Abstract
Background and Aims: Hepatitis C virus (HCV) hepatotropism is associated with intra-peripheral blood mononuclear cell (PBMC) infection that causes post-treatment relapse in RNA seronegative patients. Our understanding of the association of non-viremic hepatic fibrosis with positive anti-HCV IgG antibodies and active hepatocellular damage might be increased by PBMCs screening for intracellular infection. Thus, the goals of this study included evaluation of PBMCs PCR for diagnosing HCV infection, addressing PBMCs plus serum real-time (SRT) PCR benefits over SRT-PCR alone, studying intra-PBMCs distribution of RNA sense and antisense strands, and identifying treatment feasibility in solitary intracellular infection.
Methods: Enzyme-linked immunosorbent assay, SRT-PCR and PBMCs PCR were used to evaluate HCV infection in 401 subjects. The patients were classified into groups of negative controls (n = 30), positive controls (n = 63), non-viremia post-treatment (experienced; n = 166) and naïve (n = 49) cases, and non-viremia positive PBMCs PCR naïve (n = 35) and experienced (n = 58) patients.
Results: The diagnosis of true positive and negative by PBMCs PCR and SRT-PCR had 100% and 96.7% compatibility respectively. PBMCs PCR detected intracellular HCV infection in 49 out of 215 non-viremia patients; among them, naïve cirrhotics had significantly higher number of intracellular infection than the naïve non-cirrhotic (p < 0.001) and experienced patients (p < 0.0001). Antisense and sense strands were respectively recognized in naïve and experienced cases (p = 0.01218). Intracellular HCV strands were detected in 18.02% of experienced patients. Recognition of intracellular RNA strands showed significant decline in experienced compared to naïve patients (p < 0.05).
Conclusion: PBMCs PCR is a valid diagnostic test that can diagnose intracellular HCV when SRT-PCR is negative. Antisense and sense strands are respectively recognized more often in naïve and experienced patients. The expected overall relapsing rate in our cohort was 18.02%. Intra-PBMC infections are associated with liver cirrhosis in naïve non-viremia patients. Eradication of intracellular strands is recommended to avoid RNA seroconversion.
Ethical approval certificate: Registration number 10231.
Keywords: HCV, PBMCs, RNA, Strands
Introduction
Eradication of hepatitis C virus (HCV) infection is the goal of the direct-acting antivirals (DAAs), which otherwise would not be achieved by viremic clearance at the end of treatment. Serum real-time (SRT)-PCR has been used to monitor viremia and to evaluate HCV treatment efficacy over time, but information about status of intracellular HCV replication cannot be provided by SRT-PCR. Replication of genomic HCV-RNA inside peripheral blood mononuclear cells (PBMCs) has been confirmed1 and, in some cases, may be followed by viremia after serologic clearance.2 Therefore, dual testing for HCV by SRT-PCR and PBMCs PCR is expected to be more informative regarding concordant clearance of viral RNA from both serum and cells.
HCV is not a strictly a hepatotropic viral pathogen.3–6 The evidence of extrahepatic HCV replication by detection of RNA strands and viral proteins were shown in PBMCs7–11 and bone marrow,12 in addition to the central nervous system.13,14 All patients with positive HCV SRT-PCR are also positive on PBMCs PCR. However, some patients with positive PBMCs PCR may have negative SRT-PCR.15 Follow-up of treatment-experienced HCV patients who presented with negative SRT-PCR and positive intra-PBMCs strand infection revealed HCV RNA seroconversion within 1 year.2 It was also reported that post-treatment persistence of intracellular HCV RNA antisense strand is a predictor of relapse.16
The outcome of intracellular HCV RNA strands in treatment-naïve patients who presented with negative SRT-PCR has not been fully elucidated. However, post-treatment relapse of HCV infection may become eminent when recolonization of PBMCs, with either sense or antisense RNA genomic strands, occurs. Cryptogenic occult HCV infection (OCI) is defined by the absence of serum RNA and IgG antibodies, and the presence of genomic RNA strand(s) in PBMCs. Prevalence of OCI is around 3.3% in the healthy population.17–19 The increasing clinical demands for evaluating spontaneous and post-treatment HCV cure rates strongly argue for the need to assess cellular HCV and to determine its clinical significance. Hence, this study was designed to test for HCV by PBMCs PCR and to determine its clinical significance.
Aims of the current study
a) Evaluation of PBMCs PCR plus SRT-PCR as a diagnostic tool in delineating HCV infection as compared to the gold standard SRT-PCR test alone; b) Addressing the benefits of RNA detection inside PBMCs over SRT-PCR in recognizing HCV infection; c) Studying intra-PBMCs distribution of HCV sense and antisense RNA strands in untreated naive and post treatment-experienced patients; and d) Identifying treatment feasibility for patients who present with solitary intra-PBMCs HCV infection.
Methods
Study subjects
Ethical committee approval (Registration No. 10231) was obtained before the current study was initiated. Inclusion criteria for study participation were: age between 18 and 70 years-old; signing consent at the infectious diseases outpatient clinic (between January 2015 and September 2016); and conformity to the below-mentioned specifications of Egyptians in each of the four groups. Exclusion criteria included hepatocellular carcinoma, Child C status, and non-compliance to the antiviral therapy regimen. Sample size in each group depended upon availability of subjects that fit the inclusion criteria during a certain period of time. The 401 subjects were classified into 4 groups as summarized below in Table 1.
Table 1. Study subjects grouping.
| Study groups | Anti-HCV IgG antibodies | HCV SRT-PCR | HCV PBMCs PCR | Cases (n = 401) | |
| Naïve (n = 177) | Experienced (n = 224) | ||||
| Group I (n = 30) | Negative | Negative | TBD | 30 | 0 |
| Group II (n = 63) | Positive | Positive | TBD | 63 | 0 |
| Group III (n = 93) | Positive | Negative | Positive | 35 | 58 |
| Group IV (n = 215) | Positive | Negative | TBD | 49 | 166 |
Abbreviation: TBD, to be determined.
