, green, and black horizontal bars designate the experimental conditions meeting statistical significance. Error bars indicate SEM. For 55 kPa control, n = 11; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 6; 0.7 kPa MCAK, n = 7 (blebbistatin treated in B–D); 55 kPa MCAK, n = 8; 0.7 kPa MCAK, n = 7 (untreated in E and F). *p < 0.05. Scale bar, 20 μm.
Treatment with 20 µM blebbistatin revealed that MT growth lifetimes in control cells were reduced on stiff (55 kPa) and soft (0.7 kPa) ECMs (Figures 1D and 2C), a result similar to previously published investigations (Myers et al., 2011). In the presence of myosin-II contractility, MCAK expression resulted in a loss of compliance-mediated regulation of MT growth lifetimes (Figure 1D); however, treatment with blebbistatin in MCAK-expressing HUVECs resulted in MT growth lifetimes that were similar to controls, independent of ECM compliance, yet significantly shorter-lived on soft ECMs only in MCAK-expressing cells (Figure 2C). These results suggest that MCAK normally functions by promoting fast MT growth in response to compliance mechanosensing, and eliminates the sensitivity of MT growth lifetimes to ECM compliance (Figure 1D). In the absence of myosin-II contractility, MCAK promotes fast- and short-lived (‘’dynamic”) MT growth (Figure 2, C and F). Together these results suggest that myosin-II contractility inhibits MCAK-mediated regulation of MT dynamics, resulting in slow and long-lived MT growth.
Quantification of the number of MT growth tracks revealed that blebbistatin treatment reduced the total number of MT growth tracks as stiffness is reduced in both control and MCAK-expressing cells (2D) and resulted in an increase in growth tracks on stiff ECMs that was eliminated by MCAK in the presence of blebbistatin (compare Figures 1E and 2D). These results further suggest that myosin-II contractility inhibits MCAK and in doing so may function as an important contributor to the regulation of MT assembly. Closer comparison of the effects of blebbistatin-treated and untreated cells on MT growth speeds and growth lifetimes, specifically in MCAK-expressing cells, revealed that in the presence of myosin-II contractility (Figure 2, E and F; untreated), MCAK responds to increasing compliance by promoting increasingly fast and short-lived bouts of MT growth. However, in the absence of myosin-II contractility (Figure 2, E and F; blebbistatin), MCAK responds to increasing compliance by promoting increasingly slow and increasingly short-lived bouts of MT growth. This result further supports the conclusion that MCAK-mediated effects on MT growth dynamics are responsive to ECM compliance mechanosensing and are modulated by a myosin-II–dependent signaling pathway that promotes slow and long-lived MT growth.
MCAK-mediated regulation of MT dynamics is regionally sensitive to myosin-II contractility in branched protrusions in 2D ECMs
Substrate stiffness has been shown to modulate cell branching morphogenesis (Fischer et al., 2009). In addition, regional differences in MT dynamics within cell branches and the cell body have been characterized within ECs (Myers et al., 2011). To determine the role of MCAK in regional modulation of MT dynamics on stiff versus soft substrates, we analyzed MT dynamics in cell branches in untreated and blebbistatin-treated HUVECs.
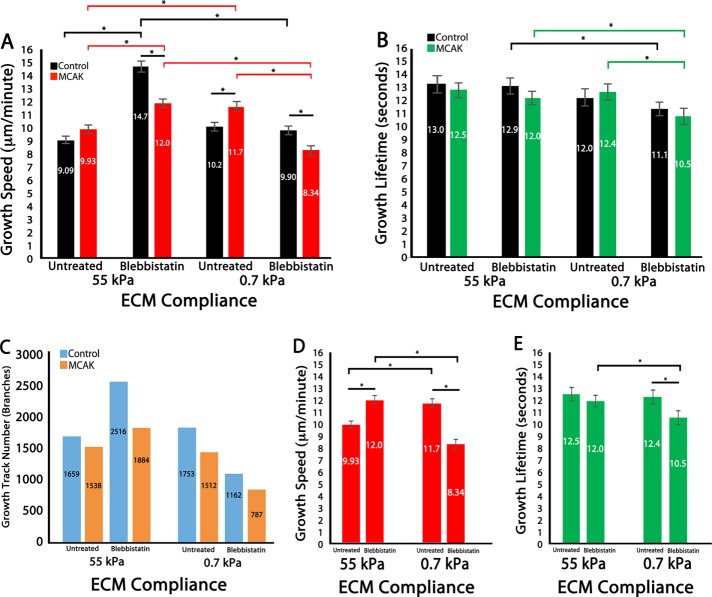
Similar to the whole-cell analysis (Figure 1), analysis of MT growth dynamics within branched regions revealed that HUVECs cultured on compliant ECMs (0.7 kPa) had significantly increased MT growth speeds compared with stiff ECMs (55 kPa) in MCAK-expressing cells (Figure 3A; untreated). Blebbistatin treatment increased MT growth speeds on stiff ECMs but not on compliant ECMs, whereas blebbistatin treatment combined with MCAK expression reduced MT growth speeds compared with MCAK expression alone on both stiff and soft ECMs. These data suggest that, within branched protrusions, MCAK-mediated regulation of MT growth speed is dependent upon myosin-II contractility.
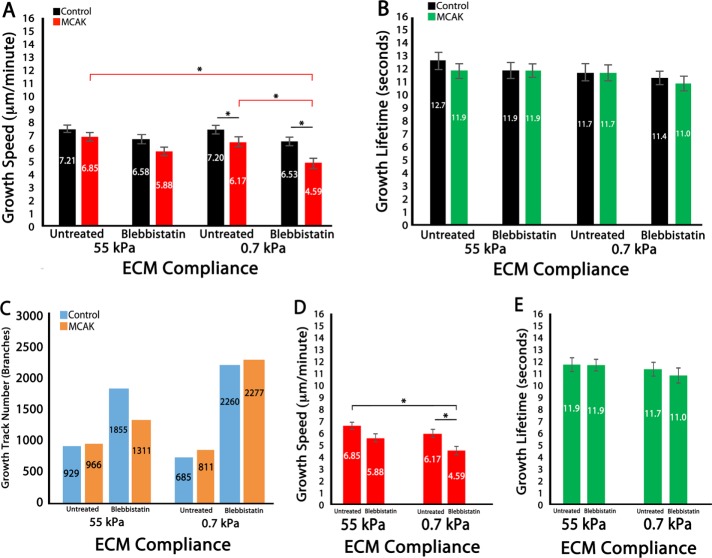
FIGURE 3:
MCAK-mediated regulation of MT dynamics is regionally sensitive to myosin-II contractility within branched protrusions in 2D ECMs. (A–E) Data analysis of HUVECs cultured on 2D polyacrylamide ECMs of varying compliance (55 and 0.7 kPa) in untreated cells or in the presence of 20 µM blebbistatin. (A, B) Mean MT growth speeds (A) and mean MT growth lifetimes (B) within branched regions of control vs. MCAK-overexpressing HUVECs. (C) MT growth track number within branched regions of HUVECs. (D, E) Comparison of MT growth speed (C) and MT growth lifetime (D) in MCAK-expressing HUVECs. Red, green, and black horizontal bars designate the experimental conditions meeting statistical significance. Error bars indicate SEM. For 55 kPa control, n = 11; MCAK, n = 8; 0.7 kPa control, n = 6; MCAK, n = 5 (blebbistatin treated); 55 kPa, control n = 9; MCAK, n = 8; 0.7 kPa control, n = 10; MCAK, n = 7 (untreated). *p < 0.05.
Analysis of MT growth lifetimes revealed that they were significantly longer lived within HUVEC branches than in the whole cell (compare Figures 1D and 3B) and also that MT growth lifetimes within branches were unaffected by ECM stiffness or MCAK expression in untreated cells. However, pharmacological inhibition of myosin-II by blebbistatin treatment caused a significant reduction in MT growth lifetimes on soft ECMs (0.7 vs. 55 kPa) that was further reduced by MCAK expression (Figure 3B). These data suggest that MCAK-mediated regulation of MT growth lifetimes in HUVEC branches is sensitive to myosin-II contractility.
Because MCAK functions as a MT-depolymerizing enzyme, it was expected that MCAK-expressing HUVECs would have a reduced number of EB3-labeled (“growing”) MTs. Analysis of total MT growth events revealed that there were fewer MT growth tracks in MCAK-expressing cells under all conditions within branched regions of the cell. In addition, total MT growth events were reduced within branches compared with the whole cell in both control and MCAK-expressing cells. This is not surprising, given that the area of cell branches is less than that of the whole cell and that MT growth events within branches are a component of the whole-cell MT growth tracks. In control HUVECs cultured on stiff ECMs, myosin-II inhibition with blebbistatin resulted in an increase in the number of growth tracks by 34.1%, whereas on soft ECMs, myosin-II inhibition reduced the number of MT growth tracks by 33.8%. Compared to untreated cells, combined myosin-II inhibition and MCAK expression increased the number of MT growth tracks on stiff ECMs (18.4%) but reduced the number of growth tracks on soft ECMs (48.1%; Figure 3C). These data are consistent with the effects of MCAK and myosin-II on whole-cell growth track number, suggesting that within EC branched protrusions, the number of MT growth events is controlled via myosin-II–dependent regulation of MT growth.
