SUMMARY
Ribosome assembly is a complex process that requires hundreds of essential assembly factors, including Rix7 (NVL2 in mammals) and Nsa1 (WDR74 in mammals). Rix7 is a type-II double ring, AAA-ATPase, which is closely related to the well-known Cdc48/p97. Previous studies in Saccharomyces cerevisiae suggest that Rix7 mediates the release of Nsa1 from nucleolar pre-60S particles however the underlying mechanisms of this release are unknown. Through multiple structural analyses we show that S. cerevisiae Nsa1 is composed of an N-terminal seven bladed-WD40 domain followed by a lysine rich C-terminus that extends away from the WD40 domain and is required for nucleolar localization. Co-immunoprecipitation assays with the mammalian homologues identified a well-conserved interface within WDR74 that is important for its association with NVL2. We further show that WDR74 associates with the D1 AAA domain of NVL2, which represents a novel mode of binding of a substrate with a type-II AAA-ATPase.
Keywords: AAA-ATPase, ribosome assembly, WD40 domain, pre-rRNA processing
eTOC Blurb
Lo et al present the 1.3Å crystal structure of the essential ribosome assembly factor Nsa1/WDR74. The N-terminal WD40 domain of WDR74 is required for binding the AAA-ATPase Rix7/NVL2, while the flexible C-terminus is required for proper nucleolar localization.
INTRODUCTION
Ribosomes are large ribonucleoprotein machines that carry out the essential role of translating mRNA into proteins. Ribosome assembly begins in the nucleolus with the transcription of the pre-ribosomal RNA (rRNA), and then proceeds through numerous carefully orchestrated steps from the nucleolus to the nucleoplasm, through the nuclear pore complex and into the cytoplasm where the final maturation steps occur (Thomson et al., 2013; Woolford and Baserga, 2013). Defects in ribosome assembly factors have been linked to a group of human diseases classified as ribosomopathies, and aberrant ribosome production has been associated with numerous cancers (McCann and Baserga, 2013; Nakhoul et al., 2014; Wang et al., 2015b; Woods et al., 2015). The assembly of ribosomes is an intricate process that relies on the aid of hundreds of ribosomal assembly factors. The ribosome assembly factors make up a large group of proteins with diverse functions including AAA-ATPases (ATPases associated with various cellular activities), endo and exo-nucleases, GTPases, RNA helicases, as well as a large number of proteins with no known enzymatic function (Kressler et al., 2010; McCann et al., 2015).
Three AAA-ATPases are specifically required for the formation of the large ribosomal subunit, including Rix7, Rea1, and Drg1 (reviewed in (Kressler et al., 2012)). The primary function of these ATPases is to catalyze the release of specific assembly factors from pre-60S ribosomal particles through the power of ATP hydrolysis (Kressler et al., 2012). Rix7 is the earliest acting AAA-ATPase and appears to be involved in driving release of the essential assembly factor Nsa1 (Nop7 associated 1) from nucleolar Nop7-containing pre-60S particles (Kressler et al., 2008). The second AAA-ATPase, Rea1 (known as Midasin in higher eukaryotes), drives release of Erb1 and Ytm1 from early nucleolar pre-60S particles and Rsa4 from later nucleoplasmic pre-60S particles (Barrio-Garcia et al., 2016; Bassler et al., 2010; Romes et al., 2016; Thoms et al., 2016; Ulbrich et al., 2009). Rea1 contains a C-terminal MIDAS (metal ion-dependent adhesion site) that is necessary for recognizing the N-terminal Ubiquitin-Like (UBL) domains on both Ytm1 and Rsa4 (Bassler et al., 2010; Bassler J, 2014; Romes et al., 2016; Ulbrich et al., 2009). Drg1 is the latest acting AAA-ATPase, and is involved in removal of Rpl24 from pre-60S particles in the cytoplasm (Kappel et al., 2012). Release and recycling of these assembly factors by Rea1, Rix7, and Drg1 is crucial for the progression and maturation of the large ribosomal subunit and important for driving large conformational changes within the maturing 60S particle. While we understand how Rea1 recognizes its substrates, little is known about how Rix7 and Drg1 recognize their proposed substrates. Rix7 and Drg1 are type-II AAA-ATPases that are closely related to the well-studied Cdc48 (p97 in mammals), whose function is to segregate proteins from cellular components such as large protein-protein complexes, chromatin, and the endoplasmic reticulum (Barthelme and Sauer, 2016). Rix7, Drg1, and Cdc48 contain unique N-terminal domains followed by tandem well-conserved AAA domains, known as the D1 and D2 domains, respectively (Kressler et al., 2012).
Nsa1 and Rix7 are functionally linked to one another and Rix7 associates with Nsa1 containing nucleolar pre-60S particles, however no direct interaction has been observed between these proteins (Kressler et al., 2008). Rix7 is essential for cell viability in yeast and is required for the maturation of the large ribosomal subunit, as temperature sensitive mutants of Rix7 lead to defects in both pre-rRNA processing and export of pre-60S particles into the cytoplasm (Gadal et al., 2001). Expression of a N-terminal truncated Rix7 lacking the first 14 residues results in Nsa1 being retained on pre-60S particles and the release of aberrant 60S particles containing Nsa1 into the cytoplasm (Kressler et al., 2008). Taken together these experiments suggest that Rix7 is involved in the recycling of Nsa1 from nucleolar pre-60S particles. Rix7 is well conserved among eukaryotes and its mammalian homologue is known as NVL2 (Nuclear valosin-containing protein (VCP)-Like 2) (Nagahama et al., 2004). Expression of a dominant negative mutant of NVL2 leads to defects in pre-rRNA processing of the large ribosomal subunit in mammalian cells, which suggests that the function of Rix7/NVL2 is well conserved across eukaryotes (Yoshikatsu et al., 2015). The mammalian homologue of Nsa1 is the nucleolar protein WDR74 (WD-repeat containing 74), which has been shown to associate with NVL2 in the presence of the nuclear Mtr4 exosome complex (Hiraishi et al., 2015). Mtr4 is a large DExH RNA helicase whose primary function is to target RNA substrates including pre-rRNA to the exosome for degradation (Thoms et al., 2015). In addition to WDR74 and Mtr4, NVL2 has also been shown to associate with numerous other macromolecules including nucleolin, ribosomal protein L5, and telomerase (Fujiwara et al., 2011; Her and Chung, 2012; Nagahama et al., 2004; Yoshikatsu et al., 2015). These observations suggest that NVL2 could play a role in the recycling of other proteins during ribosome assembly, as well as have additional roles in the cell outside of its function in ribosome assembly.
Our goals are to uncover the role of Nsa1/WDR74 during ribosome maturation and to determine how Rix7/NVL2 recognizes and dissociates its proposed substrate Nsa1/WDR74 from nucleolar pre-60S particles. As a first step in understanding the molecular mechanisms of this release reaction, we sought to determine how Rix7/NVL2 recognizes Nsa1/WDR74. Our structural analyses reveal that NVL2 recognizes a well-conserved interface along one-side of the WD40 domain of WDR74 and that WDR74 associates with the D1 domain of NVL2, which is a novel mode of interaction between a substrate and a type II-AAA-ATPase. Overall, our data suggest that the WD40 domain of WDR74 serves as a platform for recruitment of NVL2 to pre-60S particles.
RESULTS
Nsa1 is a 7-bladed β-propeller
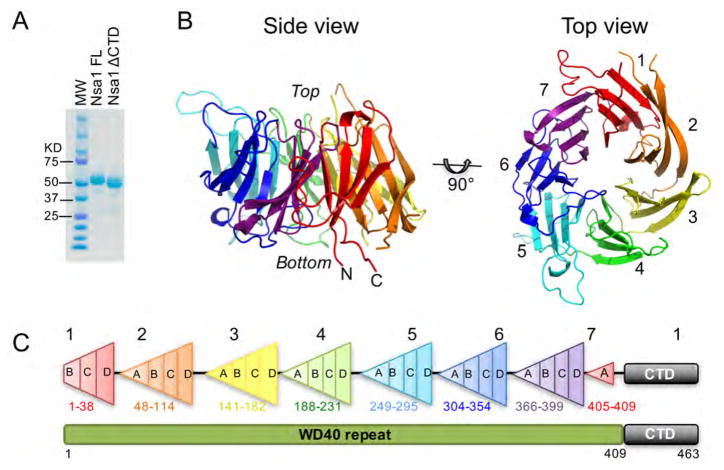
To further our understanding of Rix7 driven release of Nsa1, we first sought to solve the crystal structure of Nsa1. Crystallization trials of full-length Nsa1 from S. cerevisiae yielded large orthorhombic crystals of Nsa1 from which we were able to collect an initial data set at room temperature to 1.9 Å resolution. During optimization of these crystals we discovered that they arose as the result of proteolytic cleavage of the C-terminal region of Nsa1 (Figure 1A). Through a combination of limited proteolysis and mass spectrometry we identified the region of the C-terminus of Nsa1 that was sensitive to proteolysis (data not shown). Subsequent crystallization trials were carried out with a C-terminal truncated construct of Nsa1 containing residues 1–434.
