Summary
Much like a factory, the endoplasmic reticulum assembles simple cellular building blocks into complex molecular machines known as proteins. In order to protect the delicate protein folding process and ensure the proper cellular delivery of protein products under environmental stresses, eukaryotes have evolved a set of signaling mechanisms known as the unfolded protein response (UPR) to increase the folding capacity of the endoplasmic reticulum. This process is particularly important in plants, because their sessile nature commands adaptation for survival rather than escape from stress. As such, plants make special use of the UPR, and evidence indicates that the master regulators and downstream effectors of the UPR have distinct roles in mediating cellular processes that affect organism growth and development as well as stress responses. In this review we outline recent developments in this field that support a strong relevance of the UPR to many areas of plant life.
Keywords: Unfolded protein response, ER stress, IRE1, bZIP60, bZIP28, Arabidopsis thaliana
Introduction
As the endoplasmic reticulum (ER) is the entry point to secretory pathway (Vitale and Denecke 1999), the primary site of phospholipid synthesis (Ohlrogge and Browse 1995), a hub for critical stress and growth signaling molecules (Light et al. 2016, Ron and Walter 2007, Shore et al. 2011), and the assembly plant for a third of a cell's total proteome (Wallin and Heijne 1998), interruptions in its functions can have vast consequences in cellular health. Under physiological conditions of growth, a dedicated battery of ER-resident proteins can prevent misfolding of nascent polypeptides and facilitate acquisition of the proper tertiary structure through post-translational modification (e.g., covalent addition of oligosaccharide chains or catalysis of disulfide bond formation) (Dobson 2003, Gupta and Tuteja 2011). In plants affected by environmental stress (e.g. heat stress (Gidalevitz et al. 2011)), proper folding of secretory proteins can be impaired and misfolded proteins can accumulate in the ER igniting a potentially lethal condition known as ER stress (Dobson 2003, Hartl and Haver-Hartl 2009, Buchberger et al 2010). Indeed, under prolonged or severe levels of stress, the accumulation and aggregation of unfolded proteins can become cytotoxic and lead to death of the plant cell in a manner akin to the effect of misfolded proteins that aggregate and cause human neurodegenerative diseases such as Alzheimer's (Hoozemans et al. 2005). The unfolded protein response (UPR) is a set of signaling mechanisms designed to prevent accumulation of misfolded proteins in the ER. Specialized ER-localized membrane proteins are able to detect the buildup of unfolded proteins and activate signaling cascades that modulate the abundance of the proteins dedicated to the folding of nascent polypeptides within the ER to maintain homeostasis (Ron and Walter 2007, Ruberti and Brandizzi 2014, Ruberti et al. 2015). In metazoans the ER-localized stress sensor array consists of the Inositol Requiring Enzyme 1 (IRE1), Activating Transcription Factor 6 (ATF6) and Protein kinase R-like Endoplasmic Reticulum Kinase (PERK) (Harding et al. 1999, Shen et al. 2002, Wang et al. 1998). In plants, the functional homologs of IRE1 and ATF6 (termed IRE1a, IRE1b and bZIP28 in Arabidopsis thaliana) have been identified to date (Iwata and Koizumi 2012). In yeast, only the IRE1-dependent UPR pathway has been identified (Ron and Walter 2007, Mori 2009). A number of comparative reviews have highlighted the similarities between UPR components shared among plants, metazoans and yeast, and should be referred to for greater contexts on the depth of gene conservation shared by all eukaryotes in regards to UPR-related mechanisms (Chen and Brandizzi 2013, Liu and Howell 2016, Ruberti and Brandizzi 2014, Ruberti et al. 2015).
The study of the UPR in human and animal models largely centers on genetic defects that allow the buildup of misfolded proteins, and mitigating the cytotoxic effects of the resulting aggregates (Rao and Bredesen 2004, Stefani and Dobson 2003). In plants, UPR research also focuses on improving crop yield under adverse environmental conditions. There is a large potential for biotechnological applications for UPR-related mechanisms in ensuring plant productivity. However, considerable work must be done to understand how the UPR is integrated into intra and intercellular signaling mechanisms that are plant specific. Even though there is considerable evidence to suggest that the UPR components are required for many different aspects of plant physiology, from seed germination to meristematic maintenance (Barba-Espín et al. 2014, Chen and Brandizzi 2012,Chen et al. 2014, Deng et al. 2013, Klein et al. 2006, Deng et al. 2016, Meng et al. 2016), we are only beginning to connect the molecular activities of IRE1 and bZIP28 to the modulation of organism growth and development.
In this review, we provide a summary of causative ER stress conditions followed by an examination of UPR signaling pathways through a discussion of recent advancements in the field. Given space constraints, this review focuses mainly on UPR studies in the model dicot Arabidopsis thaliana. Throughout, we draw attention of the reader to new developments in the context of plant growth, development, and metabolic functioning. This is done in an attempt to move beyond a classical linear signal transduction paradigm, and visualize the plant UPR as a network that incorporates energy availability, plant production needs, and environmental conditions into a cohesive output governing plant life.
ER Quality Control and Important ER Client Proteins Mediate both Programmed Cell Death and Cell Cycle Control
If the plant cell was reimagined as a city, it would be easy to see how the ER could be described as the town's central factory. At the ER, shipments of raw materials in the form of amino acids and carbohydrates are reshaped and assembled into fully-functional molecular machines in the form of proteins. Properly folded proteins are then shipped out and utilized for a variety of different purposes in different places throughout or outside the cell. In order to prevent the production of faulty goods, the ER has specific machinery, collectively called ER quality control (ERQC), to survey the protein folding status, facilitate folding and ensure quality of the produced protein. The production of most secretory proteins begins with the co-translational introduction of the protein into the ER. In this process, specific peptide sequences target nascent polypeptide chains to the ER and are translocated across the membrane as they are synthesized via the Sec translocon, which is largely conserved between yeast and plants (Akopian et al. 2013, Denecke et al. 1992, Deneke et al. 1993, Schweiger and Schenkert 2013). As the polypeptide enters the ER lumen, molecular chaperones such as the luminal binding proteins (BiPs), bind to the chain of the nascent polypeptides and prevent premature folding (Carvalho et al. 2014, Foresti et al. 2003). The oligosaccharyltransferase (OST) complex (Lerouxel et al. 2005) recognizes specific amino acid sequences and transfers N-linked glycans to the peptides. In some cases, this post translational modification adds to the intrinsic stability or solubility of a protein, and importantly, it functions as a recognition beacon for major ER luminal foldase complexes (Sinclair and Elliott 2005). Nascent polypeptides undergo iterative folding cycles where they are passed between the calnexin/calreticulin complex, and UDP-glucose glycoprotein-glucosyltransferase (UGGT), which monitor protein folding and retains unfolded proteins in the ER (Totani et al., 2009) as a part of the ERQC. Other proteins participate in folding cycles under the purview of these central ER foldase complexes, such as thioredoxins (e.g protein disulfide isomerases (PDIs)), which catalyze the reduction and reformation of disulfide bonds (Bottomley et al. 2001, Wilkinson and Gilbert 2004). Properly folded proteins are then transported to the Golgi apparatus, while the unfolded or irremediably misfolded proteins are picked up by proteins like OS9 of the ER-associated protein degradation (ERAD) system, dislocated out of the ER, ubiquitinated, and finally degraded by the 26S proteasome (Huttner et al. 2012). The process is conserved across eukaryotes and, for example, it mediates the proper folding of critical client plasma membrane receptor proteins in plants, including the Arabidopsis elongation factor Tu (EF-Tu) receptor which mediates pathogen associated molecular pattern based immunity (Li et al. 2009) and brassinosteroid insensitive 1 (BRI1) receptor (Li and Chory 1997).
