Abstract
Complement factor H (CFH) is a soluble complement regulatory protein essential for the down-regulation of the alternative pathway on interaction with specific markers on the host cell surface. It recognizes the complement component 3b (C3b) and 3d (C3d) fragments in addition to self cell markers (i.e. glycosaminoglycans, sialic acid) to distinguish host cells that deserve protection from pathogens that should be eliminated. The Staphylococcus aureus surface protein serine–aspartate repeat protein E (SdrE) was previously reported to bind human CFH as an immune-evasion tactic. However, the molecular mechanism underlying SdrE–CFH-mediated immune evasion remains unknown. In the present study, we identified a novel region at CFH's C-terminus (CFH1206–1226), which binds SdrE N2 and N3 domains (SdrEN2N3) with high affinity, and determined the crystal structures of apo-SdrEN2N3 and the SdrEN2N3–CFH1206–1226 complex. Comparison of the structure of the CFH–SdrE complex with other CFH structures reveals that CFH's C-terminal tail flips from the main body to insert into the ligand-binding groove of SdrE. In addition, SdrEN2N3 adopts a ‘close’ state in the absence of CFH, which undergoes a large conformational change on CFH binding, suggesting a novel ‘close, dock, lock and latch' (CDLL) mechanism for SdrE to recognize its ligand. Our findings imply that SdrE functions as a ‘clamp' to capture CFH's C-terminal tail via a unique CDLL mechanism and sequesters CFH on the surface of S. aureus for complement evasion.
Keywords: complement factor H, crystal structure, immune evasion mechanism, ligand recognition, Staphylococcus aureus SdrE
Introduction
As the first line of immune defence for humans, the complement system plays a crucial role in pathogen recognition, destruction and elimination [1]. This powerful defence system comprises more than 30 proteins in the blood or on the cell surface, which can be activated in a cascade-dependent process via three different pathways: the classic, lectin and alternative pathways [2,3]. The three pathways converge at the generation of complement component 3 (C3) and complement component 5 (C5) convertases, which results in cleavage of C3 into fragments C3a and C3b. The C3b fragment interacts with C5 convertase to cleave C5 into fragments C5a and C5b. On formation of the convertases, anaphylatoxins (C3a/C5a), the membrane-attack complex and opsonins (C3b) are generated [3–5].
The alternative pathway is crucial for amplification of the complement cascade, because it accounts for between 80% and 90% of total complement activation [6]. To avoid damage to host tissues, complement factor H (CFH) and C3b are applied to discriminate between host and foreign cells [5,7], with C3b spontaneously deposited on the surface of all cells (host cells, as well as pathogens) exposed to the activated complement system. CFH, a complement regulator in either soluble or membrane-bound form, binds to the C3b and C3d fragments, as well as to host-cell markers [polyanions, such as glycosaminoglycans (GAGs) and sialic acid] to distinguish host cells from pathogens or altered host cells (e.g. cancer cells) and protect host cells from unintended complement-mediated injury.
CFH is composed of 20 complement-control protein (CCP) units (also known as short consensus repeats), with spacers consisting of three to eight amino acids between units. Each unit contains ∼60 amino acids, with the sequence highly conserved across units [8]. As a key regulator in the alternative pathway, CFH is abundant in serum, with a concentration of ∼500 mg/ml. However, fluctuations in serum CFH concentrations can range from 116 mg/ml to 562 mg/ml according to different environmental and genetic factors [9,10]. The important regulatory function and high concentration of CFH make it a favourable target for surface binding by pathogens, in order to disguise themselves as normal host cells and escape elimination by the complement system [11]. It has been reported that the surface-exposed lipoprotein fHbp of Neisseria meningitidis mimics the host carbohydrates to bind CFH–CCP6 [12]. The outer surface protein E (OspE) of Borrelia burgdorferi binds to CFH–CCP20 in a similar way to host cells binding CFH–CCP20 via GAGs [13]. In addition, Staphylococcus aureus surface protein SdrE, which belongs to the serine–aspartate repeat-containing protein (Sdr) subfamily within the microbial surface component recognizing adhesive matrix molecule (MSCRAMM) family, was recently reported to bind CFH as an immune-evasion tactic [14]. Sdr subfamily members adopt a similar structural pattern, including an N-terminal signal peptide (S), a functional ligand-binding region (A), a B region, a Ser–Asp repeated R region, a C-terminal cell wall membrane-spanning region (W), a hydrophobic membrane region (M) and a cytoplasmic tail (C). The N2 and N3 domains in the A region form IgG-like folds that bind ligands [15]. Although the fibrinogen (Fg)-bound structures of the N2–N3 domains of other MSCRAMM proteins, including SdrG [16], branch-point-binding protein (Bbp) [17], clumping factor A (ClfA) [18] and B (ClfB) [19], have been reported, little is known about how the newly identified ligand CFH is recognized by MSCRAMM family proteins. Furthermore, the molecular basis for SdrE–CFH recognition and SdrE–CFH-mediated immune evasion still remains to be elucidated.
