Abstract
Multiple independent adeno-associated virus (AAV) gene therapy clinical trials for hemophilia B, utilizing different AAV serotypes, have reported a vector dose-dependent loss of circulating factor IX (FIX) protein associated with capsid-specific CD8+ T cell (Cap-CD8) elimination of transduced hepatocytes. Hemophilia B patients who develop transient transaminitis and loss of FIX protein may be stabilized with the immune-suppressive (IS) drug prednisolone, but do not all recover lost FIX expression, whereas some patients fail to respond to IS. We developed the first animal model demonstrating Cap-CD8 infiltration and elimination of AAV-transduced hepatocytes of immune-deficient mice. Here, we extend this model to an immune-competent host where Cap-CD8 transfer to AAV2-F9-treated mice significantly reduced circulating and hepatocyte FIX expression. Further, we studied two high-expressing liver tropic AAV2 variants, AAV2-LiA and AAV2-LiC, obtained from a rationally designed capsid library. Unlike AAV2, Cap-CD8 did not initially reduce circulating FIX levels for either variant. However, FIX levels were significantly reduced in AAV2-LiC-F9-treated, but not AAV2-LiA-F9-treated, mice at the study endpoint. Going forward, the immune-competent model may provide an opportunity to induce immunological memory directed against a surrogate AAV capsid antigen and study recall responses following AAV gene transfer.
Keywords: AAV vector, capsid immunity, mouse model, hemophilia
Graphical Abstract
Introduction
Adeno-associated Virus Gene Therapy for Hemophilia B
Adeno-associated virus (AAV) is a nonpathogenic weakly immunogenic dependoparvovirus with a single-stranded DNA genome of ∼4.7 kb. Humans are naturally infected with AAV, often in the presence of an immunogenic helper virus, which is required for AAV replication. This dependency is thought to provide the necessary activation signals that prime anti-AAV humoral and cell-mediated immunity.1 AAV-based vectors have been developed to episomally deliver therapeutic genes into post-mitotic cells, such as hepatocytes, and provide stable long-term expression. Patients with hemophilia B lack functional coagulation factor IX (FIX) protein and are treated by infusing recombinant or plasma-derived FIX protein. Because FIX is normally produced by hepatocytes and FIX activity levels as low as 5% of normal can significantly reduce bleeding events, gene therapy was considered a promising means of restoring FIX expression. Positive outcomes in preclinical studies evaluating liver-directed AAV vectors expressing the coagulation FIX protein, in small and large animal models,2, 3, 4 prompted the first clinical trial of an AAV2 vector (expressing FIX from a hepatocyte-specific enhancer/promoter combination, AAV2-ApoE-hAAT-F9) delivered to human hepatocytes.5 A patient receiving a vector dose of 2 × 1012 vector genome (vg)/kg had transient therapeutic FIX expression levels that returned to baseline accompanied with a self-resolving transaminitis.5 It was hypothesized and later demonstrated that the activation of AAV2 capsid memory CD8+ T cells was the likely cause for the elimination of AAV2-transduced hepatocytes.5, 6, 7, 8 To avoid potential immunological responses to the AAV capsid, follow-up clinical studies have now adopted transient immune suppression with prednisolone, alternative capsid serotypes, and optimized F9 expression cassettes including codon optimization and a hyperactive FIX (R338L) variant.9, 10, 11, 12, 13, 14, 15, 16
Developing a Mouse Model for Capsid CD8+ T Cell Targeting of AAV-Transduced Hepatocytes
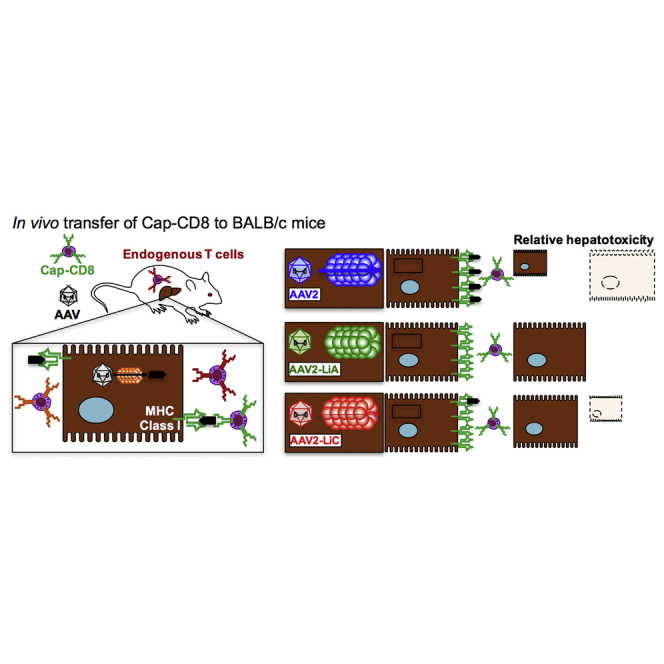
What was particularly frustrating for the AAV gene therapy field was that no preclinical studies predicted a limiting role of capsid-specific CD8+ T cells (Cap-CD8) in long-term FIX expression. Although an absence of Cap-CD8 responses in mice was anticipated because of no prior exposure to wild-type AAV,17 nonhuman primates are naturally infected with AAV and also failed to predict this outcome.18 Until recently, efforts directed toward mimicking the immunological response against AAV capsid in mouse models had been unsuccessful.19, 20, 21, 22, 23, 24 Studies conducted in mice with an AAV capsid engineered to express the CD8+ epitope for ovalbumin (ova) induced activated ova-specific CD8+ T cells, confirming that input AAV capsid was processed and presented in mice.25 Therefore, we hypothesized that previous attempts may have lacked specific and sufficient activation signals during capsid antigen exposure. To address this, we designed an immunization protocol with an in vivo prime and boost followed by in vitro expansion. In place of full-length capsid, we used the dominant capsid CD8+ T cell epitope,26 a CpG oligonucleotide activator of TLR9, and CD8+ T cell immune-stimulatory cytokines (interleukin-2 [IL-2], IL-15, and IL-21) to expand and activate Cap-CD8 followed by adoptive transfer into immune-deficient BALB/c-Rag1−/− mice.27 Although this model reflected the elimination of AAV-transduced hepatocytes, as observed in human clinical trials, immune-deficient mice come with a number of limitations. The following studies were designed to improve the utility of the model by transitioning into immune-competent mice. We observed a comparable elimination of AAV2-ApoE-hAAT-F9-transduced hepatocytes by Cap-CD8 and then proceeded to examine the immunogenicity of two engineered library-selected mouse hepatotropic AAV2 variants.
