Abstract
Prostate cancer (PCa) is the second leading cause of cancer deaths in men. A better understanding of the molecular basis of prostate cancer proliferation and metastasis should enable development of more effective treatments. In this study we focused on the lncRNA, prostate cancer associated transcript 29 (PCAT29), a putative tumor suppressive gene. Our data show that the expression of PCAT29 was reduced in prostate cancer tumors compared to paired perinormal prostate tissues. We also observed substantially lower levels of PCAT29 in DU145 and LNCaP cells compared to normal prostate (RWPE-1) cells. IL-6, a cytokine which is elevated in prostate tumors, reduced the expression of PCAT29 in both DU145 and LNCaP cells by activating signal transducer and activator of transcription 3 (STAT3). One downstream target of STAT3 is microRNA (miR)-21, inhibition of which enhanced basal PCAT29 expression. In addition, we show that resveratrol is a potent stimulator of PCAT29 expression under basal condition and reversed the down regulation of this lncRNA by IL-6. Furthermore, we show that knock down of PCAT29 expression by siRNA in DU145 and LNCaP cells increased cell viability while increasing PCAT29 expression with resveratrol decreased cell viability. Immunohistochemistry studies showed increased levels of STAT3 and IL-6, but low levels of programmed cell death protein 4 (PDCD4), in prostate tumor epithelial cells compared to adjacent perinormal prostate epithelial cells. These data show that the IL-6/STAT3/miR-21 pathway mediates tonic suppression of PCAT29 expression and function. Inhibition of this signaling pathway by resveratrol induces PCAT29 expression and tumor suppressor function.
Introduction
About 2% human genome sequences are functional genes -which coding for functional proteins- and regulatory elements, while other of the human genome sequence is considered non-coding sequence with different functions. [1] Recently, multiple studies have revealed new forms of RNA widely known as long non-coding RNA (ncRNA) transcribed from non-coding sequence in DNA [2]. These ncRNAs have different functions and can be divided according to their sizes into two groups. These include short ncRNA (less than 200 nucleotides in length), such as microRNA (miRNA) and interference RNA (siRNA) and long ncRNA (more than 200 nucleotides) such as long non-coding RNA (lncRNA) [3]. There are also other classes of RNA which have housekeeping function in protein synthesis, such as transfer RNA (tRNA) and ribosomal RNA (rRNA) [4–6]. The functions of lncRNA are still largely unknown but some studies have linked them to different cellular roles including epigenetics regulation of transcription, such as ANRIL and HOTAIR, which interact with PRC1 and PRC2 transcription complex and repress gene transcription [7,8]. Enhancer-derived RNA (eRNA) acts as a transcription enhancer which are important role in transcription activation in androgen positive prostate cancer cells [9]. In addition, MEG3 is a tumor suppresser whose activity is dependent on the p53 protein [10–12]. MALAT and NEAT lncRNAs are implicated in mRNA splicing, editing and exporting mRNA to cytoplasm [13,14]. In addition, the oncogene PCAT1 activates cell proliferation and inhibits BRCA2 [8].
Prostate cancer (PCa) is the second leading cause of cancer deaths in in men [15,16]. Several risk for developing PCa include genetic modifications in oncogenes such as STAT3 [16], modification in tumor suppressor genes such as phosphatase and tensin homolog deleted on chromosome 10 (PTEN) gene, or mutations in androgen receptors [17–19]. Epigenetic alterations, including DNA methylation in tumor suppresser genes promoters and modification in histone modifying enzymes also contribute to PCa carcinogenesis [20]. Recent studies have showed that alteration in different lncRNAs, such as PTENP1 [21,22], Linc00963 [23], PCGIM1 [24], PRNCR1 [25], CBR-3AS1 [26], CTP1 AS [27], GAS5 [28], ANRIL [29], ANRASSF1 [30] and PCAT1 [8, 31] are associated with PCa. LncRNAs, such as prostate cancer associated transcript (PCAT1 and PCAT3), increase cancer cells proliferation [8,31]. However, another LncRNA, PCAT29 exhibits tumor suppressor features in prostate cancers by decreasing the proliferation and migration of PCa [32]. Studies have reported that both LncRNA and microRNA, such as PCAT1 and miR-3667, can act synergistically to regulate PCa progression [1]. Furthermore, lncRNA and microRNA activate RNA degradation machinery and recruit STAU1 protein to enable RNA degradation [33]. While many studies have implicated PCAT lncRNAs in prostate cancer [31], their exact roles in the development of PCa remain unclear.
This study focuses on the regulation of PCAT29, a tumor suppressor lncRNA by IL-6 in prostate cancer cells. We show that IL-6 reduced PCAT29 mRNA by activating STAT3 and miR-21. Co-regulation of STAT3 and miR-21 was observed in human prostate samples. Furthermore, the chemopreventative agent, resveratrol, blocked IL-6 reduction of PCAT29, by interfering with the STAT3 and miR-21 signaling.
Materials and methods
Ethics statement
All studies involving clinical specimen were conducted in accordance with federal regulations and using a protocol authorized by The Tissue Banking Facility of Southern Illinois University School of Medicine and approved by The Springfield Committee for Research Involving Human Subjects (SCRIHS).
Materials
Resveratrol was purchased from Sigma-Aldrich (R5010) whereas, IL-6 was bought from Life Technologies (10395HNAE). Antibodies: PDCD4 rabbit monoclonal antibody (cat# 9535) and pSTAT3 mouse monoclonal antibody (cat# 4113) were purchased from Cell Signaling Technology®, while STAT3 mouse monoclonal (cat# sc-8019) and β-actin mouse monoclonal (cat# sc-69879) antibodies were obtained from Santa Cruz Biotechnology. All antibodies for Western blotting were used in the dilution of 1:1000 except β-actin, which was used in the dilution of 1:10,000. IL-6 mouse monoclonal antibody was purchased from Novus Biologicals (cat# nbp1-47810) and used in the dilution of 1:50 for immunohistochemistry. Supplies for cell culture: RPMI 1640 media and complete keratinocyte serum-free media (K-SFM) were from Gibco, penicillin/streptomycin were obtained from ThermoFisher Scientific, while fetal bovine serum (FBS) was from Atlanta Biologicals. Synthetic anti-miR-21 oligonucleotide, STAT3 siRNA and their negative control were purchased from Ambion, while PCAT29 siRNA and their negative control were purchased from Dharmacon™. The TRI reagent for RNA isolation was purchased from Sigma-Aldrich. The items for real-time RT-PCR such as iScript cDNA Synthesis Kit was purchased from Bio-Rad, while Fast SYBR™ Green Master Mix, TaqMan® MicroRNA Reverse Transcription Kit and TaqMan® Universal PCR Master Mix were purchased from and Applied Biosystems. CellTiter 96® AQueous One Solution Cell Proliferation Kit for determining cell viability was purchased from Promega. Immunohistochemistry was performed using ImmunoCruz™ ABC Staining System which was purchased from Santa Cruz Biotechnology.
