Abstract
IDH1 mutations occur in the majority of low-grade gliomas and lead to the production of the oncometabolite, D-2-hydroxyglutarate (D-2HG). To understand the effects of tumor-associated mutant IDH1 (IDH1-R132H) on both the neural stem cell (NSC) population and brain tumorigenesis, genetically faithful cell lines and mouse model systems were generated. Here, it is reported that mouse NSCs expressing Idh1-R132H displayed reduced proliferation due to p53-mediated cell cycle arrest as well as a decreased ability to undergo neuronal differentiation. In vivo, Idh1-R132H expression reduced proliferation of cells within the germinal zone of the subventricular zone (SVZ). The NSCs within this area were dispersed and disorganized in mutant animals, suggesting that Idh1-R132H perturbed the NSCs and the microenvironment from which gliomas arise. Additionally, tumor-bearing animals expressing mutant Idh1 displayed a prolonged survival and also overexpressed Olig2, features consistent with IDH1-mutated human gliomas. These data indicate that mutant Idh1 disrupts the NSC microenvironment and the candidate cell of origin for glioma; thus, altering the progression of tumorigenesis. Additionally, this study provides a mutant Idh1 brain tumor model that genetically recapitulates human disease, laying the foundation for future investigations on mutant IDH1-mediated brain tumorigenesis and targeted therapy.
Keywords: IDH1-R132H, D-2HG, glioma, brain tumor, animal model, hydrocephalus
Introduction
Gliomas are the most common primary malignant tumor of the central nervous system (CNS) and account for 80% of all malignant brain tumors. Despite decades of research, overall survival of patients with malignant glioma remains dismal (1,2). Even with aggressive treatment involving surgical resection, radiation, and chemotherapy, malignant gliomas almost invariably recur, due in large part to their invasiveness and resistance to therapy (3).
Previous studies have identified isocitrate dehydrogenase 1 (IDH1) as a major cancer gene among the low-grade gliomas, which have an inherent tendency to progress from low-grade (grade II and grade III) to high-grade (grade IV) gliomas, also known as secondary glioblastoma (GBM). One key signature among the diffuse gliomas is the co-occurrence of IDH1 mutations with other alterations, such as TP53 mutations (4). The presence of these mutations in the initial biopsies of most patients with these gliomas suggests they are early events in tumorigenesis and are important drivers of malignant progression (3–5).
Normally, IDH1 functions in the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) in an NADP+-dependent manner. The most common mutation in IDH1 is a hotspot missense mutation at arginine 132 (R132), and among gliomas, it is most commonly mutated to histidine (R132H) (4). Those cells expressing IDH1-R132H display neomorphic enzymatic activity in the NADPH-dependent reduction of α-KG to D-2-hydroxyglutarate (D-2HG) (6). Consequently, low- and high-grade IDH1-R132H-expressing gliomas produce D-2HG at levels 100-fold higher than in IDH1 wildtype tumors (6).
The IDH1 mutation and elevated D-2HG levels confer a myriad of cellular effects including inhibition of α-KG-dependent dioxygenases, aberrant histone methylation, contribution to the glioma CpG island methylation phenotype (G-CIMP), increased trimethylation of histone H3 at lysine 9 and 27, and decreased levels of 5-hydroxymethylcytosine (7–10). Additionally, mutant IDH-mediated epigenetic dysregulation is thought to inhibit normal differentiation patterns and impart significant effects on metabolism (11–16). Due to its promiscuous activity throughout a cell, mutant IDH1 likely promotes tumorigenesis via a convergence of various perturbed intracellular pathways within both the tumor cells of origin and the surrounding microenvironment.
To determine the effects of mutant IDH1 and D-2HG on the glioma cell of origin and the microenvironment, we generated a genetically faithful conditional knock-in mouse model of mutant Idh1. In vitro analyses of NSCs expressing mutant Idh1 show a reduction in proliferation and a block to neuronal differentiation, corroborating previous studies describing mutant IDH1’s effects on differentiation programs. Similarly, expression of mutant Idh1 in vivo reduces the proliferation of cells in the subventricular zone (SVZ) of the lateral ventricles and causes a disorganization to this structure which is normally very tightly organized. Together, this suggests that expression of mutant Idh1 has damaging effects on the glioma cell of origin and the microenvironment. Guided by the genetic signature of human gliomas, we used a mouse glioma model driven by Tp53 deficiency to show that expression of mutant Idh1 leads to distinct molecular and histological features, and alters the course of tumor progression by slowing tumor growth and prolonging survival. These findings provide new insight into IDH1-R132H-driven tumorigenesis, and establish a platform for future preclinical investigations focused on mutant IDH1-targeted therapy.
Materials and Methods
Generation of mutant Idh1 conditional knock-in mouse model and transgenic mice
A targeting vector based off of the genomic sequence of the 129P2/Ola strain of mouse was generated and included a 5,426-base pair 5′ homology arm encompassing exon 2 of Idh1 followed by a LoxP site, three copies of the SV40 polyadenylation signal, a neomycin-resistance gene flanked with FRT sites, another LoxP site, and a 2,416-base pair 3′ homology arm encompassing exon 3 of Idh1 and the R132H mutation. A two-base pair change from the CGA codon for arginine to the CAC codon for histidine was produced. This construct was electroporated into 129P2/Ola mouse embryonic stem cells and recombinants were selected using G418. PCR across the 3′ homology arm was performed to identify homologous recombinants. hGFAP-Cre, Nestin-CreERT2, and Tp53fl/fl animals were generously provided by Dr. Rob Wechsler-Reya and were crossed into the Idh1-R132H conditional knock-in mouse model. Animals were maintained in a barrier facility, under pathogen-free conditions according to NIH guidelines.
Genotyping
Tissue was lysed in lysis buffer (100mM Tris-HCl pH 8; 5mM EDTA pH 8; 0.2% SDS; 200mM NaCl; Proteinase K, and water) and DNA was ethanol precipitated. Pellet was resuspended in 50 μl sterile H2O and further diluted 1:50 in sterile H2O. 1.5 μl of diluted DNA were used for PCR reactions. A touchdown PCR protocol was performed: 1x (95°C 5 minutes); 3x (95°C 20 seconds, 64°C 20 seconds, 72°C 20 seconds), 3x (95°C 20 seconds, 61°C 20 seconds, 72°C 20 seconds), 3x (95°C 20 seconds, 57°C 20 seconds, 72°C 20 seconds), 30x (95°C 20 seconds, 55°C 20 seconds, 72°C 20 seconds), 1x (72°C 5 minutes). Genotyping primers can be found in Supplementary Table S1.
Analysis of D-2HG
Quantification of D-2-HG in biological media/tissues was done by LC-ESI-MS/MS method as published by Struys et al. with modifications to accommodate different equipment and sample matrices(17).
Materials
D-2-HG and Diacetyl-L-tartaric anhydride (DATAN) were from Sigma/Aldrich. Racemic mixture of L- and D-2-HG-d4 was prepared by mixing 1 mg of α-ketoglutarate-d6 (Sigma/Isotec) with 1 mg of NaBH4 (Sigma) in 0.2 mL anhydrous MeOH (Sigma) followed by 30 min incubation at 60°C. Sodium hydroxide (25%) and formic acid (98%) were from Fluka, Germany (mass spectrometry grade). Other reagents and solvents were of analytical grade.
Sample preparation/derivatization
To 20 μL of sample 10 μL of 20 μg/mL of each L/D-2-HG-d4 (internal standard) in water was added and mixture evaporated to dryness under gentle stream of nitrogen at 65°C. The dry residue was treated with 50 μL of 50 mg/mL of freshly prepared DATAN in dichloromethane/glacial acetic acid (4/1 by volume) and heated at 75°C for 30 min. After drying (65°C, 1h) the residue was dissolved in 20 μL LC mobile phase (see below) for LC/MS/MS analysis.
LC/MS/MS analysis
Instrument: Shimadzu 20A series HPLC and Sciex/Applied Biosystems API 4000 QTrap. Mobile phase (isocratic elution): water, 3% acetonitrile, 0.5% methanol, 2mM ammonium hydroxide, pH adjusted to 3.6 by formic acid. Analytical column: ACE #ACE-111-1546 (C18, 150 × 4.6mm), and #ACE-111-0103GD guard column. Column temperature: 45°C. Run time: 4 min. Injection volume: 5 μL. Mass spectrometer parameters (voltages, gas flow, and temperature) were optimized by infusion of 100 ng/mL of analytes in mobile phase at 10 μL/min using Analyst 1.6.2 software tuning module. The Q1/Q3 (m/z) transitions monitored: 363/147 (D-2-HG) and 367/151 (L/D-2-HG-d4).
