Abstract
The success of lentiviral vectors in curing fatal genetic and acquired diseases has opened a new era in human gene therapy. However, variability in the efficacy and safety of this therapeutic approach has been reported in human patients. Consequently, lentiviral-vector-based gene therapy is limited to incurable human diseases, with little understanding of the underlying causes of adverse effects and poor efficacy. To assess the role that host genetic variation has on efficacy of gene therapy, we characterized lentiviral-vector gene therapy within a set of 12 collaborative cross mouse strains. Lentiviral vectors carrying the firefly luciferase cDNA under the control of a liver-specific promoter were administered to female mice, with total-body and hepatic luciferase expression periodically monitored through 41 weeks post-vector administration. Vector copy number per diploid genome in mouse liver and spleen was determined at the end of this study. We identified major strain-specific contributions to overall success of transduction, vector biodistribution, maximum luciferase expression, and the kinetics of luciferase expression throughout the study. Our results highlight the importance of genetic variation on gene-therapeutic efficacy; provide new models with which to more rigorously assess gene therapy approaches; and suggest that redesigning preclinical studies of gene-therapy methodologies might be appropriate.
Keywords: gene therapy, lentivirus, mouse, collaborative cross, heritability
Introduction
Similar to other viral vectors, lentiviral vectors are designed to support a single round of transduction process1 comprising multiple steps, which are dependent on unique host factors and can also be restricted by retroviral-specific restriction factors as well as the host innate and adoptive immune responses.2, 3, 4, 5, 6, 7, 8 The process of a single-round vector transduction includes vector attachment to the relevant envelope receptor on the host cell membrane, uncoating,9, 10, 11, 12, 13 reverse transcription, nuclear import,14, 15, 16 integration,15, 17, 18 and transgene expression.19 Notwithstanding the central role of the host factors in the process of viral infection, current preclinical studies, which determine the efficacy and safety of viral vectors are usually based on rodent studies comprising a large cohort of genetically identical animals, followed by a large animal (e.g., primate) study using a relatively small number of outbred animals. Consequently, several clinical trials using various viral vectors resulted in major adverse effects that could not be predicted by their cognate preclinical studies.20, 21, 22, 23, 24 Furthermore, the limited numbers of individuals who are eligible for gene therapy clinical trials and the fact that these individuals already have significant health issues limit our ability to meaningfully use their outcome to determine what were the host genetic variants that put these individuals at higher risk for adverse effects or rendered them likely to benefit from gene therapy protocols. Advancements in the lentiviral vector system were followed by successful human clinical trials on either gene replacement or immunotherapy for either fatal monogenic diseases or cancer, respectively.20, 25, 26, 27, 28, 29, 30 Notwithstanding the overall successes of the above clinical trials, significant variation was observed in the efficacy and safety both between the trials as well as within patients in a specific trial. Notably, among all HIV-1 vector-treated patients, only one patient who received hematopoietic-stem-cell-directed gene replacement therapy of β-thalassemia demonstrated vector-induced insertional mutagenesis.20 The variability in the efficacy and safety of HIV-based vector clinical trials was in line with earlier preclinical studies, which demonstrated significant strain-specific differences in HIV-1 vector efficacy and safety.31, 32 However, the low numbers of animal and inbred strains employed in these studies limited the ability to assess the impact of specific genetic differences on these phenomena. Indeed, given the growing body of evidence that host genetic variants can impact antiviral responses33, 34, 35 as well as overall gene regulation within individuals,36, 37, 38 being able to accurately assess and quantify genetic variants effects on gene therapy approaches is critical for the advancement of this field.
Relevant for studies of in vivo mammalian responses, the collaborative cross (CC) panel of recombinant inbred mouse strains exists.39 This genetic reference panel of >70 inbred mouse strains were derived from a set of eight founder strains, including five classical laboratory mouse strains, and three wild-derived strains. Together, these founders represent all three Mus musculus subspecies and contain over 40 million SNPs and four million insertions and deletions segregating at high minor allele frequencies across the collaborative cross. Furthermore, across a growing number of studies, variants within the collaborative cross and related diversity outbred (DO) have been shown to impact a variety of antiviral40, 41, 42, 43, 44 and gene-regulatory processes.36, 45 Specifically, a number of these studies have identified host genetic variants impacting innate antiviral responses40, 42 and also aberrant adaptive immune responses to pathogens.41 Given the reliance of gene therapeutic efficacy assessments on one or two classic inbred strains (e.g., C57BL/6 or BALB/c), diverse populations such as the collaborative cross provide a useful tool with which to assess the impact of host genetic variation on the potential efficacy of vector-based gene therapeutic approaches and potential off-target effects and ultimately to identify polymorphic genome features driving these responses. In order to assess the impact of host genetic variation on the efficacy as well as safety of a lentivirus-based gene therapeutic regimen, we transduced 12 collaborative cross strains as well as C57BL/6J mice with a liver-targeting lentivirus vector expressing luciferase. We found evidence of strong effects of host genetic variants on the ability of the lentiviral vectors to successfully ferry genetic cargos to host cell nuclei and maintain hepatic transgene expression. This study highlights the critical need to assess the safety and efficacy of gene therapeutic approaches across a range of genetically variable backgrounds.
