The ART-Rsp5 network is regulated by the ubiquitin system. Ubp2 and Ubp15 prevent hyperubiquitination and proteasomal degradation of ARTs. Ubp2 and Ubp15 ensure efficient endocytosis by stabilizing ARTs.
Abstract
Endocytic down-regulation of cell-surface proteins is a fundamental cellular process for cell survival and adaptation to environmental stimuli. Ubiquitination of cargo proteins serves as the sorting signal for downstream trafficking and relies on the arrestin-related trafficking adaptor (ART)-Rsp5 ubiquitin ligase adaptor network in yeast. Hence proper regulation of the abundance and activity of these ligase–adaptor complexes is critical for maintenance of optimal plasma membrane protein composition. Here we report that the stability of ARTs is regulated by the deubiquitinating enzymes (DUBs) Ubp2 and Ubp15. By counteracting the E3 ubiquitin ligase Rsp5, Ubp2 and Ubp15 prevent hyperubiquitination and proteasomal degradation of ARTs. Specifically, we show that loss of both Ubp2 and Ubp15 results in a defect in Hxt6 endocytosis associated with Art4 instability. Our results uncover a novel function for DUBs in the endocytic pathway by which Ubp2 and Ubp15 positively regulate the ART-Rsp5 network.
INTRODUCTION
Ubiquitination is a reversible posttranslational modification involved in numerous cellular processes, including protein degradation, membrane trafficking, and signaling. Conjugation of ubiquitin to substrates is achieved by an enzymatic cascade involving E1 activating enzymes, E2 conjugating enzymes, and E3 ubiquitin ligases. Deubiquitinating enzymes (DUBs) remove ubiquitin from substrates and serve many cellular functions, including processing ubiquitin precursors, editing/rescuing ubiquitin conjugates, facilitating proteasomal degradation, and maintaining free ubiquitin pools (Amerik and Hochstrasser, 2004). Many DUBs and E3 ligases interact with each other and work in concert. Such DUB–E3 interactions can be used to fine-tune substrate ubiquitination, as well as regulate their own stability or activity (Nijman et al., 2005).
Proper remodeling of cell-surface receptors and transporters is critical for cell survival in response to environmental stimuli or changing nutrient conditions. Endocytic down-regulation is a major mechanism to regulate the homeostasis and composition of cell-surface proteins. Ubiquitin has emerged as a critical component of endocytosis; ubiquitin conjugation to plasma membrane (PM) proteins, or cargoes, promotes their internalization, followed by sorting into multivesicular bodies (MVBs) and delivery to the lysosome/vacuole (Henne et al., 2011; MacGurn et al., 2012; Piper et al., 2014). Failure to properly down-regulate PM receptors is associated with many human diseases. For example, inability to attenuate growth signaling by endocytosis can lead to abnormal cell proliferation and cancer (Mosesson et al., 2008).
In budding yeast, arrestin-related trafficking adaptors (ARTs) recruit the Nedd4-like ubiquitin ligase Rsp5 to the PM and mediate cargo ubiquitination (Lin et al., 2008; Nikko and Pelham, 2009; Hatakeyama et al., 2010). ARTs have similarity to human arrestin domain-containing (ARRDC) proteins, with both having N-terminal arrestin-like domains and C-terminal PY (PPXY/LPXY) motifs (Lin et al., 2008; Becuwe et al., 2012a), and they promote PM protein remodeling through endocytic down-regulation in response to nutrient availability, environmental stress, and proteotoxic stress (Lin et al., 2008; MacGurn et al., 2011; Merhi and Andre, 2012; Becuwe et al., 2012b; Zhao et al., 2013; Alvaro et al., 2014; Crapeau et al., 2014). The ART-Rsp5 network is highly regulated and influenced by intracellular signaling, such as phosphorylation/dephosphorylation (MacGurn et al., 2011; Becuwe et al., 2012b; O’Donnell et al., 2013; Herrador et al., 2015). For example, Art1/Ldb19 is phosphorylated and inhibited by Npr1, which in turn is inhibited by TORC1 (MacGurn et al., 2011). Similarly, Art4/Rod1 is subjected to inhibitory phosphorylation by yeast AMPK Snf1 (Becuwe et al., 2012b). Therefore ARTs are important regulatory hubs for ubiquitin-mediated endocytosis of PM proteins.
Of interest, ARTs are also Rsp5 substrates. In some cases, these ubiquitination events are critical for ART function in promoting PM protein turnover (Lin et al., 2008; Herrador et al., 2010; Becuwe et al., 2012b), whereas in other cases, the role of ART ubiquitination is unclear (Kee et al., 2006; Alvaro et al., 2014). Similarly, ubiquitination of endocytic machinery and endosomal sorting complexes required for transport (ESCRTs) has been reported (Stamenova et al., 2004; Gupta et al., 2007; Lu et al., 2008; Dores et al., 2010; Lauwers et al., 2010; Erpapazoglou et al., 2012; Weinberg and Drubin, 2012, 2014), but the functional significance remains to be elucidated.
The human genome encodes ∼90 DUBs with diverse functions (Nijman et al., 2005; Ye et al., 2009). Given the broad involvement of ubiquitination in endocytosis, as well as in other protein-sorting pathways, it is not surprising that DUBs have been implicated in many aspects of membrane protein trafficking (Millard and Wood, 2006; Clague et al., 2012a, b). Although DUBs are clearly important for endocytic down-regulation of PM proteins in yeast (Amerik et al., 2000b; Dupre and Haguenauer-Tsapis, 2001; Ren et al., 2007; Lam et al., 2009; Erpapazoglou et al., 2012; Tardiff et al., 2013; Weinberg and Drubin, 2014), precisely how they coordinate and interact with the ART-Rsp5 network remains unclear. Here we describe a novel function of the DUBs Ubp2 and Ubp15 in the endocytic pathway by which Ubp2 and Ubp15 protect ARTs from proteasomal degradation, and we propose deubiquitination of ARTs as a critical regulatory mechanism that contributes to the fine-tuning of ART-Rsp5 network activity.
RESULTS
Identifying potential ART-Rsp5–regulating DUBs
To investigate whether ART function is regulated by DUBs, we set out to look for changes of ART protein modifications and/or levels in strains with deletions of DUBs. We initially focused on Art1 and Art4, two of the well-characterized ARTs that regulate the ubiquitin-mediated endocytosis of several PM cargoes (Lin et al., 2008; Nikko and Pelham, 2009; Becuwe et al., 2012b; Becuwe and Leon, 2014; Alvaro et al., 2014; O’Donnell et al., 2015; Prosser et al., 2015). In addition, both Art1 and Art4 are regulated by the phosphorylation/dephosphorylation cycle (MacGurn et al., 2011; Becuwe et al., 2012b; Alvaro et al., 2014, 2016; Llopis-Torregrosa et al., 2016), yet it is unclear whether Art1 and Art4 are subject to DUB-mediated regulation despite both of them being ubiquitinated by Rsp5 (Lin et al., 2008; Becuwe et al., 2012b).
To identify DUBs that potentially regulate Art1 and Art4, we took a candidate gene/protein approach and investigated the effects of DUBs with known genetic and/or physical interactions with the ART-Rsp5 network (Supplemental Figure S1A): Ubp2 has been shown to physically/genetically interact with the E3 ligase Rsp5 (Kee et al., 2005, 2006; Lam et al., 2009), as well as to deubiquitinate Art2/Ecm21 and Art8/Csr2 in vitro and influence their ubiquitination in vivo (Kee et al., 2006). Ubp7 also physically interacts with Rsp5 (Ren et al., 2007), and overexpression of Ubp7 and its paralogue Ubp11 affects Rsp5-mediated endosomal trafficking (Tardiff et al., 2013). Ubp9 (and its paralogue Ubp13) and Ubp15 were also included in our initial consideration because they exhibit similar cellular localizations (peripheral punta or cytoplasmic punta; Huh et al., 2003) to ARTs (Lin et al., 2008; Herrador et al., 2010; O’Donnell et al., 2010; MacGurn et al., 2011; Becuwe and Leon, 2014).
Plasmid-encoded Art4 and Art1 were expressed ectopically in yeast strains with deletions of these DUBs, and protein extracts were analyzed by immunoblot. We observed an apparent decrease in steady-state Art4 (and less obviously Art1) protein level in ubp2Δ (Figure 1A and Supplemental Figure S1B), a phenotype consistent with hyperubiquitination and enhanced protein turnover. To test whether any of these DUBs play a redundant role with Ubp2, we also examined Art4 and Art1 levels in ubp2Δ ubp15Δ, ubp2Δ ubp7Δ ubp11Δ, and ubp2Δ ubp9Δ ubp13Δ mutants. We found whereas ubp2Δ ubp7Δ ubp11Δ and ubp2Δ ubp9Δ ubp13Δ exhibited similar Art4 and Art1 levels to ubp2Δ, ubp2Δ ubp15Δ showed a more prominent decrease in Art4 and Art1 levels than with ubp2Δ (Figure 1A and Supplemental Figure S1B).