Calibration and quantification
A set of calibrator samples in NeuroCult NSC base media was prepared by adding appropriate amounts of pure D-2-HG at the following concentration levels: 0, 0.032, 0.16, 0.8, 4, and 20 μg/m. The calibration samples were analyzed alongside the experimental samples and accuracy acceptance criteria was 85% for each but the lowest level (0.032 μg/mL, 80%, LLOQ). The obtained calibration curve was linear (r2=0.999). Analyst 1.6.2 software was used for integration of the chromatograms, calibration curve calculation, and quantification of the study samples.
Isolation, culturing, differentiation, transduction, and authentication of NSCs
NSCs were harvested from E14.5 embryos. Pregnant dams were euthanized via CO2 asphyxiation. NSCs were harvested and cultured from embryos in accordance with StemCell Technologies Technical Manual for In Vitro Proliferation and Differentiation of Mouse Neural Stem and Progenitor Cells Using NeuroCult (StemCell Technologies, 05704). 1-day post-harvest, cells were infected with Cre-expressing RGD fiber modified adenovirus (Vector Biolabs, 1769) or GFP-expressing adenovirus (Vector Biolabs, 1768) at an MOI of 10–100. Differentiations were performed in accordance with the aforementioned StemCell Technologies kit. Primary mouse cell lines were generated in-house from February 2014 to February 2016 and have been authenticated through PCR genotyping. Cell lines are routinely submitted for mycoplasma contamination testing. All lines tested negative for these studies.
Cell proliferation assay
Cell proliferation was assessed via CyQUANT Cell Proliferation Assay Kit (ThermoFisher-C7026) and protocol was performed as recommended.
q-RT-PCR
q-RT-PCR was performed using the KAPA SYBR FAST qPCR Master Mix (KapaBioSystems, KK4600) following a touchdown PCR protocol. Primer sequences can be found in Supplementary Table S1.
Western blot
Standard western blot protocols were followed. Antibodies included p53 (1C12) Mouse mAb (Cell Signaling Technology, 2524), IDH1-R132H (Dianova, H-09), and GAPDH (D16H11) XP Rabbit mAb (Cell Signaling Technology, 5174). Secondary antibodies included Anti-mouse IgG, HRP-linked (Cell Signaling Technology, 7076), Anti-rabbit IgG, HRP-linked (Cell Signaling Technology, 7074).
Gene expression of lower grade glioma samples, data analysis, and statistics
RNAseq data and mutation status for IDH1 of lower grade gliomas (n=534) were obtained from TCGA website. RNAseq normalized counts were utilized for comparison of gene expression between mutant IDH1 (n=416) and wild type IDH1 (n=118) samples. Gene expression comparison was shown by Boxplot with dots. Student t-test was used for statistical analysis. All graphs and statistical analyses were performed using either GraphPad Prism or R. Gene expression profiling and pathway analysis can be found in supplementary materials and methods.
Animal studies
Animals were mated and checked for the presence of a vaginal plug, which indicated 0.5 days post coitum (dpc). Tamoxifen was administered to induce activation of Cre-recombinase in those animals harboring Nestin-CreERT2. Tamoxifen (Sigma, T5648) was reconstituted in pharmaceutical grade corn oil (20 mg/ml), warmed to 37° Celsius and vortexed thoroughly until completely dissolved. 3 mgs of tamoxifen was administered to pregnant dams at 18.5 dpc via oral gavage. Randomization of animals was not necessary for these studies as genotype dictated treatment. Gender was recorded, though was not a criteria for inclusion or exclusion from treatment cohorts. Adult animals were euthanized (via CO2 asphyxiation or through a lethal dose of Urethane (10 μl/g of 20% Urethane in sterile PBS). Once unresponsive, animals underwent intracardiac perfusion with 15 mls PBS followed by 15 mls of 10% Neutral Buffered Formalin. Tissues were harvested and post-fixed in 10% Neutral Buffered Formalin for 24 hours and transferred to 70% Ethanol for paraffin embedding. Animals euthanized for the postnatal day 3 (P3) timepoint were placed on gauze and ice until deeply anesthetized followed by decapitation and tissue harvesting. Brains were placed in 10% Neutral Buffered Formalin for 24 hours, at which point they were transferred to either 70% Ethanol for paraffin embedding or in 20% Sucrose in PBS and placed at 4° Celsius until cryoprotected for OCT embedding. Paraffin blocks were cut 5–6 μM on charged slides. OCT blocks were cut at 14 μM on charged slides and stored at −80° Celsius until IF staining.
H&E and immunofluorescence
Standard H&E and immunofluorescent staining procedures were performed. The Duke University Pathology Research Histology and Immunohistochemistry Laboratory processed and stained samples for IHC. A standard protocol requiring antigen retrieval in citrate buffer (pH 6.0, 80° Celsius) was used. For IHC, the DAKO Autostainer 3400 was used. Antibodies included: β(III)Tubulin 1:1000 Covance-MMS-435P), CD31 1:100 (Abcam-ab28364), IDH1-R132H 1:100 (Dianova-H09), Ki67 1:100 (BD Pharm-550609), Olig2 1:500 (Abcam-ab136253), Sox2 1:100 (StemCell Technologies-60055). Secondary antibodies included: Alexa Fluor 488 (ThermoFisher) and Alexa Fluor 594 (ThermoFisher). Immunofluorescent staining was performed in accordance with the aforementioned StemCell Technologies kit. H&E and IHC samples were imaged using the Leica DMD 108. Immunofluorescent images were imaged on the Nikon Eclipse T2000-E. Positively stained cells were both manually counted and counted with ImageJ. Those assigned to counting were blinded to animal genotype and were given designated quadrants of specific size and magnification to count.
Global DNA Methylation analysis
Genomic DNA was extracted using the AllPrep DNA/RNA Mini Kit (Qiagen 80204). DNA was quantified and MethylFlash Global DNA Methylation (5-mC) ELISA Easy kit (Epigentek P-1030-48) colorimetric assay was performed to assess global DNA methylation in accordance with supplied handbook.
Promoter methylation analysis
EpiTect II DNA Methylation Enzyme Kit (Qiagen 335452) was used with EpiTect Methyl II PCR Assay (Qiagen 335002) to assess the methylation status of the second promoter of the mouse BCAT1. Protocol was followed in accordance to handbook using primers specific for mouse CGI 48 (human CGI 70) (Qiagen EPMM109656). Data was analyzed using the supplied Methylation Data Analysis template for single assays.
Data availability
All relevant data and reagents are available from the authors upon request and fulfillment of proper university required documentation.
Ethics statement
All animal studies were approved by the Institutional Animal Care and Use Committee of Duke University, an institution accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC), International. Protocol numbers: A109-10-04, A072-13-03 and A037-16-02.
Additional information can be found in Supplementary Materials and Methods.
Results
Generation of a mutant Idh1 conditional knock-in mouse model
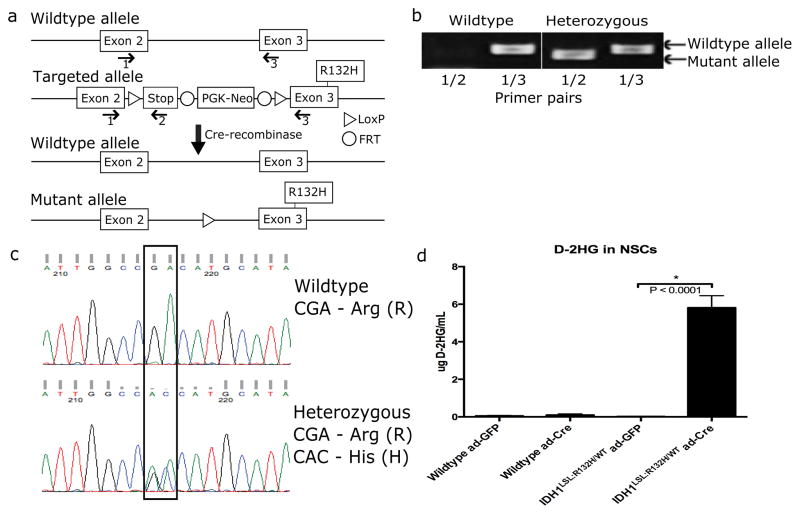
A mutant Idh1 conditional knock-in model was generated via a targeting vector containing a stop cassette flanked by LoxP sites (LSL) and the IDH1-R132H mutation (referred to as Idh1LSL:R132H) (Fig. 1a). Homologous recombination into the endogenous mouse Idh1 locus results in a mutant Idh1 allele with a CGA to CAC substitution in exon 3, encoding the arginine to histidine substitution and resembling the cancer-specific IDH1-R132H mutation. The expression of the modified allele is blocked by the preceding LoxP-flanked stop sequence, resulting in a knocked out allele that is restored through Cre-recombinase-mediated excision of the stop cassette. PCR- and sequencing-based genotyping of animals indicated that only Idh1LSL:R132H heterozygous animals are generated (Fig. 1b and c) (Primer sequences can be found in Supplementary Table S1). Sixty-two crosses between Idh1LSL:132H/WT heterozygous animals generated a total of 292 offspring, with 113 wildtype (Idh1WT/WT) and 179 heterozygous (Idh1LSL:R132H/WT) animals. Testing for the expected Mendelian ratio of 1:2:1 for wildtype:heterozygous:homozygous mutant animals, respectively, gives a p-value less than 1.0 × 10−9 (Fisher’s exact test). The failure to recover Idh1LSL:R132H/LSL:R132H homozygous animals suggested embryonic or perinatal lethality of homozygous Idh1 knockouts.
