Abstract
Routine manufacture, detonation and disposal of explosives in land and groundwater have resulted in complete pollution. Explosives are xenobiotic compounds, being toxic to biological systems, and their recalcitrance leads to persistence in the environment. The methods currently used for the remediation of explosive contaminated sites are expensive and can result in the formation of toxic products. The present study aimed to investigate the bacterial strains using the Biolog plates in the soil from the Riyadh community. The microbial strains were isolated using the spread plate technique and were identified using the Biolog method. In this study we have analyzed from bacterial families of soil samples, obtained from the different sites in 5 regions at Explosive Institute. Our results conclude that Biolog MicroPlates were developed for the rapid identification of bacterial isolates by sole-carbon source utilization and can be used for the identification of bacteria. Out of five communities, only four families of bacteria indicate that the microbial community lacks significant diversity in region one from the Riyadh community in Saudi Arabia. More studies are needed to be carried out in different regions to validate our results.
Keywords: Bacterial strains, Contaminated soil, Riyadh community, Biolog plate
1. Introduction
Bacteria use wastes for their own metabolism and finally produce some simple and useful compounds which are important for soil health, plant growth and overall to keep a good balance of the natural ecosystem (Manonmani et al., 2015). Bacteria, fungi and actinomycetes are important contributors to optimal agricultural and kitchen waste bioconversion (Barman et al., 2011, Manonmani et al., 2015). The microbial ecology of the soil is extremely important to the biogeochemical cycling of nutrients that is vital to life on our planet. Soil provides the largest reservoir of biodiversity on Earth, and sustains all other forms of terrestrial diversity while providing many ecosystem services (Hinsinger et al., 2009). Understanding the microbial community structure is important for predicting the fate of contaminants in the environment (Manonmani et al., 2015). At the most basic level, characterizing the microbial community can give an indication of whether the desired microorganisms for bioremediation are present. If there is no population capable of degradative processes, inoculation with foreign organisms may be required (Liu and Suflita, 1993). Methods used to assign strains to bacterial species, and the definition of species, have been much debated (Cohan, 2006, Gevers et al., 2005, Gevers et al., 2006, Staley, 2006). There is widespread recognition that the current methods of defining prokaryotic species are no longer adequate and there has been little progress in developing an agreed concept of species that can guide a new definition of species (Achtman and Wagner, 2008, Cohan, 2002, Fraser et al., 2007). The current approach for distinguishing species within a genus, and for defining new species, is based on polyphasic taxonomy, which incorporates all available phenotypic and genotypic data into a consensus classification (Vandamme et al., 1996). One approach is to observe the distribution of a large number of strains of closely related species in sequence space and to identify clusters of strains that are well resolved from other clusters (Bishop et al., 2009). In this study, we aimed to investigate the bacterial strains using the Biolog plate method to evaluate soil from the Riyadh community.
2. Materials and methods
2.1. Isolation of bacteria
Soil samples were collected from the Riyadh province, Saudi Arabia. The soil samples were passed through a sieve (1.7 mm mesh) to remove large pieces of debris and vegetation. The bacteria were originally isolated by plating dilutions of soils in saline solution (0.9% NaCl) on nutrient agar containing glucose, 10 g/L, NaCl, 1 g/L, peptone, 5 g/L, yeast extract, 3 g/L and agar 20 g/L and incubated at 37 °C for 48 h. The developed colonies were counted in plates and the average number of colonies per three plates was determined. The number of total bacteria (CFU) per gram dry weight of soil was determined. Individual colonies of bacteria which varied in shape and color were picked up and purified by streaking on nutrient agar. The bacterial isolates were kept on nutrient agar at 4 °C and recultured every 4 weeks. Serial dilution method for bacterial isolation and purification from explosive-contaminated soil was shown in (Figure 1, Figure 2).
Figure 1.