Analysis of branching morphology revealed that ECM compliance induced a fourfold increase in branch number and that MCAK inhibited this increase (reduced to 1.5-fold; Table 1). Inhibition of myosin II contractility resulted in a large increase in branch number in control cells, particularly on stiff ECMs (55 kPa, 7.5-fold; 0.7 kPa, 1.8-fold increase). After myosin-II inhibition, branch numbers were similar in control and MCAK cells, as well as on 55 kPa and 0.7 kPa ECMs. Branch lengths were similarly reduced by MCAK or blebbistatin treatment on stiff ECMs but were generally shorter and less influenced by either treatment on compliant ECMs. Thus branching morphology data suggest that MCAK-mediated regulation of MT dynamics in HUVEC branches is sensitive to myosin-II contractility.
TABLE 1:
Quantification of total branch number, fold change in branch number, and mean branch length of HUVECs cultured on 2D type 1 collagen ECMs.
| Untreated | Blebbistatin (20 µM) | |||
|---|---|---|---|---|
| Measurement | Control | MCAK | Control | MCAK |
| 2D 55 kPa ECM | ||||
| Total branch number | 2 | 4 | 15 | 13 |
| Fold change in branch number | 1 | 1 | 1 | 1 |
| Mean branch length (μm) (SEM) | 55.80 (13) | 36.24 (3) | 37.25 (1) | 30.48 (1) |
| 2D 0.7 kPa ECM | ||||
| Total branch number | 8 | 6 | 14 | 13 |
| Fold change in branch number | 4 | 1.5 | 0.93 | 1 |
| Mean branch length (μm) (SEM) | 20.90 (1) | 25.01 (2) | 25.71 (0.7) | 21.80 (0.7) |
Comparison of MCAK-expressing HUVECs revealed that blebbistatin treatment resulted in a significant increase in MT growth speeds on stiff ECMs but in a significant reduction in growth speeds within the branches of HUVECs cultured on compliant substrates (Figure 3D). MCAK’s effects on MT growth lifetimes were not responsive to myosin-II inhibition on stiff ECMs but were reduced on compliant ECMs (Figure 3E), resulting in the slowest-growing and shortest-lived MT growth excursions. These results suggest that within MCAK-expressing HUVEC branches on 2D ECMs, compliance mechanosensing–mediated down-regulation of myosin-II contractility promotes increased MT growth speeds without affecting MT growth lifetimes. In addition, these results suggest that the total loss of myosin-II contractility (blebbistatin) relieves MCAK inhibition, thereby enhancing MT dynamic instability and increasing the overall number of branched protrusions (Figure 3, D and E, and Table 1).
3D ECM engagement makes MT growth persistence insensitive to compliance and MCAK
MT dynamics exhibit differential behaviors in response to physical interactions with the ECM. Dimensionality (2D vs. 3D) modulates ECM compliance-mediated changes in MT dynamics and cell branching in distinct ways. For example, in 2D ECMs, myosin-II contractility regulates MT growth speed, growth lifetime, and branching, whereas in 3D ECMs, MT growth lifetime is regulated in a myosin-II–independent manner (Myers et al., 2011). We sought to determine whether ECM dimensionality mechanosensing effects on MT growth dynamics were mediated through a MCAK-dependent signaling pathway and whether this was dependent or independent of myosin-II contractility. We compared MT dynamics and branching morphogenesis in HUVECs grown in 3D “sandwich cultures” in which both dorsal and ventral surfaces of the cell are in contact with the ECM (Fischer et al., 2009). This type of cell culture system allows for high-resolution imaging of ECs when the ventral cell surface is engaged with ECM coupled to PA of varying stiffness (Fischer et al., 2009; Myers et al., 2011).
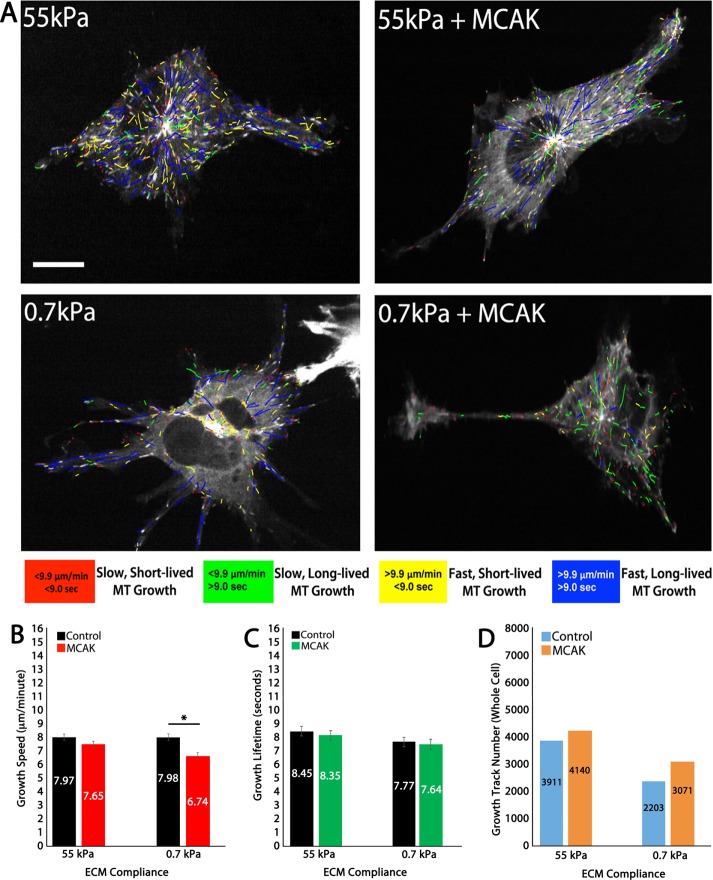
In 3D ECMs, MT growth speeds were sensitive to ECM compliance in an MCAK-dependent manner. Compared to control, on stiff (55 kPa) 3D ECMs, MCAK expression had no effect on MT growth speed or lifetime, whereas on compliant ECMs (0.7 kPa), MCAK expression resulted in significantly reduced MT growth speeds (Figure 4, A and B). MT growth lifetimes were similar on stiff and compliant ECMs in both control and MCAK-expressing ECs (Figure 4, A and C). These data support previous findings that MT growth lifetimes are insensitive to ECM compliance mechanosensing (Figure 4C; Myers et al., 2011). Here we additionally report that MT growth lifetimes are similarly insensitive to MCAK expression in 3D ECMs (Figure 4C).
FIGURE 4:
3D ECM engagement makes MT growth persistence insensitive to compliance and MCAK. (A) Color-coded MT growth track subpopulation overlays from 2-min time-lapse movies of mApple-EB3 (frame rate, 2 s) on representative cells for comparison between a cell cultured in 3D stiff (55 kPa) or soft (0.7 kPa) polyacrylamide gels in control (left) or in MCAK-overexpressing HUVECs (+MCAK; right). Color code for classification of MT growth excursion subpopulations is shown below. (B, C) Whole-cell comparisons of mean MT growth speeds (B) and mean MT growth lifetimes (C) in cells under the conditions in A. (D) Quantification of total MT growth excursion number in cells under the conditions in A. Red and black horizontal bars designate experimental conditions meeting statistical significance. Error bars indicate SEM. For 55 kPa control, n = 6; 55 kPa MCAK, n = 7; 0.7 kPa control, n = 7; 0.7 kPa MCAK, n = 10. *p < 0.05. Scale bar, 20 μm.
In control HUVECs, MT growth tracks were reduced compared with 2D results on stiff (55 kPa) and soft (0.7 kPa) ECMs, whereas in MCAK-expressing HUVECs, the number of growth tracks were similar to MCAK-expressing HUVECs in 2D (compare Figures 1D and 4D), suggesting that MCAK regulates the capacity of the cells to assemble MTs in 3D ECMs but no longer modulates MT growth lifetimes in response to ECM compliance.
