Abstract
Several fields in neuroscience have been revolutionized by the advent of optogenetics, a technique that offers the possibility to modulate neuronal physiology in response to light stimulation. This innovative and far-reaching tool provided unprecedented spatial and temporal resolution to explore the activity of neural circuits underlying cognition and behaviour. With an exponential growth in the discovery and synthesis of new photosensitive actuators capable of modulating neuronal networks function, other fields in biology are experiencing a similar re-evolution. Here, we review the various optogenetic toolboxes developed to influence cellular physiology as well as the diverse ways in which these can be engineered to precisely modulate intracellular signalling and transcription. We also explore the processes required to successfully express and stimulate these photo-actuators in vivo before discussing how such tools can enlighten our understanding of neuronal plasticity at the systems level.
Keywords: Optogenetics, neuronal plasticity, intracellular signalling, epigenetics, transcription, gene expression
Introduction
External milieu and neuronal plasticity
In mammals, the ontogeny of the central nervous system (CNS) is achieved through different genetic programmes in prenatal life, which cause the diverse types of neuronal cells that build up the brain to be produced in the right number and at the right place. Modalities of sensory experience in post-natal life, however, markedly shape the CNS architecture by modulating the structure and function of synaptic connectivity.1 The experience-induced modulation of neural circuit functionality leads to the formation of short-term and long-term forms of neuronal plasticity that alter the computational properties of sensory systems and ultimately have an impact at the level of higher brain functions and behaviour.
Environmental influences on brain development and function are mediated by the release of neurotransmitters at specific synapses between neural cells, which, following the binding to appropriate receptors on the post-synaptic neuron, set in motion a number of intracellular signalling pathways that arrive to the nucleus and regulate gene transcription. Different events can be drawn along these physiological mechanisms. First, a transient increase in calcium concentration within the post-synaptic cell promotes synapse-specific modifications that lead to changes in synaptic transmission.2,3 Second, the activation of intracellular signal transduction pathways results in the initiation of gene programmes that alter dendritic growth,4 synapse development,5 and neuronal connections.6 Thus, modifications in the structure and function of synaptic connectivity within neuronal networks arise from intracellular transduction pathways driven by electrical signals associated with neuronal activity, which in turn control the expression of plasticity genes. This underlies the capability of the brain to adjust in response to changing environmental conditions.
Aside from identifying transcription factors that regulate gene transcription in response to external stimuli, epigenetic mechanisms that exert a long-lasting control of gene expression by altering chromatin structure, rather than changing the DNA sequence itself, have recently emerged as conserved processes by which the CNS accomplishes the induction of plasticity.7,8 A hot spot in the field of neuroscience is the identification of physiological mechanisms associated with experience that promote alterations in the pattern of DNA methylation9–11 and/or posttranslational modifications of histones12–14 that control the expression of plasticity genes in the brain. A new set of optogenetic tools capable of affecting both epigenetics and transcription is emerging (detailed below), offering new ways to study intracellular mechanisms underlying neuronal plasticity.
Many mechanisms have been implicated in the occurrence of activity-dependent plasticity.15 According to Hebb’s rule,16 neurons that fire together wire together, whereas neurons that fire out of synchrony lose their link. Decades of effort in the field have elucidated the involvement of different post-synaptic receptors in this mechanism. Glutamate (Glu) receptors of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-d-aspartate (NMDA) type are key proteins in processes of long-term and short-term potentiation of synaptic transmission. Post-synaptic NMDA receptors detect synchronized neuronal activity between the pre- and post-synaptic neurons and reinforce the downstream signalling of the post-synaptic cell.17,18 The increased activity and coordination between pre- and post-synaptic receptors leads to permanent changes in synaptic connectivity, i.e., plasticity. Conversely, nerve terminals that experience weakened and unsynchronized activity will eventually lose their synaptic contacts and retract. This phenomenon seems to be mediated by the internalization of post-synaptic AMPA receptors.19 Other forms of synaptic plasticity, such as homeostatic plasticity,20 γ-aminobutyric acid (GABA)–mediated plasticity,21 neurogenesis,21 and synaptogenesis22 are also involved in the regulation of synaptic transmission.
Activity-dependent plasticity plays an important role in processes of learning and memory. Hence, it is responsible for adaptation of an individual to the environment. Until recently, optogenetic tools have been used to extrinsically modify neuronal activity using type-1 rhodopsin derivatives (described below), enabling a primary impact on the electrical state of the neuron and only a secondary impact on intracellular signalling and transcription (Figure 1). Novel optogenetic toolboxes described here have now the potential to affect both those processes directly.
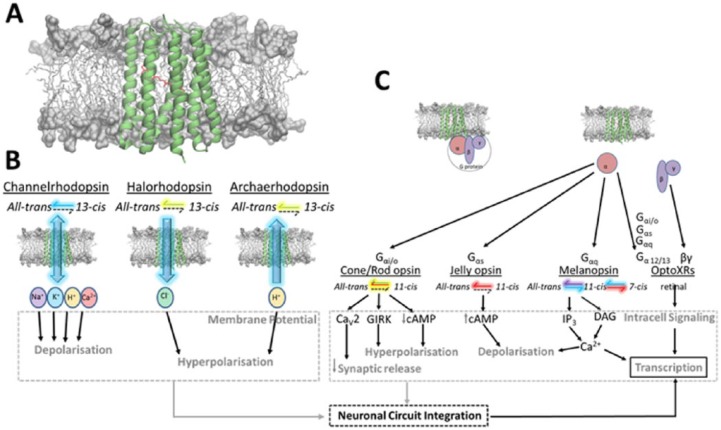
Figure 1.
Rhodopsin transmembrane photo-actuator. (A) Pictorial representation of rhodopsin structure in a membrane environment: the protein is represented as green cartoons and the retinal chromophore as red sticks. Membrane lipids are represented as grey sticks (hydrophobic region) or surface (hydrophilic region). Generated in the Visual Molecular Dynamics software.23 (B and C) Examples of (B) type I and (C) type II rhodopsins, with their activation spectra depicted as coloured arrows and retinal cycle along with their influence on intracellular function and its impact on transcription and neuronal circuitries’ connectivity.
Optogenetics and neuronal plasticity
One major challenge for modern neuroscience is to control the activity of a single type of neuron in the mammalian brain while leaving others unaltered.24 The application of molecular engineering to the field of optogenetics provided precious tools to control specific groups of neurons that underlie behaviour.24–28 Optogenetics has had a major impact in the unravelling of issues that include cross-modal plasticity,29 information processing by neuronal circuitries,30,31 hippocampal memory formation,32,33 anxiety and depression,34,35 fear conditioning,36 aggression,37 feeding behaviour,38–40 restoration of visual functions in blind animals,41–43 the Parkinson disease,44,45 and epilepsy.46,47
Interpreting the occurrence of plastic phenomena under physiological conditions requires a well-defined correlation between the activity of specific neuronal subtypes and the computational properties of neuronal networks within sensory systems. Unfortunately, the lack of a clear relationship between morphological and functional characteristics of different cell types precludes a wide view of neuronal connectivity patterns within neuronal networks. This, together with the absence of experimental approaches with high temporal and spatial resolution to address the role of specific cellular subtypes in phenomena of plasticity, is one of the major reasons by which the screening of neuronal populations underlying brain plasticity has remained elusive. Only recently, the development and application of optogenetics in the field of neuroscience has made it possible to investigate the relationship between sensory perception, plasticity phenomena, and the activity of specific neuronal subpopulations in primary sensory areas.
The subcellular targeting of optogenetic tools has opened up new avenues in neuroscience as it enables researchers to address the function of well-defined intracellular domains.48,49 High-precision spatiotemporal control of gene transcription has been recently accomplished,14 which allowed to modulate endogenous gene expression and epigenetic chromatin modifications that underlie behaviour (for review, see Moglich and Hegemann50). The fast advances in the diverse optogenetic toolboxes described in this review provide novel ways to establish causal relationships between the activity of specific neuronal networks and the related functional outcomes.
Photoactivatable Actuators
Photoreceptor protein domains respond to light by absorbing photons at a prosthetic group or chromophore. Interaction with light induces a cascade of events called the photocycle, comprising molecular transformations including chemical changes at the chromophore, and conformational and functional changes of the protein scaffold. The photocycle usually connects 2 states, the dark and lit states, separated by intermediates and is in general reversible because the photoreceptor is able to recover its dark state by thermal fluctuations. Isomerization or establishment of a covalent bond with the protein chain is among the most common chemical changes observed at the chromophores.
