Abstract
Background
The molecular profiles of tumors may inform the selection of appropriate targeted therapies. Circulating tumor cells (CTCs) reflect the real-time status of tumor genotypes. CTCs exhibit high genetic heterogeneity within a patient; accordingly, the analysis of individual CTCs, including their heterogeneity, may enable more precise treatments. We analyzed KRAS mutations in single CTCs from patients with metastatic colorectal cancer (mCRC) using a new single-cell picking system.
Methods
Blood samples were obtained from 61 patients with mCRC. CTCs were enriched and fluorescently labeled using the CellSearch® System. They were recovered using the single-cell picking system based on the fluorescence intensity of marker dyes. Single CTCs and tumor tissue samples were examined for mutations in codons 12 and 13 of the KRAS gene.
Results
CTCs were detected in 27 of 61 patients with mCRC. We isolated at least two CTCs from 15 of 27 patients. KRAS genotype was evaluated in a total of 284 CTCs from 11 patients, and 15 cells with mutations were identified in four patients. In 10 of 11 patients, the KRAS status was the same in the primary tumor and CTCs. In one patient, the KRAS status was discordant between the primary tumor and CTCs. In two patients, different KRAS mutations were found among individual CTCs.
Conclusions
We successfully isolated single CTCs and detected KRAS mutations in individual cells from clinical samples using a novel application of single-cell isolation system. Using the system, we detected CTC heterozygosity and heterogeneity in KRAS status among CTCs within a patient and between CTCs and tumor tissues.
Keywords: Circulating tumor cells, Mutation analysis, KRAS, Single cell analysis, Heterogeneity
Background
Colorectal cancer (CRC) is one of the leading causes of cancer deaths worldwide. Recently, the use of new antitumor agents for metastatic CRC (mCRC), such as epidermal growth factor receptor-targeted monoclonal antibodies (anti-EGFR), has significantly improved the treatment of colorectal disease [1, 2].
KRAS mutations are present in 30–40% of CRC patients [3]. Activating mutations in KRAS are responsible for anti-EGFR therapy resistance in mCRC; accordingly, KRAS genotyping is recommended before EGFR-targeted therapies are administered (e.g., cetuximab and panitumumab) [4]. Although KRAS is a negative predictive marker, not all patients with wild-type KRAS in tumor cells respond to EGFR-targeted therapies. KRAS genotype may not be an accurate predictor of treatment response owing to genetic differences between primary and metastatic tumors.
Several studies have shown that distant metastases can have unique genetic alterations that are different from those in the primary tumor [5, 6]. In addition, acquired resistance is partly achieved by the selection of pre-existing minor subclones harboring mutations that confer resistance to targeted therapy [7, 8]. Primary tumor specimens are not always representative of metastases, which can occur many years after resection of the primary tumor [9, 10]. Characterization of metastatic sites may provide more important information than characterization of primary tumors with respect to guiding targeted therapies [11]. However, invasive biopsies of metastatic sites are not always feasible and repeated testing for real-time surveillance is often difficult.
To overcome the abovementioned problems, circulating tumor cells (CTCs), which can be analyzed clinically by “liquid biopsy,” may be useful for the noninvasive characterization of tumors. These cells reflect subpopulations of primary and/or metastatic tumor cells and are accessible by blood collection [12]. The number of CTCs is correlated with prognosis in several tumor types, such as breast, prostate, and colorectal cancers [13–15]. Monitoring alterations in CTC number during anticancer treatment not only improves prognostic prediction, but also provides information regarding therapy response [14–20]. In addition to enumeration, the molecular characterization of CTCs is important for therapeutic decision-making [21].
Among other challenges with respect to CTC characterization, the isolation of pure CTCs that are not contaminated with leukocytes is still difficult owing to their rarity in peripheral blood [12]. Several studies have detected heterogeneity among CTCs at the single cell level [22, 23]. This suggests the importance of analyzing CTCs at the single-cell level for accurate tumor profiling. However, genetic heterogeneity has not been incorporated into clinical treatments.
Here, we demonstrated the feasibility of detecting KRAS mutations in single CTCs isolated from mCRC patients in a novel application of an automated single-cell isolation system to identify individual cancer cells. Our objective was to analyze high-purity CTCs using this cell recovery system and to evaluate the discordance in KRAS status between primary tumors and CTCs as well as variation among CTCs.
Methods
Ethics and consent statement
This study was approved by the ethical committee of Tokyo Women’s Medical University (approval number, 247) and all patients provided written informed consent prior to participation in the study. All participants in this study provided written informed consent for the publication of their clinical details.