Although not directly related to the activation of the UPR, the essential physiological relevance of these folding processes, which are monitored by the UPR regulators (via unfolded protein accumulation), are continuously being demonstrated. Beyond enabling proper function of cellular signaling pathways with receptors at the plasma membrane (like EF-Tu and BRI1), the specificity of N-linked glycosylation-bearing proteins has recently been shown to play important roles in regulating cell death. The Arabidopsis BAK1 (BRI1-associated receptor kinase 1) and SERK4 (somatic embryo receptor kinase 4) both interact with immune receptors and BRI1 and negatively regulate hypersensitive response-like programmed cell death (PCD) through yet-unknown mechanisms (Gou et al. 2012, Li et al. 2002, Nam and Li 2002, Roux et al. 2011). Intriguingly, loss of STT3a (staurosporin and temperature sensitive 3), one of the two catalytic subunits of the OST complex involved in N-glycosylation of ER proteins, is linked to the cell death phenotype observed in BAK1/SERK4 silenced plants (de Oliveira et al. 2016). However, UPR deficient mutants did not respond differently to BAK1/SERK1 silencing, which led to the conclusion that UPR regulators IRE1a, IRE1b and bZIP28 may not modulate this specific type of PCD (de Oliveira et al. 2016).
When examined under salt stress conditions, a stt3a knockout line showed UPR activation and halted cell cycle progression in a similar manner to that described in yeast and mammalian cells after being subjected to ER stress-inducing conditions (Arnold and Tanner 1982, Brewer et al. 1999, Koiwa et al. 2003). This may suggest a possible antagonistic role between STT3a and the UPR components, or may simply indicate that the proteins needed to adapt to salt stress may be glycosylated in order to fold or function properly. In both cases, although the UPR may not directly regulate the expression of BAK1/SERK4 or N-glycosylated salt protective genes, these examples underscore the importance of maintaining the ER as a fully-functional protein folding factory so a plant may adapt to various sources of stresses.
The Causes of ER Stress: Unavoidable Exogenous Threats
Enhancing the UPR appears potentially critical to efforts to maintain crop productivity by priming plants to survive under adverse environmental conditions (Valente et al. 2009, Carvalho et al. 2014, Luan et al. 2016 Xiang et al. 2016). One of the most thoroughly described environmental UPR inducer is heat stress (Deng et al. 2011, Duke and Doehlert 1996, Gao et al. 2008, Schmollinger et al. 2013, Yang et al. 2009). More conveniently than heat conditions in the lab, chemical UPR inducers such as tunicamycin, which inhibits the N-linked glycosylation in the ER lumen, are often used to investigate the UPR in many eukaryotic model organisms by mimicking the conditions associated with environmental stresses that cause the buildup of unfolded proteins. Extreme osmotic stress and heavy metals such as selenium have also shown to induce the UPR (Liu et al. 2007, Van Hoewyk 2016). The UPR also plays a role in response to pathogens and other biotic stresses (Moreno et al. 2012, Prasch and Sonnewald 2013, Zhang et al. 2015). Through an unknown mechanism, treatment of Arabidopsis plants with the biotic stress-hormone salicylic acid (SA) was shown to activate both arms of the UPR controlled by IRE1 and bZIP28 (Moreno et al. 2012, Nagashima et al. 2014). This SA-induced UPR activation was also given context by Meng et al. (2016), which showed that the SA-accumulating cpr5 (Constitutive Expression of PR genes 5) mutant is dependent on the UPR to suppress growth. Other studies have demonstrated the interesting effect of organelle sourced reactive oxygen species (ROS) (e.g. plastids and mitochondria) on the induction of the UPR (Ozgur et al. 2015) suggesting a possible functional connection between the UPR and non-secretory organelles. It was demonstrated that plastid-originated ROS production induced UPR activation suggesting that plastidial stress may be intimately linked with, and responded to, through the ER stress signal transduction mechanisms (Ozgur et al. 2015). Given the close connections between SA and ROS signaling (Torres et al. 2006, Mou et al. 2003), these observations may indicate a functional link between the two UPR activating conditions.
The Canonical Response to Unfolded Proteins: New Look at the Master Regulators IRE1, bZIP60, and bZIP28
IRE1
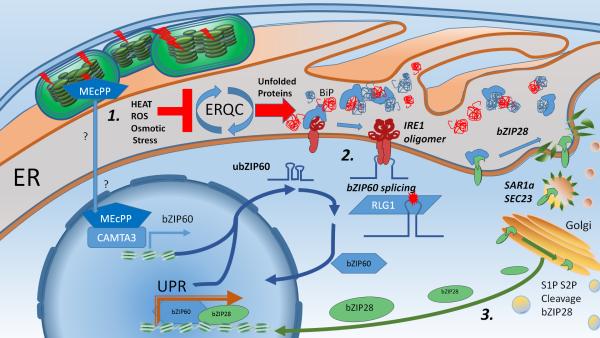
In environmentally stressful conditions, the accumulation of unfolded and irremediably misfolded proteins leads to the activation of the UPR via IRE1 and bZIP28 (Figure 1). Although the activation mechanism of IRE1 has yet to be established in plants, studies of IRE1 in yeast have indicated that BiP binds the IRE1 luminal domain in non-stressful conditions. However, upon stress, BiP chaperones preferentially bind unfolded proteins thereby freeing the ER luminal domains of IRE1 (Pincus et al. 2010). Unfolded proteins then bind to IRE1 (Gardner and Walter 2011) which oligomerizes; as a consequence, the kinase domain in the cytosolic portion of the protein is autophosphorylated (Shamu and Walter 1996, Welihinda and Kaufman 1996). These steps lead to activation of the IRE1 ribonuclease domain (Cox and Walter 1996). In mammalian cells misfolded proteins bind to BiP chaperones which keep IRE1 in an inactive monomeric state under normal conditions (Berlotti et al. 2000). After dissociation from BiP, interactions between the freed luminal domains of IRE1 bring cytoplasmic domains together allowing the necessary autophosphorylation (Credle et al. 2005, Ali et al. 2011). Although the underlying mechanisms for activation of plant IRE1 are not known, it is certain that activation of IRE1 results in the unconventional cytosolic splicing of the mRNA transcripts of bZIP60, the only UPR target of plant IRE1 known to date (Deng, et al. 2011, Nagashima et al. 2011, Hayashi et al. 2012, Lu et al. 2012, Li et al. 2012 Moreno et al. 2012). In Arabidopsis, the tRNA ligase RLG1 was recently shown to complete the splicing of bZIP60 in vitro by ligating the fragments derived from IRE1 cleavage (Nagashima et al. 2016). Whether the ligation of bZIP60 spliced transcripts is mediated by RLG1 in vivo, while likely, has yet to be experimentally demonstrated, partly due to the lethality of complete RLG1 loss-of-function mutations (Nagashima et al. 2016). In yeast and mammalian cells, the ligation of the functional equivalents of bZIP60, Hac1 and XbpI, respectively, occurs via tRNA ligases that operate in opposite fashion. In detail, the ligation of Hac1 mediated by RLG1p in yeast is completed in a 5’-3’ fashion followed by removal of 2` phosphate group by a phosphatase (Sawaya et al. 2003, Steiger et al. 2005). In mammalian cells, the ligation is mediated by RTCB, which operates in a 3’-5’ ligation (Jurkin et al. 2014). Evidence suggests that plants have evolved a 5’-3’ ligation mechanism similar to that found in yeast, and this finding is supported by observations that Arabidopsis RLG1 can also ligate the Hac1 mRNA in yeast cells (Mori et al. 2010). However, the resulting Hac1 transcripts are not efficiently translated indicating that functional differences between yeast and Arabidopsis catalytic mechanisms remain. The splicing of bZIP60 leads to a frameshift that removes a transmembrane domain from the translated transcription factor. Spliced bZIP60 subsequently translocated to the nucleus for transcriptional modulation of the downstream UPR target genes. Intriguingly, the activity of IRE1 is not limited to unconventional splicing of a transcription factor. Regulated IRE1 dependent decay (RIDD) is a process which regulates mRNA abundance and it is active during ER stress. Such IRE1 mediated cleavage of cytosolic and ER-associated mRNAs has been implicated in promoting cell death in yeast (Tam et al. 2014). In metazoans, RIDD activity under prolonged or severe ER stress also has proapoptotic effects, and can be associated with human diseases such as diabetes (Maurel et al. 2014, Hetz and Glimcher 2011). RIDD activity was also described for the Arabidopsis IRE1 homolog (Mishiba et al. 2013), although the implications related to stress outcomes are yet to be tested in vivo.
