Abstract
With the expansion of the microbiology field of research, a new genome editing tool arises from the biology of bacteria that holds the promise of achieving precise modifications in the genome with a simplicity and versatility that surpasses previous genome editing methods. This new technique, commonly named CRISPR/Cas9, led to a rapid expansion of the biomedical field; more specifically, cancer characterization and modeling have benefitted greatly from the genome editing capabilities of CRISPR/Cas9. In this paper, we briefly summarize recent improvements in CRISPR/Cas9 design meant to overcome the limitations that have arisen from the nuclease activity of Cas9 and the influence of this technology in cancer research. In addition, we present challenges that might impede the clinical applicability of CRISPR/Cas9 for cancer therapy and highlight future directions for designing CRISPR/Cas9 delivery systems that might prove useful for cancer therapeutics.
Keywords: CRISPR/Cas9, gene editing, Cas9 regulation, multiplex CRISPR/Cas9, cancer, phage-derived vectors
Main Text
The increasing burden of cancer in the human population represents a major concern for our society, and finding alternative treatments that are safe as well as efficient has become a major goal for researchers around the world. For the past decades, we have witnessed an effervescence of technologies that explore DNA structure and function, and our understanding of cancer has expanded to an extent that enables characterization of this disease at a deeper molecular level. The progression of basic research to reach clinical applications necessitates reliable pre-clinical models of cancer in which therapeutic strategies and agents can be evaluated for efficacy or efficiency.
Introducing targeted modifications in the genome for functional studies and cancer modeling or, moreover, for therapeutic purposes, requires highly efficient systems that are able to alter the existing DNA pattern with great precision. Nucleases of bacterial origin represent powerful tools that have been widely used for genome engineering purposes in an attempt to studying gene function and, ultimately, to implement new therapeutic strategies.
Among the existing nucleases with genome editing capabilities, the CRISPR system surpasses other nuclease-based systems, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), in terms of simplicity of design and versatility1, 2, 3 (Table 1). By 2013, the ability of the CRISPR/Cas9 system to engineer mammalian cell genomes has been experimentally validated, and the crystal structure of the Cas9 effector complex was resolved in 2014.4 Among the three CRISPR/Cas systems (I–III) identified in both bacteria and archaea, the type II system from Streptococcus thermophilus or Streptococcus pyogenes is the most versatile for genome engineering purposes.5
Table 1.
Comparison of ZFNs, TALENs, and CRISPR/Cas9 Nuclease Systems for Genome Editing
| Genome Editing Tool | ZFNs | TALENs | CRISPR/Cas9 |
|---|---|---|---|
| Features | ZF-FokI nuclease protein fusion | TALE-FokI nuclease protein fusion | Cas9 nuclease and sgRNA |
| Target site identification | protein-DNA interaction, capable of targeting virtually any site | protein-DNA interaction, capable of targeting virtually any site | RNA-DNA interaction, site selection restricted by the NGG motif, which occurs statistically every 8 nt in a random DNA sequence |
| Genome altering | DSBs directed by the FokI domain, repaired through NHEJ or HDR | DSBs directed by the FokI domain, repaired through NHEJ or HDR | DSBs directed by Cas9, repaired through NHEJ or HDR |
| Design | + | ++ | +++ |
| custom design based on the target sequence, labor intensive and time-consuming | custom design based on the target sequence, labor intensive, less time-consuming than ZFN | very simple design by altering the crRNA sequence of the sgRNA | |
| Efficiency | ++ | ++ | +++ |
| Biallelic targeting | ++ | ++ | +++ |
| Off-target effects | ++ | +++ | + |
| specific | more specific than ZFN | low compared with ZFN and TALEN as a result of allowed mismatches by the Cas9 nuclease between sgRNA and the DNA target sequence | |
| Multiplexing | + | + | +++ |
| rarely used | rarely used | yes, capable of targeting multiple sites simultaneously | |
| Size and delivery | +++ | ++ | + |
| the ZFN monomer is significantly smaller than Cas9 | TALEN monomers are situated in the middle of Cas9 and ZFN nucleases in terms of size | massive Cas9 protein encoded by a 4.2-kb sequence |
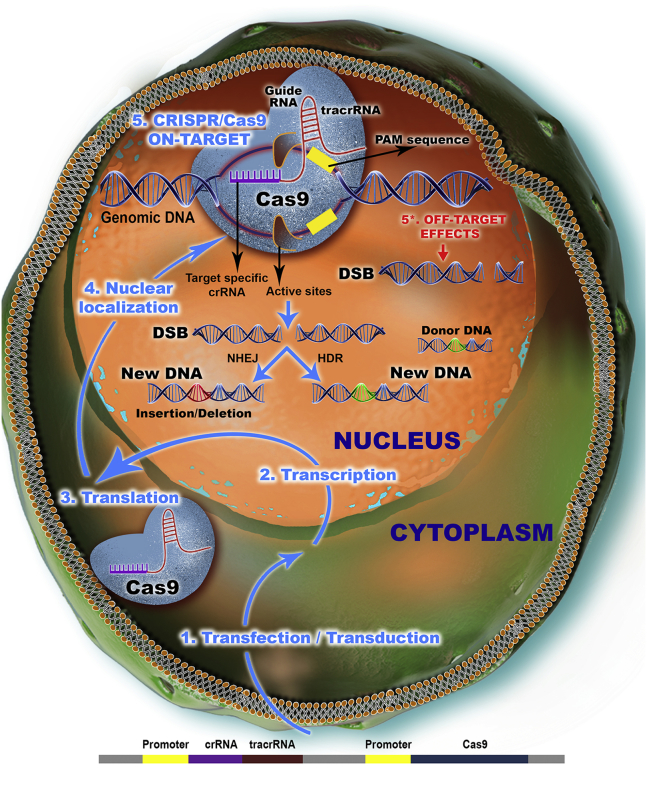
The CRISPR/Cas9 type II system consists of the Cas9 nuclease and a single guide RNA (sgRNA or gRNA), which is a fusion of a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) that binds Cas9 nuclease and directs it to a target sequence based on a complementary base-pairing rule. The target sequence must be adjacent to a protospacer-adjacent motif (PAM) consisting of a canonical NGG or NAG sequence. At the recognition site, a double-strand break (DSB) is generated that can be repaired by non-homologous end joining (NHEJ), resulting in small insertions or deletions (indels) usually associated with loss of function (knockdown/knockout) (Figure 1). In the presence of an exogenous donor DNA, by a homology-directed recombination (HDR) mechanism, precise modifications can be achieved at the targeted site, resulting in gain of function (knockin).6
Figure 1.