Cell lines
The H1975 human lung cancer cell line containing EGFR mutations was obtained from the ATCC Cell Bank (Manassas, VA, USA) and was used for cell-recovery experiments. The A549 human lung cancer cell line containing KRAS mutations was obtained from the ATCC Cell Bank and was used for blood spiking experiments. H1975 was cultured in RPMI-1640 medium containing 10% fetal bovine serum (both from Thermo fisher scientific, Waltham, MA, USA) in a humidified 5% CO2 incubator at 37 °C. A549 was cultured in F-12 K medium (Thermo fisher scientific) containing 10% fetal bovine serum in a humidified 5% CO2 incubator at 37 °C.
Tumor cell enrichment, staining, and enumeration
The enrichment and enumeration of tumor cells from whole blood were performed using the FDA-approved CellSearch® System (Janssen Diagnostics, Raritan, NJ, USA). First, 7.5 mL of the whole blood sample was processed using the CellSearch® CTC Kit (Janssen Diagnostics). In this assay, EpCAM-based immunomagnetically enriched cells were fluorescently counterlabeled with DAPI to stain nuclei, phycoerythrin (PE)-conjugated antibodies directed against cytokeratins 8/18/19, and allophycocyanin (APC)-conjugated antibodies directed against CD45 to stain the remaining WBCs. After enrichment, isolated fluorescently labeled cells were resuspended in a MagNestⓇ Cartridge Holder (Janssen Diagnostics) and analyzed (i.e., identified and enumerated) using the CellTracks Analyzer IIⓇ (Janssen Diagnostics) according to the manufacturer’s instructions.
Single CTC isolation
To isolate single cells, an automated single-cell isolation system was used, i.e., the ASONECell Picking System (ASONE, Osaka, Japan), to identify individual cancer cells based on staining [24].
Each CTC-enriched sample was recovered from the CellSearch® cartridge and manually loaded onto the microchamber array chip (84,640 wells of 30-μm diameter, 196,000 wells of 20-μm diameter). The fluorescently labeled cells were introduced into each well of the microchamber by centrifugation (2 repetitions of acceleration at 200 rpm for 1 min by plate centrifugation). After loading the microchamber array into the single-cell picking system, the fluorescence intensity of each cell was scanned and analyzed using a computer with a robot. Cells of interest were marked according to PE, APC, and DAPI fluorescence intensity. Marked cells were automatically collected with a glass capillary attached to the micromanipulator of the robot. Each cell was transferred and recovered in 10 μL of PBS in a 200-μL PCR tube. The samples were dried completely then stored in a deep freezer at −80 °C until use. Representative images obtained using the ASONECell Picking System are shown in Fig. 1.
Fig. 1.
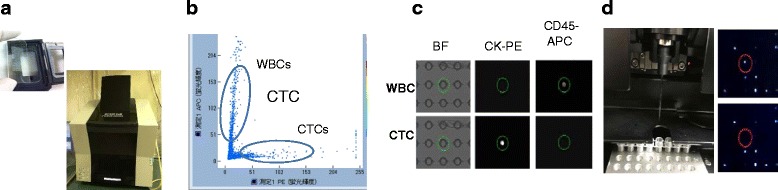
Summary of the ASONECell Picking System. a Fluorescently labeled cells are loaded in a microchamber array and sorted by the machine. b Scatter plot of mean fluorescence intensities for CK-PE (x-axis) and CD45-APC (y-axis) staining. c Bright-field, PE, and APC channel images of peripheral blood mononuclear cells (PBMCs) and circulating tumor cells (CTCs). CTCs can be distinguished from contaminated leukocytes by combining the fluorescence filters. d Cells marked with a red circle are automatically collected with a glass capillary
Evaluation of cell collection using the new single cell picking application
A solution of H1975 cells stained with Cell Tracker™ Green (Thermo fisher scientific) was loaded on the single-cell picking system and single cells were collected and added to individual wells of a 96-well microplate. The existence of a single cell in each well was confirmed by fluorescent microscopy. To quantify tumor cells identified using the single-cell picking system, approximately 1500 or a small number of (2–25) A549 cells were spiked into 7.5 mL of whole blood from a healthy donor (HD), which was collected in a CellSave Preservative Tube (Janssen Diagnostics). A549 cells spiked in HD blood were processed using the CellSearch® CTC Kit (Janssen Diagnostics), and A549 cell counts were determined using the CellTracks Analyzer II® (Janssen Diagnostics). Enriched cells were loaded onto the single-cell picking system and re-counted. CTC counts obtained by CellSearch® and the single-cell picking system were compared. When a small number of cells, i.e., A549 cells, were spiked, single cells were recovered and the recovery rate was calculated.