In 3D ECMs, MCAK-mediated regulation of MT growth speeds and the number of MT growth events is myosin-II dependent
To determine whether the effects of MCAK on MT growth speeds and growth lifetimes in 3D ECMs were dependent on myosin-II contractility, we measured MT dynamics in HUVECs cultured in 3D sandwich gels and treated with blebbistatin (20 µM). On stiff (55 kPa) and soft (0.7 kPa) 3D ECMs, inhibition of myosin-II contractility resulted in reduced MT growth speeds (compare Figures 4B and 5B). Compared to control, MT growth speeds were significantly reduced by MCAK only on soft 3D ECMs (Figure 5B). On stiff and soft 3D ECMs, MT growth lifetimes were similar in the presence and absence of myosin-II contractility (compare Figures 4C and 5C) and were insensitive to compliance mechanosensing and MCAK expression (Figure 5C).
FIGURE 5:
In 3D ECMs, MCAK-mediated regulation of MT growth speeds and number of MT growth events are myosin-II dependent. (A) Color-coded MT growth track subpopulation overlays from 2-min time-lapse movies of mApple-EB3 (frame rate, 2 s) on representative cells for comparison between a cell cultured on 3D stiff (55 kPa) or soft (0.7 kPa) polyacrylamide gels under conditions of blebbistatin treatment (20 μM) in control (+Blebb; left) or in MCAK-overexpressing HUVECs (+MCAK +Blebb; right). Color code for classification of MT growth excursion subpopulations is shown below. (B, C) Whole-cell comparisons of mean MT growth speeds (B) and mean MT growth lifetimes (C) in cells under the conditions in A. (D) Quantification of total MT growth excursion number in cells under the conditions in A. (E, F) Comparison of MT growth speeds in MCAK-expressing HUVECs cultured on stiff vs. compliant ECMs in the absence (untreated) or presence of 20 μM blebbistatin. Black horizontal bars designate experimental conditions meeting statistical significance. Error bars indicate SEM. For 55 kPa control, n = 8; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 13; 0.7 kPa MCAK, n = 9 (blebbistatin treated in B–D); 55 kPa MCAK, n = 7; 0.7 kPa MCAK, n = 10 (untreated in E and F). *p < 0.05. Scale bar, 20 μm.
Similar to 2D ECMs, myosin-II inhibition increased the number of MT growth tracks on stiff ECMs. However, unlike in 2D, on soft 3D ECMs (Figure 5D, 0.7 kPa), myosin-II inhibition resulted in an increase in the number of MT growth tracks that was similar to control and MCAK-expressing cells (control, 267% increase; MCAK, 280% increase; compare Figures 4D and 5D). These data suggest that in 3D ECMs, MCAK-mediated inhibition of MT growth track number remains sensitive to myosin-II contractility. Taken together with MT growth lifetime data, these results suggest the possibility that, in the presence of myosin-II contractility, MCAK may be actively depolymerizing nongrowing (“stable”) MTs that would not normally be detected by tracking EB3 dynamics. Further, these results show that 3D ECM engagement inhibits MCAK-mediated depolymerization of growing MTs in a myosin-II–dependent manner.
A closer comparison of the effects of MCAK expression in untreated and blebbistatin-treated cells on MT growth speeds and growth lifetimes revealed that in the absence of myosin-II contractility, MCAK was sensitive to compliant substrates (0.7 kPa), promoting slow bouts of MT growth in blebbistatin-treated HUVECs (Figure 5E), with no effect on MT growth lifetimes (Figure 5F). These results suggest that in 3D ECMs, MCAK has no effect on MT growth lifetimes but promotes slow MT growth in response to increased ECM compliance in a myosin-II–dependent manner.
MT growth speed, but not growth persistence, is regulated through an MCAK- and myosin-II–dependent signaling pathway within branched protrusions in 3D ECMs
Analysis of MT growth dynamics within branched regions of cells revealed that HUVECs cultured on 3D stiff (55 kPa) versus compliant ECMs (0.7 kPa) had similar MT growth speeds, and MCAK expression reduced MT growth speeds on compliant ECMs, resulting in the slowest MT growth speeds on 0.7 kPa ECMs (6.17 µm/min; Figure 6A). Blebbistatin treatment reduced MT growth speeds on all ECMs, whereas blebbistatin treatment combined with MCAK expression furthered this effect, resulting in MT growth speeds that were significantly reduced compared with control untreated and control blebbistatin, specifically on soft ECMs (Figure 6A). These results suggest that similar to whole-cell growth dynamics, within branched protrusions, MCAK promotes slow MT growth in response to increased ECM compliance in a myosin-II–dependent manner.
FIGURE 6:
MT growth speed, but not growth persistence, is regulated through a MCAK- and myosin-II–dependent signaling pathway within branched protrusions in 3D ECMs. (A–E) Data analysis of HUVECs cultured on 3D polyacrylamide ECMs of varying compliance (55 or 0.7 kPa) in untreated cells or in the presence of 20 mM blebbistatin. (A, B) Mean MT growth speeds (A) and mean MT growth lifetimes (B) within branched regions of control vs. MCAK-overexpressing HUVECs. (C) MT growth track number within branched regions of HUVECs. (D, E) Comparison of MT growth speed (D) and MT growth lifetime (E) in MCAK-expressing HUVECs. Red, green, and black horizontal bars designate experimental conditions meeting statistical significance. Error bars indicate SEM. For 55 kPa control, n = 8; MCAK, n = 8; 0.7 kPa control, n = 13; MCAK, n = 9 (blebbistatin treated); 55 kPa control, n = 6; MCAK n = 7; 0.7 kPa control, n = 7; MCAK, n = 10 (untreated). *p < 0.05.
Similar to results from 2D ECMs, MT growth lifetimes were significantly longer lived within branched regions of the cell than within whole-cell MTs (compare Figures 4C and 6B). However, unlike in 2D ECMs, there was no statistically significant change in MT growth lifetimes from 3D ECM compliance mechanosensing, from MCAK expression, or in response to myosin-II inhibition by blebbistatin (Figure 6B). These data suggest that, after 3D ECM engagement, MT growth lifetimes within EC branches are regulated via a myosin-II–independent pathway.
Analysis of total MT growth events revealed that blebbistatin treatment increased the number of growth tracks on all stiffnesses and that MCAK reduced the number of growth tracks in blebbistatin-treated cells on stiff but not on soft ECMs. MT growth track number was similar within the branches of HUVECs cultured on 2D versus 3D ECMs in both control and MCAK overexpression conditions (compare Figures 3C and 6C). 3D growth tracks within branched protrusions accounted for 30.3% (55 kPa) and 27.4% (0.7 kPa) of the whole-cell MT track count.
Analysis of branching morphology in HUVECs cultured on stiff (55 kPa) versus soft (0.7 kPa) 3D ECMs demonstrated that in control untreated cells, there was a twofold increase in the number of branched protrusions on soft ECMs. On stiff ECMs, MCAK expression similarly caused a twofold increase in the number of branched protrusions, but this effect was independent of compliance mechanosensing (Table 2). In addition, MCAK expression resulted in reduced mean branch length on all ECM stiffnesses (average 2.82 μm shorter; Table 2). Blebbistatin treatment dramatically increased the total number of branches (3.5-fold on 55 kPa; 2.1-fold on 0.7 kPa), whereas MCAK expression in blebbistatin-treated cells reduced branch number, with the greatest effect on compliant ECMs (threefold reduced; Table 2). These data suggest that, in 3D ECMs, MCAK functions to inhibit the number and length of branched protrusions by promoting slow MT growth, and this function is sensitive to both compliance mechanosensing and direct inhibition of myosin-II with blebbistatin.
TABLE 2:
Quantification of total branch number, fold change in branch number, and mean branch length of HUVECs cultured on 3D type 1 collagen ECMs.
| Untreated | Blebbistatin (20 µM) | |||
|---|---|---|---|---|
| Measurement | Control | MCAK | Control | MCAK |
| 3D 55 kPa ECM | ||||
| Total branch number | 16 | 29 | 57 | 48 |
| Fold change in branch number | 1 | 1 | 1 | 1 |
| Mean branch length (μm) (SEM) | 19.66 (0.6) | 15.71 (0.5) | 25.49 (0.3) | 22.55 (0.2) |
| 3D 0.7 kPa ECM | ||||
| Total branch number | 32 | 26 | 66 | 22 |
| Fold change in branch number | 2 | 0.9 | 1 | 0.5 |
| Mean branch length (μm) (SEM) | 16.45 (0.2) | 14.77 (0.4) | 25.37 (0.2) | 18.49 (0.5) |
Within HUVEC branches in 3D ECMs, comparison of MCAK-expressing HUVECs revealed that blebbistatin treatment resulted in a significant reduction in MT growth speeds on soft ECMs (Figure 6D) and that, similar to whole-cell MT dynamics, MCAK expression did not modify MT growth lifetimes within branches (Figure 6E). These data show that following 3D ECM engagement, MCAK is ineffective in regulating MT growth lifetimes, whereas MCAK-mediated effects on MT growth speeds are regulated similarly by myosin-II contractility within the whole-cell regions and within 3D branched regions of the cell.