Figure 1. Mechanisms of the plant UPR activation.
Environmental stresses (e.g. heat, oxidative stress, selenium, chemical inhibitors) negatively affect the protein folding process, leading to the buildup of unfolded proteins. Plastid stress promotes the buildup of MEcPP (2-C-Methyl-D-erythritol-2,4-cyclopyrophosphate), which through an unknown mechanism, activates CAMTA3 (Calmodulin biding transcriptional activator 3) inducing transcription of bZIP60. 2. BiP ER luminal proteins bind unfolded proteins. The buildup of unfolded proteins leads to activation of IRE1 mediated splicing of bZIP60, which is then potentially ligated by the RNA ligase RLG1. The spliced bZIP60 is a nuclear localized transcription factor which binds to promoters of downstream UPR target genes, Regulated IRE1 dependent decay (RIDD) although not depicted here, regulates mRNA abundance and it is active during ER stress. 3. bZIP28 after being freed from BiPs by the presence of unfolded proteins is trafficked to the Golgi in a SAR1a/SEC23-dependent way. S2P proteases cleave the active transcription factor from its transmembrane domain. bZIP28 enters the nucleus and acts redundantly with bZIP60 to transcriptionally activate the UPR.
Evidence that plant IRE1 assumes other roles besides the splicing of bZIP60, including RIDD and/or other yet-unknown activities of IRE1, can be found when comparing the stress responsive and vegetative phenotypic differences between IRE1 and bZIP60 loss-of-function mutants in Arabidopsis. Although no other direct splicing substrate has been found for IRE1 activity other that bZIP60 in plants, bzip60 knockout mutants are not nearly as sensitive as the ire1a/ire1b mutant to prolonged ER stress (Deng, et al. 2013, Mishiba, et al. 2013), implying diverse roles of IRE1 during the UPR. Whether these may depend on the ability of IRE1 to phosphorylate other substrates as opposed to its ribonuclease domain is a tantalizing idea that is yet to be experimentally tested. Furthermore, bzip60 plants are apparently indistinguishable from wild-type plants under normal conditions of growth, while IRE1 partial loss of function mutants (ire1a/ire1b) display shorter root growth as developmental defects, and ire1 null mutations are lethal (Chen and Brandizzi 2012, Deng et al. 2013, Lu and Christopher 2008). Comparative analyses between bzip60 and ire1a/ire1b mutants under stressed and unstressed conditions may yet yield information that could potentially explain these physiological differences and provide a better understanding of the molecular mechanisms which tie IRE1 specifically to the control of plant health and cell fate decisions.
bZIP60
Although seemingly unimportant when compared with IRE1 or redundant when compared with other UPR signal transducers like bZIP28 (Sun et al. 2013a) in conditions tested thus far, bZIP60 appears to have unique contributions to the UPR management. Indeed, a close examination of the ER molecular phenotypes associated with bzip60 knockouts and expression patterns of bZIP60 yields some interesting information. The unspliced version of bZIP60 encodes a membrane bound transcription factor, and while it is found to be transcribed under normal conditions, the accumulation of the protein encoded by unspliced bZIP60 has been debated (Iwata et al. 2008, Iwata et al. 2009, Parra-Rojas et al. 2015). Although methods of detection between studies have been different, it has been shown that the product of the unspliced bZIP60 isoform accumulates to higher levels in seedlings treated with a proteasome inhibitor compared to untreated seedlings (Parra-Rojas et al. 2015). When this is taken into context with the observation that a cellular recycling process known as macroautophagy (hereafter termed autophagy) is constitutively active in the bzip60 knockout (Liu et al. 2012), it could be hypothesized that the dynamic levels of unspliced bZIP60 products are post-translationally subject to selective protein degradation and may regulate aspects of ER homeostasis, including protection of the ER against autophagy.
bZIP60 has also been shown to be transcriptionally regulated in conditions outside of the canonical UPR as well. In chloroplasts, reductive potential is funneled into carbon fixation and the synthesis of long carbon chains through the plastid isoprenoid metabolic pathway, collectively known as the methylerythritol 4-phosphate (MEP) pathway (Banerjee and Sharkey 2014). This biosynthetic track produces numerous molecular-end products, which range in function from growth and development (e.g., gibberellin precursors) to synthesis of isoprene gas, which protects chloroplast under heat stress (Banerjee and Sharkey 2014, Hedden and Thomas 2012, Sharkey 2005). Retrograde stress signaling mechanisms from chloroplasts to nucleus utilize the buildup of the MEP pathway intermediate methylerythritol cyclodiphosphate (MEcPP) in an abiotic stress dependent manner (Xiao et al. 2012). The CEH1 (1-hydroxy-2-methyl-2-(E)-butenyl4-diphosphate synthase) enzyme, which catalyzes the conversion of MEcPP to hydroxymethylbutenyl diphosphate (HMBPP) is dependent upon reductive potential of NADPH from the light reactions and, as a result, the activity of CEH1 is known to be oxidative stress sensitive (Ostrovsky et al. 1998). Fascinatingly, the MEcPP retrograde signal, transmitted in a calcium-dependent way via the calmodulin binding transcription activator 3 (CAMTA3), leads directly to transcriptional induction of bZIP60 (Benn et al. 2016). The possibility that the UPR responds to plastid metabolic dysfunction may also indicate that, like in humans (Ron and Walter 2007), the UPR may have tissue-specific metabolic reprogramming functions that have gone unnoticed in plants. This intracellular organelle bridge deserves further scrutiny, and its connection to the observations made by Ozgur et al. (2015) in their study of UPR activation under oxidative stress cannot be understated. When sourced from different organelles, ROS had highly variable effects in stimulating expression of some UPR genes while repressing others (e.g. methyl viologen induces BiP3 expression but heavily repressed BiP2 expression). Although bZIP60 expression and splicing have yet to be tracked under these conditions, this information could help identify mechanisms for selective upregulation or downregulation of UPR genes. This is especially valuable given that current studies of the two arms of the plant UPR are most often considered in the context of coordinated upregulation of UPR genes (Nagashima et al. 2014).
bZIP28
The ER transmembrane transcription factor bZIP28 is released from binding to BiP chaperones and is trafficked to the Golgi apparatus where putative site 1 and site 2 protease (S1P and S2P)-mediated proteolytic mechanisms split the active transcription factor from its transmembrane domain and allow its relocation to the nucleus (Gao et al. 2008, Liu et al. 2007, Srivastava et al. 2013, Liu et al. 2013, Sun et al. 2013c, Sun et al. 2015). Once in the nucleus the activated bZIP28 forms protein complexes with NF-Y transcription factors, and together bind specific ER stress related cis-elements (ERSE), which consist of two consensus sequences separated by 10 nucleotides (CCAAT-N10-CACG) and are commonly found in UPR upregulated gene promoter regions (Liu et al. 2010).
