Abstract
Previous results from genome wide association studies (GWASs) in chickens divergently selected for abdominal fat content of Northeast Agricultural University (NEAUHLF) showed that many single nucleotide polymorphism (SNP) variants were associated with abdominal fat content. Of them, six top significant SNPs at the genome level were located within SRD5A3, SGCZ, DLC1, GBE1, GALNT9 and DNAJB6 genes. Here, expression levels of these six candidate genes were investigated in abdominal fat and liver tissue between fat and lean broilers from the 14th generation population of NEAUHLF. The results showed that expression levels of SRD5A3, SGCZ and DNAJB6 in the abdominal fat and SRD5A3, DLC1, GALNT9, DNAJB6 and GBE1 in the liver tissue differed significantly between the fat and lean birds, and were correlated with abdominal fat traits. The findings will provide important references for further function investigation of the six candidate genes involved in abdominal fat deposition in chickens.
Keywords: Chicken, Abdominal fat, Liver tissue, Candidate genes, Differential expression
1. Introduction
Because of the strong association with a number of diseases, including insulin resistance, type 2 diabetes mellitus, atherosclerosis and ischemic heart disease, obesity produces adverse health consequences in humans (Spiegelman and Flier, 2001, Hotamisligil, 2006, Shoelson et al., 2006). A similar problem exists in chickens. Excessive accumulation of fat in chicken abdomens does not only reduce carcass yield and feed efficiency, but is also a less desirable product for consumers. Therefore, mechanisms of obesity occurrence, genes regulating fat deposition and the development of adipose tissue are issues identified either by traditional research methods or high-throughput techniques (Wang et al., 2006, Gesta et al., 2007).
Notwithstanding increased knowledge of obesity, the genes influencing fatness remain incompletely detected. As one of the major tools, genome wide association studies (GWAS) have resulted in a dramatic increase in the identification of susceptibility variants associated with obesity in humans and domestic animals (Scherag et al., 2010, Day and Loos, 2011, Hu et al., 2013).
In recent years, many variants and genes associated with obesity in chickens have been successfully identified using GWAS. Abasht et al. revealed cryptic alleles as an important factor in heterosis for fatness in a chicken F2 population (Abasht and Lamont, 2007). Liu and Sun identified some candidate genes associated with abdominal fat traits in an F2 resource population derived from a cross between a Chinese local breed and a commercial rapid-growing broiler line (Liu et al., 2013, Sun et al., 2013).
Previously, many variants associated with abdominal fat traits have been identified using GWAS in our laboratory (unpublished data). Of them, six top significant single nucleotide polymorphisms at the genome level were located within SRD5A3 (Steroid 5α-reductase 3), SGCZ (Sarcoglycan, zeta), DLC1 (Deleted in liver cancer 1), GBE1 (Glucan (1,4-alpha-), branching enzyme 1), GALNT9 (N-acetylgalactosaminyltransferase 9) and DNAJB6 (DNAJ homology subfamily B member 6), suggesting that these genes play important roles in fat deposition in chickens. Here, we investigate whether these six genes are differentially expressed in fat and liver tissues between fat and lean broilers and the relationship between their expression levels and abdominal fat content, which would help in our understanding of the roles these genes play in chicken adipose tissues.
2. Materials and methods
2.1. Experimental animals
The broilers used in this study were derived from the Northeast Agricultural University (NEAU) broiler lines divergently selected for abdominal fat content (NEAUHLF). The NEAUHLF line has been selected since 1996 and the selection procedure and raising conditions have been described in detail previously (Wang et al., 2007, Guo et al., 2011). For each line, a total of 10 male and 6 female birds from the 14th generation population were used. Birds were slaughtered at 7 weeks of age, the average abdominal fat weight (AFW) with standard error and average abdominal fat percent (AFP) with standard error of the lean line were 12.53 ± 1.17 g and 0.59% ± 0.05%, respectively, however, for the fat line, they were 54.09 ± 1.93 g and 3.29% ± 0.13%. There were significant differences in both AFW and AFP between the two lines. Samples were collected from abdominal fat and liver tissues, then weighed and immediately frozen in liquid nitrogen, and stored at −80 °C until analysis.