Cell culture
Androgen-insensitive human prostate carcinoma DU145 cells, androgen-sensitive human prostate carcinoma LNCaP cells and immortalized human prostate epithelial RWPE-1 cells, were kindly provided by Dr. Daotai Nie (SIU School of Medicine, Springfield, IL). DU145 and LNCaP cells were cultured in RPMI 1640 media (Gibco) supplemented with 10% fetal bovine serum (Atlanta Biologicals), 50 units/ml penicillin and 50 μg/ml streptomycin (ThermoFisher Scientific). RWPE-1 cells were cultured in complete K-SFM which contains 50 μg/ml of bovine pituitary extract (BPE) and 5 ng/ml epidermal growth factor (EGF) (Gibco), plus 50 units/ml penicillin and 50 μg/ml streptomycin. All cell lines were grown at 37°C in the presence of 5%CO2 and 95% ambient air. All experiments were performed on sub-confluent monolayers.
Cell viability assay (MTS Assay)
Cell viability of DU145 and LNCaP cells was assessed using CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega), as per manufacturer’s protocol. Briefly, 2,500 cells per well were seeded in a 96-well plate. Twenty four hours after they were seeded, the cells were treated and allowed to grow for another 24 h, after which 20 μl of CellTiter 96® AQueous One Solution reagent was added to each well containing cells with100 μl media. Cell were then incubated for 2 to 3 h and absorbance was measured at 490 nm using an ELISA plate reader. The production of the colored formazan product (absorbance) is directly proportional to the number of viable cells in culture and is expressed as percent cell viability relative to control.
Western blot analysis
Protein expression was determined using Western blotting technique as previously described [34]. Briefly, cells were seeded in 6-well plates and incubated until 70–80% confluence. At the end of the treatment, cells were washed once with ice-cold 1X PBS and whole-cell lysates were prepared by homogenizing in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) containing protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitor 2 and 3 (Sigma-Aldrich). Protein concentration was determined by Bradford method and equal amount of protein was resolved by SDS polyacrylamide gel electrophoresis. The proteins were then transferred on to nitrocellulose membrane and probed with specific primary antibody. Blots were then incubated with species-specific fluorescent-tagged IgG secondary antibody and scanned and visualized using LI-COR Odyssey® imaging system (LI-COR Biosciences). Each band was normalized to the corresponding total STAT3 or β-actin bands. Densitometric analysis of the bands was performed using ImageJ software and results were plotted as percent of control where control was considered as 100%.
Oligonucleotide and short interfering (si) RNA transfection
Synthetic anti-miR-21 (30 nM) (ID# AM17000, Ambion), STAT3 siRNA (10 nM) (ID# AM16708, Ambion), PCAT29 siRNA (30 nM) (ID# SHEKJ-000001, Dharmacon™) and their negative controls (scramble) were transferred into the cells using Lipofactamine® RNAiMAX transfection reagent (Life Technologies™), according to manufacturer’s protocol. Briefly, cells were seeded in a 6-well plate and incubated until they were 60–70% confluence. When the cells were ready, anti-miR-21, STAT3 siRNA, PCAT29 siRNA and their respective negative controls were diluted in 150 μl of serum free media and incubated with 9 μl of Lipofactamine® RNAiMAX for 5 min to permit the formation of transfection complex. The transfection complex was then added to the cells which were then incubated for 24 h after which they were treated with IL-6 and resveratrol for indicated time. Cell were then collected for Western blotting or real-time PCR studies to detect various proteins or RNAs, respectively.
RNA isolation
Total RNA was extracted from the cells using 500 μl of TRI reagent (Sigma). Chloroform (100 μl) was added to the TRI reagent and the tube was shaken vigorously for 15 seconds and centrifuged at 12,000 rpm for 15 min. Top aqueous layer, which contains RNA, was extracted to which 0.5 ml ice-cold isopropanol was added and the samples were centrifuged at 12,000 rpm for 10 min. Isopropanol was carefully removed and the pellet was washed with 75% ethanol in DEPC-treated H2O. The samples where then centrifuged again at 12,000 rpm for 10 min. The ethanol was removed and the tube was air dried briefly. The RNA pellet was resuspended in nuclease free water and RNA levels were determined based on their optical density using a Nanodrop® ND-1000 Spectrophotometer.
Real-time RT-PCR
Total RNA (1 μg) was converted to cDNA using iScript cDNA Synthesis Kit (Bio-Rad). The reaction mixture was set up as follows: 1 μg of total RNA, 4 μl of iScript reaction mix, 1 μl of iScript reverse transcriptase, nuclease free water to bring the total volume to 20μl. Reverse transcription was performed at 25°C for 5 min (priming), then 42°C for 30 min (revers transcription reaction) followed by 85°C for 5 min (reverse transcription reaction inactivation). This cDNA reaction mix was used for real-time PCR StepOnePlus™ Real-Time PCR Systems (Applied Biosystems). Each reaction mixture contained 0.5 μl of cDNA, 5 μl of Fast SYBR™ Green Master Mix (Applied Biosystems), 0.3 μl from each forward and reverse primers and 3.9 μl nuclease-free water to make up the volume to 10 μl. The reactions were incubated at 95°C for 20 sec (hold), followed by 40 cycles of 95°C for 3 sec (denature) and 60°C for 30 sec (anneal/extend). Specific primer pairs for PCAT29 were used for the reactions and mRNA expression levels were normalized to the levels of GAPDH. The primer sets were purchased from Invitrogen and were as follows:
H. sapiens-PCAT29 (forward): 5’-TCTGCTGAGACCCAGTGC-3’
(reverse): 5’-TTCTCTCACATTTCATTCACC-3’,
H. sapiens-GAPDH (forward): 5’-AATCCCATCACCATCTTCCA-3’
(reverse): 5- TGGACTCCACGACGTACTCA-3’
The cycle threshold (Ct) values were used to analyze the results of real-time PCR. PCAT29 expression was calculated using the 2-ddCt method relative to GAPDH and results were reported as fold change.
TaqMan® real-Time PCR
Total RNA was isolated from the cells using a protocol previously described [35]. Total RNA (100 ng) was reverse transcribed (RT) to generate miR-21 cDNA using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). Each RT reaction contained 1 μl of 1X RT specific primer for miR-21 and U6, 1.5 μL 1X RT reaction buffer, 0.15 μl of 100 mM dNTPs, 50 U/μl MultiScribe reverse transcriptase, 3.8 U/μl RNase inhibitor, 100 ng RNA and RNase-free water to make the final volume of 15 μl. The reaction mix was then incubated for 30 min at 16°C, 30 min at 42°C and 5 min at 85°C. The real-time PCR was performed using Applied Biosystem StepOnePlus™ real-time system using TaqMan® PCR kit (Applied Biosystem). After RT step, 1 μl of cDNA was combined with 0.6 μl of 20X TaqMan® primers (forward and reverse) and 5 μl TaqMan® Universal PCR Master Mix in 10 μl final volume. The amplification was performed by incubation at 95°C for 10 min followed by 40 cycle of 95°C for 15 sec and 60°C for 1 min. The 2-ddCt was used to calculate the miR-21 expression and U6 snRNA was used as internal control.