CRISPR/Cas9 Mechanism of Action
The original bacterial CRISPR/Cas9 design has been translated into an engineered instrument for genome editing purposes and is capable of introducing specific modifications in the target cell. In this regard, the vector comprising the crRNA and tracrRNA that together constitute the RNA molecule for Cas9 guidance (gRNA) is introduced in the desired cell, where it passes the cytoplasmic milieu toward the nucleus. After delivery to the nucleus, the Cas9 gene encoded by the experimental vector is transcribed and exported into the cytoplasm for translation of Cas9 nuclease. After synthesis of the active protein, the gRNA, transcribed by its own promoter, interacts with the Cas9 nuclease, resulting in the ribonucleic-protein effector complex that is internalized back into the nucleus. The cleavage of the double-stranded genomic DNA takes place in a guided manner, where the crRNA sequence of gRNA directs Cas9 toward the specific locus, based on sequence complementarity, which is positioned adjacent to the PAM. When cleaved, the continuity of the host DNA can be restored through NHEJ, where the hanging ends join together, creating small indels, or through HDR in the presence of a donor DNA.
However, Cas9 can tolerate, to a certain extent, mismatches between the sgRNA and the target sequence in the genome, resulting in off-target effects, as some previous studies have shown.7, 8 These undesirable effects of the CRISPR/Cas9 system might impede the use of this genome editing technology for clinical applications; therefore, a great deal of effort has been made to improve the efficiency and specificity of CRISPR/Cas9.
In this review, we summarize recent improvements made in CRISPR/Cas9 design and flexibility and the applications of this genome editing technology for cancer research. At the end of the manuscript, we discuss challenges and future perspectives of CRISPR/Cas9 in cancer therapy.
Minimizing the Off-Target Effects of CRISPR/Cas9
Optimizing the sgRNA design represents one approach for reducing the off-target effects of CRISPR/Cas9. It has been shown that the base composition of the 5′ sequence of the sgRNA can have a profound effect on the efficiency of CRISPR/Cas9,9 and large-scale studies of sgRNA libraries led to design algorithms for maximizing the on-target activity and reducing off-target effects.10 Several online tools are now available to assist researchers in designing sgRNAs with a higher specificity for a desired genomic locus (Table 2).
Table 2.
Online sgRNA Design Tools
| Platform | Platform Link | Reference |
|---|---|---|
| CRISPR Design | http://crispr.mit.edu/ | 8 |
| E-CRISP | http://www.e-crisp.org/E-CRISP/ | 87 |
| CRISPR MultiTargeter | http://www.multicrispr.net/basic_input.html | 88 |
| sgRNA Designer: CRISPRko | http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design | 10, 89 |
| Off-Spoter | https://cm.jefferson.edu/Off-Spotter/ | 90 |
| CCTop | http://crispr.cos.uni-heidelberg.de/index.html | 91 |
| CHOPCHOP | http://chopchop.cbu.uib.no/index.php | 92, 93 |
The activity of Cas9 nuclease is another prime factor causing undesirable off-target effects of the CRISPR/Cas9 system; specifically, elevated levels of Cas9 has been associated with unspecific cleavage.11 Therefore, controlling the activity of Cas9 nuclease would lead to a significant reduction of unwanted “side effects.” In this regard, several approaches that use chemical or physical agents to achieve conditional expression of Cas9 have been under investigation by researchers and are illustrated in Figure 2.
Figure 2.
Strategies for Regulating Cas9 Nuclease Activity
Increased levels of Cas9 can lead to unspecific cleavage, which causes hazardous effects in the target cell, resulting in off-target effects (red 5′ in Figure 1). Different strategies have been implemented to minimize and control the activity of Cas9. (A) Introduction of a Tet-controlled promoter that allows monitoring of Cas9 expression through an on/off system dependent on Tet/Dox. (B) Fusion of Cas9 with an estrogen receptor domain (ERT2) that enables the supervision of Cas9 activity through 4-HT presence/absence. (C) Control of the enzymatic activity via intein and its splicing properties. The N-terminal and C-terminal domains of Cas9, each containing a fused intein domain, are joined together by a splicing event, and, upon expression of gRNA, the intein is auto-excised, and gRNA forms with Cas9 an active complex. (D) Holding of Cas9 activity through fusion with light-responsive elements, which allows Cas9 performance only after stimulation with blue light. (E) A self-restricted CRISPR/Cas9 system that contains, in the engineered vector, a gRNA that targets the Cas9 gene itself, resulting in an auto-regulated loop. CRY2, cryptochrome circadian clock 2; VP64, viral protein 64 transactivation domain; CIBN, N-terminal domain of CIB1; P, promoter.
Turning the activity of Cas9 on and off confers to the researcher a means for a temporal control, limiting the exposure time of the genome to Cas9 nuclease, thereby reducing the off-target effects the CRISPR system exhibits under normal conditions. Placing Cas9 under the control of a tetracycline-responsive element (TRE) promoter confers the possibility to achieve a conditional expression in the presence of tetracycline/doxycycline (Tet/Dox).12, 13 However, even in the off state, when Tet/Dox are absent, Cas9 expression still exhibits “leakiness,” as recently reported.14 Placing the sgRNA under control of a hybrid Tet-responsive promoter proved to be a more advantageous alternative to obtain a tighter control of the off-target cleavage in a Dox-dependent manner.14 Regardless whether Cas9 or sgRNA expression is regulated in a Dox-dependent manner, such inducible systems require additional trans-acting factors and promoters to be incorporated in the construction of the delivery system, which, in the case of adeno-associated virus (AAV) vectors, is quite problematic because of their limited cloning capacity.15 Therefore, inducible CRISPR/Cas9 systems that use minimal genetic elements to achieve conditional expression of Cas9 might prove to be useful in designing delivery vectors for therapeutic applications.
In one study, researchers rendered Cas9 nuclease activity dependent on 4-hydroxytamoxifen (4-HT) by fusing a hormone-binding domain of the estrogen receptor (ERT2) to Cas9.16 This fusion ensures sequestration of Cas9 nuclease to the cytoplasm in the absence of 4-HT, consequently limiting the activity of the enzyme. A nuclear import is seen within the first 6 hr after induction with 4-HT, and optimal nuclease activity occurs at a time interval of 4–8 hr at 1 μM 4-HT; therefore, the off-target effects are reduced to a minimum. In a similar manner, Davis et al.17 used the self-splicing properties of inteins to render Cas9 nuclease active in the presence of 4-HT. In this report, the ERT2 domain is fused to intein to render it sensitive to 4-HT. Upon induction, intein becomes active and is effectively spliced from an inactive variant of Cas9, in which intein was inserted, leading to an active form of Cas9. However, this 4-HT-inducible system is not reversible compared with another system proposed by Liu et al.;16 therefore, it does not offer the advantage of an on/off switch for more adjustable control of Cas9 activity. It is well worth mentioning that simple split Cas9-intein systems have been developed in which an active form of Cas9 is reconstituted post-translationally from a Cas9-C domain and a Cas9-N domain after splicing of fused intein.18 These two inducible CRISPR/Cas9 systems use minimal genetic elements to achieve conditional expression of Cas9, which might prove their usefulness in designing delivery vectors for therapeutic applications.