2.2. RNA extraction and cDNA synthesis
Total abdominal and liver RNA was isolated using TRIZOL® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The extracted RNA was dissolved in DEPC-treated water and the purity and integrity were estimated by an ultraviolet/visible spectrophotometer (Pharmacia, USA) at a 260/280 nm absorbance ratio (range 1.8–2.0 indicates a pure RNA sample) and agarose gel electrophoresis. Total RNA was reverse transcribed to cDNA in a reaction volume of 20 μL containing 1 μg total RNA, 0.5 μL of 50 pmol/L Oligo d(T)18 Primers and finally supplemented with nuclease-free water to a volume of 5 μL for the first step. This mixture was heated at 70 °C for 5 min and incubated on ice-water for 5 min. Then 5× Reverse transcription Buffer 4 μL, MgCl2 (25 mM) 2.5 μL, dNTP Mixture 1 μL, RNase Inhibitor (Promega Biotech Co. Ltd) 0.5 μL, Improm-II Reverse Transcriptase (Promega, Madison, WI, USA) 1 μL and nuclease-free dH2O were added to a final volume of 20 μL. The RT mixture was incubated at 25 °C for 5 min, then 42 °C for 60 min and finally inactivated by heating at 70 °C for 15 min. The cDNA was directly for use in quantitative real-time PCR.
2.3. Quantitative analysis of mRNA expression
Special primers for amplifications of these genes were designed spanning at least one intron to avoid genomic DNA contamination using Primer Premier 5.0 software according to Ensembl. All primers were synthesized by Invitrogen Biotechnology (Shanghai) Co., Ltd. (Table 1).
Table 1.
Primer sequences used in this study.
| Gene symbol | Forward primer (5′–3 ′) | Reverse primer (5′–3′) | Production size (bp) | Anneal temp (°C) | GenBank No. |
|---|---|---|---|---|---|
| SRD5A3 | TGGACTTGGCTATTACGTTGCTG | CATCGCAACGCCTATGATGTG | 122 | 60 | ID: 422750 |
| SGCZ | GCTCTGCGTCTGTCCCAATG | AGCTCCACAAGCAGATGTTGCTA | 92 | 60 | ID: 422739 |
| DLC1 | ATGAGAGTTCAACAGACAG | TAAAAGCATAATGGCAG | 194 | 60 | ID: 422740 |
| GBE1 | ATTTGTGGATGGTGGACT | CATACCCTTTACCCTCAA | 132 | 60 | ID: 427964 |
| GALNT9 | AGATTGGCTTGCTTGAC | TGTAGGGTTTCTTTGTGC | 153 | 60 | ID: 416796 |
| DNAJB6 | AGCCTTTGCTGAGGAGT | CTTGCTGCCTTCTTTGTAT | 211 | 60 | ID: 420448 |
| GAPDH | AGAACATCATCCCAGCGT | AGCCTTCACTACCCTCTTG | 184 | 60 | ID: 374193 |
SYBR Green real-time PCR amplifications were conducted using an AB Applied Biosystems 7500 Real Time PCR System (Life Technologies, USA). The stably expressed gene, GAPDH, served as the endogenous reference for determination of targeted mRNA profiles (Bustin, 2002). Quantitative PCR amplifications were performed in a final volume of 10 μL reaction mixture under the optimum reaction conditions including 5 μL SYBR® Permix Ex Taq™ II (TaKaRa, Japan), 0.2 μL ROX Reference Dye II (TaKaRa, Japan), 0.2 μL forward primer, 0.2 μL reverse primer (10 μmol/L) of target gene or housing genes, 3.4 μL water and 1 μL template cDNA. Amplification conditions were performed starting with 30 s template denaturation step at 94 °C, followed by 40 PCR cycles of 5 s at 95 °C, 34 s at 60 °C, where the fluorescence was acquired. Finally, a dissociation curve to test PCR specificity was generated by one cycle for 15 s at 95 °C, followed by 60 °C for 1 min and increased to 95 °C with acquired fluorescence.