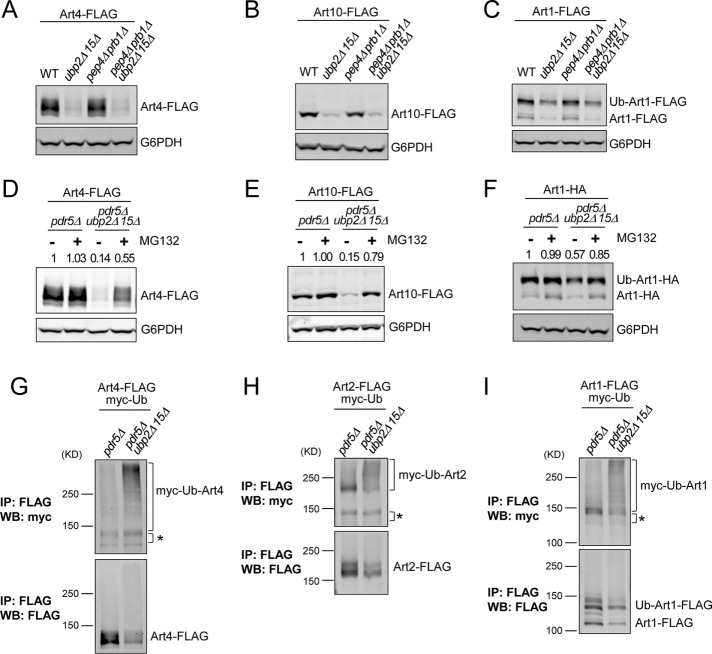
FIGURE 1:
Ubp2 and Ubp15 maintain the abundance of Rsp5 adaptor proteins. (A) Protein extracts of wild-type (WT) or the indicated yeast mutant strains expressing plasmid-encoded Art4-FLAG were analyzed by SDS–PAGE and immunoblot. (B–I) Protein extracts of WT, ubp2Δ, ubp15Δ, and ubp2Δ ubp15Δ yeast cells expressing chromosomally tagged ARTs were analyzed by SDS–PAGE and immunoblot. Ub-Art1, K486-dependent ubiquitinated form of Art1 (Lin et al., 2008). *Nonspecific band. The numbers above each lane indicate the protein abundance of ARTs determined by the LI-COR quantification and normalized with the loading control G6PDH.
Given its broad involvement in the endocytic pathway, we also tested Doa4, whose primary functions include deubiquitination in the MVB pathway, as well as maintaining cellular ubiquitin levels (Swaminathan et al., 1999; Amerik et al., 2000b). We found that doa4Δ completely suppressed the reduced abundance of Art4 and Art1 in ubp2Δ ubp15Δ (Supplemental Figure S1, C and D), indicating that Doa4 does not share a similar role to Ubp2 and Ubp15. Recently Ubp3 (as well as Ubp2) was shown to interact with Rsp5 upon heat stress and regulate the Rsp5-mediated cytosolic protein quality control pathway (Fang et al., 2016). We therefore examined the effect of ubp3Δ, and found that ubp3Δ also partially suppressed the reduced abundance of Art4 in ubp2Δ ubp15Δ (Supplemental Figure S1E). Although the reduced Art4 and Art1 protein levels in ubp2Δ ubp15Δ cells are in line with increased ubiquitination and protein degradation in which Ubp2 and Ubp15 may play a direct role, the reverse phenotypes caused by doa4Δ and ubp3Δ are likely due to indirect effects, including reduced free ubiquitin levels (see Discussion). We therefore decided to focus on investigating Ubp2 and Ubp15 as potential regulators of Rsp5 adaptor proteins.
Ubp2 and Ubp15 regulate the abundance of Rsp5 adaptor proteins, the ARTs
We first asked whether the abundance of other ART proteins is regulated by Ubp2 and Ubp15 in a similar manner. To examine the levels of ARTs more precisely, we generated WT, ubp2Δ, ubp15Δ, and ubp2Δ ubp15Δ strains with epitope-tagged ARTs at their endogenous chromosome loci. We confirmed that steady-state levels of Art4 and Art1 were indeed reduced in both ubp2Δ cells and ubp2Δ ubp15Δ cells, with ubp2Δ ubp15Δ cells showing a greater reduction (Figure 1, B and C). These results suggest that Ubp2 is the major regulator for ARTs, and Ubp15 may be partially redundant with Ubp2.
Furthermore, protein levels of many other Rsp5 adaptors, including Art2, Art6/Aly1, Art7/Rog3, Art8, Art10, and Bul1 (Figure 1, D–I), were also significantly decreased. Among these adaptors, Art4 and Art10 showed the most drastic reduction in ubp2Δ ubp15Δ. By contrast, Art3/Aly2 and Art9/Rim8 protein levels exhibited little or no decrease in ubp2Δ ubp15Δ (Supplemental Figure S1, F and G). Of importance, the abundance of Rsp5 was not decreased (Supplemental Figure S1H). These data indicate that Ubp2 and Ubp15 regulate the abundance of several Rsp5 adaptor proteins.
Ubp15 physically interacts with ARTs
Ubp2 was previously reported to physically interact with Rsp5 (Kee et al., 2005), providing a mechanism for regulation of ARTs by Ubp2. How Ubp15 is integrated into the ART-Rsp5 network, on the other hand, is not clear. To explore how Ubp15 regulates Rsp5 adaptor proteins, we performed stable isotope labeling with amino acid in culture (SILAC) analysis to identify Ubp15-interacting proteins and found Art2 as one of the top hits (Supplemental Figure S2A). We then performed coimmunoprecipitation (CoIP) experiment and confirmed the physical interaction between Art2-FLAG and GFP-Ubp15 (Supplemental Figure S2B). Consistent with the physical interactions (Rsp5-Ubp2 and Art2-Ubp15), Art2 abundance was also regulated by Ubp2 and Ubp15 (Figure 1D). The interaction between Ubp15 and Art2 raises an interesting possibility that Ubp15 might regulate other ARTs by physically associating with them as well. These interactions may not be stable enough to be detected under our experimental conditions for the Ubp15 interactome. To test directly whether Ubp15 interacts with Art4, we performed another CoIP experiment and found that indeed GFP-Ubp15 was coprecipitated with Art4-FLAG (Supplemental Figure S2C). Together these findings suggest that the genetic interaction we observed for ubp2Δ and ubp15Δ is likely to be physiologically relevant to the ART-Rsp5 network. Ubp2 and Ubp15 may regulate the ART-Rsp5 network through physical interactions: Ubp2 interacts with Rsp5, and Ubp15 interacts with ARTs. Given the greater reduction of ART protein levels in the ubp2Δ ubp15Δ double mutant compared with either single mutant, we conducted the remainder of this study by characterizing the double mutant in more detail.
Reduced abundance of ARTs in ubp2Δ ubp15Δ is due to accelerated protein turnover
To test whether the reduced abundance of ARTs in ubp2Δ ubp15Δ results from lack of deubiquitination on ARTs, we used a DUB catalytic domain (UL36) that renders its fusion proteins resistant to ubiquitination (Stringer and Piper, 2011) and assessed whether restoring deubiquitination artificially can suppress Art4 protein reduction in ubp2Δ ubp15Δ. Indeed, we found the Art4-UL36 fusion was able to maintain its abundance in ubp2Δ ubp15Δ cells, whereas fusion with catalytic-inactive UL36C40S had no effect (Figure 2A). Furthermore, expressing functional Ubp2 or Ubp15, but not catalytically inactive Ubp2C745V or Ubp15C214V, restored Art4 levels in ubp2Δ ubp15Δ cells (Figure 2B), suggesting maintaining Art4 levels requires the catalytic activity of Ubp2 and Ubp15.
FIGURE 2:
Ubp2 and Ubp15 regulate the degradation of ARTs. (A) art4Δ and art4Δ ubp2Δ ubp15Δ yeast cells carrying Art4-FLAG, Art4-UL36-FLAG, or Art4-UL36C40S-FLAG expression vector. Protein extracts were analyzed by SDS–PAGE and immunoblot. (B) Immunoblot analysis of chromosomal Art4-FLAG in WT and ubp2Δ ubp15Δ yeast cells carrying functional or catalytically dead Ubp2/Ubp15 expression vector. (C) WT and ubp2Δ ubp15Δ yeast cells expressing chromosomal Art4-FLAG or ubp2Δ ubp15Δ cells overexpressing (OE) Art4-FLAG (driven by the CYC1 promoter inserted into ART4 locus) were treated with 100 μg/ml CHX to stop protein synthesis. Protein extracts from the indicated time points were analyzed by SDS–PAGE and immunoblot. (D) Art4 levels (relative to t = 0) as in C were quantified by the LI-COR system. Data from three independent experiments are presented as mean ± SD. (E, F) similar to C and D, except that yeast strains with chromosomal Art1-FLAG were examined.
Defects in deubiquitination can lead to increased ubiquitination and subsequent protein degradation. To investigate whether the reduced abundance of ARTs in ubp2Δ ubp15Δ cells is due to protein degradation, we monitored protein turnover of Art4, as well as of Art1, in wild-type versus ubp2Δ ubp15Δ cells. The turnover of Art4 and Art1 in the presence of cycloheximide (CHX), which inhibits nascent protein synthesis, was accelerated in ubp2Δ ubp15Δ cells compared with WT cells (Figure 2, C–F). Given the drastic reduction in steady-state Art4 levels in ubp2Δ ubp15Δ cells, we further analyzed Art4 turnover after overexpression in this background and found the rate of Art4 turnover to be similar (Figure 2, C and D). These data indicate that decreased Art1 and Art4 levels are likely caused by protein degradation but not protein synthesis defects. Together our results demonstrate that the DUB activities of Ubp2 and Ubp15 prevent the degradation of ARTs.
ARTs are hyperubiquitinated and degraded by the proteasome in ubp2Δ ubp15Δ
In eukaryotic cells, ubiquitin-dependent protein degradation occurs in either the vacuole/lysosome or the proteasome. Proteins are delivered to the vacuole for degradation through either vesicle trafficking (the MVB pathway) or autophagy. Although ARTs are not transmembrane proteins and thus trafficking through the MVB pathway is unlikely, in principle they could still be degraded in the vacuole through autophagy. To test this possibility, we examined whether ART degradation in ubp2Δ ubp15Δ requires Pep4 and Prb1, two major vacuolar proteases (Hemmings et al., 1981). The degradation of Art4, Art10, and Art1 in ubp2Δ ubp15Δ was not affected by pep4Δ prb1Δ (Figure 3, A–C), suggesting that their degradation does not occur in the vacuole.