Figure 1. Generation of mutant Idh1 conditional knock-in mouse model.
(a) Schematic diagram of mutant Idh1 conditional knock-in mouse. Floxed stop cassette generates a null allele. Cre-recombinase induces excision of stop cassette and expression of Idh1-R132H. Genotyping primers (1, 2 and 3) are shown. (b) PCR-based genotyping using primers shown in (a). Primer pair (1/2) produces a 263-base pair amplicon when the mutant allele is present. Primer pair (1/3) produces a 329-base pair amplicon when the wildtype allele is present. Only wildtype and heterozygous animals were generated (p< 1.0 × 10−9 (Fisher’s exact test). (c) Sequencing-based validation of the R132H mutation among generated animals. (d) Measurement of D-2HG in wildtype and Idh1LSL:R132H/WT NSCs infected with ad-GFP or ad-Cre recombinase. Measurements reflect six independent wildtype NSC lines and six independent Idh1LSL:R132H/WT NSC lines each treated with ad-GFP or ad-Cre (unpaired Student t-test, p<0.0001, error bars indicate SEM).
To validate the faithfulness of the mutant Idh1 allele, NSCs were isolated from embryonic day 14.5 (E14.5) heterozygous mutant Idh1 conditional knock-in embryos (Idh1LSL:R132H/WT) and Idh1 wildtype embryos (Idh1WT/WT). NSC lines were transduced with adenoviral-GFP (ad-GFP) as control or adenoviral-Cre-recombinase (ad-Cre) to induce excision of the stop cassette and expression of mutant Idh1. Transient expression of Cre-recombinase led to production of D-2HG which was detected in the supernatant of mutant lines at a level 100-fold higher than in wildtype cells, suggesting D-2HG was produced at pathologically relevant levels (Fig. 1d).
Mutant Idh1 induces p21 and reduces proliferation of NSCs
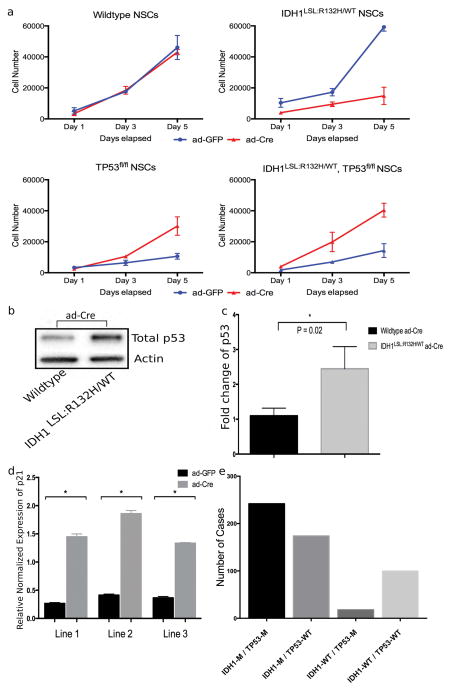
Considering the effects of mutant IDH1 are likely cell context-dependent, we sought to determine the effects of mutant Idh1 in NSCs, a candidate cell of origin for gliomas (18,19). Idh1LSL:R132H/WT NSCs displayed a nearly 4-fold reduction in proliferative capacity compared to wildtype NSC lines or ad-GFP-transduced control samples (Fig. 2a). Wildtype NSCs were transduced with ad-GFP and ad-Cre with no differences in proliferation observed. This suggested that the proliferative defects in the Idh1LSL:R132H/WT ad-Cre infected NSCs were not resultant of the generalized effects of Cre-recombinase or adenoviral transduction, and instead were mediated by expression of mutant Idh1 (Fig. 2a).
Figure 2. Mutant Idh1-induced cellular arrest is rescued in the presence of Tp53 deletion.
(a) NSCs were infected with ad-GFP or ad-Cre in vitro and expanded. Proliferation was assessed via the CyQUANT Cell Proliferation Assay at Day 1, 3, and 5. Each timepoint contains samples run in triplicate and three independent trials were performed. Figure depicts a representative trial. (b) Western blot analysis of NSCs following ad-Cre infection shows an increase in total p53 in mutant Idh1-expressing NSCs. (c) Quantification of blot intensity of total p53 western blot (unpaired Student t-test, p=0.02, error bars indicate SEM). (d) qPCR for p21 expression was performed on three Idh1LSL:R132H/WT NSC lines treated with either ad-GFP or ad-Cre (normalized to GAPDH) (unpaired Student t-test, * indicates p<0.0001, error bars indicate SEM). (e) Data was extracted from the TCGA database for lower-grade gliomas and samples were categorized based on the mutation status of IDH1 and TP53 (IDH1-M/TP53-M, n=242; IDH1-M/TP53-WT, n=174; IDH1-WT/TP53-M, n=18; IDH1-WT/TP53-WT, n=100; M=mutant, WT=wildtype).
Normal primary cells frequently respond to oncogenic insult by undergoing cell-cycle arrest or senescence via p53 activation (20,21). To determine whether the reduced proliferation of mutant Idh1-expressing NSCs was due to p53-mediated growth inhibition, we determined whether the p53 pathway was activated in NSCs following expression of mutant Idh1. Following ad-Cre transduction, total p53 was elevated in Idh1LSL:R132H/WT lines (Fig. 2b and c), which was accompanied by the upregulation of p21 (Fig. 2d). p53 mRNA was also elevated as was shown through q-RT-PCR (normalized to GAPDH) (Supplementary Fig. S1). These results suggest that NSCs respond to mutant Idh1 by activating the p53 pathway. Additionally, microarray analysis was performed on three independent Idh1LSL:R132H/WT cell lines that were transduced with ad-GFP (control) or ad-Cre (Gene Expression Omnibus Accession number: GSE88828). Hierarchical clustering of the top 1% variant genes (n=414) shows a clustering of those lines treated with ad-GFP versus those treated with ad-Cre, suggesting specific profiling following induction of mutant IDH1 expression (Supplementary Fig. S2). Pathway analysis and gene ontology was performed on those genes displaying greater than a two-fold change (Supplementary Table S2 and S3). Cell cycle and p53 signaling were among the top affected pathways involved when comparing the ad-GFP and ad-Cre cell lines. This data further corroborates our initial data as well as implicates the importance of the p53 pathway in response to induction of mutant IDH1.
To further determine the role of p53 activation, NSCs were isolated from E14.5 embryos of the following two genotypes: Idh1LSL:R132H/WT; Tp53fl/fl and Tp53fl/fl, which have Tp53 conditionally deleted (22). Mutations were induced in vitro via ad-Cre transduction and this was accompanied by loss of p53 expression and production of D-2HG in mutant IDH1 expressing NSCs (Supplementary Fig. S3a and b). As shown in Fig. 2, following deletion of Tp53, an increase in NSC proliferation was observed (Fig. 2a, bottom left). A similar increase in NSC proliferation was observed when mutant Idh1 was expressed in the context of Tp53 deletion (Fig. 2a, bottom right). However, this was an opposite effect from when mutant Idh1 was expressed in the absence of Tp53 deletion (Fig. 2a, top right). Following transduction with ad-Cre, growth of the Idh1LSL:R132H/WT; Tp53fl/fl line displayed a 2.5-fold growth increase over the Idh1-wildtype NSCs. Collectively, these results suggest that p53 activation mediates mutant Idh1-induced cell growth inhibition, and that while mutant Idh1 alone confers a growth disadvantage, the biological outcome can be different and even opposite in the presence of Tp53 deletion (Fig. 2a). This data also supports the possibility that deletion of TP53 is a cooperating alteration in mutant IDH1-driven tumorigenesis. We analyzed The Cancer Genome Atlas’ (TCGA) study on diffuse lower-grade gliomas and found that while 15% of IDH1 wildtype tumors have mutations in TP53, approximately 62% of IDH1 mutant gliomas harbor a mutation in TP53 (3). This finding suggests that while TP53 mutations promote low-grade gliomas, it is likely that it plays an even more important role in promoting IDH1-mutated low-grade gliomas (Fig. 2e).