All samples were amplified in triplicate as technical replicates and specific amplification was confirmed by single peak observation on dissociation curves. The means of Ct values were obtained for further calculations.
2.4. Statistical analysis
The expression levels of the six genes were measured using real-time PCR. The 2−ΔCt (ΔCt = Ct of the target gene – Ct of the housekeeping gene) method was used to analyze the relative quantitative data. Values were expressed as mean ± standard error of the mean. Data of expression were subjected to square root and arcsine transformation to normality distribution.
Model-based tests were carried out to evaluate the different gene expression levels on abdominal fat and liver between the two lines using Y = μ + Line + Sex + Line × Sex + e, by the GLM procedure of JMP4.0 (SAS, Chicago, IL, USA), which fitted with Line and Sex as fixed effects, Line × Sex as interaction of Line and Sex, where Y is the dependent variable for different gene expression levels of birds, μ is the overall population mean, and e is the residual random error. The Pearson coefficient of correlation between expression levels and abdominal fat traits was estimated. P < 0.05 was taken to indicate significant differences or significant correlation.
3. Results
3.1. The expression of six genes in abdominal fat tissues
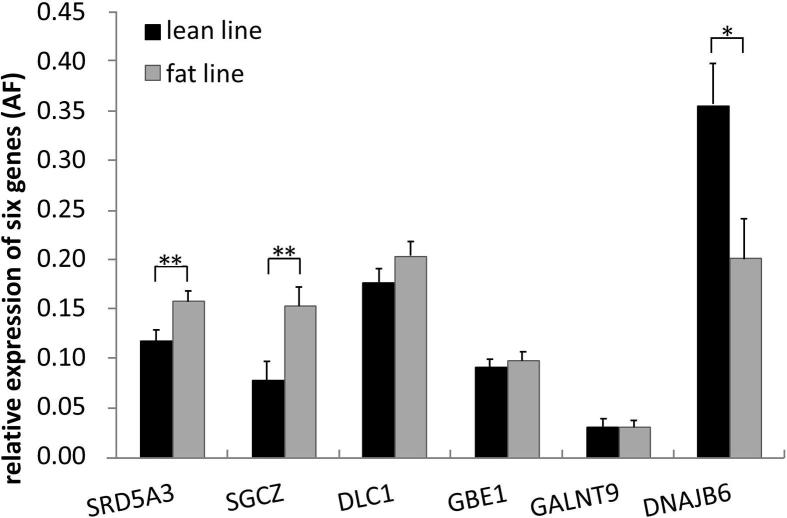
As shown in Fig. 1, all six genes were expressed in abdominal fat tissues. SRD5A3 and SGCZ were differentially expressed between the fat and lean birds (P < 0.01), and the expression levels were higher in fat birds. In contrast, DNAJB6 expression level in fat birds was significantly lower than that of lean birds (P < 0.05). For the other three genes, DLC1, GBE1 and GALNT9, no significant differences in the expression levels were observed in abdominal fat tissues between the two lines.
Figure 1.
Comparison of expression level of six genes in abdominal fat tissue between two lines.
3.2. The expression of six genes in liver tissues
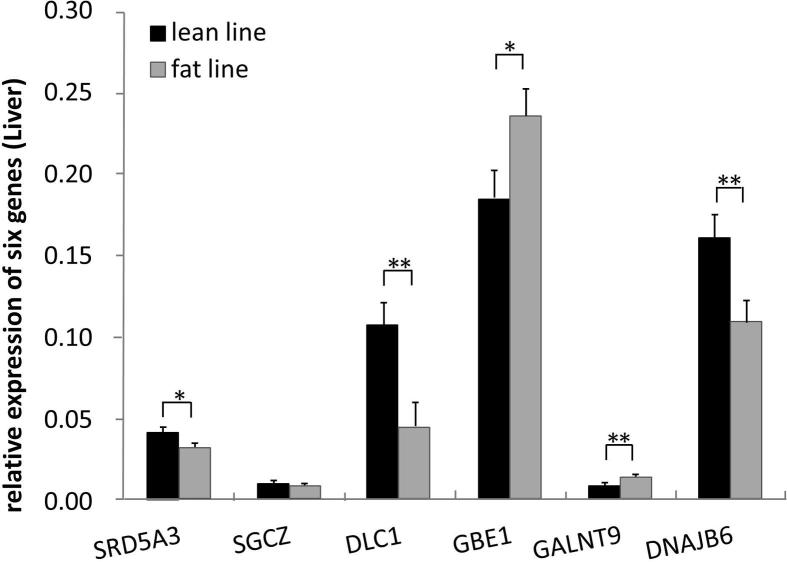
As shown in Fig. 2, all six genes were expressed in liver tissues of fat and lean birds. SRD5A3 expression levels in lean birds were significantly higher than that of fat birds (P < 0.05). Both DLC1 and DNAJB6 expression levels in lean birds were significantly higher than that of fat birds (P < 0.01). In contrast, GBE1 expression levels in lean birds were significantly (P < 0.05) lower than that of fat birds, and GALNT9 expression levels in lean birds were significantly (P < 0.01) lower than that of fat birds.