Immunohistochemistry
Formalin-fixed paraffin embedded (FFPE) human prostate tumor and their paired perinormal sections on glass slides were obtained from The Tissue Banking Facility of Simmons Cancer Institute of SIU School of Medicine. Immunohistochemistry was performed using ImmunoCruz™ ABC Staining System (Santa Cruz Biotechnology) for the detection of PDCD4, pSTAT3 and IL-6. Antigen retrieval was performed by first deparaffinizing and rehydrating the tissue sections and then immersing them in 10 mM sodium citrate buffer (pH 6) for 10 min in a 90°C water bath. After cooling down to room temperature, the tissue sections were incubated with 0.1% H2O2 for 10 min at room temperature to block the endogenous peroxidase activity. The sections were then washed twice with 1X PBS and blocked with 1.5% goat serum diluted in 1X PBS for 1 h at room temperature. The tissue sections were then incubated with 100 μl of respective primary antibodies (dilutions: 1:100 for PDCD4, 1:50 for pSTAT3 and 1:50 IL-6) overnight at 4°C. Next day, the sections were washed twice with 1X PBS and incubated with biotinylated secondary antibody for 30 min at room temperature. To identify the peroxidase activity, the section were first incubated with avidin and biotinylated horseradish peroxidase enzyme (AB reagent) for 10 min and then with peroxidase substrate until desired stain intensity develops. The sections were then washed in deionized water, counterstained with hematoxylin, dehydrated with ethanol and mounted using permanent mounting media. The tissue sections were imaged using an Olympus light microscope (Olympus imaging America Inc.) using Olympus DP controller software.
Statistical analysis
Statistical analysis were performed either using analysis of variance (ANOVA) followed by Bonferroni post hoc correction for multiple comparisons or by paired Student’s t-test using GraphPad Prism software 6.0. The value of p<0.05 was considered as statistically significant. The error bars shown in the figures represent standard error of mean (SEM).
Results
Down-regulation of PCAT29 expression in prostate cells and prostate tissues
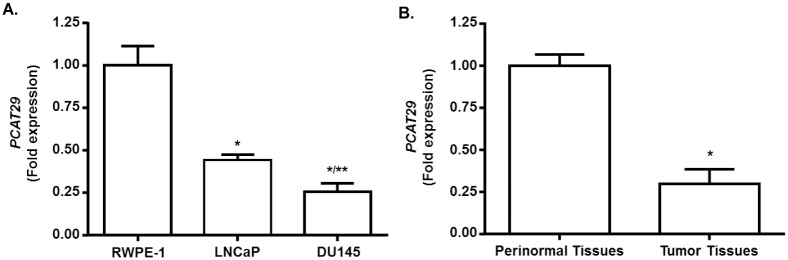
Previous studies have reported that PCAT29 was expressed at low levels in DU145 and LNCaP cells [36]. To evaluate the level of PCAT29 expression, we examined its expression in DU145, LNCaP prostate cancer cells and normal prostate cells, RWPE-1. The results showed that PCAT29 expression, normalized to that in RWPE-1 cells, were 44.0 ± 3.1% and 25.6 ± 1.8%, respectively, in LNCaP and DU145 cells, cultured under similar conditions. These results show reduced expression of PCAT29 expression in prostate cancer cells than normal prostate cells. Furthermore, the statistically significant difference in levels of PCAT29 in the more aggressive DU145 cell line, compared to LNCaP cells, implicates down-regulation of PCAT29 in oncogenesis (Fig 1A).
Fig 1. Reduced expression of PCAT29 expression in prostate cancer cells and tumor tissues.
(A) Relative mRNA levels of PCAT29 in normal human prostate epithelial cells (RWPE-1) and prostate cancer cells (LNCaP and DU145). The levels of PCAT29 were significantly reduced in prostate cancers cells compared to RWPE-1 cells. (n≥3) (B) Significant reductions in PCAT29 levels in normal prostate tissues compared to prostate cancer. Data are presented as the mean ± SEM of 4 prostate samples. Asterisks (*) indicate statistically significantly difference (p < 0.05) from RWPE-1 and from normal prostate tissue, respectively. (**) indicate statistically significantly difference (p < 0.05) from LNCaP cells.
We next examined the expression of PCAT29 in human prostate cancer tissues. These prostate tissue specimens were obtained from Tissue Banking Facility of Simmons Cancer Institute of SIU School of Medicine (Springfield, IL, USA). Fig 1B shows that PCAT29 expression was higher in normal than in tumor tissues. The relative expression was 0.29 ± 0.15 in tumor tissues as compared to normal tissues.
IL-6 down-regulates PCAT29 expression on prostate cancer cells
IL-6 plays a crucial role in the differentiation of human prostate carcinoma and benign prostatic hyperplasia. [37]. To evaluate the effect of IL-6 on PCAT29 activity, prostate DU145, LNCaP and normal prostate cell RWPE-1 cells were treated with IL-6 (10 ng/ml) for 24 h and PCAT29 expression was determined by qPCR. The results showed significant reductions in PCAT29 expression for both DU145 and LNCaP cells following IL-6 treatment for 24 h. Similar treatment of RWPE-1 cells with IL-6 did not alter PCAT29 expression. The relative expression of PCAT29 induced by IL-6 was 44.4 ± 4.0%, 47.9 ± 7.9% and 100% in DU145, LNCaP and RWPE-1 cells, respectively (Fig 2).
Fig 2. IL-6 reduced the expression of PCAT29 in prostate cancer but not normal cells.
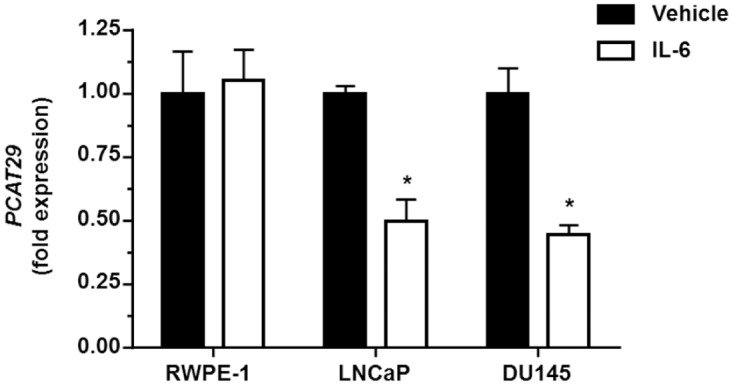
DU145 and LNCaP cells were treated with IL-6 (10 ng/ml) for 24 h, following which the expression of PCAT29 was determined by real-time RT-PCR. Both DU145 and LNCaP cells showed decreased PCAT29 expression after IL-6 treatment. There was no difference in PCAT29 expression following IL-6 treatment in RWPE-1 cells. Data are presented as the mean ± SEM of at least 3 independent samples. Asterisks (*) indicates statistically significantly difference (p < 0.05) from vehicle groups.