Results
Transitioning the Cap-CD8 Model into Immune-Competent Mice
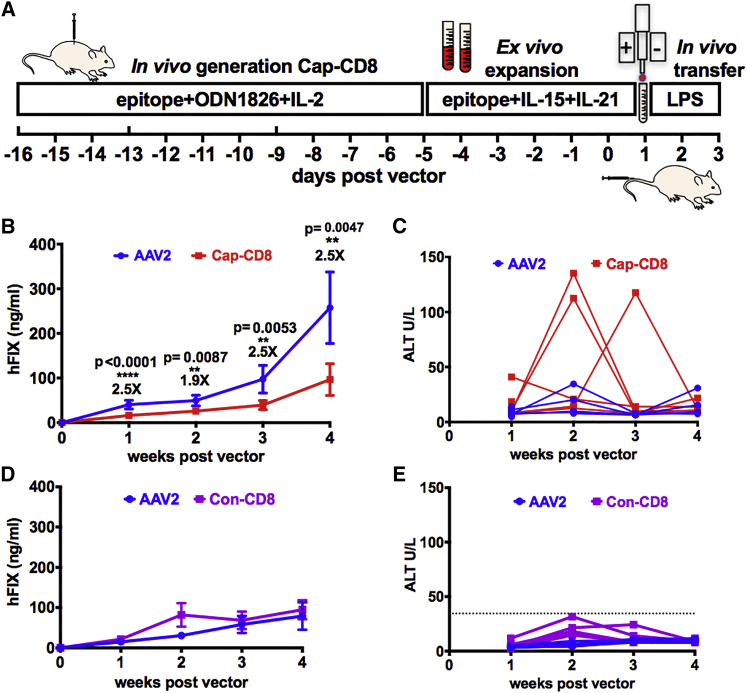
Our initial Cap-CD8 model was developed using immune-deficient Rag1−/− mice that lacked endogenous B and T cells,27 based on the speculation that a lymphopenic environment was required to support engraftment of adoptively transferred Cap-CD8. Yet, the use of an immune-deficient mouse model has inherent limitations. Therefore, we investigated whether the Cap-CD8 model would function in immune-competent mice. Using the CD90 (Thy-1) surface antigen, expressed as either CD90.1 or CD90.2 (Thy-1.1 or Thy-1.2) on T cells, allowed us to track adoptively transferred CD90.2-Cap-CD8 into congenic CD90.1-BALB/c mice (Figure 1A). For this study, we selected the AAV2-ApoE-hAAT-F9 vector at a dose of 1 × 1011 vg, where Cap-CD8 eliminated transduced hepatocytes and reduced circulating human FIX (hFIX) levels in Rag1−/− mice.27 Mice receiving Cap-CD8 compared with controls had a significant reduction in circulating hFIX levels at all time points (Figure 1B). Alanine aminotransferase (ALT) levels in plasma, a sensitive marker for liver damage, were measured at each time point (Figure 1C). Unlike the Rag1−/− mouse Cap-CD8 model in which ALT was elevated at 1 week post Cap-CD8 transfer, we observed an elevation in ALT (∼10-fold over normal levels of 9.3 ± 4.3 U/L) at 2 (in two of five mice) and 3 weeks (in one of five mice) following Cap-CD8 adoptive transfer. In all cases, ALT levels returned to baseline the following week. It is interesting to note that in the immune-competent Cap-CD8 model, the time course for transaminitis from Cap-CD8 targeting AAV2-transduced hepatocytes correlates better to that observed in a human patient. To confirm the specificity of Cap-CD8 elimination of AAV2-transduced hepatocytes in immune-competent mice, we performed a control study using an irrelevant AH1 gp70 epitope SPSYVYHQF to generate control CD8+ T cells (Con-CD8). Con-CD8 were induced, expanded, and adoptively transferred into BALB/c recipient mice following the same protocol as depicted in Figure 1A. Notably, there was no reduction in hFIX levels in Con-CD8 (Figure 1D, purple tracing) compared with AAV2 controls (Figure 1D, blue tracing) at all time points. Weekly monitoring of ALT levels failed to detect any elevation above what we have previously measured in AAV2-only-treated controls (Figures 1C and 1E). Overall, these data demonstrate that the reduction in hFIX levels was specific to Cap-CD8 transfer.
Figure 1.
Immune-Competent Mice Have a Significant Reduction in Circulating Levels of hFIX Protein after Cap-CD8 Adoptive Transfer in AAV2-F9-Treated Mice
(A) Cartoon of Cap-CD8 mouse model depicting in vivo generation and in vitro expansion of Con-CD8 and Cap-CD8 T cells followed by adoptive transfer of 2 × 106 Con-CD8 (SPSYVYHQF) cells into BALB/c or 2 × 106 Cap-CD8 (VPQYGYLTL) cells into BALB/c-CD90.1 mice receiving an AAV vector on day 0 and treated with 10 ng of LPS intraperitoneal (IP) on days 1, 2, and 3 post vector. (B) Systemic levels of hFIX protein were measured from BALB/c-CD90.1 mice injected intravenously with 1 × 1011 vg of an AAV2-ApoE-hAAT-F9 vector with adoptive transfer of Cap-CD8 (red) or controls (blue) from plasma weekly over a period of 4 weeks (n = 4–10 per group). Initial group sizes were n = 9 for AAV2 and n = 10 for Cap-CD8, and at 1 week following administration of Cap-CD8, livers were collected from five control and experimental mice. Group sizes for weeks 2–4 were n = 4 for AAV2 and n = 5 for Cap-CD8. (C) Liver function tests measuring plasma ALT were performed weekly over a period of 4 weeks following Cap-CD8 adoptive transfer. Each line represents an individual mouse over time. (D) Systemic levels of hFIX protein were measured from BALB/c mice injected intravenously with 1 × 1011 vg of an AAV2-ApoE-hAAT-F9 vector with adoptive transfer of Con-CD8 (purple) or controls (blue) from plasma weekly over a period of 4 weeks (n = 6 per group). (E) Liver function tests measuring plasma ALT were performed weekly over a period of 4 weeks following Con-CD8 adoptive transfer. Each line represents an individual mouse over time. Dotted line represents peak ALT detected in AAV2 control mouse in (C). Unpaired t tests with two-tailed analysis were used to determine significance of (B). Multiple t tests with correction for multiple comparisons using the Holm-Sidak method were used in analysis of data in (D). (B and D) Error bars indicate mean ± SD.
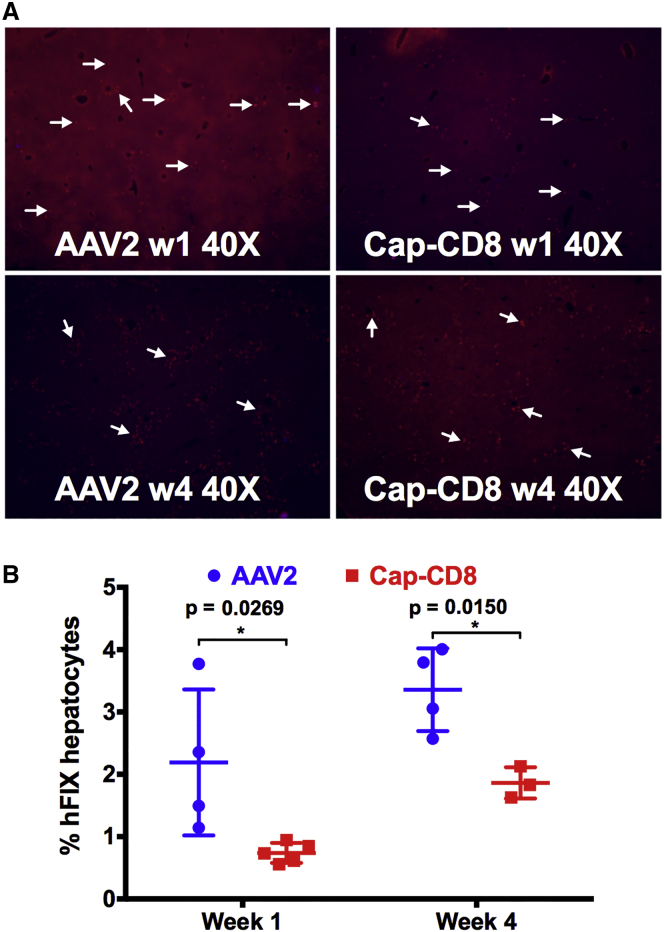
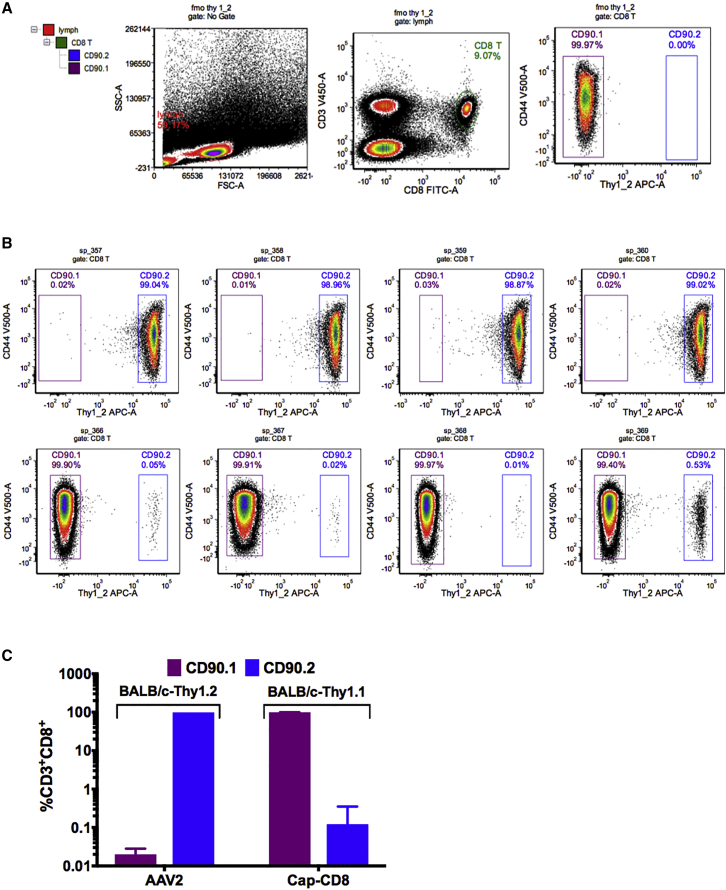