Figure 2.
Comparison of expression level of six genes in liver tissue between two lines.
3.3. The correlation analyses between these six gene expression levels and AFW and AFP
The results of a correlation analysis between these six gene expression levels and AFW are given in Table 2. SRD5A3 and SGCZ expression levels in abdominal fat tissues were significantly positively correlated with AFW and AFP (P < 0.05 or P < 0.01). DNAJB6 expression levels in abdominal fat tissues were significantly negatively correlated with AFW and AFP (P < 0.01). SRD5A3, DLC1 and DNAJB6 expression levels in liver tissues were negatively and significantly correlated with AFW and AFP (P < 0.01). GBE1 and GALNT9 expression levels in liver tissue were significantly positively correlated with AFW and AFP (P < 0.05 or P < 0.01).
Table 2.
Correlation coefficients between mRNA expression levels of six genes in abdominal fat and liver tissues and abdominal fat traits.
| SRD5A3 |
SGCZ |
DLC1 |
GBE1 |
GALNT9 |
DNAJB6 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF | Liver | AF | Liver | AF | Liver | AF | Liver | AF | Liver | AF | Liver | |
| AFW | 0.35⁎ | −0.53⁎⁎ | 0.47⁎⁎ | −0.21 | 0.13 | −0.47⁎⁎ | 0.04 | 0.35⁎ | −0.10 | 0.47⁎⁎ | −0.47⁎⁎ | −0.54⁎⁎ |
| AFP | 0.39⁎ | −0.49⁎⁎ | 0.55⁎⁎ | −0.18 | 0.24 | −0.45⁎⁎ | 0.14 | 0.38⁎ | −0.04 | 0.48⁎⁎ | −0.46⁎⁎ | −0.51⁎⁎ |
Indicates P < 0.05.
Indicates P < 0.01.
4. Discussion
Adipose and hepatic tissues play important roles in the growth and development of animals. The former not only maintains energy balance, but also functions as a crucial endocrine organ; the latter, especially in avian species, is a major site for lipid metabolism. Here, for the first time, SRD5A3, SGCZ, DLC1, GBE1, GALNT9 and DNAJB6 genes were investigated at the transcriptional level in abdominal fat and liver tissues of chicken. Significant differences in expression levels of these genes between fat and lean birds were observed for both tissues, coupled with a significant correlation of their expression levels with AFW and AFP, which suggested that these genes could be associated with abdominal fat deposition in chickens.
SRD5A3 was expressed in abdominal fat and liver tissues of chickens, and SRD5A3 expression levels in abdominal fat tissues were significantly positively correlated with AFW and AFP. It has been reported that SRD5A3 plays a crucial role in N-linked protein glycosylation, which was a process that involves many steps including the assembly of a lipid carrier for the oligosaccharide, the flip-flopping of this lipid between leaflets of the endoplasmic reticulum membrane, and multiple cycles of phosphorylation and dephosphorylation of lipids in yeast and mammals (Stiles and Russell, 2010, Cantagrel et al., 2010). In addition, SRD5A3 participates in the process of steroid hormone biosynthesis, and fatty acids had been identified as the substrates of this steroid 5α-reductase family member (Moon and Horton, 2003). Based on the above evidence, it is speculated that SRD5A3 might be essential for fat deposition.