Results
Host Genetic Variation Controls Both Levels and Kinetics of Luciferase Expression
To evaluate the effects of host genetic variation on lentiviral-vector-based gene delivery in vivo, we administered lentiviral vectors via intraperitoneal injection to 64 female mice from 12 collaborative cross strains as well as to the prototypical model C57BL/6J (n = 4 to 5/strain). These collaborative cross strains were chosen based on the availability of sufficient sized cohorts within our age window at the time of this study and were also selected to sufficiently and unbiasedly sample the genetic variation present within the entire collaborative cross population (e.g., these strains did not represent a genetically biased subset of strains). One mouse died during injection, and two other mice were excluded from the study because of post hoc observation of failure to deliver the vector to these animals. The vectors were VSV-G pseudotyped and expressed firefly luciferase under the control of a liver-specific promoter (human alpha1-antitrypsin [hAAT]). Luciferase expression in liver and whole mouse body was quantified by in vivo imaging at weeks 1, 3, 7, 15, and 41 (Figures 1 and S1). At 1 week post-infection, we identified an almost 2-log difference between mouse strains in their total-body luciferase (Log10 range of 5.78–7.27 photons/s). The classic C57BL/6J strain was intermediate in its luciferase levels (Log10 mean [SD] = 6.41 [0.58]) at this time point relative to the collaborative cross strains (Figure 2; Table 1). Differences between collaborative cross strains in total luciferase expression were highly significant (F11,44 = 9.64; p < 1 × 10−5). In order to better ascertain the percentage of phenotypic variation in this population, which can be attributed to genetic differences between strains, we estimated the broad-sense heritability46 (for more details, see Materials and Methods) to be between 46% and 63% (the more conservative coefficient of genetic determination [cgd] was 0.463, whereas the more liberal interclass correlation [icc] was 0.633). Looking across this time course, we identified significant effects of strain (F11,234 = 30.08; p < 1 × 10−10), time (F1,234 = 3.979; p = 0.047), as well as the interaction between these two factors (F11,234 = 2.9161; p = 0.001) on whole-body luciferase expression through this study. These responses can be seen clearly in Figure 2 and Table 1. Overall, modest increases in luciferase expression were seen through the experiment. However, some strains (e.g., CC008/GeniUnc and CC024/GeniUnc) showed stable luciferase expression throughout the experiment, and one strain (CC021/Unc) showed reduced luciferase expression throughout the study. Ideally, gene therapeutic products are specifically targeted to a given tissue. We were able to assess the liver-specific expression of luciferase across our set of mouse strains through this kinetic time course. In general, total and liver-specific luciferase levels were highly correlated (r2 = 0.96, Figure 3A) throughout the study, and, therefore, analysis of these data tightly mirrored the responses seen for total luciferase levels. In liver, we found evidence of strain effects (F11,234 = 40.157; p < 1 × 10−10), and also a strain-by-time point interaction (F11,234 = 3.18; p < 0.001). However, although we identified a moderate effect of time point on total luciferase levels, we did not identify an effect of time point (F1,234 = 0.0012; p = 0.97) on liver-specific luciferase expression. Furthermore, at week 1, we found that as with total luciferase, liver-specific luciferase expression heritability was high, ranging from 46% to 64% (cgd = 0.466; icc = 0.636). Examination of these data (Table 2) shows that in general, liver-specific levels were stable through the study, with some strains showing modest changes in effect. Given these lines of evidence, especially the discordance between the temporal responses for total and liver-specific luciferase, we finally asked whether the overall proportion of luciferase expression within individuals was under host genetic control. We first examined the correlation between total luciferase expression and the fraction of luciferase signal coming from the liver and found a modest correlation between these two traits (r2 = 0.579, Figure 3B). Importantly, samples having a broad range of total luciferase expression (105–107.3 photons/s) could still exhibit low levels (∼20%) of liver-specific expression, suggesting that those individual animals with low fractions of liver-specific responses were not just those with low total levels (e.g., samples difficult to assess or incorrect attribution to spillover from the liver). Therefore, we assessed how this response changed throughout the study. As we expected, we identified significant strain (F11,234 = 56.3; p < 1 × 10−10), time point (F1,234 = 27.9; p < 1 × 10−6), and an interaction between these factors (F11,234 = 2.14; p = 0.018) driving the fraction of luciferase expression coming from liver, with heritability estimates at week 1 ranging from 49% to 66% (cgd = 0.493; icc = 0.66). For example, CC033/GeniUnc and CC061/GeniUnc both maintain consistently high proportions of their total luciferase coming from liver similar to C57BL/6J, whereas CC013/GeniUnc showed a decreasing fraction of expression coming from the liver over time. Note that only C57BL/6J and CC035 showed increases in the fraction of their response coming from the liver, suggesting that although there might be fluctuations between tissues or a silencing within the liver, in most strains, there was no expansion of the luciferase expression from this tissue.
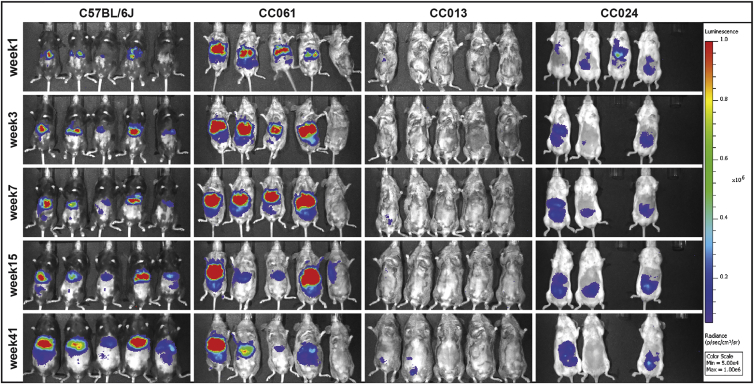
Figure 1.
In Vivo Luciferase Expression across Mouse Strains
Following introduction of a luciferase-expressing lentivirus, female mice from several strains were followed over 41 weeks, with regular assessment (weeks 1, 3, 7, 15, and 41 post-lentiviral vector administration) of in vivo luciferase expression (p/s/cm2/sr); color scale set between 5 × 104 and 1 × 106. Here, the standard laboratory strain C57BL/6J is compared to the highly expressing CC061/GeniUnc strain as well as the low/non-expressing CC013/GeniUnc and CC024/GeniUnc strains.
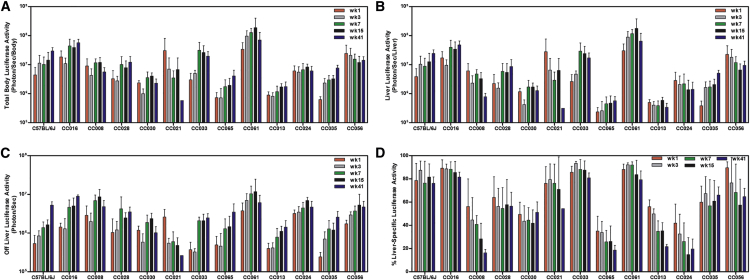
Figure 2.
Time Course of Luciferase Responses across Mouse Strains
(A–C) Whole body (A), liver-specific (B), and off-liver luciferase activity was calculated by subtracting liver-specific activity from whole-body luciferase activity (C). Log10 luciferase levels (photon/s) were assessed for each animal across a 41-week time course. (D) We also assessed the percentage of luciferase signal coming from the liver of each mouse at each time point. Figures show mean and SD of each strain at each time point measured for the appropriate trait. Time points are color coded as indicated.
Table 1.