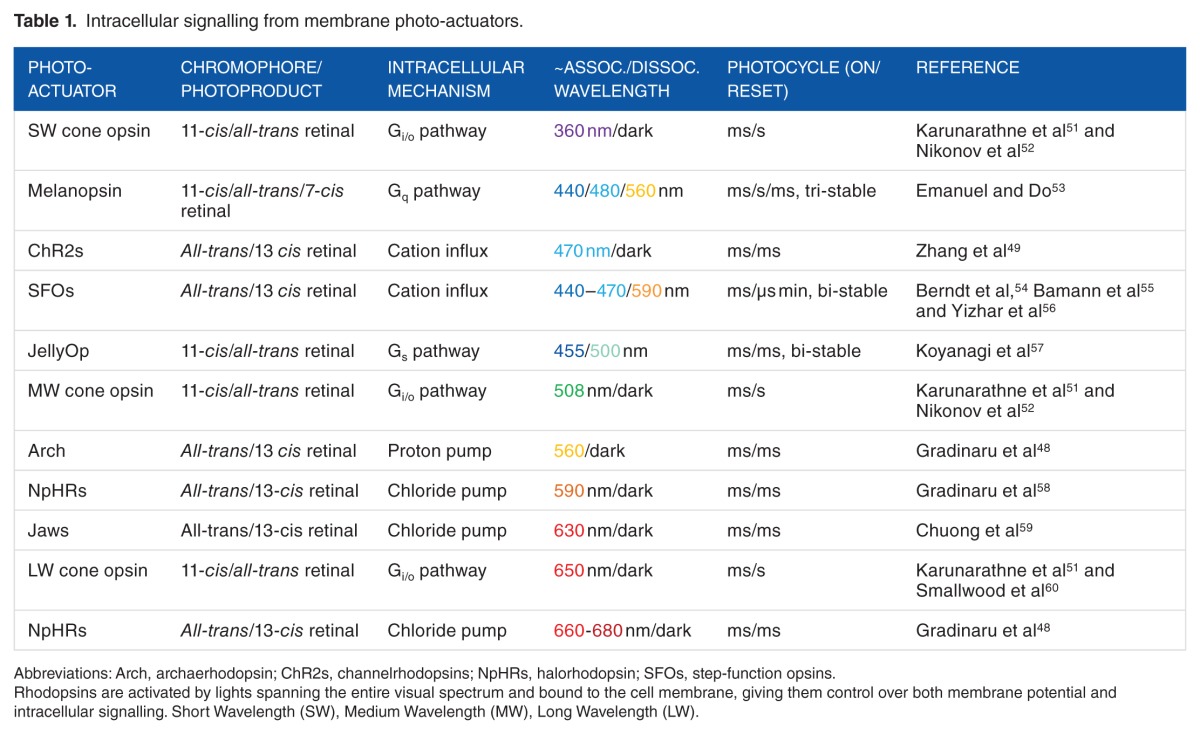
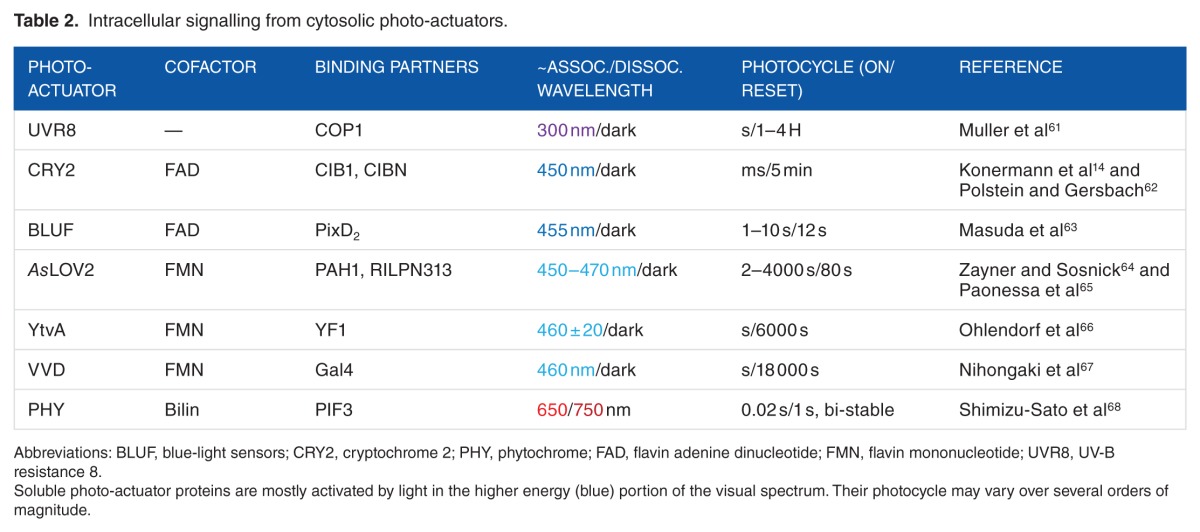
Photoreceptors are usually classified by their chromophore cofactor: rhodopsins with retinal, light-oxygen-voltage (LOV) sensors with a flavin mononucleotide (FMN), blue-light sensors (BLUF), and cryptochromes (CRYs) with a flavin adenine dinucleotide (FAD) and phytochromes (PHYs) with tetrapyrrole molecules. The UV-B receptor UV-B resistance 8 (UVR8) constitutes a separate class of photoreceptors as it does not employ a cofactor but instead absorbs UV radiation through tryptophan (Trp) residues. Although rhodopsins (Table 1) are membrane bound, the other photoreceptors (Table 2) are cytosolic, allowing for photo-transduction within any cellular compartment.
Table 1.
Intracellular signalling from membrane photo-actuators.
Table 2.
Intracellular signalling from cytosolic photo-actuators.
Rhodopsins were the first proteins to be used as optogenetic actuators in neuroscience applications.69,70 This class of light-sensitive protein is composed of 7 transmembrane α helixes (Figure 1A) combined to the photo-isomerizable chromophore retinal.71 The engineering of innovative opsins with diverse characteristics and functions originating from the two types of rhodopsins expressed in the microbial (type I) and animal (type II) kingdoms, have been extensively described.28,72,73
Type I rhodopsins
In type I rhodopsins, the transmembrane domain is generally an ionotropic effector (channel or pump) that is covalently bound to the chromophore allowing for dynamic modulation of the voltage across biological membranes at the sub-millisecond timescale69 (Figure 1B). On illumination, all-trans retinal isomerizes to 13-cis retinal, leading to a conformational change in the transmembrane domain. The most cited examples of such optogenetic tools are channelrhodopsin (ChR2),74 halorhodopsin from Natronomonas pharaonis (NpHR),75 and archaerhodopsin (Arch).76 Channelrhodopsin is permeable to sodium, potassium, protons, and, to a lesser extent, calcium on illumination with blue light (~470 nm) generating an inward current that leads to membrane depolarization and action potential firing. However, pump rhodopsins, such as NpHR, which drive chloride in the cytosol, and Arch, which pump protons out of the cell, lead to membrane hyperpolarization and a decrease in action potential firing on illumination with orange light (~590 nm). Photoactive chloride and proton pumps require constant illumination to go through their photocycle as opposed to ChR2, which only need channel opening, and bi-stable rhodopsins, which require a single pulse to move from one state to the other. Sensory rhodopsins trigger enzyme activity such as histidine kinase,77 providing the opportunity to influence intracellular signalling. Type I rhodopsins and their uses as optogenetic tools are extensively reviewed elsewhere.72
One of the most exciting features of the optogenetic revolution is the steady development of toolboxes of opsins exhibiting increasingly diverse spectra, excitation, and temporal kinetics. The NpHRs have been optimized to the point of being used in mammalian tissue58 and having red/far red sensitivity.48 Step-function opsins (SFOs) are a family of bi-stable ChR2 mutants54 designed to stabilize the active retinal isomer. They display longer inactivation time constants, allowing the activation of populations of neurons for long times (from tens of seconds54 to several55 or even tens of minutes56), together with the possibility of switching them off with a single pulse of yellow light.54 These specifications enable a myriad of uses including calcium imaging or un-tethered behavioural experiments, in which the consequences of optogenetic activation can be investigated following a single pulse, without cumbersome stimulating hardware.28 In addition to the development of such bi-stable optogenetic tools, multiplexing or even cooperative strategies are now emerging. The former allows various opsins sensitive to different wavelengths to be simultaneously but independently activated, such as the expression of ChR2 in the dendrites and the expression of NpHR in the soma of retinal ganglion cells to mimic the effect of centre-surround antagonism in the retina.43 The recently developed ‘Cl-out’ strategy uses the outward current generated by the proton pump Arch to propel the extrusion of chloride ions through the concomitant activation of a co-expressed optogenetic chloride channel.78
Type II rhodopsins
In type II rhodopsins, the 7 transmembrane domains form a G protein−coupled metabotropic receptor (GPCR), offering a broad influence over cellular physiology by tapping into secondary messenger systems. 11-cis-retinal dissociates on photo-isomerization to all-trans retinal, making the resetting kinetics considerably slower than type I rhodopsins, as well as requiring the exogenous application of 9 or 11-cis retinal anywhere other than in the retina. A large diversity of photo-transduction cascades are initiated following activation of type II rhodopsins including Gt-, Gq-, Go-, Gs-, Gi-, and Gi/o-coupled pathways. We discuss here the 3 most exploited tools (Figure 1C); for a more exhaustive review of type II rhodopsins, the reader is directed to the study by Koyanagi and Terakita.73
Cone opsins and rod opsins are expressed in the outer segment of mammalian photoreceptors, coupling to Gα transducing and activating the Gi/o pathway51 on illumination. Both cone and rod opsins present advantages, in that rod opsins are highly sensitive to light and cone opsins exist in various versions, sensitive to different wavelengths,79 thus offering the potential for polychromatic multiplexing toolboxes. A major drawback in using these opsins is the high level of bleaching due to chromophore dissociation on activation. However, this feature was successfully exploited in vivo as a light adaptation mechanism to restore vision in genetically engineered blind mice.80
Jellyfish opsins (JellyOp) have been found to signal through Gs proteins,57 inducing translocation of adenylyl cyclase, leading to an increased production of cyclic adenosine monophosphate (cAMP) followed by activation of the protein kinases, PKA and ERK pathways.81 The activation of all these proteins often leads to membrane voltage depolarization. Jellyfish opsin has a peak sensitivity of ~500 nm, and unlike visual opsins, does not bleach but is instead converted to a stable photoproduct that does not revert to its original dark state on subsequent illumination.57
Melanopsin is a photopigment expressed in intrinsically photosensitive retinal ganglion cells (ipRGCs) and is responsible for the photo-entrainment of the circadian rhythm.82 With recent evidence pointing to tri-stability,53 melanopsin activation by blue light activates Gq signalling.83 In ipRGCs, this leads to sustained firing of action potentials along axons, 80% of which (M1 ipRGCs) project to the suprachiasmatic nucleus (SCN, master clock of the CNS) leading to the daily plastic regulation of the sleep/wake cycle.82,84 Recent evidence suggests that the SCN integrates both irradiance from inner retinal ipRGCs and colour from outer retinal photoreceptors to determine the time of day.85
To target signalling pathways that do not rely solely on generic Gα subunit coupling dynamics, recent effort has been made in the development of chimeric optogenetic tools (OptoXRs) composed of extracellular loops of type II rhodopsin and the intracellular loops/C-terminal tail of GPCRs endogenously expressed in the CNS.86 The advantage of this approach consists in coupling the multiple inherent possibilities offered by type II opsins (high sensitivity, multi-chromaticity, bleach adaptation) with the quasi-infinite variety offered by endogenous GPCR systems. OptoXRs have been implemented successfully in vivo with the adrenergic,87 adenosinergic,88 opioid,89 and serotonergic90 receptor systems.
Although opsins represent powerful tools to modulate the physiology of entire cell populations, inducing or preventing plasticity in vivo through modulation of neural activity,33,51,88 they lack the spatial resolution to directly impact intracellular functions such as protein interactions or gene expression.
Soluble photoactivatable proteins
Light-oxygen-voltage (LOV) and phytochrome (PHY) protein receptors have a modular structure; ie, they are made of several adjacent globular domains originating from the same, continuous amino acid chain. Each module performs a different function. One domain is responsible for light absorption, whereas a separate one is responsible for biological activity, which can be protein binding, DNA binding, or enzymatic action. The transmission of the signal from the photo-sensor domain to the activity domain is usually a combination of structural and dynamic effects. The same principle is at the basis of engineered optogenetic probes, where the photo-sensor domain from a receptor is linked to a different protein domain exerting the biological function specifically required by the designer.
LOV domains
Light-oxygen-voltage sensor domains are found in proteins from bacteria, plants, and fungi, and they are the most widely used type of photoreceptors in optogenetic applications to influence intracellular signalling. All LOV domains show a fold typical of the so-called Per-ARNT-Sim (PAS) family,91 with a central anti-parallel 5-stranded β-sheet surrounded by α-helices, and harbour an FMN cofactor that in the dark state is non-covalently bound to the protein chain. After blue-light absorption by the flavin nucleotide, a covalent bond is established between a conserved Cys residue of the LOV domain and the isoalloxazine ring of the cofactor.92 Different LOV domains reveal different downstream effects such as dimerization Fungal photoreceptor Vivid (VVD93,94) or conformational changes Avena Sativa Light-Oxygen-Voltage domains 2 (AsLOV295). In all LOV domains, the lit state is metastable, and the dark state is recovered by thermal fluctuations in tens to thousands of seconds (Table 2). Different LOV proteins display distinct kinetics of adduct decay,96 fast (τ < 1000 seconds; AsLOV2), intermediate (1000 seconds < τ < 10 000 seconds; YtvA), and slow (10 000 seconds < τ; VVD) cycling. Several studies have shown that LOV photocycles can be tuned by acting on the photochemistry of the transition process, and several mutations have been developed to influence the time scale of the cycle, by stabilizing or destabilizing 1 of the 2 states.64
AsLOV2 is probably the most studied and employed LOV domain. Light absorption results in displacement and unfolding of the 20 residues C-terminal helix termed Jα, which in the dark state is folded and docked against the protein domain. To control the chemistry of the AsLOV2 photocycle, chemical events were investigated using a battery of mutations designed to alter side chain interactions with the FMN and the surrounding water molecules.64 For this, the authors established a library of mutations located near the chromophore able to alter photocycle times from 2 seconds to more than 2000 seconds.