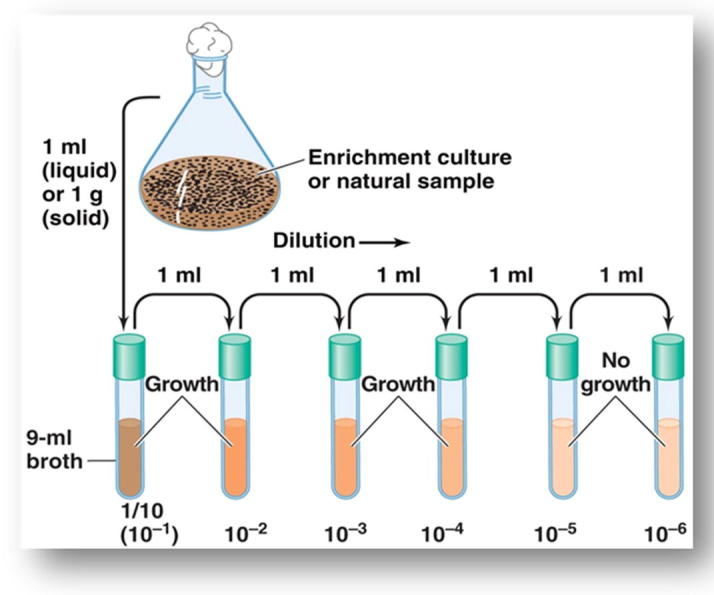
Serial dilution method for bacterial isolation from explosive-contaminated soil.
Figure 2.
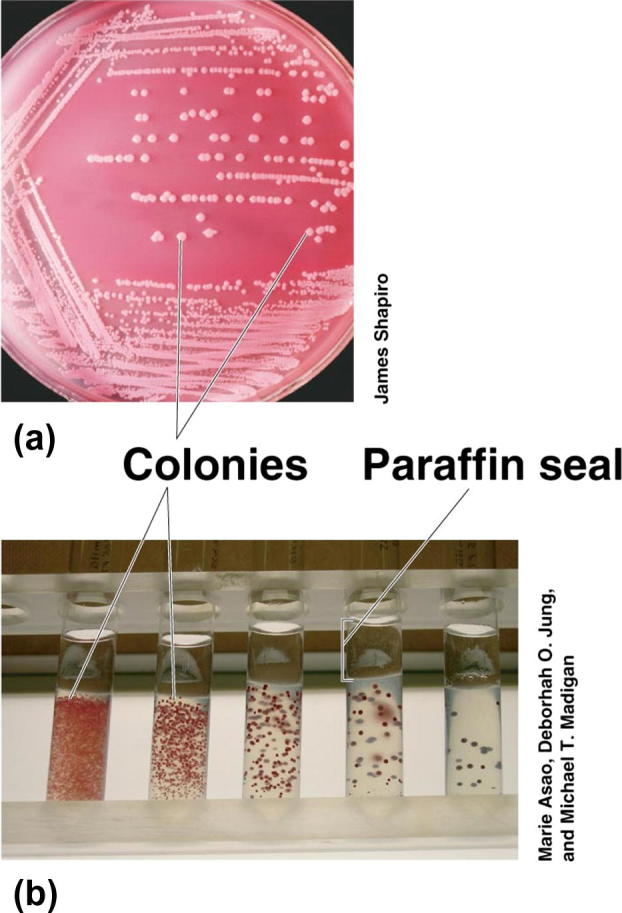
Pure culture methods from bacteria.