Mean ± SD of Log10 Total Luciferase Activity Levels Measured in Photons/s through Time Course
| Mouse Strain | Week 1 | Week 3 | Week 7 | Week 15 | Week 41 |
|---|---|---|---|---|---|
| C57BL/6J | 6.41 (0.58) | 6.88 (0.47) | 6.86 (0.41) | 7.01 (0.40) | 7.36 (0.37) |
| CC008/GeniUnc | 6.58 (0.97) | 6.38 (0.73) | 7.04 (0.17) | 7.04 (0.21) | 6.58 (0.48) |
| CC013/GeniUnc | 5.94 (0.13) | 5.89 (0.16) | 6.00 (0.27) | 6.21 (0.12) | 5.97 (0.62) |
| CC016/GeniUnc | 7.19 (0.31) | 6.96 (0.31) | 7.56 (0.33) | 7.52 (0.28) | 7.70 (0.25) |
| CC021/Unc | 7.09 (0.60) | 6.53 (0.58) | 6.38 (0.51) | 6.35 (0.82) | 5.76 (NA)a |
| CC024/GeniUnc | 6.75 (0.19) | 6.69 (0.26) | 6.77 (0.26) | 6.91 (0.12) | 6.63 (0.52) |
| CC028/GeniUnc | 6.35 (0.43) | 6.40 (0.23) | 6.87 (0.38) | 6.81 (0.31) | 6.84 (0.55) |
| CC030/GeniUnc | 6.36 (0.08) | 5.97 (0.18) | 6.41 (0.51) | 6.59 (0.14) | 6.22 (0.49) |
| CC033/GeniUnc | 6.37 (0.37) | 6.69 (0.11) | 7.34 (0.42) | 7.29 (0.39) | 7.12 (0.44) |
| CC035/Unc | 5.78 (0.11) | 6.33 (0.18) | 6.44 (0.21) | 6.49 (0.15) | 6.81 (0.29) |
| CC056/GeniUnc | 7.27 (0.33) | 7.22 (0.35) | 7.12 (0.29) | 7.00 (0.29) | 7.07 (0.32) |
| CC061/GeniUnc | 7.44 (0.33) | 7.90 (0.35) | 8.08 (0.18) | 7.66 (1.04) | 7.27 (0.87) |
| CC065/Unc | 5.82 (0.22) | 5.63 (0.51) | 6.14 (0.35) | 6.18 (0.36) | 6.39 (0.58) |
Only one animal survived to this time point.
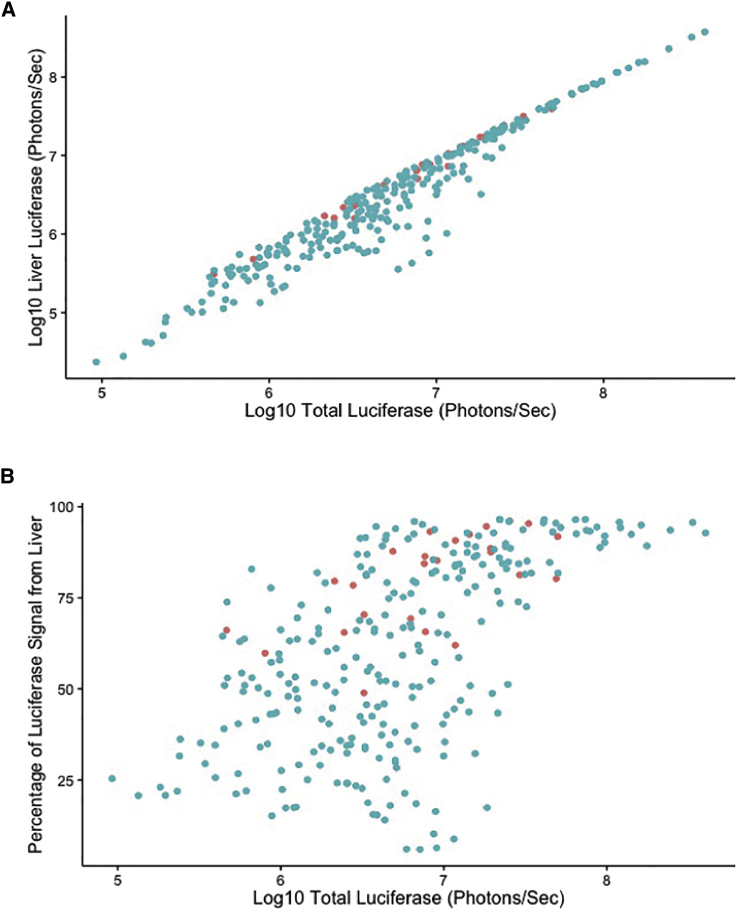
Figure 3.
Relationships between Luciferase Expression Levels
(A) A strong correlation (r2 = 0.96) was identified across all time points between the liver-specific and total-body levels of luciferase expression. (B) A moderate (r2 = 0.57) correlation was identified between total luciferase levels and the percentage of luciferase signal coming from the liver. Each point is from a single mouse at a single time point, with C57BL/6J shown as red points and collaborative cross mice shown as blue points.
Table 2.
Mean ± SD of Log10 Liver-Specific Luciferase Activity Levels Measured in Photons/s through Time Course
| Mouse Strain | Week 1 | Week 3 | Week 7 | Week 15 | Week 41 |
|---|---|---|---|---|---|
| C57BL/6J | 6.30 (0.66) | 6.82 (0.51) | 6.74 (0.52) | 6.92 (0.45) | 7.24 (0.43) |
| CC008/GeniUnc | 6.28 (1.23) | 5.99 (0.94) | 6.65 (0.19) | 6.45 (0.27) | 5.77 (0.44) |
| CC013/GeniUnc | 5.68 (0.12) | 5.58 (0.15) | 5.53 (0.25) | 5.75 (0.12) | 5.29 (0.58) |
| CC016/GeniUnc | 7.14 (0.35) | 6.91 (0.31) | 7.50 (0.33) | 7.45 (0.29) | 7.61 (0.29) |
| CC021/Unc | 6.97 (0.67) | 6.42 (0.65) | 6.26 (0.55) | 6.17 (1.02) | 5.76 (NA)a |
| CC024/GeniUnc | 6.35 (0.32) | 6.17 (0.43) | 6.13 (0.51) | 5.94 (0.50) | 5.82 (0.66) |
| CC028/GeniUnc | 6.14 (0.47) | 6.13 (0.26) | 6.59 (0.47) | 6.55 (0.47) | 6.56 (0.65) |
| CC030/GeniUnc | 6.05 (0.13) | 5.59 (0.18) | 6.04 (0.57) | 6.20 (0.18) | 5.92 (0.59) |
| CC033/GeniUnc | 6.31 (0.38) | 6.66 (0.11) | 7.29 (0.47) | 7.23 (0.43) | 7.02 (0.48) |
| CC035/Unc | 5.55 (0.18) | 6.16 (0.24) | 6.16 (0.26) | 6.26 (0.23) | 6.62 (0.32) |
| CC056/GeniUnc | 7.22 (0.35) | 7.08 (0.50) | 6.92 (0.50) | 6.73 (0.32) | 6.87 (0.34) |
| CC061/GeniUnc | 7.39 (0.34) | 7.86 (0.36) | 8.04 (0.18) | 7.58 (1.10) | 7.17 (0.92) |
| CC065/Unc | 5.34 (0.20) | 5.15 (0.58) | 5.53 (0.40) | 5.56 (0.38) | 5.64 (0.44) |
Only one animal survived to this time point.