HCV IgG antibody detection by enzyme-linked immunosorbent assay (ELISA)
The third-generation ELISA assay was used as a preliminary screening test to identify cases for enrollment in the current study. This test contained reconfigured core and NS3 antigens and, in addition, a newly incorporated antigen from the NS5 region. The procedure was performed as described.20
Quantification of HCV RNA using real-time PCR
Specimen collection and transport
Blood samples (5–10 mL) were withdrawn with standard specimen collection artus HCV RG RT-PCR Kit tubes (red cap). Whole blood was separated into plasma and cellular components by centrifugation for 20 minutes at 800–1600 × g within 6 hours of collection. A total of 200 μL of whole blood cells were transferred to a sterile polypropylene tube for PBMC studies, and the plasma was transferred into another sterile polypropylene tube. The blood samples were shipped cooled (2 to 8 °C) and the separated plasma was transported frozen (−15 to –30 °C).
RNA isolation and SRT-PCR
The QIAamp DSP Virus Kit (Cat. No. 60704; Qiagen) is validated for viral RNA purification from human plasma for use with the artus HCV RG RT-PCR Kit. The SRT-PCR was carried out after a reverse transcription step. RNA was extracted from 200 µL of plasma with the QIAamp Viral RNA Kit (Qiagen), eluted in 60 µL and stored at −80 °C. RNA (4 µL) was incubated with 1 pmol of an HCV 5NC primer RC21 for 8 minutes at 70 °C and 5 minutes at 4 °C. RNA template was then reverse transcribed at 60 °C for 1 hour with 7.5 U of Thermoscript™ Reverse Transcriptase, 20 U of RNaseOut, 10 mM DTT, 1 mM deoxyribonucleotide and cDNA buffer (50 mM Tris acetate pH 8.4, 75 mM potassium acetate, 8 mM magnesium acetate) (GibcoBRL Life Technologies) in a final volume of 10 µL. A denaturation step was performed at 95 °C for 5 minutes and was followed by a RNaseH treatment with 1 U of E. coli RNaseH (GibcoBRL Life Technologies) at 37 °C for 20 minutes. cDNA was stored at −20 °C. The primers RC1 (5′-GTCTAGCCATGGCGTTAGTA-3′) and RC21 (5′-CTCCCGGGGCACTCGCAAGC-3′) were designed to amplify a 220 bp fragment within the 5′ non-coding region of the HCV genome.
Internal control
An internal control (Hep.C Virus RG IC) was used both to control the RNA isolation procedure and to check for possible PCR inhibition. For this application, adding the internal control to the isolation at a ratio of 0.1 μL per 1 μL elution volume was performed. For example, using the QIAamp DSP Virus Kit (Qiagen), the RNA was eluted in 60 μL Elution Buffer (AVE). Hence, 6 μL of the internal control should be added initially.
Quantification
The enclosed quantification standard (Hep.C Virus RG QS1–4) was treated as the previously purified samples and the same volume was used (20 μL). To generate a standard curve on Rotor-Gene Q Instruments, all 4 quantitation standards were used. The quantitation standard values are reported as IU/μL. The following equation was applied to convert the values determined using the standard curve into IU/mL of sample material:
The initial sample volume was entered into the equation above, considering changes in sample volume.
HCV PBMCs PCR
Extraction of RNA from PBMCs
Peripheral blood (200 µL) was diluted with 10 mL freshly prepared red blood cell alkaline buffer (38.8 mmol/L NH4Cl, 2.5 mmol/L K2HCO3, 1 mmol/L EDTA pH 8.0). After 10 minutes incubation at room temperature, the nucleated cells were washed with the same buffer and lysed in 500 µL antinuclease solution (4 mol/L guanidinium isothiocyanate containing 25 mmol/L sodium citrate, 0.5 sarcosyl and 0.1 mol/L ß- mercaptoethanol). A single-step method previously described21 and subsequently modified22,23 was followed for the RNA extraction.
Retrotranscription-PCR of genomic and antigenomic strands of HCV RNA
Detection of HCV RNA strands in PBMCs was carried out as described by Lohr et al.24 in 1995. Briefly, the reaction mixture (50 µL) contained 400 ng of cellular RNA, RT-PCR bead (HVD), 50 pmol from each of the primers (1CH, P2 and 2CH) for amplification of the genomic strand, or 50 pmol from P2 and 75 pmol from 2CH for antigenomic strand and 20 U of AMV reverse transcriptase. The mixture was incubated at 42 °C for 1 hour and denatured for 15 minutes at 94 °C. Amplification of cDNA was performed in a thermal cycler for 30 cycles (94 °C × 1 minute, 55 °C × 1 minute, and 72 °C × 1 minute). Nested PCR amplification was similar to the first-round PCR except for using 10 µL from the first PCR and two nested primers (P3 and P4) and 2 U Taq DNA polymerase. To assure specificity of the assay two controls were employed: C1, ddw instead of RNA in the cDNA synthesis reaction to exclude RNA contamination; C2, PCR step with only F1 or R1 to exclude mixed primer contamination. In all conditions, the two controls provided negative amplification; special attention was paid to heat inactivate the reverse transcriptase at 95 °C for 1 hour in order to reduce false detection of the antigenomic strand before adding the forward strand. Primer sequences used included below in Table 2.