Compliance mechanosensing promotes fast MT growth in a MCAK-dependent manner in both 2D and 3D ECMs
To determine the contribution of MCAK to regulating MT growth dynamics in response to ECM dimensionality mechanosensing, we compared MT growth speeds and MT growth lifetimes in HUVECs cultured on stiff (55 kPa) versus compliant (0.7 kPa) 2D and 3D ECMs in untreated and blebbistatin treated conditions (Figure 7). Analysis of whole-cell MT dynamics revealed that MT growth speeds were significantly faster in 2D ECMs compared with 3D ECMs (Figure 7A). Inhibition of myosin-II contractility by treatment with blebbistatin enhanced the effects of ECM dimensionality mechanosensing by promoting the fastest MT growth speeds on 2D ECMs. However, comparison of 3D untreated versus 3D blebbistatin-treated HUVECs revealed that myosin-II inhibition had no effect on MT growth speeds on 3D ECMs, independent of compliance (compare 3D untreated, Figure 7A, with 3D blebbistatin, Figure 7B). Addition of MCAK promoted slow MT growth in all conditions except on compliant 2D ECMs in the presence of myosin-II contractility, where it promoted slightly faster MT growth speeds. Despite this increase, the measured values for growth speeds under these conditions were found to be statistically similar. These data suggest that MT growth speeds are sensitive to myosin-II contractility only in 2D ECMs, but are sensitive to MCAK in both 2D and 3D ECMs. Further, these data support a model in which MCAK-mediated regulation of MT growth speeds is myosin-II dependent in 2D ECMs and myosin-II independent in 3D ECMs.
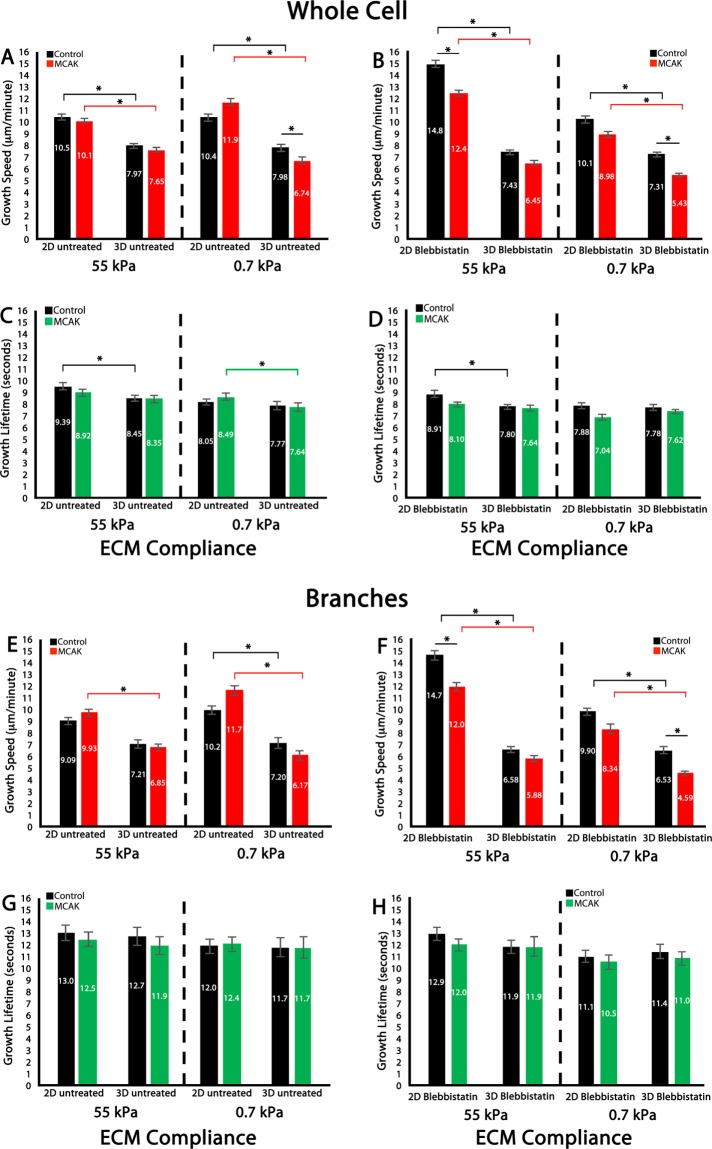
FIGURE 7:
3D ECM engagement uncouples MCAK- and myosin-II–mediated regulation of MT growth persistence within branched protrusions. (A–D) Whole-cell comparison of 2D vs. 3D MT growth speed (A, B) and MT growth lifetime (C, D) in untreated HUVECs (A, C) or HUVECs treated with 20 μM blebbistatin (B, D) in control (black bars) or in MCAK-overexpressing cells (MCAK, red or green bars). (E–H) Branched regional comparison of 2D vs. 3D MT growth speed (E, F) and MT growth lifetime (G, H) in untreated HUVECs (E, G) or HUVECs treated with 20 μM blebbistatin (F, H) in control (black bars) or MCAK overexpression (MCAK, red or green bars). Values inside the bars are mean values calculated for each condition. Red, green, and black horizontal bars designate statistical significance. Error bars indicate SEM. For 55 kPa control, n = 11; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 6; 0.7 kPa MCAK, n = 5 (whole cell, 2D, blebbistatin treatment). For 55 kPa control, n = 8; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 13, 0.7 kPa MCAK, n = 9 (whole cell, 3D blebbistatin treatment). For 55 kPa control, n = 9; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 10; 0.7 kPa MCAK, n = 7 (whole cell, 2D, untreated). For 55 kPa control, n = 6; 55 kPa MCAK, n = 7; 0.7 kPa control, n = 7; 0.7 kPa MCAK, n = 10 (whole cell, 3D, untreated). For 55 kPa control, n = 11; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 6; 0.7 kPa MCAK, n = 5 (branches, 2D, blebbistatin treatment). For 55 kPa control, n = 8; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 13; 0.7 kPa MCAK, n = 9 (branches, 3D, blebbistatin treatment). For 55 kPa control, n = 9; 55 kPa MCAK, n = 8; 0.7 kPa control, n = 10; 0.7 kPa MCAK, n = 7 (branches, 2D, untreated). For 55 kPa control, n = 6; 55 kPa MCAK, n = 7; 0.7 kPa control, n = 7; 0.7 kPa MCAK, n = 10 (branches, 3D, untreated). *p < 0.05.
Evaluation of 2D versus 3D whole-cell MT growth lifetimes revealed that growth lifetimes were the longest lived on stiff 2D ECMs and were significantly reduced via dimensionality mechanosensing on stiff 3D ECMs. This dimensionality-driven reduction in MT growth lifetimes was independent of myosin-II contractility (Figure 7, C and D). Comparison of MCAK’s effects on MT growth lifetimes in 2D and 3D ECMs revealed that MCAK expression reduced MT growth lifetimes on compliant ECMs in response to 3D ECM engagement, and this effect was myosin-II dependent (Figure 7, C and D).
3D ECM engagement uncouples MCAK-mediated regulation of MT growth persistence from myosin-II–mediated regulation of MT growth persistence within branched protrusions
Comparison of ECM compliance and dimensionality mechanosensing on EC branching morphology revealed that 3D ECM engagement resulted in a dramatic increase in total branch number (Table 3). The fold change and total branch number(s) were similarly increased in ECs on stiff ECMs (Table 3, A and B) and compliant ECMs (Table 3, C and D), and although MCAK expression did result in an increase in total branch number on stiff 3D ECMs, it did not substantially modify the fold change in branch number (Table 3, A–D, untreated). Pharmacologic inhibition of myosin-II with blebbistatin induced a large but similar relative increase in branch number in control 2D and 3D ECMs (55 kPa 2D/3D mean fold increase, 5.55; 0.7 kPa 2D/3D mean fold increase, 1.93). Blebbistatin-induced increases in branching were independent of compliance, and the combination of blebbistatin and MCAK expression had a relatively small effect of reducing branch number on 2D 55 kPa, 3D 55 kPa, or 2D 0.7 kPa ECMs (reduced 13.3, 15.8, and 7.1%, respectively). However, only on compliant 3D ECMs did the combination of blebbistatin and MCAK promote a substantial reduction in branch number (reduced 66.7%). These data suggest that EC branching is affected by MCAK only on compliant 3D ECMs in the absence of myosin-II contractility.
