Abstract
The tumor microenvironment (TME) is a dynamic tissue space in which the tumor exists, plays a significant role in tumor initiation, and is a key contributor in cancer progression; however, little is known about tumor-induced changes in the adjacent tissue stroma. Herein, tumor-induced changes in the TME were explored at the morphological and molecular level to further understand cancer progression. Tumor-adjacent mammary glands (TAGs) displayed altered branching morphology, expansion of myofibroblasts, and increased mammosphere formation, broadly suggesting a tumor-induced field effect. FACS analysis of TAGs demonstrated an increased number of Lin−CD24+/CD49+ enriched mammary gland stem cells (MaSCs), suggesting deregulated tissue homeostasis in TAGs. Comparative transcriptome analysis of TAGs and contralateral control glands coupled with meta-analysis on differentially expressed genes with two breast cancer stromal patient microarray datasets identified shared upregulation of STAT1. Knockdown of STAT1 in cancer-associated fibroblast (CAFs) co-cultured with human breast cancer cells altered cancer cell proliferation, indicating a role for STAT1 as a stromal contributor of tumorigenesis. Furthermore, depletion of STAT1 in CAFs significantly reduced periductal reactive fibrosis and delayed early breast cancer progression in vivo. Lastly, co-treatment with fludarabine, a FDA-approved STAT1 activation inhibitor and DNA synthesis inhibitor, in combination with doxorubicin, showed enhanced therapeutic efficacy in treating mouse mammary gland tumors. Taken together, these results demonstrate that stromal STAT1 expression promotes tumor progression and is a potential therapeutic target for breast cancer.
Keywords: STAT Pathway, Tumor microenvironment, Breast cancer, Field effect, Mammary gland stem cell, Combinatorial therapy
INTRODUCTION
Stepwise accumulation of mutations in key oncogenes and tumor suppressors in somatic cells typifies the classic tumor progression model(1). Cancer cells coexist with normal cells in the same tissue space, communicating with one another and constituents of their shared tumor microenvironment (TME). Seminal findings over the past decade have established the significance of the heterogeneous TME in tumor initiation and progression(2). Multi-omics analyses demonstrated that alterations within tumor-associated stroma are predictive indicators of clinical outcome(3) and targeting tumor-promoting modulators of the TME has been perceived as a promising strategy to combat the pro-tumor effects of the TME in conjunction with traditional cancer chemotherapies(4).
Breast cancer arises from a dynamic mammary gland ecosystem. A diverse array of cell types establish the context in which the majority of mammary gland development occurs(5–7). Given their fundamental role in deposition of the extracellular matrix (ECM), fibroblasts are particularly influential components of the mammary milieu(8). Reminiscent to the fibroblast contribution in wound healing (often termed ‘myofibroblasts’ in this context) are carcinoma-associated fibroblasts (CAFs) that bear molecular resemblance to activated normal fibroblasts during the wound healing(9). Through enhanced ECM production, CAFs have the capacity to alter the TME through tissue remodeling including recruitment of immune cells to TME, perturbing adipocyte differentiation, disrupting the function of epithelial stem cells, and instigating other discordant events that promote the hallmarks of cancer(10,11).
CAFs also display increased proliferation and secrete a diverse array of cytokines(12). At the core of secretory factor regulation is the Janus kinases (JAK) and signal transducers and activators of transcription (STAT) pathways (JAK-STAT). Upon phosphorylation-dependent activation, isoforms of STAT proteins function as highly-specific transcription factors independent of secondary messengers(13). Specifically, STAT1 is a central mediator of both type I (alpha and beta) and type II (gamma) IFNs, which play an essential role in cell growth, regulation, and antiviral and immune defense(14). In mouse mammary tissue, STAT1 expression is observed in the virgin mammary gland and diminishes into gestation, only to reappear during involution, potentially expressed in recruited immune cells(15). Deregulated STAT1 expression has been implicated in breast cancer development with both tumor suppressing and promoting roles of STAT1(16,17). Despite the known contributions of various STATs throughout mammary gland development, the roles of STAT1 in mammary tumor progression remain highly controversial and appear to be tumor stage- and cell type-specific(15).
In this study, we demonstrated that mammary tumors seem to induce field cancerization on the seemingly normal tissues of their microenvironment. Using a mouse-in-mouse model of breast cancer, we first observed that mammary tumors induce field cancerization on the seemingly normal tissues in the mammary gland microenvironment, including increased mammary gland branching and enriched mammary stem cells (MaSCs) and CAFs in tumor-adjacent mammary glands. Transcriptional profiling identified STAT1 as the central signaling node in the tumor-adjacent stroma. Ablation of STAT1 in CAFs decreases cancer cell proliferation through STAT1-dependent secretion of pentraxin 3 (PTX3). Depletion of STAT1 in CAFs reduces α-SMA+ periductal reactive fibroblasts and ductal carcinoma in situ (DCIS)-like lesions in a mouse model of early breast cancer progression. Finally, using an allograft model, we demonstrated that combination of a STAT1 inhibitor (fludarabine) with chemotherapy (doxorubicin) reduced stromal CAFs, STAT1 expression, and exhibited superior therapeutic efficacy.
MATERIALS AND METHODS
Cell Culture and STAT1 knockdown in CAFs
Murine mammary tumor cell line (PNA.Met1) was derived from a spontaneous mammary tumor from the MMTV-PyMT model. DCIS.COM cell line was originally purchased from Asterand, Inc. MDA-MBA-231 cell line was purchased from ATCC and has been actively passaged less than six months. The caveolin-1-deficient mouse embryonic fibroblasts were used were gifted by Dr. Zach Schafer (University of Notre Dame) (18,19). All cells lines were maintained in DMEM-F12 growth media supplemented with 10% FBS and 5% penicillin-streptomyosin at 37 °C with 5% CO2. For siRNA knockdown experiments, CAFs were treated with either the ON-TARGETplus SMARTpool siRNA targeting mouse Stat1 or ON-TARGETplus non-targeting siRNA pool (GE Healthcare Dharmacon Inc.) for six hours.
EdU Assay
Cells were seeded in serum-free media 24 hours prior to treatment to allow for cell cycle synchronization. Following designated treatment, cells were pulsed with 5-ethynyl-2- deoxyuridine (Click-iT® Plus EdU Alexa-647® Imaging Kit, Life Technologies), for two hours before fixation in 2% paraformaldehyde and subsequent EdU detection per the manufacturer’s protocol. Coverslips were mounted on slides and imaged using a Nikon A1R-MP confocal microscope. Quantification of EdU+ tumor cells were carried out using the cell counter plugin from the Image J software (NIH). The percentage of EdU+ for each field of view captured was recorded and analyzed.
Cytokine Array Analysis
Briefly, on day 0, 1×106 cells (siControl CAF or siStat1 CAF) were seeded in complete DMEM-F12. On day 3, serum-free DMEM-F12 was added and conditioned for 48 hours before cytokine array analysis by RayBiotech using the Mouse Cytokine Array Q4000 (QAM-CA-4000). After fluorescent scanning, data extraction, and data computation using array-specific scanning devices and data processing tools (Raybiotech). Concentration levels, expressed in picograms per milliliter (pg/ml), were calculated against a standard curve set for each biomarker from the positive and negative controls. Results from each of the 200 different cytokine probes were ranked by expression and subjected to bioinformatics analysis.