Preclinical validation of single cell KRAS mutation detection using the A549 cell line
To assess the feasibility of using recovered cells for downstream analyses, a known number of A549 cells was added to 7.5 mL of peripheral blood obtained from an HD, collected in a CellSave Preservative tube, and enriched using the CellSearch® system. Then, single cells were recovered into individual PCR tubes using the single-cell picking system. A total of 24 recovered A549 cells were subjected to KRAS gene-specific amplification after cell lysis with proteinase K (Takara Bio, Kusatsu, Japan) and sodium dodecyl sulfate in individual PCR tubes as previously demonstrated [25]. The DNA from single cell was subjected to KRAS gene-specific amplification and sequenced using the same protocol as that used for CTCs described below. Nine single WBCs isolated from blood samples also served as wild type control for sequencing.
Patient enrolment and tissue and sample collection
The study included 61 patients who had mCRC and underwent various anticancer therapies at the Department of Chemotherapy and Palliative Care or the Department of Surgery, Institute of Gastroenterology, Tokyo Women’s Medical University Hospital. Paraffin-embedded or fresh frozen sections collected from primary tumors were used for KRAS characterization. For each patient, two 10-mL blood samples were drawn into CellSave Preservative tubes or EDTA tubes for CTC enrichment, enumeration, and a mutation analysis. Blood samples were processed within 72 h of collection.
KRAS mutation analysis
A total of 284 single CTCs were analyzed by direct sequencing of the KRAS gene. Sequencing was performed using DNA isolated from CTCs directly or following whole-genome amplification (WGA). For the former analysis, a total of 107 single CTCs were subjected to KRAS gene-specific amplification after cell lysis with proteinase K and sodium dodecyl sulfate. The following nested PCR primers for KRAS codons 12 and 13 were designed using Primer3: outer primers, forward 5′-AAGGTACTGGTGGAGTATTTG-3′ and reverse 5′-GTACTCATGAAAATGGTGAGA-3′; inner primers, forward 5′-ATTATAAGGCCTGCTGAAAATGAGTGA-3′ and reverse 5′-ATATGCATATTAAAACAAGATTTACCTCTA-3′. The reaction was amplified for 40 cycles at 94, 59, and 72 °C for 30 s per cycle for each temperature. The remaining 177 single CTCs were first subjected to WGA using the Ampli1™ WGA Kit (Silicon Biosystems, Bologna, Italy) according to the manufacturer’s instructions. They were then subjected to KRAS gene-specific amplification using the following primers: forward 5′-CCTTATGTGTAGCATGTTCTAATATAG-3′ and reverse 5′-CTATTGTTGGATCATATTCGTCCAC-3′.
Amplified DNA from CTCs was used for direct sequencing of KRAS. PCR products were sequenced using the Big Dye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The sequencing reaction was analyzed using a 3130xl Genetic Analyzer (Applied Biosystems).
DNA from primary tumor tissue was extracted using the FFPE Tissue Kit (Qiagen, Hilden, Germany), subjected to KRAS gene-specific amplification, and sequenced using the same protocol as that used for CTCs.
Results
Evaluation of single-cell collection using the single-cell picking system
To quantify the rate of tumor cell recovery using the single-cell picking system, fluorescently labeled H1975 cells were loaded onto the single-cell picking system and collected individually in wells of a 96-well microplate. Single cells were found in 84 out of 96 wells using fluorescence microscopy, for an isolation success rate of 87.5% (Fig. 2). We next assessed the recovery rate of single CTCs from CellSearch® system. Enriched cells in CellSearch® cartridges were loaded into the single-cell picking system and analyzed. The results of eight independent experiments are summarized in Table 1. In a comparative cell identification analysis, 73.4% of the total cells detected using the CellSearch system were observed using the single-cell picking system after loading into the microchamber, on average. We examined the recovery rate using serial dilutions to obtain a more clinically relevant range (2–25 cells). The results are shown in Table 2. The recovery rate was 70.8%, on average (range 38.5–100%).
Fig. 2.
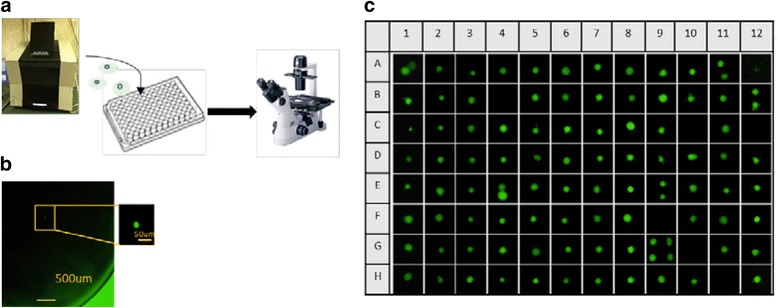
Single-cell collection. a H1975 cells stained by Cell Tracker Green were loaded onto the single-cell and collected into 96-well microplate (200uL PBS/well). b The picture of the isolated single cell confirmed by fluorescent microscopy. c Images of the recovered cells in each well of 96-well microplate. In 84 wells, isolation of single-cell was succeeded. In 12 wells, isolation was failed. In six of 12 wells, more than one cell was collected. Isolation success yield was 87.5% (84/96)
Table 1.