Figure 1.
The crystal structure of S. cerevisiae Nsa1ΔC. (A) Comparison of full-length Nsa1 and the C-terminal cleaved Nsa1 by SDS-PAGE (B) Overall structure of the Nsa1 WD40 domain shown in rainbow color. Two different views are shown, including a side and top view of the WD40 domain. The N and C-termini are labeled and they lie adjacent to one another within the first blade of the WD40 domain. The individual blades are colored as follows: Blade 1 (red), Blade 2 (orange), Blade 3 (yellow), Blade 4 (green), Blade 5 (cyan), Blade 6 (blue), and Blade 7 (purple). (C) Schematic illustration of the organization of Nsa1. The numbers correspond to S. cerevisiae Nsa1 and each blade is colored as in Fig 1B. See also Figure S1 and S2.
Secondary structure prediction analysis suggested that the N-terminus of Nsa1 contains a WD40 domain comprised of a β-propeller with 6 to 8 blades. WD40 domains are one of the most abundant protein domains and have a β-propeller architecture with varying numbers of blades (Xu and Min, 2011). The structure of Nsa1 was solved by single anomalous diffraction (SAD) from a seleno-methionine prepared derivative (Figure S1A and Table 1). The structure revealed that Nsa1 contains an N-terminal seven-bladed β-propeller (Figure 1B and 1C). The structure includes residues 1–416 of Nsa1, with the exception of two extended loops within Blade2. We followed the standard nomenclature for WD40 propellers and numbered the blades as 1 to 7. The four strands within each blade are labeled A, B, C, and D from the inside to the outside of the propeller (Figure 1C, S1B, S2).
Table 1.
X-ray data collection and refinement statistics
| Native_Nsa1FL | Native_Nsa1ΔC | SeMet_Nsa1ΔC | |
|---|---|---|---|
| Data collection | |||
|
| |||
| Wavelength (Å) | 1.00000 | 1.00000 | 0.97911 |
| Space group | P213 | P212121 | P212121 |
| Unit-cell parameters | |||
| a, b, c (Å) | a=b=c=163.1 | 54.3, 81.1, 87.4 | 54.3, 80.8, 86.7 |
| α, β, γ (°) | α=β=γ=90° | α=β=γ=90° | α=β=γ=90° |
| Resolution (Å) | 50.0 – 2.8 (2.9 – 2.8)* | 59.4–1.3 (1.35 – 1.3) | 50.0 – 1.7 (1.76 – 1.7) |
| Rmerge(%) | 10.0 (58.4) | 6.6 (54.2) | 10.1 (81.2) |
| Mosaicity | 0.5 | 0.3 | |
| I/σ(I) | 28.9 (5.6) | 29.6 (2.3) | 19.6 (3.1) |
| Completeness (%) | 96.9 (97.6) | 99.9 (99.2) | 99.9 (99.7) |
| Redundancy | 10.3 (10.0) | 7.2 (6.5) | |
| NO. molecules per ASU | 2 | 1 | 1 |
| Solvent Content (%) | 64.4 | 33.0 | 34 |
| Wilson B-factor | 47.3 | 15.7 | |
|
| |||
| Refinement | |||
|
| |||
| Resolution (Å) | 24.3 – 2.8 | 59.4 – 1.3 | |
| Unique reflections | 32680 | 93271 | |
| Rwork/Rfree (%) | 20.4/25.2 | 12.9/16.2 | |
| NO. atoms | |||
| Protein | 6144 | 3007 | |
| Water | 21 | 440 | |
| RMS bonds (Å) | 0.015 | 0.030 | |
| RMS angles (°) | 1.90 | 2.43 | |
| Ramachandran | |||
| Favored (%) | 94 | 96 | |
| Allowed (%) | 5.9 | 3.7 | |
| Outliers (%) | 0.1 | 0.3 | |
| Average B-factor | 48.5 | 22.6 | |
|
| |||
| PDB code | 5SUM | 5SUI | |
Highest resolution shell are shown in parentheses.
Like the majority of β-propellers, Nsa1 contains a velcro closure that stabilizes the radial fold. The Nsa1 velcro closure differs from the canonical WD40 velcro closure in which the final blade is composed of 3 strands from the C-terminus and the outer most strand from the N-terminus (Xu and Min, 2011). In Nsa1, the first blade is made up of 3 strands from the N-terminus (Blade 1B–1D) with the inner most strand from the C-terminus (Blade 1A) creating a velcro closure (Figure 1C). This unusual type of velcro closure was also recently observed in eIF3b (eukaryotic translation initiation factor 3 subunit b), another ribosome associated WD40 containing protein (Liu et al., 2014). A search of the DALI server (Holm and Rosenstrom, 2010) returned numerous hits of 7-bladed β-propellers with high Z-scores (>20) indicating that the WD40 domain of Nsa1 is similar to other 7-bladed β-propellers. Intriguingly the top 2 hits from the DALI search were a putative bacterial isomerase (PDB ID 1RI6) and a muconate lactonizing enzyme (PBD ID 1JOF), which both contain the same non-canonical velcro closure that is observed in Nsa1 (Figure S1B).
S. cerevisiae Nsa1 is one of the longest orthologues of the protein and its WD40 domain contains approximately 80 additional residues compared to the more compact mammalian WDR74 (Figure S2). The number of residues within each of the individual blades varies because some blades contain long extended loops (Figure S2). The overall WD40 domain is asymmetric with respect to the individual blades (Figure 1B). The root mean square deviation values across all seven blades with respect to one another ranges from 1.3 to 3.1 Å. Blades 4, 5, and 7 are most similar to one another but each contains several long loops (Figure S1C). Blades 2 and 3 are also very similar to one another and both contain elongated C and D β strands (Figure S1C). Aside from a few small positive patches along the side of the WD40 domain, Nsa1 does not appear to have an extended positively charged surface onto which RNA could bind (Figure S1D).
Nsa1 contains an extended C-terminal tail
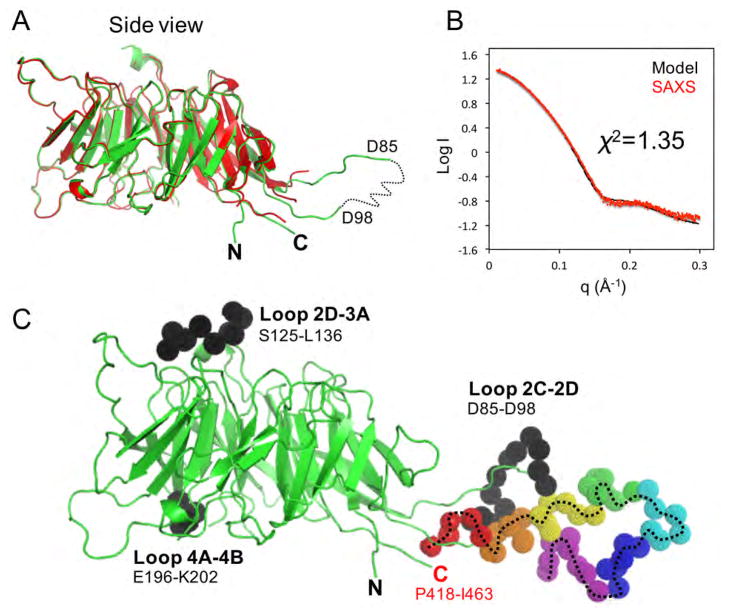
We also obtained a cubic crystal form of S. cerevisiae Nsa1 from a different crystallization condition, which could only be repeated with the full-length protein and not the C-terminal truncation. Although the crystals contained the full C-terminus of the protein we could only build a few additional residues (417–418) onto the C-terminal region of the protein suggesting that the C-terminus of Nsa1 is very flexible (Figure 2A). Overall the cubic and orthorhombic crystal forms are very similar to one another (RMSD of 0.58 Å over 372 residues). However, the cubic crystals contained a loop within the WD40 region that had improved electron density, likely because of crystal packing, which allowed us to build a more complete structure of Nsa1 (Figure 2A). There are two copies of Nsa1 in the asymmetric unit of the cubic crystal form, which are almost identical to one another (RMSD of 0.22 Å over 389 residues).