FIGURE 3:
Ubp2 and Ubp15 prevent hyperubiquitination and proteasomal degradation of ARTs. (A–C) Protein extracts of indicated yeast strains expressing plasmid-encoded Art4-FLAG (A), Art10-FLAG (B), or Art1-FLAG (C) were analyzed by SDS–PAGE and immunoblot. (D–F) pdr5Δ and pdr5Δ ubp2Δ ubp15Δ cells expressing plasmid-encoded Art4-FLAG (D), Art10-FLAG (E), or Art1-HA (F) were mock-treated (dimethyl sulfoxide [DMSO]) or treated with MG132 (25 μg/ml) for 60 min, and protein extracts were analyzed by SDS–PAGE and immunoblot. The numbers above each lane indicate the protein abundance of ARTs determined by the LI-COR quantification and normalized with the loading control G6PDH. (G–I) pdr5Δ and pdr5Δ ubp2Δ ubp15Δ cells expressing plasmid-encoded Art4-FLAG (G), Art2-FLAG (H), or Art1-FLAG (I) and myc-ubiquitin (overexpression controlled by copper-inducible promoter) were treated with MG132 (25 μg/ml) for 60 min. Samples were immunoprecipitated using anti-FLAG antibody under denaturing conditions and analyzed by SDS–PAGE and immunoblot. *Nonspecific.
We therefore investigated whether proteasomal degradation is involved. To sensitize yeast cells to proteasome inhibitor MG132, we used strains containing a deletion of the multidrug transporter gene PDR5 (Fleming et al., 2002). We found that MG132 substantially inhibited the degradation of Art4, Art10, and Art1 in pdr5Δ ubp2Δ ubp15Δ cells (Figure 3, D–F). Together with the observation that PEP4 and PRB1 are dispensable for ART degradation (Figure 3, A–C), these findings indicate that ARTs are targeted to the proteasome for degradation in the absence of Ubp2 and Ubp15.
Collectively our data demonstrate that Ubp2 and Ubp15 maintain the protein stability of ARTs by preventing their proteasomal degradation, possibly by physically interacting with Rsp5 and/or ARTs, and counteracting ubiquitination of ARTs. To explore further how loss of Ubp2 and Ubp15 affects the global ubiquitination landscape, we compared the ubiquitin-modified proteome of WT and ubp2Δ ubp15Δ cells using SILAC followed by tandem purification of ubiquitin conjugates and quantitative mass spectrometry (Supplemental Figure S3A). To improve sensitivity as well as obtain better understanding of mechanisms for Ubp2 and Ubp15 functions, we prefractionated lysates into membrane (P13) and soluble (S13) fractions for separate analysis. Our analysis revealed that several Rsp5 adaptor proteins, including Art2, Art3, Bul1, and Bul2 in the P13 fraction, as well as Art2 and Art3 in the S13 fraction, exhibited increased ubiquitination in ubp2Δ ubp15Δ cells, based on multiple peptides resolved (Supplemental Figure S3B), further implicating Ubp2 and Ubp15 in limiting ubiquitination of Rsp5 adaptor proteins, regardless of whether ARTs associate with membrane compartments.
The fact that we did not recover other ARTs such as Art4 and Art10 in this analysis may reflect the relatively low abundance of these ubiquitinated adaptors, perhaps due to their higher susceptibility to proteasomal degradation. To examine directly the ubiquitination status of Art4 in ubp2Δ ubp15Δ, we overexpressed myc-ubiquitin in pdr5Δ and pdr5Δ ubp2Δ ubp15Δ cells. After inhibition of the proteasome by MG132, Art4-FLAG was immunoprecipitated under denaturing conditions, and ubiquitin conjugates were detected by immunoblotting using anti-myc antibody. Increased high–molecular weight (MW) signals were evident in pdr5Δ ubp2Δ ubp15Δ cells compared with pdr5Δ cells (Figure 3G), indicating that Art4 is hyperubiquitinated in the absence of Ubp2 and Ubp15. Similar hyperubiquitination was observed for Art2 and Art1 in the absence of Ubp2 and Ubp15 (Figure 3, H and I). Together these results provide strong evidence that Ubp2 and Ubp15 protect at least a subset of ARTs against proteasomal degradation by preventing their hyperubiquitination.
E3 ubiquitin ligase Rsp5 mediates ART degradation in ubp2Δ ubp15Δ
Because ARTs associate with the E3 ubiquitin ligase Rsp5, we tested whether ART degradation in ubp2Δ ubp15Δ requires Rsp5 function. Rsp5 belongs to the Nedd4 ubiquitin ligase family and contains a C-terminal HECT catalytic domain and three WW domains that interact with PY motifs (PPXY or LPXY) in substrates or adaptor proteins, including ARTs. We found that disruption of Rsp5–Art4 interaction by Art4 PY-motif mutations completely suppressed Art4 degradation in ubp2Δ ubp15Δ (Figure 4A), indicating that Art4 degradation in ubp2Δ ubp15Δ requires the Art4–Rsp5 interaction. Similarly, Art1 degradation in ubp2Δ ubp15Δ was suppressed by Art1 PY-motif mutations (Figure 4B). To test whether the Rsp5 ligase activity is required, we used a hypomorphic allele of RSP5 with a mutation in the HECT domain (G747E; Oestreich et al., 2007) to monitor ART levels in ubp2Δ ubp15Δ. We found that the rsp5G747E allele suppressed the instability of both Art4 and Art1 (Figure 4, C and D), indicating that Rsp5 ligase activity is required for ART degradation in ubp2Δ ubp15Δ cells. These results suggest that Ubp2 and Ubp15 function to counteract Rsp5-mediated hyperubiquitination of ARTs.
FIGURE 4:
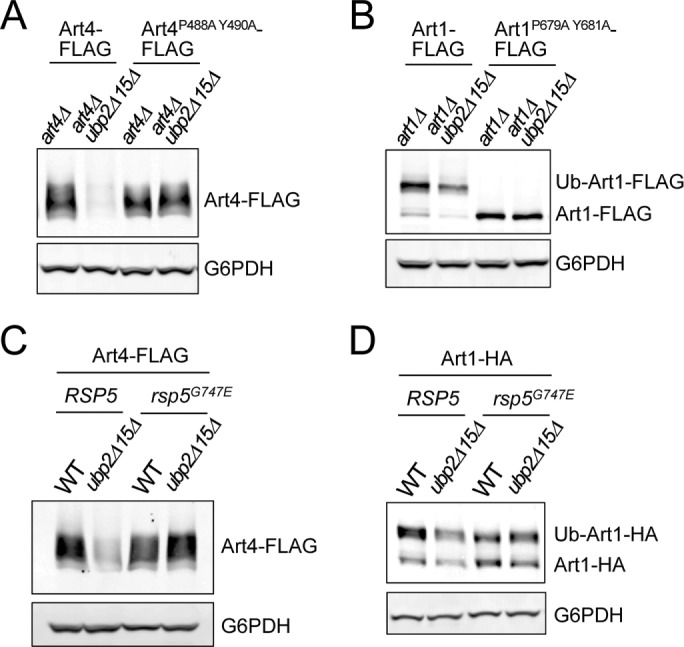
ART degradation in ubp2Δ ubp15Δ is mediated by E3 ubiquitin ligase Rsp5. (A) Protein extracts of art4Δ and art4Δ ubp2Δ ubp15Δ yeast cells expressing plasmid-encoded Art4-FLAG or Art4P488A Y490A-FLAG were analyzed by SDS–PAGE and immunoblot. (B) Protein extracts of art1Δ and art1Δ ubp2Δ ubp15Δ yeast cells expressing plasmid-encoded Art1-FLAG or Art1P679A Y681A-FLAG were analyzed by SDS–PAGE and immunoblot. (C, D) Protein extracts of RSP5, RSP5 ubp2Δ ubp15Δ, rsp5G747E, and rsp5G747E ubp2Δ ubp15Δ cells expressing plasmid-encoded Art4-FLAG (C) or Art1-HA (D) were analyzed by SDS–PAGE and immunoblot.
It was shown previously that during heat stress, Rsp5 associates with the PY motif–containing Hsp40 cochaperone Ydj1 to promote ubiquitination of misfolded proteins and target them for proteasomal degradation (Lee et al., 1996; Fang et al., 2014). Given their similarity as Rsp5-dependent proteasomal degradation processes, we tested whether ART degradation in ubp2Δ ubp15Δ is also mediated by Ydj1. We found, however, that ART degradation in ubp2Δ ubp15Δ did not require Ydj1 (Supplemental Figure S4A), indicating that it is a distinct process.
We further investigated whether other ubiquitin ligases, in addition to Rsp5, are required for ART degradation in ubp2Δ ubp15Δ by further enhancing polyubiquitination of ARTs. These ligases are sometimes referred as E4 ubiquitin chain extension enzymes (Koegl et al., 1999). Among them, Hul5 and Cul3 are particularly interesting. Hul5 has been shown to target prion-like protein Pin3/Lsb2 for proteasomal degradation (Fang et al., 2011). Pin3, like ARTs, is also a PY motif–containing protein and is ubiquitinated by Rsp5 (Chernova et al., 2011). Moreover, Rsp5 and Cul3 act sequentially to promote polyubiquitination of RNA polymerase II (Harreman et al., 2009). We therefore assessed the involvement of these E4 enzymes and found Art4 degradation in ubp2Δ ubp15Δ was largely unperturbed (Supplemental Figure S4B).
In addition, we tested other ubiquitin ligases functioning in cytoplasmic protein quality control pathways, including San1 and Ubr1 (Heck et al., 2010; Prasad et al., 2010; Khosrow-Khavar et al., 2012), as well as Doa10 (Carvalho et al., 2006; Ravid et al., 2006; Ast et al., 2014). We found none of these E3 ligases altered Art4 degradation in ubp2Δ ubp15Δ cells (Supplemental Figure S4, C and D). We cannot rule out the possibility that other ubiquitin ligases or E4 enzymes are involved in the ART degradation we observed here. Nevertheless, our results revealed a novel Rsp5-dependent mechanism targeting cytosolic proteins for proteasomal degradation.