Other models have used the propriety of light-inducible heterodimerization proteins for the modulation of Cas9 activity. These “light approaches” use a catalytically inactive form of Cas919 or a split Cas9 variant that is fused to light-responsive proteins.20 Upon stimulation with blue light, Cas9 becomes active either by transitioning from an inactive form19 or by reconstitution of the whole active Cas9 nuclease from the C domain and N domain of the enzyme.20 Although such systems are to some extent reversible and adjustable for regulating Cas9 activity, they are limited to in vitro applications, under which blue light is relatively easy to apply. For in vivo applications, stimulating Cas9 activity with blue light requires more invasive and dedicated equipment to achieve an efficient effect, a fact that might limit the use of such systems in clinical contexts.
A recent study elegantly used a self-restricted CRISPR/Cas9 system to achieve control of Cas9 nuclease to minimize off-target effects.21 In addition to a specific targeting sgRNA, the authors co-expressed an additional sgRNA that targets Cas9 itself. Using this approach, they were able to limit the expression of Cas9 to several days, even in the context of an integrative lentiviral vector. Furthermore, it has been shown that altering the energetic state of the Cas9-sgRNA-target RNA by substituting four positive amino acid residues to neutral ones in the DNA binding loop of Cas9 results in a “high-fidelity” Cas9 nuclease that displays no detectable undesired genomic alterations.22
As reviewed above, limiting undesired off-target effects is not an easy task, requiring additional genes and inducers, modifications to Cas9, or even invasive approaches to obtain spatiotemporal control of CRISPR/Cas9 so that this technology can move forward to therapeutic applications in a clinical context. In regard to “simple is beautiful,” we definitely need an easy and, at the same time, reliable system that will give us the possibility to engineer our “faulty” genomes to a healthy state and, furthermore, to an improved state of well-being.
Multiplexing CRISPR/Cas9 Editing Capabilities
Despite the fact that we currently might have a simple CRISPR/Cas9 system that is, to some extent, safe and efficient in targeting a specific genome site, there are still many other obstacles to overcome to successfully implement this strategy in clinical therapy. In some instances, targeting a single site in the genome is not sufficient to achieve a full therapeutic effect because some diseases, like cancer, have a multigenic basis and require targeting whole gene networks that sustain the pathological state of the cell. Therefore, such an approach would require the use of multiple sgRNAs able to target multiple genomic loci, adding an extra level of complexity for designing CRISPR/Cas9 systems.
Traditionally, this would mean that one must have at least two sgRNA expression cassettes on the same construct or placed on two different constructs. Co-transfection of two vectors limits the efficiency of the CRISPR/Cas9 system in vitro and, furthermore, in vivo. In addition, having multiple sgRNA expression cassettes on the same construct poses a limit of physical constrains in the number of sgRNA cassettes because of their limited cloning capacity, as seen for AAV vectors.15
Indeed, multiplexing CRISPR/Cas9 editing capabilities would offer several advantages over a conventional single sgRNA-based system for targeting multiple sites and gene networks. To date, researchers have developed two strategies for this approach. As mentioned above, one approach uses multiple sgRNA expression cassettes made of an individual RNA polymerase III promoter, sgRNA, and a transcription terminator, all imbedded in the same construct.23, 24 This means that the construct would express multiple sgRNA transcripts able to target multiple genomic loci. In the second approach, multiple sgRNAs are released from one single transcript, produced from either one RNA polymerase III promoter25 or one RNA polymerase II promoter.26 Such a polycistronic transcript offers the possibility of encoding additional exogenous factors to sgRNAs in the case of RNA polymerase II-driven promoters, although efficient processing of the transcript for releasing active sgRNAs and polyadenylated transcripts for nuclear export is rather complex in nature. First, sgRNA must be released from the primary transcript and, therefore, requires the addition of flanking sequences that are recognized by exogenous factors that must be provided in trans, such as Cys4 protein from Pseudomonas aeruginosa.26 Second, the remaining transcript must be properly processed as a translational active molecule. Additionally, a shortcoming of this system is that Csy4 has cytotoxic effects at high concentrations,26 a fact that might hinder the clinical scenario. Moreover, this number of supplemental sequences significantly hampers the efficiency of the targeting and editing capabilities of the CRISPR/Cas9 system because of the addition of extra variables.
Exploiting the endogenous RNA processing factors would be preferred, and simplistic approaches for multiplexing CRISPR/Cas9 have already been reported. In one study, the endogenous tRNA processing machinery was used for processing an sgRNA-tRNA polycistronic transcript to release multiple active sgRNAs.25 This approach is a powerful and reliable system for multiplexing CRISPR/Cas9 because it takes advantage of the abundance of endogenous cellular RNAases without heterologous expression of additional and potential cytotoxic factors that might interfere with normal cell function. This interference with normal cell function could also make the evaluation of results rather difficult and unreliable.
Having a system that can easily and efficiently target gene networks, multiplexing CRISPR/Cas9 offers the advantage of deleting genes or even inserting new genes in the genomes of choice. This provides a toolbox that opens new horizons that researchers could only dream of, with an ease never previously attained. The feasibility of CRISPR/Cas9 systems for knockin in reporter genes in different genomic sites has already been reported for different cell types, such as mouse haploid embryonic stem cells27 and human embryonic pluripotent stem cells.28 The authors used an HDR approach in which the reporter cassette was flanked by two homologous arms on the region of interest in the genome, and two sgRNAs were used for guiding Cas9 nuclease to the targeted sites. Upon cleavage, the reporter cassette is inserted in the genome by homologous recombination with an efficiency that varies depending on the targeted site.28 Using this approach, studies have shown that such a procedure permits the generation of mice harboring reporter genes up to 5 kb29 or a conditional allele30 with ease, surpassing traditional approaches in terms of simplicity, time length, and cost-effectiveness. The only requirement is the presence in the donor plasmid of two flanking homologous arms that may vary in length from 500 bp27 to 900 bp29 and two sgRNAs sequences able to direct Cas9 toward the targeted genomic fragment. In the absence of a donor DNA, large genomic deletions up to 65 kb can be easily generated,29 making CRISPR/Cas9 a preferred system for obtaining knockout mouse models for translational research.