DNAJB6 was expressed in both adipose and liver tissues, and the expression levels in both tissues differed between the two lines, and were negatively correlated with AFW and AFP. DNAJB6, named MRJ [Mammalian relative of DnaJ (HSP40)], is a member of the HSP40 family, subfamily B, owning two spliced variants, MRJ (S) (smaller isoform) and MRJ (L) (long isoform). It was found that MRJ (L) up-regulates expression of DKK1 (Mitra et al., 2010), a Wnt inhibitor, which regulates aspects of placental lipid deposition through the Wnt signaling pathway (Strakovsky and Pan, 2012). Sustained activation of the Wnt signaling pathway prevents adipogenic differentiation (Nakamura et al., 2013). Accordingly, we infer that DNAJB6 is critical for regulating abdominal fat accumulation, potentially by impacting the transcription of DKK1 involved in lipid metabolism.
SGCZ was differentially expressed between the fat and lean birds, and the expression levels in abdominal fat tissues were significantly correlated with AFW and AFP. SGCZ, a well-known gene whose protein product belongs to the sarcoglycan protein family, had been identified as a basilic factor in the pathogenesis of muscular dystrophy and is expressed mainly in vascular smooth muscle (Hack et al., 2000, Wheeler et al., 2002, Aurino et al., 2008). Vascular smooth muscle cells are the essential factor of activity and configuration of blood vessels that could be changed in cardiovascular diseases (Cannon, 2013), and obesity could induce a series of cardiovascular diseases (Lppoliti et al., 2013). Additionally, the report suggested that muscle could be seen as an important mediator for fat deposition (Brockmann et al., 2009). Therefore, we could deduce that SGCZ participates in the process of fat deposition.
Our results also showed that DLC1 was expressed in the adipose and liver tissues, which was in line with the results from human studies and proved by Durkin et al. (2002). The expression level of DLC1 in the liver was significantly different (P < 0.01) between the two lines. Coincidentally, it was negatively and significantly correlated with AFW. As a potential tumor suppressor gene in the liver (Xue et al., 2008), suppressive function of DLC1 relies on DLC1’s RhoGAP activity activated by lipid interaction and the START domain (Erlmann et al., 2009), which is typically found in lipid transfer proteins and forms a hydrophobic pocket to accommodate a single lipid molecule (Alpy and Tomasetto, 2005). When liver cancer occurs, normal lipid metabolism may be significantly influenced (Jiang et al., 2007). Therefore, it could be concluded that DLC1 indirectly influences fat metabolism in vivo.
GBE1 and GALNT9 genes were expressed in both abdominal fat and liver tissues, and the mRNA expression levels in the liver were positively significantly correlated with AFW and AFP. It was reported that a mutation on GBE1 was found causing glycogen storage disease type IV, an autosomal recessive disorder of the glycogen synthesis (Andersen, 1956). GALNT9 participates in the process of biosynthesis of O polyose and has been identified as being responsible for mitochondrial myopathy and glycobiology (Casas et al., 2004, Van der Zwaag et al., 2009). It is well known that in the liver, glycogen is converted to fat, and delivered to other tissues to be used or stored in the form of lipoprotein. Thus, these two genes could influence the synthesis of fat acid through the regulation of glycometabolism.
To sum up, our findings will provide important references for further function investigation of the six candidate genes involved in abdominal fat deposition in chickens.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgments
The authors gratefully acknowledge the members of the Poultry Breeding Group at Northeast Agricultural University for help in managing the birds. This research was supported by the National 863 Project of China (No. 2011AA100301), China Agriculture Research System (No. CARS-42) and the Program for Innovation Research Team in University of Heilongjiang Province (No. 2010td02).
Footnotes
Peer review under responsibility of King Saud University.