Mutant Idh1 perturbs the subventricular zone NSCs and blocks neuronal differentiation
Mutant Idh1 was expressed in the mouse brain under the control of different brain-specific promoters of varying expression to assess its effects on cells of the brain in vivo. First, the human glial fibrillary acidic protein promoter (hGFAP-Cre), which is broadly expressed in the CNS, was utilized. These animals display specific labeling in several neural lineages including the granule cells of the cerebellum, olfactory bulb, dentate gyrus, hippocampal pyramidal cells, the gray matter of the cerebral cortex, and in ependymal cells lining the ventricular system (23). This system was previously used to narrow the identity of the glioma cell of origin and to drive brain tumorigenesis (18,19,24,25). In order to assess the in vivo effects of mutant Idh1 when it is broadly expressed in the CNS, hGFAP-Cre mice were crossed to Idh1LSL:R132H/WT animals.
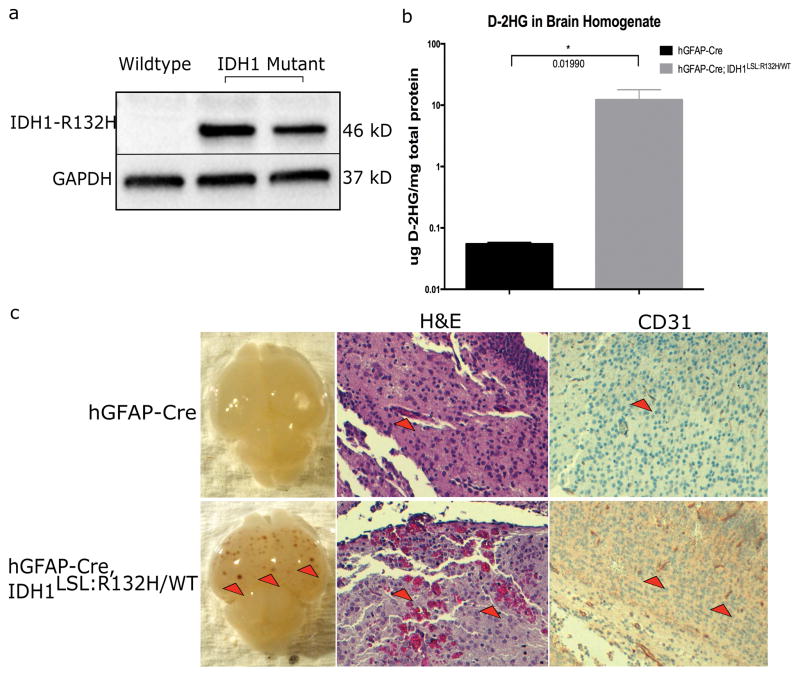
As expected, Idh1-R132H expression was detected in the brains of E15.5 animals bearing the Idh1-R132H mutation (hGFAP-Cre; Idh1LSL:R132H/WT), but not in wildtype controls (hGFAP-Cre; Idh1WT/WT) (Fig. 3a). Additionally, D-2HG was detected in the brains of mutant animals at a level 200-fold higher than in control animals (Fig. 3b). The increase of D-2HG seen in our model is consistent with the magnitude observed in human gliomas bearing the IDH1-R132H mutation, and lends credence that this system is producing biologically relevant levels of D-2HG (26).
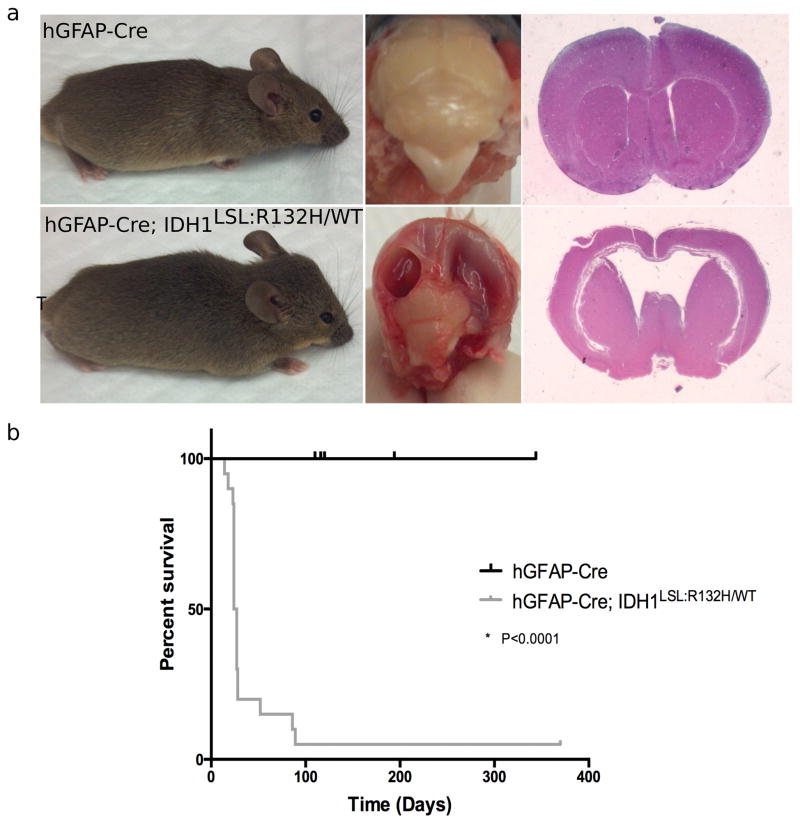
Figure 3. Idh1-R132H is expressed in hGFAP-Cre; Idh1LSL:R132H/WT brains and causes vascular abnormalities.
(a) Western blot analysis using an IDH1-R132H specific antibody on E15.5 brain lysate shows mutant Idh1 is expressed in vivo when under control of the hGFAP-Cre promoter. Blot shows wildtype brain homogenate adjacent to two Idh1-mutant brain homogenates. The production of mutant IDH1 protein was also validated through IDH1-R132H-specific IHC on hGFAP-Cre and hGFAP-Cre; Idh1LSL:R132H/WT brains (data not shown). (b) D-2HG was detected in the brains of hGFAP-Cre; Idh1LSL:R132H/WT animals (n=3 per genotype, unpaired Student t-test, p=0.019, error bars indicate SEM). (c) hGFAP-Cre; Idh1LSL:R132H/WT (bottom row) compared to hGFAP-Cre control animals (top row). Mutant brains (16/17) display areas of hemorrhagic foci (red arrowheads, left panel, bottom row) which were absent from hGFAP-Cre brains (n=20). Red blood cell extravasation was observed (red arrowheads, middle panel, bottom row, 10x) through H&E staining. CD31 staining shows thicker and more robust staining in hGFAP-Cre; Idh1LSL:R132H/WT brains (red arrowheads, right panel, 10x, indicates areas of CD31 positive staining). Phenotype was absent among control animals. Representative images are shown.
To investigate the early effects of mutant Idh1 on CNS cells, brains from postnatal day 3 (P3) hGFAP-Cre; Idh1LSL:R132H/WT and hGFAP-Cre; Idh1WT/WT animals were harvested. hGFAP-Cre; Idh1LSL:R132H/WT brains were visibly different from the control brains with greater than 80% displaying foci of hemorrhagic lesions (16/17) throughout the cortex which were absent from control animals (0/20) (Fig. 3c, left panel). H&E staining showed red blood cell extravasation in the mutant brains; however, in control brains, red blood cells were contained within vessels (Fig. 3c, middle panel). Immunohistochemistry (IHC) for CD31 was performed to visualize endothelial cells and observe any effects to their structural integrity. Interestingly, vessels among mutant brains tended to be thicker and stained more intensely with CD31 (Fig. 3c, right panel).