Art4 hyperubiquitination and degradation require the formation of K63-linked polyubiquitin chains
Proteasomal degradation is usually mediated by K48-linked polyubiquitination (Chau et al., 1989; Finley et al., 1994), although K63-linked chains have also been reported to be the targeting signal for the proteasome (Kirkpatrick et al., 2006; Saeki et al., 2009). Although Rsp5 prefers to conjugate monoubiquitin or K63-linked polyubiquitin chains (Kee et al., 2005; Kim and Huibregtse, 2009), K48-linked polyubiquitination by Rsp5 is also evident both in vitro and in vivo (French et al., 2009; Fang et al., 2014, 2016). Whereas Ubp2 appears to be K63 specific in vitro (Kee et al., 2005, 2006), evidence from in vivo studies suggests that Ubp2 may act on both K48 and K63 polyubiquitin chains (Xu et al., 2009; Anton et al., 2013). Ubp15 can target both K48 and K63 linkages in vitro, but long K63 chains are cleaved more rapidly by Ubp15 than long K48 chains (Schaefer and Morgan, 2011). To explore the linkage specificity/preference of Ubp2 and Ubp15, we used our SILAC ubiquitin proteome experiment (Supplemental Figure S3A) and analyzed linkage-specific ubiquitin peptides resolved. We found that in both membrane and soluble fractions, K63 ubiquitin linkages were increased in ubp2Δ ubp15Δ cells more prominently than with K48 linkages (Supplemental Figure S5A), consistent with the in vitro preference of both Ubp2 and Ubp15 for K63-linked polyubiquitin chains.
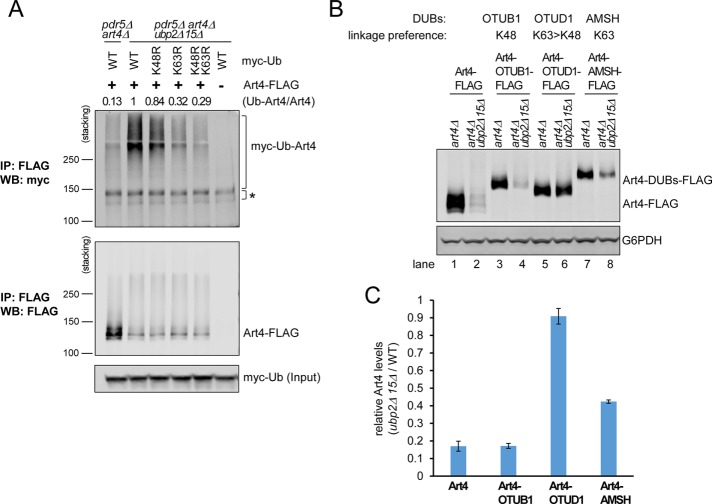
To determine directly the involvement of K63 versus K48 linkages in ART hyperubiquitination, we examined the effect of overexpressing myc-ubiquitin with K48R, K63R, or K48R K63R mutations on Art4 high-MW conjugates as described earlier (Figure 3G). We found that expressing myc-ubiquitinK63R substantially reduced Art4 hyperubiquitination in ubp2Δ ubp15Δ cells, whereas myc-ubiquitinK48R had only a mild effect (Figure 5A). In addition, the effect of the K48R K63R double mutations was similar to that of the K63R single mutation. These results suggest Art4 hyperubiquitination in ubp2Δ ubp15Δ is mediated mainly by K63-linked polyubiquitin chains, although it is possible that other linkage types, including K48, may still be involved.
FIGURE 5:
Art4 hyperubiquitination and degradation in ubp2Δ ubp15Δ depends on the formation of K63-linked polyubiquitin chains. (A) pdr5Δ art4Δ and pdr5Δ art4Δ ubp2Δ ubp15Δ yeast cells carrying Art4-FLAG or empty vector, as well as WT, K48R, K63R, or K48R K63R myc-ubiquitin expression vector. Cells were treated with MG132 (25 μg/ml) for 60 min. Samples were immunoprecipitated using anti-FLAG antibody under denaturing conditions and analyzed by SDS–PAGE and immunoblot. The numbers above each lane indicate the relative ratio of myc-Ub-Art4 vs. total Art4 determined by the LI-COR quantification. To compare more accurately the levels of myc-Ub conjugated Art4, stacking gel was included for the analysis. *Nonspecific. (B) art4Δ and art4Δ ubp2Δ ubp15Δ yeast cells carrying Art4-FLAG, Art4-OTUB1-FLAG, Art4-OTUD1-FLAG, or Art4-AMSH-FLAG expression vector. Protein extracts were analyzed by SDS–PAGE and immunoblot. (C) Relative Art4/Art4-OTUB1, Art4-OTUD1, and Art4-AMSH levels (art4Δ ubp2Δ ubp15Δ cells versus art4Δ cells) as in B were quantified using the LI-COR system. Data from at least three independent experiments are presented as mean ± SD.
To investigate further whether one or both ubiquitin linkages are responsible for Art4 degradation in ubp2Δ ubp15Δ, we used a set of mammalian DUBs exhibiting linkage specificity/preference (McCullough et al., 2006; Edelmann et al., 2009; Wang et al., 2009; Mevissen et al., 2013; Hospenthal et al., 2015), including OTUB1 (highly K48 specific), OTUD1 (prefers K63 over K48), and AMSH (highly K63 specific). We fused these DUBs to Art4 and examined their relative abundance in ubp2Δ ubp15Δ cells compared with WT cells. Similar to WT Art4, the Art4-OTUB1 fusion showed reduced abundance in ubp2Δ ubp15Δ cells (Figure 5, B, lanes 1–4, and C), suggesting that OTUB1 fusion does not affect Art4 hyperubiquitination. The level of Art4-OTUD1 fusion in ubp2Δ ubp15Δ cells, however, was nearly identical to that in WT cells (Figure 5, B, lanes 5 and 6, and C), suggesting that OTUD1 efficiently prevents Art4 hyperubiquitination. In addition, Art4-AMSH exhibited an intermediate reduction in ubp2Δ ubp15Δ cells (Figure 5, B, lanes 7 and 8, and C), suggesting partial inhibition of Art4 hyperubiquitination by AMSH. To examine whether OTUB1 and OTUD1 fusion constructs are functionally active toward K48-linked polyubiquitin chains, we also compared the effect of OTUB1 and OTUD1 fusions on CPY* degradation, which is mediated by K48 polyubiquitination (Hiller et al., 1996). We found both OTUB1 and OTUD1 delayed the turnover of CPY* (Supplemental Figure S5B), indicating that these DUB fusion constructs are indeed capable of cleaving K48-linked chains. Together these data suggest that although Art4 hyperubiquitination might be decorated by K48-linked chains in addition to K63-linked chains, K48-linked chains alone are not sufficient for Art4 degradation in ubp2Δ ubp15Δ. Instead, our results revealed that Art4 degradation in ubp2Δ ubp15Δ requires the formation of K63-linked chains. The fact that the Art4-OTUD1 fusion has a stronger effect than the Art4-AMSH fusion may suggest that either Art4 degradation in ubp2Δ ubp15Δ cells is mediated by heterotypic chains containing both K48 and K63 linkages, which are removed more efficiently by OTUD1 that cleaves both, or the degradation is mediated by very long K63 chains, which are poor substrates for AMSH without its binding partner STAM (McCullough et al., 2006; Baiady et al., 2016).
In ubp2Δ ubp15Δ, ARTs are hyperubiquitinated at residues different from the activating lysines
Next we investigated whether ART degradation in ubp2Δ ubp15Δ cells is caused by hyperubiquitination at previously identified lysine residues. Art1 K486 and Art4 4K (K235/245/264/267) are important for ubiquitination and endocytosis of several PM cargoes (Lin et al., 2008; Becuwe et al., 2012b), although they are not required for Ste2 endocytosis (Alvaro et al., 2014). To test whether the same lysine residues are also involved in ART hyperubiquitination and degradation in ubp2Δ ubp15Δ, we examined the levels of Art1K486R and Art44KR in WT versus ubp2Δ ubp15Δ cells. Note that Art1 migrates as two major species (Ub-Art1 and Art1) on the SDS gel, and the mobility shift is dependent on K486 (Lin et al., 2008). Whereas Art1K486R migrated predominantly as the fast-moving/nonubiquitinated band, the level of Art1K486R was still reduced in ubp2Δ ubp15Δ cells compared with UBP2 UBP15 cells (Figure 6A). Similarly, Art44KR did not suppress Art4 degradation in ubp2Δ ubp15Δ (Figure 6B). These results suggest that ART degradation in ubp2Δ ubp15Δ is mediated by functionally distinct ubiquitination events different from the activating ubiquitination.