Liver tissue was collected from mice in each group at 1 and 4 weeks after gene transfer, and cryosections were stained using antibodies against hFIX (red) and CD90.2+ T cells (green). Representative liver sections at 40× magnification from Cap-CD8 and control mice at 1 and 4 weeks post gene transfer are shown in Figure 2A. The percent of hFIX+-expressing hepatocytes was averaged from multiple images at 200× magnification using Volocity software. Mice receiving Cap-CD8 had a significant reduction in hFIX+ hepatocytes at both 1 and 4 weeks post gene transfer (Figure 2B), reflecting the reduction in circulating hFIX in plasma (Figure 1B). We were unable to detect infiltrating CD90.2+ cells in livers collected from Cap-CD8 mice at either 1 or 4 weeks, in contrast with our previous studies in immune-deficient mice,27 although it should be noted that Cap-CD8 were sparse in liver tissue. One likely explanation is that the absence of lymphocytes in the livers of Rag1−/− mice provides a niche for long-term residence of Cap-CD8 cells, whereas this is likely filled by endogenous CD8+ T cells in immune-competent mice. Nonetheless, flow cytometry analysis performed on splenocytes isolated from controls (CD90.2+-BALB/c mice) and Cap-CD8 adoptive transfer mice (CD90.1+-BALB/c mice) at week 4 revealed a small, but distinct fraction of CD3+ CD8+ CD90.2+ T cells in Cap-CD8-treated mice (Figure 3). Thus, the loss in both circulating and hepatocyte hFIX expression, elevation in ALT, and flow cytometry analysis demonstrates that a lymphopenic host is not required for the short-term persistence and functionality of adoptively transferred Cap-CD8.
Figure 2.
Cap-CD8 Adoptive Transfer in Immune-Competent Mice Results in a Significant Reduction in hFIX-Expressing Hepatocytes at 1 and 4 Weeks Post AAV2-ApoE-hAAT-F9 Delivery
(A) Representative liver sections collected at weeks 1 and 4 from control and Cap-CD8 mice injected intravenously with 1 × 1011 vg of AAV2-ApoE-hAAT-F9 and stained for hFIX (red) and CD90.2 (green) at 40× magnification. (B) Quantification of the % hFIX-expressing hepatocytes in Cap-CD8 adoptive transfer and control mice livers sections was performed on at least three random 200× visual fields per mouse (n = 3–5 per group) at weeks 1 and 4 after Cap-CD8. Group sizes for AAV2 were n = 4 for weeks 1 and 4 and for Cap-CD8 were n = 5 for week 1 and n = 3 for week 4. Unpaired t tests with two-tailed analysis were used to determine significance in (B). Error bars indicate mean ± SD.
Figure 3.
Cap-CD8 Persist in Immune-Competent Mice up to 4 Weeks after Adoptive Transfer
Congenic BALB/c mice, CBy.PL(B6)-Thy1a/ScrJ, were used, in which their T cells express the CD90.1 (Thy1.1) surface antigen to track adoptively transferred Cap-CD8 generated in CD90.2 (Thy1.2) donor mice. (A) Gating strategy of antibody-labeled splenocytes stained with anti-mouse CD3-BV421, B220-BV605, CD8-A488, and CD90.2-APC. CD90.2+ CD8+ T cell gating was determined using a fluorescence minus one (-CD90.2-APC)-stained sample. (B) Representative dot plots of stained splenocytes from four BALB/c-CD90.1 (CD90.2-Cap-CD8) and BALB/c-CD90.2 control mice at 4 weeks following Cap-CD8 adoptive transfer. Left box gate: purple for CD90.1+; right box gate: blue for CD90.2+ CD3+CD8+ T cells. (C) Graphical quantitation of % CD3+CD8+CD90.2− and CD3+CD8+CD90.2+ as determined by flow cytometry. Group sizes were n = 4 for AAV2 and n = 5 for Cap-CD8. Error bars indicate mean ± SD.
AAV2 Capsid Variants Selected from a Rationally Designed Capsid Library Screen for Murine Liver Tropism Have Altered Antigen Presentation
Rationally designed mutations of AAV2 with targeted substitutions of surface-exposed tyrosine residues with phenylalanine at positions 444, 500, and 730 [AAV2 (Y-F)] disrupted capsid polyubiquitination and shifted the trafficking of viral particles from the proteasome to the nucleus.28, 29 Together, these (Y-F) substitutions substantially improved vector transduction efficiency29 and reduced capsid antigen processing and presentation on hepatocyte major histocompatibility complex (MHC) class I molecules.27 We have described the design of an AAV2 capsid library incorporating two of the (Y-F) substitutions at positions 444 and 500, and following three rounds of in vivo selection on mouse liver identified two AAV2 variants, AAV2-LiA and AAV2-LiC, with equivalent transduction efficiency to AAV8.30 Sequencing of the variants revealed that AAV2-LiA had 14 aa substitutions (including Y444F and Y500F), whereas AAV2-LiC had 4 aa substitutions (including Y500F) compared with wild-type AAV2. Because our previous study focused on AAV2 (Y-F) avoidance of Cap-CD8,27 it was unclear what the relative contribution of each (Y-F) substitution had on capsid antigen presentation. Therefore, in addition to testing the relative capsid immunogenicity of two new variants with comparable transduction efficiency to AAV8, we were also able to address the requirement of (Y-F)-730 and multiple (Y-F) substitutions for attenuating capsid antigen presentation.
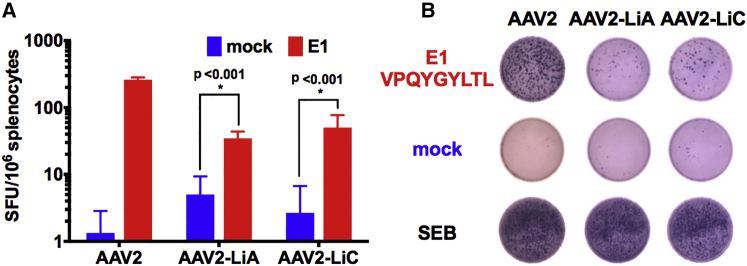
To rule out the potential generation of new Ld-restricted CD8+ T cell epitopes for AAV2-LiA and AAV2-LiC in BALB/c mice (H-2d haplotype), we performed in silico prediction of MHC class I capsid epitopes. The VPQYGYLTL epitope was similarly ranked for wild AAV2, AAV2-LiA, and AAV2-LiC capsids, suggesting that this dominant epitope may be conserved in AAV2-LiA and AAV2-LiC (Table 1). An interferon gamma (IFNγ) ELISpot assay conducted on splenocytes collected from BALB/c mice receiving an intramuscular injection of either AAV2 (n = 2), AAV2-LiA (n = 3), or AAV2-LiC (n = 3) vectors all showed an increase in IFNγ spot-forming units following stimulation with VPQYGYLTL, demonstrating that this epitope is still processed and presented by both variants (Figure 4).
Table 1.