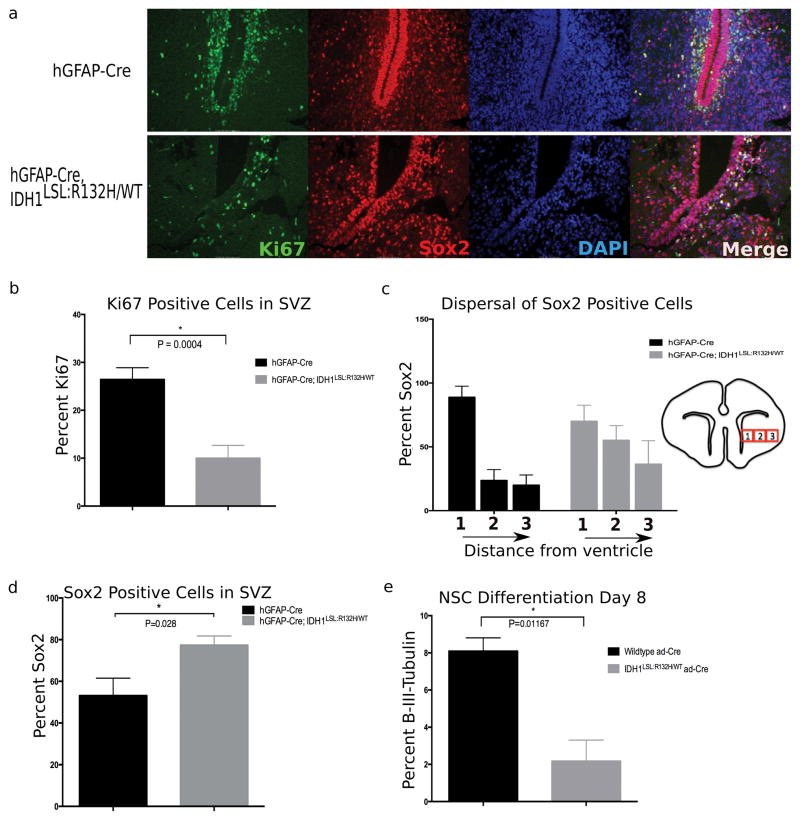
NSCs are located in the SVZ of the lateral ventricles and gives rise to other NSCs as well as differentiated progeny. Since the SVZ is a germinal zone, proliferation was assessed using the proliferation marker Ki67. As expected, robust proliferation in the SVZ of the lateral ventricles in hGFAP-Cre; Idh1WT/WT animals was observed (Fig. 4a). This was in contrast to the SVZ of hGFAP-Cre; Idh1LSL:R132H/WT animals which a 15% reduction in Ki67-positive cells, suggesting an attenuated proliferative capacity of the cells within this germinal zone, a finding consistent with our in vitro data (Fig. 2a, 4a, and 4b).
Figure 4. Broad CNS expression of mutant Idh1 disturbs the NSC microenvironment and reduces proliferation within the SVZ.
(a) Immunofluorescent staining comparing the SVZ of hGFAP-Cre (top row) and hGFAP-Cre; Idh1LSL:R132H/WT (bottom row) animals. Staining with the proliferative marker Ki67 (green) shows a decrease in the proliferative cells of this germinal zone. Sox2 (red) marks NSCs and shows a perturbed NSC niche and disorganization to the normal architecture of the region. (DAPI, nuclear stain-blue). Representative images are shown (20x). (b) Quantification of proliferative cells surrounding apex of SVZ indicates decreased proliferation within the subventricular germinal zone of mutant animals. Phenotype observed in 80% of hGFAP-Cre; Idh1LSL:R132H/WT animals (5/6 animals). Quantification derived from 2 animals per genotype, unpaired Student t-test, p=0.0004, error bars indicate SEM. (c) NSCs were quantified by taking three adjacent regions of identical size surrounding the SVZ (as indicated through boxes 1, 2, and 3-not drawn to scale). While the dispersal trend of the NSC population outwards from the SVZ is observed among mutant brains (5/6 animals), the interaction of distance from the ventricle and different genotypes is not statistically different (p=0.148, repeated measures ANOVA). Quantification derived from 2 animals per genotype, 3 regions per slide, error bars indicate SEM. (d) Sox2 positive cells were normalized to overall cell number. An accumulation of NSCs relative to total number of cells is observed in 5/6 mutant animals. Quantification derived from 2 animals per genotype, 2 fields per animal, unpaired Student t-test, p=0.02, error bars indicate SEM. (e) NSCs cultured in vitro were induced to differentiate through the neural lineage. At Day 8 of differentiation, mutant NSCs show a reduction in β-(III)-tubulin staining, a marker of mature neurons (n=3 independent NSC lines per genotype, 3 randomized fields per sample were counted, unpaired Student t-test, p=0.01, error bars indicate SEM).
To assess mutant Idh1’s effects on the NSC population, the NSC marker Sox2 was used. hGFAP-Cre; Idh1WT/WT animals displayed a tightly organized SVZ with a structured layering rich with Sox2-positive cells; in contrast, the hGFAP-Cre; Idh1LSL:R132H/WT animals displayed a perturbed SVZ and a dispersal of NSCs emanating outward from the SVZ (Fig. 4a and c). While the disorganization of this region was consistent among those animals broadly expressing mutant Idh1 in their brain (5/6), the interaction of distance from the ventricle and different genotypes was not statistically different (p=0.148, repeated measures ANOVA). However, when the NSCs of the SVZ were quantified and normalized to the overall number of cells in this region, there was a statistically significant increase in the number of ectopic Sox2 positively stained cells in the mutant brains (Fig. 4d).
The effect of mutant Idh1 on the NSCs within the SVZ lead to the question of whether mutant Idh1 also affects the NSCs in a cell-autonomous manner. To this end, Idh1LSL:R132H/WT NSCs were isolated from E14.5 embryos and expression of mutant Idh1 was induced in vitro via ad-Cre transduction. Interestingly, when cultured under conditions that promote neural differentiation, the mutant Idh1-expressing NSCs were unable to differentiate into the neuronal lineage as indicated through a reduced expression of the mature neuronal marker, β-III-tubulin (Fig. 4e). Collectively, these experiments suggest that mutant Idh1 likely affects NSCs both directly, as observed through defective differentiation, and indirectly, as observed through a disorganization of the microenvironment, two components with implications in tumor initiation and development (27). This data also corroborates a previous study investigating the role of mutant IDH1 on embryoid body induced human neural stem cells which showed that IDH1-R132H impaired the ability of these cells to differentiate, specifically into the astrocytic and neuronal lineage (28).
To determine the effects of prolonged expression of mutant Idh1 on the developing brain and assess the potential for mutant IDH1-mediated gliomagenesis, hGFAP-Cre; Idh1WT/WT and hGFAP-Cre; Idh1LSL:R132H/WT animals were aged. Physical and behavioral differences became apparent in 80% of hGFAP-Cre; Idh1LSL:R132H/WT animals shortly after weaning age. By 89 days, all hGFAP-Cre; Idh1LSL:R132H/WT animals displayed the phenotypes of head doming, squinted eyes, hunched posture, and gradual immobilization, in addition to episodes of seizure and failure to thrive (Fig. 5a). The median survival for these animals was approximately 25 days with 3 animals surviving asymptomatically for as long as 89 days before necessitating euthanasia (Fig. 5b). Dissection of the brains from mutant animals displayed large fluid-filled cavities and severely reduced cortical thickness. Histological analysis indicated that the cavities represented a severe enlargement of the lateral ventricles (Fig. 5a). These features are characteristic of severe hydrocephalus, a condition that affects 3 in 1000 births and leads to excessive accumulation of cerebrospinal fluid (CSF) in the brain, widening the ventricular spaces, and creating a build-up of pressure that could promote brain injury or even death if left untreated (29,30). An analysis of serial sections of hydrocephalic hGFAP-Cre; Idh1LSL:R132H/WT brains revealed several instances of aqueductal stenosis and an overall loss of cellularity in mutant brains.
Figure 5. Broad CNS expression of mutant Idh1 induces hydrocephalus.
(a) Comparison of hGFAP-Cre (wildtype (n=9); top row) and hGFAP-Cre; Idh1LSL:R132H/WT (mutant (n=20); bottom row) animals. Gross observation shows head doming and squinted eyes of symptomatic animal (bottom, left). Mutant brains show large, fluid-filled cavities (bottom, middle). H&E staining indicates enlargement of the lateral ventricles and hydrocephalus in mutant brains (bottom, right). (b) Kaplan-Meier survival curve comparing wildtype (hGFAP-Cre) animals (black curve, n=9) and mutant (hGFAP-Cre; Idh1LSL:R132H/WT) animals (grey curve, n=20).
The phenotypic differences observed in this hGFAP-Cre; Idh1LSL:R132H/WT model with those in the literature suggest that the timing of mutant Idh1 induction is imperative. Whereas a previously described Nestin-Cre system induced mutant Idh1 at E10.5, our hGFAP-Cre system induced it at E13.5 (31). While there was severe hemorrhage present in the former model, there were small hemorrhagic foci present in the latter model, suggesting that the severity of the mutant Idh1 phenotypes are dependent on the extent of induction. It follows that restriction of mutant Idh1 expression to a smaller population of cells may circumvent these severe phenotypes, allowing for prolonged expression and investigation of the effects of mutant Idh1.