Differential Success in Stable Vector Maintenance
Following reverse transcription, lentiviral vectors efficiently import their genetic material to the host cells’ nuclei, where it serves as a template for transcription of the relevant transgene of interest. Successful gene therapy with these vectors requires long-term persistence of vector-transduced cells. Following study termination, liver and spleen tissues were collected from these animals, and a qPCR assay was performed to identify the number of vector copies delivered into host cells’ nuclei. Vector copy number per cell (VCN) in the liver differed by over 2 logs (min = 0.001; max = 1.12; mean = 0.22 copies/cell; Figure 4A). C57BL/6J showed the highest level of VCN within the livers (0.85 ± 0.22 copies/cell), with all collaborative cross strains showing lower levels of VCN than C57BL/6J. Collaborative cross strains showed significant differences in liver VCN (F11,31 = 8.023; p = 2.1 × 10−6), and heritability estimates suggest that between 41% and 58% (cgd = 0.412; icc = 0.584) of the variation in vector copies/cell in the liver are under host genetic control. Levels of VCN within the spleen of most strains were reduced relative to liver (min < 0.001; max = 0.33; mean = 0.09 copies/cell), although there was a strong correlation between vector copies in the liver and vector copies in the spleen (r2 = 0.78). C57BL/6J showed relatively high levels of vector copies/spleen (0.13 ± 0.08 vector copies per cell), but in contrast to liver-specific VCN, C57BL/6J was not the mouse strain with the highest levels of spleen VCN (Figure 4B). As with liver levels, there was an approximately 1.5-log difference between the highest and lowest collaborative cross strain, and these differences were statistically significant (F11,29 = 4.247; p = 0.0008). Spleen levels of vector maintenance had a heritability of between 25% and 39% (cgd = 0.245; icc = 0.393) across these collaborative cross strains.
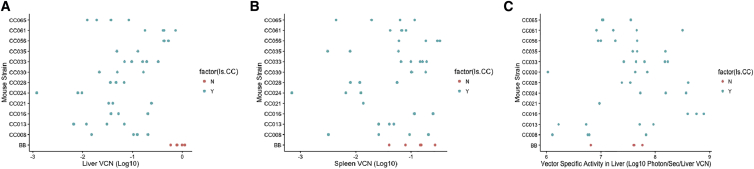
Figure 4.
Vector Copy Number and Luciferase Levels per Vector Copy Vary between Strains
(A and B) We assessed vector copies at study conclusion and found significant strain-specific differences in levels of vector copy number in liver (A) and spleen (B) at these time points. (C) We also determined luciferase levels per vector copy in liver tissues (Log10 (liver-specific luciferase/liver VCN)) based on week 41 luciferase levels and found strain-specific levels of specific activity. Each point is one animal, with C57BL/6J mice in red and collaborative cross strains in blue.
Relationship between VCN and Luciferase Expression
Given the highly significant variation in total and liver-specific luciferase expression between this collection of mouse strains, we sought to assess the extent to which host genetic differences controlled these expression levels (that is, given the strain-specific VCN, are there differences in the efficiency of transgene expression). We determined the relative expression of luciferase at week 41 within the livers of each mouse relative to liver VCN. We found that vector-specific activity in mouse livers (F11,28 = 2.81; p = 0.013) was impacted by the host strain (Figure 4C). Heritability analysis showed that between 15% and 27% (cgd = 0.153; icc = 0.266) of the variation in vector-specific activity in the liver was explained by host genetic variation. We utilized a second analysis to assess the impact of host genetic variation on luciferase expression independent of host genetic variations’ impact on vector copy number. We used a partial F-test to assess the improvement in fit when our model explaining luciferase levels included both strain and vector copy number as opposed to vector copy number alone. We found strong evidence for liver luciferase expression having other host factors driving expression (F27,38 = 3.26; p = 0.005), although the evidence for total luciferase expression was not significant (F26,36 = 1.78; p = 0.11).
Mortality and Atypical Pathological Responses across This Treated Cohort
A total of 12/61 (19%) mice within this study died during the course of this experiment. This mortality impacted seven strains (Table S1), with CC021/Unc experiencing the highest mortality (4/5 mice). After sacrifice, six mice from four strains showed what appeared to be a macroscopic abnormality, including liver (CC024/GeniUnc#1) and spleen (CC035/Unc#5) tumors. Pathological analysis suggested these were common lesion types in aging animals and unlikely to be due to the presence of vector-related issues (Figure S2; Table S2).
Discussion
Although gene therapy approaches have the power to alleviate severe congenital conditions, several aspects of viral-vector-delivered gene therapeutic regimens have raised causes for concern. Specifically, successful gene therapy requires optimal responses to a variety of processes, all with a single delivery event. These include, but are not limited to, delivery of viral vectors to specific target cells, efficient transfer of viral genomes to host cells’ nuclei, and maintenance of therapeutic levels of transgene expression without causing harmful adverse effects. Inefficiency in these processes as well as unforeseen adverse effects in some clinical trials have been the greatest impediments to broad adoption of gene therapy as a means to treat nonfatal diseases.20, 22, 23, 24 Although some studies have suggested that host genetic differences can play a role in gene therapy outcomes,31, 32 there has been little research into the role that host genetic variation plays on the efficacy, maintenance, and safety of viral-vector-driven gene therapeutic approaches. We sought to assess the magnitude of the impact that host genetic variation could have on gene therapeutic efficacy. We used a set of strains from the collaborative cross genetic reference panel, and found that host genetic variation impacted multiple aspects of the gene therapy response within hosts, and, furthermore, that these responses were not always correlated across the strains (e.g., VCN and total luciferase expression).
C57BL/6J is one of the most widely used mouse strains and is in fact the reference mouse genome. Various mouse models emulating human diseases were established on a C57BL/6J genetic background. These include several lines of C57BL/6J-factor-IX-deficient mice, which modeled hemophilia.47, 48, 49, 50 Using adenoviral- and AAV-based vectors in gene therapy preclinical studies of hemophilia B, Fields et al. and Mingozzi et al. demonstrated that induction of immune tolerance to and long-term therapeutic levels of expression of vector-delivered human factor IX were more readily achieved in C57BL/6 mice compared to BALB/c, C3H, and CD-1, although precise assessment of the genetic contributions within these studies was not characterized.51, 52 In an earlier lentiviral-vector-based preclinical study, Follenzi et al. showed superior lentiviral vector long-term expression of and less prominent adaptive immune response to vector-delivered GFP reporter gene and human factor IX in C57BL/6 mice compared to FVB/N and BALB/c mice. Not surprisingly, C57BL/6 mice are considered the most permissive model for lentiviral-vector-based gene delivery.31 In line with the Follenzi study above, we found that peak luciferase expression in the C57BL/6J strain was higher (>1 × 107.36) than in many collaborative cross strains in this study. Importantly, in this study, two collaborative cross strains (CC016/GeniUnc and CC061/GeniUnc) exhibited equivalent or higher levels of transgene expression than the C57BL/6J strain at later time points. Furthermore, our study showed differences in peak luciferase expression across strains as >100-fold. These data suggest that significant differences in the efficacy of gene therapy protocols between human patients can be anticipated solely due to genetic differences.
