Abstract
Background
miR-126 is a key regulator of oncogenic processes. It is functionally linked to cellular proliferation, survival and migration. Vascular endothelial growth factor A (VEGF-A), which is regarded as a tumorgenesis activator, could directly target miR-126 in several tumors. However, the mechanism in esophageal cancer remains unclear.
Methods and results
In this study, the expression of miR-126 and VEGF-A were assessed in esophageal cancer tissues and esophageal cancer cell lines. We found that miR-126 has significantly lower expression in esophageal cancer tissues and esophageal cancer cell lines than in healthy tissues, while the expression of VEGF-A is high. Luciferase reporter assays were performed to investigate the relationship between VEGF-A and miR-126. We confirmed that VEGF-A is a target for miR-126. Furthermore, the proliferation of esophageal cancer cells with miR-126 overexpression and miR-126 knockdown was monitored using the MTT assay. The results showed that miR-126 could inhibit esophageal cancer cell proliferation in vitro. The effect of miR-126 was also detected in BALB/c nude mice with transplanted esophageal cancer cells. In vivo study showed that tumor growth was significantly suppressed by miR-126 overexpression.
Conclusions
We believe that restoring miR-126 levels may be a promising therapeutic approach in cases of esophageal cancer.
Keywords: miR-126, Esophageal cancer, VEGF-A, Tumorgenesis, Cell proliferation, Lentivirus package, MTT assay, Xenograft model
Introduction
Over the past few decades, esophageal cancer incidence and mortality increased worldwide. Previous studies showed that microRNA (miRNA) expression and esophageal cancer are closely related. miR-203 and miR-205 expression in esophageal squamous cell carcinoma is significantly lower than in normal epithelial tissues, while the expression of miR-21 is significantly higher than in the normal epithelial tissues [1].
miRNA-126 (miR-126), an endothelial cell-restricted miRNA, was found to regulate developmental angiogenesis [2]. miR-126 is located within intron 7 of epidermal growth factor-like domain 7 (EGFL7). It acts as a tumor suppressor gene in many malignant tumor cells, inhibiting the progression of some cancers via negative control of proliferation, migration, invasionand cell survival. However, in some cases, miR-126 supports cancer progression via promotion of blood vessel formation [3].
VEGF can promote endothelial cell proliferation and play significant roles in angiogenesis and tumor growth. During tumor expansion, VEGF can increase the permeabilization of blood vessels and the production of new blood vessels [4]. VEGF-A is a target gene of miR-126. Downregulation of miR-126 increases VEGF-A activity in lung cancer [5] and breast cancer [6].
Although studies on miRNA in esophageal cancer have been reported, the underlying mechanisms have not been fully elucidated. Therefore, this study focused on the regulatory functions of miRNA-126 in the progression of esophageal cancer.
Materials and methods
Cell cultures and animal subjects
The human esophageal adenocarcinoma cell lines OE19 and OE33 were purchased from the European Collection of Cell Cultures. The 293T (ATCC CRL-3216) cell line and JH-EsoAd1 cells were purchased from the American Type Culture Collection.
Cells were cultured in RPMI 1640 medium (R0883; Sigma) supplemented with 2 mM glutamine, 10 % fetal bovine serum (FBS), 100 U/ml penicillin G and 100 mg/ml streptomycin (Imperial Laboratories) and maintained in monolayer culture at 37 °C in an incubator of humidified air with 5 % CO2.
Male BALB/c nude mice aged 8–10 weeks were obtained from the Animal House of Xi’an Jiaotong University and their conditions were approved by the Animal Care and Use Committee of the Second Affiliated Xi’an Jiaotong University.
Tissue sample collection
A set of 13 esophageal cancer tissue samples and paired adjacent normal tissue samples were obtained from patients undergoing esophageal cancer surgery at the Second Affiliated Hospital of Xi’an Jiaotong University. All of the esophageal cancer cases were pathologically confirmed by two independent experts. The collected tissues were frozen at −80 °C for further analysis. Informed consent was obtained, and this study was in accordance with the Declaration of Helsinki and approved by the Human Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University.