In Vivo Animal Experiments
Mice were housed in the Animal Facility of the Freimann Life Science at the University of Notre Dame (Notre Dame, IN) in compliance with the Institutional Animal Care and Usage Committee (IACUC) standards. Female FVB mice (12 to 16 weeks) were purchased from The Jackson Laboratories. Estrous Cycle Monitoring: Murine estrous cycle monitoring was completed by vaginal lavage according to published protocol(20). Briefly, the vaginal cavity of each animal was washed with sterile ddH2O by pipette, which was then pipetted onto clean slide glass. Dried slides were stained with 0.1% crystal violet for 1 minute before mounting. Ten fields of view were analyzed for each animal to determine estrous cycle. Mammary Fat Pad (MFP) tumor model: Two million ZsGreen-labeled PNA.Met1 cells in 100μL of serum-free DMEM-F12 was injected into one of the inguinal (fourth) MFP of female FVB mice with control injections of 100 μL of serum-free DMEM-F12 into the contralateral gland. Upon tumor formation (approximately 14–20 days post-injection), mammary glands and tumors were collected for further analysis. Ductal carcinoma in situ (DCIS) model (intraductal tumor injections): Fifty thousand cells in 8 μL HBSS were injected into the nipple of the inguinal mammary glands of mature age-matched, cage-matched mice via intraductal injection using 31 gauge RN needle, Hamilton #7803-03). Mammary tissue was collected 14 days post-injection. For the CAF co-injection experiment, 2 × 106 CAF cells were first injected to mammary fatpad. 24 hours later, 50,000 DCIS.COM cells were injected intraductially. In Vivo Drug Treatment: Following MFP injection of PNA.Met1 cells into each inguinal fat pad, we measured and recorded the calculated ellipsoidal tumor volume and randomly distributed mice into 4 treatment groups (vehicle, doxorubicin, fludarabine, doxorubicin+fludarabine). Doses of both doxorubicin and fludarabine (Selleck Chemical) were determined based on the body surface area (BSA) of each animal based on reported human doses(21–23). Drug treatments were administered every 3 days by intraperitoneal (IP) injection and tumor volume was logged. Upon sacrificing animals, tumors and adjacent mammary glands were collected for formalin fixation and paraffin embedding (FFPE) for subsequent evaluation.
Whole Mount Analysis
Mammary glands from each animal were aseptically collected, fixed, and stained according to the established protocol. Briefly, each gland was fixed for two hours in 4% paraformaldehyde prior to overnight staining with carmine alum solution. Glands were washed in an ethanol gradient (70%, 95%, and 100% for one hour each) prior to overnight clearing in xylene and mounting on glass for branching analysis in Image J.
Propagation of Primary Mammospheres
Mammary glands were collected and digested in collagenase/hyaluronidase (Stem Cell Technologies) in EpiCult™-B complete growth medium (Stem Cell Technologies). After overnight digestion, each sample was washed with 1× PBS and treated with dispase (5 mg/mL) and DNAse (0.1 mg/mL), then filtered through a sterile 40 mm mesh filter. An equal number of cells were plated in EpiCult B complete growth media on Corning Ultra-Low attachment 96-well plates (Sigma). Total number of mammospheres per well was determined after 7 days at 10× magnification.
Fluorescent Activated Cell Sorting (FACS)
Intraductal injections of PNA.Met1 cells labeled with DiR (LifeTechnologies) were carried out on age-matched, cage-matched FVB mice. Cells were stained with the APC-conjugated lineage cocktail (Lin) (BD Biosciences) containing CD45, Ter119, and CD31. Cells were also stained with PE-conjugated CD49f and FITC-conjugated CD24 (BD Biosciences).
Hemotoxylin and Eosin Staining and Immunohistochemistry (IHC)
5 μm FFPE sections were deparaffinized using standard protocol and stained with Harleco® Gill’s Modified Hematoxylin (EMD Millipore) and eosin (Sigma-Aldrich). Slides remained in xylene until mounted with Permount Mounting Media (Electron Microscopy Sciences). For IHC staining, 5 μm FFPE sections were subjected to antigen retrieval in boiling 10 mM sodium citrate (pH 6.0) buffer for 10 minutes. After blocking endogenous peroxidase with 3% H2O2 for 15 minutes, slides were washed 3 times in 1× PBS prior to blocking for 60 minutes at room temperature (Protein Block, Serum-Free; Dako), followed with primary antibody (STAT1, 1:1000, #14994S, Cell Signaling Technologies; α-SMA, 1:500, #ab124964, Abcam; 1:500. Cytokeratin-8, #ab53280, Abcam) and incubated overnight at 4 °C. After washing with PBS, the sections were incubated with secondary antibody for another 60 minutes (Polink-2 Plus HRP Rabbit with DAB Kit, GBI Labs). Each slide was counterstained with Gill’s hematoxylin. IHC quantifications were carried out using Image J (NIH).
RNA-seq, Meta-analysis and Network analysis
RNA-seq was conducted after SMART-seq2 library preparation(24). Following RNA-seq, raw reads were analyzed by DESeq2 to explore transcriptional differences between control and TAG tissue(25). Meta-analysis and network construction were conducted through the web-based bioinformatics package NetworkAnalyst (http://www.networkanalyst.ca) based on recommended protocol(26,27). Three stromal transcriptome analyses datasets used in the meta-analysis are: 1) Zhang_RNA-seq. The TAG gene expression signature was derived from comparison of the TAG with a contralateral control mammary gland using a syngeneic mouse mammary tumor transplantation model (this study). 2) GSE14548: a publically available microarray dataset of 14 patient-matched normal epithelium, normal stroma, tumor epithelium and tumor-associated stroma samples. 3) GSE26910: a publically available microarray dataset of 6 patient-matched tumor-adjacent stromal samples with 6 normal stromal samples. Prostate samples in this dataset were excluded from our studies. The top differentially expressed genes from 3 independent datasets (FDR p< 0.05, < 0.01, <0.01 for Zhang_RNA-seq, GSE14548, GSE26910 respectively) were curated for meta-analysis. Based on statistical heterogeneity estimated using Cochran’s Q tests, we applied random effects models (REM, p < 0.05) for metagene analysis. Forty-two differentially expressed metagenes were selected for NetworkAnalyst using InnateDB interactome database(26). During the network analysis, we removed UBC, ACTB and GRB2, which are commonly involved in many nonspecific interactions as suggested in NetworkAnalyst protocol.
Statistical Analysis
All quantitative data with normal distribution are compared by Student t test (two-tailed). Whole mount branch point analysis was compared by paired t-test. For all analyses, a p-value <0.05 is considered statistically significant.