Mutant Idh1 alters tumor biology and the course of tumor progression
The inducibly activated Nestin promoter (Nestin-CreERT2) displays restricted expression in NSCs of embryonic and adult animals and has been used previously to both model gliomas and to define the NSC as the candidate cell of origin for glioma (19,32). Nestin-CreERT2 was utilized to achieve spatio-temporal control over mutant Idh1 expression in the NSC population following administration of tamoxifen. This system was validated by crossing it to a conditional tdTomato reporter mouse (33). Following tamoxifen treatment of Nestin-CreERT2; tdTomatofl/fl animals at E18.5, robust recombination and subsequent expression of tdTomato was observed in the SVZ of P3 animals (Supplementary Fig. S4). This system permits the controlled and restricted expression of mutant Idh1 in the candidate cell of origin for glioma and allows for the prolonged and specific expression of mutant Idh1 in the NSCs specifically, allowing us to ask whether mutant Idh1 is capable of promoting tumorigenesis. Both Nestin-CreERT2; Idh1WT/WT and Nestin-CreERT2, Idh1LSL:R132H/WT animals were generated and tamoxifen was administered at E18.5. After one year, all animals were asymptomatic, with survival being indistinguishable between genotypes (Fig. 6a).
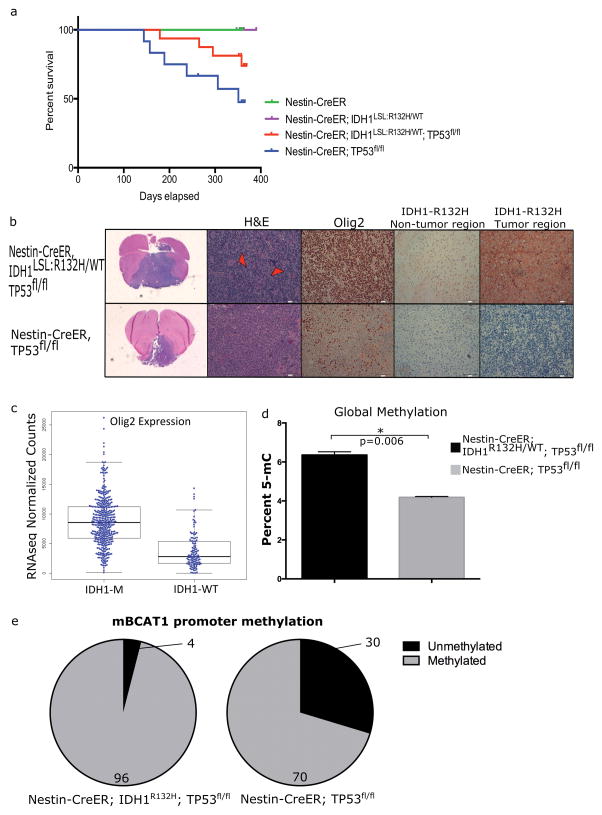
Figure 6. Mutant Idh1 alters progression of Tp53-deleted tumors and changes tumor biology.
(a) Kaplan-Meier survival of E18.5-treated animals. Tp53-deleted animals develop brain tumors and the presence of mutant Idh1 alters progression of tumor development (Nestin-CreERT2, n=6; Nestin-CreERT2; Idh1LSL:R132H/WT, n=7; Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl, n=16; Nestin-CreERT2; Tp53fl/fl, n=12) (b) Histological comparisons between tumors that developed in Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl (n=2) (top row) and Nestin-CreERT2; Tp53fl/fl (n=2) (bottom row). H&E staining shows the presence of giant cells (red arrowheads) in the mutant Idh1 cohort. Nearly 100% of tumor cells stained positive for Olig2 in Idh1 mutant tumors compared to 20%–60% in Tp53-deleted tumors. An IDH1-R132H-specific antibody shows the presence of Idh1 mutant cells in both non-tumor brain tissue as well as tumor tissue from tumor-bearing Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl animals. This is in contrast to tumors from Nestin-CreERT2; Tp53fl/fl animals which show no IDH1-R132H-positive staining. (c) Data was extracted from the TCGA dataset for lower-grade gliomas. Olig2 expression was assessed in IDH1 mutant (n=416) and IDH1 wildtype (n=118) human glioma samples. Data shows a significant increase in Olig2 expression among the IDH1 mutant gliomas (p<0.05). (d) Global methylation was assessed in tumor-derived cell lines that developed in Nestin-CreERT2; Tp53fl/fl and Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl animals. (e) The methylation status of the second promoter of mouse Bcat1 was assessed in aforementioned mouse brain tumor-derived cell lines. Greater promoter methylation is observed in the Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl compared to the Nestin-CreERT2; Tp53fl/fl lines. Percent of the loci that is methylated and unmethylated is shown.
We next considered the effects of mutant Idh1 in the context of tumor biology and tumor progression. Considering the prevalence of co-occurring mutations in IDH1 and TP53 (Fig. 2e), coupled with the in vitro and in vivo effects of mutant Idh1 on NSCs (Fig. 2a, 4b and 4d), conditionally deleted Tp53 animals were crossed into the mutant Idh1 conditional knock-in animals, leading to a compound genetic mouse model harboring the most common mutations found among the low-grade gliomas (22). Two genotypes, Nestin-CreERT2; Tp53fl/fl and Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl were administered tamoxifen at E18.5 to assess the impact of mutant Idh1 on brain tumorigenesis. Consistent with previous neural-specific, Tp53-deleted tumor models, tumors developed in Tp53-deleted animals (Nestin-CreERT2; Tp53fl/fl) at a penetrance of 50% (6/12) (34). Among animals that harbored both the Idh1-R132H mutation and Tp53 deletion (Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl), the penetrance was reduced to 15% (4/26).
An IDH1-R132H-specific antibody was tested on tumor-bearing brains from Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl and Nestin-CreERT2; Tp53fl/fl animals (Fig. 6b). Tumors that developed in Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl animals stained positively with the IDH1-R132H antibody. This was in contrast to those tumors that developed in animals of the genotype Nestin-CreERT2; Tp53fl/fl, which showed a lack of IDH1-R132H staining, suggesting that tumors were initiated from cells with mutations in both genes (Fig. 6b).
While the overall tumor penetrance was low, several histological observations include that Idh1 mutant tumors tended to be sarcomatoid in nature, with more perivascular infiltration into surrounding brain parenchyma rather than single-cell infiltration. This is in contrast to the Idh1 wildtype tumors, which were more epithelioid and displayed both single-cell and perivascular infiltration. Pseudopalisading necrosis was present in several of the Idh1 mutant tumors. Additionally, there was a prevalence of giant cells among the Idh1 mutant tumors (Fig. 6b, arrowhead). The presence of giant cells among glioma patients is associated with a prolonged survival, a finding consistent among our mutant Idh1 tumor-bearing animals (35).
IHC characterization of tumors was performed using common markers for clinical diagnosis, including markers for astrocytes (GFAP), oligodendrocytes (Olig2), and proliferative cells (Ki67). The majority of tumors stained positively for GFAP; however, this was determined to be reactive astrocytosis and spanned genotypes. Additionally, 100% of the tumor cells in the Idh1 mutant cases stained positively for Olig2, in contrast to 20% to 60% of the tumor cells in the Idh1 wildtype tumors (Fig. 6b). Among human diffuse glioma samples, those harboring IDH1 mutations display an elevated expression of Olig2 compared to wildtype samples, suggesting a cooperation between mutant IDH1 and Olig2 expression in promoting tumorigenesis (Fig. 6c) (36,37). This is a feature specific for the proneural group of gliomas, as characterized by TCGA (36,37).
When comparing overall survival of experimental animals, mutant Idh1 is shown to confer a prolonged survival among Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl tumor-bearing animals compared to Nestin-CreERT2; Tp53fl/fl tumor-bearing animals (Fig. 6a). Though the median survival was unable to be determined due to a low number of affected animals in the Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl cohort, the average time for onset of symptoms was 233 days compared to 274 days for the Nestin-CreERT2; Tp53fl/fl versus Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl animals, respectively. The survival trend corroborates the clinical data, which shows a prolonged survival among brain tumor patients with mutations in IDH1 compared to patients with wildtype IDH1 tumors. For instance, among patients with Grade III anaplastic astrocytoma, survival was 65 months for those with IDH1 mutations compared to 20 months for those with wildtype IDH1 tumors. Similarly, among patients with Grade IV GBM, those with IDH1-mutant tumors displayed a 31-month survival compared to 15 months for those with IDH1-wildtype tumors (4,38).