The mechanism of LOV domains photoexcitation has been widely used to engineer chimeric proteins whose action can be finely modulated by light. Remarkable examples realized using AsLOV2 include the control of gene transcription by modulating the action of the Trp repressor,97 the binding of tetracycline to Tet repressor,98 the activity of endogenous transcription factors,65 the control of small GTPases’ activity and actin cytoskeletal dynamics,99 and the caging of peptides.100 Other LOV-based optogenetic probes successfully provided control over light-repressed or light-induced gene expression using YtvA,66 caspase-9 activation to regulate apoptosis with VVD,67 and gene activation by engineered zinc finger transcription factors.101 Recently, LOV sensors were employed to modulate the action of RNA-guided programmable DNA endonuclease Cas9102 and the structural disorder of diverse proteins, thus generating robust allosteric switches.103
BLUF proteins
The Blue-light sensor (BLUF) proteins are found in bacteria, and they are unique among photoreceptors because the light-induced structural changes at the chromophore are very limited.104 The BLUF structures that have been determined so far show 5 anti-parallel β-strands faced by 2 α-helices running parallel to the β-sheet that dock the FAD chromophore between them.105,106 Although the complete structural features of BLUF photocycle and signal cascade have not been clarified yet, it is known from spectroscopic and structural studies that, after light absorption, new hydrogen bonds are formed between the chromophore and a conserved Met or Trp residue in the protein. These changes trigger modifications in one of the strands (β5) that, in turn, induce structural modifications in the C-terminal α-helices.104 Optogenetic applications include use of BLUF proteins to cluster transcription factors, thus regulating their activity,63 and to control the conversion of adenylyl cyclases into guanylyl cyclases.107
Cryptochromes
Cryptochromes (CRY) are blue light–absorbing photoreceptors found in all kingdoms of life as components of circadian clocks.108 Several crystal structures of CRYs are available, which show binding of a FAD cofactor in a conserved α-helical domain. Although the fine details of CRYs’ photochemistry are still under debate,109 it is known that after interaction with light, Arabidopsis thaliana cryptochrome 2 (CRY2) undergoes both homo- and hetero-dimerization, the latter occurring via association with its interacting partner calcium and integrin–binding protein 1 (CIB1).110 The association with CIB1 takes place within milliseconds after illumination in mammalian cells, and the dimer shows a half-life of about 5 minutes in the dark. Although homo-dimerization has been observed only in plant cells, the property has recently started to be used also in applications to modulate functions of mammalian cells. Indeed, recent analysis revealed that the 2 processes can happen concomitantly in such cells, and that the use of specific CIB1 fusion proteins can suppress homo-oligomerization. Examples of CRY uses in optogenetics include the regulation of gene transcription in cultured neurons and in vivo14 or the activation of the Raf/MEK/ERK pathway to induce neurite outgrowth.111
Phytochromes
Phytochromes (PHYs) are found in fungi, plants, and bacteria and they bind covalently the red light–and far-red light–absorbing bilin tetrapyrrole chromophores.112 They are made of 3 individual domains called PAS, cGMP phosphodiesterase/adenylcyclase/FhlA (GAF), and PHY, the last two being also structurally similar to PAS. The chromophore makes the larger number of contacts with residues in the GAF domain, but the presence of the PAS and PHY domains is also required for photo-activity, in all but a few limited exceptions.113 In PHYs, the photo-cycle consists of conversions between spectral states that absorb far-red and red light. The 2 states are associated with dark and light conditions differently in diverse PHYs. Hence, apart from thermal fluctuations, the dark state can also be recovered by photon absorption. In both cases, the transition involves isomerization of the bilin cofactor. The fundamental details of signal transduction in PHYs are not yet known. Some crystal structures show parallel dimers with the individual domains linearly disposed around a central bundle of α-helices, but the effect of light signal propagation is still to be uncovered.114 Bacterial PHYs have been engineered into several types of probes including fluorescent proteins,115,116 protein-protein interaction reporters,117 and regulators of cAMP levels in zebra fish and mammalian cells.118
UVR8
The UVR8 photoreceptor, mostly found in plants, forms a homodimer stabilized by salt bridges between facing Arg and Glu/Asp residues.119 UV light absorption occurs at Trp residues clustered in proximity to the salt bridges and induces the dissociation of the dimer, most likely because of perturbations in the interaction pattern between monomers. The UVR8 receptor reverts back to its ground state by homo-dimerization and restores light responsiveness when UV light is switched off.120 The UVR8 dimer restoration appears to be faster in vivo (1–2 hours) than in vitro (24–48 hours).120,121 It has been used to control gene expression,61 protein recruitment to chromatin,122 and protein secretion from mammalian cells.123
Photo-labile protecting groups
The concept of caging bio-active compounds with a photo-labile construct to render them inactive until ‘un-caged’ by light was first developed to improve the spatial resolution of protein synthesis at the transcriptional level.124 The technology rapidly evolved to enable control of both gene promotion and repression.125 Systems neuroscience then greatly benefited from the technique, which allowed the spatiotemporally precise ‘uncaging’ of the amino acid Glu with light126 to map out glutamatergic circuitry. One of the major drawbacks of this approach was the need to exogenously apply the caged compounds. However, recent advances have made it possible to genetically encode caging groups for lysines, tyrosines, and cysteines inside the protein sequences, enabling photo-control over protein localization,127 signal transduction,128 and gene expression.129
Chemical optogenetics
Photoswitchable tethered ligands undergo conformational changes on illumination at a specific wavelength, therefore activating or antagonizing receptor activity on demand while preserving their physiological function. This approach was pioneered by the Isacoff group, who created a light-sensitive ionotropic Glu receptor (LiGluR) by tethering an agonist to its receptor via the azobenzene-photosensitive linker MAG (cysteine-reactive Maleimide/Azobenzene photoswitch/Glu head group). The conjugation of azobenzene to the receptor was made possible by a single cysteine substitution in the receptor sequence.130,131 The MAG linker isomerizes from trans to cis on illumination at 380 nm, presenting the agonist to the receptor, and reverts back to the trans conformation on 500 nm illumination. The photoswitch is extremely rapid, on the time scale of milliseconds, and could efficiently modulate firing and synaptic transmission in cultured neurons,132,133 allowing the activation of plasticity with single-synapse specificity.134
Besides cultured neurons, this system was successfully used in entire organisms such as zebra fish132,135 and Drosophila larvae.136 By following a similar approach, light-agonized and light-antagonized metabotropic Glu receptors (LimGluRs),137 a light-regulated GABAA receptor (LiGABAR),138 and a chimera of LiGluR and K+ channel (Hylighter)139 were created. The 380 nm, near-UV wavelength required to activate the switch, however, was not convenient for biological experiments, which prompted further optimization of the initial LiGluR molecule140 to achieve a red-shifted photoswitch (L-MAG0460), activated by 400 to 520 nm light, which undergoes spontaneous relaxation in the dark, thus displaying an interesting single-wavelength behaviour.141 More recently, a wider panel of LiGlu/MAG variants was applied in vivo to control the activity of mouse brain when injected in the visual cortex.142 Some of the abovementioned engineered variants have been used in mouse and canine models of blindness: when expressed by adeno-associated virus (AAV) injection and activated by subsequent MAG administration, they were able to restore responses to visible light in mouse and canine retinal explants and to restore light-dependent behaviour in rodents.143,144 Thus, despite the main disadvantage of being a 2-component, non-entirely genetically coded system, chemical optogenetic constructs hold a high potential in biomedical sciences.
Photo-actuated Transcription
Photo-transcription
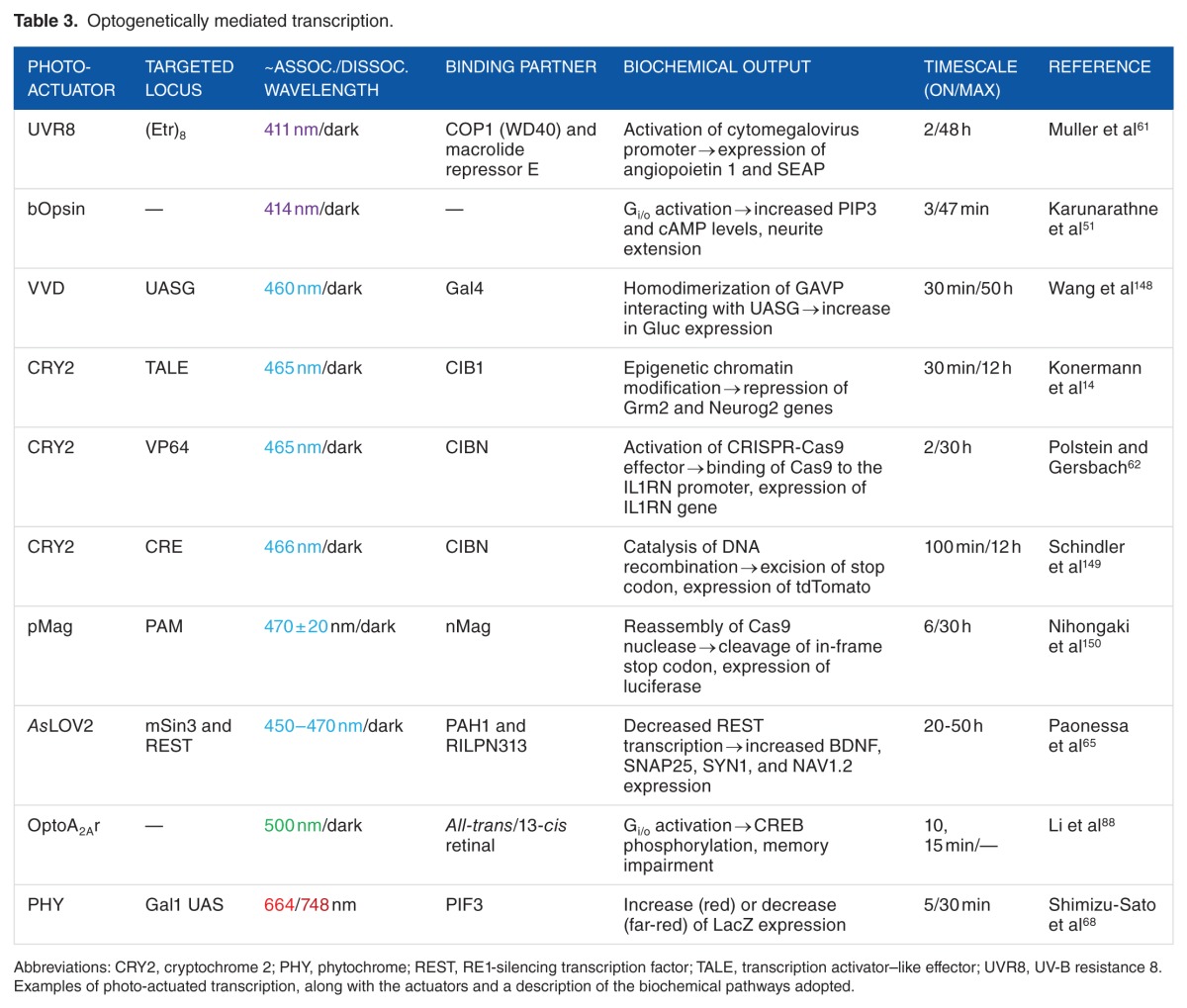
The modulation of gene expression is crucial for long-term synaptic plasticity, and thus, the possibility to control protein levels by optogenetics is attracting increasing attention. The control of protein expression has been achieved through several strategies acting both at transcriptional and translational levels. Among the several available light-sensitive proteins, researchers have mostly exploited few plant-derived, light-dependent systems, such as (1) the blue light−dependent interaction between the photoreceptor CRY2 and the basic helix-loop-helix (bHLH) transcription factor CRY-interacting bHLH 1 (CIB1),145 (2) the red/far-red–dependent association of Phy with the PHY interaction factor,146 (3) the UVR8,147 all derived from A thaliana, and (4) the LOV domain from Avena sativa and its variants (ie, the fungal circadian clock photoreceptor VVD or the 222 amino acid LOV-transcription factor from Erythrobacter litoralis EL222), which all undergo conformational changes in response to blue light96 (Table 3).