RESULTS
Early mammary tumor development deregulates the mammary gland microenvironment and alters stromal cells
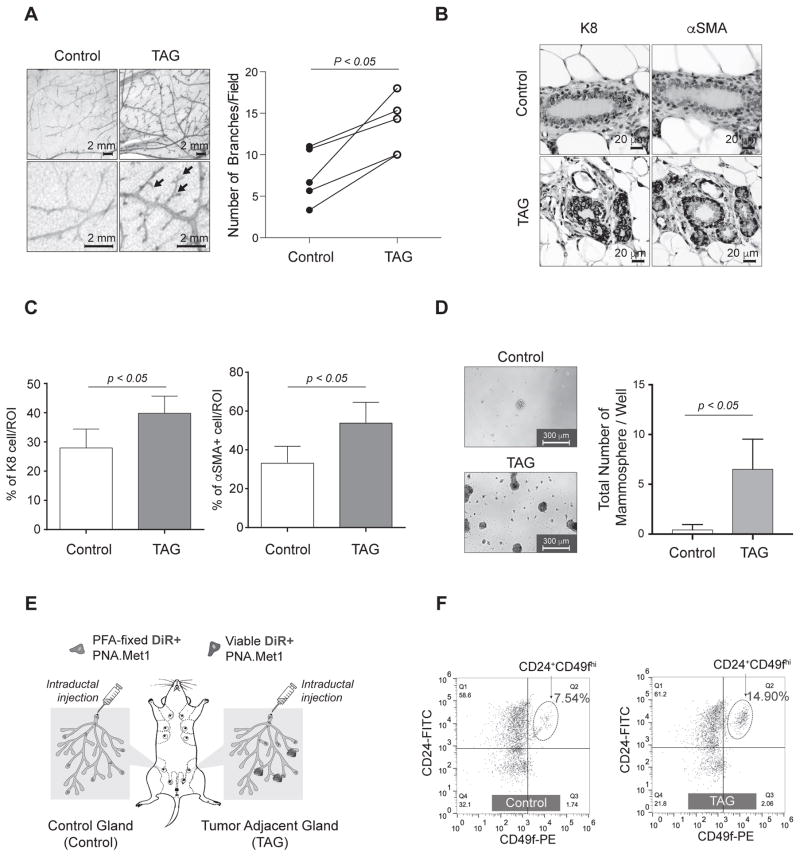
We began by exploring whether tumors induce either physical or biochemical changes on ostensibly healthy, non-transformed mammary tissues. Considering mammary gland tissue homeostasis is under the tight regulation of local and systemic signals (e.g. ovarian hormones)(5), we age-matched and cage-matched experimental mice and confirmed the estrous cycle of each animal by vaginal lavage staining, as indicated by the presence of cornified squamous epithelial cells (Supplementary Fig. 1A). We then performed an acute mammary fat pad (MFP) transplantation of syngeneic PNA.Met1 tumor cells, an aggressive breast cancer cell line derived from a mammary tumor collected from the MMTV-PyMT transgenic mouse, to study changes in tumor-adjacent tissues (Supplementary Fig. 1B)(28). We assayed branching morphology in both the vehicle-injected control and contralateral tumor-adjacent glands (TAGs) by mammary whole mount(29). We observed a marked increase in the number of branch points per duct (p < 0.05) in each TAG compared to control glands 21 days post-injection (Fig. 1A). To further assess tumor-induced changes in the mammary gland, we performed immunohistochemistry (IHC) on TAGs and control glands for cytokeratin 8 (K8 - luminal epithelial cells) and α-SMA (myoepithelial and myofibroblasts) (Fig. 1B). Quantification showed an increase in the K8+ luminal epithelial compartment of TAG tissue compared to control (K8, Fig. 1C, left; α-SMA, Fig. 1C, right). Compared to vehicle injected control glands, TAG tissue, while visibly undisturbed, also exhibited an increase in the number of α-SMA+ stromal and myoepithelial cells (Fig. 1C). Given prior knowledge that fibroblast-secreted factors can alter the mammary epithelial hierarchy(30,31), we next examined the impact of tumor burden on the MaSC niche via mammosphere formation of cells derived from TAGs. We observed a significant increase (p < 0.05) in the number mammospheres derived from TAGs compared to tissue from the contralateral control gland, strongly suggesting an increased MaSC population in TAG (Fig. 1D).
Figure 1.
Tumor development deregulates the mammary gland microenvironment and alters stromal cells. A, Left, whole mount analysis of control (left) and tumor-adjacent (TAG, right) mammary glands. Arrows indicate branch points. Right, branch point quantification of matched control and tumor-adjacent glands, with analysis from paired t-test. B, Representative images of IHC staining of cytokeratin 8 (K8) and α-smooth muscle actin (α-SMA) in control (top) and TAG (bottom). C, Quantification of IHC (shown in B) of percent of K8+ and α-SMA+ cells per 40× region of interest (ROI) in both control and TAG. D, Representative images of both control and TAG mammospheres with corresponding quantifications at day 10 post-plating. E, Schematic depicting experimental strategy for intraductal injection experiments. F, FACS plot of MaSCs from both control (left) and TAG (right) mammary glands.
We next asked whether the MaSC expansion occurs at a critical transition stage of tumor progression, such as the transition from ductal carcinoma in situ (DCIS) to invasive ductal carcinoma (IDC). To faithfully mimic the DCIS to IDC transition, we used an intraductal injection model that provides an accurate recapitulation of DCIS and its progression to IDC in vivo (Fig. 1E)(32). Intraductal injection of PNA.Met1 cells gave rise to DCIS-like lesions in approximately 3 weeks (Supplementary Fig. 1C). We labeled the tumor cells with DiR to facilitate tumor cell tracking and injected paraformaldehyde (PFA)-fixed tumor cells in the contralateral gland to control the potential impact of the physical stress imposed to mammary gland tissue during intraductal injection (Fig. 1E) and characterize the enriched MaSC population in mammary tissue using FACS (Supplementary Fig. 1D). It has been widely accepted that lineage-negative (Lin−) CD24+/CD49flow cells are enriched for luminal epithelial cells, and the CD24+CD49hi basal epithelial population is enriched for MaSCs(33,34). Therefore, we examined TAG tissue for CD24+CD49hi cells after gating out Lin+ cells as well as DiR+ cells (tumor cells). Quantification of the Lin−/CD24+/CD49fhi population in both tumor and control glands showed that the enriched MaSC population comprised 14.9% of the total cell population in the TAGs, compared to 7.58% in the control glands (Fig. 1F). Taken together, these results suggest that early stage tumors may impose a cancer field effect on adjacent, ostensibly normal tissue by stimulating the stromal myoepithelial and myofibroblast population and inducing an expansion of MaSC population(35).