In addition to peak levels of transgene expression, long-term maintenance of therapeutic levels of vector-delivered transgenes is a required characteristic of in vivo gene transfer procedures and directly affects the success of in vivo gene therapy protocols. An early study by Zhang et al. associated the development of humoral immune response to human factor IX delivered by AAV-based vectors with slow kinetics of in vivo transduction.53 In our study, the kinetics of lentiviral-vector-mediated luciferase expression throughout the study differed significantly between mouse strains. Although there were modest increases in luciferase expression in most strains, a subset of strains showed reduction of luciferase expression throughout the study. Most notably, this was seen in the strain CC021/Unc, which showed decreasing levels of luciferase expression throughout the study and also showed high mortality throughout the study. Overall, these results were in line with earlier studies showing stable long-term expression of luciferase and human factor IX from integrating lentiviral vectors in immune-competent C57BL/6J mice.54, 55 However, other earlier lentiviral-vector-based preclinical studies reported on dramatic reduction in transgene expression between weeks 2 and 4 post-injection.31, 56 This phenomenon was alleviated in a transgene-dependent manner by inhibiting off-target transgene expression in antigen presenting cells.56, 57, 58 Several mechanisms can contribute to reduction in vector-delivered transgene expression and thus limit the efficacy and safety of gene therapy protocols. These mechanisms include adaptive immune response directed to vector-transduced cells, adaptive immune responses to the vector-delivered gene products themselves,23, 31, 56, 59, 60 transgene toxicity, and silencing of the vector expression cassette.61 Previous clinical trials and other studies have suggested that immune responses against targeted cells can lead to adverse effects.23, 24, 60, 62 Although our study was not able to conclusively investigate whether this transgene silencing and mortality were connected and if they were the results of immune responses, these data suggest that there are host genotype-specific responses that can lead to suboptimal and adverse gene therapeutic responses.
Vector biodistribution is a key characteristic of all gene-delivery systems. Lentiviral vector biodistribution is controlled by the efficiency at which reverse-transcribed vector genomes are nuclear imported and expressed in various host organs following systemic administration. The ability of HIV-1 to transduce non-dividing cells, which facilitates in vivo gene delivery, is the hallmark of lentiviruses and lentiviral vector systems.63, 64 Surprisingly, to date, the effects of host genetic variation on vector biodistribution has not been characterized. We were able to identify a difference in the success of VSV-G-pseudotyped lentiviral vectors to deliver reverse-transcribed genomes into both desired target (liver) and bystander (spleen) tissues (expressed as VCN) between the collaborative cross strains we assessed. With the exception of three mouse strains (C57BL/6J, CC056/GeniUnc, and CC061/GeniUnc), in which VCN levels in liver tissues were remarkably high, all other collaborative cross mouse strains exhibited comparable levels of VCNs in liver and spleen tissues. Furthermore, there was a strong correlation across all animals in the level of copies between tissues. These findings are in line with an earlier report by Pan et al. showing comparable VCN levels in liver and spleen tissues following intravenous administration of VSV-G-pseudotyped lentiviral vectors to BALB/c mice.65 These results suggest there are some variant host factors, which impact the efficacy of delivery or maintenance of cells transformed by the vector. Nuclear import of vector DNA is a prerequisite for vector transcriptional activity and directly affects biodistribution of transgene expression. However, vector design and host factors also have major effects on transgene expression in vector-transduced cells. In this study, a liver-specific promoter (hAAT) controlled vector-mediated luciferase expression. Indeed, most mouse strains exhibited predominantly hepatic expression. However, several mouse strains demonstrated relatively high extra-hepatic transgene expression. Furthermore, an earlier study reported on extra-hepatic transgene expression in non-hepatic cell lines in vitro and extra hepatic tissues in vivo and from lentiviral vectors carrying a liver-specific promoter.59 Although the mechanism involved in this phenomenon has not been completely elucidated, it was generally attributed to enhancer/promoter effects of the host chromatin on liver-specific promoters in integrated lentiviral vectors.56, 66 In addition to VCN levels, specific activity of a lentiviral vector expression cassette as determined by total transgene activity per vector genome directly affects the therapeutic efficacy of a lentiviral vector as a therapeutic agent. Although vector-specific activity was comparable across the collaborative cross mouse strains, significant high specific activity was exhibited by strain CC016/GeniUnc. Furthermore, and strikingly, both the prototypical C57BL/6J and CC061/GeniUnc strains showed much more modest specific activity than their overall luciferase expression levels would indicate, further highlighting the need for assessment of both transduction efficiency as well as transgene expression in considering the success of gene therapy. This phenomenon can be attributed to a single or a combination of strain-specific transcriptional and post-translational host factors, which enhanced luciferase activity in the above mouse strain. This observation is highly important in the gene therapy field because it strongly suggests that therapeutic vector loads should be patient specific. This approach could potentially reduce transgene cytotoxicity on one hand and optimize overall vector genomes required to achieve therapeutic transgene expression with minimal adverse effects on the other hand. Indeed, insertional mutagenesis is a major biosafety inherent to all integrating vectors, including lentiviral vectors. A previous study found some evidence for differential oncogenic potential.32 We identified mice from four of the 12 collaborative cross strains used in this study (Table S2), which had macroscopic liver abnormalities following termination. The presence of these abnormalities did not appear to be tied to the lentiviral transduction, and the propensity to develop specific cancer types is known to be under genetic control.67, 68 However, more in-depth future studies in a more robust case-control framework will be required to both understand any potential exacerbation of tumorigenesis driven by lentiviral vectors as well as to assess the distribution of integration into oncogenes and the extent to which host genetic differences directly (e.g., integration site biases) or indirectly (e.g., epigenetic and chromatin differences) might alter this. Here, we have demonstrated the strong effects of host genetic variation on the levels and kinetics of transgene expression, levels of VCN, and vector-specific activity. A larger number of collaborative cross strains should facilitate identification of putative genetic loci involved in the above strain-specific characteristics of in vivo lentiviral vector gene delivery. To avoid gender-specific effects on hepatic transduction, this study was premised on female mice only. Although more resource demanding, additional studies comprising male mice are needed to identify gender-specific effects on lentiviral vector transduction efficiency.