Comparison of High to Low Binding H2Ld Predicted Epitopes Using In Silico Prediction
| AAV2 |
AAV2-LiA |
AAV2-LiC |
||||||
|---|---|---|---|---|---|---|---|---|
| Start | End | Peptide | Start | End | Peptide | Start | End | Peptide |
| 534 | 542 | FPQSGVLIF | 534 | 542 | FPQSGVLIF | 534 | 542 | FPQSGVLIF |
| 631 | 639 | SPLMGGFGL | 631 | 639 | SPLMGGFGL | 631 | 639 | SPLMGGFGL |
| 363 | 371 | PPFPADVFM | 363 | 371 | PPFPADVFM | 363 | 371 | PPFPADVFM |
| 372 | 380 | VPQYGYLTLa | 372 | 380 | VPQYGYLTLa | 372 | 380 | VPQYGYLTLa |
| 307 | 315 | RPKRLNFKL | 307 | 315 | RPKRLNFKL | 502 | 510 | WPGATTYHLa |
| 165 | 173 | QPARKRLNF | 165 | 173 | QPARKRLNF | 307 | 315 | RPKRLNFKL |
| 621 | 629 | IPHTDGHFH | 621 | 629 | IPHTDGHFH | 165 | 173 | QPARKRLNF |
| 437 | 445 | LIDQYLYYL | 437 | 445 | LIDQYLYFL | 621 | 629 | IPHTDGHFH |
| 651 | 659 | TPVPANPST | 651 | 659 | TPVPANPST | 437 | 445 | LIDQYLYYL |
| 724 | 732 | RPIGTRYLT | 724 | 732 | RPIGTRYLT | 651 | 659 | TPVPANPST |
| 45 | 53 | LVLPGYKYL | 45 | 53 | LVLPGYKYL | 724 | 732 | RPIGTRYLT |
| 653 | 661 | VPANPSTTF | 653 | 661 | VPANPSTTF | 45 | 53 | LVLPGYKYL |
| 276 | 284 | STPWGYFDF | 276 | 284 | STPWGYFDF | 653 | 661 | VPANPSTTF |
| 310 | 318 | RLNFKLFNI | 310 | 318 | RLNFKLFNI | 276 | 284 | STPWGYFDF |
| 190 | 198 | QPPAAPSGL | 190 | 198 | QPPAAPSGL | 310 | 318 | RLNFKLFNI |
| 515 | 523 | SLVNPGPAM | 515 | 523 | SLVNPGPAM | 190 | 198 | QPPAAPSGL |
| 639 | 647 | LKHPPPQIL | 639 | 647 | LKHPPPQIL | 515 | 523 | SLVNPGPAM |
| 407 | 415 | NNFTFSYTF | 407 | 415 | NNFTFSYTF | 639 | 647 | LKHPPPQIL |
| 462 | 470 | FSQAGASDI | 462 | 470 | FSQAGASDI | 407 | 415 | NNFTFSYTF |
| 357 | 365 | AHQGCLPPF | 357 | 365 | AHQGCLPPF | 462 | 470 | FSQAGASDI |
| 346 | 354 | SEYQLPYVL | 346 | 354 | SEYQLPYVL | 357 | 365 | AHQGCLPPF |
| 186 | 194 | QPLGQPPAA | 186 | 194 | QPLGQPPAA | 346 | 354 | SEYQLPYVL |
| 403 | 411 | LRTGNNFTF | 403 | 411 | LRTGNNFTF | 186 | 194 | QPLGQPPAA |
| 370 | 378 | FMVPQYGYL | 370 | 378 | FMVPQYGYL | 403 | 411 | LRTGNNFTF |
| 502 | 510 | WTGATKYHL | 502 | 510 | WTGATKYHL | 370 | 378 | FMVPQYGYL |
| 362 | 370 | LPPFPADVF | 362 | 370 | LPPFPADVF | 362 | 370 | LPPFPADVF |
| 435 | 443 | NPLIDQYLY | 435 | 443 | NPLIDQYLY | 435 | 443 | NPLIDQYLY |
| 508 | 516 | YHLNGRDSL | 508 | 516 | YHLNGRDSL | 508 | 516 | YHLNGRDSL |
| 401 | 409 | QMLRTGNNF | 401 | 409 | QMLRTGNNF | 401 | 409 | QMLRTGNNF |
| 7 | 15 | LPDWLEDTL | 7 | 15 | LPDWLEDTL | 7 | 15 | LPDWLEDTL |
| 298 | 306 | RLINNNWGF | 298 | 306 | RLINNNWGF | 298 | 306 | RLINNNWGF |
| 629 | 637 | HPSPLMGGF | 629 | 637 | HPSPLMGGF | 629 | 637 | HPSPLMGGF |
| 704 | 712 | YNKSVNVDF | 704 | 712 | YNKSVNVDF | 704 | 712 | YNKSVNVDF |
The dominant epitope used in this study is in bold. The new predicted epitope for AAV-LiC is underlined.
Figure 4.
AAV2 Capsids Variant AAV2-LiA and AAV2-LiC, Selected for Enhanced Murine Hepatocyte Tropism from an AAV2 Capsid Library, Retain the Dominant MHC Class I Epitope VPQYGYLTL of Wild-Type AAV2
(A) Frequency of interferon gamma (IFNγ)-secreting cells per 1 × 106 cells upon in vitro stimulation of splenocytes isolated from BALB/c mice (n = 2–3 per vector) 7 days following intramuscular (IM) injection of 1 × 1011 vg of AAV2 (n = 2), AAV2-LiA (n = 3), or AAV2-LiC (n = 3) vectors. Cells were stimulated in triplicate with media alone (blue) or with media containing the dominant CD8+ T cell epitope VPQYGYLTL, designated E1 (red). As an additional control, all splenocytes were stimulated with the bacterial superantigen SEB. The data are presented as an average ± SD. (B) Representative ELISpot wells for each vector AAV2, AAV2-LiA, and AAV2-LiC (left to right) with either VPQYGYLTL (E1), mock, or SEB stimulation (top to bottom). Multiple t tests with correction for multiple comparisons using the Holm-Sidak method were used in analysis of data in (A).