Comparison of tumor cell counts obtained using CellSearch and the ASONECell Picking System
| n = 8 | CellSearch® (cells) |
ASONECell Picking System (cells) |
Re-identification rate (%) ASONECell/ CellSearch® |
|---|---|---|---|
| 1 | 1634 | 1072 | 65.6% |
| 2 | 1692 | 1258 | 74.3% |
| 3 | 1674 | 1430 | 85.4% |
| 4 | 1827 | 1463 | 80.1% |
| 5 | 1874 | 1335 | 71.2% |
| 6 | 1927 | 1369 | 71% |
| 7 | 1964 | 1324 | 67.4% |
| 8 | 1783 | 1289 | 72.3% |
| Average | 1797 | 1318 | 73.4% |
Table 2.
Re-identification rate and recovery rate for a small number of cells (2–25 cells)
| n = 9 | CellSearch® Count (cells) | ASONECell | Re-identification ratea (%) | Recovery rateb (%) | |
|---|---|---|---|---|---|
| Count (cells) | Pick up (cells) | ||||
| 1 | 2 | 1 | 1 | 50% | 50% |
| 2 | 2 | 2 | 2 | 100% | 100% |
| 3 | 4 | 4 | 4 | 100% | 100% |
| 4 | 8 | 7 | 5 | 87.5% | 62.5% |
| 5 | 13 | 6 | 5 | 46.2% | 38.5% |
| 6 | 19 | 15 | 14 | 78.9% | 73.7% |
| 7 | 21 | 13 | 11 | 61.9% | 52.4% |
| 8 | 25 | 21 | 18 | 84% | 72% |
| 9 | 25 | 23 | 22 | 92% | 88% |
| Average | 13.2 | 9.7 | 9.1 | 77.8% | 70.8% |
aRe-identification rate, the number of cells counted using CellSearch® divided by the number of cells re-counted using ASONECell Picking system
bRecovery rate, the number of cells counted using CellSearch® divided by the number of cells picked up using ASONEcell Picking system
Preclinical validation of single cell KRAS mutation detection using the A549 cell line
After CellSearch® enrichment, 24 single A549 cells were recovered by the single-cell picking system and subjected to KRAS gene-specific amplification. The A549 cell line harbors homozygous KRAS mutation (G12S). Codons 12 and 13 of the KRAS gene were sequenced in all sorted cells. In all recovered single A549 cells, KRAS mutation of codon 12 was detected. In 21 of the 24 single A549 cells, the known original homozygous mutation was detected. In the remaining three single A549 cells, the wild-type KRAS allele was detected by sequencing, in addition to the mutant allele (i.e., the samples were heterozygous). This may be explained by contamination with HD blood. Nine single WBCs isolated from HD blood sample were confirmed the expected wild-type genotype.
Patient characteristics
Sixty-one mCRC patients were enrolled in the study. The patient characteristics, including the number of CTCs based on CellSearch®, are listed in Table 3. CTCs (≥1) were detected in 27 out of 61 (44.3%) patients. The range of CTC counts in the CTC-positive patient group (CTC ≥ 1) was 1 to 105 cells.
Table 3.
Patient characteristics according to CTC number assessed by CellSearch
| Patients’ characteristics | CTC = 0 | CTC ≥ 1 | Total (%) |
|---|---|---|---|
| n = 34 | n = 27 | n = 61 | |
| Age | |||
| Median | 69 | 63 | 67 |
| (range) | (34–80) | (36–82) | (34–82) |
| Site of primary tumor | |||
| Right hemicolon | 14 | 9 | 23 (38%) |
| Left hemicolon | 7 | 7 | 14 (23%) |
| Rectum | 13 | 10 | 23 (38%) |
| Other | 0 | 1 | 1 (1%) |
| Site of metastasis | |||
| Liver only | 14 | 10 | 24 (40%) |
| Others | 20 | 17 | 37 (60%) |
| Disease status | |||
| Primary | 11 | 12 | 23 (38%) |
| Recurrence | 23 | 15 | 38 (62%) |
| KRAS status in primary tissue | |||
| Wild-type | 21 | 14 | 35 (57%) |
| Mutant | 9 | 10 | 19 (31%) |
| Unknown | 4 | 3 | 7 (12%) |
In the CTC-negative patient group (CTC = 0), a KRAS mutation was found in 9 out of 34 (25%) patients. In the CTC-positive patient group, the mutation was found in 10 out of 27 (37%) patients. The presence of CTCs was not related to clinical characteristics.