RNA isolation and quantitative RT-PCR
RNA was extracted from the 13 human esophageal cancer tissues and cultured cells using Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. It was then reverse transcribed into cDNA using aPrimeScript RT Reagent Kit (TaKaRa). The cDNA underwent PCR with an TaKaRaBio SYBR Green Master Mix kit using the miR-126 primer set as follows: sense, 5’-CAA CAG AAG GGG CAG GTT GCC CGG AGC-3’; and antisense, 5’-ATT CTG ATC ACG CCT AAG TAC GTC GGG GC-3’. The U6 primers set was: sense, 5’-ATC CGC AAA GAC CTG T-3’; and antisense, 5’-GGG TGT AAC ACT AAG-3’ (Sangon). The relative levels of miR-126 transcripts were normalized to the control U6 mRNA. The cycle number at which the reaction crossed an arbitrarily placed threshold (Ct) was determined for each gene, and the relative amount of each miRNA to U6 snRNA was calculated using the equation 2-ΔΔCt, where ΔΔCt = (C tmiRNA - C tU6 snRNA) transfected - (C tmiRNA - C tU6 snRNA)control. Each experiment was performed in triplicate.
Plasmid construction and lentivirus package
The lentiviral vector pCDH-CMV (pLV, System Biosciences, SBI) was used to construct the pLV-mir-126 plasmid. The template was the human genome DNA of miR-126 (Accession number: NCBI Reference Sequence: NR_029695.1). Lentiviral constructs contained miR-126 (LV-miR-126) and anti-miR-126 (LV-anti-miR-126) along with the miR negative control (LV-miR-NC). To produce the lentivirus, three plasmid DNAs were individually transfected into 29 cells using psPAX2, a pMD2G packaging construct, and lipofectamin plus reagent (Invitrogen) according to the manufacturer’s instructions. After 8 h, the original medium was replaced with fresh medium, and the lentiviral supernatant was collected 48 h later. The expression level of green fluorescent protein (GFP) was measured to establish the titer of the virus, according to the manufacturer’s instruction.
Luciferase reporter assay
293 cells were seeded in 96-well plates and grown in RPMI 1640 containing 10 % FBS at 37 °C in a humidified atmosphere containing 5 % CO2. After 24 h, luciferase reporter plasmids were transfected separately or co-transfected with VEGF-A wild-type 3’-UTR and VEGF-A mutation-type 3’-UTR. Firefly and renilla luciferase activities were measured 48 h after transfection using the Dual-Glo Luciferase Assay System (Promega). Firefly luciferase was normalized to renilla luciferase activity.
In vivo tumor xenograft model
1 × 107 cells stably transfected with LV-miR-126, LV-anti-miR-126 and lenti-miR-NC were injected subcutaneously into the right foreleg armpit of three groups of 18–26 g male BALB/c nude mice (8 mice/group) to establish a cancer model. The tumor size was detected every week using a caliper. At 42 days after inoculation, all of the mice were killed and the tumor masses were successively excised, weighed, photographed and subjected to western blot and luciferase reporter assays.
Western blotting analysis
Immunoblotting was performed to detect the expression of VEGF-A in the cancer tissues and in the human esophageal adenocarcinoma cell lines OE33 and JH-EsoAd1, which had been infected with the recombinant lentivirus. For western blot analysis, the samples were lysed using the RIPA buffer (Pierce) in the presence of a protease inhibitor cocktail (Pierce). Samples with 25 μg of total protein were resolved by SDS-PAGE on a 10 % gel and transferred onto an NC membrane. The blot was incubated with TBST with 1:500 goat primary antibodies against human VEGF-A (sc-152), followed the secondary HRP-conjugated anti-goat antibody (both from Santa-Cruz Biotechnology). After washing, the bands were detected using chemiluminescence (ECL detection kit) and imaged with Kodak film. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous protein for normalization.
MTT assay
Cell proliferation was analyzed using the3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) assay. Cells were seeded into 96-well plates (5 × 103 cells/well) directly or at 1 day after stable transfection, and incubated for 1, 3,5, 7, 9 and 11 days. After incubation with 25 μl of MTT (5 mg/ml, Sigma) at 37 °C for 4 h, the supernatants were removed, and 150 μl of dimethylsulfoxide (DMSO, Sigma) was added to each well. The absorbance value (OD) of each well was measured at 490 nm. All experiments were performed three times and the average results were calculated.