Figure 2.
SAXS analysis of full-length S. cerevisiae Nsa1. (A) Superposition of the orthorhombic (red) and cubic (green) crystal forms of Nsa1. (B) Comparison of the experimental SAXS scattering curve (red circles) with the scattering curve derived from the Bunch model of Nsa1 (black line) shown in C. (C) A combination of rigid body and ab initio modelling of Nsa1 determined by Bunch. The WD40 domain is shown in cartoon colored in green (crystal structure Nsa1FL: PDB 5SUM), the flexible C-terminus is shown as rainbow color spheres and the three missing loops are shown as black spheres. See also Figure S3, S4 and S5, and Table S1.
Because we could not observe density for the proteolytic sensitive C-terminus of Nsa1 by crystallography we used another structural biology technique, SAXS (small angle X-ray scattering) to determine the position of the C-terminal tail of Nsa1 in solution. SAXS data were recorded for full-length Nsa1 across a concentration series. The Nsa1 samples did not reveal the presence of radiation damage or concentration-dependent inter-particle interactions (Figure S3). The data set with the highest quality estimate determined by AutoRg and AutoGNOM from the ATSAS software suite was used for subsequent analysis (Petoukhov et al., 2012). The calculated molecular weight of Nsa1 (51.9 kDa) was determined using the volume of correlation and corresponds well with the expected molecular weight of a monomer (52.3 kDa) (Table S1). To determine the position of the C-terminus of Nsa1, we used the Nsa1 crystal structure (PDB 5SUM) in the program Bunch from the ATSAS software suite to carry out a combination of rigid body and ab initio modeling (Petoukhov et al., 2012; Petoukhov and Svergun, 2005). The WD40 domain of Nsa1 was treated as a rigid body, while the C-terminus of Nsa1 along with the three missing loops were modeled to fit the experimental SAXS data. The WD40 domain of Nsa1 alone (PDB ID 5SUI or 5SUM) is not a good fit of the experimental SAXS data, giving the chi2 values of 33.32 and 19.62 respectively (Figure S4). The discrepancy of the ab initio model generated by Bunch and the experimental SAXS data resulted in a chi2 value of 1.35, which is a significant improvement over the crystal structure alone (Figure 2B). The ab initio model positions the C-terminus of Nsa1 as extending away from the WD40 domain (Figure 2C and Figure S5). Overall our SAXS data indicate that in solution S. cerevisiae Nsa1 has a compact globular WD40 domain with a C-terminal tail that extends outward from the WD40 domain.
The lysine-rich C-terminus of Nsa1/WDR74 is required for nucleolar localization
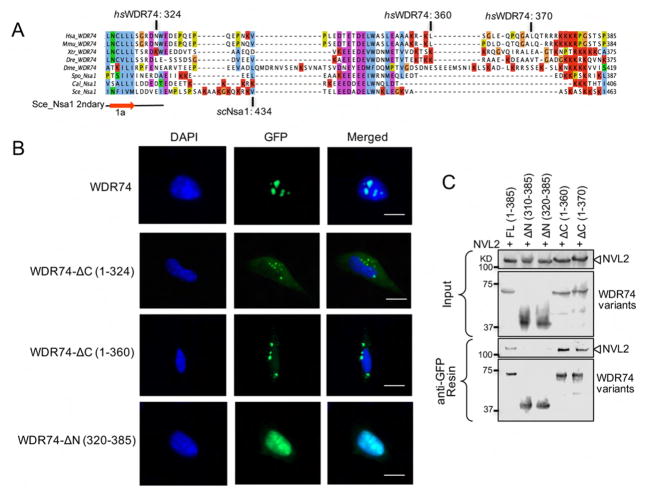
After determining the solution structure of the C-terminus of Nsa1 we wanted to understand its function within the cell. A multiple sequence alignment of the C-terminal region of Nsa1/WDR74 revealed that, aside from a small acidic patch in the middle, it is not well conserved (Figure 3A). However, one striking feature is common amongst homologues: the presence of numerous lysine residues. The incidence of multiple patches of basic residues is often indicative of a nucleolar localization signal (NoLS), which the C-terminus of WDR74 is predicted to contain (Scott et al., 2010). We fused full-length and C-terminal truncations of WDR74 to GFP and then observed subcellular localization in U2OS cells. Full-length WDR74-GFP displays prominent nucleolar localization as expected (Figure 3B). In contrast both C-terminal truncations of WDR74 (residues 1–324 and 1–360), were unable to localize to the nucleolus or nucleus, and are instead found in punctate structures within the cytoplasm (Figure 3B). We also fused the C-terminal tail of WDR74 (residues 320–385) to GFP and found that this region alone could localize to both the nucleus and nucleolus (Figure 3B). The C-terminal tail does not display exclusive nucleolar localization indicating that the WD40 domain is also required for proper targeting of WDR74 to the nucleolus. Our localization studies with mammalian WDR74 confirmed that at least one function of this lysine-rich C-terminal tail is targeting of WDR74 to the nucleus and nucleolus. This suggests that the C-terminus of WDR74 could be important for binding to a nucleolar transporter protein and/or for interacting directly with the nucleolar pre-rRNA (Scott et al., 2010).
Figure 3.
The WDR74 C-terminus is required for nucleolar localization (A) Multiple sequence alignment of the C-terminus of Nsa1/WDR74 from different homologues of Homo sapiens (Hsa), Mus musculus (Mmu), Xenopus tropicalis (Xtr), Danio rerio (Dre) Drosophila melanogaster (Dme), Schizosaccharomyces pombe (Spo), Candida albicans (Cal), and Saccharomyces cerevisiae (Sce). The alignment was done in Tcoffee and colored with Jalview. All of the lysine residues are highlighted in red. The final β strand from the WD40 domain is labeled. The sequence boundaries used for immunofluorescence are marked. (B) U2OS cells were transfected with plasmids harboring WDR74-GFP, WDR74ΔC (1–324)-GFP, WDR74ΔC (1–360)-GFP, or WDR74ΔN (320–385)-GFP, and then fixed and stained with DAPI (blue). The 20 μm scale bar is representative for all panels. (C) Western blot analysis of the interaction between NVL2 with various truncations of WDR74. See also Figure S6.
WDR74 interacts with NVL2
Once we determined that the C-terminal tail of WDR74 was essential for nucleolar localization we wanted to resolve which region(s) of Nsa1/WDR74 were important for association with Rix7/NVL2. We first attempted to characterize the direct interaction between recombinant S. cerevisiae Nsa1 and S. cerevisiae Rix7, however we could not detect binding between recombinant Nsa1 purified from E. coli and recombinant Rix7 purified from yeast or E. coli, suggesting that the interaction may require post-translational modifications or an additional cofactor such as Mtr4, RNA, or pre-60S particles to bridge the interaction. Therefore, we carried out co-immunoprecipitation assays of transiently expressed NVL2 with various truncations of WDR74. Full-length WDR74 (residues 1–385) as well as C-terminal truncations (residues 1–360 and 1–370) were all able to associate with NVL2 (Figure 3C). However, N-terminal truncations of WDR74 (310–385 and 320–385) could not associate with NVL2. Therefore, the WD40 domain of WDR74 is essential for interacting with NVL2. While the C-terminal tail is not important for association with NVL2 in vitro, it still plays a critical role in vivo for facilitating proper nucleolar localization (Figure 3B).