FIGURE 6:
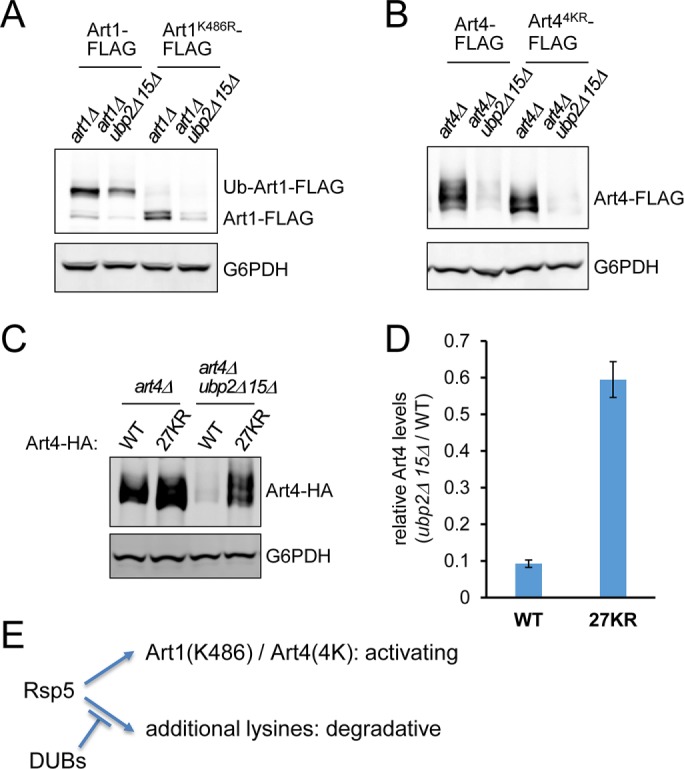
ART degradation in ubp2Δ ubp15Δ is not mediated by ubiquitination at the activating lysine residues. (A) Protein extracts of art1Δ and art1Δ ubp2Δ ubp15Δ yeast cells expressing plasmid-encoded Art1-FLAG or Art1K486R-FLAG were analyzed by SDS–PAGE and immunoblot. (B) Protein extracts of art4Δ and art4Δ ubp2Δ ubp15Δ yeast cells expressing plasmid-encoded Art4-FLAG or Art4K(235/245/264/267)R-FLAG were analyzed by SDS–PAGE and immunoblot. (C) Protein extracts of art4Δ and art4Δ ubp2Δ ubp15Δ cells expressing plasmid-encoded Art4-HA or Art427KR-HA were analyzed by SDS–PAGE and immunoblot. (D) Relative Art4 levels (art4Δ ubp2Δ ubp15Δ cells vs. art4Δ cells) as in C were quantified using the LI-COR system. Data from three independent experiments are presented as mean ± SD. (E) Model for functionally distinct ubiquitinations of ARTs. Rsp5 ubiquitinates Art1 at K486 and Art4 at K235/K245/K264/K267, which are required for their activation. In the absence of Ubp2 and Ubp15, additional lysines on ARTs are ubiquitinated, causing their degradation.
The prediction of Art4 ubiquitination sites using the online resource UbPred (Radivojac et al., 2010) revealed a cluster of lysine residues at the C-terminus and a few N-terminal lysine residues as potential sites of ubiquitination. To test whether Art4 degradation in ubp2Δ ubp15Δ is caused by ubiquitination at these sites, we generated Art4 mutants with different combinations of lysine-to-arginine (KR) substitutions (Supplemental Figure S6A). We found that the mutants with most of the N-terminal and C-terminal lysine sites mutated (27KR/30KR/44KR) prominently restored Art4 levels in ubp2Δ ubp15Δ cells (Figure 6, C and D, and Supplemental Figure S6B). We infer from these findings that ARTs can be regulated by two different types of ubiquitination: activating ubiquitination (such as at Art1 K486 or Art4 4K) and degradative ubiquitination, which is counteracted by Ubp2 and Ubp15 (Figure 6E).
Ubp2 and Ubp15 ensure efficient Hxt6 endocytosis by stabilizing Art4
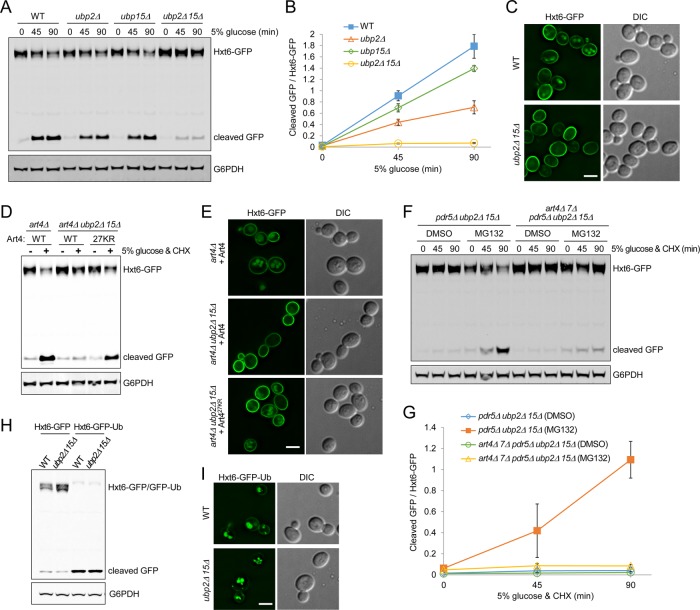
Art4 promotes endocytic trafficking of the PM hexose transporter Hxt6 in response to high concentrations of glucose (Nikko and Pelham, 2009; Llopis-Torregrosa et al., 2016). Given the prominent proteasomal degradation of Art4 in ubp2Δ ubp15Δ, we reasoned that Hxt6 endocytosis would be affected as well. Indeed, we found that ubp2Δ ubp15Δ cells exhibited a strong defect in glucose-induced Hxt6 endocytosis (Figure 7, A–C). The accumulation of Hxt6 at the PM in ubp2Δ ubp15Δ cells (Figure 7C) suggests that Ubp2 and Ubp15 affect Hxt6 internalization, likely the ubiquitination step mediated by Rsp5-ARTs. Endocytic trafficking of Jen1, another Art4-dependent PM cargo (Becuwe et al., 2012b), was also impaired in ubp2Δ ubp15Δ cells (Supplemental Figure S7A). To confirm that the Hxt6 endocytosis defect in ubp2Δ ubp15Δ is caused by Art4 hyperubiquitination and degradation, we examined the effect of Art427KR on Hxt6 endocytosis in ubp2Δ ubp15Δ cells. We used CHX as an additional inducer for Hxt6 endocytosis (Nikko and Pelham, 2009), as well as to inhibit protein synthesis. Therefore only the preexisting, PM-localized pool of Hxt6 can be delivered to the vacuole. We found that Art427KR substantially suppressed the Hxt6 trafficking defect in ubp2Δ ubp15Δ (Figure 7, D and E). We also tested whether MG132 treatment, which partially suppressed Art4 degradation (Figure 3D), can also alleviate the Hxt6 trafficking defect. We found that MG132 treatment considerably restored Hxt6 endocytic degradation in ubp2Δ ubp15Δ cells (Figure 7, F and G). Furthermore, Hxt6 endocytic degradation upon MG132 treatment was still dependent on Art4 and its paralogue Art7 (Figure 7, F and G), suggesting that upon proteasome inhibition, Hxt6 still undergoes physiological ubiquitination mediated by the ART-Rsp5 network.
FIGURE 7:
Ubp2 and Ubp15 promote Hxt6 endocytic down-regulation by stabilizing Art4. (A–C) The indicated strains expressing chromosomal Hxt6-GFP were grown to mid log phase in medium containing 0.05% glucose. Hxt6 endocytosis was stimulated with 5% glucose. (A) Protein extracts at the indicated time points were analyzed by SDS–PAGE and immunoblot. The cleaved GFP signals indicate the product of Hxt6-GFP degradation in the vacuole. (B) The ratio of cleaved GFP vs. full-length Hxt6-GFP as in A was determined by the quantification using the LI-COR system. Data from three independent experiments are presented as mean ± SD. (C) Samples were analyzed by fluorescence microscopy after 90 min of stimulation. Scale bar, 2.5 μm. (D, E) art4Δ and art4Δ ubp2Δ ubp15Δ cells expressing chromosomal Hxt6-GFP and plasmid-encoded Art4 or Art427KR were grown in medium containing 0.05% glucose. Hxt6 endocytosis was stimulated with 5% glucose and 50 μg/ml CHX. (D) Protein extracts after 3.5 h of stimulation were analyzed by SDS–PAGE and immunoblot. (E) Samples were analyzed by fluorescence microscopy after 3 h of stimulation. Scale bar, 2.5 μm. (F) pdr5Δ ubp2Δ ubp15Δ and art4Δ art7Δ pdr5Δ ubp2Δ ubp15Δ yeast cells expressing chromosomal Hxt6-GFP were grown to early log phase in medium containing 0.05% glucose and then mock-treated (DMSO) or treated with MG132 (25 μg/ml) for 90 min. Hxt6 endocytosis was stimulated with 5% glucose and 50 μg/ml CHX. Protein extracts from the indicated time points were analyzed by SDS–PAGE and immunoblot. (G) The ratio of cleaved GFP vs. full-length Hxt6-GFP as in F was determined by the quantification using the LI-COR system. Data from three independent experiments are presented as mean ± SD. (H) WT and ubp2Δ ubp15Δ cells expressing plasmid-encoded Hxt6-GFP or Hxt6-GFP-Ub were grown to mid log phase in medium containing 0.05% glucose. Protein extracts were analyzed by SDS–PAGE and immunoblot. (I) Similar to H, WT and ubp2Δ ubp15Δ cells expressing plasmid-encoded Hxt6-GFP-Ub were analyzed by fluorescence microscopy. Scale bar, 2.5 μm.
To further confirm that impaired Hxt6 trafficking in ubp2Δ ubp15Δ cells is not caused by other processes such as defects in MVB sorting, we used a single-ubiquitin fusion that bypasses Rsp5/ART-dependent ubiquitination and constitutively sorts cargo proteins into the vacuole (Stringer and Piper, 2011). We found that Hxt6-GFP-Ub was sorted to the vacuole and degraded in both WT and ubp2Δ ubp15Δ cells (Figure 7, H and I), suggesting that there is no gross defect in the ESCRT machinery or MVB formation. Moreover, we found that whereas Art2-dependent Lyp1 endocytosis (Lin et al., 2008) is partially defective (Supplemental Figure S7B), Art3-dependent Dip5 endocytosis (Hatakeyama et al., 2010) was nearly unchanged in ubp2Δ ubp15Δ cells (Supplemental Figure S7C). These trafficking phenotypes are consistent with the reduced abundance of Art2 (Figure 1D) and unaltered levels of Art3 (Supplemental Figure S1F) in ubp2Δ ubp15Δ. Of importance, these data also demonstrate that endocytic processes are not generally impaired. We therefore conclude that Ubp2 and Ubp15 ensure efficient endocytic down-regulation of Hxt6, at least in part by preventing Art4 hyperubiquitination and subsequent proteasomal degradation.