Statistical analysis
Results were analyzed statistically using Student’s t-test for comparisons between two groups. Data are presented as the means ± SD. Correlation parameters were submitted to Pearson and non-parametric Spearman correlations. A P value less than 0.05 was considered to indicate statistical significance. Analyses were performed using SPSS 17.0 for Windows (SPSS).
Result
Expression of miR-126 and VEGF-A in esophageal cancer tissue samples and cells
The expression levels of miR-126 in 13 esophageal cancer tissue samples and paired adjacent normal tissue samples were determined using quantitative RT-PCR. The results showed that the expression level of miR-126 was approximately 7 times lowerin all 13 esophageal cancer tissue samples than that in the normal tissues (Fig. 1a).
Fig. 1.
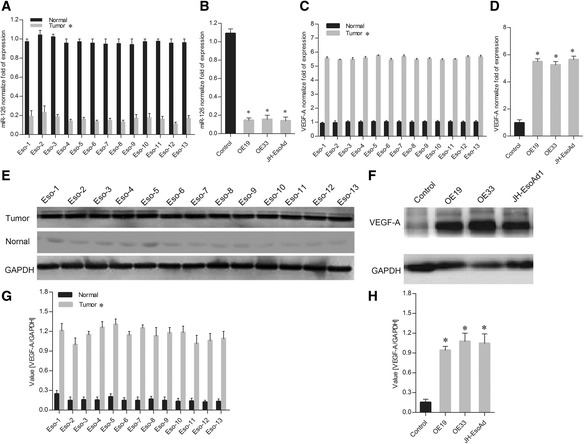
Expression of miR-126 and VEGF-A in esophageal cancer tissues and esophageal carcinoma cell lines. a qRT-PCR analysis of miR-126 expression in 13 esophageal cancer tissues and paired adjacent normal tissues. b qRT-PCR analysis of miR-126 expression in three esophageal carcinoma cell lines and a control cell line. c qRT-PCR analysis of VEGF-A expression in the esophageal cancer tissues and paired adjacent normal tissues. d qRT-PCR analysis of VEGF-A expression in three esophageal carcinoma cell lines and a control cell line. e Western blot analysis of VEGF-A expression in esophageal cancer tissues and paired adjacent normal tissues. f Western blot analysis of VEGF-A expression in three esophageal carcinoma cell lines and a control cell line. g Quantitation of protein expression for Fig. 1e. h Quantitation of protein expression for Fig. 1f. The data is shown as the means ± SD. *p <0.01, compared to the control group
Furthermore, we also tested the expression of miR-126 in the OE19, OE33 and JH-EsoAd1 esophageal carcinoma cell lines. Compared with the control cell lines, the expression levels of miR-126 in the esophageal carcinoma cell lines were also significantly lower (Fig. 1b). This indicts that miR-126 might play a negative role in esophageal carcinogenesis.
The expression of VEGF-A in all 13 cancer and normal tissue samples and the carcinoma cell lines was also assessed using qRT-PCR and western blot. Compared with the control, the VEGF-A expression levels were significant upregulated (Fig. 1) at both the mRNA and protein levels.
Inhibition of VEGF-A expression in esophageal cancer cells with miR-126 overexpression
To find direct targets of miR-126, luciferase activity was measured by the co-transfection of the VEGF-A wild-type 3’-UTR with miR-126. The miRNA decreased luciferase activity, whereas this effect was completely ablated by deletion of the miR-126-binding site in the VEGF-A 3’-UTR (Fig. 2a). These results suggest that VEGF-A was the direct target of miR-126.
Fig. 2.