2.2. Biolog fingerprint: bacterial species identification system
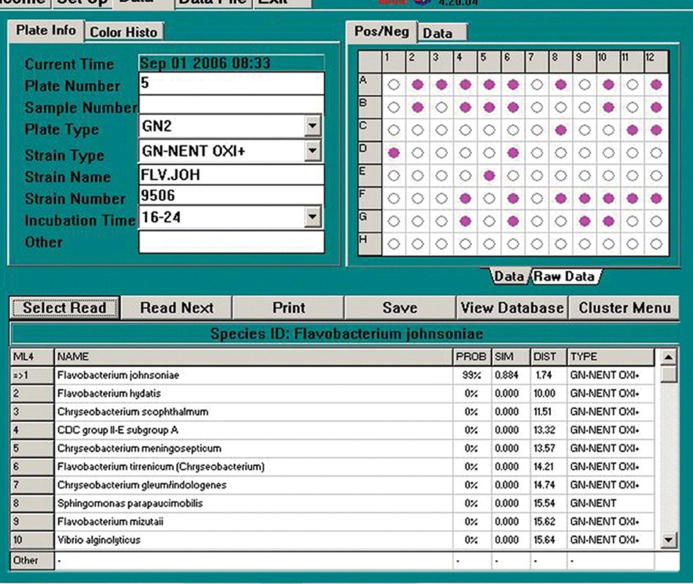
The Biolog (Biolog Inc., Hayward, CA, USA; http://www.biolog.com/microID.html) utilizes and identification test panel (YT Micro Plate) consisting of a matrix of 8–12 wells (Fig. 3). The first three rows contain 35 carbon source oxidation tests using tetrazolium violet as an indicator of oxidation. The next five rows contain carbon assimilation tests which are scored turbidometrically against a negative control panel containing only water. The last row contains two carbon sources and tests for the co- utilization of various carbon sources with D-xylose. The hardware (Biolog MicroStation Reader) consists of an automated plate reader coupled with a computer, which interprets the results and compares them with the resident database which currently includes 267 species.
Figure 3.

The Biolog microbial ID system.
The biochemical composition of bacterial isolates was determined by standard determinative tests as outlined in the BACTID system, in which pre-selected media were incubated after inoculation with single bacterial colonies (Fig. 4).
Figure 4.
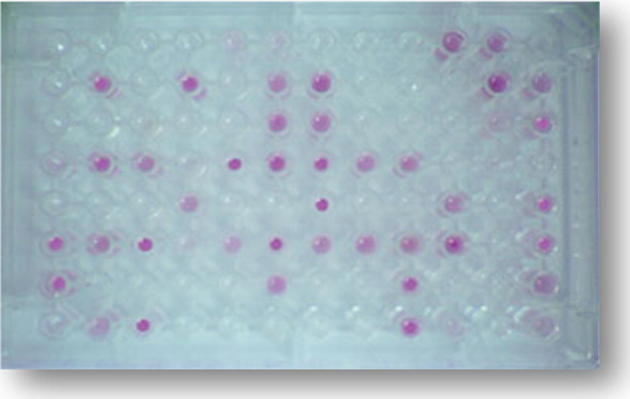
This automated identification system uses a 96-well microliter plate with 95 different carbon sources. The microorganism of interest is applied to each well. Each microorganism has a unique capacity to oxidize some of the various carbon sources. When these carbon sources are oxidized by the microorganism, a purple dye develops visible patterns of positive (purple) and negative (clear) wells which provide a metabolic signature of the organism. The system’s computer examines the pattern signature with its database to determine bacterial species identification.
2.3. Identification of bacteria
Isolates exhibiting distinct colonial morphologies were isolated by repeated sub culturing into basal salt medium and solidified basal salt medium until purified strains were obtained. Identification at species level was performed by using Biolog GN microplate (Biolog, Hayward, CA, USA) according to the manufacturer’s instructions. Briefly, a pure culture of the bacterium was grown on a Biolog Universal Growth agar plate. The bacteria were swabbed from the surface of the agar plate, and suspended to a specified density in GN/GP inoculating fluid. Hundred and fifty microlitre of a bacterial suspension was pipetted into each well of the micro-plate. The micro-plate was incubated at 30 °C or 35 °C depending upon the nature of the organism for 4–24 h according to manufacturer’s specification. The micro-plate was read with the Biolog MicroStation TM system and compared to the database. Databases include aerobic and anaerobic bacteria, yeast, and fungi (Fig. 5).
Figure 5.