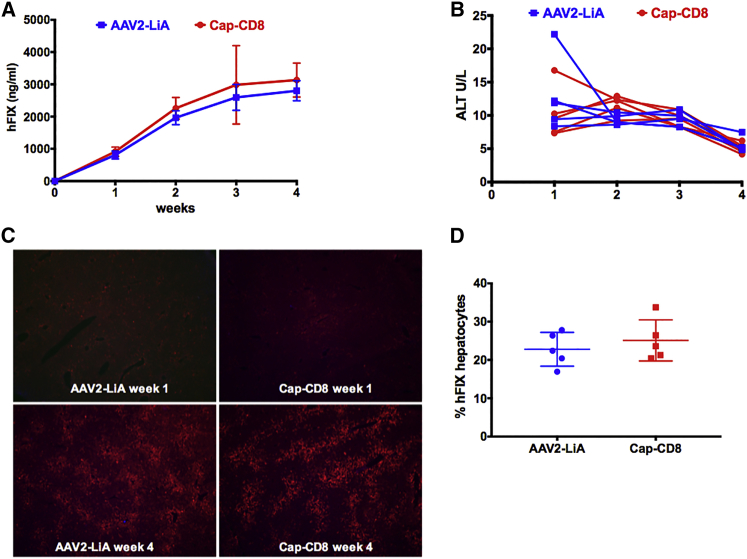
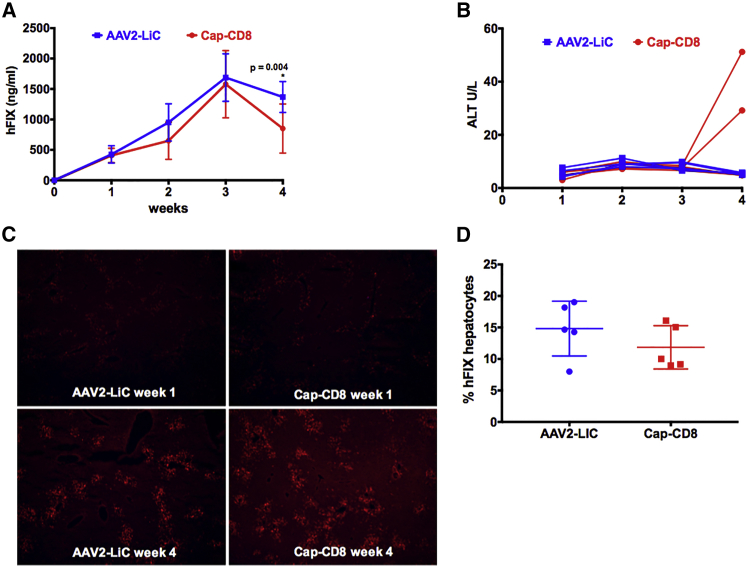
AAV2-LiA and AAV2-LiC ApoE-hAAT-F9 vectors were evaluated at a dose of 1 × 1011 vg in the Cap-CD8 immune-competent mouse model. Mice transduced with either AAV2-LiA or AAV-LiC vectors expressed elevated levels of hFIX protein as compared with AAV2 (Figures 5A and 6A). Yet, contrary to wild-type AAV2 (Figure 1B), no initial loss in hFIX expression was observed in Cap-CD8-treated mice compared with controls (Figures 5A and 6A). At study endpoint, a small but significant decrease in circulating hFIX levels was observed in AAV2-LiC-injected, but not AAV2-LiA-injected, animals. Plasma ALT levels were unchanged over time in AAV2-LiA mice receiving Cap-CD8 (Figure 5B), whereas a rise in ALT levels (∼5-fold over normal levels of 9.3 ± 4.3 U/L) was detected in two of five AAV2-LiC mice receiving Cap-CD8 at 4 weeks (Figure 6B). Liver tissue from each group was collected at 1 and 4 weeks post vector administration, and immunostaining for hFIX (red) and CD90.2+ T cells (green) was performed on liver sections from control and Cap-CD8 mice. Representative sections at 40× magnification from each vector (AAV2-LiA and AAV2-LiC) and group at 1 and 4 weeks are provided in Figures 5C and 6C, respectively. Despite a moderate decrease in circulating hFIX protein detected in AAV2-LiC Cap-CD8 mice, quantitation of hFIX+ hepatocytes at 4 weeks did not reveal a significant decrease in hFIX+ hepatocytes in Cap-CD8 compared with control mice in both AAV2-LiA- and AAV2-LiC-treated mice (Figures 5D and 6D). As with AAV2-injected mice, no CD90.2+ Cap-CD8 cell infiltration was observed in liver sections, and flow cytometry staining of splenocytes at 4 weeks did not detect CD90.2+ Cap-CD8 in either AAV2-LiA or AAV2-LiC Cap-CD8 mice (data not shown).
Figure 5.
AAV2-LiA Mutant Capsid Avoids Elimination by Cap-CD8 in Immune-Competent Mice
Control and Cap-CD8 congenic CD90.1 mice were injected intravenously with 1 × 1011 vg of an AAV2-LiA-ApoE-hAAT-F9 vector, and experimental mice were given 2 × 106 Cap-CD8 cells the following day. (A) Systemic levels of hFIX protein were measured from plasma weekly over a period of 4 weeks (n = 5–8 per group). Starting group sizes for AAV2-LiA and Cap-CD8 were n = 8. For weeks 2–4, group sizes for AAV2-LiA and Cap-CD8 were n = 5. One and 4 weeks following administration of Cap-CD8 to AAV2-LiA mice, livers were collected from control and Cap-CD8 mice. (B) Liver function tests measuring plasma ALT were performed weekly over a period of 4 weeks following Cap-CD8 adoptive transfer. Each line represents an individual mouse over time. (C) Representative liver sections at 1 and 4 weeks from control and Cap-CD8 mice stained for hFIX (red) and CD90.2 (green) at 40× magnification. (D) Quantification of the % hFIX-expressing hepatocytes (AAV2-LiA) was performed on controls and Cap-CD8 liver sections on at least three random 200× visual fields per mouse collected 4 weeks post vector at study endpoint (n = 5 per group). Multiple t tests with correction for multiple comparisons using the Holm-Sidak method were used in analysis of data in (A) and (D). (A and D) Error bars indicate mean ± SD.
Figure 6.
AAV2-LiC Mutant Capsid Has a Delayed Elimination of Vector-Transduced Hepatocytes by Cap-CD8 in Immune-Competent Mice
Control and Cap-CD8 congenic CD90.1 mice were injected intravenously with 1 × 1011 vg of an AAV2-LiC-ApoE-hAAT-F9 vector, and experimental mice were given 2 × 106 Cap-CD8 cells the following day. (A) Systemic levels of hFIX protein were measured from plasma weekly over a period of 4 weeks (n = 5–8 per group). Starting group sizes for AAV2-LiA and Cap-CD8 were n = 8. For weeks 2–4, group sizes for AAV2-LiA and Cap-CD8 were n = 5. One and 4 weeks following administration of Cap-CD8 to AAV2-LiC mice, livers were collected from control and Cap-CD8 mice. (B) Liver function tests measuring plasma ALT were performed weekly over a period of 4 weeks following Cap-CD8 adoptive transfer. Each line represents an individual mouse over time. (C) Representative liver sections at 1 and 4 weeks from control and Cap-CD8 mice stained for hFIX (red) and CD90.2 (green) at 40× magnification. (D) Quantification of the % hFIX-expressing hepatocytes (AAV2-LiC) was performed on controls and Cap-CD8 liver sections on at least three random 200× visual fields per mouse collected 4 weeks post vector at study endpoint (n = 5 per group). Multiple t tests with correction for multiple comparisons using the Holm-Sidak method were used in analysis of data in (A) and (D).
Discussion
Clinical Studies Show that Vector Dose Strongly Correlates with Risk of Capsid Immunity
No animal model predicted that anti-AAV immunity would limit FIX expression levels in human clinical trials. Data from three independent clinical studies evaluating different AAV-based gene therapy vectors for hemophilia B now show that cell-mediated immune responses targeting transduced hepatocytes presenting AAV capsid-derived epitopes has compromised FIX expression in some patients with a strong correlation to administered vector dose.5, 10, 31, 32 Thus, we developed the first animal model of capsid-reactive CD8+ T cells directed against a native capsid epitope that specifically targeted AAV-ApoE-hAAT-F9-transduced hepatocytes.27
Advantages and Limitations of the Two Cap-CD8 Models
In the initial design of our Cap-CD8 model, several factors were considered in the selection of immune-deficient mice. Adoptive transfer of lymphocytes into immunodeficient mice has aided in defining the subpopulations of lymphocytes involved in protective immunity33, 34 and autoimmune disease.35, 36 Engraftment and persistence of adoptively transferred T cells is enhanced by lymphodepletion,37, 38 and the lack of endogenous effector CD8+ T cells reduces competition for cytokines and survival signals, whereas absence of CD4+CD25+FoxP3+ regulatory T cells eliminates the risk of bystander suppression. The success of our model was likely due to both in vivo activation and in vitro expansion of Cap-CD8 using the dominant Ld-restricted VPQYGYLTL capsid epitope, as opposed to previous approaches that used an immunogenic virus (adenovirus) and delivery route (intramuscular) to express the full-length capsid.23 With this model, we have addressed the relative risk of cell-mediated immunity directed against AAV-transduced mouse hepatocytes in terms of serotype, novel capsid mutants (Figures 5 and 6), and drugs, such as proteasome inhibitors, that interfere with MHC class I presentation.27 However, the use of an immune-deficient host does not reflect the clinical experience and has several limitations. Expanding the Cap-CD8 model to immune-competent mice may now provide an opportunity to address some of these limitations.