STAT1 is a Central Node of Tumor-Stimulated Stromal Signature
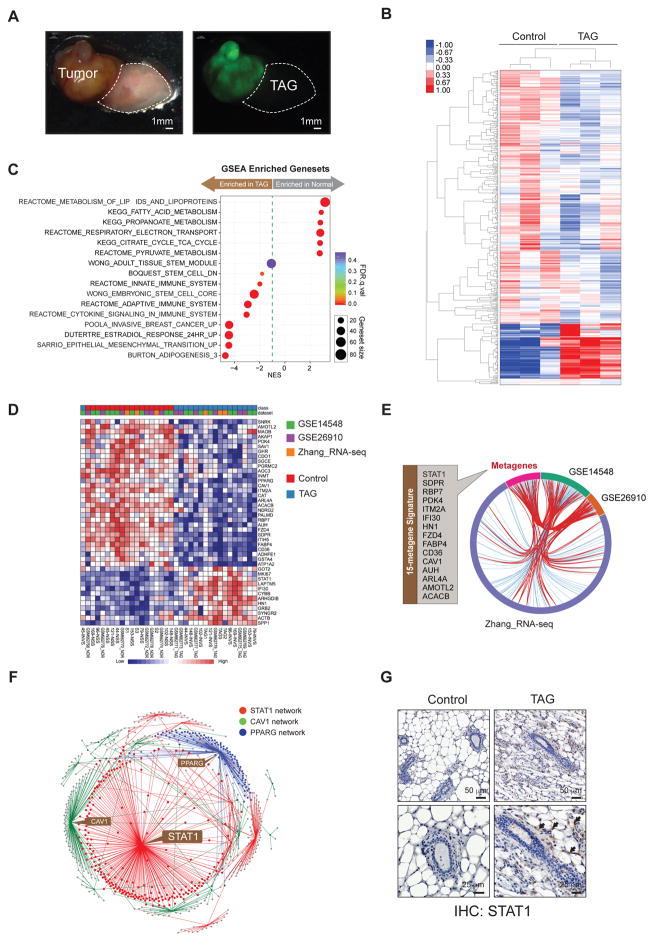
To explore the molecular mechanisms driving both the shift in tissue morphology as well as the impact on the mammary epithelial hierarchy, we conducted comparative transcriptome analysis (RNA-seq) of control and TAG tissue (Fig. 2A). A diagnostic MA plot shows evenly distributed reads around zero (zero-fold change) across read count values (Supplementary Fig. 2A). In total, we detected 17,877 differentially expressed genes (DEGs) in TAGs after stimulation by the presence of a tumor. Among the top DEGs (p < 0.05), approximately 20% of the highest ranked DEGs were upregulated in the TAGs, whereas approximately 80% were upregulated in the control glands (Fig. 2B). To reveal the functional implications of DEGs, we performed gene set enrichment analysis (GSEA) using all DEGs (p < 0.05, q < 0.25) based on curated gene sets (C2) from the Molecular Signature Database (MSigDB). Interestingly, our TAG samples showed significant enrichment of gene sets that represent mammary gland/stem cell development, aggressive tumor phenotypes, and immune activation (Fig. 2C, Supplementary Fig. 2C, Supplementary Table 1). To validate the clinical relevance of our findings and identify potential candidates that drive the phenotype observed in tumor-adjacent tissue, we performed meta-analysis based on a random effect model across our RNA-seq dataset as well as two publically available clinical breast cancer stromal microarray datasets (GSE14548 and GSE26910) using NetworkAnalyst (http://www.networkanalyst.ca)(26,36). We identified a total of 42 DE meta-genes (FDR < 0.05) across our RNA-seq dataset and two breast cancer stromal patient microarray datasets (Fig. 2D, Supplementary Table 2). Moreover, of the 42 differentially expressed meta-genes, 15 of these meta-genes are shared among all three datasets as depicted in the chord diagram, which we refer to as our 15 meta-gene signature (Fig. 2E). Downregulated genes of interest included cluster of differentiation 36 (CD36), fatty acid binding protein 4 (FABP4), and caveolin-1 (CAV1). Upregulated genes of interest in the tumor-associated included signal transducers and activators 1 (STAT1) and hematological and neurological expressed 1 (HN1). Next, we created a nodal network from the 15-gene meta-signature in order to identify functionally important drivers of tumorigenesis. Using the Module Explorer panel in NetworkAnalyst, we isolated the network into densely connected modules whose node genes are likely to work toward a common biological function. A search algorithm is performed on each seed node that ranks each node gene based on the number of its first-degree interactions. Interestingly, NetworkAnalyst distinctly revealed 3 top-ranked key nodes: 1) STAT1, an upregulated node in TAG tissue (centrality degree=222, betweeness=15442.82); 2) PPARG, a downregulated node (centrality degree=123, betweeness=7616.63); and 3) CAV1, a downregulated node (centrality degree=117, betweeness=7629.22) (Fig. 2F). The loss of CAV1 expression is a reported feature of CAFs, a major constituent of the tumor stroma(8,37). Most importantly, the highest degree of centrality in our 15 meta-gene signature is STAT1, which indicates that the presence of a tumor in the same tissue space promotes upregulation of STAT1 in tumor-adjacent tissue, which could play a central role in the deregulated TAG. Since the mammary gland is highly heterogeneous and compartmentalized, we sought to confirm the expression of STAT1 in tumor-adjacent tissue. We confirmed our RNA-seq findings by staining for STAT1 by IHC and found that expression is negligible in the control gland, while more prominently expressed in tumor-adjacent mammary tissue, primarily in stromal fibroblasts (Fig. 2G).
Figure 2.
STAT1 is a Central Node of Tumor Stimulated Stromal Signature. A, Bright field (left) and fluorescent (right) images of ZsGreen PNA.Met1 xenograft tumor and tumor associated gland (TAG). Dashed line indicates tumor-adjacent tissue extracted for RNA-sequencing. B, Clustering analysis showing the top DE genes in control and TAG groups. C, Cleveland plot of gene set enrichment analysis (GSEA) from all DEGs (p < 0.05, q < 0.25) with curated gene sets (C2) from the Molecular Signature Database (MSigDB). D, Heat map of top 42 DEGs (FDR p < 0.05) from meta-analysis of Zhang_RNA-seq with GSE14548 and GSE26910. E, Chord diagram of 15 meta-genes (red line) shared among all three datasets (Zhang_RNAseq, GSE14548, and GSE26910). F, Nodal network generated from the 15 meta-gene signature showing the top three ranked nodes. G, Representative images of IHC staining for STAT1 in control (left) and TAG (right).
Fibroblast STAT1-mediated Pentraxin 3 (PTX3) Secretion Promotes Tumor Cell Proliferation
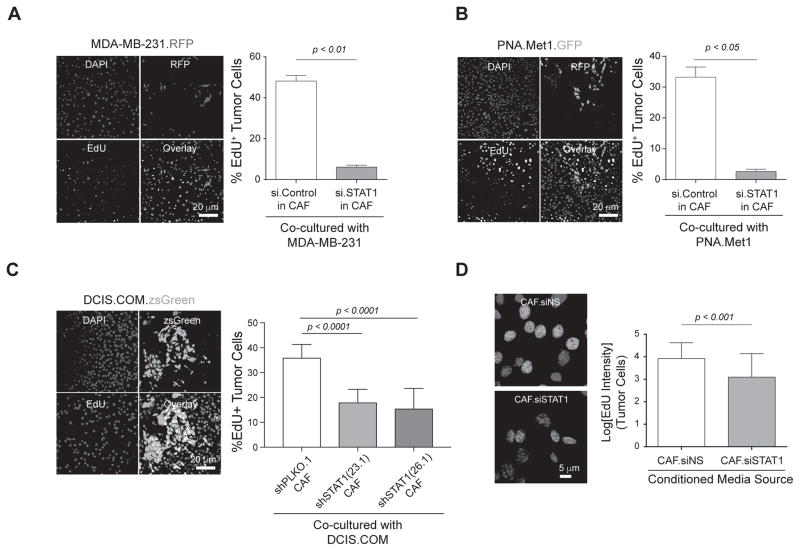
Our immunohistochemical analyses indicate the upregulation of STAT1 occurs predominantly in the stromal fibroblast compartment of the tumor-adjacent mammary glands, not the epithelial compartment (Fig. 2G). Therefore, to examine the functional importance of fibroblast-derived STAT1 in tumor-adjacent mammary tissue on tumor proliferation, we co-cultured tumor cells with CAV1-deficient MEFs (herein referred to as CAFs), a well-established model cell line with similar transcriptome and secretome profile as human CAFs(18,19). Transient depletion of STAT1 in CAFs by siRNA resulted in a decrease of proliferation as evidenced by less EdU incorporation (Supplementary Fig. 3A, 3B). We co-cultured the same number of CAFs, transfected with either control siRNA or STAT1 siRNA, with MDA-MB-231 breast cancer cells. Interestingly, coculture with siSTAT1 CAFs resulted in decreased proliferation as evidenced by significantly less EdU incorporation in the tdTomato-red-positive MDA-MB-231 cancer cells (Fig. 3A). Similar results were observed from CAFs co-cultured with ZsGreen-labeled PNA.Met1 breast cancer cells (Fig. 3B) or DCIS.COM cells, a well-characterized early stage human breast cancer line (Fig. 3C). To exclude the possible contact-dependent role of CAFs, we treated MDA-MB-231 tumor cells with conditioned media collected from either siSTAT1 CAFs or non-silencing control CAFs for 48 hours. Treatment of conditioned media collected from siSTAT1 CAFs led to significantly weaker EdU fluorescent intensity in tumor cells, which is a direct result of decrease of EdU incorporation (Fig. 3D).