CRISPR/Cas9 Libraries
Taking advantage of the CRISPR/Cas9 system’s ability to target almost any genetic loci within a target cell, researchers have created entire libraries of sgRNAs that are directed toward specific genes to examine the meaning of a “custom-made” phenotype in different experimental setups.31 By introducing into cells sgRNA libraries via delivery vectors that are most often lentiviral particles, there is the possibility to repress, activate, or even knock out different target genes at once to elucidate their function within a pathologic or homeostatic context. The extensiveness of the library is also a critical factor, where there is the possibility to target the whole genome with the use of a single, large “pooled” library of sgRNAs or to target specific subsets of genes that are thought to be interconnected, known as sub-pooled libraries.32, 33 Moreover, the permanent nature of the genetic modifications mediated by CRISPR/Cas9 technology permits the evaluation of the modified cells for extended periods of time, where there is the possibility that a certain phenotype will manifest only after several cell divisions.
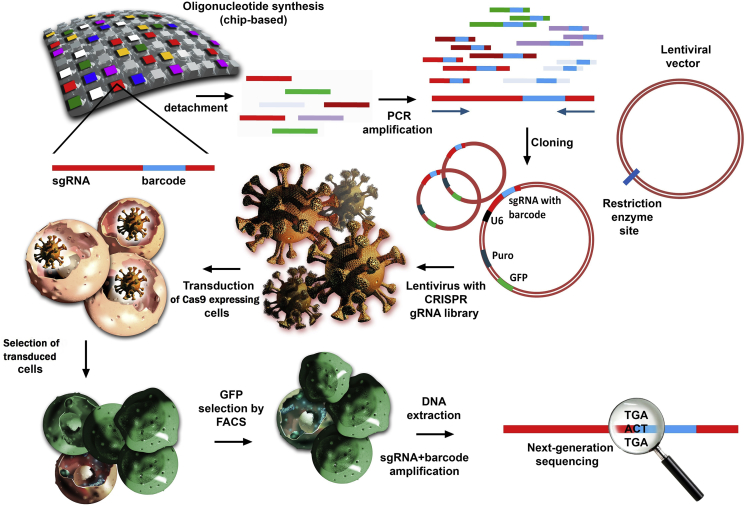
The first step when performing a screening investigation is viral administration of the selected sgRNAs in cells that consistently expresses Cas9 enzyme or in combination with the Cas9-encoded sequence under the construct. After selection of the transfected cells, further selective agents can be added that will enrich the resistant phenotype population. The final step consists of sequencing arrays that can reveal the difference between the experimental and control groups in terms of enriched sgRNAs and depleted ones, which can be seen in Figure 3. When these data are overlapped with the original sgRNA library and screened for the target genes, statistical analysis can reveal the key genes within the phenotype of concern.31, 34.
Figure 3.
Generation and Quality Control of sgRNA Libraries for CRISPR/Cas9-Mediated Phenotypic Screening
The first step illustrates the synthesis of sgRNA libraries consisting of a heterogeneous population of sequences approximately 100 bases in length with a reduced number of mutations. Quality control is significantly facilitated when a barcode of a known sequence is incorporated into each sgRNA and analyzed at the final stages through next-generation sequencing (NGS). After detachment of oligonucleotides from the solid support, the targeted sequences are amplified through PCR and then cloned in lentiviral vectors that contain a selection marker for antibiotic resistance (e.g., puromycin) and also an enrichment sequence that can be detected with the help of fluorescence-activated cell sorting (FACS) (e.g., GFP). Every sgRNA is under the activity of an U6 promoter that will facilitate its expression. The entire complex is packed in viruses that are further used for the transduction of the target cells. The first selection consists of the capacity of cells to survive in an antibiotic-enriched medium because of their integrated gene for puromycin or any other antibiotic that is used in the experiment. The second selection consists of the enrichment of the transduced cell population through GFP selection by FACS. The remaining cells that contain the viral construct are then collected for DNA extraction and analyzed through NGS, which detects individual sgRNAs because of the inserted barcode.
Shalem et al.35 and Wang et al.36 pioneered the genome-wide screening of cells using CRISPR/Cas9 libraries and presented the advantages of this method in contrast to small interfering RNA. Although their approaches were quite different, where Shalem et al.35 transfected cells with a vector containing both sgRNAs and a Cas9-expressing sequence, and Wang et al.36 administered viral particles containing only sgRNAs molecules in cells that already stably expressed Cas9 enzyme, the results proved encouraging for both groups. Thus, even though this approach for the identification of entire genes set functions in living cells is still in its infancy, CRISPR/Cas9 libraries hold significant advantages over the preceding technologies (e.g., small interfering RNA [siRNA libraries]), allowing the generation of a permanent “mutated” phenotype that can be studied across several cell division steps under different experimental conditions.
The Big Step of CRISPR/Cas9 in Cancer Research
Cancer is one of the most investigated fields in medicine because of its high prevalence in the human population, the complexity and heterogeneity of the disease, which makes current treatments rather unspecific, raising the death toll of the affected individuals.37, 38, 39 Understanding the molecular mechanisms that underline the proliferative process is a prerequisite for the development of reliable experimental models in which the altered cellular pathways can be reproduced and novel targeted therapies can be implemented. Hence, it did not take long until the genome editing capabilities of CRISPR/Cas9 proved its utility in cancer research and development of therapeutic strategies. Such applicability ranges from functional validation of genes implicated in cancer development and progression to cancer modeling and therapeutic concepts, as described in Table 3. Therefore, CRISPR/Cas9 has spread to every aspect of cancer research in a very short period of time, making it a versatile technology that could leave an important footprint with regard to redefining the meaning of cancer biology and its treatment. From small-scale studies40, 41, 42 to high-throughput screens of gene function,43, 44, 45 CRISPR/Cas9 has brought new insights into cancer progression and metastasis and led to the identification of new potential therapeutic targets. Cancer modeling has an opportunity with CRISPR/Cas9 for both in vitro and in vivo models because the traditional Cre/LoxP recombination technology for animal models has shortcomings in terms of labor, time length, and costs for producing stable in vivo lines of cancer models. In addition, this might bypass tumor xenograft models, where the risk of immune incompatibility can interfere with the production of reliable in vivo models. The use of immunocompromised animals requires special conditions for maintenance of the lines, and lack of an immune response might interfere with the results obtained in preclinical studies, a crucial step for evaluating the safety profile of novel therapeutic agents.
Table 3.