To further test the faithfulness of this brain tumor model, brain tumors were harvested from symptomatic animals and cultured in vitro. Tumor cultures were derived from animals of the genotype Nestin-CreERT2; Tp53fl/fl and Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl. These lines did not express p53 (Supplementary Fig. S3b) and D-2HG was elevated in the mutant IDH1-expressing brain tumor line (Supplementary Fig. S5). This observation coupled with the observation that non-tamoxifen treated animals failed to develop tumors (Figure 6a), suggests that these tumors were derived from those cells that recombined following tamoxifen administration and further implicates the NSC as the cell of origin for these tumors.
Mutant IDH1 has been shown to promote G-CIMP (7,39). To determine whether mutant IDH1 confers the expected hypermethylation phenotype, global methylation was assessed in cell lines derived from tumor-bearing animals. An increase in global methylation in the mutant IDH1-expressing brain tumor cell line was observed (Figure 6d). Additionally, the second promoter of BCAT1, a loci known to be differentially methylated in response to mutant IDH1 was investigated (40). The homologous CpG island in mouse was identified and the methylation status was differentially methylated as it is in human IDH1-mutated glioma, with the Nestin-CreERT2; Idh1LSL:R132H/WT; Tp53fl/fl line showing greater methylation than the Nestin-CreERT2; Tp53fl/fl line at the second promoter of Bcat1 (Figure 6e). Combined, these data suggest that the tumors derived from our genetic model recapitulate many of the features of mutant IDH1 gliomas, including expression of Olig2, a prolonged survival among mutant IDH1 patients, elevated global DNA methylation, and hypermethylation of the Bcat1 gene.
Discussion
Despite the prevalence of the IDH1 mutation among gliomas, its effects on the NSC population (which are a candidate cell of origin for glioma), the microenvironment from which gliomas arise, or how this mutation affects the course of tumor development are incompletely understood. We address these three aspects using conditional mutant Idh1 in vitro and in vivo mouse models.
Considering the large body of evidence implicating the NSC as the cell of origin for glioma, our studies have focused on the effects of mutant IDH1 on the NSC in vitro and in vivo (18,19,24,27). However, there are other candidate cells of origin, including the oligodendroglial progenitor cell (OPC) for which there is impressive data suggesting it as the cell of origin (41). Further investigations are required in order to assess the differential effects of mutant IDH1 on different candidate cells of origin.
We show that mutant Idh1 attenuates proliferation of NSCs, suggesting that mutant Idh1 acts as an oncogenic insult to which cells respond by undergoing cell-cycle arrest. This in vitro growth reduction as a result of mutant Idh1 corroborates both of our in vivo models: the first which shows that the Idh1 mutation reduces proliferation in the SVZ of the lateral ventricle among hGFAP-Cre; Idh1LSL:R132H/WT animals, and the second model which shows a failure of mutant Idh1 to impart a tumorigenic phenotype among Nestin-CreERT2; Idh1LSL:R132H/WT animals. We show that this cellular response is mediated by p53 activation, as evidenced by increased p53 mRNA and protein and the upregulation of p21. Most importantly, deletion of Tp53 rescues the proliferative defect attributed to mutant Idh1, suggesting the potential cooperative effects between Idh1 and Tp53 mutations, and explaining the co-occurring nature of these mutations among glioma patients (3,5,37).
Mutant IDH1’s effects on cellular behavior are cell context-dependent, reducing proliferation in some cases, and increasing proliferation and promoting tumorigenesis in other cases (42,43). The finding linking IDH1 and TP53 mutations presents clear genetic evidence that in addition to cellular context, additional genetic components play integral roles in dictating the distinct effects of mutant IDH1, a notion that is supported by previous findings. For example, Koivunen et al. discovered that immortalized human astrocytes infected with mutant IDH1 display a proliferative advantage and an enhanced ability for colony formation (42). These lines were immortalized and transformed through the expression of the human papillomavirus 16 E6, which inactivates p53 (44). Based on the status of TP53, a proliferative advantage would be expected in these cells, as was the case. Another study by Bralten et al. shows that mutant IDH1 decreases the proliferation of the established glioma cell line U87 and a human embryonic kidney (HEK) cell line, both of which have TP53 intact (43). While these findings do not provide direct evidence that TP53 is the critical factor in deciding the cellular effects of mutant IDH1, they do support the notion that additional genetic aberrations alter the effects of mutant IDH1.
It is well accepted that the microenvironment plays critical roles in driving tumorigenesis and dictating tumor progression (45,46). This is especially true for the IDH1 mutation, as one main feature of mutant IDH1 is the generation of the neometabolite D-2HG, which can be secreted into the extracellular compartment (6). The broadly expressed hGFAP-Cre promoter, which has been used previously to successfully generate glioma models (19,25), was used to drive mutant Idh1 expression in the brain. This model revealed the striking effects that mutant Idh1 has on the CNS generally, and on the SVZ specifically. Perinatal hGFAP-Cre; Idh1LSL:R132H/WT animals presented with areas of hemorrhagic foci throughout the cortex, a phenotype reminiscent of a model utilizing a Nestin-Cre driver that showed severe hemorrhage and perinatal lethality (31). This phenotypic variation can likely be explained by the timing and extent of mutant Idh1 induction; while Cre-recombinase begins to be expressed at E10.5 in the Nestin-Cre system, it begins at E13.5 in the hGFAP-Cre system (23,32). At P3, hGFAP-Cre; Idh1LSL:R132H/WT animals displayed reduced cortical thickness, loss of cellularity, and a disrupted SVZ, as indicated through a dispersal of Sox2-positive NSCs emanating outwards from the SVZ of the lateral ventricles. The SVZ and the NSC microenvironment are critical for the regulation of proliferation and progenitor cell differentiation; therefore, this disorganization likely perturbs the normal differentiation patterns of the resident NSCs.
While further studies are needed to reveal the mechanism behind mutant Idh1’s effects on the microenvironment, we speculate that the effects are at least partially mediated by D-2HG. There is an interesting pathological similarity between hGFAP-Cre; Idh1LSL:R132H/WT animals and patients with D-2-hydroxyglutaric aciduria, a neurometabolic disorder characterized by systemic elevation of D-2HG (47). Though a rare disorder, an international survey of patients with D-2-hydroxyglutaric aciduria described that all patients (17/17) exhibited ventriculomegaly, an enlargement of the lateral ventricles, as well as delayed cerebral maturation (48). These features suggest that D-2HG has a significant effect on the candidate cell of origin for gliomas by altering the signaling pathways within the niche and disrupting the microenvironment of the resident NSCs; these occurrences are likely imperative for tumor initiation.
The genetic data confers an unambiguous causal role of mutant IDH1 in gliomagenesis. Paradoxically, glioma patients with mutations in IDH1 tend to have a better survival clinically (3–5,37). Together, this suggests that the IDH1 mutation is an effective promoter for clinically less aggressive gliomas. There are two likely underlying mechanisms for this phenomenon: (i) It is possible that the IDH1 mutation can only target and transform a unique cellular population; a population distinct from those cells that give rise to non-IDH1-mutant gliomas, and a population that intrinsically gives rise to less aggressive tumors; or (ii) While the IDH1 mutation promotes cellular transformation initially, its continuous presence in tumor cells may attenuate tumor progression and aggressiveness. Our experiments using the inducible Nestin-CreERT2 model indicate that in tumors driven by Tp53 deletion, mutant Idh1 is expressed in all tumor cells, supporting the notion that the two mutations target the same population of cells that give rise to tumors. The prolonged survival among tumor-bearing animals with both mutant Idh1 and Tp53 mutations suggests that mutant Idh1 alters tumor biology as well as the course of tumor progression. Our in vivo findings, coupled with previous in vitro models (43,49), support the latter mechanism and stress an important point for consideration when devising glioma treatments on the basis of mutant IDH1 inhibition. Notably, a recent study has shown that the response of cells to mutant IDH1 inhibition is dependent on whether treatment is administered before, at the same time, or after mutant IDH1 is expressed(50). This study raises an important question regarding the timing of therapeutic intervention and the potential reversibility of mutant IDH1-mediated epigenetic effects. Our models provide a valuable tool for addressing this question in a relevant pathological setting.
Collectively, our findings provide new insight into the effects of mutant Idh1 on a candidate cell of origin for glioma, mutant Idh1’s role in disrupting the microenvironment from which gliomas arise, and mutant Idh1’s effect on the course of glioma progression. Additionally, this study highlights the challenges involved in modeling mutant IDH1, including (i) the impact of broad expression of mutant IDH1 on the CNS, which hinders long-term investigations of mutant IDH1 in contributing to brain tumorigenesis; (ii) the necessity to test other cooperating genetic alterations; (iii) the unclear identity of the cells of origin that are most susceptible to mutant IDH1-mediated transformation, and (iv) the unknown contribution of cell-autonomous and non-cell-autonomous effects on gliomagenesis. Further studies involving alternative approaches, such as orthotopic transplantation of other candidate glioma cells of origin, will help overcome these challenges and will provide a faithful foundation for understanding mutant Idh1’s role in tumorigenesis and for investigating preclinical targeted therapies.