Figure 3.
Fibroblast STAT1-mediated Pentraxin 3 (PTX3) secretion promotes tumor cell proliferation. A, Left: Representative images of EdU assay from tumor cells grown in co-culture with CAFs (siSTAT1). Blue: DAPI, Red: MDA-MB-231, Yellow: EdU+ cells, Pink: EdU+ cells in merged image. Right: Quantification of Percent EdU+ cells. B, Left: Representative images of EdU assay ZsGreen PNA.Met1 tumor cells in co-culture with CAFs as mentioned in A. Blue: DAPI, Green: ZsGreen PNA.Met1, Yellow: EdU+ cells. Right: Quantification of Percent EdU+ cells, Pink: EdU+ cells in merged image. C, Left: Representative images of EdU assay from DCIS.COM tumor cells cocultured with either shPLKO.1 (control) CAFs or with CAFs containing either construct of shSTAT1 (23.1 or 26.1). Blue: DAPI, Green: DCIS.COM, Red: EdU+ cells, Yellow: EdU+ cells in merged image. Right: Quantification of percent EdU+ cells. D, Left: Representative merged images of DAPI-stained DCIS.COM cells after treatment for 48 hours with conditioned media collected from siSTAT1 CAFs and non-silencing control (siNS) CAFs. Blue: DAPI, Pink: EdU+ cells. Right: Quantification represents log[EdU intensity] of PNA.Met1 cells.
PTX3 is secreted by STAT1-expressing CAFs and promotes tumor cell viability and proliferation
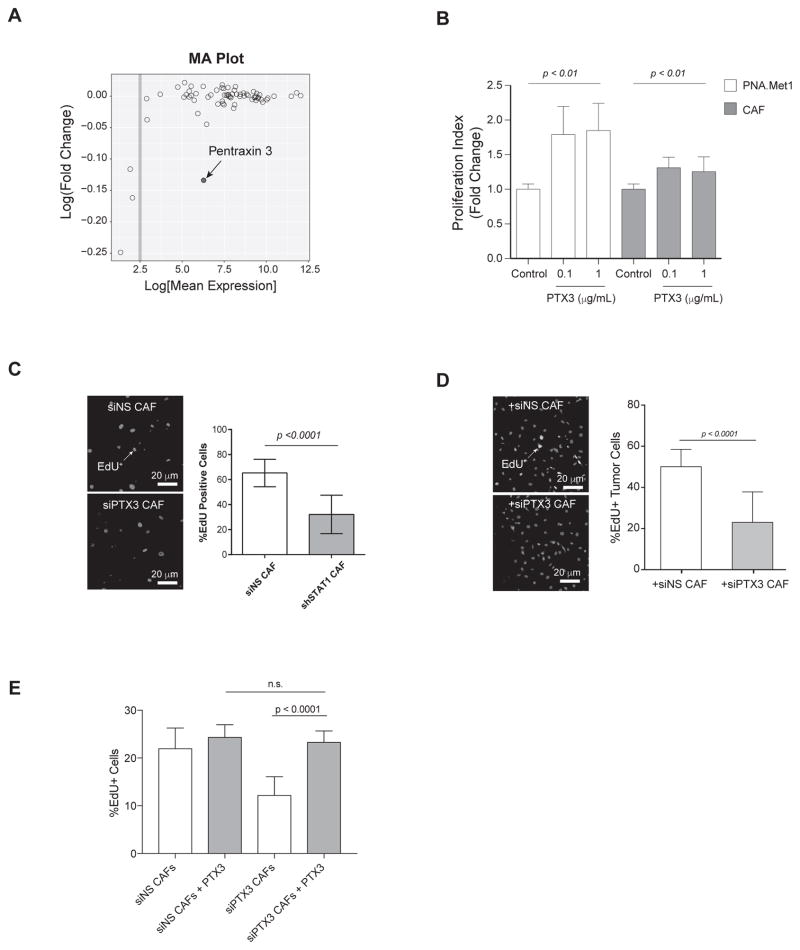
To further elucidate the STAT1-downstream secretory factors released in CAF-conditioned media, we performed unbiased cytokine array analysis to probe for 200 different secretory proteins in the conditioned media. Principle component analysis of 68 total detectable cytokines showed a 39% total variance of the first component (PC1) and 22% total variance of the second component (PC2), suggesting differentially regulated cytokine profiles between the si.Control and si.STAT1 group (Supplementary Fig. 3C). As shown in the heat map of hierarchical clustering, there was a consistent pattern of down-regulation of pentraxin 3 (PTX3) in siSTAT1-derived conditioned media compared with si.Control group (Supplementary Figure 3D). Further analysis using MA plot identified PTX3 as the only abundant cytokine (log[expression] > 2.5) significantly down-regulated in the media from siSTAT CAFs (Fig. 4A and Supplementary Fig. 3D). To further explore the role of CAF-derived PTX3 in tumor progression, we treated PNA.Met1 tumor cells with purified PTX3 protein. Treating PNA.Met1 tumor cells with PTX3 showed a dose-dependent increase of cell proliferation compared to vehicle-treated control cells (Fig. 4B). Moreover, coculture MDA-MB-231 breast tumor cells with PTX3-depleted CAFs led to a significant decrease of EdU incorporation in tumor cells (23.25% in siPTX3 group vs. 50.3% in siNS group) (Fig. 4C and Supplementary Fig. 3E). Similar decrease of tumor cell proliferation was observed when tumor cells cocultured with siPTX3 CAF cells (Fig. 4D). In addition, overnight treatment of siPTX3 CAFs with 1 μg purified PTX3 restored CAF cell proliferation (23.34% PTX3 treated vs. 12.24% EdU incorporation in vehicle-treated group (Fig. 4E), suggesting a self-sustained proliferation of CAF via PTX3 autocrine. Collectively, our data demonstrated that STAT1-dependent secretion of PTX3 from CAFs is one of the facilitators of tumor cell proliferation.
Figure 4. PTX3 is secreted by STAT1-expressing CAFs and promotes tumor cell viability and proliferation.
A, MA plot showing the cytokine array analysis. Log fold change is plotted as a function of log mean expression. Each circle represents one probe. B, Proliferation index of PNA.Met1 tumor cells and CAFs 24 hours after treatment with recombinant human pentraxin 3 in serum-free DMEM- F12. C, Left: Representative images of non-silencing control (siNS) CAFs (top) or PTX3-depleted CAFs (bottom). Blue: DAPI, Red: Red: EdU+ cells. Right: Quantification of percent EdU+ cells. D, Left: Representative images of MDA-MB-231 cells cocultured with non-silencing control (siNS) CAFs (top) or PTX3-depleted CAFs (bottom). Blue: DAPI, Red: Tumor Cells, Yellow: EdU+ cells. Right: Quantification of percent EdU+ cells. E, Quantification of percent EdU+ cells of non-silencing control (siNS) or PTX3-depleted (siPTX3) CAFs treated with vehicle or 1 μg PTX3 overnight.