List of CRISPR/Cas9 Applications in Cancer Research Organized by In Vitro and In Vivo Studies of Gene Function, Cancer Modeling, and Therapy
| Cell Line/Organism | Delivery System | Cancer Type | Targeted Gene | Local/Systemic | Reference |
|---|---|---|---|---|---|
| Gene Function Studies In Vitro | |||||
| A375 melanoma cell line (human) | LV | melanoma | SAM library | 45 | |
| A375 melanoma cell line (human) | LV | melanoma | GeCKO library | 43 | |
| Non-small-cell lung cancer cell line | LV | non-small-cell lung cancer | GeCKO library | 44 | |
| MDA-MB-231, MCF-7 (human) | LV | triple-negative breast cancer | Shcbp1 | 42 | |
| T24, J82, 5637, SW-780 bladder cancer cell lines (human) | plasmid transfection | bladder cancer | p21, E-cadherin, hBax | 63 | |
| HEK293T (human) | plasmid transfection | non-small-cell lung cancer | Cd74-Ros1 | 64 | |
| Eml4-Alk | |||||
| Ki5b-Ret | |||||
| HEK293 (human) | plasmid transfection | non-small-cell lung cancer | Met | 65 | |
| Dld-1 (human) | plasmid transfection | colon cancer | Pkc | 66 | |
| B16-F10, Ret melanoma cell lines (mouse) | plasmid transfection | melanoma | Id1, Id3 | 67 | |
| BT-474, SKBR-3, MCF-7 (human) | RV/plasmid transfection | breast cancer | Her2 | 68 | |
| In Vivo | |||||
| Mouse | LV | prostate cancer | TGFBRII | ex vivo | 69 |
| DU145 cells (human) | |||||
| Mouse | LV | prostate cancer | Nanog1 | ex vivo | 40 |
| Nanoggp8 | DU145 cells (human) | ||||
| Mouse | LV | triple-negative breast cancer | Cripto-1 | ex vivo | 41 |
| JygMC(A) (mouse) | |||||
| Mouse | LV | triple-negative breast cancer | Ctbp1 | ex vivo | 70 |
| MDA-Mb-231 (human) | |||||
| Mouse | plasmid transfection | cervical cancer | HPV E6, E7 | ex vivo | 71 |
| SiHa, C33-A (human) | |||||
| Mouse | LV | Burkitt lymphoma | Mcl-1, TP53 | ex vivo | 72 |
| HSC (mouse) | |||||
| Mouse |
plasmid transduction |
acute myeloid leukemia |
Mll3 |
ex vivo |
73 |
| HSC (mouse) | |||||
| Cancer Modeling In Vitro | |||||
| Myoblast cells (mouse) | LV | alveolar rhabdomyosarcoma | Pax3, Foxo1 | 74 | |
| HEK293A, hMSCs, PBMCs, HL-60 (human) | plasmid transduction/EP | Ewing sarcoma, acute myeloid leukemia | Ewsr1, Fli1, Runx1, Eto | 75 | |
| In Vivo | |||||
| Mouse | LV | non-small-cell lung cancer | Eml4-Alk | local | 76 |
| Mouse | LV | non-small-cell lung cancer | Nkx2.1, Pten, Apc | local | 77 |
| Mouse | LV/Ad | pancreatic ductal adenocarcinoma | Lkb1 | local | 78 |
| Mouse | Ad | non-small-cell lung cancer | Eml4-Alk | local | 79 |
| Mouse | AAV | lung adenocarcinoma | TP53, Lkb1, Kras | local | 80 |
| Mouse | HI | hepatocellular carcinoma | multiple | systemic | 81 |
| intrahepatic cholangiocarcinoma | |||||
| Mouse | HI | hepatocellular carcinoma | Pten, TP53 | systemic | 82 |
| Mouse | EP | glioblastoma | Pten, Apc, Nf1 | local | 83 |
| Mouse | PEI/EP | medulloblastoma | Ptch1 | local | 83 |
| Mouse | EP | pancreatic cancer | multiple | local | 84 |
| Mouse | LV | acute myeloid leukemia | Tet2, Dnmt3a, Nf1, Ezh2 | ex vivo | 85 |
| HSCs (mouse) | |||||
| Mouse |
LV |
colorectal cancer |
Apc, TP53, Kras, Smad4, PIK3CA |
ex vivo |
86 |
| intestinal stem cells (human) | |||||
| Therapeutic Concepts In Vitro | |||||
| Osteosarcoma cell lines (KHOS), u-20 s (human) | EP | osteosarcoma | Cdk11 | 46 | |
| Primary T cells (human) | EP | tumor immunotherapy | Cxcr4, PD-1 | 47 | |
| Primary T cells (human) | EP | tumor immunotherapy | PD-1 | 48 | |
| In Vivo | |||||
| Mouse | LV | pancreatic ductal adenocarcinoma | p57 | local | 49 |
| Mouse | LV/EP | prostate cancer | TCR, B2M, PD-1 | ex vivo | 50 |
| leukemia | CAR-T cells (human) | ||||
RV, retrovirus; Ad, adenovirus; HI, hydrodynamic injection; EP, electroporation; PEI, polyethylenimine; HSC, hematopoietic stem cell; hMSC, human mesenchymal stem cells; PBMC, peripheral blood mononuclear cells; CAR, chimeric antigen receptor
A few studies published recently on in vitro46, 47, 48 and in vivo49, 50 cancer models have paved the way for CRISPR/Cas9 in therapeutic applications for the oncology niche. In this regard, cancer immunotherapy gained special attention by reprograming T cells through disruption of the programmed death-1 receptor (PD-1) over conventional PD-1 antibody therapy.48 Because the PD-1 ligand is a negative regulator of T cell activity and expressed on dendritic cells as well as in some tumors, knocking down PD-1 with CRISPR/Cas9 could reprogram T cell activity toward PD-1 ligand-expressing tumors. This approach is already in phase I clinical trials for castration-resistant prostate cancer, muscle-invasive bladder cancer, metastatic non-small-cell lung cancer, and metastatic renal cell carcinoma (https://clinicaltrials.gov).
Challenges and Future Perspectives of CRISPR/Cas9 for Cancer Therapy
The simplicity that resides in the genome editing capabilities of CRISPR/Cas9 attracted the attention of many researchers around the world, as confirmed by the overwhelming numbers of studies that have been published on the subject for the past 2 years. This newly characterized RNA-guided Cas9 nuclease system opened the era of “molecular surgery,” and, as reviewed above, cancer research has and will continue to benefit from this new genome editing technology. In particular, modeling oncogenesis in mice using CRISPR/Cas9 surpassed previous genome editing tools in terms of time length and cost effectiveness without the need of multiplying colonies of mice.