TABLE 3:
Comparison of total branch number, fold change in branch number, and mean branch length of HUVECs cultured on 2D (A, C) versus 3D (B, D) type 1 collagen ECMs.
| Untreated | Blebbistatin (20 µM) | |||
|---|---|---|---|---|
| Measurement | Control | MCAK | Control | MCAK |
| 55 kPa: 2D versus 3D | ||||
| A. 2D 55 kPa ECM | ||||
| Total branch number | 2 | 4 | 15 | 13 |
| Fold change in branch number | 1 | 1 | 1 | 1 |
| Mean branch length (μm) (SEM) | 55.80 (13) | 36.24 (3) | 37.25 (1) | 30.48 (1) |
| B. 3D 55 kPa ECM | ||||
| Total branch number | 16 | 29 | 57 | 48 |
| Fold change in branch number | 8 | 7 | 3.8 | 3.7 |
| Mean branch length (μm) (SEM) | 19.66 (0.6) | 15.71 (0.5) | 25.49 (0.3) | 22.55 (0.2) |
| 0.7 kPa: 2D versus 3D | ||||
| C. 2D 0.7 kPa ECM | ||||
| Total branch number | 8 | 6 | 14 | 13 |
| Fold change in branch number | 1 | 1 | 1 | 1 |
| Mean branch length (μm) (SEM) | 20.90 (1) | 25.01 (2) | 25.71 (0.7) | 21.80 (0.7) |
| D. 3D 0.7 kPa ECM | ||||
| Total branch number | 32 | 26 | 66 | 22 |
| Fold change in branch number | 4 | 4.3 | 4.7 | 1.7 |
| Mean branch length (μm) (SEM) | 16.45 (0.2) | 14.77 (0.4) | 25.37 (0.2) | 18.49 (0.5) |
Regional examination of MT growth lifetimes in EC branches revealed that the effects of ECM dimensionality mechanosensing were similar on comparing the whole cell with HUVEC branched regions (Figure 7). Within branches, MT growth speeds were reduced by engagement of stiff or soft 3D ECMs. In 2D branches, MCAK expression promoted faster MT growth speeds, whereas in 3D branches, MCAK expression had no effect on MT growth speeds (Figure 7E). Treatment with blebbistatin caused MT growth speeds to increase by an average of 38.1% in 2D ECMs, and MCAK expression inhibited the blebbistatin-induced increase in MT growth speeds (Figure 7, E and F), a result that mimicked the whole-cell MT growth speed data.
MT growth lifetimes within branches were extremely long and uniform under all conditions measured (Figure 7, G and H). This result was unlike the whole-cell result, where 3D ECM engagement reduced MT growth lifetimes in an ECM compliance–dependent manner. Also dissimilar to whole-cell analysis of MT growth lifetimes was the finding that MCAK-expressing cells were insensitive to myosin-II inhibition within branched protrusions (Figure 7H). Together these results suggest that within EC branched protrusions, the regulation of MT growth lifetimes is independent of MCAK and myosin-II in both 2D and 3D ECMs, whereas the speed of MT assembly depends on both MCAK and myosin-II in 2D ECMs but is independent of myosin-II on 3D ECM engagement.
DISCUSSION
The physical and mechanical attributes of the ECM affect cellular morphology and cytoskeletal behaviors. Substrate compliance and dimensionality are known to induce changes in MT dynamic instability that, in turn, modify cell-branching morphology and EC migration (Myers et al., 2011). Control of cell shape is mediated by both the MT cytoskeleton and actin-myosin contractility (Wang, 1998; Dent and Kalil, 2001; Rodriguez et al., 2003; Hasaka et al., 2004; Myers et al., 2006). Global inhibition of myosin-II promotes excessive cell branching and inhibits directional migration (Connolly et al., 2002; Myers et al., 2011; Elliott et al., 2015), whereas localized myosin-II inhibition is sufficient to promote branch initiation (Fischer et al., 2009). In addition, myosin-II is down-regulated in response to 3D ECM engagement, as well as in response to increased ECM compliance, such that on compliant 3D ECMs, cells have the greatest number of branched protrusions (Fischer et al., 2009; Myers et al., 2011; Doyle et al., 2015; Petrie and Yamada, 2015). Additionally, MT growth speeds and growth lifetimes are sensitive to myosin-II when HUVECs engage 2D ECMs; however, engagement of a 3D ECM results in MT growth lifetimes that are myosin-II independent (Myers et al., 2011).
It is well understood that MT dynamics are controlled by association with MAPs that function to modulate MT growth and disassembly through a wide array of activities, the majority of which have been characterized and have allowed the organization of MAPs into distinct functional families (Avila et al., 1994; Mandelkow and Mandelkow, 1995; Marx et al., 2006; Howard and Hyman, 2007; Yu et al., 2015; Nogales and Zhang, 2016). MCAK, a kinesin-13 MAP responsible for inducing MT disassembly, associates with growing MT plus ends, leading to questions about how MCAK’s MT-depolymerizing activity is regulated (Kline-Smith and Walczak, 2002; Ovechkina et al., 2002; Andrews et al., 2004; Moore and Wordeman, 2004; Moore et al., 2005; Moores et al., 2006; Manning et al., 2007; Zhang et al., 2007, 2008; Hertzer and Walczak, 2008; Braun et al., 2014). Recent evidence shows that the depolymerase activity of MCAK is spatiotemporally inhibited within the leading edge but not within the trailing edge of polarized wound-edge HUVECs in order to drive polarized MT growth toward the leading edge (Braun et al., 2014). The finding that MCAK is locally inhibited to establish polarized regulation of MT growth dynamics led us to investigate the effects of MCAK on MT dynamics as they engage ECMs of varying compliance and dimensionality.
By evaluating MT growth dynamics and HUVEC branching morphology on stiff (55 kPa) versus compliant (0.7 kPa) 2D and 3D collagen-coupled PA ECMs, we sought to determine whether ECM mechanosensing-induced control of MT growth dynamics is regulated by the MT-depolymerizing enzyme MCAK and to determine whether MCAK-dependent regulation of MT growth dynamics functions through a myosin-II–dependent or –independent mechanism. First, we demonstrated that MCAK is a target of ECM mechanosensing-mediated signaling in response to both ECM compliance and ECM dimensionality. Second, we identified that MCAK is a MAP that is responsible for distinguishing between the previously identified myosin-II–dependent and myosin-II–independent regulation of MT growth lifetimes (Myers et al., 2011). Third, we showed that MT growth speeds, but not growth lifetimes, are regulated through an MCAK- and myosin-II–dependent signaling pathway within EC branched protrusions in 3D ECMs. Fourth, we found that compliance mechanosensing promotes fast MT growth and is MCAK-dependent in both 2D and 3D ECMs. Finally, we demonstrated that 3D ECM engagement uncouples MCAK-mediated regulation of MT growth persistence from myosin-II–mediated regulation of MT growth persistence within branched protrusions. Thus, one important mechanism of compliance and dimensionality mechanosensing–mediated regulation of MT growth dynamics is achieved through myosin-II–dependent regulation of MCAK, which functions to guide branching morphogenesis in response to HUVEC engagement of physically complex ECMs.
These results provide insight into how ECM engagement is able to transmit physical, mechanosensitive signaling from the actin-myosin–sensing components of the cell to at least one component of the MT-associated regulatory machinery—in this case, MCAK. On 2D ECMs, compliance mechanosensing in the presence of MCAK results in fast MT growth, with no effect on MT growth persistence. There is also a large reduction in MT growth track number, suggesting that in addition to regulating MT growth speeds, MCAK may be working to prevent new MT growth. Inhibition of myosin-II with blebbistatin results in slow and short-lived MT growth that is MCAK dependent, suggesting that myosin-II contractility is important for MCAK’s effects on MT growth dynamics during EC compliance mechanosensing in 2D collagen ECMs.
Comparison of compliance mechanosensing in ECs cultured in 2D ECMs and in ECs cultured in 3D ECMs reveals that MCAK promotes compliance-dependent changes in MT growth speed but does not significantly affect MT growth lifetimes, suggesting that MCAK-mediated regulation of MT growth speed is sensitive to ECM compliance, but MCAK-mediated regulation of MT growth persistence is insensitive to ECM compliance. Because MCAK is a MT-depolymerizing enzyme, we expected that the primary effects of MCAK manipulation would be effects on the lifetime of MT growth rather than effects on the speed of MT growth. Indeed, using in vitro reconstitution assays, previous studies showed that titration of MCAK expression does not dramatically influence MT growth speed (Montenegro Gouveia et al., 2010; Gardner et al., 2011). Similar to these in vitro findings, we find that in living cells cultured on the stiffest 2D ECMs (55 kPa), MCAK expression does not modify MT growth speed (Figure 1, A and C). However, when cells are forced to adapt to ECMs of increasing compliance and/or dimensionality, we find that MT growth speeds are dramatically altered by MCAK expression. These data support the notion that ECM compliance mechanosensing controls MT growth dynamics through a MCAK-dependent signaling pathway.