To determine if RNA or pre-60S particles are a requirement to bridge the WDR74-NVL2 interaction we repeated our co-immunoprecipitation experiments with the addition of benzonase and RNase A to the lysis and wash buffer and found that RNA is not a requirement for the WDR74-NVL2 interaction (Figure S6). Treatment of cells with Actinomycin D for 2 hours prior to harvest, to halt transcription of pre-rRNA also had no effect on the WDR74-NVL2 interaction (Figure S6). Moreover, a large scale co-immunoprecipitation was resolved by SDS-PAGE and coomassie staining and the two most prominent bands on the gel were those corresponding to WDR74 and NVL2 (Figure S6). We could detect some ribosomal proteins and ribosome associated factors by whole-lane mass spec analysis but they were not visible by coomassie staining. Taken together these results suggest that the WDR74-NVL2 interaction is not dependent upon RNA or pre-60S ribosome particles. Previous studies have suggested that WDR74 has a greater affinity for binding to a mutant of NVL2 that is deficient in ATP hydrolysis, by mutation of the Walker B motif glutamate (E) residue in both the D1 and D2 domains (Hiraishi et al., 2015). We also mutated the Walker B residues within NVL2 (E364Q/E681Q) and observed an enhanced interaction of WDR74 and NVL2 with the Walker B mutation in our large-scale co-immunoprecipitation (Figure 6S). These studies suggest that WDR74 interacts more strongly with NVL2 in a pre ATP-hydrolysis state.
Subcellular localization of WDR74
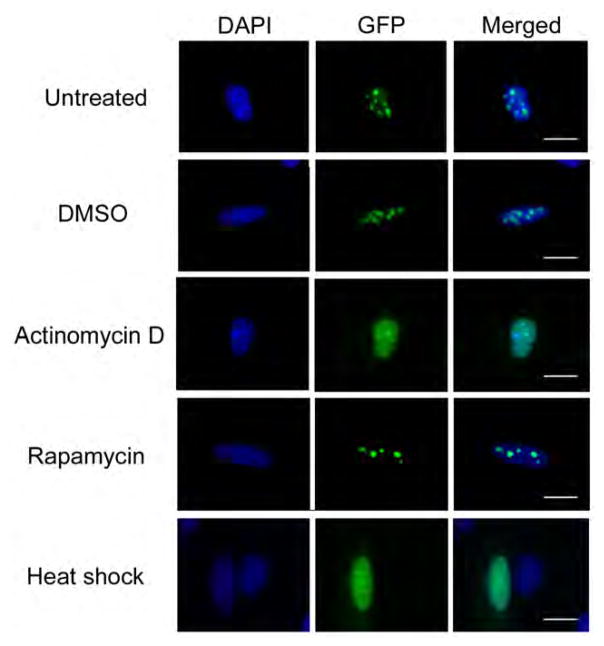
After establishing that the lysine-rich C-terminus of WDR74 is required for nucleolar localization, we wanted to determine if other factors also influence the localization of WDR74. Many steps of ribosome assembly take place within the nucleolus, including pre-rRNA transcription and early pre-rRNA processing and assembly steps (Thomson et al., 2013). Inhibition of Pol I pre-rRNA transcription has been shown to influence the localization of numerous ribosome assembly factors including NVL2 and WDR12 (Nagahama et al., 2004; Romes et al., 2016). Treatment of cells with low concentrations of actinomycin D, which inhibits Pol I transcription, led to a loss of the nucleolar localization of WDR74 and its redistribution to the nucleoplasm (Figure 4). Thus, localization of WDR74 is dependent on active Pol I transcription of pre-rRNA. The mammalian target of rapamycin, complex I (mTORC1) is a positive regulator of ribosome assembly, and inhibition of mTORC1 affects both pre-rRNA transcription and processing (Iadevaia et al., 2012). Inhibition of mTORC1 by rapamycin does not cause a disruption of the nucleolus, but it has been shown to have a selective effect on the localization of certain nucleolar proteins (Iadevaia et al., 2012). Upon treatment of cells with rapamycin for 2 hours, we did not observe any significant difference in the nucleolar localization of WDR74, indicating that mTORC1 activity does not significantly influence localization of WDR74 (Figure 4). Environmental stress has also been shown to impact ribosome assembly so we looked at the effects of heat shock on WDR74 localization (Jacob et al., 2013). Heat shock at 43°C for 3 hours also led to a redistribution of WDR74 into the nucleoplasm mimicking the effects of actinomycin D treatment (Figure 4).
Figure 4.
Localization of WDR74. U2OS cells were transfected with a plasmid containing WDR74-GFP and treated with DMSO, actinomycin D, or rapamycin for 2 hours prior to fixing. Heat shock was induced for 3 hours at 43°C prior to fixing. The nucleus was visualized by staining with DAPI (blue) and the 20 μm scale bar is representative for all panels.
WDR74 associates with NVL2 through a conserved interface in the WD40 domain
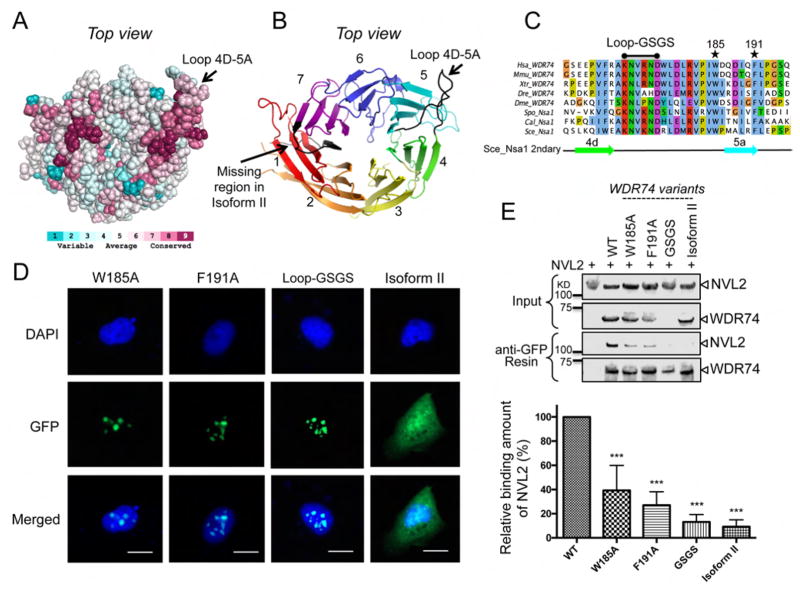
WD40 domains are important scaffold molecules that facilitate protein-protein interactions and WD40 domains have three distinct surfaces (top, bottom, or circumference) for promoting these protein-protein interactions (Stirnimann et al., 2010). In order to understand which region of the WD40 domain of Nsa1/WDR74 was important for association with Rix7/NVL2 a multiple sequence alignment was generated followed by consurf conservation analysis. This analysis revealed a cluster of conserved residues along the top interface of the WD40 domain (Figure 5A and 5B) indicating that the top surface of the propeller could be an important interface for interacting with Rix7/NVL2. Included in this interface is a long-extended loop between blades 4 and 5 (Loop 4D-5A) that is well conserved and ordered in our Nsa1 crystal structure (Figure 5A). Loop 4D-5A is comprised of 16 residues and extends several angstroms away from the central WD40 domain (Figure 5B and 5C). Previous studies in yeast have hinted at the importance of this loop as the W230R mutant in S. cerevisiae Nsa1 was identified in a yeast Rix7 complementation assay (Kressler et al., 2008). W230 (F168 in WDR74) is the last residue in the β strand preceding loop 4D-5A and this mutation confers a slow-growth phenotype in yeast (Kressler et al., 2008). To determine if the top interface of Nsa1/WDR74 is important for interacting with Rix7/NVL2 we carried out co-immunoprecipitation assays with transiently expressed WDR74 and NVL2. Replacement of part of loop 4D-5A (residues 171–176 of WDR74) with alternating glycine and serine residues (Loop-GSGS) greatly reduces the amount of NVL2 that can be pulled down by WDR74, suggesting that this loop is critical for the association of NVL2 and WDR74 (Figure 5D and 5E). The transfection efficiency of the GSGS loop mutant is lower than the wild type protein, making a direct comparison with the wild-type protein impossible. However, it can still be pulled down with affinity resin and maintains proper nucleolar localization, suggesting that it is properly folded and unable to associate with NVL2 under the experimental conditions tested (Figure 5D and 5E). We made additional point mutations to residues, W185A and F191A, which are near loop 4D-5A and these mutations also impacted binding of NVL2 (Figure 5C, 5D, and 5E).
Figure 5.