Functional studies of multiple genes in cancer cell lines with CRISPR/Cas9 (Table 3), especially large-scale studies with sgRNAs libraries (Table 3),43, 44, 45 again stressed the complexity of this insidious killer that makes cancer particularly difficult to treat. The multigenic and multi-mutated status gives cancer an unique heterogeneity characteristic; this heterogeneity is even seen in different individuals affected by the same cancer type,51, 52 and this truly represents the major challenge for clinical therapy. Although surgery and chemotherapy/radiation therapy are currently in clinical use for the treatment and palliative care of cancer patients, the lifespan, because of tumor relapse, is quite short, and the quality of life because of adverse effects of therapy is rather poor. All of this gave researchers the impulse to find alternative treatments that are efficient for targeting the diseased cells within our body, and gene therapy emerged as a vison for treatment of human diseases that would be safe and specific.
Most of the biotherapies are based on oncolytic vectors that selectively replicate and destroy cancer cells, releasing tumor-specific antigens, resulting in a second immune-humoral response against distant metastatic tumors.53 However, these therapies are limited to local administration, a fact that imposes several limitations, such as tumor localization, that require invasive approaches that are not feasible in certain cancer types. For the time being, CRISPR/Cas9 has been investigated as a potential therapeutic strategy in ex vivo preclinical setups for cancer immunotherapy in both hematological and solid tumors by targeting the PD-1 gene in T cells (Table 3), reprograming these cells of the immune system to recognize and attack cancer cells. To date, there are four clinical trials under investigation based on the strategy of targeting PD-1 by CRISPR/Cas9 in four different carcinomas (https://clinicaltrials.gov). Both the preclinical and clinical studies are using lentiviruses (LVs) as delivery systems in T cells of the CRISPR/Cas9 components. For ex vivo therapeutic setups, transduction of primary T cells from cancer patients, characterization, and expansion in the laboratory of the transduced cells are quite labor-intensive and expensive. Although lentiviruses have been reported to be efficient for tumor regression on in vivo models, with CRISPR/Cas949 or other experimental setups, like utilizing antagomirs,54 a valuable lesson has been learned in terms of the tumorigenic potential of integrating vectors in clinical trials.55
Having a technology that gives us the possibility to knock out or knock in single or multiple genes with an ease that surpasses other genome editing tools is clearly a major achievement. Furthermore, researchers have engineered Cas9 nuclease into an RNA-guided transcriptional activator by fusing the VP64 transcription activation domain to a catalytically inactive Cas9 and linking a MS2-p65-HSF1 activation complex to sgRNA via a hairpin aptamer.45 Rendering sgRNA from 20 bp to 11–15 bp, an RNA-guided transcriptional activation complex, Cas9-MS2-p65-HSF1, was obtained with an active Cas9.56 This system offers the possibility of simultaneous orthogonal activation of gene expression with gene knockout by “classical” Cas9-sgRNA system in a single cell population. The specificity of such an approach can be further enhanced by substituting the Streptococcus pyogenes Cas9 (spCas9) with the Streptococcus aureus Cas9 (saCas9); this allows for recognition of the PAM of the consensus sequence NNGRR(T), which is more complex than the NGG spCas9 PAM.57 SaCas9 also has a significantly smaller size than spCas9, a particularity that might be useful when physical constrains are imposed by the type of vector that is used as the delivery vehicle. However, such a complex PAM reduces the number of genomic loci that can be targeted.
Despite the numerous efforts that have been made to improve the specificity and efficiency of CRISPR/Cas9, a major hurdle experienced with other gene therapy approaches lies before us. Delivery into the body and targeting the diseased cells without side effects is not something new, and each type of delivery vector has its own advantages and limitations.58 Therefore, a safe and efficient delivery vector for systemic administration of CRISPR/Cas9 would be highly desirable. An emerging class of gene delivery vectors is derived from an M13 filamentous bacteriophage in which a tumor-targeting ligand is expressed on the bacteriophage capsid.59, 60 Because bacteriophages do not have natural receptors on human cells, attaching a ligand like Arg-Gly-Asp (RGD) confers an improved specificity for tumors over conventional vectors derived from human viruses, which need complex engineering of the capsid to limit the tropism for their cognate receptors. In addition, bacteriophage-derived vectors could bypass the immune response, commonly experienced with adenoviral vectors, in which a pre-existing immunity against adenoviral strains can result in severe life-threatening allergic side effects, as previously reported in the classic case of the Jesse Gelsinger gene therapy trial.61 Furthermore, M13 bacteriophage-derived vectors can accommodate into their genome exogenous genetic material up to 13 kb without affecting their 3D structure, an important aspect to consider when designing an all-in-one vector; the physical constraint experienced with AAV delivery vectors limits the cloning capacity to 4 kb.15 Differential microRNA expression patterns between the tumor and normal tissue62 can be exploited to restrict Cas9 activity to cancer cells where a specific miRNA is downregulated by the inclusion of miRNA targeting sequences in the 3′ UTR of Cas9, further increasing the specificity of the bacteriophage-derived vector for cancer cells. Likewise, using a tumor-specific promoter to drive Cas9 expression can lead to an increase of CRISPR/Cas9 specificity for cancer cells.60
Is the bacteriophage-derived vector the vehicle that will move CRISPR/Cas9 to clinical applications in a simple and efficient manner without the potential genotoxic effects of integrating vectors? Perhaps, but we cannot envision such a scenario at this time because more preclinical studies must be implemented to test its clinical significance and evaluate the pharmacokinetic properties of phage-derived nanoparticles. However, we must keep in mind that the multigenic and heterogeneous nature of cancer is definitively the major challenge for an efficient therapy. Even with a systemic delivery vector that can efficiently deliver CRISPR/Cas9 to cancer cells without side effects, there is no guarantee that a full therapeutic effect will be achieved. A thoughtful understanding of the proliferative process and connections between different cellular pathways in cancer cells is mandatory for developing efficient therapeutics, especially considering that each type of cancer has its own genomic and phenotypic profiles. Whether CRISPR/Cas9 technology will make a difference in cancer therapeutics is hard to tell, but it is definitely worth the effort. And the knowledge that will arise from this process of “trial and error” will pave the way for CRISPR/Cas9 in our daily medical practice and, if not for cancer, then perhaps for other diseases that are threatening our own existence and well-being.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Mihail Buse for English proofreading of the manuscript. This work was supported by national grants Nr.128,014-PN-II-PT-PCCA-2013-4-2166 (GenCanD) and POC Nr.35/01.09.2016, ID 37_796 (CANTEMIR).
Contributor Information
Sergiu Chira, Email: sergiu.chira@umfcluj.ro.
Pierre Cordelier, Email: pierre.cordelier@inserm.fr.