Looking beyond the functions of bZIP28 in the nucleus, recent work has elucidated the mechanisms by which bZIP28 is shuttled to the Golgi apparatus in an ER stress dependent fashion. In exploring the functional diversity of the COPII components involved in ER export, Zeng et al. (2015) mapped the biochemical interactions necessary for proper transfer of bZIP28 to the Golgi apparatus under ER stress. ER-to-Golgi transport is mediated by a specialized protein machinery collectively known COPII (Brandizzi and Barlowe, 2014). Assembly of this machinery requires SAR1, a GTPase that in the active form recruits the COPII coat components, SEC23/Sec24 and SEC13/SEC31. These components are required for cargo selection at the ER and shuttling to the Golgi. Although SAR1a shares high sequence identity to other secretion associated RAS-related GTPase homologs of the SAR1-family in Arabidopsis (Hanton et al. 2008), a single cysteine residue substitution at the amino acid position 84 was found to be required for interaction with SEC23a (Zeng et al. 2015). A dominant negative allele of SAR1a impaired the export of bZIP28 from the ER to the Golgi under tunicamycin-induced stress in Arabidopsis protoplasts. In contrast with other SAR1 and SEC23 homologs in Arabidopsis, the SAR1a/SEC23a pair is transcriptionally induced under ER stress conditions (Song et al. 2013). As such they may play an important role in promoting and sustaining the activity of bZIP28 during ER stress. This also opens up avenues to explore other conditions which could promote condition-specific trafficking of bZIP28, and subsequently potential activation of bZIP28 in developmental or growth contexts without activation of the canonical UPR cascade. This may be especially impactful with respect to bZIP28 functions in promoting brassinosteroid (BR) sensitivity (Che et al. 2010). S1P-S2P mediated cleavage of bZIP28 and another bZIP-transcription factor (bZIP17) under ER stress promote active BR signaling in adaptation to abiotic stress responses (Che et al. 2010). Although this phenomenon is presumed to be result of ERQC upregulation and increased delivery of functional client protein BRI1 sensor to the cell surface (Gendron and Wang 2007, Jin et al. 2007, Hong et al. 2008, Jin et al. 2009, Che et al. 2010), the exact mechanism through which bZIP28 and bZIP17 positively impact BR signaling remains to be discovered.
Future Exploration: bZIP60 and bZIP28 mediate Epigenetic Modifications with the COMPASS like Complex
In contrast with the single bzip60 and bzip28 mutants, the bzip60 bzip28 double mutant shows enhanced susceptibility to prolonged ER stress (Deng et al. 2013), indicating both functional redundancy and potential cooperativity. Under ER stress conditions both bZIP60 and bZIP28 localize to the nucleus and have been shown to interact with each other to modulate the expression of UPR genes (Song et al. 2015). The recent findings implicating bZIP60 and bZIP28 in the direction of histone methylation activities of the COMPASS complex presents interesting opportunities for further study. In plants the COMPASS complex is responsible for increasing the frequency of trimethylation of histone 3 in the promoter region of actively expressed genes through association with an unidentified histone methyltransferase (Jiang et al. 2011). Previously, although there was strong evidence supporting the correlation between H3K4 histone trimethylation and gene transcription, no mechanisms were known to direct these epigenetic modifications in sequence-specific ways in plants (Li et al. 2008, Zhang et al. 2009). However, Song et al. (2015) showed that bZIP60 and bZIP28 interact with the proteins Ash2 (absent, small, homeotic like factor 2) and WDR5a (WD40 containing repeat 5a), which form the core elements of the COMPASS complex in plants (Jiang et al. 2011). Furthermore, the inducible expression of a subset of UPR responsive genes during ER stress was dramatically compromised in the wdr5a and ash2 mutants (Song et al. 2015). The lasting effects of these epigenetic marks after ER stress are not clear, but H3K4 trimethylation marks accumulate on genes involved in heat stress memory (Lämke et al. 2015). This may indicate that the UPR plays an important role in priming plants against future environmental stresses. Furthermore, H3K4 methylation and the COMPASS complex also play important roles in mediating temperature sensitive transitions in plant development (Jiang et al. 2011, Kumar and Wigge 2010, Zilberman et al. 2008). Exploring how UPR stress signaling and the constitutive functions of UPR related genes integrate with these epigenetic regulating mechanisms through bZIP28 and particularly bZIP60 might also provide useful insights into the developmental phenotypes found in ire1a/ire1b (Chen and Brandizzi 2012, Deng et al. 2013), which are deficient in bZIP60 splicing.
Send in the Cavalry: Downstream Effectors during ER Stress
Nuclear translocation of spliced bZIP60 and cleaved bZIP28 leads to an increase in the transcription of genes coding for ER luminal proteins (BiP/Hsp70, ERdj/HSP40, HSP90), which (i) prevent aggregation of misfolded protein and newly translated polypeptides (Gupta et al. 2011), (ii) limit uncontrolled folding of nascent polypeptides through ERQC, and (iii) translocate terminally misfolded proteins across the ER membrane to the 26S proteasome by a group of membrane bound complexes in a processes collectively termed ER associated degradation (ERAD-L, -M and –C for luminal, membrane, and cytoplasmic ERAD, respectively (Olzmann et al. 2013). These downstream effectors, although largely conserved across eukaryotes, have been found to have specific significance in plants during normal growth and in response to ER stress (Klein et al. 2006, Liu et al. 2015, Yang et al. 2009). A noteworthy example of this is provided in the study of the plant specific properties of the conserved ER luminal chaperone HSP90.7, also known in Arabidopsis as SHEPHERD (SHD) (Ishiguro et al. 2002). When compared to the non-selective foldase activities of its mammalian counterpart GRP94, the designation of HSP90.7 as a general ER chaperone in plants (like BiP/HSP70) has been questioned (Klein et al. 2006, Marzec et al. 2012). In particular, Arabidopsis HSP90.7 which is highly upregulated under ER stress conditions was also found to have specific functions in proliferating tissues (Ishiguro et al. 2002, Klein et al. 2006). The shd (hsp90.7) knockout mutant is phenotypically identical to clv a mutant defective in CLAVATA signaling, a critical negative modulator of shoot apical meristem activity, indicating it may be required for plant specific production of the CLAVATA peptide (Aichinger et al. 2012, Miwa et al. 2009). Additional plant specific activities of HSP90.7 were demonstrated by Chong et al.(2014) who found that a short sequence of highly charged amino acids present only in plant ER-localized homologs drastically affected the survival rates of Arabidopsis seedlings under tunicamycin - and high calcium- induced ER stress. As expected overexpression of the HSP90.7 chaperone conferred significant resistance to tunicamycin, heat and high calcium induced ER stress; conversely overexpression of an HSP90.7 mutant with the highly charged 22 residue sequence deleted (HSP90.7Δ22 ), while still more resistant to heat stress than wild type, showed a marked increase in lethality in response to tunicamicyn compared to both native HSP90.7 overexpressor plants and even wild-type plants. These observations beyond illustrating plant specific chaperone functions also provide an important example which implies that in plants tunicamycin-induced stress may be responded to in a manner that is separate from heat induced ER stresses. Furthermore, it may be possible that the UPR directly mediates meristematic growth within undifferentiated tissues themselves through the HSP90.7/ CLAVATA relationship, perhaps in combination with transmission of signals through secondary messengers from distant tissues.