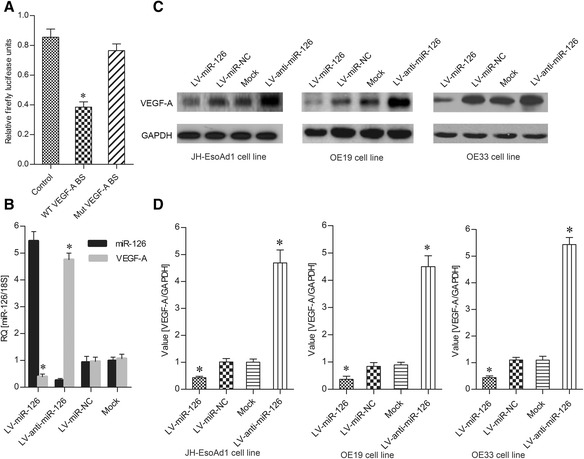
The expression of VEGF-A is regulated by miR-126. a Relative firefly luciferase units in 293 cells transfected with WT VEGF-A or Mut VEGF-A. b Expression of miR-126 and VEGF-A in 293 cells treated with different vectors. c Western blot analysis of VEGF-A expression in three esophageal carcinoma cell lines treated with different vectors. d Quantitation of protein expression for Fig. 2c. The data is shown as the means ± SD. *p <0.01, compared to the controls LV-miR-NC and Mock
To investigate the regulatory role of miR-126 on VEGF-A expression in esophageal cancer in vitro, VEGF-A expression was detected in esophageal cancer cell lines with miR-126 overexpression and knockdown. Compared to the lentivirus miR negative control (LV-miR-NC) and the vector without lentivirus (Mock), the expression of miR-126 in cells infected with recombinant lentivirus miR-126 was significantly increased, whereas with the treatment with anti-miR-126 inhibitor (LV-anti-miR-126), the miR-126 expression decreased significantly (Fig. 2b). Furthermore, the overexpression and inhibition of miR-126 in esophageal cancer cells could downregulate or upregulate the expression of VEGF-A both at the mRNA and protein levels (Fig. 2b–d). These results demonstrate that overexpression of miR-126 could inhibit VEGF-A expression.
miR-126 inhibits cell proliferation in esophageal cancer
The MTT assay was employed to investigate the effects of miR-126 on cell proliferation in esophageal cancer. LV-miR-126 was transfected into esophageal cancer cells for this assay. The results revealed that exogenous expression of miR-126 could inhibit the proliferation of JH-EsoAd1, OE19 and OE33 cells (Fig. 3). Statistical analysis suggested similar effects of miR-126 on cell proliferation. These results suggest that miR-126 could inhibit VEGF-A expression and then inhibit esophageal cancer cell proliferation in vitro.
Fig. 3.
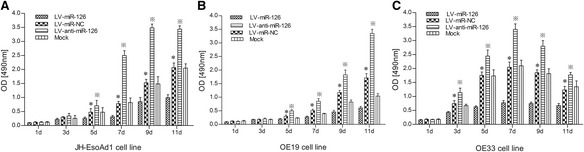
The proliferation of esophageal cancer cells as determined using the MTT assay. a Effects of LV-miR-126 on the cell line JH-EsoAd1. b Effects of LV-miR-126 on OE19. c Effects of LV-miR-126 on OE33. The data is shown as the means ± SD. *p <0.01, compared to LV-miR-NC and Mock
Effect of miR-126 expression on the progression of esophageal cancer in vivo
To figure out whether changes in miR-126 expression could influence the growth of tumorsin vivo, three groups of nude mice were inoculated with OE33 cells that had been stably transfected with recombinant lentivirus miR-126 (LV-miR-126), anti-miR-126 inhibitor (LV-anti-miR-126) and lentivirus miR negative control (LV-miR-NC). Tumor formation was observed and tumor weight was measured in these three groups (Fig. 4). These results showed that the ectopic expression of miR-126 inhibited tumorigenesis in vivo. The average tumor weight of mice inoculated with lenti-miR-126-transfected OE33 cells was significantly lower on day 42 (p < 0.01) than that for mice inoculated with anti-miR-126 transfected OE33 cells and the miR-NC negative control group. These results revealed that miR-126 could inhibit esophageal cancer growth by downregulating VEGF-A expression.
Fig. 4.