Nsa1/WDR74 associates with Rix7/NVL2 (A) A surface representation of Nsa1 illustrating conserved (dark pink) to not-conserved residues (cyan). The figure was produced using the consurf website http://consurf-hssp.tau.ac.il (B) Cartoon of top view of Nsa1 highlighting the missing region of WDR74 isoform II (black) and the conserved loop (black) between blade 4D and blade 5A. (C) Selected alignment of Nsa1/WDR74 between blade 4D and blade 5A (see Figure S2 for full alignment) from different homologues of Homo sapiens (Hsa), Mus musculus (Mmu), Xenopus tropicalis (Xtr), Danio rerio (Dre) Drosophila melanogaster (Dme), Schizosaccharomyces pombe (Spo), Candida albicans (Cal), and Saccharomyces cerevisiae (Sce). The asterisk indicate the position of the point mutations in Hsa WDR74 and the black line indicates the position of the loop-GSGS (D) U2OS cells were transfected with plasmids harboring WDR74-GFP mutants and then fixed and stained with DAPI (blue). The 20 μm scale bar is representative for all panels. (E) Western blot analysis of the interaction between NVL2 with various mutations of WDR74. The relative amount of NVL2 bound to the resin is quantified below (39.3% for W185A, 27.1% for F191A, 13.2% for Loop-GSGS, and 9.2% for Isoform II) from 5 independent co-immunoprecipitations (*P < 0.05; **P < 0.01; ***P < 0.001).
There are two known isoforms of WDR74 that arise because of alternative splicing. Isoform I, which is predicted to be the principal isoform, contains an additional 19 residues (308–326) that correspond to β strands 7D and 1A in our S. cerevisiae Nsa1 crystal structure (Figure 5B and S2). Based on our sequence alignment, the WD40 domain of isoform II likely has a destabilized radial WD40 fold. Previous studies revealed that NVL2 interacts with isoform I of WDR74 (Hiraishi et al., 2015). We confirmed this interaction with our co-immunoprecipitations and did not observe any interaction between NVL2 and isoform II of WDR74 (Figure 5E). Surprisingly, despite the absence of two β strands, isoform II of WDR74 expresses well, however the protein does not properly localize to the nucleolus or nucleus (Figure 5D).
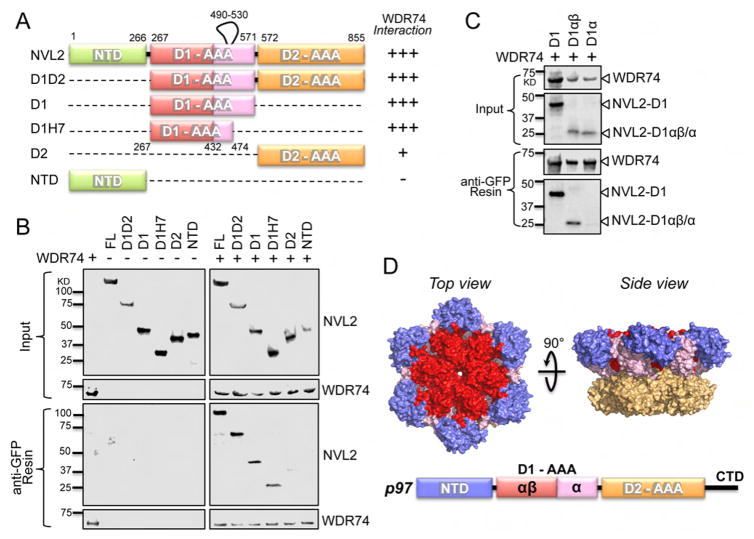
NVL2 associates with WDR74 through its D1 AAA domain
To uncover the structural organization of the NVL2-WDR74 complex, we carried out a series of co-immunoprecipitation assays using various truncations of transiently expressed NVL2 with full-length WDR74 (Figure 6A). Our initial hypothesis was that the N-terminal domain of NVL2 would be responsible for its interaction with WDR74 because Cdc48/p97 has numerous cofactors that interact with its N-terminal domain (Buchberger et al., 2015). However, we did not observe any interaction between the N-terminal region of NVL2 and WDR74 (Figure 6B). WDR74 instead binds to the tandem D1 and D2 AAA domains and the isolated D1 AAA domain (Figure 6B). This is in agreement with previously published data from yeast, which showed a strong yeast two-hybrid interaction between Nsa1 and residues 166–557 of Rix7, which includes a portion of the N-terminal region, the entire D1 domain and a small portion of the D2 domain (Kressler et al., 2008). The D1 domain of Rix7/NVL2 contains an insertion of ~50 amino acids that is not well conserved following Helix 7 (H7) of the AAA module and is absent in Cdc48/p97 (Kressler et al., 2012). Truncation of the D1 domain within H7 (residues 267–474) (Figure 6A) does not effect binding to WDR74 (Figure 6B). AAA domains can be subdivided into two subdomains known as the αβ-domain which contains the Walker A, Walker B, Sensor 1 and 3, and arginine finger motifs and the α-helical lid domain which contains the sensor 2 motif (Erzberger and Berger, 2006). ATP binds at the interface between these two subdomains and the neighboring AAA domain and the ATP hydrolysis state determines the position of the lid domain with respect to the αβ-domain (Erzberger and Berger, 2006). Only the αβ-domain of NVL2 is required for association with WDR74 (Figure 6C). Based upon the high sequence similarity with p97, the αβ-domain of NVL2, is accessible along the top surface of the D1 domain, suggesting that is where WDR74 binds (Figure 6D).
Figure 6.
WDR74 binds the D1 domain of NVL2. (A) Schematic illustration of NVL2 and its truncations. The numbering is based on Mus musculus NVL2, which was used for the pull downs. (B) Western blot analysis of the domain mapping of the interaction between WDR74 and the individual domains of NVL2. The constructs of NVL2 used for the pull downs are depicted in figure 6A and contained the following residues FL (1–855), D1D2 (267–855), D1 (267–571), D1H7 (267–474), D2 (572–855) and NTD (1–266). (C) Western blot analysis of the mapping of the interaction between WDR74 and the D1-AAA domain of NVL2. The constructs of NVL2 used for the pull downs contained the following residues D1 (267–571), D1αβ (267–432), and D1α (432–571). (D) Surface representation of p97 taken from PDB ID 5C18. The NTD is shown in purple, D1αβ is shown in red, D1α is shown in pink and the D2 domain is shown in orange.
DISCUSSION
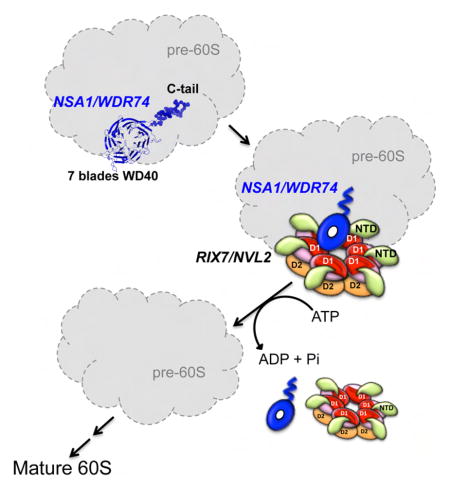
Eukaryotic ribosome assembly is a complex process that relies on hundreds of non-ribosomal proteins, including Nsa1 and Rix7 to perform diverse roles during the assembly process. Nsa1 is essential for cell viability in yeast and WDR74 was recently shown to be required for blastocyst formation in mice however the function of Nsa1/WDR74 in ribosome assembly remains unclear (Kressler et al., 2008; Maserati et al., 2011). Our structural and functional analyses reveal important functions for the N and C terminus of Nsa1/WDR74. The N-terminal WD40 domain of Nsa1/WDR74 contains a seven-bladed β-propeller, which provides a platform for protein-protein interactions. The top surface of the β-propeller within Nsa1/WDR74 is well conserved and important for the association of WDR74 with NVL2. In contrast to the WD40 domain, the C-terminus of Nsa1 is not structured, extends away from the WD40 domain and harbors numerous lysine residues, which are important for targeting the protein to the nucleus and nucleolus.