Table 3.
Optogenetically mediated transcription.
Using various combinations of the abovementioned tools, modulation of transcription of exogenous genes has been achieved in lower organisms such as yeast and bacteria.68,151–154 Some of these tools have been successfully applied to the modulation of exogenous genes containing the appropriate consensus binding sites also in live organisms such as zebra fish155,156 and Drosophila melanogaster.157 Two of these opto-tools have been tested in living rodents: the VVD-GAL4 construct, which can upregulate the expression of a reporter gene in the mouse liver,148 and the ‘LITE’ system, which combined the customizable transcription activator–like effector (TALE) DNA-binding domain with the light-sensitive CRY2-CIB1 dimerizing system,14 and could efficiently boost gene expression in the mouse brain. Among all these strategies, the LITE system was actually the only one that could target endogenous genes, rather than exogenous constructs artificially introduced into the animal.
Another challenging task, which goes beyond the ‘simple’ switching on and off of gene transcription, is to modify the epigenetic landscape of cells, to achieve a more physiologic variation of gene expression.158 To our knowledge, only 2 attempts have been made to address this point. The abovementioned ‘LITE’ system was coupled with several histone effectors to induce light-dependent epigenetic modification in primary neurons.14 Following a different approach, the activity of the RE1-silencing transcription factor (REST), a transcriptional repressor that assembles a chromatin-modifying complex on the promoter of its target genes, could undergo light-dependent inhibition by the expression of inhibitory peptides fused to the LOV domain (Figure 2). Interestingly, when such probes were expressed in primary neurons, illumination triggered upregulation of selected neuronal genes and the consequent alteration of cell excitability.65 On the same line, the inhibition of the activity of another pleiotropic transcription factor, cAMP response element binding protein (CREB), was achieved, in this case by the expression of a dominant negative CREB protein fused to the photoactive yellow protein.159
Figure 2.
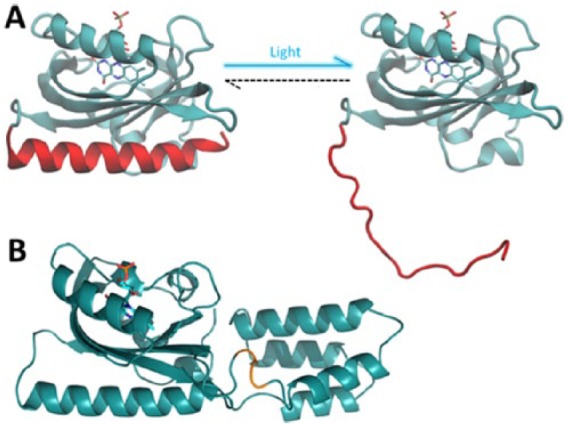
The AsLOV2 domain and AsLOV2-PAH1 chimera. (A) Pictorial representation of the structure of LOV2 domain from Avena sativa phototropin 1 in the dark (left) and lit state (right); the protein is represented as cyan cartoons. The C-terminal Jα helix is coloured in red, and the FMN chromophore is represented as sticks. On illumination, the Jα helix undocks from the protein core and unfolds. (B) Pictorial representation of the AsLOV2-PAH1 chimera engineered in Paonessa et al65 and obtained by attaching the LOV2 domain of A sativa to the REST-binding PAH1 domain of the mSin3a cofactor. The backbone of AsLOV2 and PAH1 domains is represented as cyan cartoons and the FMN chromophore as sticks. The amino acids linking the 2 domains are highlighted in orange. Generated in the Visual Molecular Dynamics software.23
Opto-CRE/CRISPR
Besides targeting the genome, today’s molecular biology makes routine use of tools capable of modifying gene sequences, ie, the lox/Cre recombinase and the CRISPR/Cas9 systems. Both systems have been rendered light-sensitive through various optogenetic strategies. Photoactivatable Cre recombinases based on the CRY2-CIB1 system149 and on the ‘magnet’ interacting proteins160 were tested in primary neurons and live animals. Alternatively, a photo-caged Cre-ONBY has also been created, although its use has been so far limited to cells.161 Similar to what has been described for Cre recombinase, also the CRISPR/Cas9 system has been combined with CRY2-CIBN62 and with the magnet proteins150 to achieve light-dependent genome editing.
A more subtle control on protein expression can be achieved by manipulating endogenous messenger RNA (mRNA) levels. This has proved to be a more complex issue, and only few attempts have been made to engineer mRNA-specific optoprobes. The CRY2-CIBN system was used to boost translation, by exploiting its ability to bind to specific sequences on target RNAs.162 In addition, the cis-trans photo-isomerization of the photoresponsive 7-methyl-8-styrylguanosine (8ST) cap was exploited to engineer an 8ST-capped translation initiator.163 However, despite these attempts, the optogenetic manipulation of RNA levels is still far from being satisfactorily achieved, and efficient RNA-directed optogenetic systems are still missing.164 In this context, proteins of the PUF (Pumilio and FBF) family of RNA-binding proteins have been the tools of choice to make sequence-specific probes165 and represent promising candidates to build mRNA-targeted, light-sensitive optogenetic proteins.
Applying Optogenetic Tools to In Vivo Systems
Targeted expression of photo-actuators
The design of effective optogenetic experiments relies heavily on the targeted expression of the photoactive actuators. As described above, these may be expressed in specific compartments (membrane, cytosol, or nucleus) of distinct cell populations (Figure 3). Targeting appropriate cell populations may be achieved by promoting gene expression through tagging to a specific portion of the genome that is exclusively expressed in a particular cell type. For example, ON-bipolar cells of the retina, which express the mGluR6 metabotropic Glu receptor, can be targeted using the Grm6 promoter,80 excitatory pyramidal cells and inhibitory interneurons can be targeted by their respective expression of CaMKIIα and parvalbumin,56 or adenosinergic neurons can be targeted by the combined expression of A2Ar and CaMKIIα promoters.88 Recent studies have highlighted the higher specificity of the short CaMKIIα promoter compared with the longer one (which has a widespread use in the scientific community) in both mice166 and monkeys.167 The above examples rely both on the genetic construct and the specific locations in which they are injected.
Figure 3.
Subcellular targeting of opto-tools. Various light-sensitive systems localize to different intracellular compartments and as such are able to target a specific domain of signalling molecules. For instance, the Phytochromes and the OptoXR system are both membrane associated and have been used to modulate, among other phenomena, peripheral actin dynamics and G-protein signalling. LOV domains are soluble proteins that have been coupled with a number of cytoplasmic effectors. In the LITE system, the transcription activator–like effector (TALE) modules were exploited to target the CRY2-CIB system to the nucleus, thus rendering gene transcription light sensitive. Much like sensory inputs, all the optogenetic tools effectively trigger long-term modifications of neuronal physiology (for specific references, see main text).
Expressing optogenetic actuators in a specific location requires initially the delivery of genetic materials to the nucleus of the correct cell types. Viral transduction is the most common strategy employed in optogenetic studies, intrinsically limiting expression to the injection site.168 Controlled injection procedures and incubation periods allow anterograde transfection169 for lentiviruses and AAV or retrograde transport for herpes simplex or rabies viruses,170 yielding a complete mapping of neural circuits. High infection efficacy in human cells and the absence of associated pathological effects make AAVs the preferred viral vectors for biologists and clinical therapists.171 AAVs have been used successfully in the delivery of genes56,88 as well as genome editing DNA-binding domains such as zinc finger endonucleases,172 RNA-guided Cas9 enzymes,14 or transcription activator–like effector nucleases.173 Unfortunately, some transcription promoter sequences are quite large, whereas the amount of genetic material packaged in viruses is limited, with AAVs’ cargo limited to 4800 base pairs. To overcome this limitation, scientists make use of the wide variety of CRE recombinase driver lines available to the scientific community in combination with CRE-dependent optogenetic vectors56,88,168 because the CRE promoter sequences are much smaller than selective promoter sequences. Additional tools such as FLEX174 or lox sites, which are also very short sequences, can be introduced into the viral genome, flanking the gene of interest, to minimize expression leakage into unwanted cell types.168,174
Nanotechnological development has enabled significant advances in synthetic genetic vector systems such as liposomes,175 nanotubes,176 and polymer scaffolds.177 Although these systems allow the packaging of far greater amounts of nucleic acid than viruses, their naturally evolved counterparts remain, for now, the most efficient systems for gene delivery.178,179 A more promising avenue may be the further optimization of virus functionality, such as external receptor targeting180 or even optogenetic modulation of nuclear translocation, as demonstrated through the insertion of light-sensitive PHY proteins within the capside.181
Challenges of complex stimulation in vivo
Scattering problem
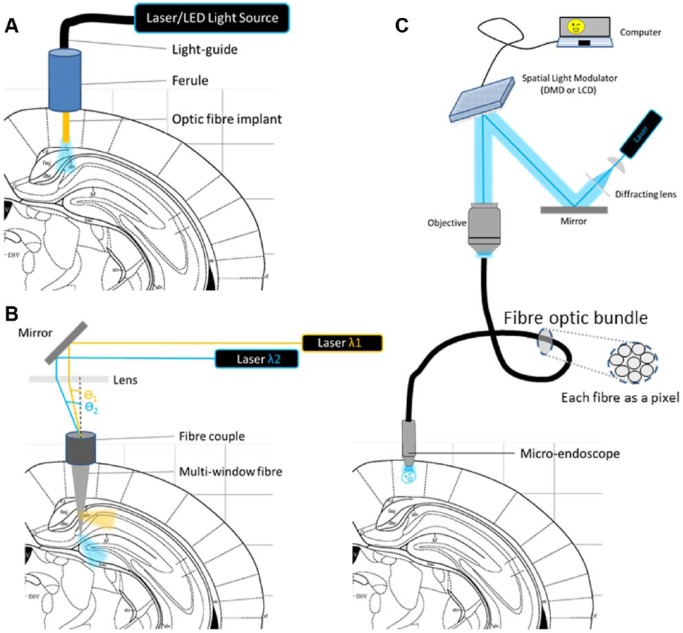
The scattering of incident light through live tissue is a fundamental problem for non-invasive photo-stimulation of CNS targets. To reach cortical neurons, light has to penetrate through skin, bone, dura, pia, and blood vessels. Deep brain structures are found several millimeters (mice) to tens of centimeters (humans) deeper. The depth of light penetration is proportional to the wavelength, with infrared radiation achieving depths of 2.5 mm, near-infrared (NIR) of 1.4 mm and red of 0.9 mm in explanted human brain tissue.182 More powerful illumination with NIR has been demonstrated to penetrate through bone and skin samples183 as deep as 40 mm within the brain.184 (As an additional tool, the reader is directed to the Deisseroth lab’s tissue penetration calculator: http://web.stanford.edu/group/dlab/cgi-bin/graph/chart.php). Furthermore, the photons’ energy content is inversely proportional to the wavelength, with UV radiation capable of inducing cell death185,186 and DNA damage187 on its way to the targeted photo-actuators. As such, red and far-red–sensitive proteins such as PHYs188 or the red-shifted cruxhalorhodopsin Jaws59 have the potential to be activated non-invasively. Alternatively, blue photo-actuators can be activated distally with 2-photon excitation (2PE). The 2PE technique relies on the convergence of 2 low-energy infrared photons onto a target molecule into a higher energy electronic state189 and has been shown to work, with some limitations, also on optogenetic proteins.190 However, this requires a moving objective and a femtosecond laser (both bulky and expensive), so most in vivo studies to date have been performed with blue-sensitive ChR2 and have had to rely on implanted cannulae bearing light guides connected to an external laser (Figure 4A). Considerable resources have now been dedicated to the development of implantable wireless light-emitting diode (LED) optrodes,191–193 but these are still subject to physical (heat dissipation, power requirements) and biomedical (tissue damage, immune response, surgical implantation, discomfort) constraints.