Further examples of UPR effectors which have plant-specific roles in growth and development can also be found with respect to ERdj3 (ER resident J domain 3) protein function during gametophyte development (Yamamoto et al. 2008). J domain proteins (Hsp40) found in the ER lumen bind BiP proteins and stabilize their interactions with client unfolded proteins (Misselwitz et al. 1998, Yamamoto et al. 2008). ERdj3A, which is induced under ER stress, contains a C-terminal protein disulfide isomerase domain that has reductive capabilities on substrates in vitro (Yang et al. 2009), in addition to a HSP40 ATPase activity (Ma et al. 2015). This suggests that ERdj3A may act on a specific subset of client proteins. Further in vivo analysis of ERdj3A and its homologs ERdj3B and P58IPK support this possibility by demonstrating their importance in development (Maruyama et al. 2014b). Indeed, genetic analysis of the mutant Thermosensitive male sterile 1 (tms1) revealed a nonfunctional allele of ERdj3A that under elevated temperatures was defective in pollen tube growth (Yang et al. 2009). Under normal conditions, in conjunction with P58IPK and ERdj3b, ERdj3A was also shown to mediate polar haploid nuclei fusion in female gametophytes (Maruyama et al. 2014b) prior to double fertilization. During this nuclear fusion process, the perinuclear ER fuses with the outer nuclear envelope and creates a continuous outer membrane around the two haploid nuclei, and it is followed by a second fusion of the inner nuclear membranes (Jensen 1964, Maruyama et al. 2010). Recently it was demonstrated that ERdj3A and P58IPK are required for the fusion of the ER membrane with the outer nuclear membranes. A double knockout (erdj3a p58IPK) resulted in seed abortion after fertilization due to aberrant endosperm proliferation, similar to that found in bip1 bip2 double mutants (Maruyama et al. 2010, Maruyama et al. 2014b). The inner membrane fusion requires the ERdj3B/ P58IPK pair, and although the erdj3b p58 IPK double mutants had unfused haploid nuclei in close proximity, unlike erdj3a p58IPK no aborted seeds were found (Maruyama et al. 2014a). The developmental defects found in plants with mutant alleles of UPR induced ER resident proteins (e.g., ERdj, BiP, SHD) are consistent with the evidence that pollen development in an ire1a ire1b double mutant is highly vulnerable to heat stress (Deng et al. 2016, Fragkostefanakis et al. 2016). However, the observed rescue of male fertility through the overexpression of a single COPII coat component SEC31a is intriguing given the large gene list shown to be regulated by IRE1 in these conditions (Deng et al. 2016). This underscores the need to fully understand the detailed functional mechanisms of downstream UPR components. Although studies exploring the similarities between yeast, mammalian, and plant UPR have led to significant advances in plant ER stress research, in order to fully understand the mechanisms connecting the UPR to plant specific physiology it will also be important to look at the contrasting characteristics. These plant specific cases, such as the single amino acid substitution in the case of SAR1a important for bZIP28 shuttling (Zeng et al. 2015), or the small charged region in the case of HSP90.7, which drastically alters stress responsive phenotypes compared to the deletion mutant (Chong et al. 2014), are potent reminders to expect the unexpected, even in evolutionarily conserved contexts.
NAC Membrane Transcription Factors: A Second Set of UPR activators?
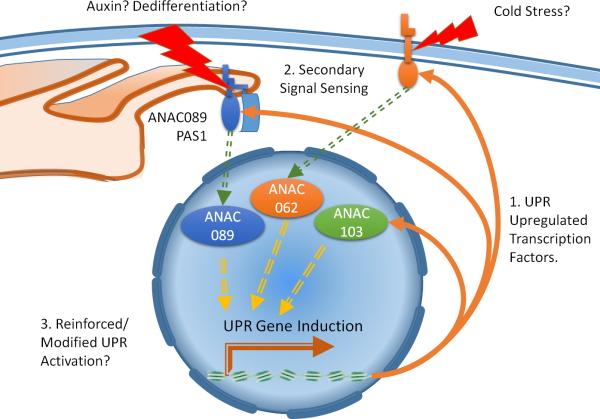
In addition to the transcriptional activities of bZIP60 and bZIP28, a veritable menagerie of plant-specific transcription factors is involved in the response to ER stress (Figure 2) (Sun et al. 2013b, Yang et al. 2014a, Yang et al. 2014b). Thus far however, their mechanistic involvement in the UPR is yet to be fully elucidated. ANAC062 a plasma membrane-bound transcription factor, which is proteolytically cleaved and nuclear localized in response to cold stress, has been linked to the expression of multiple pathogenesis-related genes in a salicylic acid independent fashion and has also been shown to be upregulated under ER stress (Seo et al. 2010). Overexpression of a truncated form lacking the C-terminal membrane domain was shown to induce canonical UPR responsive chaperones and improve prolonged ER stress outcomes (Yang et al. 2014b), supporting a functional connection between ANAC062 and the UPR. Furthermore, ANAC103, a soluble nuclear transcription factor was shown to be transcriptionally induced under ER stress (Sun et al. 2013b). Overexpression of ANAC103 with a small C-terminal deletion to enhance stability of the expressed protein was also shown to induce UPR genes, although true knockouts showed no appreciable ER stress phenotype suggesting a functional redundancy between ANAC103 and other UPR related transcription factors in response to tunicamycin (Sun et al. 2013b). A third transcription factor, the membrane bound ANAC089, has been implicated in promoting programmed cell death in Arabidopsis. Inducible overexpression of truncated ANAC089 ignited mammalian apoptotic-like symptoms in roots (Yang et al. 2014a). RNAi lines targeting ANAC089 also displayed resistance to tunicamycin induced ER stress (Yang et al. 2014a). These results support earlier findings implicating ANAC089 in the control of ER-related homeostatic mechanisms. Smyczynsk et al. (2006) in their study of a conserved immunophilin peptidylprolyl cis-trans-isomerase PASTICCINO1 (PAS1) demonstrated through a series of biochemical experiments that PAS1 and ANAC089 (which they termed FAN, for FKBP associated NAC) interact in vivo and in vitro (Smyczynski et al. 2006). PAS1 is important in maintaining proper morphology during embryo development in Arabidopsis (Vittorioso et al. 1998). Expressed in response to cytokinin, ER-localized PAS1 associates with the VLCFA (very long chain fatty acid) synthesis complex and promotes sphingolipid biosynthesis and subsequently organ polarity through the lipids effect on the localization of the PIN1 efflux carrier (Roudier et al. 2010). A C-terminal domain of PAS1 is required for interaction and co-localization with ANAC089 (Smyczynski et al. 2006). That same domain is also required for nuclear exclusion of the PAS1. Interestingly, both PAS1 and ANAC089 are highly expressed in the mature embryo and were found to relocate from ER to the nucleus dependent upon exogenous 1-naphthaleneacetic acid treatment during cellular dedifferentiation in Arabidopsis root tips (Smyczynski et al. 2006). When overexpressed in the pas1 loss of function allele, the full-length ANAC089 resulted in partial complementation of the deformed growth phenotype (Smyczynski et al. 2006). When this is taken into account in the context of the RNAi knockdowns of ANAC089 under ER stress, it may be possible that the reduction of endogenous ANAC89 may perturb cytokinin-auxin balance in a manner that promotes cell divisions in spite of continued stress signals. Taken together, these ANAC transcription factors may indeed play important roles in the secondary activation of the UPR cascade; they might also play important roles in activation of the UPR in response to other stress or hormone signaling mechanisms. For example, in a manner opposite STT3a which seems to prevent the activation of the UPR in salt stress conditions (Koiwa et al. 2003), perhaps plants treated with tunicamycin in cold conditions may require NAC062 for full UPR gene expression regulation as cold signaling mechanisms may interfere with canonical activation of IRE1 or bZIP28. Expansion of combinatorial stress experiments incorporating tunicamycin-induced stress with other environmental stressors may give a more accurate picture of regulation, and downstream targets of these transcription factors. This may consequently lead to more information regarding the in vivo function of the unmodified transcription factors in relation to the unfolded protein response.
Figure 2. UPR-regulated Transcription Factors may Tailor UPR Activation to Specific Stress Combinations.
Several plant-specific transcription factors are upregulated under ER stress in an IRE1/bZIP28 dependent way. Two membrane-bound transcription factors, ANAC062 and ANAC089, have been shown to relocate to the nucleus under ER stress. ANAC062 also is nuclear localized under cold stress conditions. ANAC089 along with interactor PAS1 (PASTICCINO1) were also shown to relocate to the nucleus in roots treated with 1-naphthaleneacetic acid to induce dedifferentiation. ANAC103, a soluble transcription factor, was also shown to upregulate UPR genes upon over expression. Although truncated forms of these transcription factors were shown to upregulate UPR responsive genes, the molecular mechanisms of action of native transcription factors are still unknown. After upregulation by IRE1/bZIP28 dependent mechanisms, these transcription factors may respond to secondary signals which reinforce or alter UPR gene expression.