Stromal STAT1 influences progression of early stage breast cancer in vivo
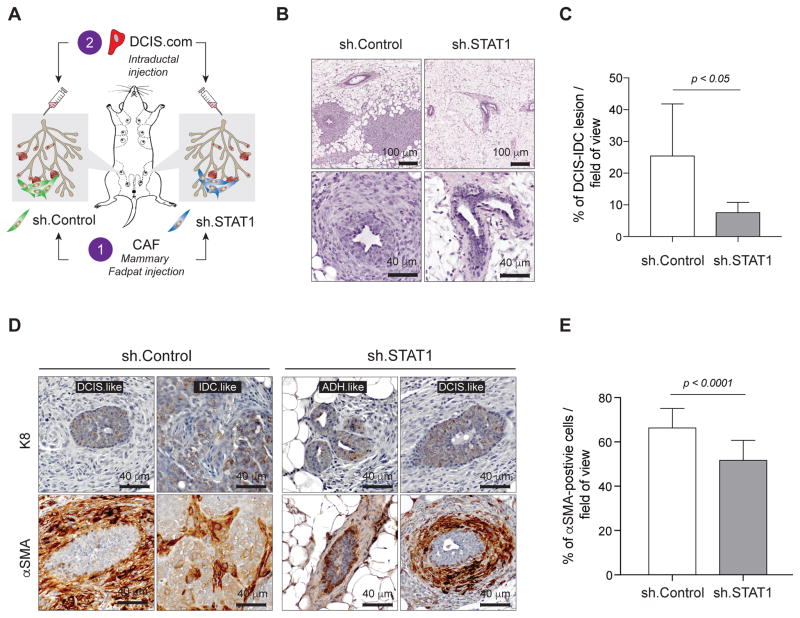
In order to assess the importance of stromal STAT1 in tumor progression, we first manipulated the mammary gland microenvironment by mammary fat pad injection of either STAT1-depleted CAFs (sh.STAT1) or control CAFs (sh.Control) in the contralateral site. We then performed intraductal injections of DCIS.COM cells to model the clinical progression of DCIS (Fig. 5A)(32). Over the four weeks of DCIS formation, we noticed a palpable increase in mammary gland stiffness in the control glands compared to sh.STAT1 glands. The significant increase of periductal fibrosis in the control glands was evidenced in H&E staining (Fig. 5B). Importantly, on average, 25.53% of each field of view was composed of a DCIS or IDC-like lesion in the control glands, compared to only 7.65% in the sh.STAT1 injected glands (Fig. 5C), suggesting a less aggressive mammary tumor development conferred by deprivation of STAT1 in CAFs. To reveal the cellular composition of the abnormal mammary tissue, we stained K8 for epithelial cells and α-SMA for myoepithelial and reactive fibroblasts. In sh.Control CAF-injected gland, we observed K8+ DCIS-like and IDC-like tumors, whereas only ADH/DCIS-like lesions were observed in the shSTAT1.CAF-injected glands (Fig. 5D). Moreover, we observed a significant decrease of α-SMA+ cells surrounding mammary ducts in the sh.STAT1 CAF-injected glands (Fig. 5D and E), suggesting a reduction in tumor-induced myoepithelial cells as well as decreased periductal fibrosis (38).
Figure 5. Stromal STAT1 influences progression of early stage breast cancer lesions in vivo.
A, Schematic depicting experimental strategy for intraductal injection experiments. Control CAFs were injected into one mammary fat pad (n = 5), while STAT1-depleted CAFs (shSTAT1) were injected into the contralateral fat pad (n = 5). After 24 hours, DCIS.COM cells were injected into the nipple of each mammary gland. B, Representative H&E images of tumor progress in glands injected with control CAFs (left) and shSTAT1 CAFs (right) at both 10× (top) and 40× (bottom). C, Mean percent of DCIS/IDC-like lesion per field of view in glands injected with control CAFs (n = 5) compared to glands injected with shSTAT1 CAFs (n = 5). D, Representative 40× images showing tumor lesions in glands injected with control CAFs (left) and shSTAT1 CAFs (right) stained with either human K8 (top row) or α-SMA (bottom row). DCIS: ductal carcinoma in situ, IDC: invasive ductal carcinoma, ADH: advanced ductal hyperplasia. E, Quantification of percent α-SMA+ cells per 40× field of view in 5 μm sections from mammary glands injected with sh.Control CAFs (left) or sh.STAT1 CAFs (right).
Inhibition of stromal STAT1 enhances chemotherapeutic efficacy
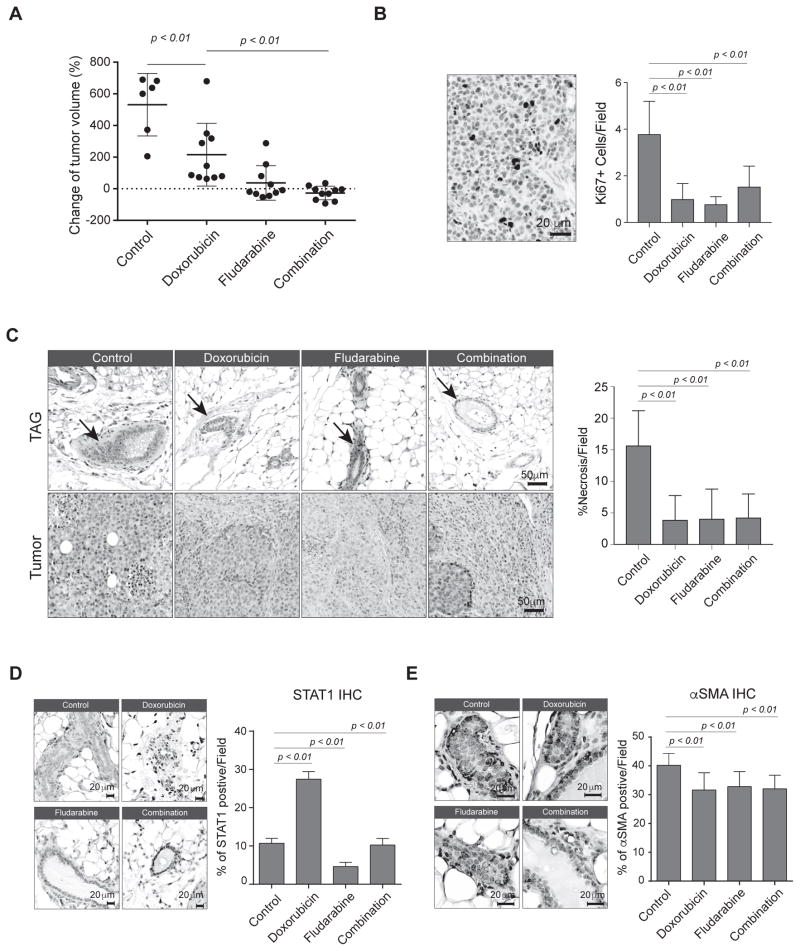
Given the prominent role of stromal STAT1 in facilitating breast cancer progression, we hypothesized that combined treatment of STAT1-inhibiting agents with doxorubicin, a conventional chemotherapeutic agent used to treat breast cancer, could enhance chemotherapeutic efficacy in breast cancer treatment. Fludarabine is a clinically approved chemotherapy that inhibits cytokine-induced activation of STAT1 and is often used for the treatment of chronic lymphocytic leukemia and other hematological malignancies(22). In this proof-of-concept study, we used our mouse-in-mouse allograft model to evaluate the therapeutic efficacy of fludarabine in combination with doxorubicin. Combination treatment of fludarabine and doxorubicin further decreased tumor volume (p <0.01) compared with single-treatment alone (Fig. 6A). Consistent with reduction of tumor size, we observed decreased proliferation (Ki67 IHC staining) and decreased necrosis in residual tumors either single-drug treatment or combination treatment (Fig. 6B–C). Interestingly, single-treatment of fludarabine showed evidence of increased fibrosis within the residual tumor mass (Fig. 6C, lower panel). In addition, fludarabine treatment normalized the TAG tissue structure. In both the single-treatment fludarabine and combination-treatment group, TAG glands exhibited regular adipocyte morphology and, most importantly, an evident decrease of resident fibroblasts (Fig. 6C, upper panel), suggesting fludarabine inhibits the abnormal expansion of CAFs. Doxorubicin-treated tumor-adjacent tissue showed an increase in STAT1 expression, likely due to drug-induced immune cell infiltration, whereas either fludarabine- or combination-treated tumor-adjacent tissue showed a complete inhibition of STAT1 expression (Fig. 6D)(11). Consistently, fludarabine or combination-treatment significantly suppressed STAT1 expression in residual tumor tissues while doxorubicin treatment showed a moderate effect on STAT1 expression in tumor tissues (Supplementary Fig. 4). Lastly, all treatment groups showed a mild decrease of α-SMA expression in tumor-adjacent glands (Fig. 6E). Taken together, these findings suggest that targeting the STAT1 pathway could further improve the therapeutic efficacy of conventional chemotherapy.