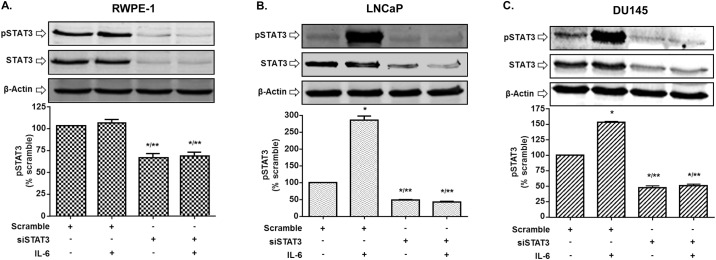
Since IL-6 signaling pathway includes STAT3, we performed Western blot studies to assess the status of p-STAT3 in the different prostate cancer cell lines following IL-6 treatments. Significant increases in p-STAT3 levels (normalized to STAT3) were observed following treatment with IL-6 (10 ng/ml) in DU145 and LNCaP cells. However, IL-6 treatment did not significantly alter STAT3 phosphorylation in RWPE-1 cells. Knockdown of STAT3 using siRNA, not only blunted the responses of IL-6 in DU145 and LNCaP cells but reduced basal pSTAT3 activities in all three cell lines. These data suggest that IL-6 activates STAT3 in DU145 and LNCaP cells but not in the normal RWPE-1 cell line (Fig 3A, 3B and 3C). These results suggest a positive correlation between p-STAT3 and IL-6 and suggest that pSTAT3 is involved in down-regulation of PCAT29.
Fig 3. IL-6 induces STAT3 phosphorylation in prostate cancer cells.
(A) RWPE-1 cells were transfected with either scramble or siSTAT3 (10 mM) for 24 h then subjected to IL-6 treatment (10 ng/ml) for 30 min, then cells lysate were used for Western blotting analysis. IL-6 showed no significant effect on STAT3 phosphorylation on this cell. (B, C) LNCaP and DU145 cells were transfected with a scrambled siRNA sequence or siSTAT3 (10 nM) for 24 h then treated with vehicle or IL-6 (10 ng/ml) for 5 min (DU145) or 30 min (LNCaP). Cells were then lysed and used for Western blotting analysis. Bar graph represents mean ± SEM of 3 independent experiments. Asterisks (*) (**) indicate statistically significantly difference (p < 0.05) from scramble or IL-6, respectively.
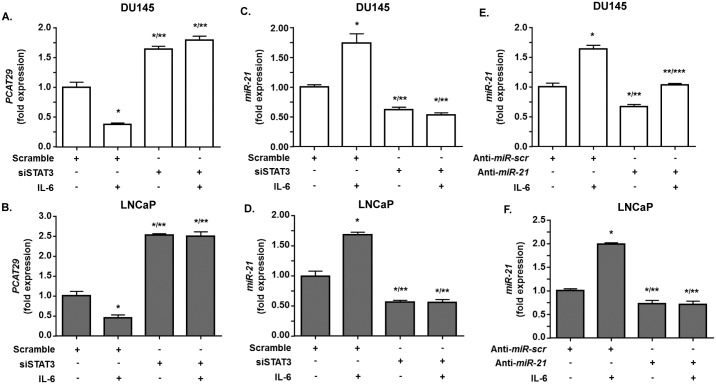
Additional studies were performed to confirm a role of STAT3 in regulation of PCAT29 expression. Knockdown of STAT3 using siRNA resulted in increased levels of PCAT29 in both DU145 and LNCaP cells. In addition, knockdown of STAT3 abolished the ability of IL-6 to decrease PCAT29 (Fig 4A and 4B). The relative expression of PCAT29 following STAT3 knockdown and treatment with vehicle or IL-6 were 1.64 ± 0.05-fold and 1.79 ± 0.07-fold over scramble siRNA, respectively, in DU145 cells. In LNCaP cells, the responses of IL-6 following STAT3 knockdown were 2.25 ± 0.03-fold and 2.47 ± 0.11-fold over scramble siRNA, respectively.
Fig 4. STAT3 knockdown increase the level of PCAT29 expression.
(A, B) DU145 and LNCaP cells were transfected with either scramble siRNA sequence or siSTAT3 (10 nM) for 24 h then treated with IL-6 (10 ng/ml) for another 24 h. Cells were then used to determine PCAT29 level by real-time RT-PCR. IL-6 suppressed PCAT29 expression in both DU145 and LNCaP cells which were pretreated with scrambled siRNA but not following transfection with siSTAT3. (C, D) DU145 and LNCaP cells were transfected with either scramble or siSTAT3 (10 nM) for 24 h, then treated with IL-6 (10 ng/ml) for 24 h, and used to determine miR-21 levels by TaqMan® real-time PCR. The levels of miR-21 were increased by IL-6 in the cells treated with scrambled siRNAs but not after transfection with siSTAT3. In fact, siSTAT3 reduced both basal and IL-6 stimulated miR-21 expression. (E, F) DU145 and LNCaP cells were transfected with a scrambled sequence (anti-miR-scr) or anti-miR-21 (30ng/ml) for 24 h and then treated with IL6 (10 ng/ml) for 24 h. The expression of miR-21 was the determined by TaqMan® real-time PCR. IL-6 increased miR-21 in the group treated with the scrambled sequence but its effects were significantly reduced by anti-miR-21 in DU145 cells and completely reversed in LNCaP cells. Data represent the mean ± SEM of at least 3 independent experiments. Asterisks (*), (**), and (***) indicate statistically significantly difference (p < 0.05) from scramble, from scramble +IL-6 and from anti-miR-21, respectively.
Our results also showed that IL-6 significantly increased miR-21 levels. The relative increases in expression of miR-21 were 1.84 ± 0.09 and 1.72 ± 0.04 over the control in DU145 and LNCaP cells, respectively. However, knockdown of STAT3 led to significantly decreased IL-6 stimulated miR-21 levels in both DU145 and LNCaP cells as compare to control. The relative expression of miR-21 following STAT3 knockdown were 0.61 ± 0.11 and 0.61 ± 0.03 in DU145 and LNCaP cells, respectively. In addition to relative expression of miR-21 in STAT3 transfected groups treated with IL-6 were 0.54 ± 0.08 and 0.51 ± 0.13 in DU145 and LNCaP cells, respectively. (Fig 4C and 4D).