Supplementary Material
Implications.
Through the use of a conditional mutant mouse model that confers a less aggressive tumor phenotype, this study reveals that mutant Idh1 impacts the candidate cell of origin for gliomas.
Acknowledgments
Financial support:
This work was supported by the James S. McDonnell Foundation Research Award and R01-CA140316 (HY).
We thank Dr. Rob Wechsler-Reya at the Sandford-Burnham Medical Research Institute for sharing reagents and Dr. Darell Bigner at Duke’s Preston Robert Tisch Brain Tumor Center for providing access to resources. We thank Ms. Jenna Lewis, Mr. Bill Diplas, Dr. Xuhui Bao, and Dr. Vidya Chandramohan for editorial revisions to the manuscript. We thank the UNC Animal Model Core for the initial animal generation. We thank the Pathology Research Histology and Immunohistochemistry Laboratory of Duke University for IHC staining. This work was supported by the James S. McDonnell Foundation Research Award and R01-CA140316. YH was supported by an AACR-Aflac Career Development Award, the National Comprehensive Cancer Network Young Investigator Award and the Circle of Service Foundation.
Footnotes
Conflicts of interest:
H.Y. receives royalties from Agios Pharmaceuticals and Personal Genome Diagnostics. H.Y. is a co-founder of Genetron Health (Beijing) Co. Ltd.
References
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al. SEER Cancer Statistics Review, 1975–2011. National Cancer Institute; Bethesda, MD: Apr, 2014. http://seer.cancer.gov/csr/1975_2011/, based on November 2013 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Ostrom QT, Gittleman H, de Blank PM, Finlay JL, Gurney JG, McKean-Cowdin R, et al. American Brain Tumor Association Adolescent and Young Adult Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol. 2016;18(Suppl 1):i1–i50. doi: 10.1093/neuonc/nov297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. New Engl J Med. 2015;372(26):2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. New Engl J Med. 2009;360(8):765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Killela PJ, Pirozzi CJ, Reitman ZJ, Jones S, Rasheed BA, Lipp E, et al. The genetic landscape of anaplastic astrocytoma. Oncotarget. 2014;5(6):1452–7. doi: 10.18632/oncotarget.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–44. doi: 10.1038/nature08617. nature08617 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–9. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–7. doi: 10.1158/0008-5472.can-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108(8):3270–5. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izquierdo-Garcia JL, Viswanath P, Eriksson P, Chaumeil MM, Pieper RO, Phillips JJ, et al. Metabolic reprogramming in mutant IDH1 glioma cells. PloS One. 2015;10(2):e0118781. doi: 10.1371/journal.pone.0118781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, Nishimura MC, Kharbanda S, Peale F, Deng Y, Daemen A, et al. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci USA. 2014;111(39):14217–22. doi: 10.1073/pnas.1409653111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reitman ZJ, Duncan CG, Poteet E, Winters A, Yan LJ, Gooden DM, et al. Cancer-associated isocitrate dehydrogenase 1 (IDH1) R132H mutation and d-2-hydroxyglutarate stimulate glutamine metabolism under hypoxia. J Biol Chem. 2014;289(34):23318–28. doi: 10.1074/jbc.M114.575183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Struys EA, Jansen EE, Verhoeven NM, Jakobs C. Measurement of urinary D- and L-2-hydroxyglutarate enantiomers by stable-isotope-dilution liquid chromatography-tandem mass spectrometry after derivatization with diacetyl-L-tartaric anhydride. Clin Chem. 2004;50(8):1391–5. doi: 10.1373/clinchem.2004.033399. [DOI] [PubMed] [Google Scholar]
- 18.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25(1):55–7. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 19.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 21.Elbendary AA, Cirisano FD, Evans AC, Jr, Davis PL, Iglehart JD, Marks JR, et al. Relationship between p21 expression and mutation of the p53 tumor suppressor gene in normal and malignant ovarian epithelial cells. Clin Cancer Res. 1996;2(9):1571–5. [PubMed] [Google Scholar]
- 22.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 23.Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31(2):85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 24.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. New Engl J Med. 2005;353(8):811–22. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 25.Abel TW, Clark C, Bierie B, Chytil A, Aakre M, Gorska A, et al. GFAP-Cre-mediated activation of oncogenic K-ras results in expansion of the subventricular zone and infiltrating glioma. Mol Cancer Res. 2009;7(5):645–53. doi: 10.1158/1541-7786.mcr-08-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin G, Reitman ZJ, Spasojevic I, Batinic-Haberle I, Yang J, Schmidt-Kittler O, et al. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PloS One. 2011;6(2):e16812. doi: 10.1371/journal.pone.0016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim DA, Alvarez-Buylla A. Adult neural stem cells stake their ground. Trends Neurosci. 2014;37(10):563–71. doi: 10.1016/j.tins.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosiak K, Smolarz M, Stec WJ, Peciak J, Grzela D, Winiecka-Klimek M, et al. IDH1R132H in Neural Stem Cells: Differentiation Impaired by Increased Apoptosis. PloS One. 2016;11(5):e0154726. doi: 10.1371/journal.pone.0154726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter CS, Vogel TW, Zhang Q, Seo S, Swiderski RE, Moninger TO, et al. Abnormal development of NG2+PDGFR-alpha+ neural progenitor cells leads to neonatal hydrocephalus in a ciliopathy mouse model. Nat Med. 2012;18(12):1797–804. doi: 10.1038/nm.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kousi M, Katsanis N. The Genetic Basis of Hydrocephalus. Annual Review of Neuroscience. 2016 doi: 10.1146/annurev-neuro-070815-014023. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26(18):2038–49. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmerman L, Parr B, Lendahl U, Cunningham M, McKay R, Gavin B, et al. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron. 1994;12(1):11–24. doi: 10.1016/0896-6273(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 33.Arenkiel BR, Hasegawa H, Yi JJ, Larsen RS, Wallace ML, Philpot BD, et al. Activity-induced remodeling of olfactory bulb microcircuits revealed by monosynaptic tracing. PloS One. 2011;6(12):e29423. doi: 10.1371/journal.pone.0029423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15(6):514–26. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deb P, Sharma MC, Mahapatra AK, Agarwal D, Sarkar C. Glioblastoma multiforme with long term survival. Neurol India. 2005;53(3):329–32. [PubMed] [Google Scholar]
- 36.Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63(5):499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 37.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Killela PJ, Pirozzi CJ, Healy P, Reitman ZJ, Lipp E, Rasheed AB, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–25. doi: 10.18632/oncotarget.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19(7):901–8. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zong H, Parada LF, Baker SJ. Cell of origin for malignant gliomas and its implication in therapeutic development. Cold Spring Harbor Persp Biol. 2015;7(5) doi: 10.1101/cshperspect.a020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–8. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bralten LB, Kloosterhof NK, Balvers R, Sacchetti A, Lapre L, Lamfers M, et al. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol. 2011;69(3):455–63. doi: 10.1002/ana.22390. [DOI] [PubMed] [Google Scholar]
- 44.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61(13):4956–60. [PubMed] [Google Scholar]
- 45.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes & development. 2015;29(12):1203–17. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaacoub K, Pedeux R, Tarte K, Guillaudeux T. Role of the tumor microenvironment in regulating apoptosis and cancer progression. Cancer Lett. 2016;378(2):150–9. doi: 10.1016/j.canlet.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C. Progress in understanding 2-hydroxyglutaric acidurias. J Inherit Metab Dis. 2012;35(4):571–87. doi: 10.1007/s10545-012-9462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Knaap MS, Jakobs C, Hoffmann GF, Nyhan WL, Renier WO, Smeitink JA, et al. D-2-Hydroxyglutaric aciduria: biochemical marker or clinical disease entity? Ann. Neurol. 1999;45(1):111–9. doi: 10.1002/1531-8249(199901)45:1<111::aid-art17>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 49.Piaskowski S, Bienkowski M, Stoczynska-Fidelus E, Stawski R, Sieruta M, Szybka M, et al. Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer. 2011;104(6):968–70. doi: 10.1038/bjc.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johannessen TA, Mukherjee J, Viswanath P, Ohba S, Ronen SM, Bjerkvig R, et al. Rapid Conversion of Mutant IDH1 from Driver to Passenger in a Model of Human Gliomagenesis. Mol Cancer Res. 2016;14(10):976–83. doi: 10.1158/1541-7786.mcr-16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data and reagents are available from the authors upon request and fulfillment of proper university required documentation.