Evaluation of CTCs in clinical samples
Eighty-eight blood samples from 61 patients were analyzed using CellSearch®; the full analysis is summarized in the sample flowchart shown in Fig. 3. Samples obtained from 27 patients (44.3%) for whom at least one CTC was detected using CellSearch® were selected for sorting by the single-cell picking system. For 15 (24.6%) of these 27 patients, at least two single CTCs were recovered by the single-cell picking system.
Fig. 3.
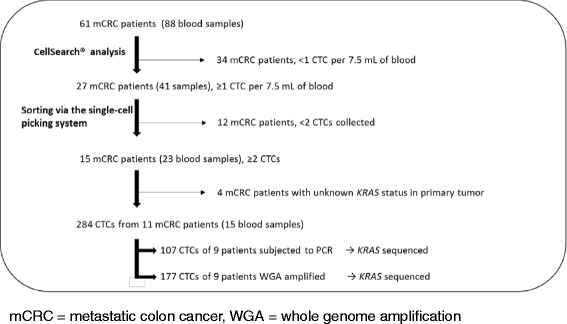
Sample flowchart
Single CTCs recovered from 11 (18%) patients from whom primary tumor samples were available were evaluated to determine the KRAS genotype; primary tumor samples were also sequenced in these cases.
KRAS mutational status of single CTCs determined by PCR
A total of 284 single CTCs were recovered from 11 mCRC patients; 107 single CTCs from nine patients were subjected to direct KRAS gene-specific amplification and 77 were successfully sequenced (median percentage of sequenced CTCs per patient, 70%; range, 20–100%; Table 4, left panel). Sequencing failure may reflect cell loss during sample manipulation or PCR amplification failure.
Table 4.
KRAS mutation analysis in single CTCs and primary tissue
| Pt ID | Blood Sample ID | CTC | Primary tumor | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | WGA | |||||||||||||
| Number of samples analyzed | Number of successful sequences | Sequence success rate (%) | KRAS status | Number of samples analyzed | Number of successful sequences | Sequence success rate (%) | KRAS status | |||||||
| Number of wild-type cells | Number of mutant cells | Number of wild-type cells | Number of mutant cells | |||||||||||
| I | #43 | 12 | 12 | 100% | 12 | 0 | wild | 9 | 7 | 77.8% | 7 | 0 | wild | wild |
| #46 | 12 | 12 | 100% | 12 | 0 | wild | 18 | 16 | 88.9% | 16 | 0 | wild | ||
| II | #44 | 2 | 2 | 100% | 2 | 0 | wild | - | - | - | - | - | - | wild |
| III | #82 | 3 | 3 | 100% | 1 | 2 | p.G12A | - | - | - | - | - | p.G12A | |
| VI | #92 | 10 | 7 | 70% | 7 | 0 | wild | - | - | - | - | - | - | wild |
| #96 | 12 | 8 | 66.7% | 8 | 0 | wild | 1 | 1 | 100% | 1 | 0 | wild | ||
| V | #95 | - | - | - | - | - | - | 6 | 6 | 100% | 6 | 0 | wild | wild |
| #97 | 10 | 6 | 60% | 6 | 0 | wild | 16 | 14 | 87.5% | 14 | 0 | wild | ||
| #142 | - | - | - | - | - | - | 4 | 4 | 100% | 4 | 0 | wild | ||
| VI | #98 | 10 | 5 | 50% | 3 | 2 | p.G12D | 8 | 2 | 25% | 2 | 0 | wild | p.G12D |
| #99 | 12 | 8 | 66.7% | 4 | 4 | p.G12D | 74 | 73 | 98.6% | 70 | 3 | p.G12D | ||
| VII | #114 | 5 | 1 | 20% | 1 | 0 | wild | 5 | 3 | 60% | 3 | 0 | wild | wild |
| VIII | #126 | - | - | - | - | - | - | 4 | 3 | 75% | 3 | 0 | wild | wild |
| #129 | - | - | - | - | - | - | 7 | 6 | 85.7% | 6 | 0 | wild | ||
| IX | #130 | 6 | 5 | 83.3% | 5 | 0 | wild | 8 | 8 | 100% | 7 | 1 | p.G13D | wild |
| #131 | 6 | 2 | 33.3% | 1 | 1 | p.G13D | 13 | 7 | 53.8% | 6 | 1 | p.G12D | ||
| X | #135 | - | - | - | - | - | - | 2 | 2 | 100% | 2 | 0 | wild | wild |
| XI | #137 | 7 | 6 | 85.7% | 5 | 1 | p.G12D | 2 | 1 | 50% | 1 | 0 | wild | p.G12D |
| Total | 107 | 77 | 72% | 177 | 153 | 85.9% | ||||||||
CTCs from five of nine patients had wild-type KRAS at codons 12 and 13. Ten CTCs from the remaining four patients (Patients III, VI, IX, and XI) contained mutations in the KRAS gene.