Table 2. Primers used for PCR.
| Primer | Sequences |
| 1CH | 5′-ggtgcacggtctacgagacctc-3′ |
| 2CH | 5′-aactcatgtcttcacgcagaa-3′ |
| P2 | 5′-tgctcatggtgcacggtcta-3′ |
| P3 | 5′-ctttcgcgacccaacactac-3′ |
| P4 | 5′-agagccatagtggtctgcgg-3′ |
Medicine administration
The anti-HCV drugs were administered according to guidelines regarding doses, routes of administration, and duration of therapy. The antiviral therapeutic regimens included: 1) sofosbuvir at 400 mg + ladepasvir at 90 mg once daily for 24 weeks; 2) sofosbuvir at 400 mg + weight-based ribavirin daily for 24 weeks; 3) sofosbuvir at 400 mg + daclatasvir at 60 mg + weight-based ribavirin daily for 12 weeks; 4) sofosbuvir at 400 mg + simeprevir at 150 mg daily for 12 weeks; 5) sofosbuvir at 400 mg + ribavirin at 800 mg daily + peg-interferon alfa 2b1 at 5 mg/kg weekly for 12 weeks.
Statistical analysis
A proposed diagnostic procedure (PBMCs PCR) was compared to the gold standard test (SRT-PCR) in this case-control study that contained four groups of selected cases. All available cases, which fulfilled the inclusion criteria, were included during the 21 consequent month period. The statistical parameters of the proposed test were studied. Data analysis was performed using SSPS version 9.0 for Windows. Normality and homoscedasticity of continuous variables were tested by the Kolmogorov-Smirnov and Levene’s tests, respectively. The mean was compared by using the Student’s t test (for variables with normal distribution) or the Mann-Whitney U test (for variables with non-normal distribution). Categorical variables were compared using the chi-squared or Fisher’s exact tests. Pearson’s (for variables with Gaussian distribution) or Spearman’s (for non-normally distributed variables) correlation coefficients were determined to study the existence of correlation between the variables. All tests performed were two-sided and statistical significance was considered at a p value of 0.05.
Results
Matching of PBMCs HCV PCR
As demonstrated in Table 3, all of the 63 (100%) HCV-infected patients who were diagnosed by SRT-PCR had sense and anti-sense strands in their PBMCs. The same Table shows that HCV RNA strands were detectable in 3.3% of subjects who tested negative by SRT-PCR. These findings validated the PBMCs PCR as a diagnostic test in HCV infection because of the 100% compatibility with the gold standard SRT-PCR test and its unique advantage in diagnosing OCI.
Table 3. Concordance between HCV SRT-PCR and PBMCs PCR.
| Serum real-time HCV PCR result | PBMCs HCV PCR | |||
| RNA sense strand, n (%) | RNA antisense strand, n (%) | |||
| Positive | Negative | Positive | Negative | |
| Positive SRT-PCR and IgG-Abs (n = 63) | 63 (100) | 0 (0) | 63 (100) | 0 (0) |
| Negative SRT-PCR and IgG-Abs (n = 30) | 1 (3.3) | 29 (96.7) | 0 (0) | 30 (100) |
| p | 0.0 | 0.0 | 0.0 | 0.0 |
Anti-sense and sense RNA strands were equally detected in PBMCs of all HCV patients who were diagnosed by SRT-PCR. None of the HCV RNA genomic materials were recognized by either SRT-PCR or PBMCs PCR for 96.7% of the negative controls.
Benefits of PBMCs PCR over SRT-PCR in HCV RNA detection
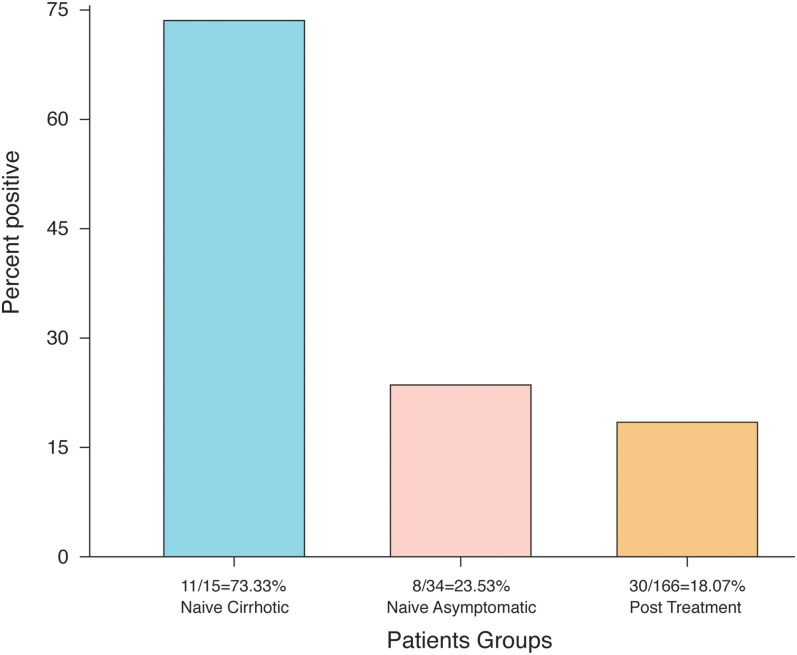
Table 4 illustrates recognition of HCV strands inside PBMCs of patients who presented with positive serum IgG antibodies and negative SRT-PCR (group III). It was found that PBMCs PCR had the benefit of HCV RNA strands detection in 49 out of 215 (22.79%) patients who tested negative for HCV infection by SRT-PCR. Nineteen out of 49 naïve patients (38.78%) and 30 out of 166 post-treatment patients (18.07%) had detectable intracellular HCV RNA (p = 0.00197). Further classification of the naïve patients into asymptomatic and cirrhotic is demonstrated in Fig. 1. The 15 naïve cirrhotic patients had a significantly higher intracellular HCV infection (11/15, 73.33%), compared to the 8 asymptomatic naïve patients (8/34, 23.73%; p = 0.00079) and the 30 experienced cases (30/166, 18.07%; p = 0.000009). We also found insignificant differences in intracellular HCV infection of asymptomatic naïve (8/34, 23.73%) as compared to experienced patients (30/166, 18.07%; p = 0.23179).