In the present study, we mapped the minimal binding region [amino acids 1206–1226 (CFH1206–1226)] in CFH–CCP20 for interaction with SdrEN2N3, and solved the structures of apo-SdrEN2N3 (PDB code: 5WTA) and the SdrEN2N3–CFH1206–1226 complex (PDB: 5WTB). Structural analysis and comparison with other MSCRAMM proteins revealed a novel closed state of SdrE, wherein the ligand-binding groove formed by the N2 and N3 domains is occupied by LoopA–B in the absence of ligand. To capture CFH's C-terminal tail, LoopA–B of SdrEN2N3 rotates ∼180° to facilitate CFH binding. This indicates that SdrE recognizes its ligands by using a unique ‘close, dock, lock and latch' (CDLL) mechanism observed in the MSCRAMM protein family for the first time. Compared with the structures of CFH in complex with C3b, fHbp or OspE, a large conformational change of the C-terminal tail of CFH was observed on SdrE binding, demonstrating a novel recognition mechanism between CFH and its ligands. All these findings illustrate that SdrE binds to a unique region in CFH's C-terminal region with a high affinity for capturing CFH for complement evasion. Our findings suggest that SdrE functions as a ‘clamp' to capture the CFH C-terminal tail via a novel ‘CDLL' mechanism and sequesters CFH on the surface of S. aureus to evade the human immune system.
Experimental
Cloning, expression and purification
DNA fragments encoding residues 270–599 of wild-type SdrE or its mutants were obtained by PCR and ligated to pET-22b(+) (Novagen) vector with a C-terminal 6×His tag. The construct was transformed into Escherichia coli BL21 (DE3) strain and grown with shaking overnight at 37°C in a 20-ml starter culture of Luria–Bertani (LB) medium containing 100 µg/ml of ampicillin. The overnight starter culture was then transferred into 1 l of LB medium and incubated at 37°C with shaking until reaching an absorbance at 600 nm of ∼0.6. The culture was then induced with 0.4 mM IPTG and incubated at 16°C for 18–24 h. The cells were harvested by centrifugation (277 K, 6000g, 8 min) and suspended in lysis buffer [50 mM Tris-HCl, pH 7.8, 500 mM NaCl and 5%(v/v) glycerol]. The cells were then homogenized by sonication and the lysate was centrifuged for 30 min at 12 000g and 277 K. The supernatant was loaded onto a Ni/NTA nickel-chelating column (Qiagen) pre-equilibrated with lysis buffer. The column was washed by approximately 20 column volumes of lysis buffer with 40 mM imidazole to remove contaminants. The eluted target protein was concentrated by centrifugal ultrafiltration (Millipore, 10-kDa cut-off) and further purified using a Superdex 75 16/60 size exclusion column (GE Healthcare) equilibrated with buffer consisting of 20 mM Tris-HCl, pH 7.8, 400 mM NaCl, 5% (v/v) glycerol and 1 mM DTT. Various truncated fragments and mutants of human CFH were amplified by PCR and cloned to the pGEX-6P-1 vector. The recombinant proteins were expressed in E. coli BL21 (DE3) strain and purified by GST affinity chromatography.