For Patient III, a c.35G > C (p.G12A) mutation in codon 12 of the KRAS gene was detected in two of three CTCs. In one CTC, the mutation was homozygous, while it was heterozygous in the other (Fig. 4). Thus, CTCs exhibited genetic heterogeneity at the single-cell level and showed the potential for loss of heterozygosity of the wild-type allele.
Fig. 4.
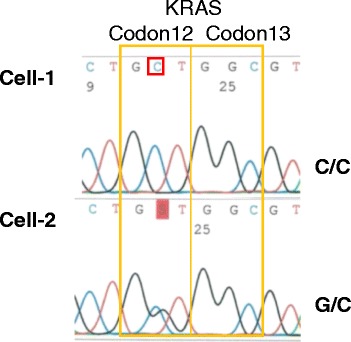
KRAS mutations in single CTCs from Patient III. Direct sequencing results for KRAS codons 12 and 13; the mutation in codon 12 was homozygosis in Cell-1 and heterozygosis in Cell-2
For Patient VI, sample #98 contained a c.35G > A (p.G12D) mutation in codon 12 of the KRAS gene in two of five CTCs and sample #99 had the same mutation in four of eight CTCs. For Patient IX, sample #130 had the wild-type KRAS genotype for all five analyzed CTCs and sample #131 contained a c.38G > A (p.G13D) mutation in codon 12 in one of two CTCs. For Patient XI, the c.35G > A (p.G12D) mutation in codon12 was detected in one of six CTCs.
KRAS mutational status of single CTCs subjected to WGA
The remaining 177 single CTCs from nine patients were subjected to WGA. KRAS of 153 CTCs was successfully sequenced (median percentage of sequenced CTCs per patient, 85.9%; range, 25–100%; Table 4, right panel). Sequencing failure may have been caused by cell loss during sample manipulation, the WGA reaction, or PCR amplification failure.
CTCs from seven of nine patients were wild type for KRAS codons 12 and 13. Five CTCs from the remaining two patients (Patient VI and IX) contained mutations in the KRAS gene.
For patient VI, sample #98 did not have a mutation in KRAS codons 12 and 13 in the two analyzed CTCs and sample #99 contained a c.35G > A (p.G12D) mutation in codon 12 in three of 73 CTCs. For patient IX, two serial blood samples contained different mutations. Sample #130 showed a c.38G > A (p.G13D) mutation in codon 13 in one of eight CTCs and sample #131 contained a c.35G > A (p.G12D) mutation in codon 12 in one of seven CTCs.
KRAS mutational status of primary tissues compared with CTCs
Primary tumor tissues were available for 11 patients. The KRAS mutation status for each of these samples is summarized in Table 4. Wild-type KRAS was detected in eight of 11 samples, while mutant KRAS was detected in three primary tumor samples. In seven of 11 patients, both CTCs and primary tissues were wild type for codons 12 and 13 of the KRAS gene. In one patient (Patient III), both CTCs and primary tissues showed the same mutation in the KRAS gene. In the remaining three patients (Patient VI, IX, and XI), there was discordance between the KRAS mutational status of primary tumor tissues and CTCs.
Discussion
In this study, we evaluated the feasibility of detecting KRAS mutations in single CTCs isolated from mCRC patients using the ASONECell Picking System. This system is an automated single-cell isolation system that allows the isolation of rare cells from a large number of candidate cells via the analysis of immunofluorescence signals. This is the first report indicating that the new cell picking system can be used to isolate CTCs in clinical samples. We performed a comparative analysis of cells obtained using the CellSearch® system and the single-cell picking system. The new system resulted in 26.6% cell loss, on average, relative to the number of cells obtained using the CellSearch® system. The lower cell counts may reflect manual processing issues, such as pipetting errors. The re-identification rate observed using the single-cell picking system is comparable to that of another previously reported device, the DEPArray™ system (Silicon Biosystems, Bologna, Italy) [26, 27]. The recovery rate in a small number of cells was 70.8%, on average (range 38.5–100%). This result demonstrated the feasibility of this application in a more clinically relevant range.
In a preclinical validation of the KRAS mutation analysis of single cells, known mutations were confirmed in 87.5% of samples. The other 12.5% of samples showed the wild-type allele, which may indicate contamination with normal cells during CTC selection. In this examination, no wild-type KRAS cells were found. Thus, loss of mutant-type allele was not occurred and false negativity was not detected. This result indicated that the system is feasible for the detection of KRAS mutations by liquid biopsy.
In our study, we demonstrated analyses of KRAS mutations using two different DNA amplification methods, direct PCR and WGA. We showed the feasibility of KRAS mutation analyses using both methods. Direct PCR is more convenient with respect to time and cost compared with WGA, but few mutations can be analyzed. If information for a single mutation is needed (i.e., EGFR T790 M for targeted therapy in lung cancer) for treatment choices, direct PCR might be suitable. WGA can be used for multi-locus molecular profiling. In the colorectal cancer field, information for several mutations is required for treatment decisions, therefore the WGA method is appropriate.