Table 4. HCV sense and antisense strands detection in PBMCs of RNA-seronegative patients who presented with positivity for serum IgG antibodies.
| Patient group (n) | PBMCs PCR results | |
| Positive, n (%) | Negative, n (%) | |
| Naïve untreated (49) | 19 (38.78) | 30 (61.22) |
| Experienced (166) | 30 (18.07) | 136 (81.93) |
| Total (215) | 49 (22.79) | 166 (77.21) |
| p: naïve vs. post-treatment | 0.00197 | |
Fig. 1. HCV RNA strands’ prevalence in PBMCs of naïve and post-treatment patients who presented with negative SRT-PCR.
Naïve cirrhotic patients had a significantly increased intracellular HCV infection, compared to naïve asymptomatic patients (p = 0.00079) and experienced patients (p < 0.000009); no significant changes were observed upon comparing naïve asymptomatic with experienced patients (p = 0.23179).
PBMCs PCR recognized 49 out of 215 HCV infections that were missed by SRT-PCR. Naïve patients had a significantly higher frequency of intracellular HCV infection as compared to experienced patients (p = 0.00197). Clearance of intracellular HCV infection in experienced patients is significantly higher than that of naïve patients (p = 0.00197).
Intracellular distribution of sense and antisense HCV RNA strands
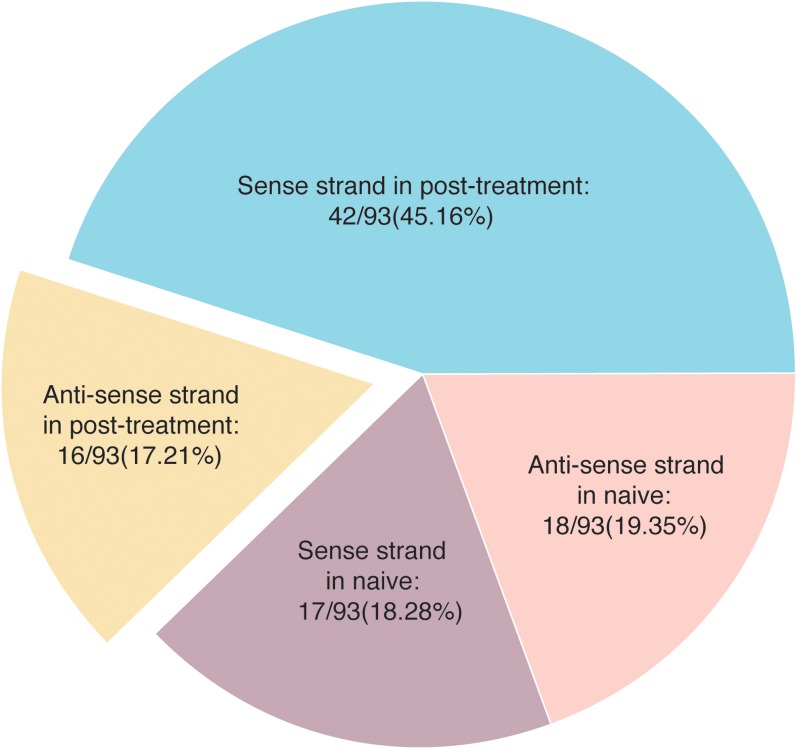
Fig. 2 represents data of the total 93 HCV cases in group III. All had positive PBMCs PCR and serum anti-HCV IgG antibodies, but none had viremia. The relative distribution of intra-PBMCs sense and antisense RNA strands are illustrated in the same figure. As shown, naïve patients had 18 out of 93 (19.35%) and 17 out of 93 (18.28%) positive values for the sense and antisense strands respectively (p = 0.40824). On the other hand, the relative distribution of the sense and antisense strands differed significantly (p < 0.0001) in post-treatment patients, with the sense strand detected in 42 out of 93 (45.16%) as compared to the antisense strand detection in 16 out of 93 (17.2%). Subgrouping of these 93 patients into naïve (n = 35) and experienced (n = 58) patients is shown in Table 5. The results show that the antisense strand is more detectable in naïve (18/35, 51.43%) than in post-treatment patients (16/58, 27.59%; p = 0.01218). The RNA sense strand was more detectable in post-treatment than in naïve patients (42/58, 72.41% vs. 17/35, 48.57%; p = 0.01218).
Fig. 2. Distribution of HCV RNA sense and antisense strands in PBMCs of naïve and post-treatment patients who had negative SRT-PCR.
In naïve patients, both sense and antisense strands were almost equally identified (p = 0.40824), while in experienced patients, the sense strand was more frequently recognized than the antisense strand (p = 0.00000)
Table 5. Verification of HCV RNA strand type in infected PBMCs of naïve patients than in experienced patients who presented with negative SRT-PCR.
| Study group (n) | Type of HCV strands in PBMCs | |
| Antisense strand, n (%) | Sense strand, n (%) | |
| Naïve untreated (35) | 18 (51.43) | 17 (48.57) |
| Experienced (58) | 16 (27.59) | 42 (72.41) |
| Total (93) | 34 (36.56) | 59 (63.44) |
| p | 0.01218 | |
Solitary sense strand was more frequently detected in experienced patients as compared to naïve patients (p > 0.05). Solitary antisense strand was more frequently detected in naïve patients as compared to experienced patients (p < 0.5).