In the present study, we observed a reduction in the fold change in hFIX levels and the timing and magnitude of elevated ALT levels in Cap-CD8 immune-competent mice compared with immune-deficient mice.27 These changes are likely mediated by both inherent biological differences and technical differences in sample analysis for ALT. The adoptive transfer of effector T cells into immune-deficient mice has become a standard model for studying inflammatory bowel disease.39 In contrast with genetic knockout models, adoptive transfer of effector T cells into an immune-deficient host induces synchronized disease,40 similar to what we have observed in our two different Cap-CD8 models. Additionally, endogenous lymphocytes may directly or indirectly impact Cap-CD8 engraftment, activity, and migration into the liver. This is in part supported by our data showing an asynchronous elevation in ALT levels in Cap-CD8- and AAV2-treated immune-competent mice (Figure 1C, red tracings) and difficulties with detecting infiltrating Cap-CD8 in the liver. And although the overall ALT levels were not as high as previously reported in immune-deficient mice, the fold increase over normal ALT levels was comparable with or greater than what we have previously reported in immune-deficient mice. Further, we have found that AAV murine liver gene transfer efficiency differs between immune-deficient and -competent mice.27, 41 Taken together, the asynchronous response of Cap-CD8 and altered transduction efficiency likely both contribute to the lower reduction in hFIX levels seen at earlier time points. Note that at 4 weeks we observed a 3-fold reduction in hFIX in the immune-deficient model, which is comparable with the 2.5-fold reduction in the immune-competent model (Figure 1B).
Blending Rational Design with Library Diversity Generates a Superior AAV Vector
Capsid libraries provide a high starting level of complexity and allow for the identification of capsid variants that meet a user-defined selection criterion.30, 42, 43, 44, 45 Taking a rationalized approach, we generated a highly complex AAV2-based capsid library selectively substituting naturally occurring amino acids in conserved surface-exposed loops, as well as including targeted (Y-F) substitutions at positions 444 and 500.28, 29, 30 AAV2-LiA and AAV2-LiC ApoE-hAAT-F9 vector-injected mice given Cap-CD8 did not initially lose hFIX expression, yet at study endpoint, AAV2-LiC-injected mice had a moderate, but significant drop in circulating hFIX levels. It is possible that alternative Ld-restricted dominant capsid epitopes may exist for AAV2-LiA or AAV2-LiC, such as WPGATTYHL for AAV2-LiC (Table 1), to explain the observed delay in loss of hFIX levels and ALT elevation. Follow-up studies are planned to address whether this alternative AAV2-LiC epitope is presented on transduced hepatocytes. Surprisingly, a new epitope was predicted for AAV2-LiC that differs from wild-type AAV2 by only 4 aa, whereas AAV2-LiA, which differs by 14 aa, shares the top 30 ranked epitopes with wild-type AAV2. We have previously demonstrated an additive effect of combining AAV2 (Y-F) mutations at positions 444, 500, and 730 on transgene expression, but it was unclear whether this impacted capsid antigen presentation.27, 28, 29 Here, we show that (Y-F)-730, the best-performing single (Y-F) substitution,28 does not appear to be critical for reducing capsid antigen presentation, although our data do support an additive effect of having at least two (Y-F) substitutions. Some caution is justified in this observation because it is unclear how the other amino acid substitutions in AAV2-LiA and AAV2-LiC may impact capsid processing and presentation. Follow-up studies with AAV2 (Y-F)-444 and -500 and AAV2 (Y-F)-500 capsid variants will aid in addressing this. Another interesting observation, overlooked with whole-body imaging in our original description of AAV2-LiA and AAV2-LiC,30 is that despite the higher levels of hFIX expression, the transduction profile in murine liver still resembles that of wild-type AAV2, with clustering of transduced hepatocytes around vessels (Figures 5C and 6C). How these variants provide for substantially higher levels of transgene expression, with transgene expression restricted to approximately 25% of total hepatocytes, will be addressed in future studies.
Future Directions of the Cap-CD8 Model
Although conservation of MHC class I-restricted capsid epitopes has been observed in humans,46 this study was limited to just three serotypes, and it is unclear whether this will be true for other naturally occurring serotypes, as well as library-selected and engineered capsid serotypes. To improve the flexibility of the model, we plan to optimize Cap-CD8 induction using full-length VP1 in place of the dominant epitope. Recently, there has been increased interest in identifying capsid serotypes with an improved affinity for human hepatocytes.41, 42 Such serotypes may have limited affinity for murine hepatocytes and pose a challenge to evaluate capsid immunity in our current model. A potential approach to address this is the use of chimeric immune-deficient mice reconstituted with human hepatocytes41 and a human immune system.47 A major difference between our model and human patients is that in humans, AAV gene delivery likely triggers the activation and expansion of capsid-specific memory CD8+ T cells. Using immune-competent mice, it is now possible to mimic this scenario, allowing induction of Cap-CD8 and subsequent AAV vector delivery into the same animal. We have recently identified an important link between innate sensing of viral DNA and capsid antigen presentation in professional antigen-presenting cells (plasmacytoid dendritic cells and conventional dendritic cells) in mice,48 which will aid in the design of an effective immunization protocol. Induction of an immunological memory response to capsid will also provide an opportunity to test and optimize prophylactic immune-suppressive (IS) protocols.
Each of our Cap-CD8 models has its respective strengths and limitations, briefly discussed above. Depending on the specific question to answer, one can select the appropriate system to use. The present study revealed a variable delay in Cap-CD8 elimination of AAV2-transduced hepatocytes, as measured by elevation in plasma ALT, compared with Rag 1−/− mice. Because this has been observed only with a single serotype, we do not have enough data to support this holding as true for other serotypes. However, it would be important that investigators empirically determine optimal follow-up times in each model when studying different AAV serotypes. Although no model is perfect, these mouse models provide a unique opportunity to assess the potential risks of cell-mediated capsid immunity in terms of serotype, vector dose, transgene, and route of administration in vivo. Understanding these potential risks will help to improve clinical trial design and the chances for achieving sustained therapeutic transgene expression in patients.
Materials and Methods
This study was approved under University of Florida Institutional Animal Care and Use Committee protocols 201307941 and 201503182.
AAV Vectors
AAV vectors expressing hFIX from the liver-specific ApoE enhancer and α1-antitrypsin promoter were packaged into AAV2, AAV2-LiA, or AAV2-LiC capsids as previously published.27 Vectors were administered by tail-vein injection at 1 × 1011 vg.
Generation of Murine Control CD8+ T Cells and Capsid-Specific CD8+ T Cells
Con-CD8 and Cap-CD8 T cells were generated as previously published27 with the following modifications. Male BALB/c mice (6–8 weeks old; Jackson Laboratory) received three subcutaneous injections every 4 days consisting of 1,000 IU mouse IL-2 (R&D Systems), 25 μg of TLR9 activator ODN 1826 (InvivoGen), and 10 μg of the control AH1 gp70 epitope SPSYVYHQF or dominant capsid CD8+ T cell epitope VPQYGYLTL emulsified in Sigma Adjuvant System (Sigma). Splenocytes were isolated 12 days following the first immunization, and Cap-CD8 were expanded in vitro. A total of 2 × 107 splenocytes was cultured in 6 mL of stimulation media over a 6-day period at 37°C in 15-mL Falcon tubes (BD Biosciences) with media exchange on day 3. Stimulation medium was RPMI-1640 (Invitrogen) supplemented with 10% fetal bovine serum, 1% penicillin and streptomycin, 10 μg/mL SPSYVYHQF or VPQYGYLTL, 25 ng/mL IL-15, and 10 ng/mL IL-21. Following in vitro expansion, CD8+ T cells were magnetically purified using a mouse CD8− selection kit (Miltenyi) and used for in vivo studies.
In Silico Prediction of MHC Class I Capsid Epitopes
The MHC class I binding predictions were made on April 15, 2015, using the IEDB analysis resource consensus tool,49 which combines predictions from artificial neural network (ANN),50, 51 stabilized matrix method (SMM),52 and comblib.53
Antibodies for Flow Cytometry
Purified anti-CD16/32 (Fc Block) and anti-CD8 (A488) antibodies were purchased from BD Biosciences; anti-CD3 (BV421) and anti-CD90.2 (antigen presenting cell [APC]) antibodies were purchased from BioLegend.
IFNγ ELISpot
Splenocytes were isolated 7 days following an intramuscular injection of 1 × 1011 vg of AAV2, AAV2-LiA, or AAV2-LiC vectors and cultured for ELISpot as previously published.27, 54 Stimulation media was supplemented with 5 μg/mL of the dominant AAV2 MHC class I epitope (VPQYGYLTL). Stimulation of splenocytes with staphylococcal enterotoxin B (SEB) was used as a positive control. IFNγ-producing cells were quantitated using the CTL-ImmunoSpot system.