DISCUSSION
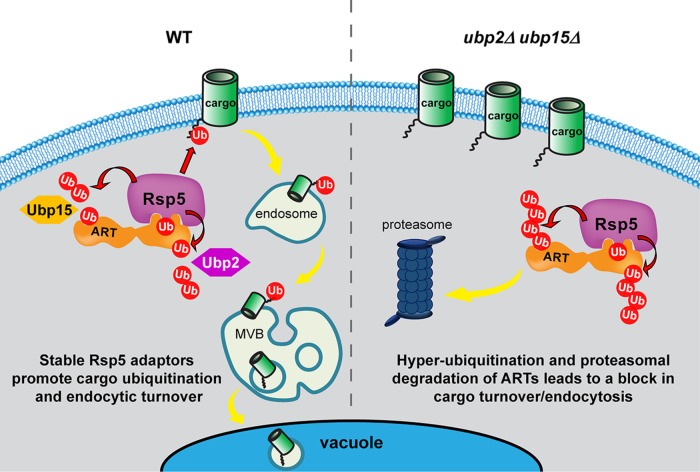
Endocytic down-regulation is crucial for maintaining PM protein homeostasis. By regulating the composition of integral membrane proteins at the PM, cells are able to adapt to various environmental stimuli and challenges, such as nutrient availability and heat stress. The ART-Rsp5 network ubiquitinates cargoes and promotes their internalization at the PM. Therefore proper regulation of ART-Rsp5 is of great importance for cells to survive in response to the changing environment. In this study, we identified two deubiquitinating enzymes, Ubp2 and Ubp15, as novel regulators of the ART-Rsp5 network. We demonstrate that Ubp2 and Ubp15 maintain the stability of several ARTs by counteracting Rsp5-mediated hyperubiquitination and subsequent proteasomal degradation of ARTs. Loss of Ubp2 and Ubp15 causes decreased steady-state levels of ARTs, leading to defects in endocytic turnover of PM cargoes (Figure 8). Our results revealed another layer of regulatory complexity in the ART-Rsp5 network: besides inhibitory phosphorylation/activating dephosphorylation by kinases/phosphatases, ARTs are also subject to ubiquitination/deubiquitination regulation imposed by E3/DUBs so that cells can maintain appropriate levels of Rsp5 adaptor proteins for proper control of endocytosis and PM composition.
FIGURE 8:
Model for Ubp2 and Ubp15 preventing Rsp5-mediated ART hyperubiquitination. In WT cells, Ubp2 and Ubp15 counteract Rsp5 activity and prevent ART hyperubiquitination. This is achieved most likely by physical interactions between Ubp2 and Rsp5, as well as between Ubp15 and ARTs. We propose that ART-Rsp5 complexes are direct targets of Ubp2 and/or Ubp15. Ubp2 and Ubp15 may regulate different steps of an ART-Rsp5 loading cycle, with Ubp2 engaging ART-Rsp5 complexes by binding to Rsp5, and Ubp15 engaging ARTs. Whether Ubp15 interacts with ARTs in complexes with Rsp5 or with ARTs that have dissociated from Rsp5 is unclear. ART-Rsp5 promotes PM cargo ubiquitination, leading to endocytic down-regulation. In ubp2Δ ubp15Δ cells, ARTs are hyperubiquitinated by Rsp5. At least several ARTs exhibit enhanced protein degradation through the proteasome, leading to reduced ART abundance and defective cargo turnover. The degradative hyperubiquitination of Art4 occurs most likely at N- and C-terminal regions. Regions of degradative ubiquitination on other ARTs have not been determined. Although the model illustrates that Ubp2 and Ubp15 target different residues/regions, it is unknown how these DUBs recognize substrates or whether motif specificity exists. Ub, ubiquitin; red arrow, Rsp5 E3 ligase activity.
The ART-Rsp5 network is regulated by the ubiquitin system
It is evident that many components of the ubiquitin system are also regulated by ubiquitination (Weissman et al., 2011). Several ARTs are ubiquitinated, but functions of these modifications are not always clear (Kee et al., 2006; Lin et al., 2008; Herrador et al., 2010; Becuwe et al., 2012b; Alvaro et al., 2014). Here we show Ubp2 and Ubp15 counteract ART hyperubiquitination, thereby preventing degradation of several ARTs. On the basis of our findings and those from other studies, we propose that Ubp2 and Ubp15 are crucial regulators for the ART-Rsp5 network. Ubp2 operates on ARTs by binding to Rsp5 (Kee et al., 2005), and Ubp15 may engage ARTs directly or indirectly through other interacting partners (Supplemental Figure S2, A–C, and Figure 8). Additional work is required to determine mechanistic details of ART-Ubp15 interactions.
In contrast to the ubp2Δ ubp15Δ mutant, deletion of DOA4 or UBP3 inhibited the degradation of ARTs (Supplemental Figure S1, C–E). It is known that Doa4 maintains free ubiquitin levels by recycling ubiquitin from the MVB pathway (Swaminathan et al., 1999; Amerik et al., 2000b). Hence ubiquitin deficiency in doa4Δ mutants may in turn limit ART degradation in the absence of Ubp2 and Ubp15. How ubp3Δ affects ART degradation in ubp2Δ ubp15Δ is less clear, as there is no obvious defect in free ubiquitin levels in ubp3Δ (Amerik et al., 2000a). Recently Ubp3 and Ubp2 were shown to regulate the Rsp5-mediated cytosolic protein quality control pathway upon heat stress (Fang et al., 2016). In this pathway, Ubp3 and Ubp2 cleave Rsp5-conjugated K63 chains, which, together with the enhanced K48-catalyzing activity of Rsp5 upon heat shock, promotes proteasomal degradation of cytosolic misfolded proteins. Although this function of Ubp3 could potentially explain the suppression of the ART degradation observed here, we suspect that it is unlikely for the following reasons: 1) Ubp2 plays an opposite role and inhibits the degradation of ARTs; 2) the degradation of ARTs in ubp2Δ ubp15Δ cells does not require heat shock; and 3) K48-linked polyubiquitination does not play a major role in ART degradation in ubp2Δ ubp15Δ.
Proteasomal degradation of Art4 requires K63-linked polyubiquitin chains
We show in this study that Art4 hyperubiquitination in ubp2Δ ubp15Δ is mediated mainly by K63-linked chains (Figure 5A), consistent with the preferences of Rsp5, Ubp2, and Ubp15 toward K63 linkages (Kee et al., 2005, 2006; Kim and Huibregtse, 2009; Schaefer and Morgan, 2011). Given that we find no evidence that Art4 (as well as Art10 and Art1) is degraded in the vacuole (Figure 3, A–C), and the proteasome inhibitor MG132 can significantly suppress ART degradation in ubp2Δ ubp15Δ cells (Figure 3, D–F), it is likely that hyperubiquitination of ARTs leads to protein degradation by the 26S proteasome. Collectively these findings suggest that ART degradation in ubp2Δ ubp15Δ cells is via unconventional K63-mediated proteasomal targeting, because proteasomal degradation is widely considered to be mediated by non-K63 linkages (Chau et al., 1989; Finley et al., 1994; Hershko and Ciechanover, 1998; Pickart and Fushman, 2004; Xu et al., 2009). It is also evident, however, that K63-linked polyubiquitin chains can be used as the proteasome targeting signal in certain circumstances (Saeki et al., 2009; Isasa et al., 2016), and longer chain lengths can potentially increase the affinity with the proteasome (Saeki et al., 2009). Furthermore, proteasome recruitment of a human E3 ligase, TRIM21, involved in virus degradation is preceded by the formation of K63-linked polyubiquitination (Fletcher et al., 2015). Hyperubiquitinated ARTs may therefore represent another example of the complexity of the ubiquitin-proteasome pathway and highlight the importance of K63-specific DUBs in preserving nonproteolytic ubiquitinated proteins.
Alternatively, the degradation of ARTs in ubp2Δ ubp15Δ can be the consequence of heterotypic ubiquitin chains containing both K63 and K48 linkages, which could have higher affinity for the proteasome, as described previously for K11/K48 mixed ubiquitin chains (Grice et al., 2015). We demonstrate here that the OTUB1 fusion can delay CPY* turnover mediated by K48-linked polyubiquitination (Supplemental Figure S5B) but has no effect on Art4 degradation in ubp2Δ ubp15Δ (Figure 5, B and C). These data suggest that Art4 degradation in ubp2Δ ubp15Δ is unlikely caused solely by K48 linkages. We cannot, however, rule out the possibility that in addition to K63 linkages, some K48-linked chains are present and enhance the delivery to the proteasome. Such K48 linkages could potentially be catalyzed by Rsp5 itself, as evident by the considerable (though weaker than K63) K48-conjugating activity of Rsp5 in vitro (French et al., 2009; Kim and Huibregtse, 2009), or they could result from activities of other E3 ligases or E4 enzymes that require ubiquitination events primed by Rsp5. Although we show here that several E3s/E4s, including San1, Ubr1, Doa10, Cul3, and Hul5, are not involved (Supplemental Figure S4, A and D), it remains possible that there are other ligase activities conjugating these K48 linkages.
Modulation of the ART-Rsp5 network by deubiquitinating enzymes Ubp2 and Ubp15
Of the 10 Rsp5 adaptors we tested, steady-state levels of 8—Art1, Art2, Art4, Art6, Art7, Art8, Art10, and Bul1—are reduced in ubp2Δ ubp15Δ cells (Figure 1, B–I). It is intriguing that Ubp2 and Ubp15 have little or no effect on the levels of Art3 and Art9 (Supplemental Figure S1, F and G), and yet Art3 ubiquitination is still increased in the absence of Ubp2 and Ubp15 (Supplemental Figure S3C). It therefore seems that Ubp2 and Ubp15 are general regulators for preventing hyperubiquitination of most, if not all, Rsp5 adaptor proteins, but the outcomes (degraded or not) of hyperubiquitination are further dictated by other factors, such as protein–protein interactions, posttranslational modifications, or subcellular localizations. Further studies are required to elucidate mechanisms determining the fates of individual Rsp5 adaptor proteins upon hyperubiquitination.