Evaluation of HCV treatment on the basis of intracellular strands’ detection in absence of detectable serum RNA
Table 6 demonstrates results for the total 215 patients in group IV. All were screened by nested PCR for detection of intra-PBMCs HCV sense and antisense strands. We found that 30 out of 166 (18.02%) experienced patients, compared to 19 out of 49 (38.78%) naïve cases, had intracellular infection (p = 0.00197), despite absence of HCV-RNA in their sera. In the same table, experienced patients are classified according to treatment regimens. The sense and antisense RNA strands were almost equally distributed within PBMCs in the PCR-positive cases for each regimen (p > 0.05), except for the sofosbuvir + ribavirin regimen which had more antisense than sense strands (p = 0.0281). As illustrated in Table 7, the 49 HCV PBMCs PCR-positive patients were segregated and further studied. The experienced patients in most of the regimens studied showed significantly decreased intracellular HCV RNA antisense (except for sofosbuvir + ribavirin) and sense (except for the INF-containing regimens) strands, compared to naïve cases (p < 0.05). This finding validates treatment of those who present with negative SRT-PCR but positive intra-PBMCs antisense strand with one of the following regimens: sofosbuvir + daclatasvir and ribavirin (p = 0.01051), sofosbuvir + ladipasvir (p = 0.01051), sofosbuvir + simeprevir (p = 0.00280), and finally INF-containing regimens (p = 0.0284). Sofosbuvir + ribavirin might be contraindicated (p = 0.3878). Regarding treatment of patients who presented with negative SRT-PCR and positivity for intra-PBMCs sense strand, one of the following regimens can be used: sofosbuvir + daclatasvir + ribavirin (p = 0.03375), sofos buvir + ribavirin (p = 0.00478), sofosbuvir + ladipasvir (p = 0.00115), and sofosbuvir + simeprevir (p = 0.00115). Regimens that might be contraindicated in this group are INF-containing ones (p = 0.12083).
Table 6. HCV-RNA strands detection inside PBMCs of experienced and naïve patients in absence of serum viral particles.
| PBMCs PCR | Dacla Sofo Riba (n = 36) | Sofo Riba (n = 64) | INF-Cont Regim (n = 45) | Sofo Ladi (n = 11) | Sofo Simep (n = 10) | Naïve Asympt (n = 34) | Naïve Cirrh (n = 15) |
| Patients with antisense strand, n (%) | 2 (5.56) | 8 (12.5) | 3 (6.67) | 2 (18.18) | 1 (10.0) | 3 (8.82) | 6 (40.0) |
| Patients with sense strand, n (%) | 4 (11.11) | 2 (3.125) | 6 (13.33) | 1 (9.091) | 1 (10.0) | 5 (14.70) | 5 (33.33) |
| p | 0.2181 | 0.0281 | 0.1604 | 0.3036 | 0.5000 | 0.2418 | 0.3624 |
Table 7. Distribution of sense and antisense HCV RNA strands inside PBMCs in experienced and naïve patients with non-viremia.
| Treatment regimens | HCV PBMCs PCR (n = 49) | |||
| Antisense strand (n = 25) | Sense strand (n = 24) | |||
| n (% positive) | p-value on comparison with naïve | n (% positive) | p-value on comparison with naïve | |
| Dacla + Sofo + Riba | 2 (8.00) | 0.0105 | 4 (16.67) | 0.03375 |
| Sofo + Riba | 8 (32.0) | 0.3878 | 2 (8.330) | 0.00478 |
| INF Cont. Regim. | 3 (12.0) | 0.0284 | 6 (25.00) | 0.12083 |
| Sofo + Ladi | 2 (8.00) | 0.0105 | 1 (4.170) | 0.00115 |
| Sofo + Simep | 1 (4.00) | 0.0028 | 1 (4.170) | 0.00115 |
| None (naïve patients) | 9 (36.00) | 10 (41.67) | ||
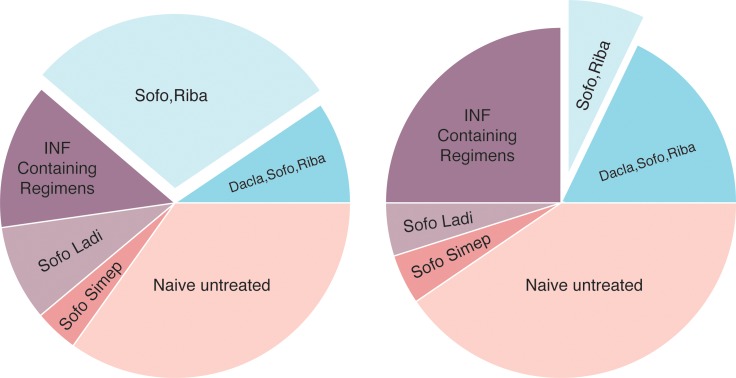
Comparison of percent positive values of HCV RNA antisense with sense strands in the 49 patients that were further subdivided into 19 naïve and 30 experienced patients who completed the various anti-HCV regimens is demonstrated in Fig. 3. Naïve patients had insignificant changes in persistence of intracellular HCV RNA antisense compared to sense strands in their PBMCs (p = 0.34839). There were insignificant differences in the frequency of HCV RNA antisense detected in comparison to the sense strands for the following therapeutic regimens: sofosbuvir + daclatasvir + ribavirin (p = 0.19959), sofosbuvir + ladipasvir (p = 0.32023), and sofosbuvir + simeprevir (p = 0.48979), and INF-containing regimens (p = 0.13547). The HCV RNA antisense strand detection was significantly associated with the sofosbuvir + ribavirin regimen as compared to sense strand (p = 0.02452).
Fig. 3. HCV antisense strand (left) and sense strand (right) detected by PBMCs PCR in post-treatment compared to naïve untreated patients.