Our data also reveal that the consequences of MCAK expression on MT growth speeds are antagonistically modified when ECM compliance is modulated in 3D ECMs. When HUVECs engage a 2D collagen ECM, for example, MT growth speeds increase in response to increasing ECM compliance in a MCAK-dependent manner (Figure 1). However, when HUVECs engage a 3D collagen ECM, MCAK causes reduced MT growth speeds in response to increasing ECM compliance (Figure 4). This trend is consistent when comparing 2D versus 3D MT dynamics within the whole cell and within cell branches, despite branches having significantly longer-lived MT growth under all conditions of ECM compliance and dimensionality (Figures 3, 6, and 7).
Comparison of dimensionality mechanosensing in 2D versus 3D ECMs (Figure 7) reveals that MCAK is sufficient to reduce MT growth lifetimes after 3D ECM engagement on compliant substrates and that this effect is eliminated by pharmacologic inhibition of myosin-II. The finding that the MCAK-dependent effect is specific to the cell body and is eliminated within HUVEC branches in either the presence or absence of myosin-II contractility (Figure 7, C and G vs. D and H) suggests that 3D ECM engagement uncouples MCAK-mediated regulation of MT growth persistence from myosin-II–mediated regulation of MT growth persistence within branched protrusions. These data support the notion that MCAK is a critical regulator of regional MT dynamic instability. Further, these data suggest that MCAK-mediated regulation of MT dynamics distinguishes MTs within the cell body and within the cell branches and uniquely regulates MT dynamic instability within these subcellular locations in response to cell engagement of the ECM.
Regional analysis of MT growth persistence also revealed that MT growth lifetimes are significantly longer lived within cell branches under all conditions of compliance, dimensionality, or myosin-II inhibition. These data support previous studies of MT growth lifetimes within the branches of ECs (Myers et al., 2011), but also suggest that persistent MT growth within EC branches is not solely dependent on myosin-II contractility, nor is it entirely dependent on MCAK-mediated regulation of MT growth speeds or growth lifetimes. We posit that additional MAPs are likely involved in mediating ECM mechanosensing-induced regulation of MT growth lifetimes during endothelial cell angiogenesis. An important study recently identified that the effects of MT-destabilizing enzymes (MCAK and Kif18A) have opposite time-dependent effects on MT growth dynamics in short-term versus longer-term inhibition experiments, and these differences result primarily from feedback responses on tubulin autoregulation (Wordeman et al., 2016). These findings suggest that in the absence (knockdown) of MT destabilizers, cells are able to adapt MT assembly dynamics through modified regulation of tubulin synthesis, which is likely to have effects on any number of MAPs that contribute to the regulation of MT dynamic instability. Because the experiments described in the present study fall within the short-term time frame of MCAK modulation, they should not be influenced by modified tubulin synthesis. Nevertheless, it remains possible and perhaps even likely that other MAPs and their effects on MT dynamic instability may be modulated via ECM compliance and dimensionality mechanosensing, as well as by short-term modulation of MCAK and myosin-II. Future investigations are necessary to determine whether and which other MAPs are involved in modulating MT growth dynamics with regional specificity and determine how upstream signaling from myosin-II is controlled to coordinate additional MAP-mediated regulation of MT dynamics.
MATERIALS AND METHODS
Cells and DNA expression constructs
HUVECs were cultured in endothelial cell basal medium (EBM) supplemented with EGM-MV Single Quots (Lonza) and penicillin-streptomycin (Fisher) and maintained at 37°C in 5% CO2. Transfection with GFP-WT-MCAK and mApple-EB3 cDNAs (final concentration, 1 µg/µl) was completed using Amaxa Cell Line Nucleofector Kit V for HUVECs (Lonza), setting A-034, and experiments were performed 3–4 h later. Transfected cells (300,000–400,000 cells) were seeded in a 35-mm Petri dish (Corning) and incubated for 1.5 h to select healthy living cells from cellular debris and dead cells that were the product of the transfection procedure. Cells were treated with (−)-blebbistatin (20 µM; Cayman Chemicals) or (+)-blebbistatin (control) in dimethyl sulfoxide (DMSO; 0.001%) for 60 min before imaging and maintained in blebbistatin-containing medium plus 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) throughout imaging. Blebbistatin-containing medium and blebbistatin-treated samples were protected from light to maintain the pharmacological activity of blebbistatin and avoid phototoxicity from light-induced toxic byproducts (Kolega, 2004; Sakamoto et al., 2005).
2D and 3D cell culture
Preactivation of coverslips and polyacrylamide conjugation.
Square glass coverslips, 22 × 22 mm (No. 1.5; Corning), were first activated with 0.5% 3-aminopropyltrimethoxysilane (Acros-organics) in double-distilled (dd) H2O for 10 min, washed by immersion in ddH20 (6×), and allowed to sit in ddH2O for 10 min. Coverslips were dried in a 37°C incubator and allowed to cool to room temperature. Coverslips were then incubated in 0.5% glutaraldehyde aqueous solution (Fisher) in Dulbecco’s phosphate-buffered saline (DPBS; Life Technologies) for 30 min, washed in ddH2O (3×, 10 min/wash), and dried at room temperature.
Coverslips were then cross-linked to PA of stiff or compliant elastic moduli (55 or 0.7 kPa, respectively). The various PA elastic moduli were prepared by varying concentrations of 40% acrylamide (Bio-Rad) and 2% Bis (Bio-Rad) in ddH2O before the addition of 10% ammonium persulfate (0.1 g/ml ddH2O; Sigma) and N,N,N’,N’-tetramethylethylenediamine (Sigma-Aldrich) as detailed in Tables 4 and 5.
TABLE 4:
Recipe 1: polyacrylamide stock solution.
| Reagent | 55 kPa (stiff) polyacrylamide | 0.7 kPa (soft) polyacrylamide |
|---|---|---|
| ddH2O (ml) | 2.25 | 3.75 |
| Acrylamide 40% (ml) | 2.25 | 0.300 |
| Bis-acrylamide 2% (ml) | 0.500 | 0.950 |
| Total volume (ml) | 5.00 | 5.00 |
TABLE 5:
Recipe 2: polyacrylamide polymerization solution.
| Reagent | 55 kPa (stiff) polyacrylamide | 0.7 kPa (soft) polyacrylamide |
|---|---|---|
| ddH2O (ml) | 54.5 | 124 |
| Polyacrylamide stock solution (ml) | 111.1 | 41.6 |
| Ammonium persulfate 10% (ml) | 0.830 | 0.830 |
| Tetramethylethylenediamine (ml) | 0.250 | 0.250 |
| Total volume (ml) | 166.68 | 166.68 |
The polyacrylamide stock solution used in Recipe 2 is the final solution produced in Recipe 1.
Immediately after preparation of the PA solution, 15 µl was pipetted onto a Rain-X–washed glass slide and covered with an activated coverslip for 20 min to allow for polymerization. After 20 min, the coverslip was carefully removed from the glass slide with a razor blade by insertion under the edge of the coverslip and with gentle peeling upward. The coverslip was washed with DPBS (three times) and stored in DPBS at 4°C for ≤2 wk as previously described (Myers et al., 2011; Fischer et al., 2012).
Preparation of 2D substrates.
To generate 2D ECMs of controlled compliance, coverslips with PA were activated with 2 mM Sulfo-SANPAH (Pierce; 1 mg/ml DMSO). Sulfo-SANPAH (250 µl) was added to the top of the coverslip and exposed to ultraviolet (UV) light (7500 J) at ∼3.5 in. from five 365-nm bulbs in a Stratagene UV Strata Linker 1800. The activated PA-coated coverslip was rinsed (three times) by dipping in a beaker of ddH2O by holding the corner of the coverslip with tweezers. The excess ddH2O on the non-PA side was dried with a Kimwipe.
Immediately after activation of the PA-coated coverslip, 15 µl of 300 µg/ml rat-tail collagen type I solution (final concentration 90 µg/ml; Corning) was pipetted onto a Rain-X–washed glass slide, covered with the activated coverslip, and allowed to polymerize for 3–4 h at 37°C in 5% CO2. The rat-tail collagen solution was prepared immediately after addition to the glass slide, stored on ice, and composed of glutamine-free 10× MEM (9.6 µl; Life Technologies), 7.5% sodium bicarbonate solution (7 µl; Sigma), and ddH2O (54 µl). After polymerization, the coverslip was carefully removed from the glass slide with a razor blade by insertion under the edge of the coverslip and with gentle peeling upward. The coverslip was washed with DPBS (10 times) and rinsed overnight by rotation at room temperature on a shaker. The next day, before seeding of transfected cells, the PA coverslips conjugated to collagen were rinsed three times with DBPS.
Preparation of 3D substrates.