MiR-21 regulates PCAT29 in prostate cancer cells
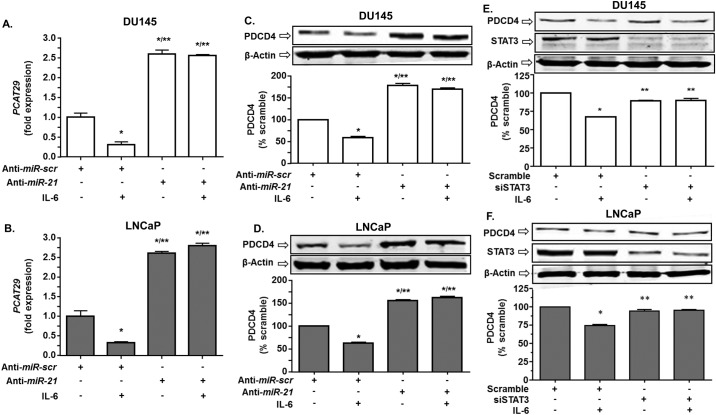
Previous studies have shown that miR-21 is a relevant target of STAT3, which mediates growth factor receptors regulation of cell proliferation. Additional studies show that lncRNAs can be regulated by microRNA [3]. We first investigated the efficiency of miR-21 knocking down using anti-miR-21 oligonucleotides. This resulted in significantly reduced miR-21 expression in both DU145 and LNCaP by 33.4 ± 3.8% and 27.3 ± 3.5%, respectively. IL-6 treatment increased miR-21 levels by 1.64 ± 0.04 and 1.98 ± 0.07-fold over vehicle control in DU145 and LNCaP cells, respectively. However, the responses of these cells to IL-6 were reduced (in DU145) or completely abrogated (in LNCaP) following partial knockdown of miR-21 (Fig 4E and 4F). Measurement of PCAT29 expression in these cells showed that IL-6 suppressed PCAT29 expression in DU145 and LNCaP cells and the relative expression were reduced to 0.30 ± 0.01-fold and 0.32 ± 0.03-fold, respectively. Partial knockdown of miR-21 resulted in significant increases in the basal levels of PCAT29 and blunted its reduction by IL-6 in DU145 and LNCaP cells. The relative expressions of PCAT29 were 2.59 ± 0.03-fold and 2.6 ± 0.05-fold over control following partial knockdown of miR-21 and treatment with IL-6 in DU145 and LNCaP cells, respectively. There were no significant differences in PCAT29 levels in the miR-21 knockdown groups treated with either vehicle or IL-6 (Fig 5A and 5B). These results provide good evidence that miR-21 is an important regulator of PCAT29 expression. The almost complete abolition of the IL-6 response following partial knockdown of miR-21 suggests that PCAT29 expression is very sensitive to changes in miR-21 levels in prostate cancer cells.
Fig 5. Knockdown of miR-21 increases the levels of PCAT29 and PDCD4.
(A, B) DU145 and LNCaP cells were transfected with a scrambled oligo sequence (anti-miR-scr) or anti-miR-21 (30 nM) for 24 h. Cells were then treated with vehicle or IL6 (10 ng/ml) for 24 h and the expression of PCAT29 were determined by real-time RT-PCR. IL-6 significantly decreased the expression of PCAT29 in the anti-miR-scr. Knockdown of miR-21 enhanced the expression of PCAT29 and abolished the effect of IL-6 in both cell lines. (C, D) DU145 and LNCaP cells were transfected with anti-miR-21 (30 nM) for 24 h and then treated with IL-6 (10 ng/ml) for another 24 h. Cell lysates were prepared and used for Western blotting analysis. IL-6 significantly decreased the expression of PDCD4. Knockdown of miR-21 increased basal PDCD4 levels and abolished the reduction of this protein by IL-6. (E, F) DU145 and LNCaP cells were transfected with siRNA against STAT3 (10 nM) for 24 h. Cells were then treated with IL-6 (10 ng/ml) for 24 h and lysates were used for Western blotting analysis. IL-6 reduced PDCD4 levels, but this effect was abolished by siSTAT3. Data are presented as the mean ± SEM of at least 3 independent experiments. Asterisks (*) and (**) indicate statistically significantly difference (p < 0.05) from scramble and from scramble +IL-6 treatment groups, respectively.
We next examined the levels of PDCD4, a downstream target negatively regulated by miR-21. Our results also show a negative correlation between miR-21 and PDCD4. IL-6 treatment reduced PDCD4, as anticipated due to the ability of this cytokine to increase miR-21 levels. Knockdown of miR-21 resulted in a significant increase of PDCD4 protein levels in both DU145 and LNCaP cells. IL-6 treatment showed decrease in PDCD4 in both DU145 and LNCaP cells (Fig 5C and 5D). In addition, partial knockdown of STAT3 resulted in increased levels of PDCD4 and abrogated the effect of IL-6 (Fig 5E and 5F). Overall, these data support the above finding that miR-21 could regulate the expression of PCAT29 in prostate cancer cells.
Resveratrol blocks IL-6 effects on prostate cancer cells
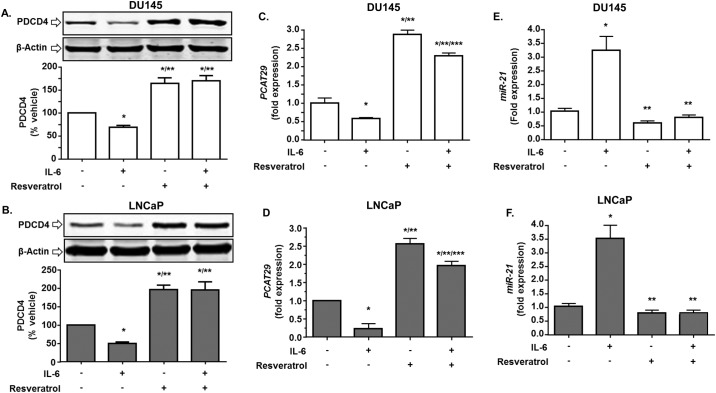
We have previously shown that the anti-tumor actions of resveratrol are mediated, in part, by inhibiting the miR-21 signaling pathway in prostate cancer [34]. Since IL-6 regulation of PCAT29 appears dependent on miR-21, we tested the effect of resveratrol on PCAT29 expression. One important gene which is negatively regulated by miR-21 is the tumor suppressor programmed cell death protein 4 (PDCD4) [16] which is induced by resveratrol [34]. As shown above, IL-6 decreased the levels of PDCD4 following 24 h treatment in DU145 and LNCaP cells, while knockdown of miR-21 increased PDCD4 levels in both vehicle- and IL-6 treated cells and abolish IL-6 effect (Fig 5C and 5D). Additional studies show that the reductions in PDCD4 produced by IL-6 treatment was reversed by resveratrol (Fig 6A and 6B). In fact, resveratrol significantly increased PDCD4 levels above control levels. In addition, in presence of resveratrol, IL-6 was unable to decrease PCAT29 expression in both DU145 and LNCaP cells. The fold expression of PCAT29 for the resveratrol-pretreated groups in absence and presence of IL-6 were 2.87 ± 0.12 and 2.2 ± 0.08, respectively for DU145 cells. In LNCaP cells the fold expression of PCAT29 were 2.56 ± 0.15 and 1.9 ± 0.11 in vehicle and IL-6 treatment groups, respectively (Fig 6C and 6D). Furthermore, we show that while IL-6 increased miR-21 in these cells, the addition of resveratrol (25μM) for 24 h led to significant reductions in both basal and IL-6 induced miR-21 levels (Fig 6E and 6F). Thus, resveratrol induction of PCAT29 expression involves suppression of IL-6/STAT3/miR-21 signaling.