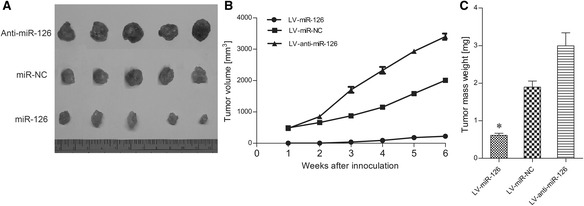
The effects of miR-126 on tumor growth in nude mice inoculated with esophageal cancer cells. a The measurement of tumor volumes in each group. b The tumor volumes over the experimental period. c The tumor mass weights for each group. The data is shown as the means ± SD. *p <0.01, compared to LV-miR-NC and Mock
Discussion
It has been demonstrated that abnormal expression of miR-126 correlates with human tumorigenesis and that miR-126 has roles in cancers of the gastrointestinal tract, genital tracts, breasts, thyroid, lungs and some other tissues and organs [7]. Li found that miR-126 expression was downregulated in gastric carcinoma tissues compared with matched non-cancer tissues when assessed with qRT-PCR [8]. In addition, miR-126 was identified as a tumor suppressor in gastric cancer and as an inhibitor of V-crk avian sarcoma virus CT10 oncogene homolog-like (CRKL)thanks to its targeting of the 3’ UTR region of CRKL mRNA [9] or VEGF-A mRNA [10]. It has also been determined that miR-126 expression is deregulated in colorectal cancer [11–14]. For the mechanism of its repression of colon cancer proliferation and invasion, Li reported that miR-126 negatively regulates the expression of CXCR4 and inhibits the RhoA/ROCK (Rho-associated proteinkinase) signalling pathway [15]. In oral cancer, Sasahira has reported that low miR-126 expression is associated with tumour progression due to upregulation of the VEGF-A signal [16]. In lung carcinoma, miR-126 expression was found to be reduced [17].
Crawford discovered that miR-126 can suppress lung cancer invasion by directly targeting CRK [18], and Liu reported that miR-126 suppresses the expression of VEGF-A, inhibiting cancer cell growth [19]. Zhu suggests that enhanced expression of miR-126 elevates the sensitivity of non-small cell lung cancer cells to anticancer therapy via negative regulation of the VEGF/PI3K/Akt/MRP1 signalling pathway [20]. These studies suggest that miR-126 is a potential tumor suppressor gene. Based on these reports, we postulate that miR-126 plays a critical but as-yet-unknown role in human esophageal cancer.
Recently, Koumangoye et al. reported that miR-31 is significantly decreased in esophageal cancer cells, while upregulation of miR-31 inhibits growth, migration and invasion of esophageal adenocarcinoma (EAC) and squamous cell carcinoma (ESCC) cell lines [21]. Yu et al. suggested that miR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells [22].
Liu et al. discovered that microRNA126-3p (miR-126) was significantly downregulated in esophageal squamous cell carcinoma (ESCC), and its downregulation correlated with poor ESCC prognosis. Furthermore, they found downregulation of miR-126 was due to promoter hypermethylation of its host gene, Egfl7 [23]. In another study, Liu et al. reported that when miR-126 in the esophageal cancer tissues was compared with its levels in matched normal tissues using miRNA microarrays, it was found to be reduced [24].
However, the effect of miR-126 in esophageal cancer remains unclear. In this study, we showed that miR-126 was bound to VEGF-A 3’-UTR, which resulted in reduced expression of VEGF-A in esophageal cancer cells, which inhibited esophageal cancer growth. We found miR-126 had lower expression in esophageal cancer samples compared to adjacent normal samples around the cancer, which is consistent with earlier results [25].
However, there were some controversies for miR-126 in esophageal cancer. Hu discovered that miRNA-126 failed to show a relationship to the outcome of patients with esophageal adenocarcinoma [26]. These contradictory results might due to the distinct biology and clinical features of esophageal squamous carcinoma in our study and esophageal adenocarcinoma in the previous study. The differences in the subject populations (Asian vs. American) might also contribute to this.