Biolog MicroStationTM system and compared to database.
3. Results and discussion
3.1. Identification of bacterial strains using Biolog plates for analysis of bacterial community function
Biolog MicroPlates were originally developed for the rapid identification of bacterial isolates by sole-carbon source utilization, through the inoculation of 95 individual carbon sources plus a water control on a 96 well plate. The plates are read between 24 and 72 h following inoculation with a pre-grown isolate. Metabolism of the substrate in particular wells results in formazan production, producing a color change in the tetrazolium dye. Individual species may be identified by the specific pattern of color change on the plate, providing an identifiable metabolic fingerprint. There is a Biolog database enabling the rapid identification of 1449 bacterial and yeast species/taxa (Biolog sales information) mainly of medical importance. However, the tetrazolium dye immediately introduces some bias since not all bacteria are able to reduce it nor are fungi, hence the plates do not necessarily give a complete picture. A range of different types of Biolog MicroPlates are available containing different substrates for which we have compiled a single table to allow direct and rapid comparison of the substrates available. Bacterial families of soil samples, obtained from the different sites in 5 regions at Explosive Institute (Ryiadh) is shown as follows:
3.1.1. Bacterial community from region 1
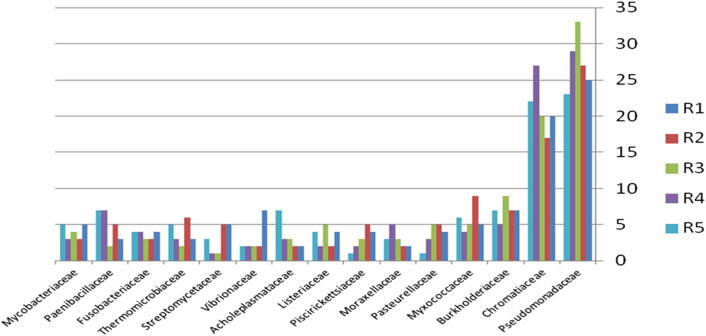
The relative abundance of the microbial community present in soil contaminated with explosive materials at the family and phylum taxonomic level is provided in Fig. 6. There were 4 phyla, 6 classes, 9 orders, and 15 families classifications obtained from the different sites in region 1 at Explosive Institute (Ryiadh). The dominant families in region 1 (R1) are Burkholderiaceae and Chromatiaceae which represent 45% of the relative abundance at the family level. The dominant phylum present was proteobacteria which represented 55% of the relative abundance at the phylum level. Moraxellaceae and Acholeplasmataceae had a low percentage occurrence frequency of 2%, respectively. Nearly 59% of the relative abundance was represented by only four families of bacteria, indicating that the microbial community lacks significant diversity in region one.
Figure 6.
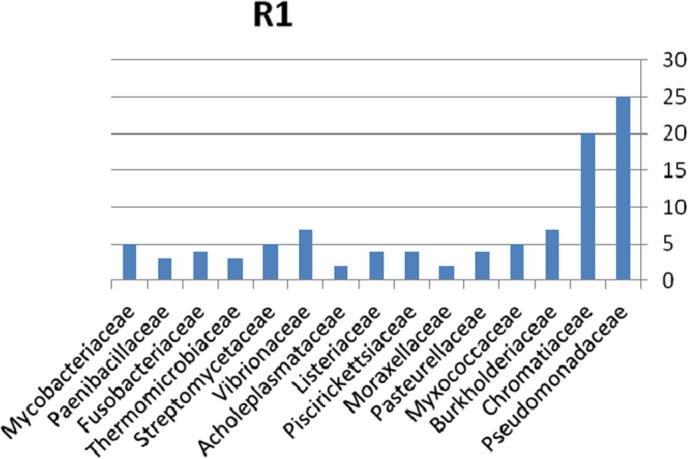
Bacterial community at the family classification level collected from region 1 in Explosive Institute (Ryiadh).