Nsa1 associates with late nucleolar pre-60S particles and Rix7 exclusively associates with the Nsa1 pre-60s particle and is not bound to other pre-60S intermediate particles (Kressler et al., 2008). The exclusive interaction of Rix7 with the Nsa1 pre-60S particle coupled with our co-immunoprecipitation analysis with the mammalian homologues strongly suggests that one of the functions of Nsa1/WDR74 is the recruitment of Rix7/NVL2 to late nucleolar pre-60S particles. This recruitment is driven in part through an extended loop between the 4th and 5th blades of the WD40 domain of Nsa1/WDR74 that is well conserved amongst eukaryotes. Recent work on Rea1, another AAA-ATPase involved in ribosome assembly, suggests that Rea1 is also involved in Rix7 recruitment, as inhibition of Rea1 in vivo, leads to a reduced association of Rix7 with Nsa1 pre-60S particles (Kawashima et al., 2016). The function of Rea1 at this early stage in ribosome assembly is unknown but it could be important for driving conformational changes within Nsa1 pre-60S particles to allow for Rix7 binding.
NVL2 is a closely related homologue of the well-studied and highly abundant protein p97 that functions in numerous cellular processes. The major difference between NVL2 and p97 lies in the N-terminal and C-terminal regions of the proteins. Both proteins contain unique N-terminal regions and p97 includes a C-terminal extension following the D2 domain, which is missing in NVL2. The N-terminal domain and C-terminal extension of p97 are the platforms on which all of the diverse p97 adapter proteins bind. There are at least 40 known cofactors of mammalian p97, the bulk of which interact with p97 through small conserved binding modules (Buchberger et al., 2015). Here we demonstrate that WDR74 associates with the αβ region of the D1 domain of NVL2. Previous mutagenesis studies in yeast also revealed that ATP binding but not ATP hydrolysis within the D1 domain of Rix7 is essential for viability (Pratte et al., 2013). The D1 domain of NVL2 likely then has 3 functions, ATP binding and oligomerization, which are functions in common with p97 and it also carries out the unique function of associating with WDR74. Our large scale co-immunoprecipitations with WDR74 and NVL2 (wild type and Walker B mutant) agree with previous results that suggest that WDR74 has a higher affinity for the Apo or ATP-bound state of NVL2 (Hiraishi et al., 2015). ATP hydrolysis by NVL2 could therefore drive release of WDR74 from NVL2 and pre-60S particles. p97 is thought to undergo dramatic conformational changes during the ATP hydrolysis cycle and structural studies have revealed at least two known positions of the NTD (Xia et al., 2016). Therefore, as is the case in p97, the nucleotide bound state of the D2 domain of NVL2 likely contributes to the positioning of the NTD and/or conformational changes within the D1 domain. Re-positioning of these domains likely influences the ability of WDR74 to interact with NVL2 through its D1 domain.
In addition to interacting with WDR74, NVL2 has also been found to associate with Mtr4 and the exosome complex (Hiraishi et al., 2015; Nagahama et al., 2006; Yoshikatsu et al., 2015). Mtr4 is an RNA helicase that directs the exosome complex to specific pre-ribosome particles to mediate pre-rRNA processing and degradation (Thoms et al., 2015). The function of the Mtr4-exosome, NVL2, and WDR74 complex is unknown. Mtr4 is specifically recruited to Nop53 and Utp18 containing pre-ribosome particles where it functions to target regions of the 5′ETS and ITS2 for exosome mediated degradation (Thoms et al., 2015). Therefore, one possibility is that WDR74 and NVL2 aid in the recruitment of Mtr4 to pre-60S particles to target the exosome for the degradation of parts of the pre-rRNA. Another possibility is that Mtr4 functions as a cofactor for NVL2 and through the interaction with NVL2, Mtr4 could aid in mediating release of WDR74 from pre-60S particles. Mtr4, NVL2, and WDR74 could also be involved in a pre-60S quality control mechanism, whereby NVL2 is able to sense misassembled pre-60S particles and target the pre-rRNA to the exosome for degradation. Previous work in yeast has shown that over expression of Rix7 enhances the 60S assembly defects of mutations of two other assembly factors Ebp2, and Mak5 suggesting that Rix7 may somehow sense damaged pre-60S particles (Pratte et al., 2013).
Finally, WDR74 and NVL2 have been linked to human disease. WDR74 has been previously shown to be unregulated in mammary gland cancer cells and it was recently reported that the WDR74 promoter is frequently mutated in cancer cells (Krol et al., 2010; Weinhold et al., 2014). NVL, the gene which encodes for NVL2, has been identified as a new prognostic outlier gene for prostate cancer and siRNA knockdown of NVL had a mild inhibitory effect on cell proliferation suggesting a role for NVL2 in cancer progression (Zhao et al., 2016). NVL has also been shown to confer a risk in the Han Chinese population to mental illness, including major depressive disorder and schizophrenia (Wang et al., 2015a). Further studies are needed to understand what roles WDR74 and NVL2 play in human disease but here we reveal the molecular details about the association of these essential proteins, which play important roles in the assembly of the large ribosomal subunit.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Robin Stanley (robin.stanley@nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
S. cerevisiae Nsa1 for crystallization studies was expressed in in BL21(DE3) star cells (Life Technologies) supplemented with 100 μg/ml ampicillin and grown at 37°C. U-2 OS and HEK 293-F cells were supplied by the NIEHS protein expression core. U2OS cells were grown in DMEM/F-12 medium (Gibco by Life Technologies) supplemented with 10% (v/v) fetal bovine serum, antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin), and 2 mM L-glutamine. Cells were incubated at 37°C in 5% CO2.
METHOD DETAILS
Nsa1 Protein Expression and Purification
Full-length Nsa1 (Nsa1FL, residues 1–463) and C-terminal truncated Nsa1 (Nsa1ΔC, residues 1–434) were generated by PCR amplification of genomic DNA isolated from S. cerevisiae and inserted into the pHMBP vector between the BamHI and NotI restriction sites for expression of N-terminal MBP-tagged Nsa1. The plasmids were transformed into BL21(DE3) star cells (Life Technologies) for protein expression. Expression of Nsa1 was induced when the OD600 reached 0.8 by addition of IPTG to 1 mM followed by incubation at 25°C overnight. Cells were harvested by centrifugation and then resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% glycerol, 10 mM MgCl2) containing one EDTA-free protease inhibitor tablet (Roche Applied Science) and lysed by sonication at 4°C. Clarified lysate was applied to a gravity flow column loaded with TALON metal affinity resin (Clontech). Unbound protein was removed by successive washes with lysis buffer, and MBP-Nsa1 was eluted with buffer containing 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 5 mM MgCl2, 5% glycerol and 200 mM imidazole. The MBP tag was removed by TEV protease overnight and dialyzed against Buffer A containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM MgCl2, 5% glycerol and 1 mM β-ME at 4°C. Cleaved Nsa1 was further purified by size exclusion chromatography on a Superdex 200 column (GE Healthcare) pre-equilibrated with Buffer A. Column fractions were analyzed by SDS-PAGE, and those containing Nsa1 were concentrated to 8 mg/ml and used immediately for crystallization. Selenomethionyl (SeMet) incorporation was achieved by expression of Nsa1 in BL21(DE3) star cells grown in M9 minimal medium that was supplemented with SeMet (Molecular Dimensions). The SeMet Nsa1 was purified as described above, and incorporation of SeMet was verified by mass spectrometry.
Nsa1 Crystallization
Two different crystal forms were observed in crystallization trials of Nsa1. Cubic crystals were grown by hanging-drop vapor diffusion method at 25°C, by mixing 1 μl of protein with 1 μl of well solution containing 1.6 M sodium citrate tribasic. Orthorhombic crystals were also grown using the hanging-drop vapor diffusion method at 25°C by mixing 1 μl of protein solution with 1 μl of well solution containing 20%(v/v) PEG 1500, 13%(v/v) PEG 400, 0.1 M HEPES/sodium hydroxide pH 7.5. During crystallization optimization, we discovered that the orthorhombic crystals arose as a result of proteolytic cleavage of Nsa1. Using a combination of limited proteolysis coupled with mass spectrometry, we determined that the C-terminal 29 residues of Nsa1 were sensitive to proteolysis and removal of the C-terminal tail was required for production of the orthorhombic crystal form (Nsa1ΔC). Nsa1ΔC crystals were cryo-protected with cryo-solution supplemented with 22.5%(v/v) PEG 1500 and 15%(v/v) PEG 400 before flash freezing in liquid nitrogen. Cubic crystals were directly flash frozen in 1.6 M sodium citrate tribasic.