We anticipate that using the collaborative cross mouse system in an in vitro setting would further elucidate the molecular mechanisms by which host genetic variation affects specific steps in the HIV-1 life cycle. However, establishing the ability to genetically identify host factors affecting the efficacy of lentiviral vectors ferrying therapeutic genetic cargos in the setting of a genetic disease model will be more challenging.
Materials and Methods
Lentiviral Particles Production, Concentration, and Titration
Lentiviral vector (pTK979) harboring the firefly luciferase cDNA under the control of a liver-specific promoter (hAAT) was constructed and used as described earlier.54 Lentiviral vector particles were packaged with the packaging cassette, ΔNRF, in 293T cells using three-plasmid transient transfection as previously described.69 Vector titer was determined by measuring the p24 capsid concentration using ELISA as previously described.70 All vector preps were tested for the absence of replication-competent retrovirus as described earlier.71
Animal Studies
All procedures involving animal study were performed in accordance with the Guide for the Care and Use of Laboratory Animals. The animal study protocol was approved by the University of North Carolina (UNC) Institutional Animal Care and Usage Committee. All mice were purchased from Jackson Laboratories (C57BL/6J) or the UNC Systems Genetics Core Facility (collaborative cross strains listed in Table 1; http://csbio.unc.edu/CCstatus/index.py?run=AvailableLines). Female mice were acquired at 5 to 6 weeks of age, and acclimated for 1 week in our experimental facility. Female animals (n = 4 to 5/strain) were intraperitoneally injected with 50 μg of p24gag lentiviral vectors (in 250 μL of phosphate-buffered solution). In vivo expression of vector-delivered firefly luciferase in live animals was determined at weeks 1, 3, 7, 15, and 41 using the IVIS Lumina optical imaging system (PerkinElmer). To this end, animals were intraperitoneally injected with 200 μL (5 mg) of D-luciferin potassium salt reconstituted in phosphate-buffered solution (Regis Technologies). Imaging was initiated 10 min after D-luciferin injection using a 5-min exposure time. The relative light counts obtained through a charge-coupled device (CCD) camera were converted to physical units of surface radiance (p/s/cm2/sr) and displayed in luminescence radiance mode. All in vivo luciferase activities shown in this study were analyzed using Living Image Software (PerkinElmer) and reported in total flux (photon/s). To measure whole-body and liver-specific luciferase activity, identical regions of interest (ROIs) were measured. These comprised either the whole animal body (excluding head and tail) or the liver, respectively (Figure S1). Luciferase activity in the respective ROIs in each animal was measured as described above. Off-liver luciferase activity was calculated by subtracting liver-specific activity from whole-body luciferase activity.
Mouse Tissue DNA Preparation for Quantification of Vector Copy Number
At the end of the experiment, the viral copy number of liver and spleen tissues was determined. Genomic DNAs from tissues were isolated using the Blood & Tissue DNeasy kit (QIAGEN) according to the manufacturer’s instructions. RNAs were removed using RNase A (Fermentas). All samples were treated with DpnI (New England Biolabs). Total vector copy number of each sample was quantified using multiplex qPCR, as previously described.55
Histopathology
Upon tissue harvesting, liver and spleen tissues with physical abnormalities in either size or appearance observed in six animals from four strains (Figure S2; Table S2) were sent to the UNC Animal Histopathology Core Facility for a histopathological examination. To this end, tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned with a microtome, and stained with H&E. A board-certified veterinary pathologist analyzed these H&E-stained sections.
Statistical Analysis
All data were analyzed in the R programming environment (cran.r-project.org). Broad-sense heritability was assessed as either the interclass correlation or the coefficient of genetic determination.46 Briefly, each of these approaches uses the ANOVA model fit and then assesses the relative ratios of mean square explained by strain differences relative to the mean square error in the data. Both approaches then apply a scaling factor relative to the number of samples (n) in each treatment class as follows:
-
(1)
Interclass correlation: (MSstrain − MSerror)/(MSstrain + (n − 1)MSerror)
-
(2)
Coefficient of genetic determination:
In order to assess the significance of strain differences on given phenotypic traits, data were normalized and ANOVA was used to identify significance. For the model fitting assessing vector and strain effects driving luciferase expression, we used a partial F-test framework.
Data Availability
All data from this study, including calculated luciferase, vector copy number levels, and raw images for luciferase calculation, are available upon request. The 12 collaborative cross strains used within this study are available from the Systems Genetics Core Facility (http://csbio.unc.edu/CCstatus/index.py?run=AvailableLines), and both genotype files and haplotype reconstructions are available for these strains on that site.
Author Contributions
Conceptualization and design of the study, T.K., and F.P.-M.d.V.; Acquisition of data, T.S., P.H., S.A.M., and T.G.; Analysis and interpretation of data, T.K., M.T.F., and F.P.-M.d.V.; Writing – Original Draft, T.K., M.T.F., and T.S.; Statistical analysis, M.T.F.; Critical revision of the manuscript, T.K., M.T.F., F.P.-M.d.V., and T.S.
Conflicts of Interest
T.K. is an inventor of a technology which was licensed by UNC to a commercial entity.
Acknowledgments
The following reagent was obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Division of AIDS, the National Institute of Allergy and Infectious Diseases: the HIV-1 p24 monoclonal antibody (183-H12-5C) from Bruce Chesebro and Kathy Wehrly. The study was supported by the UNC Center for AIDS Research and by the NIH grants R01-HL128119 (to T.S., M.T.F., P.H., T.G., F.P.-M.d.V., and T.K.), R01-DK058702 (to T.S., P.H., T.G., and T.K.), and U19-AI100625 (to M.T.F. and F.P.-M.d.V.). Histopathology analysis was performed in the UNC Animal Histopathology Core Facility, which is supported in part by an NCI Center Core Support Grant (2P30CA016086-40) to the UNC Lineberger Comprehensive Cancer Center. This study is dedicated to the U.S. Marine Corps and the Gold Star families. In memory of Boaz Kofman, the epitome of a mensch.
Footnotes
Supplemental Information includes two figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2017.03.009.