Figure 4.
Different approaches to in vivo optical stimulation. Depiction of a rodent brain implanted with a chronic fibre and tethered to stimulation hardware. A coronal section of the brain is used to illustrate in subsequent sub-figures the illumination strategy. (A) Single core fibre optic implant. Here, the light is delivered to an implantable fibre connected extrinsically in the manner of a cannula. (B) Multi-point optical stimulation using an implantable fibre with windows etched along its length, allowing it to deliver light of different spectral compositions (λ) at multiple depths depending on the incident angle (θ) of the light as it penetrates the coupler.194 (C) 2-dimensional optical stimulation using a spatial light modulator (SLM) which is generally a transmissive liquid crystal display (LCD) or a digital micro-mirror device (DMD). The video signal is focused through a microscope objective onto a fibre optic bundle where each of the fibres transmits a single pixel to an implanted micro-endoscope.195 The depictions in this figure are neither to scale nor exhaustive.
2-dimensional/3-dimensional light stimulation
The studies described above rely on wide-field excitation, which allows the selective targeting of genetically analogous neuronal populations within an illuminated volume. Spatially selective activation can be achieved in 2 dimensions (2D) with the use of a spatial light modulator (SLM)196,197 (Figure 4C) or a multi-LED array.198 An elegant example of 2D digital holography performed in vivo in tethered behaving mice consists of a fibrescope set-up combining distinct coherent blue (modulated with a liquid crystal SLM for holographic stimulation of ChR2) and green (modulated with a digital micro-mirror for confocal calcium imaging of genetically expressed GCaMP5-G) light paths.195
The 2D spatial modulation can be useful when targeting a 2D structure such as the retina, but dissecting neuronal circuits in the brain requires the 3-dimensional sculpting of light into complex volumes. This has been achieved by combining digital holography with 2PE (reviewed thoroughly for their use in optogenetic stimulation in Oron et al199) in vitro, initially for Glu uncaging200 and more recently for optogenetic activation of fast-acting201 and red-shifted202 optogenetic probes. Modulation of the stimulating light in the z dimension in vivo is technically challenging as it requires a moveable objective, making its use in freely behaving animals virtually impossible. In vivo multi-photon imaging of genetically encoded calcium indicators has generated ground-breaking studies in the last decade, relying on 2PE microscopes imaging brain tissue through a cranial window of head-fixed animals.203–205 Although their head is fixed, animals can perform in behavioural experiments based on their navigation atop a 2D treadmill in a projected virtual environment.206,207
Deep brain stimulation
Cranial windows do not allow for deep-brain light stimulation, although this could be achieved with the insertion of a micro-objective or a smaller gradient index lens–based objective.208 Another possible method for achieving complex light stimulation in deeper structures, without requiring complex lenses, is through the use of single-core, micro-fabricated multimode fibres.209 This approach is based on digital phase conjugation requiring a calculation of the appropriate wavefront bringing light into defined patterns at the fibre outputs. Further applications of such fibres include advanced microscopy210 as well as the possibility to deliver spectrally distinct radiations at different depths194 (Figure 4B). However, there may be complications due to fibre bending, making this approach inappropriate for tethered animals.
Future Perspectives
Chemiluminescence
Optogenetics is an extremely powerful tool to control neuronal excitability and signalling; however, illuminating the cells, a relatively easy task in vitro, becomes the limiting step when trying to apply the same probes to living and behaving animals. Although successful attempts have been made to control animal behaviour by conveying the light through implanted cannulae,34,35,37–40 it is difficult to envisage this method to be the technique of choice for future applications in biomedicine.
In recent years, scientists have started to turn back to what had become an almost obsolete tool, i.e., bioluminescence-emitting luciferases. With the aim of in vivo applications, several groups have tried to engineer red-shifted variants of both luciferases and their substrates.211,212 Interestingly, luciferase-emitted bioluminescence can be used to activate optogenetic probes, provided the emission of the luciferases and the excitation spectra of the optogenetic probes substantially overlap, thus potentially bypassing the need of surgical implants to deliver light from an external source and using systemic injections instead. The use of luciferase to activate optogenetic probes differs from that of chemogenetic modulatory systems such as Designer Receptor Exclusively Activated by Designer Drugs (DREADDs). The former can specifically affect protein function at the intracellular and extracellular level, whereas the later has to make use of signalling pathways originating from membrane-bound receptors.213 Firefly luciferase could activate Natronomonas NpHR when co-expressed in neurons.214 Taking a step forward, the bioluminescence resonance energy transfer–based nanolantern probe, formed by the fluorescent protein Venus and Renilla luciferase,215,216 was fused to Natronomonas NpHR. The resulting construct, named inhibitory luminopsin or ‘iLMO’, could silence neuronal firing on coelenterazine (CTZ) administration and modify mouse behaviour when expressed into the striatum and globus pallidum.217 Similarly, a small luciferase from the sea shrimp Gaussia princeps (Gluc) was fused to ChR2.218 Using enhanced versions of Gluc, more efficient probes have been recently created by fusing the luciferase with either ChR2 (excitatory luminopsins) or proton pumps (iLMOs). These constructs proved to work also in vivo, as they were delivered to the substantia nigra of living mice and could be activated by both direct and systemic delivery of CTZ.219
Thus, there is an increasing interest towards the use of bioluminescence as an internal light source. This undoubtedly provides several advantages compared with the ‘classic’ optogenetic approach, chiefly the opportunity to avoid surgery, effectively representing a hardware-free opto-system. This approach is, however, still in its infancy and awaits the availability of a wider panel of luciferases, with different emission kinetics and spectra, as well as of chemically modified and/or caged substrates, which could render luciferase activation more specific in time and space. The combinatorial use of these tools could bring optogenetics a step closer to routine and user-friendly biomedical applications.
Optimization of optogenetic probes
A commonly adopted strategy to tune the photoreceptor photocycle, and thus to adapt it to the specific activity of the effector domain, is to engineer mutations that alter the photocycle time scale by modifying either the chemistry of the cofactor or the properties of the downstream structural moiety. Another possibility is to act directly on the effector domain, for example, by designing mutations that can enhance the interaction with its target. As a speculative example, let us discuss one of the chimeras realized by our group, the AsLOV2-PAH1 construct,65 where, as effector domain we used the minimal REST-interacting sequence of the corepressor mSin3a. By inspection of the atomic structure obtained by nuclear magnetic resonance studies, it is possible to observe that the REST/PAH1-interacting surface is composed mostly of hydrophobic residues, except for a charged Lys amino acid (K155) on PAH1, which is buried among neutral partners. By performing a computational analysis using a dedicated software,220 we predicted that changing this residue into a hydrophobic one, such as Phe, would increase the binding free energy of the REST/PAH1 complex by 1.5 kcal/mol. Thus, a chimeric construct carrying the K155F-mutated effector domain should compete more effectively with the endogenous PAH1 binding to REST and give rise to an enhanced binding of the photo-excited chimera.
Recent developments in the field of developmental biology have also yielded new optogenetic tools capable of activating differentiation and developmental features. Examples include the photo-induction of ectopic endodermal cells in zebra fish,221 an ectopic tail-like structure in Xenopus222 and the triggering of embryogenesis in Drosophila.223
Conclusions
In this review, we have tried to summarize the most significant aspects of optogenetics applied to neuronal plasticity. When trying to engineer a novel light-sensitive actuator, but also when ‘simply’ using one of the many tools already available, several challenges have to be faced. This includes the choice of the most appropriate light-sensitive system, of the most efficient way to target it to the cell/brain region under study and of the hardware that could most effectively deliver light while minimizing off-target effects. This may seem an insidious and daunting task; however, despite such complexity, several elegant studies have demonstrated the feasibility of these approaches, showing that synaptic plasticity can indeed be interrogated and/or manipulated using light. The scientific community is putting a continuous effort in optimizing available systems and engineering novel probes, which makes optogenetics a constantly evolving field. Such strive is especially needed to devise optogenetic-based biomedical applications, for which ease-of-use, low-cost, and simple equipment are highly desirable. With the amount of technical and practical knowledge currently in our hands, the advent of such user-friendly tools may not be very far ahead.
Footnotes
PEER REVIEW: Seven peer reviewers contributed to the peer review report. Reviewers’ reports totalled 1461 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) received no financial support for the research, authorship, and/or publication of this article.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
CE, FC, LM, FB, and JFMV jointly developed the structure and arguments of the manuscript. All authors reviewed and approved the final version of the script.