Figure 6.
Inhibition of stromal STAT1 enhances chemotherapeutic efficacy. A, Percent change of tumor volume at end point compared to day zero for each tumor in each treatment group: vehicle control (n = 6), doxorubicin (n = 10), fludarabine (n = 10), or combination of doxorubicin and fludarabine (n = 10). B, Left, representative image showing IHC for Ki67+ tumor cells in control treatment group. Right, mean number of Ki67+ cells per 40× field of view in each group. C, Hematoxylin and eosin (H&E) staining of tumors from each treatment group. Left, representative images showing the tissue morphology after the treatments. Right, bar graph showing the percent of necrosis in each 10× field of view. D, IHC analysis of STAT1 in TAG. Left, representative images of IHC STAT1. Right, STAT1+ cells per 40× field of view. E, IHC analysis of STAT1 in TAG a-SMA. Left, representative images of IHC α-SMA. Right, α-SMA+ cells per 40× field of view.
DISCUSSION
Breast cancer development is an evolutionary process with extensive interplay between tumor cells and their stroma. It is indisputable that a ‘tumor-centric’ approach limits our understanding of the tumor as a component of a dynamic ecosystem. While previous studies primarily focus on the pathological consequences of tumor-associated stroma on tumorigenesis, our data collectively demonstrated a field cancerization effect instigated by early tumor development on normal adjacent tissue, which facilitates a vicious cycle of the tumor development (Supplementary Fig. 5).
The first focal point of our study is the tissue niche surrounding the developing tumor. As a consequence of tumor initiation, a cancer field effect is induced, reflected in altered branching morphogenesis, an increased capacity for mammosphere formation, and an increasing MaSC population (Fig. 1). Mechanistically, analysis of tumor-associated stroma using a systems biology approach revealed upregulation of STAT1 specific to CAFs (Fig. 2). Transient depletion of STAT1 in CAFs suggested a tumor-promoting role for STAT1 mediated by secretion of PTX3 (Fig. 4). In addition, in vivo depletion of STAT1 in CAFs reduced periductal α-SMA+ myoepithelial/myofibroblasts and delayed mammary tumor progression, suggesting a stroma-specific role for STAT1 in promoting invasion of early stage lesions (Fig. 5). Finally, we showed that treatment with the STAT1 inhibitor fludarabine in combination with doxorubicin significantly reduced tumor burden compared to mice treated with doxorubicin alone (Fig. 6).
Our results demonstrated a context- and tissue compartment-specific role of STAT1 in breast cancer progression. STAT1 activity in tumor cells is often ascribed a tumor suppressor function; however, in the TME context, recent evidence has emerged suggesting the transcription factor may also play a tumor-promoting role, particularly in a breast cancer. Hix et al. used a syngeneic mouse injection model to demonstrate a pro-tumor role for STAT1 in tumor cells due to expression of STAT1 and recruitment of CD33+ myeloid cells. Similarly, Tymoszuk et al. found that high levels of STAT1 mRNA and increased levels of STAT1 target genes in tumor cells are associated with macrophage infiltration and poor prognosis(39). These findings imply that, in addition to its cell-autonomous role in controlling tumor cell proliferation, tumor cell-derived STAT1 might also play a pivotal role in TME immune recruitment. Distinct from the aforementioned tumor cell-focused studies, in our study, we provide compelling evidence for the role of STAT1 within the mammary TME. Specifically, tumor cell-instigated upregulation of STAT1 in CAFs alters the proliferation of tumor cells through paracrine and autocrine signaling of PTX3 (Fig. 3 and Fig. 4). We further demonstrated that CAF-secreted PTX3 promotes tumor cell viability and proliferation. Bonavita et al. published evidence suggesting the opposite, showing increased susceptibility for epithelial and mesenchymal tumorigenesis and Trp53 mutations in PTX3−/− mice(40). Despite compelling analyses suggesting PTX3 promoter methylation in certain subtypes of cancer, we strongly believe that the role of PTX3 in regulating the tumor niche is complex and entirely context dependent. For example, Chi et al. determined that PTX3 is a CCAAT/enhancer binding protein delta (CEBPD)-responsive gene and displays a pro-tumor phenotype(41), including acquired drug resistance, enhanced migration, and stem-like characteristics. Given these recent findings, we believe our results further support a pro-tumor role for PTX3 in a tissue niche with activated CAFs.
Furthermore, our novel observation underscores the importance of studying cancer progression as cohesive and evolving ecosystem(42), which will provide mechanistic insight of perplexing clinical observation of field cancerization in mammary tissue(35). Although we have proposed that tumors impose field cancerization on their niche constituents resulting in upregulation of STAT1, the means by which tumors initially activate fibroblasts is multilayered and complex. One potential explanation is that tumor cells secrete pro-inflammatory factors that promote resident fibroblast activation and maintain fibroblasts in this reactive state (38,43). In addition, evidence suggests that bone marrow-derived circulating myofibroblasts home to tumors by a systemic response reminiscent of wound healing (44,45). This phenomenon could possibly occur in parallel with the activation of tissue resident fibroblasts, producing a rapid supply of myofibroblasts in the tumor microenvironment.
Lastly, from a translational perspective, identifying molecular targets in the tumor-associated stroma provides new therapeutic opportunities to ‘normalize’ the tumor-promoting TME. It is increasingly acknowledged that targeting tumor stroma as a neoadjuvant therapy or in combination with chemotherapy is a promising new avenue to enhance therapeutic efficacy and attenuate drug resistance(46). While a clinically applicable target of STAT1 for breast cancer is currently unavailable, we conducted a proof-of-concept study of chemotherapy with fludarabine, a FDA approved chemotherapy that has been show to significantly inhibit STAT1 signaling(24). Our results demonstrated that combinatorial treatment of fludarabine with doxorubicin enhanced the overall therapeutic efficacy and reduced stroma abnormalities (Fig. 6). Further development of a STAT1-specific agent could ultimately facilitate the clinical proposition of co-targeting the tumor and stroma as a strategy to reduce tumor burden.
In summary, our study highlighted the tumor-imposed insults on the microenvironment and mammary gland homeostasis and subsequent induction of a molecular clutch (STAT1) driving a vicious cycle of tumor progression and shed light on future design of rationalized combinatorial therapy targeting both cancer cell as well as its co-evolved tumor microenvironment.
Supplementary Material
IMPLICATIONS.