References
- Abasht B., Lamont S.J. Genome-wide association analysis reveals cryptic alleles as an important factor in heterosis for fatness in chicken F2 population. Anim. Genet. 2007;38:491–498. doi: 10.1111/j.1365-2052.2007.01642.x. [DOI] [PubMed] [Google Scholar]
- Alpy F., Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- Andersen D.H. Familial cirrhosis of the liver with storage of abnormal glycogen. Lab. Invest. 1956;5:11–20. [PubMed] [Google Scholar]
- Aurino S., Piluso G., Saccone V., Cacciottolo M., Amico F.D., Dionisi M., Totaro A., Belsito A., Vivino U.D., Nigro V. Candidate-gene testing for orphan limb-girdle muscular dystrophies. Acta Myol. 2008;27:90–97. [PMC free article] [PubMed] [Google Scholar]
- Brockmann G.A., Tsaih S.W., Nuschi C., Churchill G.A., Li R. Genetic factors contributing to obesity and body weight can act through mechanisms affecting muscle weight, fat weight, or both. Physiol. Genomics. 2009;36:114–126. doi: 10.1152/physiolgenomics.90277.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Cannon B. Cardiovascular disease: biochemistry to behaviour. Nature. 2013;493:S2–S3. doi: 10.1038/493S2a. [DOI] [PubMed] [Google Scholar]
- Cantagrel V., Lefeber D.J., Ng B.G., Guan Z., Silhavy J.L., Bielas S.L., Lehle L., Hombauer H., Adamowicz M., Swiezewska E., De Brouwer A.P., Blümel P., Sykut-Cegielska J., Houliston S., Swistun D., Ali B.R., Dobyns W.B., Babovic-Vuksanovic D., van Bokhoven H., Wevers R.A., Raetz C.R., Freeze H.H., Morava E., Al-Gazali L., Gleeson J.G. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell. 2010;142:203–217. doi: 10.1016/j.cell.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas K., Bykhovskaya Y., Mengesha E., Wang D., Yang H., Taylor K., Inbal A., Fischel-Ghodsian N. Gene responsible for mitochondrial myopathy and sideroblastic anemia (MSA) maps to chromosome 12q24.33. Am. J. Med. Genet. A. 2004;127A:44–49. doi: 10.1002/ajmg.a.20652. [DOI] [PubMed] [Google Scholar]
- Day F.R., Loos R.J.E. Developments in obesity genetics in the era of genome-wide association studies. J. Nutrigenet. Nutrigenomics. 2011;4:222–238. doi: 10.1159/000332158. [DOI] [PubMed] [Google Scholar]
- Durkin M.E., Yuan B.Z., Thorgeirsson S.S., Popescu N.C. Gene structure, tissue expression, and linkage mapping of the mouse DLC-1 gene. Gene. 2002;288:119–127. doi: 10.1016/s0378-1119(02)00462-6. [DOI] [PubMed] [Google Scholar]
- Erlmann P., Schmid S., Horenkamp F.A., Geyer M., Pomorski T.G., Olayioye M.A. DLC1 activation requires lipid interaction through a polybasic region preceding the RhoGAP domain. Mol. Biol. Cell. 2009;20:4400–4411. doi: 10.1091/mbc.E09-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S., Tseng Y.H., Kahn C.R. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Guo L., Sun B., Shang Z.C., Leng L., Wang Y.X., Wang N., Li H. Comparison of adipose tissue cellularity in chicken lines divergently selected for fatness. Poult. Sci. 2011;90:2024–2034. doi: 10.3382/ps.2010-00863. [DOI] [PubMed] [Google Scholar]
- Hack A.A., Groh M.E., McNally E.M. Sarcoglycans in muscular dystrophy. Microsc. Res. Tech. 2000;48:167–180. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<167::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Hu Z.L., Park C.A., Wu X.L., Reecy J.M. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41:871–879. doi: 10.1093/nar/gks1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.T., Xu N., Zhang X.Y., Wu C.P. Lipids changes in liver cancer. J. Zhejiang Univ. Sci. B. 2007;8:398–409. doi: 10.1631/jzus.2007.B0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.R., Sun Y.F., Zhao G.P., Wang F.J., Wu D., Zheng M.Q., Chen J.L., Zhang L., Hu Y.D., Wen J. Genome-wide association study identifies loci and candidate genes for body composition and meat quality traits in Beijing-You chickens. PLoS ONE. 2013;8:e61172. doi: 10.1371/journal.pone.0061172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lppoliti F., Canitano N., Businaro R. Stress and obesity as risk factors in cardiovascular diseases: a neuroimmune perspective. J. Neuroimmune Pharmacol. 2013;8:212–226. doi: 10.1007/s11481-012-9432-6. [DOI] [PubMed] [Google Scholar]