Crystallization and data collection
The apo-SdrEN2N3 protein was concentrated to 20 mg/ml in 20 mM Tris-HCl, pH 7.8, 400 mM NaCl, 5% (v/v) glycerol and 1 mM DTT. The synthesized CFH1206–1226 peptide and SdrEN2N3 were mixed at a molar ratio of 5:1 and incubated at 277 K overnight. Initial crystallization screening was carried out at 298 K using a sitting-drop, vapour-diffusion method with commercial screen kits from Hampton Research (Crystal Screen, Crystal Screen 2, SaltRx 1, SaltRx 2, PEGRx 1, PEGRx 2 and Index). The apo-SdrEN2N3 crystals were grown in 0.1 M citric acid, pH 3.5, 16% (w/v) PEG-3350 and 2% (v/v) 2-methyl-2,4-pentanediol (MPD). The complex crystals were obtained from 0.1 M Hepes, pH 7.5, 0.2 M magnesium chloride and 30% (v/v) PEG-400. X-ray diffraction data for apo-SdrEN2N3 and the SdrEN2N3–CFH1206–1226 complex were collected at 100 K at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1 and BL18U1, respectively. Diffraction data were indexed, integrated and scaled using the programs iMOSFLM [20], POINTLESS [21] and SCALA [21] in the CCP4i suite [22].
Structure determination, refinement and analysis
The crystal structure of apo-SdrEN2N3 was determined by molecular replacement using Phaser [23] in the CCP4i suite with SdrGN2N3 (PDB: 1R17) as the search model. After several runs of structure refinement using the programs REFMAC5 [24] and COOT [25], the final model was refined to 2.3 Å (1 Å = 0.1 nm) resolution with an Rwork of 19.4% and an Rfree of 25.2%. The SdrEN2N3–CFH1206–1226 complex structure was solved by molecular replacement using apo-SdrEN2N3 as the search model. The final model was refined to 3.3 Å resolution with an Rwork of 22.2% and an Rfree of 28.1%. The quality of the final models was analysed using the program MolProbity [26]. Data collection and model refinement statistics are shown in Table 1.
Table 1. Data collection and structure refinement statistics.
| Apo-SdrEN2N3 | SdrEN2N3–CFH1206–1226 | |
|---|---|---|
| Data collection | ||
| Space group | P1 | P212121 |
| PDB ID | 5WTA | 5WTB |
| Unit cell parameters | ||
| a, b, c (Å) | 41.81, 61.73, 139.61 | 117.50, 117.50, 154.39 |
| α, β, γ (°) | 80.89, 89.83, 73.34 | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 40.01–2.30 (2.42–2.30)1 | 49.75–3.30 (3.48–3.30) |
| Wavelength (Å) | 0.9792 | 0.9779 |
| Unique reflections | 55 992 | 31 291 |
| Completeness (%) | 95.5 (95.2) | 96.1 (97.9) |
| Overall I/σ(I) | 7.8 (3.4) | 7.0 (1.9) |
| Multiplicity | 2.2 (2.3) | 2.8 (2.8) |
| Rsym (%)2 | 8.1 (26.4) | 10.4 (56.1) |
| Refinement | ||
| Resolution range (Å) | 40.00–2.30 | 49.75–3.30 |
| Rwork3 | 0.194 | 0.222 |
| Rfree4 | 0.252 | 0.281 |
| RMSD bond lengths (Å) | 0.013 | 0.010 |
| RMSD bond angles | 1.408 | 1.371 |
| Average B-factors (Å2) | ||
| Protein | 22.2 | 89.5 |
| Water | 26.4 | |
| Ramachandran plot5 | ||
| Most favoured regions (%) | 97.1 | 90.4 |
| Allowed regions (%) | 2.9 | 9.6 |
| Generously allowed regions (%) | 0.0 | 0.0 |
| MolProbity | ||
| Clash score6/percentile | 8.89/96 | 24.63/89 |
| Overall score/percentile | 2.20/81 | 3.31/80 |
The values in parentheses refer to statistics in the highest shell.