Fig 6. Resveratrol blocks IL-6 suppression of PDCD4 and PCAT29 and its induction of miR-21.
(A, B) Western blotting analysis of DU145 and LNCaP cells showed that IL-6 reduced the levels of PDCD4. Resveratrol (25 μM) treatment for 24 h blocked this reduction and increased the levels of PDCD4 above that of control. This effect of resveratrol persisted even in the presence of IL-6. (C, D) Resveratrol also increased PCAT29 by greater than 2-fold and this effect was partly reduced by concurrent IL-6 treatment. (E, F) Resveratrol restored IL-6 stimulated expression of miR-21 in LNCaP and DU145 cells. Data are presented as the mean ± SEM of at least 3 independent experiments. Asterisks (*), (**), and (***) indicate statistically significantly difference (p < 0.05) from vehicle, from vehicle+IL-6 and from resveratrol, respectively.
PCAT29 regulates oncogenic phenotypes in vitro
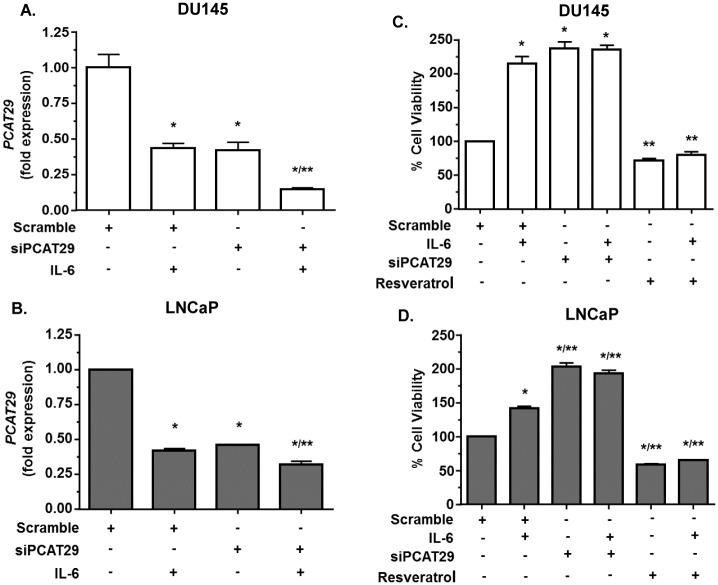
Previous studies showed PCAT29 is an important gene for inhibiting prostate cancer [32]. Since PCAT29 is negatively regulated by IL-6 and positively regulated by resveratrol, we decided to investigate the functional role of PCAT29. First siRNA was designed to knockdown the expression of PCAT29 in prostate cancer cells. Using siRNA, we shown a significant reduction in PCAT29 level in both DU145 and LNCaP cells, by 60 ± 3% and 54 ± 2%, respectively. IL-6 treatment decreased PCAT29 levels by 26 ± 1% and 41 ± 1% in DU145 and LNCaP cells, respectively (Fig 7A and 7B). Knockdown of PCAT29 led to increased proliferation in both DU145 and LNCaP cells compared to increased cell proliferation obtained with IL-6 in normal cells. Resveratrol abolished both basal and IL-6 dependent cell proliferation in both DU145 and LNCaP cells (Fig 7C and 7D).
Fig 7. PCAT29 tonically suppressed prostate cancer cell proliferation.
(A, B) DU145 and LNCaP cells were transfected with scrambled siRNA or siRNA against PCAT29 (siPCAT29, 30 nM) for 24 h, followed by treatment with IL6 (10 ng/ml) for 24 h. Cells were then used to determine PCAT29 levels. (C, D) DU145 and LNCaP cells were transfected with scrambled siRNA or siPCAT29 (30 ng/ml) for 24 h, followed by treatment with IL-6 for 24 h and cells proliferation was determined by MTS assay. IL-6 treatment significantly increased cells proliferation, which was mimicked by siPCAT29. Knockdown of PCAT29 did not increase cell proliferation above that observed with IL-6 alone in DU145 but not in LNCaP cells. Resveratrol treatment significantly reduced cell proliferation, even in presence of IL-6. Data are presented as the mean ± SEM of at least 3 independent experiments. Asterisks (*) and (**) indicate statistically significantly difference (p < 0.05) from scramble and from scramble + IL-6, respectively.
Human prostate cancer specimen demonstrate increase in pSTAT3
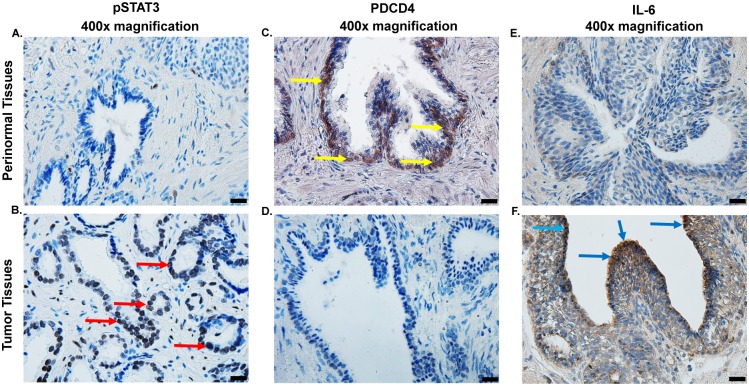
STAT3 acts as an oncogenic tumor marker in prostate and colon cancer tissues, where it mediates hyperplasia and neoplastic transformation [37]. The role of STAT3 as a downstream conduit of IL-6 signaling and as a regulator of miR-21 prompted us to determine whether pSTAT3 levels was inversely correlated with PDCD4 in human prostate specimens. Immunohistochemical comparisons were made between prostate cancer and its paired perinormal tissue using paraffin embedded sections. We observed a higher degree of pSTAT3 staining in prostate cancer than those in perinormal tissues (Fig 8A). Immunolabeling for PDCD4 showed high expression of this protein in normal specimens, compared to lower levels of this protein in prostate cancer (Fig 8B). In addition, we observed localization of PDCD4 mainly in nuclei of epithelial cells in normal but not in these cells in prostate cancer specimens. In addition, immunohistochemical comparison between normal and prostate cancer using paraffin embedded sections show a higher degree of IL-6 staining in prostate cancer than those in perinormal tissue sections (Fig 8C). Overall, data from human prostate specimens indicate a reciprocal relationship between the levels of pSTAT3 and IL-6 versus those of PDCD4.