To generate 3D ECMs of controlled compliance, coverslips with PA were activated two times with 2 mM Sulfo-SANPAH (1 mg/ml DMSO; Pierce). Sulfo-SANPAH (250 µl) was added to the top of the coverslip and exposed to UV light (7500 J) at ∼3.5 in. from five 365-nm bulbs in a Stratagene UV Strata Linker 1800. The activated PA-coated coverslip was rinsed (three times) by dipping in a beaker of ddH2O by holding the corner of the coverslip with tweezers. The excess ddH2O on the non-PA side was dried with a Kimwipe.
Immediately after activation of the PA-coated coverslip, 40 µl of 5.3 mg/ml rat-tail collagen type I solution (final concentration 1.6 mg/ml; Corning) was pipetted onto a Rain-X–washed glass slide, covered with the activated coverslip, and allowed to polymerize for 3–4 h at 37°C. The rat-tail collagen solution was prepared immediately before addition to the glass slide, stored on ice, and composed of glutamine-free 10× MEM (9.6 µl; Life Technologies), 7.5% sodium bicarbonate solution (7 µl; Sigma-Aldrich), and ddH2O (54 µl). After polymerization, the coverslip was carefully removed from the glass slide with a razor blade by insertion under the edge of the coverslip and with gentle peeling upward. The coverslip was washed with DPBS (10 times) and rinsed overnight by rotation at room temperature on a shaker. The next day, before seeding of transfected cells, the collagen-conjugated PA coverslips were rinsed three times with DBPS. The coverslip was assembled in a Rose Chamber, and transfected cells were seeded onto the coverslip and allowed to adhere for 2 h. One hour before imaging, collagen was applied on top of the cells and polymerized at 37°C/5% CO2. For blebbistatin treatments, HUVECs were exposed to blebbistatin (20 µM), when they were seeded to PA after the 1.5-h intermediate seeding period in 35-mm Petri dishes.
Preparation of Rain-X–washed slides.
Both sides of a glass slide were wiped with Rain-X wipes to make the surfaces hydrophobic. Once dry, the slides were wiped down with 100% ethanol (200 proof; Pharmco-AAPER) on a Kimwipe to remove visible Rain-X. The slides were placed in a slide holder and rinsed in running ddH2O for 20 min. After the ddH2O rinsing, the slides were soaked in 100% ethanol (200 proof; Pharmco-AAPER) for 20 min and then rinsed one final time in ddH2O (two or three times) and dried at 37°C.
Live-cell imaging
Time-lapse images were acquired using a high-resolution spinning-disk confocal microscope equipped with 488- and 561-nm lasers and a 60× oil immersion objective lens (1.4 numerical aperture) for 2 min every 2 s using a 700-ms exposure time. For blebbistatin-treated samples, time-lapse images were acquired with only the 561-nm laser for 2 min every 2 s using a 700-ms exposure time. After time-lapse imaging, a single 700-ms exposure with the 488-nm laser was obtained to verify MCAK expression. Because multiple cells from a single coverslip were imaged, a different field of view was imaged to avoid photoinactivation and phototoxicity from this single 488-nm exposure of blebbistatin-treated samples.
MT dynamics analysis and branching quantification.
MT dynamics were visualized by tracking mApple-EB3, a plus-end growth marker, and analyzed using the automated MATLAB-based software package plusTipTracker (Matov et al., 2010; Applegate et al., 2011). To identify and distinguish MCAK-overexpressing cells from control cells, we defined MCAK overexpression as GFP-WT-MCAK expression that exceeded the median grayscale fluorescence intensity of GFP-WT-MCAK expression within the entire experimental cell population. The number of MT growth tracks was calculated using plusTipTracker software and is presented as the sum of the growth tracks counted for n = 5, which represented the minimum number of cells evaluated in any one representative group or subgroup region of interest (whole-cell, branches, control, experimental, untreated, or blebbistatin treated). For cell-branching quantification, branches were defined as protrusions extending from the cell >10 µm in length, and branch origin was designated by the angle of greatest curvature on each side of the branch where it protruded from the cell membrane (Myers et al., 2011).
Immunofluorescence
Fixation and processing of samples for immunofluorescence labeling were performed using a paraformaldehyde/glutaraldehyde coextraction/fixation buffer (PGF-PHEM; 4% paraformaldehyde, 0.15% glutaraldehyde, and 0.2% Triton X-100 in 60 mM PIPES, 27.3 mM HEPES, 10 mM EGTA, and 8.2 mM MgSO4, pH 7.0). PGF-PHEM was added to a coverslip with bound HUVECs at room temperature for 10 min, followed by rinsing (3 × 5 min) with 1× PBS (PBS minus Ca2 and Mg2; HyClone), treated with 0.01 g/ml NaBH4 in 1× PBS (2 × 15 min) to quench reactive aldehydes, rinsed once with 1× PBS, and finally blocked with 5% fat-free milk in 1× PBS (1 h, room temperature). Primary antibodies diluted in 1× PBS and were added and incubated on a rocking platform overnight at 4°C (mouse anti-MCAK, 1:1000; ab50778; Abcam). The next morning, the primary antibody was removed, and the cells were rinsed 3 × 5 min in 1× PBS, followed by incubation in a secondary antibody in 5% fat-free milk (2 h, 37°C; Cy3 donkey anti-mouse, 1:1000; 715-225-150; Jackson ImmunoResearch). Cells were then mounted in Dako mounting medium and imaged on the TiE confocal spinning-disk microscope described earlier. For image analysis, a mask was manually traced around the cell boundary, and the measured mean fluorescence intensity within the masked boundary was recorded (Nikon Elements software).
Statistical analysis
MT growth tracks were pooled per cell, and the average growth speeds and lifetimes from each cell were taken collectively as one event for each experimental group (n = number of cells). All MT dynamics data were collected from three separate experiments and are displayed as mean ± SEM. Using GraphPad Prism software, one-way analysis of variance tests with post-hoc Tukey analysis was performed to determine statistical significance between cells within each experimental group and between each experimental group separately for MT growth speed and MT growth lifetime. For statistical significance, α was set to 0.05, yielding 95% confidence level.
Supplementary Material
Acknowledgments
We thank members of the Myers laboratory and the Department of Biological Sciences for their feedback, support, and helpful discussions. In particular we thank former lab members Kyvan Dang, Tam Linh Nyugen, Patrick Jones, and Felinah Buslig for their contributions. This work was supported by Academic Career Award funding to K.A.M. from the National Institutes of Health/NHLBI (grant number HL113069) and by funding from the W.W. Smith Charitable Trust (grant number H1306). L.D. and N.M.M. were supported by National Institutes of Health grant HL113069 and by the Department of Biological Sciences, University of the Sciences in Philadelphia.
Abbreviations used:
- EC
endothelial cell
- ECM
extracellular matrix
- HUVEC
human umbilical vein endothelial cell
- MCAK
mitotic centromere–associated kinesin
- MT
microtubule
- PA
polyacrylamide.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0306) on March 15, 2017.