3.1.2. Bacterial community from region 2
The relative abundance of the microbial community present in soil contaminated with explosive materials at the family and phylum taxonomic level is provided in Fig. 7. There were 4 phyla, 6 classes, 9 orders, and 15 families classified obtained from the different sites in region 1 at Explosive Institute (Ryiadh). The dominant families in region 2 (R2) are Burkholderiaceae, Chromatiaceae and Myxococcaceae which represent 48% of the relative abundance at the family level. The dominant phylum present was proteobacteria which represented 60% of the relative abundance at the phylum level. Listeriaceae, Acholeplasmataceae and Vibrionaceae had a low percentage occurrence frequency of 2%, respectively. Nearly 66% of the relative abundance was represented by only four families of bacteria, indicating that the microbial community lacks significant diversity in region one.
Figure 7.
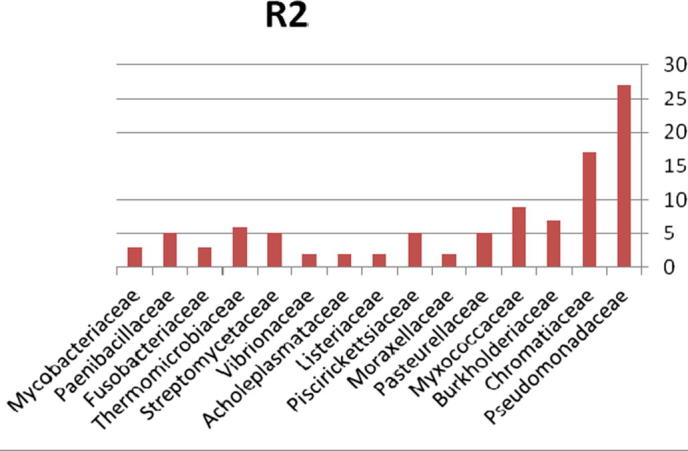
Bacterial community at the family classification level collected from region 2 in Explosive Institute (Ryiadh).
3.1.3. Bacterial community from region 3
The relative abundance of the microbial community present in soil contaminated with explosive materials at the family and phylum taxonomic levels. There were 4 phyla, 6 classes, 9 orders, and 15 families classified obtained from the different sites in region 1 at Explosive Institute (Ryiadh). As shown in Fig. 8, the dominant families in region 3 (R3) are Burkholderiaceae, Chromatiaceae and Pseudomonadaceae which represent 61% of the relative abundance at the family level. The dominant phylum present was proteobacteria which represented 62% of the relative abundance at the phylum level. Listeriaceae, Paenibacillaceae and Vibrionaceae had a low percentage occurrence frequency of 2%, respectively. Nearly 67% of the relative abundance was represented by only four families of bacteria, indicating that the microbial community lacks significant diversity in region one.
Figure 8.
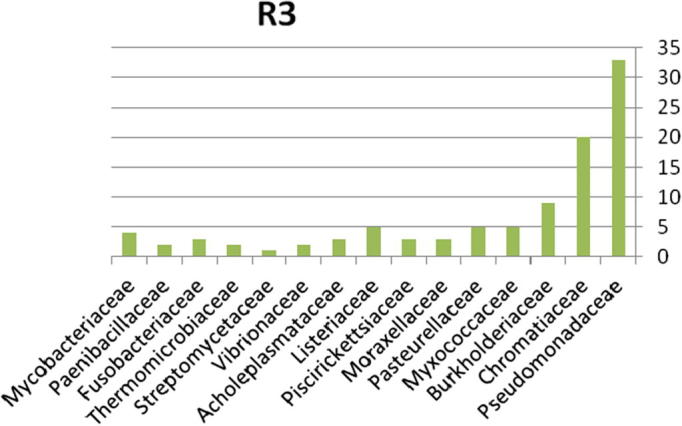
Bacterial community at the family classification level collected from region 3 in Explosive Institute (Ryiadh).