Tumors induce stromal STAT1-dependent cytokine secretion that promotes tumor cell proliferation and can be targeted using clinically-approved inhibitors of STAT1.
Acknowledgments
We’d like to thank Dr. Zonggao Shi, HCRI Biorepository Director, Dr. Charles Tessier, and the Notre Dame Genome Core Facility for their technical support. This works is partially supported by Department of Defense W81XWH-15-1-0021 (S.Z.), NIH 1R01CA194697-01 (S.Z.), Walther Cancer Foundation Advancing Basic Cancer Research Grant II (S.Z.), Indiana CTSI core pilot fund (S.Z.), HCRI Notre Dame Day Pilot Fund (S.Z.). S.Z. is a Nancy Dee Assistant Professor in Cancer Research at the University of Notre Dame and Harper Cancer Research Institute.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–27. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X-J, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Research. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nat Rev Cancer. 2007;7:139–47. doi: 10.1038/nrc2067. [DOI] [PubMed] [Google Scholar]
- 5.Sternlicht MD. Key stages in mammary gland development: The cues that regulate ductal branching morphogenesis. Breast Cancer Research. 2005;8:201. doi: 10.1186/bcr1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142:1028–42. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- 7.Polyak K, Kalluri R. The Role of the Microenvironment in Mammary Gland Development and Cancer. Cold Spring Harb Perspect Biol. 2010;2:a003244. doi: 10.1101/cshperspect.a003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing F, Saidou J, Watabe K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci. 2010;15:166–79. doi: 10.2741/3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Augsten M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front Oncol. 2014;4:62. doi: 10.3389/fonc.2014.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Coussens LM. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell. 2012;21:309–22. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Kessenbrock K, Dijkgraaf GJP, Lawson DA, Littlepage LE, Shahi P, Pieper U, et al. A Role for Matrix Metalloproteinases in Regulating Mammary Stem Cell Function via the Wnt Signaling Pathway. Cell Stem Cell. 2013;13:300–13. doi: 10.1016/j.stem.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 13.Aaronson DS, Horvath CM. A Road Map for Those Who Don’t Know JAK-STAT. Science. 2002;296:1653–5. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 14.Stark GR, Darnell JE., Jr The JAK-STAT Pathway at Twenty. Immunity. 2012;36:503–14. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haricharan S, Li Y. STAT signaling in mammary gland differentiation, cell survival and tumorigenesis. Mol Cell Endocrinol. 2014;382:560–9. doi: 10.1016/j.mce.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L. Loss of STAT1 from Mouse Mammary Epithelium Results in an Increased Neu-Induced Tumor Burden. Neoplasia. 2010;12:899–905. doi: 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khodarev N, Ahmad R, Rajabi H, Pitroda S, Kufe T, McClary C, et al. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene. 2009;29:920–9. doi: 10.1038/onc.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sotgia F, Del Galdo F, Casimiro MC, Bonuccelli G, Mercier I, Whitaker-Menezes D, et al. Caveolin-1−/− Null Mammary Stromal Fibroblasts Share Characteristics with Human Breast Cancer-Associated Fibroblasts. The American Journal of Pathology. 2009;174:746–61. doi: 10.2353/ajpath.2009.080658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigel KJ, Jakimenko A, Conti BA, Chapman SE, Kaliney WJ, Leevy WM, et al. CAF-Secreted IGFBPs Regulate Breast Cancer Cell Anoikis. Mol Cancer Res. 2014;12:855–66. doi: 10.1158/1541-7786.MCR-14-0090. [DOI] [PubMed] [Google Scholar]
- 20.McLean AC, Valenzuela N, Fai S, Bennett SAL. Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification. J Vis Exp. 2012:e4389. doi: 10.3791/4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staedler D, Idrizi E, Kenzaoui BH, Juillerat-Jeanneret L. Drug combinations with quercetin: doxorubicin plus quercetin in human breast cancer cells. Cancer Chemother Pharmacol. 2011;68:1161–72. doi: 10.1007/s00280-011-1596-x. [DOI] [PubMed] [Google Scholar]
- 22.Frank DA, Mahajan S, Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat Med. 1999;5:444–7. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 23.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–61. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 24.Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols. 2014;9:171–81. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 25.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia J, Benner MJ, Hancock REW. NetworkAnalyst - integrative approaches for protein–protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–74. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia J, Gill EE, Hancock REW. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protocols. 2015;10:823–44. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 28.Borowsky AD, Namba R, Young LJT, Hunter KW, Hodgson JG, Tepper CG, et al. Syngeneic mouse mammary carcinoma cell lines: Two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis. 2005;22:47–59. doi: 10.1007/s10585-005-2908-5. [DOI] [PubMed] [Google Scholar]
- 29.Plante I, Stewart MKG, Laird DW. Evaluation of mammary gland development and function in mouse models. J Vis Exp. 2011 doi: 10.3791/2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiseman BS, Werb Z. Stromal Effects on Mammary Gland Development and Breast Cancer. Science. 2002;296:1046–9. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaenel P, Hahnewald S, Wotzkow C, Strange R, Andres A-C. Overexpression of EphB4 in the mammary epithelium shifts the differentiation pathway of progenitor cells and promotes branching activity and vascularization. Develop Growth Differ. 2014;56:255–75. doi: 10.1111/dgd.12126. [DOI] [PubMed] [Google Scholar]
- 32.Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Research. 2009;11:R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 34.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M-L, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 35.Heaphy CM, Griffith JK, Bisoffi M. Mammary field cancerization: molecular evidence and clinical importance. Breast Cancer Res Treat. 2009;118:229–39. doi: 10.1007/s10549-009-0504-0. [DOI] [PubMed] [Google Scholar]
- 36.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucl Acids Res. 2013;41:D1228–33. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2012;32:303–15. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 39.Tymoszuk P, Charoentong P, Hackl H, Spilka R, Müller-Holzner E, Trajanoski Z, et al. High STAT1 mRNA levels but not its tyrosine phosphorylation are associated with macrophage infiltration and bad prognosis in breast cancer. BMC Cancer. 2014;14:257. doi: 10.1186/1471-2407-14-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, et al. PTX3 Is an Extrinsic Oncosuppressor Regulating Complement-Dependent Inflammation in Cancer. Cell. 2015;160:700–14. doi: 10.1016/j.cell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Chi J-Y, Hsiao Y-W, Li C-F, Lo Y-C, Lin Z-Y, Hong J-Y, et al. Targeting chemotherapy-induced PTX3 in tumor stroma to prevent the progression of drug-resistant cancers. Oncotarget. 2015;6:23987–4001. doi: 10.18632/oncotarget.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat R, Bissell MJ. Of plasticity and specificity: dialectics of the microenvironment and macroenvironment and the organ phenotype: Of plasticity and specificity. Wiley Interdisciplinary Reviews: Developmental Biology. 2014;3:147–63. doi: 10.1002/wdev.130. [DOI] [PubMed] [Google Scholar]
- 43.Östman A, Augsten M. Cancer-associated fibroblasts and tumor growth – bystanders turning into key players. Current Opinion in Genetics & Development. 2009;19:67–73. doi: 10.1016/j.gde.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Direkze NC, Hodivala-Dilke K, Jeffery R, Hunt T, Poulsom R, Oukrif D, et al. Bone Marrow Contribution to Tumor-Associated Myofibroblasts and Fibroblasts. Cancer Res. 2004;64:8492–5. doi: 10.1158/0008-5472.CAN-04-1708. [DOI] [PubMed] [Google Scholar]
- 45.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes Dev. 2008;22:559–74. doi: 10.1101/gad.1636908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.