X-ray Data Collection and Structure Determination
X-ray diffraction data were collected at 100 K on the SER-CAT beam lines 22-ID and 22-BM of the Advanced Photon Source at Argonne National Laboratory (Chicago, IL). Diffraction images were processed and scaled with the HKL2000 package (Otwinowski and Minor, 1997). The Nsa1ΔC structure was solved by SeMet-SAD (single-wavelength anomalous dispersion) followed by automated density modification and building in AutoSolve (Terwilliger et al., 2009). The cubic crystal form (Nsa1FL) was solved by molecular replacement using the Nsa1ΔC structure as the search model in Phaser (Adams et al., 2010). Coot was used to examine electron density maps and for model building (Emsley et al., 2010). The refinement and final analysis were performed with Refmac with no NCS and geometric restraints (Murshudov et al., 2011; Winn et al., 2011). Unit cell parameters and crystallographic statistics are presented in Table 1.
SAXS Data Collection and Processing
SAXS data were recorded for full length S. cerevisiae Nsa1 at the Advanced Light Source, SIBYLS beamline 12.3.1 at Lawrence Berkeley National Laboratory (Berkeley, CA) (Dyer et al., 2014). Twenty-four hours prior to shipment of Nsa1 to the beamline, it was resolved over a Superdex 200 gel filtration column pre-equilibrated in buffer A. The samples were then added to a microplate, sealed, and shipped overnight at 4°C. The plate was stored at 4°C until data collection. Prior to data collection the plate was spun at 3700 rpm for 10 minutes at 4°C. SAXS data were recorded for the buffer before and after each protein concentration series. Thirty-three consecutive scans of 0.3 seconds were collected for Nsa1 over a concentration series of 6.2, 4.5 and 3 mg/ml at 10°C. Data quality was assessed by comparing the solution scattering across all thirty-three frames and averaged to generate the final scattering curve. The radius of gyration and pair-distance distribution functions were determined using Primus and Gnom from the ATSAS 2.7.2 suite and compared across all protein concentrations (Petoukhov et al., 2012) (see Table S1 and Figure S3). We compared the experimental scattering of Nsa1 to the theoretical scattering curve, generated from the atomic structure of Nsa1 (PDB 5SUM), using Crysol (Svergun et al., 1995). A rigid body model of full-length Nsa1 was generated with the cubic crystal structure using Bunch (Petoukhov and Svergun, 2005).
HEK293 Mammalian Transfections
DNA encoding mammalian NVL2 was PCR amplified from mouse NVL2 cDNA (GeneScript), and DNA fragments encoding full-length and truncated NVL2 were subcloned into the pCAG vector that encodes an N-terminal OSF (One-Strep-Flag) tag (Morita et al., 2007). Mammalian WDR74, (which corresponded to isoform II), was PCR amplified from WDR74 cDNA (Open Biosystems), and subcloned into the pLexM vector that encodes a C-terminal GFP tag (Aricescu et al., 2006). Isoform I of WDR74, which includes an additional 19 residues near the C-terminus, was generated by adding the additional cDNA sequences through successive rounds of PCR. Mutations to WDR74 and NVL2 were generated using the Q5 site directed mutagenesis kit (NEB) and verified by DNA sequencing. Transfections were carried out in 40 ml cultures of HEK293 cells that had been adapted to grow in suspension. Cells were transfected with 1 μg/ml of purified plasmid DNA and 2 μg/ml of PEI, and harvested 72 hours after transfection, and then frozen at −80°C until used.
Co-immunoprecipitation Assays
Transfected HEK293 cells were lysed in 50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 10% (v/v) glycerol, 0.5% (v/v) Nonidet P40, EDTA-free protease inhibitors (Roche Applied Science), and 10 units of Benzonase (Sigma) with gentle rocking at 4°C for 1 h. Clarified lysate was incubated with anti-GFP conjugated resin prepared using NHS-activated Sepharose™ Fast Flow (GE Healthcare) for 1 h at 4°C. Resin was washed four times with 500 μl lysis buffer (without benzonase). Resin-bound protein in 2× beta-mercaptoethanol and 2× SDS loading dye (Bio-Rad) was boiled and then loaded onto SDS-PAGE gels for identification by western blot.
Western Blots
Proteins were separated on 4–15% mini-Protean TGX Stain Free gels (BioRad) and then transferred to Immuno-Blot LF-PVDF membranes (Bio-Rad), which were blocked for one hour in Odyssey Blocking Buffer TBS (Li-COR). Membranes were washed 3 times with TBS, then incubated one hour at room temperature in TBS Buffer containing both anti-GFP mouse IgG monoclonal primary antibody (Roche; 1:1000) and anti-Flag antibody produced in rabbit (Sigma; 1:2000 dilution). After incubation, membranes were washed 3 times with TBS and then incubated with IRDye 680LT Donkey anti-mouse (Li-COR; 1:4000 dilution) and IRDye 800CW Donkey anti-rabbit (Li-COR; 1:4000 dilution) for 1 hour at room temperature. Membranes were washed three more times with TBST and once with TBS, and then the bands were visualized on a Li-COR Odyssey Fc Imaging System (Image Studio Lite).
Fluorescence Microscopy
U2OS cells were grown on 35-mm glass bottom culture dishes (MatTek Corp.) until 70–80% confluent. Plasmid DNA (6 μg) was delivered into cells with FuGENE 6 (Promega) according to manufacturer’s instructions. Treated cells were incubated with DMSO (0.5% (v/v)), actinomycin D (10 μM), or Rapamycin (200 nM) for 2 hours at 37°C in 5% CO2 where indicated. Heat shock cells were incubated for 3 hours at 43°C in 5% CO2. Cells were fixed in 1× PBS containing 4% paraformaldehyde for 1 hour. U2OS cells were rinsed with 1× PBS and permeabilized with 0.5% Triton X-100 in 1× PBS for 1 hour. Cells were incubated with 300 nM DAPI for 5 minutes and rinsed in 1× PBS before collecting images using a Zeiss Axio Observer Z1 Epifluorescence Microscope with a Zeiss EC Plan-Neofluar 40×/0.9 objective and aperture lens. WDR74-GFP constructs were imaged at room temperature on the Zeiss AxioCam MRm camera with Zen Pro 2012 (Blue Edition) software. Images were prepared in ImageJ (Schneider et al., 2012).
QUANTIFICATION AND STATISTICAL ANALYSIS
All co-immunoprecipitation analyses were repeated at least three times and representative results were shown.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The atomic coordinates of both Nsa1 crystal forms been deposited in the Protein Data Bank under accession codes 5SUI and 5SUM (see Table 1).
Supplementary Material
HIGHLIGHTS.
The N-terminal WD40 domain of Nsa1/WDR74 is a seven-bladed β-propeller
The flexible C-terminus of WDR74 is required for nucleolar localization
A conserved loop within the WDR74 WD40 domain is required for association with NVL2
WDR74 associates with the D1-AAA domain of the AAA-ATPase NVL2
Acknowledgments
We would like to thank the NIEHS Protein Expression Core for their help with mammalian cell culture and the NIEHS X-ray Crystallography Core for their help with x-ray data collection and analysis. Diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID and 22-BM beamlines at the Advanced Photon Source (APS), Argonne National Laboratory. The SAXS data was collected on the SIBYLS beamline at the Advance Light Source (ALS), Lawrence Berkeley National Laboratory. We would like to thank the staff at the SIBYLS beamline for their help with remote data collection and processing. We would also like to thank the NIEHS Fluorescence Microscopy and Imaging Center for their help with the immunofluorescence and the NIEHS Mass Spectrometry Core for help with identifying the limited proteolysis boundaries. We thank Dr. Traci Hall and Dr. Thomas Kunkel for their critical reading of this manuscript. This work was supported by the US National Institute of Health Intramural Research Program; US National Institute of Environmental Health Sciences (NIEHS) (ZIA ES103247 to R. E. S.). Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. Use of the Advanced Light Source (ALS) was supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Additional support for the SIBYLS SAXS beamline comes from the National Institute of Health project MINOS (R01GM105404) and a High-End Instrumentation Grant S10OD018483.