REFERENCES
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. plusTipTracker: quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung A, Seo YN, Lu S, Wang Y, Jamora C, del Alamo JC, Varghese S. 3D traction stresses activate protease-dependent invasion of cancer cells. Biophys J. 2014;107:2528–2537. doi: 10.1016/j.bpj.2014.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila J, Dominguez J, Diaz-Nido J. Regulation of microtubule dynamics by microtubule-associated protein expression and phosphorylation during neuronal development. Int J Dev Biol. 1994;38:13–25. [PubMed] [Google Scholar]
- Bae YH, Mui KL, Hsu BY, Liu SL, Cretu A, Razinia Z, Xu T, Pure E, Assoian RK. A FAK-Cas-Rac-lamellipodin signaling module transduces extracellular matrix stiffness into mechanosensitive cell cycling. Sci Signal. 2014;7:ra57. doi: 10.1126/scisignal.2004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JL, Critser PJ, Whittington C, Kuske JL, Yoder MC, Voytik-Harbin SL. Collagen oligomers modulate physical and biological properties of three-dimensional self-assembled matrices. Biopolymers. 2011;95:77–93. doi: 10.1002/bip.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelemi S, Robinet J, Garnotel R, Antonicelli F, Schittly E, Hornebeck W, Lorimier S. Mechanical forces-induced human osteoblasts differentiation involves MMP-2/MMP-13/MT1-MMP proteolytic cascade. J Cell Biochem. 2012;113:760–772. doi: 10.1002/jcb.23401. [DOI] [PubMed] [Google Scholar]
- Beningo KA, Dembo M, Wang YL. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci USA. 2004;101:18024–18029. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A, Dang K, Buslig F, Baird MA, Davidson MW, Waterman CM, Myers KA. Rac1 and Aurora A regulate MCAK to polarize microtubule growth in migrating endothelial cells. J Cell Biol. 2014;206:97–112. doi: 10.1083/jcb.201401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey SP, Rahman A, Kraning-Rush CM, Romero B, Somasegar S, Torre OM, Williams RM, Reinhart-King CA. Comparative mechanisms of cancer cell migration through 3D matrix and physiological microtracks. Am J Physiol Cell Physiol. 2015;308:C436–C447. doi: 10.1152/ajpcell.00225.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JO, Simpson N, Hewlett L, Hall A. Rac regulates endothelial morphogenesis and capillary assembly. Mol Biol Cell. 2002;13:2474–2485. doi: 10.1091/mbc.E02-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Kalil K. Axon branching requires interactions between dynamic microtubules and actin filaments. J Neurosci. 2001;21:9757–9769. doi: 10.1523/JNEUROSCI.21-24-09757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat Commun. 2015;6:8720. doi: 10.1038/ncomms9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Yamada KM. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp Cell Res 343, 60–66. 2015 doi: 10.1016/j.yexcr.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar LT, Hoying JB, Utzinger U, Underwood CJ, Krishnan L, Baggett BK, Maas SA, Guilkey JE, Weiss JA. Mechanical interaction of angiogenic microvessels with the extracellular matrix. J Biomech Eng. 2014a;136:021001. doi: 10.1115/1.4026471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar LT, Underwood CJ, Guilkey JE, Hoying JB, Weiss JA. Extracellular matrix density regulates the rate of neovessel growth and branching in sprouting angiogenesis. PLoS One. 2014b;9:e85178. doi: 10.1371/journal.pone.0085178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott H, Fischer RS, Myers KA, Desai RA, Gao L, Chen CS, Adelstein RS, Waterman CM, Danuser G. Myosin II controls cellular branching morphogenesis and migration in three dimensions by minimizing cell-surface curvature. Nat Cell Biol. 2015;17:137–147. doi: 10.1038/ncb3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Pérez-González C, Castro N, Zhu C, Trepat X, Roca-Cusachs P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. 2016;18:540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19:260–265. doi: 10.1016/j.cub.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RS, Myers KA, Gardel ML, Waterman CM. Stiffness-controlled three-dimensional extracellular matrices for high-resolution imaging of cell behavior. Nat Protoc. 2012;7:2056–2066. doi: 10.1038/nprot.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Ghibaudo M, Di Meglio JM, Hersen P, Ladoux B. Mechanics of cell spreading within 3D-micropatterned environments. Lab Chip. 2011;11:805–812. doi: 10.1039/c0lc00221f. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ. Mitosis: MCAK under the aura of Aurora B. Curr Biol. 2004;14:R346–R348. doi: 10.1016/j.cub.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Guo WH, Frey MT, Burnham NA, Wang YL. Substrate rigidity regulates the formation and maintenance of tissues. Biophys J. 2006;90:2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasaka TP, Myers KA, Baas PW. Role of actin filaments in the axonal transport of microtubules. J Neurosci. 2004;24:11291–11301. doi: 10.1523/JNEUROSCI.3443-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Hertzer KM, Walczak CE. The C-termini of tubulin and the specific geometry of tubulin substrates influence the depolymerization activity of MCAK. Cell Cycle. 2008;7:2727–2737. doi: 10.4161/cc.7.17.6590. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hu YL, Li S, Miao H, Tsou TC, del Pozo MA, Chien S. Roles of microtubule dynamics and small GTPase Rac in endothelial cell migration and lamellipodium formation under flow. J Vasc Res. 2002;39:465–476. doi: 10.1159/000067202. [DOI] [PubMed] [Google Scholar]
- Kline-Smith SL, Walczak CE. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol Biol Cell. 2002;13:2718–2731. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva E, Putnam AJ. Endothelial cell traction and ECM density influence both capillary morphogenesis and maintenance in 3-D. Am J Physiol Cell Physiol. 2009;297:C179–C187. doi: 10.1152/ajpcell.00018.2009. [DOI] [PubMed] [Google Scholar]
- Knowlton AL, Vorozhko VV, Lan W, Gorbsky GJ, Stukenberg PT. ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr Biol. 2009;19:758–763. doi: 10.1016/j.cub.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun. 2004;320:1020–1025. doi: 10.1016/j.bbrc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Kutys ML, Yamada KM. Rho GEFs and GAPs: emerging integrators of extracellular matrix signaling. Small GTPases. 2015;6:21–24. doi: 10.4161/21541248.2014.989792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Hong J, Lee J. Cell motility regulation on a stepped micro pillar array device (SMPAD) with a discrete stiffness gradient. Soft Matter 12, 2325–2333. 2016 doi: 10.1039/c5sm00649j. [DOI] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E, Mandelkow EM. Microtubules and microtubule-associated proteins. Curr Opin Cell Biol. 1995;7:72–81. doi: 10.1016/0955-0674(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A, Muller J, Mandelkow EM, Hoenger A, Mandelkow E. Interaction of kinesin motors, microtubules, and MAPs. J Muscle Res Cell Motil. 2006;27:125–137. doi: 10.1007/s10974-005-9051-4. [DOI] [PubMed] [Google Scholar]
- Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat Methods. 2010;7:761–768. doi: 10.1038/nmeth.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro Gouveia S, Leslie K, Kapitein LC, Buey RM, Grigoriev I, Wagenbach M, Smal I, Meijering E, Hoogenraad CC, et al. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr Biol. 2010;20:1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Moore A, Wordeman L. C-terminus of mitotic centromere-associated kinesin (MCAK) inhibits its lattice-stimulated ATPase activity. Biochem J. 2004;383:227–235. doi: 10.1042/BJ20040736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, Andrieux A, Job D, Wordeman L. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Cooper J, Wagenbach M, Ovechkina Y, Wordeman L, Milligan RA. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5:1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- Mousavi SJ, Doweidar MH, Doblare M. Cell migration and cell-cell interaction in the presence of mechano-chemo-thermotaxis. Mol Cell Biomech. 2013;10:1–25. [PubMed] [Google Scholar]
- Myers KA, Applegate KT, Danuser G, Fischer RS, Waterman CM. Distinct ECM mechanosensing pathways regulate microtubule dynamics to control endothelial cell branching morphogenesis. J Cell Biol. 2011;192:321–334. doi: 10.1083/jcb.201006009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Tint I, Nadar CV, He Y, Black MM, Baas PW. Antagonistic forces generated by cytoplasmic dynein and myosin-II during growth cone turning and axonal retraction. Traffic. 2006;7:1333–1351. doi: 10.1111/j.1600-0854.2006.00476.x. [DOI] [PubMed] [Google Scholar]
- Nogales E, Zhang R. Visualizing microtubule structural transitions and interactions with associated proteins. Curr Opin Struct Biol. 2016;37:90–96. doi: 10.1016/j.sbi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina Y, Wagenbach M, Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J Cell Biol. 2002;159:557–562. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Yamada KM. Fibroblasts lead the way: a unified view of 3D cell motility. Trends Cell Biol. 2015;25:666–674. doi: 10.1016/j.tcb.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riching KM, Keely PJ. Rho family GTPases: making it to the third dimension. Int J Biochem Cell Biol. 2015;59:111–115. doi: 10.1016/j.biocel.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Limouze J, Combs CA, Straight AF, Sellers JR. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry. 2005;44:584–588. doi: 10.1021/bi0483357. [DOI] [PubMed] [Google Scholar]
- Thievessen I, Fakhri N, Steinwachs J, Kraus V, McIsaac RS, Gao L, Chen BC, Baird MA, Davidson MW, Betzig E, et al. Vinculin is required for cell polarization, migration, and extracellular matrix remodeling in 3D collagen. FASEB J. 2015;29:4555–4567. doi: 10.1096/fj.14-268235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood CJ, Edgar LT, Hoying JB, Weiss JA. Cell-generated traction forces and the resulting matrix deformation modulate microvascular alignment and growth during angiogenesis. Am J Physiol Heart Circ Physiol. 2014;307:H152–H164. doi: 10.1152/ajpheart.00995.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N. Mechanical interactions among cytoskeletal filaments. Hypertension. 1998;32:162–165. doi: 10.1161/01.hyp.32.1.162. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Decarreau J, Vicente JJ, Wagenbach M. Divergent microtubule assembly rates after short- versus long-term loss of end-modulating kinesins. Mol Biol Cell. 2016;27:1300–1309. doi: 10.1091/mbc.E15-11-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I, Garnham CP, Roll-Mecak A. Writing and reading the tubulin code. J Biol Chem. 2015;290:17163–17172. doi: 10.1074/jbc.R115.637447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ems-McClung SC, Walczak CE. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol Biol Cell. 2008;19:2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.