Furthermore, in ourqRT-PCR and western blot assays, we discovered that the VEGF-A expression levels were upregulated in human esophageal cancer cells and negatively correlated with miR-126. These results suggest that miR-126 as a possible negative regulatory role in VEGF-A expression. It has been demonstrated that downregulated miR-126 increases VEGF-A activity in oral cancer, lung cancer and breast cancer [16, 19, 27] and that miR-126 may act as a tumor suppressor by regulating VEGF-A expression in esophageal cancer.
To understand the molecular mechanisms of miR-126 suppressing esophageal cancer cells, we searched for putative miR-126 targets. VEGF-A is an important invasion and metastasis factor [28, 29]. This has led to the hypothesis that VEGF-A is atarget for miR-126. Based on luciferase activity assays, we found that VEGF-A is a direct target of miR-126and confirmed that miR-126 negatively regulates VEGF-A expression in overexpressed miR-126 esophageal cancer cells from the lines JH-EsoAd1, OE19 and OE33. To further clarify this, the MTT assay was carried out, and the results showed that miR-126 could inhibit esophageal cancer cells proliferation in vitro. More importantly, ectopic expression of miR-126 in nude mouse inhibited tumorigenesis.
Our result was consistent with a previous study, which demonstrated that downregulation of miR-126 was reverse correlated with VEGF-A expression in gastric cancer. Furthermore, Chen et al. also suggested that the downregulation of miR-126 inhibited gastric tumor growth and tumor angiogenesis through activation ofAkt, mTORand Erk1/2 of VEGF-A signaling downstream genes [9, 30].
In conclusion, miR-126 was naturally complementary to the VEGF-A 3’-UTR and could downregulate overexpression of VEGF-A in esophageal cancer cells, thus inhibiting the esophageal cancer growth.
Acknowledgement
We would like to thank Dr. Xijing He and Dr. Bin Zhou for their insightful comments and suggestions.
Funding
This study had no funding support.
Availability of data and materials
The dataset of human genome DNA of miR-126 supporting the conclusions of this article is available in the Genbank (NCBI) repository (Assession number: NR_029695.1, Gene ID: 262205369, http://www.ncbi.nlm.nih.gov/nuccore/NR_029695.1).
The dataset of human genome DNA of VEGF-A supporting the conclusions of this article is available in the Genbank (NCBI) repository (Assession number: NM_001317010.1, Gene ID: 949394727, http://www.ncbi.nlm.nih.gov/nuccore/NM_001317010.1).
The dataset of OE19 cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA4186904, ID: 4729602, http://www.ncbi.nlm.nih.gov/biosample/?term=OE19).
The dataset of OE33 cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA4186711, ID: 4728790, http://www.ncbi.nlm.nih.gov/biosample/?term=OE33).
The dataset of JH-EsoAd1 cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA4446651, ID: 4776544, http://www.ncbi.nlm.nih.gov/biosample/?term=JH-EsoAd1).
The dataset of 293T cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA3782549, ID: 4856025, http://www.ncbi.nlm.nih.gov/biosample/?term=293T).
The dataset of plasmid pCDH-CMV supporting the conclusions of this article is available in the Addgene repository (Assession number: Plasmid # 72265, http://www.addgene.org/search/advanced/?q=pCDH-CMV).
Authors’ contributions
BZ contributed to design the experiment. YFM and XPY collected the clinical tissues samples; JF and SML contributed to the analysis and interpretation of the data. RRK, and JTJ performed the experiments and drafted the manuscript. ZQ and JZ contributed to analysis of the data. WZ contributed to revise the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Studies involving human tissue:
The informed consents were obtained from the involved patients, this study was in accordance with the Declaration of Helsinki and approved by the Human Ethics Committee of Second Affiliated Hospital of Xi’an Jiaotong University, xi’an, China.
Studies involving animals:
The animal experiment was approved by the Animal Care and Use Committee of the Second Affiliated Xi’an Jiaotong University, Xi’an, China.