Fig 8. Levels of pSTAT3, PDCD4 and IL-6 in human prostate specimens.
(A, B) Immunohistochemistry studies were performed to determine the levels of pSTAT3 in formalin-fixed paraffin embedded prostate tumor specimens, as compared to adjacent normal prostate specimen. Diaminobenzidine tetrahydrochloride (DAB) staining revealed high expression of pSTAT3 (B) as dark-brown labeling in epithelial cells of tumor samples (marked by red arrows) while low expression was observed in paired perinormal prostate tissues (A). In contrast, high expression of PDCD4 was observed in perinormal prostate tissues (as evidenced by dark-brown labeling, indicated by yellow arrows) (C), as compared to prostate tumors (D). Immunolabling of prostate tumor revealed high expression of IL-6 (F), compared to perinormal prostate specimens. Representative images show immunohistochemical studies performed in four different specimens obtained from three different patients. Pictures magnification is 400x. Scale bar is 20 μm.
Discussion
There is relationship between inflammation and cancer development, as pro-inflammatory cytokines participate in the progression of cancer cells to a more aggressive form [38]. In particular, the IL-6/STAT3 signaling pathway appears to be crucial for the progression of prostate cancer. High levels of IL-6 expression is detected in patients with prostate cancer [16,37] which could activate IL-6 receptor, leading to increased STAT3 activation [39]. The levels of STAT3 have also been found to be elevated in many cancers where they stimulate cell proliferation (via cyclin D1) and induction of anti-apoptotic proteins (such as Bcl2) [40]. Our findings show that IL-6 is a negative regulator of PCAT29 expression which is mediated by activation of STAT3. Activated STAT3 promoted down-regulation of PCAT29 expression in both androgen dependent and independent prostate cancer cells by induction of miR-21. Moreover, we showed reduced expression of LncPCAT29 in Grade 4 prostate tumor samples compared to prostate samples from normal. These studies identify PCAT29 as a novel target of miR-21, which could contribute to its pro-oncogenic properties. The current study shows another target of IL-6, namely LncPCAT29, whose down-regulation requires an interaction between STAT3 and miR-21. Down-regulation of PCAT29 was associated with increased STAT3 and decreased levels of PDCD4 (a downstream target of miR-21) in Grade 4 prostate cancer. Previous studies have shown IL-6 can regulate the expression of LncTCF7 in hepatocellular carcinoma via a STAT3 pathway [41]. Similarly, the IL-6/STAT3 pathway is implicated in the induction of HOTAIR LncRNA in human bronchial epithelial cells [42]. The expression of HOTAIR LncRNA was found to be regulated by a direct interaction of miR-34a with this gene [43]. MiR-21 regulates prostate cancer progression by down-regulating tumor suppressor genes, such as PTEN and PDCD4 [44]. This is the first study to show that PCAT29 is also a relevant target of miR-21 in prostate cancer cells, as inhibition of miR-21 expression leads to stimulation of PCAT29.
Resveratrol is generally regarded as a chemo-preventive agent for various diseases. This property is derived from the interaction of resveratrol with multiple targets and many molecular pathways [34,45]. The structure of resveratrol is similar to androgen and estrogen, suggesting that its might produce its beneficial actions via these hormone receptors [46]. We have previously shown that resveratrol reduces prostate cancer growth and metastasis by inhibiting the AKT/miR-21 pathway [34]. In the current study, we show that targeting miR-21 could also contribute to the induction of PCAT29 by resveratrol. As such, resveratrol was shown to induce miR-21 targets such as PDCD4 in both LNCaP and DU145. These findings suggest that miR-21 could provide tonic suppression of PCAT29 expression in prostate cancer cells which could be reversed by resveratrol.
MiR-21 is an important cancer risk factor whose expression is induced by IL-6 [47,48]. Overexpression of miR-21 is observed in many types of cancers, including, pancreas, lung, breast and prostate cancer [49]. This microRNA possesses oncogenic properties as it induces many proteins associated with cell proliferation [50,51]. Previous studies have shown that STAT3 binds directly to the miR-21 promoter to induce its expression [52]. Our qPCR resulted showed that resveratrol down regulate miR-21 expression in prostate cancers cells, presumably by suppressing STAT3. Thus, by interfering with the STAT3/miR-21 pathway, resveratrol could block IL-6 mediated down-regulation of PCAT29 and reduce tumorigenesis.
The overall impact of this study is significant as is provide strong evidence for the suppression of PCAT29 expression by the IL-6 signaling pathway. This study identifies STAT3 and miR-21 as important players in the regulation of PCAT29. Furthermore, it provides a clinically relevant drug, resveratrol, which could abrogate IL-6 signaling to boost the expression of PCAT29 and facilitate its anti-tumor action.