- Mitra A., Menezes M.E., Shevde L.A., Samant R.S. DNAJB6 induces degradation of beta-catenin and causes partial reversal of mesenchymal phenotype. J. Biol. Chem. 2010;285:24686–24694. doi: 10.1074/jbc.M109.094847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon Y.A., Horton J.D. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J. Biol. Chem. 2003;278:7335–7343. doi: 10.1074/jbc.M211684200. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Hinoi E., Lezaki T., Takada S., Hashizume S., Takahata Y., Tsuruta E., Takahashi S., Yoneda Y. Repression of adipogenesis through promotion of Wnt/β-catenin signaling by TIS7 up-regulated in adipocytes under hypoxia. BBA-Mol. Basis Dis. 2013;1832:1117–1128. doi: 10.1016/j.bbadis.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Scherag A., Jarick I., Grothe J., Biebermann H., Scherag S., Volchmar A., Vogel C.I.G., Grene B., Hebebrand J., Hinney A. Investigation of a genome wide association signal for obesity: synthetic association and haplotype analyses at the melanocortin 4 receptor gene locus. PLoS ONE. 2010;5:e13967. doi: 10.1371/journal.pone.0013967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoelson S.E., Lee J., Goldfine A.B. Inflammation and insulin resistance. J. Clin. Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman B.M., Flier J.S. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- Stiles A.R., Russell D.W. SRD5A3: a surprising role in glycosylation. Cell. 2010;142:196–198. doi: 10.1016/j.cell.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakovsky R.S., Pan Y.X. A decrease in DKK1, a WNT inhibitor, contributes to placental lipid accumulation in an obesity-prone rat model. Biol. Reprod. 2012;86:1–11. doi: 10.1095/biolreprod.111.094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.F., Zhao G.P., Liu R.R., Zheng M.Q., Hu Y.D., Wu D., Zhang L., Li P., Wen J. The identification of 14 new genes for meat quality traits in chicken using a genome-wide association study. BMC Genomics. 2013;14:458. doi: 10.1186/1471-2164-14-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Zwaag B., Franke L., Poot M., Hochstenbach R., Spierenburg H.A., Vorstman J.A., van Daalen E., de Jonge M.V., Verbeek N.E., Brilstra E.H., van ’t Slot R., Ophoff R.A., van Es M.A., Blauw H.M., Veldink J.H., Buizer-Voskamp J.E., Beemer F.A., van den Berg L.H., Wijmenga C., van Amstel H.K., van Engeland H., Burbach J.P., Staal W.G. Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS ONE. 2009;4:e5324. doi: 10.1371/journal.pone.0005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.B., Li H., Wang Q.G., Wang Y.X., Han H.B., Shi H. Microarray analysis of adipose tissue gene expression profiles between two chicken breeds. J. Biosci. 2006;31:565–573. doi: 10.1007/BF02708408. [DOI] [PubMed] [Google Scholar]
- Wang Q.G., Li H., Li N., Leng L., Wang Y.X., Tang Z.Q. Polymorphism of heart fatty acid-binding protein gene associated with fatness traits in the chicken. Anim. Biotechnol. 2007;18:91–99. doi: 10.1080/10495390601038900. [DOI] [PubMed] [Google Scholar]
- Wheeler M.T., Zarnegar S., McNally E.M. Zeta-sarcoglycan, a novel component of the sarcoglycan complex, is reduced in muscular dystrophy. Hum. Mol. Genet. 2002;11:2147–2154. doi: 10.1093/hmg/11.18.2147. [DOI] [PubMed] [Google Scholar]
- Xue W., Krasnitz A., Lucito R., Sordella R., Vanaelst L., Cordon-Cardo C., Singer S., Kuehnel F., Wigler M., Powers S., Zender L., Lowe S.W. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]