In Vivo Murine Cap-CD8 T Cell Model for the Elimination of AAV-Transduced Hepatocytes
CBy.PL(B6)-Thy1a/ScrJ (005443) mice (BALB/c-Thy1.1 male, 6–8 weeks old) were purchased from Jackson Laboratories. AAV vectors (1 × 1011 vg) and Cap-CD8 cells (2 × 106 cells per mouse) were delivered by tail-vein injection into BALB/c congenic CBy.PL(B6)-Thy1a/ScrJ (005443) mice (BALB/c-CD90.1). Mice received Cap-CD8 cells 1 day post AAV gene delivery. In order to augment innate activation signals in AAV-transduced hepatocytes and upregulate MHC class I expression, mice received three intraperitoneal injections of 10 ng of lipopolysaccharide (LPS; InvivoGen) 30 min prior to and 24 and 48 hr following cap-CD8 adoptive transfer. Plasma was isolated from weekly retro-orbital bleeds into heparinized capillaries for measuring circulating FIX protein levels and ALT levels. Liver tissue was collected 7 or 28 days after Cap-CD8 transfer and flash frozen in liquid nitrogen using Optimal Cutting Temperature tissue-freezing medium.
Measurement of Plasma FIX and ALT
Plasma levels of FIX protein were measured by ELISA as previously published.27 Plasma ALT levels were determined by an ALT Activity Assay (MAK052; Sigma) per manufacturer’s instructions.
Immunostaining of Liver Sections
Immunostaining for hFIX, mouse CD8 T cells, and mouse CD90.2 T cells was performed on liver cryosections with the following antibodies: goat anti-hFIX (Affinity Biologicals), rat anti-mouse CD8 (BD Pharmingen), rat anti-mouse CD90.2 (eBioscience), donkey anti-goat IgG-Alexa 568, and donkey anti-rat IgG-Alexa 488. Images were captured with a Nikon Eclipse 80i fluorescence microscope (Nikon) and Retiga 2000R digital camera (QImaging). The percent of hFIX-expressing murine hepatocytes in liver sections was determined with Volocity software (Perkin Elmer). Images were post-processed with Volocity 3D Image Analysis Software v6.3 (Perkin-Elmer). Quantitation of hFIX-stained hepatocytes was performed using multiple liver sections, and multiple images per section were captured at 200× per animal. Statistical analysis and graphical presentation were carried out with GraphPad Prism 7 (GraphPad Software).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7 software using multiple t tests with correction for multiple comparisons using the Holm-Sidak method with two-tailed analysis or unpaired t tests with two-tailed analysis when group sizes were not matched.
Author Contributions
B.P. and D.M.M. performed experiments. D.M.M. and R.W.H. designed experiments. B.P. and D.M.M. interpreted the data. D.M. and S.Z. provided reagents. D.M.M., R.W.H., and S.Z. supervised and coordinated the study. D.M.M., B.P., D.M., R.W.H., and S.Z. wrote the manuscript.
Conflicts of Interest
R.W.H. has been receiving royalty payments from Genzyme Corp. for license of AAV-FIX technology.
Acknowledgments
This work was supported by grants from the NIH (grant R01 HL097088 to R.W.H. and S.Z.), the Bayer Hemophilia Awards Program (D.M.M.), and the Children’s Miracle Network-UF Department of Pediatrics (B.P. and D.M.).
References
- 1.Zaiss A.K., Liu Q., Bowen G.P., Wong N.C., Bartlett J.S., Muruve D.A. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 2002;76:4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mingozzi F., Liu Y.L., Dobrzynski E., Kaufhold A., Liu J.H., Wang Y., Arruda V.R., High K.A., Herzog R.W. Induction of immune tolerance to coagulation factor IX antigen by in vivo hepatic gene transfer. J. Clin. Invest. 2003;111:1347–1356. doi: 10.1172/JCI16887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mount J.D., Herzog R.W., Tillson D.M., Goodman S.A., Robinson N., McCleland M.L., Bellinger D., Nichols T.C., Arruda V.R., Lothrop C.D., Jr., High K.A. Sustained phenotypic correction of hemophilia B dogs with a factor IX null mutation by liver-directed gene therapy. Blood. 2002;99:2670–2676. doi: 10.1182/blood.v99.8.2670. [DOI] [PubMed] [Google Scholar]
- 4.Rogers G.L., Herzog R.W. Gene therapy for hemophilia. Front. Biosci. (Landmark Ed.) 2015;20:556–603. doi: 10.2741/4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 6.Mingozzi F., Maus M.V., Hui D.J., Sabatino D.E., Murphy S.L., Rasko J.E., Ragni M.V., Manno C.S., Sommer J., Jiang H. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 7.Pien G.C., Basner-Tschakarjan E., Hui D.J., Mentlik A.N., Finn J.D., Hasbrouck N.C., Zhou S., Murphy S.L., Maus M.V., Mingozzi F. Capsid antigen presentation flags human hepatocytes for destruction after transduction by adeno-associated viral vectors. J. Clin. Invest. 2009;119:1688–1695. doi: 10.1172/JCI36891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn J.D., Hui D., Downey H.D., Dunn D., Pien G.C., Mingozzi F., Zhou S., High K.A. Proteasome inhibitors decrease AAV2 capsid derived peptide epitope presentation on MHC class I following transduction. Mol. Ther. 2010;18:135–142. doi: 10.1038/mt.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathwani A.C., Rosales C., McIntosh J., Rastegarlari G., Nathwani D., Raj D., Nawathe S., Waddington S.N., Bronson R., Jackson S. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 2011;19:876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monahan P., Walsh C.E., Powell J.S., Konkle B.A., Josephson N.C., Escobar M., McPhee S.J., Litchev B., Cecerle M., Ewenstein B.M. Update on a phase 1/2 open-label trial of BAX335, an adeno-associated virus 8 (AAV8) vector-based gene therapy program for hemophilia B. J. Thromb. Haemost. 2015;13:87. [Google Scholar]
- 11.UniQure Biopharma BV, and Chiesi Farmaceutici, SpA. Trial of AAV5-hFIX in severe or moderately severe hemophilia B. National Library of Medicine. 2015, NCT02396342. https://clinicaltrials.gov/show/NCT02396342.
- 12.Spark Therapeutics, Children’s Hospital of Philadelphia, University of Pittsburgh, Royal Prince Alfred Hospital, and St. James’s Hospital. Hemophilia B gene therapy - Spark. National Library of Medicine. 2012, NCT01620801. https://clinicaltrials.gov/show/NCT01620801.
- 13.Spark Therapeutics, Children’s Hospital of Philadelphia, The Hemophilia Center of Western Pennsylvania, and Royal Prince Alfred Hospital. LTFU for gene transfer subjects with hemophilia B. National Library of Medicine. 2007, NCT00515710. https://clinicaltrials.gov/show/NCT00515710.
- 14.Spark Therapeutics and Pfizer. A gene therapy study for hemophilia B. National Library of Medicine. 2015, NCT02484092. https://clinicaltrials.gov/show/NCT02484092.
- 15.St. Jude Children’s Research Hospital, National Heart, Lung, and Blood Institute, Hemophilia of Georgia, Inc., and Children’s Hospital of Philadelphia. Dose-escalation study of a self complementary adeno-associated viral vector for gene transfer in hemophilia B. National Library of Medicine. 2009, NCT00979238. https://clinicaltrials.gov/show/NCT00979238.
- 16.Baxalta US Inc. Open-label single ascending dose of adeno-associated virus serotype 8 factor IX gene therapy in adults with hemophilia B. National Library of Medicine. 2012, NCT01687608. https://clinicaltrials.gov/show/NCT01687608.