References
- 1.Gupta R.M., Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Invest. 2014;124:4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan F.A., Pandupuspitasari N.S., Chun-Jie H., Ao Z., Jamal M., Zohaib A., Khan F.A., Hakim M.R., ShuJun Z. CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases. Oncotarget. 2016;7:52541–52552. doi: 10.18632/oncotarget.9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sontheimer E.J., Barrangou R. The Bacterial Origins of the CRISPR Genome-Editing Revolution. Hum. Gene Ther. 2015;26:413–424. doi: 10.1089/hum.2015.091. [DOI] [PubMed] [Google Scholar]
- 6.Sánchez-Rivera F.J., Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H., Xiao T., Chen C.H., Li W., Meyer C.A., Wu Q., Wu D., Cong L., Zhang F., Liu J.S. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dow L.E., Fisher J., O’Rourke K.P., Muley A., Kastenhuber E.R., Livshits G., Tschaharganeh D.F., Socci N.D., Lowe S.W. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J., Wu L., Zhang S.M., Lu M., Cheung W.K., Cai W., Gale M., Xu Q., Yan Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44:e149. doi: 10.1093/nar/gkw660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Solis C.A., Ho A., Holehonnur R., Ploski J.E. The Development of a Viral Mediated CRISPR/Cas9 System with Doxycycline Dependent gRNA Expression for Inducible In vitro and In vivo Genome Editing. Front. Mol. Neurosci. 2016;9:70. doi: 10.3389/fnmol.2016.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh A., Yue Y., Lai Y., Duan D. A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol. Ther. 2008;16:124–130. doi: 10.1038/sj.mt.6300322. [DOI] [PubMed] [Google Scholar]
- 16.Liu K.I., Ramli M.N., Woo C.W., Wang Y., Zhao T., Zhang X., Yim G.R., Chong B.Y., Gowher A., Chua M.Z. A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat. Chem. Biol. 2016;12:980–987. doi: 10.1038/nchembio.2179. [DOI] [PubMed] [Google Scholar]
- 17.Davis K.M., Pattanayak V., Thompson D.B., Zuris J.A., Liu D.R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truong D.J., Kühner K., Kühn R., Werfel S., Engelhardt S., Wurst W., Ortiz O. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polstein L.R., Gersbach C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nihongaki Y., Kawano F., Nakajima T., Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y., Liu X., Zhang Y., Wang H., Ying H., Liu M. A Self-restricted CRISPR System to Reduce Off-target Effects. Mol. Ther. 2016;24:1508–1510. doi: 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabadi A.M., Ousterout D.G., Hilton I.B., Gersbach C.A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakuma T., Nishikawa A., Kume S., Chayama K., Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie K., Minkenberg B., Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura Y., Oda M., Nakatani T., Sekita Y., Monfort A., Wutz A., Mochizuki H., Nakano T. CRISPR/Cas9-mediated reporter knock-in in mouse haploid embryonic stem cells. Sci. Rep. 2015;5:10710. doi: 10.1038/srep10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkle F.T., Neuhausser W.M., Santos D., Valen E., Gagnon J.A., Maas K., Sandoe J., Schier A.F., Eggan K. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11:875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., Jia R., Palange N.J., Satheka A.C., Togo J., An Y., Humphrey M., Ban L., Ji Y., Jin H. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS ONE. 2015;10:e0120396. doi: 10.1371/journal.pone.0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrotis A., Ketteler R. A new age in functional genomics using CRISPR/Cas9 in arrayed library screening. Front. Genet. 2015;6:300. doi: 10.3389/fgene.2015.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassett A.R., Kong L., Liu J.L. A genome-wide CRISPR library for high-throughput genetic screening in Drosophila cells. J. Genet. Genomics. 2015;42:301–309. doi: 10.1016/j.jgg.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heigwer F., Zhan T., Breinig M., Winter J., Brügemann D., Leible S., Boutros M. CRISPR library designer (CLD): software for multispecies design of single guide RNA libraries. Genome Biol. 2016;17:55. doi: 10.1186/s13059-016-0915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 35.Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braicu C., Catana C., Calin G.A., Berindan-Neagoe I. NCRNA combined therapy as future treatment option for cancer. Curr. Pharm. Des. 2014;20:6565–6574. doi: 10.2174/1381612820666140826153529. [DOI] [PubMed] [Google Scholar]
- 38.Braicu C., Chiorean R., Irimie A., Chira S., Tomuleasa C., Neagoe E., Paradiso A., Achimas-Cadariu P., Lazar V., Berindan-Neagoe I. Novel insight into triple-negative breast cancers, the emerging role of angiogenesis, and antiangiogenic therapy. Expert Rev. Mol. Med. 2016;18:e18. doi: 10.1017/erm.2016.17. [DOI] [PubMed] [Google Scholar]
- 39.Berindan-Neagoe I., Calin G.A. Molecular pathways: microRNAs, cancer cells, and microenvironment. Clin. Cancer Res. 2014;20:6247–6253. doi: 10.1158/1078-0432.CCR-13-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawamura N., Nimura K., Nagano H., Yamaguchi S., Nonomura N., Kaneda Y. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015;6:22361–22374. doi: 10.18632/oncotarget.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castro N.P., Fedorova-Abrams N.D., Merchant A.S., Rangel M.C., Nagaoka T., Karasawa H., Klauzinska M., Hewitt S.M., Biswas K., Sharan S.K., Salomon D.S. Cripto-1 as a novel therapeutic target for triple negative breast cancer. Oncotarget. 2015;6:11910–11929. doi: 10.18632/oncotarget.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng W., Li H.C., Xu K., Chen Y.F., Pan L.Y., Mei Y., Cai H., Jiang Y.M., Chen T., Feng D.X. SHCBP1 is over-expressed in breast cancer and is important in the proliferation and apoptosis of the human malignant breast cancer cell line. Gene. 2016;587:91–97. doi: 10.1016/j.gene.2016.04.046. [DOI] [PubMed] [Google Scholar]