Antisense strand detection was significantly associated with the Sofo + Riba regimen compared to sense strand (p = 0.02452). There were insignificant differences in frequency of the detection of antisense compared to that of sense strands in naïve patients (p = 34839) and in the following therapeutic regimens: Sofo + Dacla + Riba (p = 0.19959), Sofo + Ladi (p = 0.32023), and Sofo + Simep (p = 0.48979), and INF-containing regimens (p = 0.13547).
Thirty out of 166 (18.02%) experienced patients, compared to 19 out of 49 (38.78%) naïve patients, had intracellular infection (p = 0.00197). Naïve patients had insignificant changes in frequency of detection of intracellular HCV infection with antisense as compared to sense strands (p = 0.3624). The antisense strand was significantly recognized more than the sense strand in the sofo + riba regimen (p = 0.0281), with insignificant changes in the other regimens (p > 0.15). Sofo: Sofosbuvir; Riba: Ribavirin; INF Cont. Regim.: Interferon-containing regimens; Dacla: Daclatasvir; Simep: Simeprevir; Ladi: Ladipasvir; Asympt.: Asymptomatic; Cirrh.; Cirrhotic.
The experienced patients in most regimens studied had significantly decreased intracellular solitary HCV RNA antisense and sense strands, compared to naïve cases (p < 0.05). This finding validated the treatment of those who presented with negative SRT but positive PBMCs PCR with any of the above-mentioned regimens except for the Sofo + Riba regimen for antisense strand and the INF-containing regimens for sense strand.
Discussion
Most of the study subjects (n = 371), except for the negative controls (n = 30), tested positive for anti-HCV IgG antibodies at baseline, and all of them (n = 401) were subjected to serum and PBMCs HCV PCR. The 100% sensitivity and 96.7% specificity found for the PBMCs PCR validated this test as a potential diagnostic tool for HCV infection. These results are in full agreement with previous reports.15 The 3.3% of negative subjects who tested positive by PBMCs PCR might be considered cryptogenic OCI as previously reported by other investigators.17–19 Recognition of OCI by detecting HCV strands within PBMCs has an extra advantage of cellular over SRT-PCR. The current results indicated that all SRT-PCR-positive patients had both HCV RNA strands within PBMCs. However, elimination of serum viremia does not exclude intra-PBMCs infection. These findings support the recommendation of the use of both SRT-PCR and PBMCs PCR during follow-up of HCV treatment and during validation of the SVR, because intracellular actively-replicating HCV genomes are confirmed.1,25,26 Occasionally, naïve cirrhotic and non-cirrhotic patients test negative by SRT-PCR, despite high anti-HCV IgG antibody titer and a positive PBMCs PCR. Evidence that supports treatment of solitary intracellular HCV infection in both naïve cirrhotic and experienced cases who presented with active hepatocellular damage are insufficient. Establishment of a clinically applied HCV PCR test, which recognizes the dormant intracellular RNA genomic materials, would add tremendous benefits in management of cirrhotic naïve cases, post-treatment potential relapsers, and OCI cases who have negative SRT-PCR.
Our results are compatible with the previously reported data15 regarding confirmation of HCV diagnosis by PBMCs PCR in the absence of viremia. The presence of anti-HCV IgG antibodies without viremia will not be helpful in predicting the actual etiology that led to post-hepatitis cirrhosis. Identification of HCV RNA inside cells in the above-mentioned situation, however, will confirm post-HCV liver cirrhosis, particularly in naïve patients. So, our study concluded for the first time that elimination of intracellular HCV strands might lead to prevention of high prevalence of post-hepatitis cirrhosis, even in the absence of viremia. Post-treatment persistence of overt intracellular HCV infection is related to the serologic relapse that can occur over time.2 Considering the advantage of higher efficacy of the new DAAs for treating HCV, eradication of intracellular infection might help in reducing and/or preventing both HCV liver cirrhosis and relapse. Regarding response to anti-HCV treatment regimens, the experienced patients in our study showed a significant reduction of intracellular HCV infection as compared to both naïve cirrhotic and naïve asymptomatic patients (p = 0.00197). This finding sheds more light on the objectives of therapeutic values of DAAs in achieving SVR.
The antiviral drugs evaluated in this study succeeded in elimination of the HCV antisense strand, but they showed significantly weaker effects on the sense strand. The pathophysiological impact of this finding is not fully understood; however, it might reflect the favorable effects of each antiviral regimen. Regardless, the recognition of negative strand in PBMCs before treatment may be suggested as a potential marker of good treatment response, while detection of negative strand at the end of therapy is a predictor of relapse.16 Our findings are supported by results from a small-scale study that evaluated sofosbuvir + ladipasvir treatment outcomes, in which 14 out of 75 negative viremia patients (18.7%) tested positive for intra-PBMCs HCV sense strand at the end of 6 months of therapy. Moreover, 9 out of these 14 (64.29%) showed seroconversion of HCV RNA after 3 months (unpublished data). Most naïve cirrhotic patients who present with intra-PBMCs HCV sense strand infection, in the absence of antisense strand as well as of viremia, have actual hepatocellular damage, as indicated by impaired liver function (clinical observations). It would be predicted then that solitary presence of HCV RNA sense strand is associated with active hepatic tissue damage. It would not, however, be easy to knock-out solitary RNA sense strands from the infected cells because DAAs act mainly on the replication complex.