3.1.4. Bacterial community from region 4
The relative abundance of the microbial community present in soil contaminated with explosive materials at the family and phylum taxonomic levels. There were 4 phyla, 6 classes, 9 orders, and 15 families classified obtained from the different sites in region 1 at Explosive Institute (Ryiadh). As shown in Fig. 9, the dominant families in region 4 (R4) are Burkholderiaceae, Chromatiaceae and Paenibacillaceae which represent 61% of the relative abundance at the family level. The dominant phylum present was proteobacteria which represented 61% of the relative abundance at the phylum level. Listeriaceae, and Listeriaceae, Vibrionaceae and Streptomycetaceae had low percentage occurrence frequencies of 2% and 1%, respectively. Nearly 67% of the relative abundance was represented by only four families of bacteria, indicating that the microbial community lacks significant diversity in region one.
Figure 9.
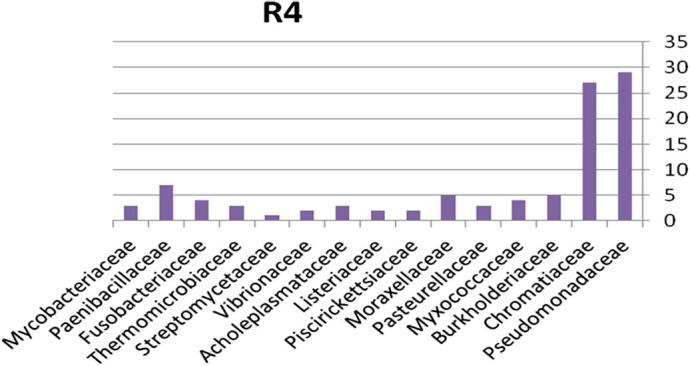
Bacterial community at the family classification level collected from region 4 in Explosive Institute (Ryiadh).
3.1.5. Bacterial community from region 5
The relative abundance of the microbial community present in soil contaminated with explosive materials at the family and phylum taxonomic levels. There were 4 phyla, 6 classes, 9 orders, and 15 families classified obtained from the different sites in region 1 at Explosive Institute (Ryiadh). As shown in Fig. 10, the dominant families in region 5 (R5) are Burkholderiaceae, Chromatiaceae and Pseudomonadaceae which represent 49% of the relative abundance at the family level. The dominant phylum present was proteobacteria which represented 52% of the relative abundance at the phylum level. Vibrionaceae and Piscirickettsiaceae had low percentage occurrence frequencies of 2% and 1%, respectively. Nearly 59% of the relative abundance was represented by only four families of bacteria, indicating that the microbial community lacks significant diversity in region one.
Figure 10.
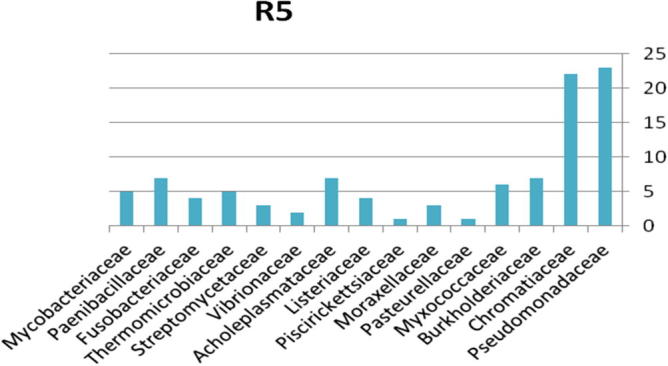
Bacterial community at the family classification level collected from region 5 in Explosive Institute (Ryiadh).
3.2. Bacterial community from combined regions
As shown in Fig. 11, the dominant families in five regions were Burkholderiaceae, Chromatiaceae and Pseudomonadaceae which represent 49% of the relative abundance at the family level. The dominant phylum present was proteobacteria which ranged from 52% to 67% of the relative abundance at the phylum level. Vibrionaceae and Piscirickettsiaceae had a low percentage occurrence frequency of 2%, respectively. Nearly 59–67% of the relative abundance was represented by only four families of bacteria, indicating that the microbial community lacks significant diversity in region one.