We analyzed KRAS mutations in single CTCs and matched primary tumors from patients with mCRC. In total, 36.4% of patients had KRAS mutations in CTCs, whereas 27.3% of patients had mutations in primary tumors. In 10 of 11 (90.9%) cases, the KRAS status of the primary tumor matched that of CTCs by either direct PCR or WGA methods. In one patient (Patient IX, Table 4), we found discordant results between the KRAS status of single CTCs and the primary tumor. In this case, the mutation was found in the CTC and wild-type KRAS was found in the primary tumor. The mutation may be present in only a minor subclone of the primary tumor. Although a number of reports have examined the concordance between KRAS mutations in primary tumors and metastatic lesions in mCRC, the significance of observed cases of discordance has only recently been considered [28–30]. Several studies have shown discrepancies between the genetic profiles of CTCs and primary tumors [31, 32] and heterogeneity among individual CTCs [27]. Because single-CTC analyses by liquid biopsy provide information regarding the real-time status of existing tumors, these data might provide more accurate information for personalized therapy.
In one patient (Patient IX), KRAS mutations in CTCs differed among blood samples obtained at different time periods. One CTC had a p.G13D mutation, and the other had p.G12D. In another patient (Patient III), the mutation was homozygous in one CTC, but heterozygous in another CTC. In these cases, either more than one subclone was present in a tumor at a given time or a mutation was acquired during the clinical course of the disease. These results are consistent with the growing number of studies reporting high heterogeneity among CTCs within a patient [18, 33–35]. Our results raise several clinical questions about the real value and significance of CTC analyses. One question is which status is appropriate for treatment decisions if the CTC mutational status was different from that of the primary tumor. Another question is which mutational status is the most clinically significant if CTCs show genetic heterogeneity. Although heterogeneity among single CTCs has been observed at several loci that are drug targets (e.g., EGF receptor inhibitors) or associated with drug resistance (e.g., PIK3CA and KRAS), the clinical relevance of this variation is unknown. To address these questions, clinical studies are needed to monitor changes in the mutational status of CTCs and primary and/or metastatic tumors during treatment as well as to identify indicators of the treatment response.
Conclusions
We examined the molecular profiles of single CTCs using the ASONECell Picking System, a new cell sorter that enables the isolation of single or small groups of cells from mixed-cell suspensions. We demonstrated that the isolation and molecular characterization of single CTCs is feasible in mCRC patients. We detected CTC heterozygosity as well as differences between primary tumors and CTCs with respect to KRAS status. This system may facilitate future analyses of the clinical significance of CTC heterogeneity.
Acknowledgements
We thank Ms. Sayaka Kinoshita, Mr. Takeshi Watabe, Ms. Ayano Kanazawa, Mr. Masatoshi Mori and Mr. Gen Fujii for excellent technical assistance. We thank Mr. Hajime Sugisaki, Ms. Hiroko Higashimoto, and Mr. Masao Oomura for helpful scientific discussions.
Funding
No funding was provided for this research.
Availability of data and materials
The datasets supporting the conclusions of this article are available from the corresponding author on reasonable request.
Authors’ contributions
YK, HH, KH, KK and MY designed the study. YK, KK and KH contributed patient samples. HH and YM developed the technology of the single-cell picking system. YK and YM performed experiments and analyzed the sequencing assays. YK drafted the manuscript. All authors have read and approved the final manuscript.
Competing interests
All authors report that they have no competing interest associated with this study.
Consent for publication
All participants in this study gave us written informed consent for publication of their clinical details.
Ethics approval and consent to participate
This study was approved by the ethical committee of Tokyo Women’s Medical University (approval number, 247) and all patients provided written informed consent prior to participation in the study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- APC
Allophycocyanin
- CRC
Colorectal cancer
- CTC
Circulating tumor cell
- HD
Healthy donor
- mCRC
metastatic colorectal cancer
- PE
Phycoerythrin
- WGA
Whole-genome amplification
Contributor Information
Yuurin Kondo, Phone: +81-(0)3-3353-8111, Email: kondo.yurin@twmu.ac.jp.
Kazuhiko Hayashi, Email: hayashi@chemo.jp.
Kazuyuki Kawakami, Email: kawakami.kazuyuki@twmu.ac.jp.
Yukari Miwa, Email: ino_yuka@srl.srl-inc.co.jp.
Hiroshi Hayashi, Email: hayash@srl.srl-inc.co.jp.
Masakazu Yamamoto, Email: yamamoto.ige@twmu.ac.jp.