Supplemental Information
References
- 1.Cockrell A.S., Kafri T. Gene delivery by lentivirus vectors. Mol. Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz P.D. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 3.Borsotti C., Borroni E., Follenzi A. Lentiviral vector interactions with the host cell. Curr. Opin. Virol. 2016;21:102–108. doi: 10.1016/j.coviro.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Goff S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 5.Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y., Craigie R. The road to chromatin - nuclear entry of retroviruses. Nat. Rev. Microbiol. 2007;5:187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- 7.Taltynov O., Desimmie B.A., Demeulemeester J., Christ F., Debyser Z. Cellular cofactors of lentiviral integrase: from target validation to drug discovery. Mol. Biol. Int. 2012;2012:863405. doi: 10.1155/2012/863405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Towers G.J., Hatziioannou T., Cowan S., Goff S.P., Luban J., Bieniasz P.D. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 9.Dochi T., Nakano T., Inoue M., Takamune N., Shoji S., Sano K., Misumi S. Phosphorylation of human immunodeficiency virus type 1 capsid protein at serine 16, required for peptidyl-prolyl isomerase-dependent uncoating, is mediated by virion-incorporated extracellular signal-regulated kinase 2. J. Gen. Virol. 2014;95:1156–1166. doi: 10.1099/vir.0.060053-0. [DOI] [PubMed] [Google Scholar]
- 10.Fricke T., White T.E., Schulte B., de Souza Aranha Vieira D.A., Dharan A., Campbell E.M., Brandariz-Nuñez A., Diaz-Griffero F. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology. 2014;11:68. doi: 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayah D.M., Sokolskaja E., Berthoux L., Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 12.Shah V.B., Shi J., Hout D.R., Oztop I., Krishnan L., Ahn J., Shotwell M.S., Engelman A., Aiken C. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 2013;87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan T., Yao W., Tokunaga K., Yang R., Sun B. An HIV-1 capsid binding protein TRIM11 accelerates viral uncoating. Retrovirology. 2016;13:72. doi: 10.1186/s12977-016-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrose Z., Aiken C. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology. 2014;454-455:371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borrenberghs D., Dirix L., De Wit F., Rocha S., Blokken J., De Houwer S., Gijsbers R., Christ F., Hofkens J., Hendrix J. Dynamic oligomerization of integrase orchestrates HIV nuclear entry. Sci. Rep. 2016;6:36485. doi: 10.1038/srep36485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilditch L., Towers G.J. A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr. Opin. Virol. 2014;4:32–36. doi: 10.1016/j.coviro.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delelis O., Carayon K., Saïb A., Deprez E., Mouscadet J.F. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology. 2008;5:114. doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poeschla E.M. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 2008;65:1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mbonye U., Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454-455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaspar H.B., Cooray S., Gilmour K.C., Parsley K.L., Adams S., Howe S.J., Al Ghonaium A., Bayford J., Brown L., Davies E.G. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 22.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 23.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 24.Raper S.E., Yudkoff M., Chirmule N., Gao G.P., Nunes F., Haskal Z.J., Furth E.E., Propert K.J., Robinson M.B., Magosin S. A pilot study of in vivo liver-directed gene transfer with an adenoviral vector in partial ornithine transcarbamylase deficiency. Hum. Gene Ther. 2002;13:163–175. doi: 10.1089/10430340152712719. [DOI] [PubMed] [Google Scholar]
- 25.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 27.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 28.Frey N.V., Porter D.L. CAR T-cells merge into the fast lane of cancer care. Am. J. Hematol. 2016;91:146–150. doi: 10.1002/ajh.24238. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S., Thrasher A.J., Gaspar H.B. Gene therapy for monogenic disorders of the bone marrow. Br. J. Haematol. 2015 doi: 10.1111/bjh.13520. Published online June 5, 2015. [DOI] [PubMed] [Google Scholar]
- 30.Kalos M., Levine B.L., Porter D.L., Katz S., Grupp S.A., Bagg A., June C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Follenzi A., Battaglia M., Lombardo A., Annoni A., Roncarolo M.G., Naldini L. Targeting lentiviral vector expression to hepatocytes limits transgene-specific immune response and establishes long-term expression of human antihemophilic factor IX in mice. Blood. 2004;103:3700–3709. doi: 10.1182/blood-2003-09-3217. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y., Keating K., Thorpe R. Comparison of toxicogenomic profiles of two murine strains treated with HIV-1-based vectors for gene therapy. Toxicol. Appl. Pharmacol. 2007;225:189–197. doi: 10.1016/j.taap.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Duggal P., Thio C.L., Wojcik G.L., Goedert J.J., Mangia A., Latanich R., Kim A.Y., Lauer G.M., Chung R.T., Peters M.G. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann. Intern. Med. 2013;158:235–245. doi: 10.7326/0003-4819-158-4-201302190-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., GenISIS Investigators. MOSAIC Investigators IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferris M.T., Heise M.T. Quantitative genetics in the study of virus-induced disease. Adv. Virus Res. 2014;88:193–225. doi: 10.1016/B978-0-12-800098-4.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chick J.M., Munger S.C., Simecek P., Huttlin E.L., Choi K., Gatti D.M., Raghupathy N., Svenson K.L., Churchill G.A., Gygi S.P. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elbahesh H., Schughart K. Genetically diverse CC-founder mouse strains replicate the human influenza gene expression signature. Sci. Rep. 2016;6:26437. doi: 10.1038/srep26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong H., Morrison J., Ferris M.T., Gralinski L.E., Whitmore A.C., Green R., Thomas M.J., Tisoncik-Go J., Schroth G.P., Pardo-Manuel de Villena F. Genomic profiling of collaborative cross founder mice infected with respiratory viruses reveals novel transcripts and infection-related strain-specific gene and isoform expression. G3 (Bethesda) 2014;4:1429–1444. doi: 10.1534/g3.114.011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collaborative Cross Consortium The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferris M.T., Aylor D.L., Bottomly D., Whitmore A.C., Aicher L.D., Bell T.A., Bradel-Tretheway B., Bryan J.T., Buus R.J., Gralinski L.E. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013;9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham J.B., Swarts J.L., Wilkins C., Thomas S., Green R., Sekine A., Voss K.M., Ireton R.C., Mooney M., Choonoo G. A mouse model of chronic West Nile virus disease. PLoS Pathog. 