It is an emerging theme that many DUBs physically interact with E3 ubiquitin ligases and in many cases protect E3s from autoubiquitination and degradation. Whereas Rsp5 remains stable in the absence of Ubp2 and Ubp15 (Supplemental Figure S1H), we found in this study that ART adaptor proteins are susceptible to Rsp5-mediated hyperubiquitination and degradation, and therefore require DUB activities to maintain their stability and trafficking function. ARRDC1, one of the mammalian counterparts of ARTs, has been shown to form heterodimers with β-arrestins and recruit the Nedd4 ubiquitin ligase Itch to down-regulate Notch receptor at the PM (Puca et al., 2013). Of interest, the mammalian DUB USP9X associates with Itch and protects Itch from proteasomal degradation (Mouchantaf et al., 2006). It is conceivable that USP9X or other DUBs may also function in receptor down-regulation by protecting ARRDCs or other ligase–adaptor complexes. The human orthologue of Ubp15, USP7, is an integral component of an endosomal E3 ligase complex and promotes endosomal protein recycling by preventing autoubiquitination and degradation of the ligase (Hao et al., 2015). It remains to be seen whether USP7 is also involved in regulating PM receptors by associating with other E3 ligases and/or adaptors.
Ubp2 and Ubp15 affect cargo trafficking by regulating ART stability
This work unveiled a novel role for Ubp2 and Ubp15 in maintaining the stability of ARTs by preventing their hyperubiquitination and subsequent proteasomal degradation. Given that ARTs recognize PM cargoes and promote their endocytic turnover, Ubp2 and Ubp15 may modulate the endocytic pathway by stabilizing ARTs. We demonstrate that ubp2Δ ubp15Δ cells exhibit defects in Hxt6 and Jen1 endocytic turnover (Figure 7, A–C, and Supplemental Figure S7A), consistent with the instability of Art4 (Figure 1B). These findings imply that the endocytic phenotypes in ubp2Δ ubp15Δ cells are likely caused by the degradation of ARTs. In support of this idea, suppressing Art4 degradation by either proteasome inhibition (Figure 3D) or 27KR mutations (Figure 6, C and D) considerably restores Hxt6 trafficking to the vacuole in ubp2Δ ubp15Δ (Figure 7, D–G).
Of note, partial suppression of the Hxt6 trafficking defect by proteasome inhibition reveals that ARTs reside at a point of cross-talk between vacuolar and proteasomal protein degradation pathways. Recently expression of nonpolymerizable (lysine-less/K0-Ub) ubiquitin has been used to identify polyubiquitination-dependent proteasomal substrates (stabilized upon K0-Ub expression) and monoubiquitination-dependent proteasomal substrates (further degraded upon K0-Ub expression; Braten et al., 2016). Jen1 and Hxt6 are among the proteins exhibiting enhanced degradation upon expression of K0-Ub. Although it is possible that Jen1 and Hxt6 are direct substrates of the proteasome under certain conditions, as shown by the increased proteasomal targeting of other PM transporters at low temperature (Isasa et al., 2016), this finding can also be explained by increased vacuolar degradation due to stabilization of Art4 upon K0 expression, consistent with the present observations that Art4 is a proteasomal substrate and proteasome inhibition improves Hxt6 trafficking to the vacuole in ubp2Δ ubp15Δ.
Although our findings strongly support a model in which Ubp2 and Ubp15 affect endocytic trafficking by regulating ART stability, they do not exclude the possibility that Ubp2 and Ubp15 play additional roles in the endocytic pathway. Ubp2 has been implicated in efficient MVB sorting of Fur4 and Cps1 (Ren et al., 2007; Lam et al., 2009). Ubp2, along with Ubp7, has also been shown to regulate formation and stability of the endocytic coat by deubiquitinating the early endocytic protein Ede1 (Weinberg and Drubin, 2014). It remains to be investigated how certain DUBs, such as Ubp2, can coordinate various steps, including cargo ubiquitination, internalization, and MVB sorting, to achieve successful endocytic down-regulation of PM cargoes.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions
All Saccharomyces cerevisiae strains and plasmids used in this study are listed in Supplemental Tables S1 and S2. Standard procedures were used for manipulation of yeast. Homologous recombination was used to tag or delete genes (Longtine et al., 1998). All integrations and deletions were verified by PCR analysis. For protein extractions and fluorescence microscopy, yeast liquid cultures were grown to mid log phase and analyzed. All yeast cultures were grown at 30°C unless otherwise indicated. To induce Hxt6 expression and PM localization, synthetic medium with 0.05% glucose (instead of regular 2% glucose) was used. Under this condition, the culture density is able to reach OD600 ≈ 1.2. For the Hxt6-GFP degradation assay, yeast cells in medium with 0.05% glucose were grown to OD600 ≈ 0.4–0.5 before addition of 5% glucose, or 5% glucose plus 50 μg/ml CHX to induce Hxt6 endocytic turnover.
Cellular protein levels and antibodies
Denaturing whole-cell lysates were prepared by pelleting three to five OD600 equivalents of cells and precipitating with 10% trichloroacetic acid (TCA). Samples were then washed in acetone, dried, resuspended in cracking buffer (6 M urea, 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% SDS), and mechanically disrupted by bead beating. Urea sample buffer (6 M urea, 150 mM Tris, pH 6.8, 6% SDS, and 10% β-mercaptoethanol [2-ME] with bromophenol blue) was added, and extracts were analyzed by SDS–PAGE and immunoblotting. For analysis of PM cargoes (Hxt6-GFP, Lyp1-GFP, Dip5-GFP), 8 M urea and 8% SDS were included in both cracking buffer and sample buffer, with replacement of 2-ME with 100 mM dithiothreitol in sample buffer. Standard SDS gels were used (6% for ARTs; 9.5% for PM cargoes). For Hxt6-GFP analysis, stacking gel was omitted to reduce protein aggregation. Quantitative fluorescence imaging of immunoblots was performed using an Odyssey infrared imaging system (LI-COR Biosciences). Protein levels were quantified by fluorescence intensity of immunoblots using the Odyssey software or Image Studio Lite software. For presentations, immunoblots were linearly adjusted for brightness and contrast in Odyssey or Image Studio Lite (which does not affect their quantifications) and cropped in Photoshop (Adobe). Antibodies used in this study include anti-FLAG (M2; Sigma-Aldrich), anti-hemagglutinin (12CA5; Roche), anti-GFP (B2; Santa Cruz Biotechnology), anti–glucose-6-phosphate dehydrogenase (G6PDH; Sigma-Aldrich), anti-myc (9E10; Sigma-Aldrich), anti-GFP (TP401; Torrey Pines Biolabs), and anti-FLAG (F7425; Sigma-Aldrich).
Immunoprecipitation and detection of polyubiquitinated ARTs
To facilitate the detection of proteasome-targeting polyubiquitin conjugates, MG132 (25 μg/ml; ApexBio) was used to inhibit the proteasome in the pdr5Δ strain background. Myc-ubiquitin was overexpressed under the control of the copper-inducible CUP1 promoter. Cells were grown to early log phase, treated with 100 μM CuSO4 for 2–3 h, and then treated with MG132 for 1 h. Between 20 and 25 OD600 equivalents of cells were harvested and precipitated with 10% TCA. Acetone-washed samples were disrupted by bead beating in 200 μl of cracking buffer (with 6 M urea), cleared by centrifugation at 21,130 × g for 3 min (room temperature), and then 10-times diluted in IP buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.2% NP-40, 5 mM EDTA, protease inhibitors, phosphatase inhibitors, 2.5 mM N-ethylmaleimide, and 25 μg/ml bortezomib). Art1/2/4-FLAG was immunoprecipitated with M2 anti-FLAG beads (Sigma-Aldrich) at 4°C for 2.5 h. Beads were then washed three times with IP buffer and resuspended in IP buffer plus urea sample buffer (1:1 ratio). Elutes were analyzed by SDS–PAGE and immunoblotting. To enhance the transfer efficiency of high-MW polyubiquitinated proteins, methanol was completely omitted from the transfer buffer. Ubiquitinated ARTs were detected by α-myc (9E10) antibody.
Microscopy
All microscopy was performed using the DeltaVision RT (Applied Precision, Issaquah, WA) equipped with a 100× objective and fluorescein isothiocyanate and rhodamine filters. Images were captured with a digital camera (CoolSNAP HQ; Photometrics) and deconvolved using the softWoRx software (Applied Precision). Images were linearly adjusted for brightness and contrast in softWoRx and cropped in Photoshop (Adobe).
Supplementary Material
Acknowledgments
We thank Shih-Chi Hsu and Sho Suzuki for critical reading of the manuscript, members of the Emr lab and Jeremy Thorner for helpful discussions, Shih-Chi Hsu and Ludovic Giloteaux for reagents and antibodies, and Marcus Smolka for assistance with mass spectrometry. J.A.M. is supported by a National Institutes of Health Pathway to Independence Award (R00 GM101077). This work was supported by a Cornell University Research Grant to S.D.E.
Abbreviations used:
- ART
arrestin-related trafficking adaptor
- DUB
deubiquitinating enzyme
- ESCRT
endosomal sorting complexes required for transport
- MVB
multivesicular body
- PM
plasma membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0008) on March 15, 2017.