- 17.Herzog R.W. Immune responses to AAV capsid: are mice not humans after all? Mol. Ther. 2007;15:649–650. doi: 10.1038/sj.mt.6300123. [DOI] [PubMed] [Google Scholar]
- 18.Li H., Lasaro M.O., Jia B., Lin S.W., Haut L.H., High K.A., Ertl H.C. Capsid-specific T-cell responses to natural infections with adeno-associated viruses in humans differ from those of nonhuman primates. Mol. Ther. 2011;19:2021–2030. doi: 10.1038/mt.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somanathan S., Breous E., Bell P., Wilson J.M. AAV vectors avoid inflammatory signals necessary to render transduced hepatocyte targets for destructive T cells. Mol. Ther. 2010;18:977–982. doi: 10.1038/mt.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siders W.M., Shields J., Kaplan J., Lukason M., Woodworth L., Wadsworth S., Scaria A. Cytotoxic T lymphocyte responses to transgene product, not adeno-associated viral capsid protein, limit transgene expression in mice. Hum. Gene Ther. 2009;20:11–20. doi: 10.1089/hum.2008.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Lin S.W., Giles-Davis W., Li Y., Zhou D., Xiang Z.Q., High K.A., Ertl H.C. A preclinical animal model to assess the effect of pre-existing immunity on AAV-mediated gene transfer. Mol. Ther. 2009;17:1215–1224. doi: 10.1038/mt.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Figueredo J., Calcedo R., Lin J., Wilson J.M. Cross-presentation of adeno-associated virus serotype 2 capsids activates cytotoxic T cells but does not render hepatocytes effective cytolytic targets. Hum. Gene Ther. 2007;18:185–194. doi: 10.1089/hum.2007.001. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Murphy S.L., Giles-Davis W., Edmonson S., Xiang Z., Li Y., Lasaro M.O., High K.A., Ertl H.C. Pre-existing AAV capsid-specific CD8+ T cells are unable to eliminate AAV-transduced hepatocytes. Mol. Ther. 2007;15:792–800. doi: 10.1038/sj.mt.6300090. [DOI] [PubMed] [Google Scholar]
- 24.Li C., Hirsch M., Asokan A., Zeithaml B., Ma H., Kafri T., Samulski R.J. Adeno-associated virus type 2 (AAV2) capsid-specific cytotoxic T lymphocytes eliminate only vector-transduced cells coexpressing the AAV2 capsid in vivo. J. Virol. 2007;81:7540–7547. doi: 10.1128/JVI.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Hirsch M., DiPrimio N., Asokan A., Goudy K., Tisch R., Samulski R.J. Cytotoxic-T-lymphocyte-mediated elimination of target cells transduced with engineered adeno-associated virus type 2 vector in vivo. J. Virol. 2009;83:6817–6824. doi: 10.1128/JVI.00278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabatino D.E., Mingozzi F., Hui D.J., Chen H., Colosi P., Ertl H.C., High K.A. Identification of mouse AAV capsid-specific CD8+ T cell epitopes. Mol. Ther. 2005;12:1023–1033. doi: 10.1016/j.ymthe.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Martino A.T., Basner-Tschakarjan E., Markusic D.M., Finn J.D., Hinderer C., Zhou S., Ostrov D.A., Srivastava A., Ertl H.C., Terhorst C. Engineered AAV vector minimizes in vivo targeting of transduced hepatocytes by capsid-specific CD8+ T cells. Blood. 2013;121:2224–2233. doi: 10.1182/blood-2012-10-460733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Jr., Weigel-Van Aken K.A. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markusic D.M., Herzog R.W., Aslanidi G.V., Hoffman B.E., Li B., Li M., Jayandharan G.R., Ling C., Zolotukhin I., Ma W. High-efficiency transduction and correction of murine hemophilia B using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 2010;18:2048–2056. doi: 10.1038/mt.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsic D., Govindasamy L., Currlin S., Markusic D.M., Tseng Y.S., Herzog R.W., Agbandje-McKenna M., Zolotukhin S. Vector design Tour de Force: integrating combinatorial and rational approaches to derive novel adeno-associated virus variants. Mol. Ther. 2014;22:1900–1909. doi: 10.1038/mt.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzog R.W. Hemophilia gene therapy: caught between a cure and an immune response. Mol. Ther. 2015;23:1411–1412. doi: 10.1038/mt.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harvill E.T., Osorio M., Loving C.L., Lee G.M., Kelly V.K., Merkel T.J. Anamnestic protective immunity to Bacillus anthracis is antibody mediated but independent of complement and Fc receptors. Infect. Immun. 2008;76:2177–2182. doi: 10.1128/IAI.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNeal M.M., VanCott J.L., Choi A.H., Basu M., Flint J.A., Stone S.C., Clements J.D., Ward R.L. CD4 T cells are the only lymphocytes needed to protect mice against rotavirus shedding after intranasal immunization with a chimeric VP6 protein and the adjuvant LT(R192G) J. Virol. 2002;76:560–568. doi: 10.1128/JVI.76.2.560-568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao G., Detre C., Berger S.B., Engel P., de Waal Malefyt R., Herzog R.W., Bhan A.K., Terhorst C. Glucocorticoid-induced tumor necrosis factor receptor family-related protein regulates CD4(+)T cell-mediated colitis in mice. Gastroenterology. 2012;142:582–591.e8. doi: 10.1053/j.gastro.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christianson S.W., Shultz L.D., Leiter E.H. Adoptive transfer of diabetes into immunodeficient NOD-scid/scid mice. Relative contributions of CD4+ and CD8+ T-cells from diabetic versus prediabetic NOD.NON-Thy-1a donors. Diabetes. 1993;42:44–55. doi: 10.2337/diab.42.1.44. [DOI] [PubMed] [Google Scholar]
- 37.King C., Ilic A., Koelsch K., Sarvetnick N. Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell. 2004;117:265–277. doi: 10.1016/s0092-8674(04)00335-6. [DOI] [PubMed] [Google Scholar]
- 38.Kalos M., June C.H. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity. 2013;39:49–60. doi: 10.1016/j.immuni.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson S.J., de Jong Y.P., Comiskey M., Terhorst C. Pathways of T cell pathology in models of chronic intestinal inflammation. Int. Rev. Immunol. 2000;19:1–37. doi: 10.3109/08830180009048387. [DOI] [PubMed] [Google Scholar]
- 40.Ostanin D.V., Bao J., Koboziev I., Gray L., Robinson-Jackson S.A., Kosloski-Davidson M., Price V.H., Grisham M.B. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G135–G146. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vercauteren K., Hoffman B.E., Zolotukhin I., Keeler G.D., Xiao J.W., Basner-Tschakarjan E., High K.A., Ertl H.C., Rice C.M., Srivastava A. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol. Ther. 2016;24:1042–1049. doi: 10.1038/mt.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W., Asokan A., Wu Z., Van Dyke T., DiPrimio N., Johnson J.S., Govindaswamy L., Agbandje-McKenna M., Leichtle S., Eugene Redmond D., Jr. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol. Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- 45.Maheshri N., Koerber J.T., Kaspar B.K., Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 46.Hui D.J., Edmonson S.C., Podsakoff G.M., Pien G.C., Ivanciu L., Camire R.M., Ertl H., Mingozzi F., High K.A., Basner-Tschakarjan E. AAV capsid CD8+ T-cell epitopes are highly conserved across AAV serotypes. Mol. Ther. Methods Clin. Dev. 2015;2:15029. doi: 10.1038/mtm.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strick-Marchand H., Dusséaux M., Darche S., Huntington N.D., Legrand N., Masse-Ranson G., Corcuff E., Ahodantin J., Weijer K., Spits H. A novel mouse model for stable engraftment of a human immune system and human hepatocytes. PLoS ONE. 2015;10:e0119820. doi: 10.1371/journal.pone.0119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers G.L., Zolotukhin I., Herzog R.W. Unique role of the TLR9-MyD88 signaling pathway in dendritic cells in AAV capsid-specific CD8+ T cell activation. Mol. Ther. 2015;23:S208. [Google Scholar]
- 49.Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P.E. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen M., Lundegaard C., Worning P., Lauemøller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sidney J., Assarsson E., Moore C., Ngo S., Pinilla C., Sette A., Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martino A.T., Herzog R.W., Anegon I., Adjali O. Measuring immune responses to recombinant AAV gene transfer. Methods Mol. Biol. 2011;807:259–272. doi: 10.1007/978-1-61779-370-7_11. [DOI] [PMC free article] [PubMed] [Google Scholar]