REFERENCES
- 1.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catterall WA, Few AP. Calcium channel regulation and presynaptic plasticity. Neuron. 2008;59:882–901. doi: 10.1016/j.neuron.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Higley MJ, Sabatini BL. Calcium signaling in dendrites and spines: practical and functional considerations. Neuron. 2008;59:902–913. doi: 10.1016/j.neuron.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 6.Singer W. Development and plasticity of cortical processing architectures. Science. 1995;270:758–764. doi: 10.1126/science.270.5237.758. [DOI] [PubMed] [Google Scholar]
- 7.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zocchi L, Sassone-Corsi P. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol. 2010;20:432–440. doi: 10.1016/j.conb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tognini P, Napoli D, Tola J, et al. Experience-dependent DNA methylation regulates plasticity in the developing visual cortex. Nat Neurosci. 2015;18:956–958. doi: 10.1038/nn.4026. [DOI] [PubMed] [Google Scholar]
- 12.Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 13.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 14.Konermann S, Brigham MD, Trevino AE, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Roux N, Amar M, Fossier P. Acquiring new information in a neuronal network: from Hebb’s concept to homeostatic plasticity. J Soc Biol. 2008;202:143–160. doi: 10.1051/jbio:2008018. [DOI] [PubMed] [Google Scholar]
- 17.Kampa BM, Clements J, Jonas P, Stuart GJ. Kinetics of Mg2+ unblock of NMDA receptors: implications for spike-timing dependent synaptic plasticity. J Physiol. 2004;556:337–345. doi: 10.1113/jphysiol.2003.058842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kauer JA, Malenka RC, Nicoll RA. NMDA application potentiates synaptic transmission in the hippocampus. Nature. 1988;334:250–252. doi: 10.1038/334250a0. [DOI] [PubMed] [Google Scholar]
- 19.Lei S, Pelkey KA, Topolnik L, Congar P, Lacaille JC, McBain CJ. Depolarization-induced long-term depression at hippocampal mossy fiber-CA3 pyramidal neuron synapses. J Neurosci. 2003;23:9786–9795. doi: 10.1523/JNEUROSCI.23-30-09786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- 21.Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- 22.Yates D. Synaptogenesis: a synaptic bridge. Nat Rev Neurosci. 2016;17:135. doi: 10.1038/nrn.2016.12. [DOI] [PubMed] [Google Scholar]
- 23.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Tye KM, Deisseroth K. Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat Rev Neurosci. 2012;13:251–266. doi: 10.1038/nrn3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deisseroth K, Feng G, Majewska AK, Miesenböck G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iurilli G, Ghezzi D, Olcese U, et al. Sound-driven synaptic inhibition in primary visual cortex. Neuron. 2012;73:814–828. doi: 10.1016/j.neuron.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson NR, Runyan CA, Wang FL, Sur M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature. 2012;488:343–348. doi: 10.1038/nature11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Ramirez S, Pang PT, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez S, Liu X, Lin P-A, et al. Creating a false memory in the hippocampus. Science. 2013;341:387–391. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- 34.Tye KM, Prakash R, Kim S-Y, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tye KM, Mirzabekov JJ, Warden MR, et al. Dopamine neurons modulate neural encoding and expression of depression-related behaviour. Nature. 2013;493:537–541. doi: 10.1038/nature11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haubensak W, Kunwar P, Cai H, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin D, Boyle MP, Dollar P, et al. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470:221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atasoy D, Betley JN, Su HH, Sternson SM. Deconstruction of a neural circuit for hunger. Nature. 2012;488:172–177. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jennings JH, Rizzi G, Stamatakis AM, Ung RL, Stuber GD. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science. 2013;341:1517–1521. doi: 10.1126/science.1241812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busskamp V, Duebel J, Balya D, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 42.Lagali PS, Balya D, Awatramani GB, et al. Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration. Nat Neurosci. 2008;11:667–675. doi: 10.1038/nn.2117. [DOI] [PubMed] [Google Scholar]
- 43.Greenberg KP, Pham A, Werblin FS. Differential targeting of optical neuromodulators to ganglion cell soma and dendrites allows dynamic control of center-surround antagonism. Neuron. 2011;69:713–720. doi: 10.1016/j.neuron.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 44.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kravitz AV, Freeze BS, Parker PRL, et al. Regulation of parkinsonian motor behaviors by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paz JT, Davidson TJ, Frechette ES, et al. Closed-loop optogenetic control of thalamus as a tool for interrupting seizures after cortical injury. Nat Neurosci. 2013;16:64–70. doi: 10.1038/nn.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I. On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy. Nat Commun. 2013;4:1376. doi: 10.1038/ncomms2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gradinaru V, Zhang F, Ramakrishnan C, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F, Wang LP, Brauner M, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 50.Moglich A, Hegemann P. Biotechnology: programming genomes with light. Nature. 2013;500:406–408. doi: 10.1038/500406a. [DOI] [PubMed] [Google Scholar]
- 51.Karunarathne WK, Giri L, Kalyanaraman V, Gautam N. Optically triggering spatiotemporally confined GPCR activity in a cell and programming neurite initiation and extension. Proc Natl Acad Sci U S A. 2013;110:E1565–E1574. doi: 10.1073/pnas.1220697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikonov SS, Kholodenko R, Lem J, Pugh EN., Jr Physiological features of the S- and M-cone photoreceptors of wild-type mice from single-cell recordings. J Gen Physiol. 2006;127:359–374. doi: 10.1085/jgp.200609490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emanuel AJ, Do MT. Melanopsin tristability for sustained and broadband phototransduction. Neuron. 2015;85:1043–1055. doi: 10.1016/j.neuron.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 55.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 56.Yizhar O, Fenno LE, Prigge M, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koyanagi M, Takano K, Tsukamoto H, Ohtsu K, Tokunaga F, Terakita A. Jellyfish vision starts with cAMP signaling mediated by opsin-Gs cascade. Proc Natl Acad Sci U S A. 2008;105:15576–15580. doi: 10.1073/pnas.0806215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chuong AS, Miri ML, Busskamp V, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17:1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smallwood PM, Olveczky BP, Williams GL, et al. Genetically engineered mice with an additional class of cone photoreceptors: implications for the evolution of color vision. Proc Natl Acad Sci U S A. 2003;100:11706–11711. doi: 10.1073/pnas.1934712100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muller K, Engesser R, Schulz S, et al. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masuda S, Nakatani Y, Ren S, Tanaka M. Blue light-mediated manipulation of transcription factor activity in vivo. ACS Chem Biol. 2013;8:2649–2653. doi: 10.1021/cb400174d. [DOI] [PubMed] [Google Scholar]
- 64.Zayner JP, Sosnick TR. Factors that control the chemistry of the LOV domain photocycle. PLoS ONE. 2014;9:e87074. doi: 10.1371/journal.pone.0087074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paonessa F, Criscuolo S, Sacchetti S, et al. Regulation of neural gene transcription by optogenetic inhibition of the RE1-silencing transcription factor. Proc Natl Acad Sci U S A. 2016;113:E91–E100. doi: 10.1073/pnas.1507355112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Moglich A. From dusk till dawn: one-plasmid systems for light-regulated gene expression. J Mol Biol. 2012;416:534–542. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 67.Nihongaki Y, Suzuki H, Kawano F, Sato M. Genetically engineered photoinducible homodimerization system with improved dimer-forming efficiency. ACS Chem Biol. 2014;9:617–621. doi: 10.1021/cb400836k. [DOI] [PubMed] [Google Scholar]
- 68.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 69.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 70.Aravanis AM, Wang LP, Zhang F, et al. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–S156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 71.Haupts U, Tittor J, Bamberg E, Oesterhelt D. General concept for ion translocation by halobacterial retinal proteins: the isomerization/switch/transfer (IST) model. Biochemistry. 1997;36:2–7. doi: 10.1021/bi962014g. [DOI] [PubMed] [Google Scholar]
- 72.Zhang F, Vierock J, Yizhar O, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koyanagi M, Terakita A. Diversity of animal opsin-based pigments and their optogenetic potential. Biochim Biophys Acta. 2014;1837:710–716. doi: 10.1016/j.bbabio.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 75.Chow BY, Han X, Dobry AS, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- 77.Chen X, Spudich JL. Demonstration of 2:2 stoichiometry in the functional SRI-HtrI signaling complex in Halobacterium membranes by gene fusion analysis. Biochemistry. 2002;41:3891–3896. doi: 10.1021/bi015966h. [DOI] [PubMed] [Google Scholar]
- 78.Alfonsa H, Lakey JH, Lightowlers RN, Trevelyan AJ. Cl-out is a novel cooperative optogenetic tool for extruding chloride from neurons. Nat Commun. 2016;7:13495. doi: 10.1038/ncomms13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Masseck OA, Spoida K, Dalkara D, et al. Vertebrate cone opsins enable sustained and highly sensitive rapid control of Gi/o signaling in anxiety circuitry. Neuron. 2014;81:1263–1273. doi: 10.1016/j.neuron.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 80.Cehajic-Kapetanovic J, Eleftheriou C, Allen AE, et al. Restoration of vision with ectopic expression of human rod opsin. Curr Biol. 2015;25:2111–2122. doi: 10.1016/j.cub.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bailes HJ, Zhuang LY, Lucas RJ. Reproducible and sustained regulation of Gαs signalling using a metazoan opsin as an optogenetic tool. PLoS ONE. 2012;7:e30774. doi: 10.1371/journal.pone.0030774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terakita A, Tsukamoto H, Koyanagi M, Sugahara M, Yamashita T, Shichida Y. Expression and comparative characterization of Gq-coupled invertebrate visual pigments and melanopsin. J Neurochem. 2008;105:883–890. doi: 10.1111/j.1471-4159.2007.05184.x. [DOI] [PubMed] [Google Scholar]
- 84.Bailes HJ, Lucas RJ. Melanopsin and inner retinal photoreception. Cell Mol Life Sci. 2010;67:99–111. doi: 10.1007/s00018-009-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walmsley L, Hanna L, Mouland J, et al. Colour as a signal for entraining the mammalian circadian clock. PLoS Biol. 2015;13:e1002127. doi: 10.1371/journal.pbio.1002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spangler SM, Bruchas MR. Optogenetic approaches for dissecting neuromodulation and GPCR signaling in neural circuits. Curr Opin Pharmacol. 2016;32:56–70. doi: 10.1016/j.coph.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 88.Li P, Rial D, Canas PM, et al. Optogenetic activation of intracellular adenosine A2A receptor signaling in the hippocampus is sufficient to trigger CREB phosphorylation and impair memory. Mol Psychiatry. 2015;20:1339–1349. doi: 10.1038/mp.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siuda ER, Copits BA, Schmidt MJ, et al. Spatiotemporal control of opioid signaling and behavior. Neuron. 2015;86:923–935. doi: 10.1016/j.neuron.2015.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spoida K, Masseck OA, Deneris ES, Herlitze S. Gq/5-HT2c receptor signals activate a local GABAergic inhibitory feedback circuit to modulate serotonergic firing and anxiety in mice. Proc Natl Acad Sci U S A. 2014;111:6479–6484. doi: 10.1073/pnas.1321576111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salomon M, Eisenreich W, Dürr H, et al. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc Natl Acad Sci U S A. 2001;98:12357–12361. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zoltowski BD, Schwerdtfeger C, Widom J, et al. Conformational switching in the fungal light sensor vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 96.Pudasaini A, El-Arab KK, Zoltowski BD. LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Front Mol Biosci. 2015;2:18. doi: 10.3389/fmolb.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci U S A. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moon J, Gam J, Lee SG, Suh YG, Lee J. Light-regulated tetracycline binding to the Tet repressor. Chemistry. 2014;20:2508–2514. doi: 10.1002/chem.201304027. [DOI] [PubMed] [Google Scholar]