Supporting information
(PDF)
Acknowledgments
This project was supported in part by an NIH grant, R01 CA166907, to VR. We would like to thank Dr. Daotai Nie (Department of Medical Microbiology, Immunology and Cell Biology, Southern Illinois University School of Medicine, Springfield, IL) for providing us RWPE-1, DU145 and LNCaP cells. We also like to thank The Tissue Banking Facility of Simmons Cancer Institute of SIU School of Medicine for providing us the FFPE and RNA samples of prostate tissues.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received funding from National Cancer Institute (R01 CA166907). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pickard MR, Mourtada-Maarabouni M, Williams GT (2013) Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta 1832: 1613–1623. 10.1016/j.bbadis.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 2.Weiss M, Plass C, Gerhauser C (2014) Role of lncRNAs in prostate cancer development and progression. Biol Chem 395: 1275–1290. 10.1515/hsz-2014-0201 [DOI] [PubMed] [Google Scholar]
- 3.Yang G, Lu X, Yuan L (2014) LncRNA: a link between RNA and cancer. Biochim Biophys Acta 1839: 1097–1109. 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15 Spec No 1: R17–29. [DOI] [PubMed] [Google Scholar]
- 5.Gibb EA, Brown CJ, Lam WL (2011) The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10: 38 10.1186/1476-4598-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams GT, Farzaneh F (2012) Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer 12: 84–88. 10.1038/nrc3195 [DOI] [PubMed] [Google Scholar]
- 7.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, et al. (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962. 10.1038/onc.2010.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prensner JR, Chinnaiyan AM (2011) The emergence of lncRNAs in cancer biology. Cancer Discov 1: 391–407. 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, et al. (2011) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394. 10.1038/nature10006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, et al. (2007) Activation of p53 by MEG3 non-coding RNA. J Biol Chem 282: 24731–24742. 10.1074/jbc.M702029200 [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, et al. (2010) Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res 70: 2350–2358. 10.1158/0008-5472.CAN-09-3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, et al. (2011) microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30: 4750–4756. 10.1038/onc.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, et al. (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093. 10.1038/emboj.2010.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bray F, Ren JS, Masuyer E, Ferlay J (2013) Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 132: 1133–1145. 10.1002/ijc.27711 [DOI] [PubMed] [Google Scholar]
- 16.Dong B, Shi Z, Wang J, Wu J, Yang Z, et al. (2015) IL-6 Inhibits the Targeted Modulation of PDCD4 by miR-21 in Prostate Cancer. PLoS One 10: e0134366 10.1371/journal.pone.0134366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, et al. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22. 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, et al. (2012) The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239–243. 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, et al. (2013) Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23: 159–170. 10.1016/j.ccr.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 20.Jeronimo C, Bastian PJ, Bjartell A, Carbone GM, Catto JW, et al. (2011) Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol 60: 753–766. 10.1016/j.eururo.2011.06.035 [DOI] [PubMed] [Google Scholar]
- 21.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, et al. (2010) Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 3: ra29 10.1126/scisignal.2000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, et al. (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038. 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Han S, Jin G, Zhou X, Li M, et al. (2014) Linc00963: a novel, long non-coding RNA involved in the transition of prostate cancer from androgen-dependence to androgen-independence. Int J Oncol 44: 2041–2049. 10.3892/ijo.2014.2363 [DOI] [PubMed] [Google Scholar]
- 24.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, et al. (2000) PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A 97: 12216–12221. 10.1073/pnas.97.22.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, et al. (2011) Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci 102: 245–252. 10.1111/j.1349-7006.2010.01737.x [DOI] [PubMed] [Google Scholar]
- 26.Cui Z, Ren S, Lu J, Wang F, Xu W, et al. (2013) The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol 31: 1117–1123. 10.1016/j.urolonc.2011.11.030 [DOI] [PubMed] [Google Scholar]
- 27.Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, et al. (2013) Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J 32: 1665–1680. 10.1038/emboj.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT (2009) GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28: 195–208. 10.1038/onc.2008.373 [DOI] [PubMed] [Google Scholar]
- 29.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, et al. (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674. 10.1016/j.molcel.2010.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, et al. (2013) The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet 9: e1003705 10.1371/journal.pgen.1003705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, et al. (2011) Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29: 742–749. 10.1038/nbt.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik R, Patel L, Prensner JR, Shi Y, Iyer MK, et al. (2014) The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res 12: 1081–1087. 10.1158/1541-7786.MCR-14-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong C, Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 470: 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheth S, Jajoo S, Kaur T, Mukherjea D, Sheehan K, et al. (2012) Resveratrol reduces prostate cancer growth and metastasis by inhibiting the Akt/MicroRNA-21 pathway. PLoS One 7: e51655 10.1371/journal.pone.0051655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jajoo S, Mukherjea D, Kaur T, Sheehan KE, Sheth S, et al. (2013) Essential role of NADPH oxidase-dependent reactive oxygen species generation in regulating microRNA-21 expression and function in prostate cancer. Antioxid Redox Signal 19: 1863–1876. 10.1089/ars.2012.4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakurai K, Reon BJ, Anaya J, Dutta A (2015) The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol Cancer Res 13: 828–838. 10.1158/1541-7786.MCR-15-0016-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lou W, Ni Z, Dyer K, Tweardy DJ, Gao AC (2000) Interleukin-6 induces prostate cancer cell growth accompanied by activation of stat3 signaling pathway. Prostate 42: 239–242. [DOI] [PubMed] [Google Scholar]
- 38.Michalaki V, Syrigos K, Charles P, Waxman J (2004) Serum levels of IL-6 and TNF-alpha correlate with clinicopathological features and patient survival in patients with prostate cancer. Br J Cancer 90: 2312–2316. 10.1038/sj.bjc.6601814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegsmund MJ, Yamazaki H, Pastan I (1994) Interleukin 6 receptor mRNA in prostate carcinomas and benign prostate hyperplasia. J Urol 151: 1396–1399. [DOI] [PubMed] [Google Scholar]
- 40.Carpenter RL, Lo HW (2014) STAT3 Target Genes Relevant to Human Cancers. Cancers (Basel) 6: 897–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Zhang J, Shen B, Yin K, Xu J, et al. (2015) Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res 34: 116 10.1186/s13046-015-0229-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Wang B, Liu X, Lu L, Luo F, et al. (2016) Epigenetic silencing of p21 by long non-coding RNA HOTAIR is involved in the cell cycle disorder induced by cigarette smoke extract. Toxicol Lett 240: 60–67. 10.1016/j.toxlet.2015.10.016 [DOI] [PubMed] [Google Scholar]
- 43.Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, et al. (2016) Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. Int J Mol Sci 17: 356 10.3390/ijms17030356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, et al. (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27: 2128–2136. [DOI] [PubMed] [Google Scholar]
- 45.Yang Q, Xu E, Dai J, Liu B, Han Z, et al. (2015) A novel long noncoding RNA AK001796 acts as an oncogene and is involved in cell growth inhibition by resveratrol in lung cancer. Toxicol Appl Pharmacol 285: 79–88. 10.1016/j.taap.2015.04.003 [DOI] [PubMed] [Google Scholar]
- 46.Benitez DA, Pozo-Guisado E, Alvarez-Barrientos A, Fernandez-Salguero PM, Castellon EA (2007) Mechanisms involved in resveratrol-induced apoptosis and cell cycle arrest in prostate cancer-derived cell lines. J Androl 28: 282–293. [DOI] [PubMed] [Google Scholar]
- 47.Hussain SP, Harris CC (2007) Inflammation and cancer: an ancient link with novel potentials. Int J Cancer 121: 2373–2380. [DOI] [PubMed] [Google Scholar]
- 48.Schetter AJ, Heegaard NH, Harris CC (2010) Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis 31: 37–49. 10.1093/carcin/bgp272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krichevsky AM, Gabriely G (2009) miR-21: a small multi-faceted RNA. J Cell Mol Med 13: 39–53. 10.1111/j.1582-4934.2008.00556.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103: 2257–2261. 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, et al. (2007) Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72: 397–402. 10.1159/000113489 [DOI] [PubMed] [Google Scholar]
- 52.Ou H, Li Y, Kang M (2014) Activation of miR-21 by STAT3 induces proliferation and suppresses apoptosis in nasopharyngeal carcinoma by targeting PTEN gene. PLoS One 9: e109929 10.1371/journal.pone.0109929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.