Exposure of intracellular HCV RNA strands to the stress of antivirals is associated with disappearance of the antisense strand that leads to interruption of the virus life cycle. Concordant clearance of cellular HCV RNA strands and whole virus particles from serum is a promising sign of SVR, while intra-PBMCs reappearance or non-responding antisense HCV RNA strand to antiviral therapy is a predictor of relapse. Post-treatment persistence of HCV RNA strands might be ominous because of relapse probability, the necessity of further exposure to treatment, and the inevitable hepatic fibrosis activity. In the present study, the efficacy of using DAAs in eradication of intracellular HCV sense and antisense strands was obvious in experienced cases, compared to naïve patients. Thus, our study strongly recommends treatment of naïve cirrhotic patients who have intra-PBMCs HCV strands infection and present with negative SRT-PCR. Also, OCI cases who present with only intracellular infection might be considered suitable candidates for treatment with DAAs upon recognition of the comorbid liver cirrhosis and impaired liver functions. The benefits of treating these groups are not restricted to prevention of post-treatment relapse, but include elimination of the sequelae that result from intracellular harboring of RNA strands in individuals who present with negative SRT-PCR. The improved sensitivity of HCV PBMCs PCR over SRT-PCR mandates the use of both tests at baseline and during follow-up in naïve cases and experienced patients who present with positive anti-HCV IgG-antibodies and negative SRT-PCR respectively. Our study determined that HCV patients who were treated with the sofosbuvir + ribavirin regimen are more liable to relapse, likely because of post-treatment persistence of the antisense strand.16 So eradication of both HCV RNA strands is almost equally accomplished by any of the evaluated therapeutic regimens with the exception of sofosbuvir + ribavirin.
To our knowledge, this is the first report that declares the following conclusions: a) HCV PBMCs PCR is a valid diagnostic test that has the major advantage of diagnosing intracellular infection when SRT-PCR is negative. b) HCV antisense strands are detected more often in naïve patients and sense stands are detected more often in post-treatment patients. c) The overall expected relapse rate for the current treatment regimens is 18.02%, d) Intracellular infection of PBMCs is associated with post-HCV liver cirrhosis in naïve patients who show positivity for serum anti-HCV IgG-antibodies but negative results by SRT-PCR. e) Eradication of intracellular HCV antisense and sense strands is recommended to prohibit sequelae of RNA seroconversion.
Acknowledgments
Financial support for this study was provided by the Faculty of Medicine at Al-Azhar University, National Research Center (Cairo, Egypt) and by the Academy of Scientific Research and Technology Development Fund (Grant No. 3365, to Mostafa K El Awady). The authors are grateful to Professor El-Azhary Shabaan Salah for providing the unpublished data of PBMCs PCR screening after treatment with sofosbuvir + ladipasvir which was cited in this work.
Abbreviations
- HCV
hepatitis C virus
- DAAs
direct-acting antivirals
- SRT
serum real-time
- PBMCs
peripheral blood mononuclear cells
- SVR
sustained virologic response
- RAVs
resistance-associated variants
- OCI
occult HCV infection
References
- 1.Pawełczyk A, Kubisa N, Jabłońska J, Bukowska-Ośko I, Caraballo Cortes K, Fic M, et al. Detection of hepatitis C virus (HCV) negative strand RNA and NS3 protein in peripheral blood mononuclear cells (PBMC): CD3+, CD14+ and CD19+ Virol J. 2013;10:346. doi: 10.1186/1743-422X-10-346. doi: 10.1186/1743-422X-10-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanno AFF, Mohiedeen KM, Alshayeb AF, Deghedy A. HCV RNA in peripheral blood mononuclear cells (PBMCs) as a predictor of the response to antiviral therapy in chronic hepatitis C. Alexandria Journal of Medicine. 2014;50:317–322. doi: 10.1016/j.ajme.2013.05.004. [Google Scholar]
- 3.Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Search for hepatitis C virus extrahepatic replication sites in patients with acquired immunodeficiency syndrome: specific detection of negative-strand viral RNA in various tissues. Hepatology. 1998;28:1398–1401. doi: 10.1002/hep.510280531. doi: 10.1002/hep.510280531. [DOI] [PubMed] [Google Scholar]
- 4.Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. The presence of active hepatitis C virus replication in lymphoid tissue in patients coinfected with human immunodeficiency virus type 1. J Infect Dis. 1998;178:1189–1192. doi: 10.1086/515682. [DOI] [PubMed] [Google Scholar]
- 5.Pal S, Sullivan DG, Kim S, Lai KK, Kae J, Cotler SJ, Carithers RL, Jr, et al. Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology. 2006;130:1107–1116. doi: 10.1053/j.gastro.2005.12.039. doi: 10.1053/j.gastro.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 6.Stamataki Z. Hepatitis C infection of B lymphocytes: more tools to address pending questions. Expert Rev Anti Infect Ther. 2010;8:977–980. doi: 10.1586/eri.10.86. doi: 10.1586/eri.10.86. [DOI] [PubMed] [Google Scholar]
- 7.El-Awady MK, Ismail SM, El-Sagheer M, Sabour YA, Amr KS, Zaki EA. Assay for hepatitis C virus in peripheral blood mononuclear cells enhances sensitivity of diagnosis and monitoring of HCV-associated hepatitis. Clin Chim Acta. 1999;283:1–14. doi: 10.1016/s0009-8981(99)00007-8. [DOI] [PubMed] [Google Scholar]