- 99.Wu YI, Wang X, He L, Montell D, Hahn KM. Spatiotemporal control of small GTPases with light using the LOV domain. Methods Enzymol. 2011;497:393–407. doi: 10.1016/B978-0-12-385075-1.00016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Strickland D, Lin Y, Wagner E, et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc. 2012;134:16480–16483. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richter F, Fonfara I, Bouazza B, et al. Engineering of temperature- and light-switchable Cas9 variants. Nucleic Acids Res. 2016;44:10003–10014. doi: 10.1093/nar/gkw930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dagliyan O, Tarnawski M, Chu PH, et al. Engineering extrinsic disorder to control protein activity in living cells. Science. 2016;354:1441–1444. doi: 10.1126/science.aah3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Masuda S. Light detection and signal transduction in the BLUF photoreceptors. Plant Cell Physiol. 2013;54:171–179. doi: 10.1093/pcp/pcs173. [DOI] [PubMed] [Google Scholar]
- 105.Anderson S, Dragnea V, Masuda S, Ybe J, Moffat K, Bauer C. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44:7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu Q, Gardner KH. Structure and insight into blue light-induced changes in the BlrP1 BLUF domain. Biochemistry. 2009;48:2620–2629. doi: 10.1021/bi802237r. [DOI] [PubMed] [Google Scholar]
- 107.Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem. 2010;285:41501–41508. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 109.Liu B, Liu H, Zhong D, Lin C. Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol. 2010;13:578–586. doi: 10.1016/j.pbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Che DL, Duan L, Zhang K, Cui B. The dual characteristics of light-induced cryptochrome 2, homo-oligomerization and heterodimerization, for optogenetic manipulation in mammalian cells. ACS Synth Biol. 2015;4:1124–1135. doi: 10.1021/acssynbio.5b00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang K, Duan L, Ong Q, et al. Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in C12 cell neurite outgrowth. PLoS ONE. 2014;9:e92917. doi: 10.1371/journal.pone.0092917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rockwell NC, Lagarias JC. A brief history of phytochromes. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci U S A. 2008;105:14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Piatkevich KD, Subach FV, Verkhusha VV. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat Commun. 2013;4:2153. doi: 10.1038/ncomms3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Filonov GS, Verkhusha VV. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol. 2013;20:1078–1086. doi: 10.1016/j.chembiol.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gasser C, Taiber S, Yeh CM, et al. Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci U S A. 2014;111:8803–8808. doi: 10.1073/pnas.1321600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R. The UVR8 UV-B photoreceptor: perception, signaling and response. Arabidopsis Book. 2013;11:e0164. doi: 10.1199/tab.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Heilmann M, Jenkins GI. Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 2013;161:547–555. doi: 10.1104/pp.112.206805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci U S A. 2013;110:1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Crefcoeur RP, Yin R, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 123.Chen D, Gibson ES, Kennedy MJ. A light-triggered protein secretion system. J Cell Biol. 2013;201:631–640. doi: 10.1083/jcb.201210119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lin W, Albanese C, Pestell RG, Lawrence DS. Spatially discrete, light-driven protein expression. Chem Biol. 2002;9:1347–1353. doi: 10.1016/s1074-5521(02)00288-0. [DOI] [PubMed] [Google Scholar]
- 125.Shi Y, Koh JT. Light-activated transcription and repression by using photocaged SERMs. Chembiochem. 2004;5:788–796. doi: 10.1002/cbic.200300823. [DOI] [PubMed] [Google Scholar]
- 126.Kandler K, Katz LC, Kauer JA. Focal photolysis of caged glutamate produces long-term depression of hippocampal glutamate receptors. Nat Neurosci. 1998;1:119–123. doi: 10.1038/368. [DOI] [PubMed] [Google Scholar]
- 127.Gautier A, Nguyen DP, Lusic H, An W, Deiters A, Chin JW. Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 128.Gautier A, Deiters A, Chin JW. Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hemphill J, Chou C, Chin JW, Deiters A. Genetically encoded light-activated transcription for spatiotemporal control of gene expression and gene silencing in mammalian cells. J Am Chem Soc. 2013;135:13433–13439. doi: 10.1021/ja4051026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Bio. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gorostiza P, Volgraf M, Numano R, Szobota S, Trauner D, Isacoff EY. Mechanisms of photoswitch conjugation and light activation of an ionotropic glutamate receptor. Proc Natl Acad Sci U S A. 2007;104:10865–10870. doi: 10.1073/pnas.0701274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Szobota S, Gorostiza P, Del Bene F, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 133.Izquierdo-Serra M, Trauner D, Llobet A, Gorostiza P. Optical modulation of neurotransmission using calcium photocurrents through the ion channel LiGluR. Front Mol Neurosci. 2013;6:3. doi: 10.3389/fnmol.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hou Q, Gilbert J, Man HY. Homeostatic regulation of AMPA receptor trafficking and degradation by light-controlled single-synaptic activation. Neuron. 2011;72:806–818. doi: 10.1016/j.neuron.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wyart C, Del Bene F, Warp E, et al. Optogenetic dissection of a behavioral module in the vertebrate spinal cord. Nature. 2009;461:407–410. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kauwe G, Isacoff EY. Rapid feedback regulation of synaptic efficacy during high-frequency activity at the Drosophila larval neuromuscular junction. Proc Natl Acad Sci U S A. 2013;110:9142–9147. doi: 10.1073/pnas.1221314110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Levitz J, Pantoja C, Gaub B, et al. Optical control of metabotropic glutamate receptors. Nat Neurosci. 2013;16:507–516. doi: 10.1038/nn.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin WC, Davenport CM, Mourot A, et al. Engineering a light-regulated GABAA receptor for optical control of neural inhibition. ACS Chem Biol. 2014;9:1414–1419. doi: 10.1021/cb500167u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Janovjak H, Szobota S, Wyart C, Trauner D, Isacoff EY. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat Neurosci. 2010;13:1027–1032. doi: 10.1038/nn.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Numano R, Szobota S, Lau AY, et al. Nanosculpting reversed wavelength sensitivity into a photoswitchable iGluR. Proc Natl Acad Sci U S A. 2009;106:6814–6819. doi: 10.1073/pnas.0811899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kienzler MA, Reiner A, Trautman E, Yoo S, Trauner D, Isacoff EY. A red-shifted, fast-relaxing azobenzene photoswitch for visible light control of an ionotropic glutamate receptor. J Am Chem Soc. 2013;135:17683–17686. doi: 10.1021/ja408104w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Levitz J, Popescu AT, Reiner A, Isacoff EY. A toolkit for orthogonal and in vivo optical manipulation of ionotropic glutamate receptors. Front Mol Neurosci. 2016;9:2. doi: 10.3389/fnmol.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Caporale N, Kolstad KD, Lee T, et al. LiGluR restores visual responses in rodent models of inherited blindness. Mol Ther. 2011;19:1212–1219. doi: 10.1038/mt.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gaub BM, Berry MH, Holt AE, et al. Restoration of visual function by expression of a light-gated mammalian ion channel in retinal ganglion cells or ON-bipolar cells. Proc Natl Acad Sci U S A. 2014;111:E5574–E5583. doi: 10.1073/pnas.1414162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kianianmomeni A. UVB-based optogenetic tools. Trends Biotechnol. 2015;33:59–61. doi: 10.1016/j.tibtech.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 148.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 149.Schindler SE, McCall JG, Yan P, et al. Photo-activatable Cre recombinase regulates gene expression in vivo. Sci Rep. 2015;5:13627. doi: 10.1038/srep13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 151.Chen X, Liu R, Ma Z, et al. An extraordinary stringent and sensitive light-switchable gene expression system for bacterial cells. Cell Res. 2016;26:854–857. doi: 10.1038/cr.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jayaraman P, Devarajan K, Chua TK, Zhang H, Gunawan E, Poh CL. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Res. 2016;44:6994–7005. doi: 10.1093/nar/gkw548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Levskaya A, Chevalier AA, Tabor JJ, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 154.Tabor JJ, Levskaya A, Voigt CA. Multichromatic control of gene expression in Escherichia coli. J Mol Biol. 2011;405:315–324. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu H, Gomez G, Lin S, Lin S, Lin C. Optogenetic control of transcription in zebrafish. PLoS ONE. 2012;7:e50738. doi: 10.1371/journal.pone.0050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Motta-Mena LB, Reade A, Mallory MJ, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chan YB, Alekseyenko OV, Kravitz EA. Optogenetic control of gene expression in Drosophila. PLoS ONE. 2015;10:e0138181. doi: 10.1371/journal.pone.0138181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Day JJ. New approaches to manipulating the epigenome. Dialogues Clin Neurosci. 2014;16:345–357. doi: 10.31887/DCNS.2014.16.3/jday. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ali AM, Reis JM, Xia Y, et al. Optogenetic inhibitor of the transcription factor CREB. Chem Biol. 2015;22:1531–1539. doi: 10.1016/j.chembiol.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kawano F, Okazaki R, Yazawa M, Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat Chem Biol. 2016;12:1059–1064. doi: 10.1038/nchembio.2205. [DOI] [PubMed] [Google Scholar]
- 161.Edwards WF, Young DD, Deiters A. Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem Biol. 2009;4:441–445. doi: 10.1021/cb900041s. [DOI] [PubMed] [Google Scholar]
- 162.Cao J, Arha M, Sudrik C, Bugaj LJ, Schaffer DV, Kane RS. Light-inducible activation of target mRNA translation in mammalian cells. Chem Commun. 2013;49:8338–8340. doi: 10.1039/c3cc44866e. [DOI] [PubMed] [Google Scholar]
- 163.Ogasawara S. Control of cellular function by reversible photoregulation of translation. Chembiochem. 2014;15:2652–2655. doi: 10.1002/cbic.201402495. [DOI] [PubMed] [Google Scholar]
- 164.You M, Jaffrey SR. Designing optogenetically controlled RNA for regulating biological systems. Ann N Y Acad Sci. 2015;1352:13–19. doi: 10.1111/nyas.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 166.Scheyltjens I, Laramée M-E, Van den Haute C, et al. Evaluation of the expression pattern of rAAV2/1, 2/5, 2/7, 2/8, and 2/9 serotypes with different promoters in the mouse visual cortex. J Comp Neurol. 2015;523:2019–2042. doi: 10.1002/cne.23819. [DOI] [PubMed] [Google Scholar]