REFERENCES
- Alvaro CG, Aindow A, Thorner J. Differential phosphorylation provides a switch to control how alpha-arrestin rod1 down-regulates mating pheromone response in saccharomyces cerevisiae. Genetics. 2016;203:299–317. doi: 10.1534/genetics.115.186122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro CG, O’Donnell AF, Prosser DC, Augustine AA, Goldman A, Brodsky JL, Cyert MS, Wendland B, Thorner J. Specific alpha-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol Cell Biol. 2014;34:2660–2681. doi: 10.1128/MCB.00230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000a;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000b;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton F, Dittmar G, Langer T, Escobar-Henriques M. Two deubiquitylases act on mitofusin and regulate mitochondrial fusion along independent pathways. Mol Cell. 2013;49:487–498. doi: 10.1016/j.molcel.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Ast T, Aviram N, Chuartzman SG, Schuldiner M. A cytosolic degradation pathway, prERAD, monitors pre-inserted secretory pathway proteins. J Cell Sci. 2014;127:3017–3023. doi: 10.1242/jcs.144386. [DOI] [PubMed] [Google Scholar]
- Baiady N, Padala P, Mashahreh B, Cohen-Kfir E, Todd EA, Du Pont KE, Berndsen CE, Wiener R. The Vps27/Hrs/STAM (VHS) domain of the signal-transducing adaptor molecule (STAM) directs associated molecule with the SH3 domain of STAM (AMSH) specificity to longer ubiquitin chains and dictates the position of cleavage. J Biol Chem. 2016;291:2033–2042. doi: 10.1074/jbc.M115.689869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Herrador A, Haguenauer-Tsapis R, Vincent O, Leon S. Ubiquitin-mediated regulation of endocytosis by proteins of the arrestin family. Biochem Res Int. 2012a;2012:242764. doi: 10.1155/2012/242764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Leon S. Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and the ubiquitin ligase Rsp5. Elife. 2014;3 doi: 10.7554/eLife.03307. doi: 10.7554/eLife.03307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Leon S. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol. 2012b;196:247–259. doi: 10.1083/jcb.201109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braten O, Livneh I, Ziv T, Admon A, Kehat I, Caspi LH, Gonen H, Bercovich B, Godzik A, Jahandideh S, et al. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci USA. 2016;113:E4639–E4647. doi: 10.1073/pnas.1608644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, Howie RL, O’Dell A, McNally JG, Liebman SW, et al. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell. 2011;43:242–252. doi: 10.1016/j.molcel.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. J Cell Sci. 2012a;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Liu H, Urbe S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell. 2012b;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Crapeau M, Merhi A, Andre B. Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J Biol Chem. 2014;289:22103–22116. doi: 10.1074/jbc.M114.582320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Schnell JD, Maldonado-Baez L, Wendland B, Hicke L. The function of yeast epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic. 2010;11:151–160. doi: 10.1111/j.1600-0854.2009.01003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre S, Haguenauer-Tsapis R. Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol Cell Biol. 2001;21:4482–4494. doi: 10.1128/MCB.21.14.4482-4494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann MJ, Iphofer A, Akutsu M, Altun M, di Gleria K, Kramer HB, Fiebiger E, Dhe-Paganon S, Kessler BM. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- Erpapazoglou Z, Dhaoui M, Pantazopoulou M, Giordano F, Mari M, Leon S, Raposo G, Reggiori F, Haguenauer-Tsapis R. A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol Biol Cell. 2012;23:2170–2183. doi: 10.1091/mbc.E11-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang NN, Chan GT, Zhu M, Comyn SA, Persaud A, Deshaies RJ, Rotin D, Gsponer J, Mayor T. Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat Cell Biol. 2014;16:1227–1237. doi: 10.1038/ncb3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang NN, Zhu M, Rose A, Wu KP, Mayor T. Deubiquitinase activity is required for the proteasomal degradation of misfolded cytosolic proteins upon heat-stress. Nat Commun. 2016;7:12907. doi: 10.1038/ncomms12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JA, Lightcap ES, Sadis S, Thoroddsen V, Bulawa CE, Blackman RK. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc Natl Acad Sci USA. 2002;99:1461–1466. doi: 10.1073/pnas.032516399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AJ, Mallery DL, Watkinson RE, Dickson CF, James LC. Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proc Natl Acad Sci USA. 2015;112:10014–10019. doi: 10.1073/pnas.1507534112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French ME, Kretzmann BR, Hicke L. Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem. 2009;284:12071–12079. doi: 10.1074/jbc.M901106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice GL, Lobb IT, Weekes MP, Gygi SP, Antrobus R, Nathan JA. The proteasome distinguishes between heterotypic and homotypic lysine-11-linked polyubiquitin chains. Cell Rep. 2015;12:545–553. doi: 10.1016/j.celrep.2015.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3:116. doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YH, Fountain MD, Jr, Fon Tacer K, Xia F, Bi W, Kang SH, Patel A, Rosenfeld JA, Le Caignec C, Isidor B, et al. USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol Cell. 2015;59:956–969. doi: 10.1016/j.molcel.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CA, Conaway RC, Conaway JW, Svejstrup JQ. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci USA. 2009;106:20705–20710. doi: 10.1073/pnas.0907052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama R, Kamiya M, Takahara T, Maeda T. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol Cell Biol. 2010;30:5598–5607. doi: 10.1128/MCB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings BA, Zubenko GS, Hasilik A, Jones EW. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:435–439. doi: 10.1073/pnas.78.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Herrador A, Herranz S, Lara D, Vincent O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol Cell Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrador A, Livas D, Soletto L, Becuwe M, Leon S, Vincent O. Casein kinase 1 controls the activation threshold of an alpha-arrestin by multisite phosphorylation of the interdomain hinge. Mol Biol Cell. 2015;26:2128–2138. doi: 10.1091/mbc.E14-11-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hospenthal MK, Mevissen TE, Komander D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest) Nat Protoc. 2015;10:349–361. doi: 10.1038/nprot.2015.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Isasa M, Suner C, Diaz M, Puig-Sarries P, Zuin A, Bichman A, Gygi SP, Rebollo E, Crosas B. Cold temperature induces the reprogramming of proteolytic pathways in yeast. J Biol Chem. 2016;291:1664–1675. doi: 10.1074/jbc.M115.698662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Munoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem. 2006;281:36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- Khosrow-Khavar F, Fang NN, Ng AH, Winget JM, Comyn SA, Mayor T. The yeast ubr1 ubiquitin ligase participates in a prominent pathway that targets cytosolic thermosensitive mutants for degradation. G3 (Bethesda) 2012;2:619–628. doi: 10.1534/g3.111.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29:3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Lam MH, Urban-Grimal D, Bugnicourt A, Greenblatt JF, Haguenauer-Tsapis R, Emili A. Interaction of the deubiquitinating enzyme Ubp2 and the e3 ligase Rsp5 is required for transporter/receptor sorting in the multivesicular body pathway. PLoS One. 2009;4:e4259. doi: 10.1371/journal.pone.0004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010;20:196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Llopis-Torregrosa V, Ferri-Blazquez A, Adam-Artigues A, Deffontaines E, van Heusden GP, Yenush L. Regulation of the yeast Hxt6 hexose transporter by the Rod1 alpha-arrestin, the Snf1 protein kinase, and the Bmh2 14-3-3 protein. J Biol Chem. 2016;291:14973–14985. doi: 10.1074/jbc.M116.733923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lu JY, Lin YY, Qian J, Tao SC, Zhu J, Pickart C, Zhu H. Functional dissection of a HECT ubiquitin E3 ligase. Mol Cell Proteomics. 2008;7:35–45. doi: 10.1074/mcp.M700353-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Emr SD. Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem. 2012;81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2011;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- Merhi A, Andre B. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol Cell Biol. 2012;32:4510–4522. doi: 10.1128/MCB.00463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard SM, Wood SA. Riding the DUBway: regulation of protein trafficking by deubiquitylating enzymes. J Cell Biol. 2006;173:463–468. doi: 10.1083/jcb.200602082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, Angers A. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 2006;281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Nikko E, Pelham HR. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF, Apffel A, Gardner RG, Cyert MS. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol Biol Cell. 2010;21:3552–3566. doi: 10.1091/mbc.E10-07-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF, Huang L, Thorner J, Cyert MS. A calcineurin-dependent switch controls the trafficking function of alpha-arrestin Aly1/Art6. J Biol Chem. 2013;288:24063–24080. doi: 10.1074/jbc.M113.478511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF, McCartney RR, Chandrashekarappa DG, Zhang BB, Thorner J, Schmidt MC. 2-Deoxyglucose impairs Saccharomyces cerevisiae growth by stimulating Snf1-regulated and alpha-arrestin-mediated trafficking of hexose transporters 1 and 3. Mol Cell Biol. 2015;35:939–955. doi: 10.1128/MCB.01183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ. Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell. 2007;18:707–720. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Piper RC, Dikic I, Lukacs GL. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb Perspect Biol. 2014;6:a16808. doi: 10.1101/cshperspect.a016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DC, Pannunzio AE, Brodsky JL, Thorner J, Wendland B, O’Donnell AF. alpha-Arrestins participate in cargo selection for both clathrin-independent and clathrin-mediated endocytosis. J Cell Sci. 2015;128:4220–4234. doi: 10.1242/jcs.175372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca L, Chastagner P, Meas-Yedid V, Israel A, Brou C. Alpha-arrestin 1 (ARRDC1) and beta-arrestins cooperate to mediate Notch degradation in mammals. J Cell Sci. 2013;126:4457–4468. doi: 10.1242/jcs.130500. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Kee Y, Huibregtse JM, Piper RC. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol Biol Cell. 2007;18:324–335. doi: 10.1091/mbc.E06-06-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer JB, Morgan DO. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem. 2011;286:45186–45196. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenova SD, Dunn R, Adler AS, Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J Biol Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol. 2011;192:229–242. doi: 10.1083/jcb.201008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, et al. Yeast reveal a “druggable” Rsp5/Nedd4 network that ameliorates alpha-synuclein toxicity in neurons. Science. 2013;342:979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JS, Drubin DG. Regulation of clathrin-mediated endocytosis by dynamic ubiquitination and deubiquitination. Curr Biol. 2014;24:951–959. doi: 10.1016/j.cub.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM, Shabek N, Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol. 2011;12:605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Scheel H, Hofmann K, Komander D. Dissection of USP catalytic domains reveals five common insertion points. Mol Biosyst. 2009;5:1797–1808. doi: 10.1039/b907669g. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Macgurn JA, Liu M, Emr S. The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. Elife. 2013;2:e00459. doi: 10.7554/eLife.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.