Footnotes
AUTHOR CONTRIBUTIONS
YHL purified Nsa1, crystallized Nsa1 and solved the crystal structure. YHL and MCP processed and analyzed SAXS data. YHL, EMR, and MS designed and constructed vectors for pull-downs and analyzed pull downs by western blot. MCP and YHL carried out the immunofluorescence experiments. RES outlined the study and experiments. YHL, EMR, MCP, MS and RES analyzed data, prepared figures and the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio-Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Bassler J, Beckmann R, Hurt E. Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nature structural & molecular biology. 2016;23:37–44. doi: 10.1038/nsmb.3132. [DOI] [PubMed] [Google Scholar]
- Barthelme D, Sauer RT. Origin and Functional Evolution of the Cdc48/p97/VCP AAA+ Protein Unfolding and Remodeling Machine. J Mol Biol. 2016;428:1861–1869. doi: 10.1016/j.jmb.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Molecular cell. 2010;38:712–721. doi: 10.1016/j.molcel.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler JPH, Holdermann I, Thoms M, Granneman S, Barrio-Garcia C, Nyarko A, Stier G, Clark SA, Schraivogel D, Kallas M, Beckmann R, Tollervey D, Barbar E, Sinning I, Hurt E. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J Cell Biol. 2014;207:481–498. doi: 10.1083/jcb.201408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Schindelin H, Hanzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015;589:2578–2589. doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Dyer KN, Hammel M, Rambo RP, Tsutakawa SE, Rodic I, Classen S, Tainer JA, Hura GL. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol Biol. 2014;1091:245–258. doi: 10.1007/978-1-62703-691-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA plus proteins. Annu Rev Bioph Biom. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Fujiwara K, Goda N, Iwaya N, Tenno T, Shirakawa M, Hiroaki H. Structure and function of the N-terminal nucleolin binding domain of nuclear valosin-containing protein-like 2 (NVL2) harboring a nucleolar localization signal. J Biol Chem. 2011;286:21732–21741. doi: 10.1074/jbc.M110.174680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Braspenning J, Hoepfner D, Petfalski E, Philippsen P, Tollervey D, Hurt E. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 2001;20:3695–3704. doi: 10.1093/emboj/20.14.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her J, Chung IK. The AAA-ATPase NVL2 is a telomerase component essential for holoenzyme assembly. Biochemical and biophysical research communications. 2012;417:1086–1092. doi: 10.1016/j.bbrc.2011.12.101. [DOI] [PubMed] [Google Scholar]
- Hiraishi N, Ishida Y, Nagahama M. AAA-ATPase NVL2 acts on MTR4-exosome complex to dissociate the nucleolar protein WDR74. Biochemical and biophysical research communications. 2015;467:534–540. doi: 10.1016/j.bbrc.2015.09.160. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V, Zhang Z, Jan E, Proud CG. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res. 2012;40:2527–2539. doi: 10.1093/nar/gkr1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob MD, Audas TE, Uniacke J, Trinkle-Mulcahy L, Lee S. Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol Biol Cell. 2013;24:2943–2953. doi: 10.1091/mbc.E13-04-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel L, Loibl M, Zisser G, Klein I, Fruhmann G, Gruber C, Unterweger S, Rechberger G, Pertschy B, Bergler H. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J Cell Biol. 2012;199:771–782. doi: 10.1083/jcb.201205021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Chen Z, Aoi Y, Patgiri A, Kobayashi Y, Nurse P, Kapoor TM. Potent, Reversible, and Specific Chemical Inhibitors of Eukaryotic Ribosome Biogenesis. Cell. 2016;167:512–524. e514. doi: 10.1016/j.cell.2016.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochimica et biophysica acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bergler H, Bassler J. The power of AAA-ATPases on the road of pre-60S ribosome maturation--molecular machines that strip pre-ribosomal particles. Biochimica et biophysica acta. 2012;1823:92–100. doi: 10.1016/j.bbamcr.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Roser D, Pertschy B, Hurt E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J Cell Biol. 2008;181:935–944. doi: 10.1083/jcb.200801181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol M, Polanska J, Pawlowski KM, Turowski P, Skierski J, Majewska A, Ugorski M, Morty RE, Motyl T. Transcriptomic signature of cell lines isolated from canine mammary adenocarcinoma metastases to lungs. J Appl Genet. 2010;51:37–50. doi: 10.1007/BF03195709. [DOI] [PubMed] [Google Scholar]
- Liu Y, Neumann P, Kuhle B, Monecke T, Schell S, Chari A, Ficner R. Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit. Structure. 2014;22:923–930. doi: 10.1016/j.str.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Maserati M, Walentuk M, Dai X, Holston O, Adams D, Mager J. Wdr74 is required for blastocyst formation in the mouse. PLoS One. 2011;6:e22516. doi: 10.1371/journal.pone.0022516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KL, Baserga SJ. Genetics. Mysterious ribosomopathies. Science. 2013;341:849–850. doi: 10.1126/science.1244156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KL, Charette JM, Vincent NG, Baserga SJ. A protein interaction map of the LSU processome. Genes Dev. 2015;29:862–875. doi: 10.1101/gad.256370.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Alam SL, Eckert DM, Gygi SP, Sundquist WI. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama M, Hara Y, Seki A, Yamazoe T, Kawate Y, Shinohara T, Hatsuzawa K, Tani K, Tagaya M. NVL2 is a nucleolar AAA-ATPase that interacts with ribosomal protein L5 through its nucleolar localization sequence. Mol Biol Cell. 2004;15:5712–5723. doi: 10.1091/mbc.E04-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama M, Yamazoe T, Hara Y, Tani K, Tsuji A, Tagaya M. The AAA-ATPase NVL2 is a component of pre-ribosomal particles that interacts with the DExD/H-box RNA helicase DOB1. Biochemical and biophysical research communications. 2006;346:1075–1082. doi: 10.1016/j.bbrc.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Nakhoul H, Ke J, Zhou X, Liao W, Zeng SX, Lu H. Ribosomopathies: mechanisms of disease. Clinical medicine insights Blood disorders. 2014;7:7–16. doi: 10.4137/CMBD.S16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratte D, Singh U, Murat G, Kressler D. Mak5 and Ebp2 act together on early pre-60S particles and their reduced functionality bypasses the requirement for the essential pre-60S factor Nsa1. PLoS One. 2013;8:e82741. doi: 10.1371/journal.pone.0082741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romes EM, Sobhany M, Stanley RE. The Crystal Structure of the Ubiquitin-like Domain of Ribosome Assembly Factor Ytm1 and Characterization of Its Interaction with the AAA-ATPase Midasin. J Biol Chem. 2016;291:882–893. doi: 10.1074/jbc.M115.693259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 2010;38:7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, Mccoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M, Ahmed YL, Maddi K, Hurt E, Sinning I. Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res. 2016;44:926–939. doi: 10.1093/nar/gkv1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M, Thomson E, Bassler J, Gnadig M, Griesel S, Hurt E. The Exosome Is Recruited to RNA Substrates through Specific Adaptor Proteins. Cell. 2015;162:1029–1038. doi: 10.1016/j.cell.2015.07.060. [DOI] [PubMed] [Google Scholar]
- Thomson E, Ferreira-Cerca S, Hurt E. Eukaryotic ribosome biogenesis at a glance. J Cell Sci. 2013;126:4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Bottcher B, Hurt E. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- Wang M, Chen J, He K, Wang Q, Li Z, Shen J, Wen Z, Song Z, Xu Y, Shi Y. The NVL gene confers risk for both major depressive disorder and schizophrenia in the Han Chinese population. Prog Neuropsychopharmacol Biol Psychiatry. 2015a;62:7–13. doi: 10.1016/j.pnpbp.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Wang W, Nag S, Zhang X, Wang MH, Wang H, Zhou J, Zhang R. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Medicinal research reviews. 2015b;35:225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods SJ, Hannan KM, Pearson RB, Hannan RD. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochimica et biophysica acta. 2015;1849:821–829. doi: 10.1016/j.bbagrm.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Woolford JL, Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Tang WK, Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene. 2016;583:64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikatsu Y, Ishida Y, Sudo H, Yuasa K, Tsuji A, Nagahama M. NVL2, a nucleolar AAA-ATPase, is associated with the nuclear exosome and is involved in pre-rRNA processing. Biochemical and biophysical research communications. 2015;464:780–786. doi: 10.1016/j.bbrc.2015.07.032. [DOI] [PubMed] [Google Scholar]
- Zhao SG, Evans JR, Kothari V, Sun G, Larm A, Mondine V, Schaeffer EM, Ross AE, Klein EA, Den RB, et al. The Landscape of Prognostic Outlier Genes in High-Risk Prostate Cancer. Clin Cancer Res. 2016;22:1777–1786. doi: 10.1158/1078-0432.CCR-15-1250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.