- 167.Gerits A, Vancraeyenest P, Vreysen S, et al. Serotype-dependent transduction efficiencies of recombinant adeno-associated viral vectors in monkey neocortex. Neurophotonics. 2015;2:031209. doi: 10.1117/1.NPh.2.3.031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tsai HC, Zhang F, Adamantidis A, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gerfen CR, Sawchenko PE. An anterograde neuroanatomical tracing method that shows the detailed morphology of neurons, their axons and terminals: immunohistochemical localization of an axonally transported plant lectin, Phaseolus vulgaris leucoagglutinin (PHA-L) Brain Res. 2016;1645:42–45. doi: 10.1016/j.brainres.2015.12.040. [DOI] [PubMed] [Google Scholar]
- 170.Kato S, Inoue K, Kobayashi K, et al. Efficient gene transfer via retrograde transport in rodent and primate brains using a human immunodeficiency virus type 1-based vector pseudotyped with rabies virus glycoprotein. Hum Gene Ther. 2007;18:1141–1151. doi: 10.1089/hum.2007.082. [DOI] [PubMed] [Google Scholar]
- 171.Mueller C, Flotte TR. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- 172.Ellis BL, Hirsch ML, Porter SN, Samulski RJ, Porteus MH. Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by food and drug administration-approved drugs. Gene Ther. 2013;20:35–42. doi: 10.1038/gt.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Smith C, Gore A, Yan W, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Al-Jamal WT, Kostarelos K. Liposomes: from a clinically established drug delivery system to a nanoparticle platform for theranostic nanomedicine. Acc Chem Res. 2011;44:1094–1104. doi: 10.1021/ar200105p. [DOI] [PubMed] [Google Scholar]
- 176.Pantarotto D, Singh R, McCarthy D, et al. Functionalized carbon nanotubes for plasmid DNA gene delivery. Angew Chem Int Ed Engl. 2004;43:5242–5246. doi: 10.1002/anie.200460437. [DOI] [PubMed] [Google Scholar]
- 177.Curtin CM, Cunniffe GM, Lyons FG, et al. Innovative collagen nano-hydroxyapatite scaffolds offer a highly efficient non-viral gene delivery platform for stem cell-mediated bone formation. Adv Mater. 2012;24:749–754. doi: 10.1002/adma.201103828. [DOI] [PubMed] [Google Scholar]
- 178.Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer BA. Overcoming nonviral gene delivery barriers: perspective and future. Mol Pharm. 2013;10:4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Miyata K, Nishiyama N, Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem Soc Rev. 2012;41:2562–2574. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- 180.Choi J, Young JA, Callaway EM. Selective viral vector transduction of ErbB4 expressing cortical interneurons in vivo with a viral receptor-ligand bridge protein. Proc Natl Acad Sci U S A. 2010;107:16703–16708. doi: 10.1073/pnas.1006233107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gomez EJ, Gerhardt K, Judd J, Tabor JJ, Suh J. Light-activated nuclear translocation of adeno-associated virus nanoparticles using phytochrome B for enhanced, tunable, and spatially programmable gene delivery. ACS Nano. 2016;10:225–237. doi: 10.1021/acsnano.5b05558. [DOI] [PubMed] [Google Scholar]
- 182.Stolik S, Delgado JA, Pérez A, Anasagasti L. Measurement of the penetration depths of red and near infrared light in human ‘ex vivo’ tissues. J Photochem Photobiol B. 2000;57:90–93. doi: 10.1016/s1011-1344(00)00082-8. [DOI] [PubMed] [Google Scholar]
- 183.Henderson TA, Morries LD. Near-infrared photonic energy penetration: can infrared phototherapy effectively reach the human brain? Neuropsychiatr Dis Treat. 2015;11:2191–2208. doi: 10.2147/NDT.S78182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tedford CE, DeLapp S, Jacques S, Anders J. Quantitative analysis of transcranial and intraparenchymal light penetration in human cadaver brain tissue. Lasers Surg Med. 2015;47:312–322. doi: 10.1002/lsm.22343. [DOI] [PubMed] [Google Scholar]
- 185.Zanke BW, Boudreau K, Rubie E, et al. The stress-activated protein kinase pathway mediates cell death following injury induced by cis-platinum, UV irradiation or heat. Curr Biol. 1996;6:606–613. doi: 10.1016/s0960-9822(02)00547-x. [DOI] [PubMed] [Google Scholar]
- 186.Kulms D, Schwarz T. Molecular mechanisms involved in UV-induced apoptotic cell death. Skin Pharmacol Appl Skin Physiol. 2002;15:342–347. doi: 10.1159/000064539. [DOI] [PubMed] [Google Scholar]
- 187.Sinha RP, Häder DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 188.Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem Soc Rev. 2013;42:3441–3452. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Göppert-Mayer M. Über Elementarakte mit zwei Quantensprüngen. Annalen der Physik. 1931;401:273–294. doi: 10.1002/andp.19314010303. [DOI] [Google Scholar]
- 190.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci U S A. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kwon KY, Lee HM, Ghovanloo M, Weber A, Li W. Design, fabrication, and packaging of an integrated, wirelessly-powered optrode array for optogenetics application. Front Syst Neurosci. 2015;9:69. doi: 10.3389/fnsys.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kwon KY, Lee HM, Ghovanloo M, Weber A, Li W. A wireless slanted optrode array with integrated micro leds for optogenetics; 2014 IEEE 27th International Conference on Micro Electro Mechanical Systems (MEMS); 26–30 Jan. 2014; 2014. pp. 813–816. [Google Scholar]
- 193.Clements IP, Gnade AG, Rush AD, Patten CD, Twomey MC, Kravitz AV. Miniaturized LED sources for in vivo optogenetic experimentation. 2013. p. 85860X-X-9. [Google Scholar]
- 194.Pisanello F, Sileo L, Oldenburg IA, et al. Multipoint-emitting optical fibers for spatially addressable in vivo optogenetics. Neuron. 2014;82:1245–1254. doi: 10.1016/j.neuron.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V. Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron. 2014;84:1157–1169. doi: 10.1016/j.neuron.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 196.Procyk CA, Eleftheriou CG, Storchi R, et al. Spatial receptive fields in the retina and dorsal lateral geniculate nucleus of mice lacking rods and cones. J Neurophysiol. 2015;114:1321–1330. doi: 10.1152/jn.00368.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Barrett JM, Hilgen G, Sernagor E. Dampening spontaneous activity improves the light sensitivity and spatial acuity of optogenetic retinal prosthetic responses. Sci Rep. 2016;6:33565. doi: 10.1038/srep33565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Barrett JM, Degenaar P, Sernagor E. Blockade of pathological retinal ganglion cell hyperactivity improves optogenetically evoked light responses in rd1 mice. Front Cell Neurosci. 2015;9:330. doi: 10.3389/fncel.2015.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Oron D, Papagiakoumou E, Anselmi F, Emiliani V. Two-photon optogenetics. Prog Brain Res. 2012;196:119–143. doi: 10.1016/B978-0-444-59426-6.00007-0. [DOI] [PubMed] [Google Scholar]
- 200.Dal Maschio M, Difato F, Beltramo R, Blau A, Benfenati F, Fellin T. Simultaneous two-photon imaging and photo-stimulation with structured light illumination. Opt Express. 2010;18:18720–18731. doi: 10.1364/OE.18.018720. [DOI] [PubMed] [Google Scholar]
- 201.Ronzitti E, Conti R, Papagiakoumou E, et al. Sub-millisecond optogenetic control of neuronal firing with two-photon holographic photoactivation of Chronos. bioRxiv. 2016 doi: 10.1101/062182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chaigneau E, Ronzitti E, Gajowa MA, et al. Two-photon holographic stimulation of ReaChR. Front Cell Neurosci. 2016;10:234. doi: 10.3389/fncel.2016.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Horton NG, Wang K, Kobat D, et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics. 2013:7. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Holtmaat A, Randall J, Cane M. Optical imaging of structural and functional synaptic plasticity in vivo. Eur J Pharmacol. 2013;719:128–136. doi: 10.1016/j.ejphar.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 205.Holtmaat A, Bonhoeffer T, Chow DK, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Leinweber M, Zmarz P, Buchmann P, et al. Two-photon calcium imaging in mice navigating a virtual reality environment. J Vis Exp. 2014:e50885. doi: 10.3791/50885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Aronov D, Tank DW. Engagement of neural circuits underlying 2D spatial navigation in a rodent virtual reality system. Neuron. 2014;84:442–456. doi: 10.1016/j.neuron.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ziv Y, Burns LD, Cocker ED, et al. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bianchi S, Rajamanickam VP, Ferrara L, Di Fabrizio E, Liberale C, Di Leonardo R. Focusing and imaging with increased numerical apertures through multimode fibers with micro-fabricated optics. Opt Lett. 2013;38:4935–4938. doi: 10.1364/OL.38.004935. [DOI] [PubMed] [Google Scholar]
- 210.Cižmár T, Dholakia K. Exploiting multimode waveguides for pure fibre-based imaging. Nat Commun. 2012;3:1027. doi: 10.1038/ncomms2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 212.Giuliani G, Molinarib P, Ferretti G, et al. New red-shifted coelenterazine analogues with an extended electronic conjugation. Tetrahedron Lett. 2012;53:5114–5118. [Google Scholar]
- 213.Storchi R, Milosavljevic N, Eleftheriou CG, et al. Melanopsin-driven increases in maintained activity enhance thalamic visual response reliability across a simulated dawn. Proc Natl Acad Sci U S A. 2015;112:E5734–E5743. doi: 10.1073/pnas.1505274112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Land BB, Brayton CE, Furman KE, Lapalombara Z, Dileone RJ. Optogenetic inhibition of neurons by internal light production. Front Behav Neurosci. 2014;8:108. doi: 10.3389/fnbeh.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Saito K, Chang YF, Horikawa K, et al. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat Commun. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Takai A, Nakano M, Saito K, et al. Expanded palette of nano-lanterns for realtime multicolor luminescence imaging. Proc Natl Acad Sci U S A. 2015;112:4352–4356. doi: 10.1073/pnas.1418468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tung JK, Gutekunst CA, Gross RE. Inhibitory luminopsins: genetically-encoded bioluminescent opsins for versatile, scalable, and hardware-independent optogenetic inhibition. Sci Rep. 2015;5:14366. doi: 10.1038/srep14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Berglund K, Birkner E, Augustine GJ, Hochgeschwender U. Light-emitting channelrhodopsins for combined optogenetic and chemical-genetic control of neurons. PLoS ONE. 2013;8:e59759. doi: 10.1371/journal.pone.0059759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Berglund K, Clissold K, Li HE, et al. Luminopsins integrate opto- and chemogenetics by using physical and biological light sources for opsin activation. Proc Natl Acad Sci U S A. 2016;113:E358–E367. doi: 10.1073/pnas.1510899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dehouck Y, Kwasigroch JM, Rooman M, Gilis D. BeAtMuSiC: prediction of changes in protein-protein binding affinity on mutations. Nucleic Acids Res. 2013;41:W333–W339. doi: 10.1093/nar/gkt450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Reade A, Motta-Mena LB, Gardner KH, et al. TAEL: a zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development. 2017;144:345–355. doi: 10.1242/dev.139238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Krishnamurthy VV, Khamo JS, Mei W, et al. Reversible optogenetic control of kinase activity during differentiation and embryonic development. Development. 2016;143:4085–4094. doi: 10.1242/dev.140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Johnson HE, Goyal Y, Pannucci L, et al. The spatiotemporal limits of developmental Erk signaling. Dev Cell. 2017;40:185–192. doi: 10.1016/j.devcel.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]