Concluding Remarks: The Case for an Expanding UPR
The molecular products assembled inside the ER have an ever expanding relevance to plants under environmental stress. Although many open questions still plague the study of the UPR in plants, including the identity of the molecular mechanisms for the activation and de-activation of the master regulator IRE1, the general relevance of the UPR maintaining ER homeostasis is clear. The ERQC and UPR maintain the folding capacity of the ER, and in doing so, enable a wide range of downstream processes from proper heat stress adaptation to defense against pathogens. Specifically, the downstream effectors of the UPR have been implicated in transcriptional and post transcriptional regulation of both ER homeostatic genes, and developmental processes. However, new oddities arising in research focusing upstream and downstream of the UPR offer ever expanding possibilities where the UPR may play a defining role in plant physiology. UPR activation in response to plastid metabolic dysfunction, and oxidative stress implicates the potential for the UPR to respond in many different signal transduction cascades that utilize reactive oxygen species as a secondary messenger. Further inquiry exploring the canonical UPR, in non-canonical and tissue-specific contexts may help elucidate hidden functions and better integrate our understanding of UPR functionality in plant life.
Significance Statement.
The endoplasmic reticulum (ER) is the entry point to the secretory pathway, the primary site of phospholipid synthesis, a hub for critical stress and growth signaling molecules and for the assembly a third of the proteome. The unfolded protein response (UPR) increases the protein folding capacity of the ER in response to stresses and through unknown means exerts control over plant growth and development. Here we review recent and exciting findings that explore potential molecular mechanisms that support efficient UPR in plants. We visualize the plant UPR as a network that incorporates energy availability, plant production needs, and environmental conditions into a cohesive output governing plant life.
Acknowledgements
We acknowledge support by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (award number DE-FG02-91ER20021) for infrastructure, NASA (award NNX12AN71G), NIH R01-GM101038, the DOE Great Lakes Bioenergy Research Center [DOE Office of Science BER DE-FC02-07ER64494], AgBioResearch, and a fellowship from Michigan State University under the Training Program in Plant Biotechnology for Health and Sustainability (T32-GM110523).
References
- Aichinger E, Kornet N, Friedrich T, Laux T. Plant stem cell niches. Annual review of plant biology. 2012;63:615–636. doi: 10.1146/annurev-arplant-042811-105555. [DOI] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X, Shan S.-o. Signal recognition particle: An essential protein targeting machine. Annual review of biochemistry. 2013;82:693. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MM, Bagratuni T, Davenport EL, Nowak PR, Silva-Santisteban MC, Hardcastle A, McAndrews C, Rowlands MG, Morgan GJ, Aherne W. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. The EMBO journal. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold E, Tanner W. An obligatory role of protein glycosylation in the life cycle of yeast cells. FEBS letters. 1982;148:49–53. doi: 10.1016/0014-5793(82)81240-4. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Sharkey T. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Natural product reports. 2014;31:1043–1055. doi: 10.1039/c3np70124g. [DOI] [PubMed] [Google Scholar]
- Barba-Espín G, Dedvisitsakul P, Hägglund P, Svensson B, Finnie C. Gibberellic acid-induced aleurone layers responding to heat shock or tunicamycin provide insight into the N-glycoproteome, protein secretion, and endoplasmic reticulum stress. Plant physiology. 2014;164:951–965. doi: 10.1104/pp.113.233163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn G, Bjornson M, Ke H, De Souza A, Balmond EI, Shaw JT, Dehesh K. Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proceedings of the National Academy of Sciences. 2016:201602582. doi: 10.1073/pnas.1602582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nature cell biology. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Barlowe C. Organization of the ER–Golgi interface for membrane traffic control. Nature reviews Molecular cell biology. 2013;14(6):382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JW, Hendershot LM, Sherr CJ, Diehl JA. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proceedings of the National Academy of Sciences. 1999;96:8505–8510. doi: 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Molecular cell. 2010;40(2):238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Carvalho HH, Silva PA, Mendes GC, Brustolini OJ, Pimenta MR, Gouveia BC, Valente MA, Ramos HJ, Soares-Ramos JR, Fontes EP. The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 2014;164:654–670. doi: 10.1104/pp.113.231928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Bussell JD, Zhou W, Estavillo GM, Pogson BJ, Smith SM. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 2010;3:ra69–ra69. doi: 10.1126/scisignal.2001140. [DOI] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F. AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J. 2012;69:266–277. doi: 10.1111/j.1365-313X.2011.04788.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013;23:547–555. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Yani, Aung Kyaw, Rolčík Jakub, Walicki Kathryn, Friml Jiří, Brandizzi Federica. Inter-regulation of the unfolded protein response and auxin signaling. The Plant Journal. 2014;77(1):97–107. doi: 10.1111/tpj.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong LP, Wang Y, Gad N, Anderson N, Shah B, Zhao R. A highly charged region in the middle domain of plant endoplasmic reticulum (ER)-localized heat-shock protein 90 is required for resistance to tunicamycin or high calcium-induced ER stresses. Journal of experimental botany. 2014:eru403. doi: 10.1093/jxb/eru403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87(3):391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira MV, Xu G, Li B, de Souza Vespoli L, Meng X, Chen X, Yu X, de Souza SA, Intorne AC, de AMAM, Musinsky AL, Koiwa H, de Souza Filho GA, Shan L, He P. Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nat Plants. 2016;2:15218. doi: 10.1038/nplants.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, De Rycke R, Botterman J. Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. The EMBO journal. 1992;11:2345. doi: 10.1002/j.1460-2075.1992.tb05294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneke J, EK B, Caspers M, Sinjorgo KM, Palva ET. Analysis of sorting signals responsible for the accumulation of soluble reticuloplasmins in the plant endoplasmic reticulum. Journal of experimental botany. 1993:213–221. [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:7247–7252. doi: 10.1073/pnas.1102117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH. Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci U S A. 2013;110:19633–19638. doi: 10.1073/pnas.1314749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Quilichini TD, Dong H, Bao Y, Horner HT, Howell SH. IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. The Plant Journal. 2016 doi: 10.1111/tpj.13239. [DOI] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Duke ER, Doehlert DC. Effects of heat stress on enzyme activities and transcript levels in developing maize kernels grown in culture. Environmental and experimental botany. 1996;36:199–208. [Google Scholar]
- Foresti O, Frigerio L, Holkeri H, de Virgilio M, Vavassori S, Vitale A. A phaseolin domain involved directly in trimer assembly is a determinant for binding by the chaperone BiP. Plant Cell. 2003;15:2464–2475. doi: 10.1105/tpc.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S, Mesihovic A, Hu Y, Schleiff E. Unfolded protein response in pollen development and heat stress tolerance. Plant Reprod. 2016;29:81–91. doi: 10.1007/s00497-016-0276-8. [DOI] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Wang ZY. Multiple mechanisms modulate brassinosteroid signaling. Current opinion in plant biology. 2007;10(5):436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Prahlad V, Morimoto RI. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harbor perspectives in biology. 2011;3:a009704. doi: 10.1101/cshperspect.a009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012;8:e1002452. doi: 10.1371/journal.pgen.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D, Tuteja N. Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal Behav. 2011;6:232–236. doi: 10.4161/psb.6.2.15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton SL, Chatre L, Matheson LA, Rossi M, Held MA, Brandizzi F. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant molecular biology. 2008;67(3):283–294. doi: 10.1007/s11103-008-9317-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nature structural & molecular biology. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochemical Journal. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Protein homeostasis networks in physiology and disease. Current opinion in cell biology. 2011;23(2):123. doi: 10.1016/j.ceb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism–mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. The Plant Cell. 2008;20(12):3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoozemans J, Veerhuis R, Van Haastert E, Rozemuller J, Baas F, Eikelenboom P, Scheper W. The unfolded protein response is activated in Alzheimer's disease. Acta neuropathologica. 2005;110:165–172. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- Hüttner S, Veit C, Schoberer J, Grass J, Strasser R. Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins. Plant molecular biology. 2012;79(1-2):21–33. doi: 10.1007/s11103-012-9891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Watanabe Y, Ito N, Nonaka H, Takeda N, Sakai T, Kanaya H, Okada K. SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. The EMBO Journal. 2002;21:898–908. doi: 10.1093/emboj/21.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. The Plant Cell. 2008;20:3107–3121. doi: 10.1105/tpc.108.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. The Arabidopsis membrane-bound transcription factor AtbZIP60 is a novel plant-specific endoplasmic reticulum stress transducer. Plant signaling & behavior. 2009;4:514–516. doi: 10.4161/psb.4.6.8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. Plant transducers of the endoplasmic reticulum unfolded protein response. Trends in plant science. 2012;17:720–727. doi: 10.1016/j.tplants.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Jensen WA. Observations on the fusion of nuclei in plants. The Journal of cell biology. 1964;23:669–672. doi: 10.1083/jcb.23.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011;7:e1001330. doi: 10.1371/journal.pgen.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Hong Z, Su W, Li J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proceedings of the National Academy of Sciences. 2009;106(32):13612–13617. doi: 10.1073/pnas.0906144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Molecular cell. 2007;26(6):821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, Martinez J. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. The EMBO journal. 2014;33:2922–2936. doi: 10.15252/embj.201490332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EM, Mascheroni L, Pompa A, Ragni L, Weimar T, Lilley KS, Dupree P, Vitale A. Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J. 2006;48:657–673. doi: 10.1111/j.1365-313X.2006.02904.x. [DOI] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J, Rus A, Pardo JM, Bressan RA, Hasegawa PM. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15:2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 2001;127:949–962. [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Lämke J, Brzezinka K, Altmann S, Bäurle I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. The EMBO journal. 2015:e201592593. doi: 10.15252/embj.201592593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Mouille G, Andème-Onzighi C, Bruyant MP, Séveno M, Loutelier-Bourhis C, Driouich A, Höfte H, Lerouge P. Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. The Plant Journal. 2005;42:455–468. doi: 10.1111/j.1365-313X.2005.02392.x. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, Zipfel C, Jones JD. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci U S A. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, He K, Ma Y, Su N, He H, Stolc V, Tongprasit W, Jin W, Jiang J. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. The Plant Cell. 2008;20:259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light KM, Wisniewski JA, Vinyard WA, Kieber-Emmons MT. Perception of the plant hormone ethylene: known-knowns and known-unknowns. JBIC Journal of Biological Inorganic Chemistry. 2016;21:715–728. doi: 10.1007/s00775-016-1378-3. [DOI] [PubMed] [Google Scholar]
- Liu JX, Howell SH. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016;211:418–428. doi: 10.1111/nph.13915. [DOI] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. The Plant Journal. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012;24:4635–4651. doi: 10.1105/tpc.112.101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang C, Wang D, Su W, Liu L, Wang M, Li J. EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc Natl Acad Sci U S A. 2015;112:12205–12210. doi: 10.1073/pnas.1511724112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DP, Christopher DA. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Molecular Genetics and Genomics. 2008;280(3):199–210. doi: 10.1007/s00438-008-0356-z. [DOI] [PubMed] [Google Scholar]
- Luan H, Shine MB, Cui X, Chen X, Ma N, Kachroo P, Zhi H, Kachroo A. The potyviral P3 protein targets EF1A to promote the unfolded protein response and viral pathogenesis. Plant Physiology. 2016:00505. doi: 10.1104/pp.16.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z-X, Leng Y-J, Chen G-X, Zhou P-M, Ye D, Chen L-Q. The THERMOSENSITIVE MALE STERILE 1 Interacts with the BiPs via DnaJ Domain and Stimulates Their ATPase Enzyme Activities in Arabidopsis. PloS one. 2015;10:e0132500. doi: 10.1371/journal.pone.0132500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Endo T, Nishikawa S.-i. BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2010;107:1684–1689. doi: 10.1073/pnas.0905795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Sugiyama T, Endo T, Nishikawa S.-i. Multiple BiP genes of Arabidopsis thaliana are required for male gametogenesis and pollen competitiveness. Plant and Cell Physiology. 2014a:pcu018. doi: 10.1093/pcp/pcu018. [DOI] [PubMed] [Google Scholar]
- Maruyama D, Yamamoto M, Endo T, Nishikawa S.-i. Different Sets of ER-Resident J-Proteins Regulate Distinct Polar Nuclear-Membrane Fusion Events in Arabidopsis thaliana. Plant and Cell Physiology. 2014b;55:1937–1944. doi: 10.1093/pcp/pcu120. [DOI] [PubMed] [Google Scholar]
- Marzec M, Eletto D, Argon Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2012;1823:774–787. doi: 10.1016/j.bbamcr.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends in biochemical sciences. 2014;39(5):245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Meng Z, Ruberti C, Gong Z, Brandizzi F. CPR5 modulates salicylic acid and unfolded protein response to manage tradeoffs between plant growth and stress responses. The Plant Journal. 2016 doi: 10.1111/tpj.13397. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K, Nagashima Y, Suzuki E, Hayashi N, Ogata Y, Shimada Y, Koizumi N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc Natl Acad Sci U S A. 2013;110:5713–5718. doi: 10.1073/pnas.1219047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Staeck O, Rapoport TA. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Molecular cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- Miwa H, Kinoshita A, Fukuda H, Sawa S. Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem. Journal of plant research. 2009;122:31–39. doi: 10.1007/s10265-008-0207-3. [DOI] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One. 2012;7:e31944. doi: 10.1371/journal.pone.0031944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. Journal of biochemistry. 2009;146(6):743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- Mori T, Ogasawara C, Inada T, Englert M, Beier H, Takezawa M, Yoshihisa T. Dual functions of yeast tRNA ligase in the unfolded protein response: unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Molecular biology of the cell. 2010;21(21):3722–3734. doi: 10.1091/mbc.E10-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113(7):935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Iwata Y, Ashida M, Mishiba K.-i., Koizumi N. Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant and Cell Physiology. 2014;55:1772–1778. doi: 10.1093/pcp/pcu108. [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Iwata Y, Mishiba K.-i., Koizumi N. Arabidopsis tRNA ligase completes the cytoplasmic splicing of bZIP60 mRNA in the unfolded protein response. Biochemical and biophysical research communications. 2016;470:941–946. doi: 10.1016/j.bbrc.2016.01.145. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. The Plant Cell. 1995;7(7):957. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, Christianson JC. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harbor perspectives in biology. 2013;5:a013185. doi: 10.1101/cshperspect.a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky D, Diomina G, Lysak E, Matveeva E, Ogrel O, Trutko S. Effect of oxidative stress on the biosynthesis of 2-C-methyl-D-erythritol-2, 4-cyclopyrophosphate and isoprenoids by several bacterial strains. Archives of microbiology. 1998;171:69–72. doi: 10.1007/s002030050680. [DOI] [PubMed] [Google Scholar]