- 43.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S., Sanjana N.E., Zheng K., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y., Sassi S., Shen J.K., Yang X., Gao Y., Osaka E. Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system. J. Orthop. Res. 2015;33:199–207. doi: 10.1002/jor.22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su S., Hu B., Shao J., Shen B., Du J., Du Y., Zhou J., Yu L., Zhang L., Chen F. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016;6:20070. doi: 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazur P.K., Herner A., Mello S.S., Wirth M., Hausmann S., Sánchez-Rivera F.J., Lofgren S.M., Kuschma T., Hahn S.A., Vangala D. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 2015;21:1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-1300. Published online November 4, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J., Fujimoto J., Zhang J., Wedge D.C., Song X., Zhang J., Seth S., Chow C.W., Cao Y., Gumbs C. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Bruin E.C., McGranahan N., Swanton C. Analysis of intratumor heterogeneity unravels lung cancer evolution. Mol. Cell. Oncol. 2015;2:e985549. doi: 10.4161/23723556.2014.985549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heo J., Reid T., Ruo L., Breitbach C.J., Rose S., Bloomston M., Cho M., Lim H.Y., Chung H.C., Kim C.W. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat. Med. 2013;19:329–336. doi: 10.1038/nm.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sicard F., Gayral M., Lulka H., Buscail L., Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol. Ther. 2013;21:986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 56.Dahlman J.E., Abudayyeh O.O., Joung J., Gootenberg J.S., Zhang F., Konermann S. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishimasu H., Cong L., Yan W.X., Ran F.A., Zetsche B., Li Y., Kurabayashi A., Ishitani R., Zhang F., Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chira S., Jackson C.S., Oprea I., Ozturk F., Pepper M.S., Diaconu I., Braicu C., Raduly L.Z., Calin G.A., Berindan-Neagoe I. Progresses towards safe and efficient gene therapy vectors. Oncotarget. 2015;6:30675–30703. doi: 10.18632/oncotarget.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hajitou A., Trepel M., Lilley C.E., Soghomonyan S., Alauddin M.M., Marini F.C., 3rd, Restel B.H., Ozawa M.G., Moya C.A., Rangel R. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 60.Kia A., Przystal J.M., Nianiaris N., Mazarakis N.D., Mintz P.J., Hajitou A. Dual systemic tumor targeting with ligand-directed phage and Grp78 promoter induces tumor regression. Mol. Cancer Ther. 2012;11:2566–2577. doi: 10.1158/1535-7163.MCT-12-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raper S.E., Chirmule N., Lee F.S., Wivel N.A., Bagg A., Gao G.P., Wilson J.M., Batshaw M.L. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 62.Berindan-Neagoe I., Monroig Pdel.C., Pasculli B., Calin G.A. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y., Zeng Y., Liu L., Zhuang C., Fu X., Huang W., Cai Z. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun. 2014;5:5393. doi: 10.1038/ncomms6393. [DOI] [PubMed] [Google Scholar]
- 64.Choi P.S., Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Togashi Y., Mizuuchi H., Tomida S., Terashima M., Hayashi H., Nishio K., Mitsudomi T. MET gene exon 14 deletion created using the CRISPR/Cas9 system enhances cellular growth and sensitivity to a MET inhibitor. Lung Cancer. 2015;90:590–597. doi: 10.1016/j.lungcan.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 66.Antal C.E., Hudson A.M., Kang E., Zanca C., Wirth C., Stephenson N.L., Trotter E.W., Gallegos L.L., Miller C.J., Furnari F.B. Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell. 2015;160:489–502. doi: 10.1016/j.cell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krachulec J.M., Sedlmeier G., Thiele W., Sleeman J.P. Footprintless disruption of prosurvival genes in aneuploid cancer cells using CRISPR/Cas9 technology. Biochem, Cell Biol. 2016;94:289–296. doi: 10.1139/bcb-2015-0150. [DOI] [PubMed] [Google Scholar]
- 68.Wang H., Sun W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett. 2017;385:137–143. doi: 10.1016/j.canlet.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 69.Huang G., Osmulski P.A., Bouamar H., Mahalingam D., Lin C.L., Liss M.A., Kumar A.P., Chen C.L., Thompson I.M., Sun L.Z. TGF-β signal rewiring sustains epithelial-mesenchymal transition of circulating tumor cells in prostate cancer xenograft hosts. Oncotarget. 2016;7:77124–77137. doi: 10.18632/oncotarget.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raza U., Saatci Ö., Uhlmann S., Ansari S.A., Eyüpoğlu E., Yurdusev E., Mutlu M., Ersan P.G., Altundağ M.K., Zhang J.D. The miR-644a/CTBP1/p53 axis suppresses drug resistance by simultaneous inhibition of cell survival and epithelial-mesenchymal transition in breast cancer. Oncotarget. 2016;7:49859–49877. doi: 10.18632/oncotarget.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 72.Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O’Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 73.Chen C., Liu Y., Rappaport A.R., Kitzing T., Schultz N., Zhao Z., Shroff A.S., Dickins R.A., Vakoc C.R., Bradner J.E. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lagutina I.V., Valentine V., Picchione F., Harwood F., Valentine M.B., Villarejo-Balcells B., Carvajal J.J., Grosveld G.C. Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR-Cas9 nuclease. PLoS Genet. 2015;11:e1004951. doi: 10.1371/journal.pgen.1004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torres R., Martin M.C., Garcia A., Cigudosa J.C., Ramirez J.C., Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat. Commun. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- 76.Blasco R.B., Karaca E., Ambrogio C., Cheong T.C., Karayol E., Minero V.G., Voena C., Chiarle R. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 77.Sánchez-Rivera F.J., Papagiannakopoulos T., Romero R., Tammela T., Bauer M.R., Bhutkar A., Joshi N.S., Subbaraj L., Bronson R.T., Xue W., Jacks T. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiou S.H., Winters I.P., Wang J., Naranjo S., Dudgeon C., Tamburini F.B., Brady J.J., Yang D., Grüner B.M., Chuang C.H. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–1585. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maddalo D., Manchado E., Concepcion C.P., Bonetti C., Vidigal J.A., Han Y.C., Ogrodowski P., Crippa A., Rekhtman N., de Stanchina E. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weber J., Öllinger R., Friedrich M., Ehmer U., Barenboim M., Steiger K., Heid I., Mueller S., Maresch R., Engleitner T. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc. Natl. Acad. Sci. USA. 2015;112:13982–13987. doi: 10.1073/pnas.1512392112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N.S., Cai W., Yang G., Bronson R., Crowley D.G. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuckermann M., Hovestadt V., Knobbe-Thomsen C.B., Zapatka M., Northcott P.A., Schramm K., Belic J., Jones D.T., Tschida B., Moriarity B. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat. Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maresch R., Mueller S., Veltkamp C., Öllinger R., Friedrich M., Heid I., Steiger K., Weber J., Engleitner T., Barenboim M. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 2016;7:10770. doi: 10.1038/ncomms10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 87.Heigwer F., Kerr G., Boutros M. E-CRISP: fast CRISPR target site identification. Nat. Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 88.Prykhozhij S.V., Rajan V., Gaston D., Berman J.N. CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS ONE. 2015;10:e0119372. doi: 10.1371/journal.pone.0119372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pliatsika V., Rigoutsos I. “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biol. Direct. 2015;10:4. doi: 10.1186/s13062-015-0035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]