2016;12:e1005996. doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gralinski L.E., Ferris M.T., Aylor D.L., Whitmore A.C., Green R., Frieman M.B., Deming D., Menachery V.D., Miller D.R., Buus R.J. Genome wide identification of SARS-CoV susceptibility loci using the collaborative cross. PLoS Genet. 2015;11:e1005504. doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leist S.R., Pilzner C., van den Brand J.M., Dengler L., Geffers R., Kuiken T., Balling R., Kollmus H., Schughart K. Influenza H3N2 infection of the collaborative cross founder strains reveals highly divergent host responses and identifies a unique phenotype in CAST/EiJ mice. BMC Genomics. 2016;17:143. doi: 10.1186/s12864-016-2483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen A.L., Okumura A., Ferris M.T., Green R., Feldmann F., Kelly S.M., Scott D.P., Safronetz D., Haddock E., LaCasse R. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng C.L., Wilmot B., Walter N.A., Oberbeck D., Kawane S., Searles R.P., McWeeney S.K., Hitzemann R. Splicing landscape of the eight collaborative cross founder strains. BMC Genomics. 2015;16:52. doi: 10.1186/s12864-015-1267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petkova S.B., Yuan R., Tsaih S.W., Schott W., Roopenian D.C., Paigen B. Genetic influence on immune phenotype revealed strain-specific variations in peripheral blood lineages. Physiol. Genomics. 2008;34:304–314. doi: 10.1152/physiolgenomics.00185.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin D.Y., Zhang T.P., Gui T., Stafford D.W., Monahan P.E. Creation of a mouse expressing defective human factor IX. Blood. 2004;104:1733–1739. doi: 10.1182/blood-2004-01-0138. [DOI] [PubMed] [Google Scholar]
- 48.Kundu R.K., Sangiorgi F., Wu L.Y., Kurachi K., Anderson W.F., Maxson R., Gordon E.M. Targeted inactivation of the coagulation factor IX gene causes hemophilia B in mice. Blood. 1998;92:168–174. [PubMed] [Google Scholar]
- 49.Lin H.F., Maeda N., Smithies O., Straight D.L., Stafford D.W. A coagulation factor IX-deficient mouse model for human hemophilia B. Blood. 1997;90:3962–3966. [PubMed] [Google Scholar]
- 50.Wang L., Zoppè M., Hackeng T.M., Griffin J.H., Lee K.F., Verma I.M. A factor IX-deficient mouse model for hemophilia B gene therapy. Proc. Natl. Acad. Sci. USA. 1997;94:11563–11566. doi: 10.1073/pnas.94.21.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fields P.A., Armstrong E., Hagstrom J.N., Arruda V.R., Murphy M.L., Farrell J.P., High K.A., Herzog R.W. Intravenous administration of an E1/E3-deleted adenoviral vector induces tolerance to factor IX in C57BL/6 mice. Gene Ther. 2001;8:354–361. doi: 10.1038/sj.gt.3301409. [DOI] [PubMed] [Google Scholar]
- 52.Mingozzi F., Liu Y.L., Dobrzynski E., Kaufhold A., Liu J.H., Wang Y., Arruda V.R., High K.A., Herzog R.W. Induction of immune tolerance to coagulation factor IX antigen by in vivo hepatic gene transfer. J. Clin. Invest. 2003;111:1347–1356. doi: 10.1172/JCI16887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang T.P., Jin D.Y., Wardrop R.M., 3rd, Gui T., Maile R., Frelinger J.A., Stafford D.W., Monahan P.E. Transgene expression levels and kinetics determine risk of humoral immune response modeled in factor IX knockout and missense mutant mice. Gene Ther. 2007;14:429–440. doi: 10.1038/sj.gt.3302881. [DOI] [PubMed] [Google Scholar]
- 54.Bayer M., Kantor B., Cockrell A., Ma H., Zeithaml B., Li X., McCown T., Kafri T. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol. Ther. 2008;16:1968–1976. doi: 10.1038/mt.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suwanmanee T., Hu G., Gui T., Bartholomae C.C., Kutschera I., von Kalle C., Schmidt M., Monahan P.E., Kafri T. Integration-deficient lentiviral vectors expressing codon-optimized R338LhFIX restore normal hemostasis in hemophilia B mice. Mol. Ther. 2014;22:567–574. doi: 10.1038/mt.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown B.D., Venneri M.A., Zingale A., Sergi Sergi L., Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 57.Brown B.D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 58.Matsui H., Hegadorn C., Ozelo M., Burnett E., Tuttle A., Labelle A., McCray P.B., Jr., Naldini L., Brown B., Hough C. A microRNA-regulated and GP64-pseudotyped lentiviral vector mediates stable expression of FVIII in a murine model of Hemophilia A. Mol. Ther. 2011;19:723–730. doi: 10.1038/mt.2010.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Follenzi A., Sabatino G., Lombardo A., Boccaccio C., Naldini L. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum. Gene Ther. 2002;13:243–260. doi: 10.1089/10430340252769770. [DOI] [PubMed] [Google Scholar]
- 60.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 62.Raper S.E., Chirmule N., Lee F.S., Wivel N.A., Bagg A., Gao G.P., Wilson J.M., Batshaw M.L. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 63.Lewis P., Hensel M., Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 65.Pan D., Gunther R., Duan W., Wendell S., Kaemmerer W., Kafri T., Verma I.M., Whitley C.B. Biodistribution and toxicity studies of VSVG-pseudotyped lentiviral vector after intravenous administration in mice with the observation of in vivo transduction of bone marrow. Mol. Ther. 2002;6:19–29. doi: 10.1006/mthe.2002.0630. [DOI] [PubMed] [Google Scholar]
- 66.De Palma M., Montini E., Santoni de Sio F.R., Benedicenti F., Gentile A., Medico E., Naldini L. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- 67.Bai L., Yang H.H., Hu Y., Shukla A., Ha N.H., Doran A., Faraji F., Goldberger N., Lee M.P., Keane T. An integrated genome-wide systems genetics screen for breast cancer metastasis susceptibility genes. PLoS Genet. 2016;12:e1005989. doi: 10.1371/journal.pgen.1005989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winter J.M., Gildea D.E., Andreas J.P., Gatti D.M., Williams K.A., Lee M., Hu Y., Zhang S., NISC Comparative Sequencing Program. Mullikin J.C. Mapping complex traits in a diversity outbred F1 mouse population identifies germline modifiers of metastasis in human prostate cancer. Cell Syst. 2016;4:31–45.e6. doi: 10.1016/j.cels.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu K., Ma H., McCown T.J., Verma I.M., Kafri T. Generation of a stable cell line producing high-titer self-inactivating lentiviral vectors. Mol. Ther. 2001;3:97–104. doi: 10.1006/mthe.2000.0238. [DOI] [PubMed] [Google Scholar]
- 70.Kantor B., Ma H., Webster-Cyriaque J., Monahan P.E., Kafri T. Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc. Natl. Acad. Sci. USA. 2009;106:18786–18791. doi: 10.1073/pnas.0905859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kafri T., van Praag H., Gage F.H., Verma I.M. Lentiviral vectors: regulated gene expression. Mol. Ther. 2000;1:516–521. doi: 10.1006/mthe.2000.0083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from this study, including calculated luciferase, vector copy number levels, and raw images for luciferase calculation, are available upon request. The 12 collaborative cross strains used within this study are available from the Systems Genetics Core Facility (http://csbio.unc.edu/CCstatus/index.py?run=AvailableLines), and both genotype files and haplotype reconstructions are available for these strains on that site.