- 8.Laskus T, Operskalski EA, Radkowski M, Wilkinson J, Mack WJ, deGiacomo M, et al. Negative-strand hepatitis C virus (HCV) RNA in peripheral blood mononuclear cells from anti-HCV-positive/HIV-infected women. J Infect Dis. 2007;195:124–133. doi: 10.1086/509897. doi: 10.1086/509897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laskus T, Radkowski M, Piasek A, Nowicki M, Horban A, Cianciara J, et al. Hepatitis C virus in lymphoid cells of patients coinfected with human immunodeficiency virus type 1: evidence of active replication in monocytes/macrophages and lymphocytes. J Infect Dis. 2000;181:442–448. doi: 10.1086/315283. doi: 10.1086/315283. [DOI] [PubMed] [Google Scholar]
- 10.Müller HM, Pfaff E, Goeser T, Kallinowski B, Solbach C, Theilmann L. Peripheral blood leukocytes serve as a possible extrahepatic site for hepatitis C virus replication. J Gen Virol. 1993;74:669–676. doi: 10.1099/0022-1317-74-4-669. doi: 10.1099/0022-1317-74-4-669. [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez-Iñigo E, Casqueiro M, Navas S, Bartolomé J, Pardo M, Carreño V. Fluorescent “in situ” hybridization of hepatitis C virus RNA in peripheral blood mononuclear cells from patients with chronic hepatitis C. J Med Virol. 2000;60:269–274. doi: 10.1002/(sici)1096-9071(200003)60:3<269::aid-jmv4>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 12.Radkowski M, Kubicka J, Kisiel E, Cianciara J, Nowicki M, Rakela J, et al. Detection of active hepatitis C virus and hepatitis G virus/GB virus C replication in bone marrow in human subjects. Blood. 2000;95:3986–3989. [PubMed] [Google Scholar]
- 13.Bürgel B, Friesland M, Koch A, Manns MP, Wedemeyer H, Weissenborn K, et al. Hepatitis C virus enters human peripheral neuroblastoma cells - evidence for extra-hepatic cells sustaining hepatitis C virus penetration. J Viral Hepat. 2011;18:562–570. doi: 10.1111/j.1365-2893.2010.01339.x. doi: 10.1111/j.1365-2893.2010.01339.x. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson J, Radkowski M, Laskus T. Hepatitis C virus neuroinvasion: identification of infected cells. J Virol. 2009;83:1312–1319. doi: 10.1128/JVI.01890-08. doi: 10.1128/JVI.01890-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalheiro Nde P, Filgueiras TC, Melo CE, Morimitsu SR, de Araújo ES, Tengan FM, et al. Detection of HCV by PCR in serum and PBMC of patients with hepatitis C after treatment. Braz J Infect Dis. 2007;11:471–474. doi: 10.1590/s1413-86702007000500006. [DOI] [PubMed] [Google Scholar]
- 16.Inglot M, Pawlowski T, Szymczak A, Malyszczak K, Zalewska M, Radkowski M. Replication of hepatitis C virus in peripheral blood mononuclear cells in patients with chronic hepatitis C treated with pegylated interferon alpha and ribavirin. Postepy Hig Med Dosw (Online) 2013;67:186–191. doi: 10.5604/17322693.1038785. [DOI] [PubMed] [Google Scholar]
- 17.De Marco L, Gillio-Tos A, Fiano V, Ronco G, Krogh V, Palli D, et al. Occult HCV infection: an unexpected finding in a population unselected for hepatic disease. PLoS One. 2009;4:e8128. doi: 10.1371/journal.pone.0008128. doi: 10.1371/journal.pone.0008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rho J, Ryu JS, Hur W, Kim CW, Jang JW, Bae SH, et al. Hepatitis C virus (HCV) genotyping by annealing reverse transcription-PCR products with genotype-specific capture probes. J Microbiol. 2008;46:81–87. doi: 10.1007/s12275-007-0121-8. doi: 10.1007/s12275-007-0121-8. [DOI] [PubMed] [Google Scholar]
- 19.Bastani MN, Bokharaei-Salim F, Keyvani H, Esghaei M, Monavari SH, Ebrahimi M, et al. Prevalence of occult hepatitis C virus infection in Iranian patients with beta thalassemia major. Arch Virol. 2016;161:1899–1906. doi: 10.1007/s00705-016-2862-3. doi: 10.1007/s00705-016-2862-3. [DOI] [PubMed] [Google Scholar]
- 20.Colin C, Lanoir D, Touzet S, Meyaud-Kraemer L, Bailly F, Trepo C, et al. Sensitivity and specificity of third-generation hepatitis C virus antibody detection assays: an analysis of the literature. J Viral Hepat. 2001;8:87–95. doi: 10.1046/j.1365-2893.2001.00280.x. [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Fong TL, Shindo M, Feinstone SM, Hoofnagle JH, Di Bisceglie AM. Detection of replicative intermediates of hepatitis C viral RNA in liver and serum of patients with chronic hepatitis C. J Clin Invest. 1991;88:1058–1060. doi: 10.1172/JCI115368. doi: 10.1172/JCI115368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goergen B, Jakobs S, Symmons P, Hornes E, Meyer zum Büschenfelde KH, Gerken G. Quantitation of HCV-replication using one-step competitive reverse transcription-polymerase chain reaction and a solid phase, colorimetric detection method. J Hepatol. 1994;21:678–682. doi: 10.1016/s0168-8278(94)80118-5. [DOI] [PubMed] [Google Scholar]
- 24.Löhr HF, Goergen B, Meyer zum Büschenfelde KH, Gerken G. HCV replication in mononuclear cells stimulates anti-HCV-secreting B cells and reflects nonresponsiveness to interferon-alpha. J Med Virol. 1995;46:314–320. doi: 10.1002/jmv.1890460405. [DOI] [PubMed] [Google Scholar]
- 25.Bokharaei Salim F, Keyvani H, Amiri A, Jahanbakhsh Sefidi F, Shakeri R, Zamani F. Distribution of different hepatitis C virus genotypes in patients with hepatitis C virus infection. World J Gastroenterol. 2010;16:2005–2009. doi: 10.3748/wjg.v16.i16.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu DZ, Xie Y, Li ZQ. Clearance of HCV RNA in peripheral blood mononuclear cell as a predictor of response to antiviral therapy in patients with chronic hepatitis C. Hepatobiliary Pancreat Dis Int. 2005;4:550–553. [PubMed] [Google Scholar]