Abbreviations
- Akt
Protein kinase B
- CRKL
V-crk avian sarcoma virus CT10 oncogene homolog-like
- CXCR4
c-x-c chemokine receptor type 4
- EGFL7
Epidermal growth factor-like domain 7
- Erk1/2
Extracellular regulated protein kinases 1/2
- ESCC
Esophageal squamous cell carcinoma
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GFP
Green fluorescent protein
- HRP
Horse reddish peroxidase
- LV-miR-126
Lentiviral vector-miR-126
- LV-miR-NC
Lentiviral vector-miR- negative control
- miR-126
microRNA 126
- miRNA
microRNA
- MRP1
Multi-drug resistant associate protein
- mTOR
Mechanistic Target of Rapamycin
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
- OD
Absorbance value
- PI3K
Phosphatidylinositol 3 kinase
- RhoA
Ras homolog gene family member A
- ROCK
Rho-associated protein kinase
- VEGF-A
Vascular endothelial growth factor A
Contributor Information
Ranran Kong, Email: wanglierfuyuan@163.com.
Yuefeng Ma, Email: shilipingsyy@163.com.
Jie Feng, Email: leipengsyy@163.com.
Shaomin Li, Email: xiaozhu2009@yeah.net.
Wei Zhang, Email: 1966951059@qq.com.
Jiantao Jiang, Email: zhangyurenmin@163.com.
Jin Zhang, Email: spectacularview123@163.com.
Zhe Qiao, Email: dedicatedperson@163.com.
Xiaoping Yang, Email: qibaozhu@yeah.net.
Bin Zhou, Email: zhoubineryuan@163.com.
References
- 1.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135(2):255–60. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madanecki P, Kapoor N, Bebok Z, Ochocka R, Collawn JF, Bartoszewski R. Regulation of angiogenesis by hypoxia: the role of microRNA. Cell Mol Biol Lett. 2013;18(1):47–57. doi: 10.2478/s11658-012-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebrahimi F, Gopalan V, Smith RA, Lam AK. miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol. 2014;96(1):98–107. doi: 10.1016/j.yexmp.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Connolly DT, Olander JV, Heuvelman D, Nelson R, Monsell R, Siegel N, Haymore BL, Leimgruber R, Feder J. Human vascular permeability factor. Isolation from U937 cells. J Biol Chem. 1989;264(33):20017–24. [PubMed] [Google Scholar]
- 5.Alevizakos M, Kaltsas S, Syrigos KN. The VEGF pathway in lung cancer. Cancer Chemother Pharmacol. 2013;72(6):1169–81. doi: 10.1007/s00280-013-2298-3. [DOI] [PubMed] [Google Scholar]
- 6.Arias-Pulido H, Chaher N, Gong Y, Qualls C, Vargas J, Royce M. Tumor stromal vascular endothelial growth factor A is predictive of poor outcome in inflammatory breast cancer. BMC Cancer. 2012;12(298):1471–2407. doi: 10.1186/1471-2407-12-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meister J, Schmidt MH. miR-126 and miR-126*: new players in cancer. ScientificWorldJournal. 2010;10:2090–100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59(5):579–85. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Chen X, Li P, Su L, Yu B, Cai Q, Li J, Yu Y, Liu B, Zhu Z. CRKL promotes cell proliferation in gastric cancer and is negatively regulated by miR-126. Chem Biol Interact. 2013;206(2):230–8. doi: 10.1016/j.cbi.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N, Zhou X, Chen C. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget. 2014;5(23):11873–85. doi: 10.18632/oncotarget.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, Naziri W, Marcuard SP. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6(5):281–95. [PubMed] [Google Scholar]
- 12.Diaz R, Silva J, Garcia JM, Lorenzo Y, Garcia V, Pena C, Rodriguez R, Munoz C, Garcia F, Bonilla F, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47(9):794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 13.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47(11):939–46. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen TF, Sorensen FB, Lindebjerg J, Jakobsen A. The predictive value of microRNA-126 in relation to first line treatment with capecitabine and oxaliplatin in patients with metastatic colorectal cancer. BMC Cancer. 2012;12(83):1471–2407. doi: 10.1186/1471-2407-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Li N, Wu M, Li X, Luo Z, Wang X. Expression of miR-126 suppresses migration and invasion of colon cancer cells by targeting CXCR4. Mol Cell Biochem. 2013;381(1-2):233–42. doi: 10.1007/s11010-013-1707-6. [DOI] [PubMed] [Google Scholar]