- Ozgur R, Uzilday B, Sekmen AH, Turkan I. The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in arabidopsis. Ann Bot. 2015;116:541–553. doi: 10.1093/aob/mcv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Rojas J, Moreno AA, Mitina I, Orellana A. The dynamic of the splicing of bZIP60 and the proteins encoded by the spliced and unspliced mRNAs reveals some unique features during the activation of UPR in Arabidopsis thaliana. PLoS One. 2015;10:e0122936. doi: 10.1371/journal.pone.0122936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragón T, Van Anken E, Vidal SE, El-Samad H, Walter P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch CM, Sonnewald U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 2013;162:1849–1866. doi: 10.1104/pp.113.221044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RV, Bredesen DE. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Current opinion in cell biology. 2004;16:653–662. doi: 10.1016/j.ceb.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews Molecular cell biology. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Roudier F, Gissot L, Beaudoin F, Haslam R, Michaelson L, Marion J, Molino D, Lima A, Bach L, Morin H. Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. The Plant Cell. 2010;22:364–375. doi: 10.1105/tpc.109.071209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. The Arabidopsis leucine-rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. The Plant Cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti C, Brandizzi F. Conserved and plant-unique strategies for overcoming endoplasmic reticulum stress. Front Plant Sci. 2014;5:69. doi: 10.3389/fpls.2014.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti C, Kim SJ, Stefano G, Brandizzi F. Unfolded protein response in plants: one master, many questions. Curr Opin Plant Biol. 2015;27:59–66. doi: 10.1016/j.pbi.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R, Schwer B, Shuman S. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. Journal of Biological Chemistry. 2003;278:43928–43938. doi: 10.1074/jbc.M307839200. [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Schulz-Raffelt M, Strenkert D, Veyel D, Vallon O, Schroda M. Dissecting the heat stress response in Chlamydomonas by pharmaceutical and RNAi approaches reveals conserved and novel aspects. Mol Plant. 2013;6:1795–1813. doi: 10.1093/mp/sst086. [DOI] [PubMed] [Google Scholar]
- Schweiger R, Schwenkert S. AtTPR7 as part of the Arabidopsis Sec post-translocon. Plant signaling & behavior. 2013;8(8):e25286. doi: 10.4161/psb.25286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. The Plant Journal. 2010;61:661–671. doi: 10.1111/j.1365-313X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. The EMBO journal. 1996;15(12):3028. [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD. Effects of moderate heat stress on photosynthesis: importance of thylakoid reactions, rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant, Cell & Environment. 2005;28:269–277. [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Developmental cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Current opinion in cell biology. 2011;23:143–149. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AM, Elliott S. Glycoengineering: the effect of glycosylation on the properties of therapeutic proteins. Journal of pharmaceutical sciences. 2005;94:1626–1635. doi: 10.1002/jps.20319. [DOI] [PubMed] [Google Scholar]
- Smyczynski C, Roudier F, Gissot L, Vaillant E, Grandjean O, Morin H, Masson T, Bellec Y, Geelen D, Faure J-D. The C terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. Journal of Biological Chemistry. 2006;281:25475–25484. doi: 10.1074/jbc.M601815200. [DOI] [PubMed] [Google Scholar]
- Song Z-T, Sun L, Lu S-J, Tian Y, Ding Y, Liu J-X. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proceedings of the National Academy of Sciences. 2015;112:2900–2905. doi: 10.1073/pnas.1419703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH. BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. The Plant Cell. 2013;25:1416–1429. doi: 10.1105/tpc.113.110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. Journal of molecular medicine. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- Steiger MA, Jackman JE, Phizicky EM. Analysis of 2′-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: Evidence for a conserved two-step reaction mechanism. RNA. 2005;11:99–106. doi: 10.1261/rna.7194605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Lu S-J, Zhang S-S, Zhou S-F, Sun L, Liu J-X. The lumen-facing domain is important for the biological function and organelle-to-organelle movement of bZIP28 during ER stress in Arabidopsis. Molecular plant. 2013a;6:1605–1615. doi: 10.1093/mp/sst059. [DOI] [PubMed] [Google Scholar]
- Sun L, Yang ZT, Song ZT, Wang MJ, Sun L, Lu SJ, Liu JX. The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. The Plant Journal. 2013b;76:274–286. doi: 10.1111/tpj.12287. [DOI] [PubMed] [Google Scholar]
- Sun L, Lu SJ, Zhang SS, Zhou SF, Sun L, Liu JX. The lumen-facing domain is important for the biological function and organelle-to-organelle movement of bZIP28 during ER stress in Arabidopsis. Molecular plant. 2013c;6(5):1605–1615. doi: 10.1093/mp/sst059. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang SS, Lu SJ, Liu JX. Site-1 protease cleavage site is important for the ER stress-induced activation of membrane-associated transcription factor bZIP28 in Arabidopsis. Science China Life Sciences. 2015;58(3):270–275. doi: 10.1007/s11427-015-4807-6. [DOI] [PubMed] [Google Scholar]
- Tam AB, Koong AC, Niwa M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell reports. 2014;9:850–858. doi: 10.1016/j.celrep.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant physiology. 2006;141(2):373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totani K, Ihara Y, Tsujimoto T, Matsuo I, Ito Y. The Recognition Motif of the Glycoprotein-Folding Sensor Enzyme UDP-Glc: Glycoprotein Glucosyltransferase†. Biochemistry. 2009;48(13):2933–2940. doi: 10.1021/bi8020586. [DOI] [PubMed] [Google Scholar]
- Valente MAS, Faria JA, Soares-Ramos JR, Reis PA, Pinheiro GL, Piovesan ND, Morais AT, Menezes CC, Cano MA, Fietto LG, Loureiro ME. The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. Journal of experimental botany. 2009;60(2):533–546. doi: 10.1093/jxb/ern296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D. Defects in endoplasmic reticulum-associated degradation (ERAD) increase selenate sensitivity in Arabidopsis. Plant Signal Behav. 2016;0 doi: 10.1080/15592324.2016.1171451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Denecke J. The endoplasmic reticulum—Gateway of the secretory pathway. The Plant Cell. 1999;11:615–628. doi: 10.1105/tpc.11.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittorioso P, Cowling R, Faure J-D, Caboche M, Bellini C. Mutation in the Arabidopsis PASTICCINO1Gene, Which Encodes a New FK506-Binding Protein-Like Protein, Has a Dramatic Effect on Plant Development. Molecular and cellular biology. 1998;18:3034–3043. doi: 10.1128/mcb.18.5.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E, Heijne GV. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Science. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. The EMBO journal. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ. The Unfolded Protein Response Pathway in Saccharomyces cerevisiae oligomerization and trans-phosphorylation of Ire1P (Ern1p) are required for kinase activation. Journal of Biological Chemistry. 1996;271(30):18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2004;1699(1):35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149:1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Sun X, Gao S, Qin F, Dai M. Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Molecular Plant. 2016 doi: 10.1016/j.molp.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Maruyama D, Endo T, Nishikawa S.-i. Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant and cell physiology. 2008;49:1547–1562. doi: 10.1093/pcp/pcn119. [DOI] [PubMed] [Google Scholar]
- Yang KZ, Xia C, Liu XL, Dou XY, Wang W, Chen LQ, Zhang XQ, Xie LF, He L, Ma X, Ye D. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 2009;57:870–882. doi: 10.1111/j.1365-313X.2008.03732.x. [DOI] [PubMed] [Google Scholar]
- Yang Z-T, Wang M-J, Sun L, Lu S-J, Bi D-L, Sun L, Song Z-T, Zhang S-S, Zhou S-F, Liu J-X. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014a;10:e1004243. doi: 10.1371/journal.pgen.1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZT, Lu SJ, Wang MJ, Bi DL, Sun L, Zhou SF, Song ZT, Liu JX. A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. The Plant Journal. 2014b;79:1033–1043. doi: 10.1111/tpj.12604. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Chung KP, Li B, Lai CM, Lam SK, Wang X, Cui Y, Gao C, Luo M, Wong K-B. Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2015;112:14360–14365. doi: 10.1073/pnas.1519333112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen H, Brandizzi F, Verchot J, Wang A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 2015;11:e1005164. doi: 10.1371/journal.pgen.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di-and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome biology. 2009;10:1. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]