References
- 1.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 3.Barault L, Veyrie N, Jooste V, Lecorre D, Chapusot C, Ferraz JM, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–2259. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 4.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 5.Dupont Jensen J, Laenkholm AV, Knoop A, Ewertz M, Bandaru R, Liu W, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic. Clin Cancer Res. 2011;17:667–677. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe T, Kobunai T, Yamamoto Y, Matsuda K, Ishihara S, Nozawa K, et al. Heterogeneity of KRAS status may explain the subset of discordant KRAS status between primary and metastatic colorectal cancer. Dis Colon Rectum. 2011;54:1170–1178. doi: 10.1097/DCR.0b013e31821d37a3. [DOI] [PubMed] [Google Scholar]
- 7.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108:479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen LC, Yeh YS, Chen CW, Wang HM, Tsai HL, Lu CY, et al. Detection of KRAS oncogene in peripheral blood as a predictor of the response to cetuximab plus chemotherapy in patients with metastatic colorectal cancer. Clin Cancer Res. 2009;15:4508–4513. doi: 10.1158/1078-0432.CCR-08-3179. [DOI] [PubMed] [Google Scholar]
- 10.Buim ME, Fanelli MF, Souza VS, Romero J, Abdallah EA, Mello CA, et al. Detection of KRAS mutations in circulating tumor cells from patients with metastatic colorectal cancer. Cancer Biol Ther. 2015;16:1289–1295. doi: 10.1080/15384047.2015.1070991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 12.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 13.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 14.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 15.Allen JE, El-Deiry WS. Circulating tumor cells and colorectal cancer. Curr Colorectal Cancer Rep. 2010;6:212–220. doi: 10.1007/s11888-010-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danila DC, Fleisher M, Scher H. Circulating tumor cells as biomarkers in prostate cancer. Clin Cancer Res. 2011;17:3903–3912. doi: 10.1158/1078-0432.CCR-10-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldkorn A, Ely B, Quinn DI, Tangen CM, Fink LM, Xu T, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32:1136–1142. doi: 10.1200/JCO.2013.51.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 19.Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, et al. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27:5153–5159. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Gao P, Song Y, Sun J, Chen X, Zhao J, et al. Relationship between circulating tumor cells and tumor response in colorectal cancer patients treated with chemotherapy: a meta-analysis. BMC Cancer. 2014;14:976. doi: 10.1186/1471-2407-14-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 23.Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2015;9:749–757. doi: 10.1016/j.molonc.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimoto N, Kida A, Jie X, Kurokawa M, Iijima M, Niimi T, et al. An automated system for high-throughput single cell-based breeding. Sci Rep. 2013;3:1191. doi: 10.1038/srep01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutterlin M, Sleiman PA, Onadim Z, Delhanty J. Single cell detection of inherited retinoblastoma predisposition. Prenat Diagn. 1999;19:1231–1236. doi: 10.1002/(SICI)1097-0223(199912)19:13<1231::AID-PD725>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 26.Peeters DJ, De Laere B, Van den Eynden GG, Van Laere SJ, Rothé F, Ignatiadis M, et al. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting. Br J Cancer. 2013;108:1358–1367. doi: 10.1038/bjc.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francesco F, Silvia C, Wainer Z, Paola U, Giluia G, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett. 2013;335:225–231. doi: 10.1016/j.canlet.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Jin K, Lan H, Teng L. Heterogeneity in primary colorectal cancer and its corresponding metastases: a potential reason of EGFR-targeted therapy failure? Hepato-Gastroenterology. 2011;58:411–416. [PubMed] [Google Scholar]
- 29.Mao C, Wu XY, Yang ZY, Threapleton DE, Yuan JQ, Yu YY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep. 2015;5:8065. doi: 10.1038/srep08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasch C, Bauernhofer T, Pichler M, Langer-Freitag S, Reeh M, Seifert AM, et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem. 2013;59:252–260. doi: 10.1373/clinchem.2012.188557. [DOI] [PubMed] [Google Scholar]
- 31.Lyberopoulou A, Aravantinos G, Efstathopoulos EP, Nikiteas N, Bouziotis P, Isaakidou A, et al. Mutational analysis of circulating tumor cells from colorectal cancer patients and correlation with primary tumor tissue. PLoS One. 2015;10:e0123902. doi: 10.1371/journal.pone.0123902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mostert B, Jiang Y, Sieuwerts AM, Wang H, Bolt-de Vries J, Biermann K, et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int J Cancer. 2013;133:130–141. doi: 10.1002/ijc.27987. [DOI] [PubMed] [Google Scholar]
- 33.Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6:1371–1386. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng G, Krishnakumar S, Powell AA, Zhang H, Mindrinos MN, Telli ML, et al. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer. 2014;14:456. doi: 10.1186/1471-2407-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are available from the corresponding author on reasonable request.