- 16.Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K, Kirita T, Kuniyasu H. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107(4):700–6. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373(4):607–12. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 19.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169–75. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X, Li H, Long L, Hui L, Chen H, Wang X, Shen H, Xu W. miR-126 enhances the sensitivity of non-small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A. Acta Biochim Biophys Sin. 2012;44(6):519–26. doi: 10.1093/abbs/gms026. [DOI] [PubMed] [Google Scholar]
- 21.Koumangoye RB, Andl T, Taubenslag KJ, Zilberman ST, Taylor CJ, Loomans HA, Andl CD. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer. 2015;14(1):24. doi: 10.1186/s12943-014-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu T, Cao R, Li S, Fu M, Ren L, Chen W, Zhu H, Zhan Q, Shi R. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer. 2015;15(1):29. doi: 10.1186/s12885-015-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Gu J, Jiang P, Zheng Y, Liu X, Jiang X, Huang E, Xiong S, Xu F, Liu G, et al. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21(4):854–63. doi: 10.1158/1078-0432.CCR-14-1740. [DOI] [PubMed] [Google Scholar]
- 24.Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ, Wang TY, Li HC, Wu XN. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett. 2013;5(5):1639–42. doi: 10.3892/ol.2013.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie YF, Shu R, Jiang SY, Liu DL, Zhang XL. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int J Oral Sci. 2011;3(3):125–34. doi: 10.4248/IJOS11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Correa AM, Hoque A, Guan B, Ye F, Huang J, Swisher SG, Wu TT, Ajani JA, Xu XC. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2011;128(1):132–43. doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351(1-2):157–64. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 28.Mulkeen AL, Silva T, Yoo PS, Schmitz JC, Uchio E, Chu E, Cha C. Short interfering RNA-mediated gene silencing of vascular endothelial growth factor: effects on cellular proliferation in colon cancer cells. Arch Surg. 2006;141(4):367–74. doi: 10.1001/archsurg.141.4.367. [DOI] [PubMed] [Google Scholar]
- 29.Petit AM, Rak J, Hung MC, Rockwell P, Goldstein N, Fendly B, Kerbel RS. Neutralizing antibodies against epidermal growth factor and ErbB-2/neu receptor tyrosine kinases down-regulate vascular endothelial growth factor production by tumor cells in vitro and in vivo: angiogenic implications for signal transduction therapy of solid tumors. Am J Pathol. 1997;151(6):1523–30. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Huang Y, Zhang J. Molecularly targeting the PI3K-Akt-mTOR pathway can sensitize cancer cells to radiotherapy and chemotherapy. Cell Mol Biol Lett. 2014;19(2):233–42. doi: 10.2478/s11658-014-0191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset of human genome DNA of miR-126 supporting the conclusions of this article is available in the Genbank (NCBI) repository (Assession number: NR_029695.1, Gene ID: 262205369, http://www.ncbi.nlm.nih.gov/nuccore/NR_029695.1).
The dataset of human genome DNA of VEGF-A supporting the conclusions of this article is available in the Genbank (NCBI) repository (Assession number: NM_001317010.1, Gene ID: 949394727, http://www.ncbi.nlm.nih.gov/nuccore/NM_001317010.1).
The dataset of OE19 cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA4186904, ID: 4729602, http://www.ncbi.nlm.nih.gov/biosample/?term=OE19).
The dataset of OE33 cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA4186711, ID: 4728790, http://www.ncbi.nlm.nih.gov/biosample/?term=OE33).
The dataset of JH-EsoAd1 cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA4446651, ID: 4776544, http://www.ncbi.nlm.nih.gov/biosample/?term=JH-EsoAd1).
The dataset of 293T cell line supporting the conclusions of this article is available in the BioSample database (Assession number: SAMEA3782549, ID: 4856025, http://www.ncbi.nlm.nih.gov/biosample/?term=293T).
The dataset of plasmid pCDH-CMV supporting the conclusions of this article is available in the Addgene repository (Assession number: Plasmid # 72265, http://www.addgene.org/search/advanced/?q=pCDH-CMV).