Figure 11.
Bacterial community at the family classification level collected from five regions in Explosive Institute (Ryiadh).
The present study reports the identification of bacterial strains by the microplate analysis from the Riyadh community. From this current study Biolog MicroPlates were developed for the rapid identification of bacterial isolates by sole-carbon source utilization, through the inoculation of 95 individual carbon sources plus a water control on a 96 well plate. The plates are read between 24 and 72 h following inoculation with a pre-grown isolate. The relative abundance of the microbial community present in soil contaminated with explosive materials at the family and phylum taxonomic level is provided. There were 4 phyla, 6 classes, 9 orders, and 15 families classification obtained from the different sites in region 1 at Explosive Institute (Ryiadh). The dominant families in five regions were Burkholderiaceae, Chromatiaceae and Pseudomonadaceae which represent 49% of the relative abundance at the family level. The dominant phylum present was proteobacteria which ranged from 52% to 67% of the relative abundance at the phylum level. Vibrionaceae and Piscirickettsiaceae had a low percentage occurrence frequency of 2%, respectively. Nearly 59–67% of the relative abundance was represented by only four families of bacteria, indicating that the microbial community lacks significant diversity in region one.
4. Conclusion
The present study concluded that Biolog MicroPlates methods were identical for the rapid identification of bacterial isolates by sole-carbon source utilization and can be used for the identification of bacteria. Out of five communities, only four families of bacteria indicate that the microbial community lacks significant diversity in region one from the Riyadh community in Saudi Arabia. More studies are needed to be carried out in different regions to validate our results.
Acknowledgement
Gratitude is conveyed to the King Abdul-Aziz city of science and technology, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
References
- Achtman M., Wagner M. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- Barman D., Saud Z.A., Habib M.R., Islam M.F., Hossain K., Yeasmin T. Isolation of cellulytic bacterial strains from soil for effective and efficient bioconversion of solid waste. Life Sci. Med. Res. LSMR. 2011;25 [Google Scholar]
- Bishop C.J., Aanensen D.M., Jordan G.E., Kilian M., Hanage W.P., Spratt B.G. Assigning strains to bacterial species via the internet. BMC Biol. 2009;7:3. doi: 10.1186/1741-7007-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan F.M. What are bacterial species? Ann. Rev. Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Cohan F.M. Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philos. Trans. R. Soc. B. 2006;361:1985–1996. doi: 10.1098/rstb.2006.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C., Hanage W.P., Spratt B.G. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D., Cohan F., Lawrence J., Spratt B.G., Coenye T., Feil E.J., Stackebrandt E., Manfio G., Peer Van de Y., Nesme X., Thompson F., Swings J. Re-evaluating bacterial species. Nat. Microbiol. Rev. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Gevers D., Dawyndt P., Vandamme P., Willems A., Vancanneyt M., Swings J., de Vos P. Stepping stones to a new prokaryotic taxonomy. Philos. Trans. R. Soc. B. 2006;361:1911–1916. doi: 10.1098/rstb.2006.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsinger P., Bengough A.G., Vetterlein D., Young I.M. Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil. 2009;321(1–2):117–152. [Google Scholar]
- Liu S., Suflita J.M. Ecology and evolution of microbial-populations for bioremediation. Trends Biotechnol. 1993;11(8):344–352. doi: 10.1016/0167-7799(93)90157-5. (Review) [DOI] [PubMed] [Google Scholar]
- Manonmani P., Raj S.P., Ramar M., Erusan R.R. Load of infectious microorganisms in hospital effluent treatment plant in Madurai. South Ind. J. Biol. Sci. 2015;1(1):30–33. [Google Scholar]
- Staley J.T. The bacterial species dilemma and the genomic phylogenetic species concept. Philos. Trans. R. Soc. B. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Pot B., Gillis M., de Vos P., Kersters K., Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]