Rsym = |Ii−|/|Ii| where Ii is the intensity of the ith measurement, and is the mean intensity for that reflection.
Rwork = Σhkl||Fobs| − k|Fcal| |/Σhkl |Fobs| where Fobs and Fcal are observed and calculated structure factors, respectively, calculated over all reflections used in the refinement.
Rfree, is similar to Rwork but calculated over a subset of reflections (5%) excluded from all stages of refinement.
Statistics for the Ramachandran plot from an analysis using MolProbity.
The MolProbity clash score indicates the number of steric overlaps >0.4 Å per 1000 atoms.
GST pull-down assay
Various truncated fragments and mutants of human CFH were overexpressed in E. coli BL21 (DE3) cells. The cells were suspended in lysis buffer [50 mM Tris-HCl, pH 7.8, 400 mM NaCl, 5% (v/v) glycerol and 1 mM DTT] and then homogenized by sonication. After centrifugation of the lysates, the supernatant was incubated with 30 µl of GST beads (pre-equilibrated in lysis buffer) at 4°C for 1 h. The beads were washed with 1 ml of lysis buffer three times to remove the impurities. Then GST beads immobilized with GST–CFH were incubated with purified wild-type SdrEN2N3 or its mutant at 4°C for 1 h. The beads were washed with 1 ml of lysis buffer five times to remove proteins that were non-specific binding. Beads were boiled with SDS/sample buffer and proteins retained on the GST beads were analysed using SDS/PAGE (Figure 1B–D and see Figure 3B and D).
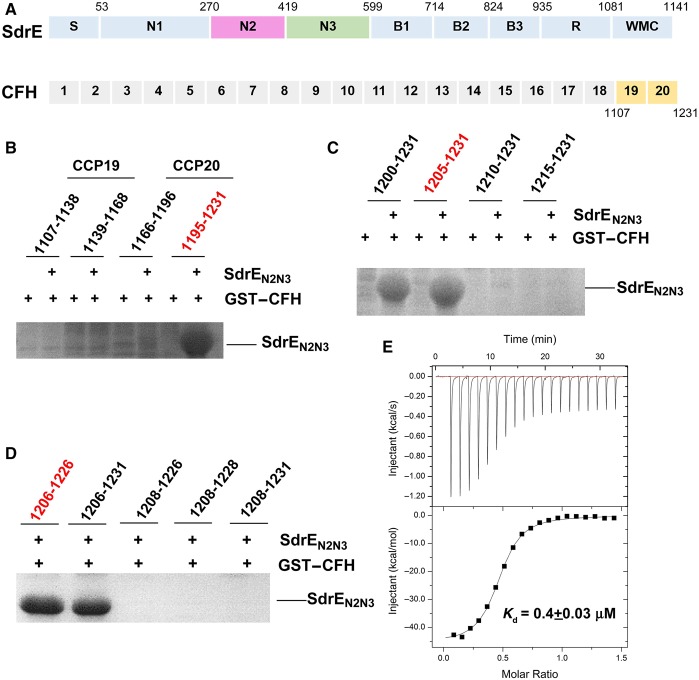
Figure 1. CFH–CCP20 is responsible for SdrEN2N3 binding.
(A) Schematic representation of SdrE and CFH domain organization. S, signal sequence; N1–N3, ligand-binding region; B1–B3, B repeats region; R, serine–aspartate repeat region; W, wall-spanning region; M, membrane-spanning region; C, cytoplasmic, positively charged tail. The amino acid residue number identifying the boundary between each subdomain is indicated above. (B–D) GST pull-down assays of SdrEN2N3 with different fragments of the CFH fused to a GST tag. The retaining amino acid residues of each CFH fragment are indicated in each panel. (E) The ITC fitting curves of GST–CFH1206–1226 to SdrEN2N3.
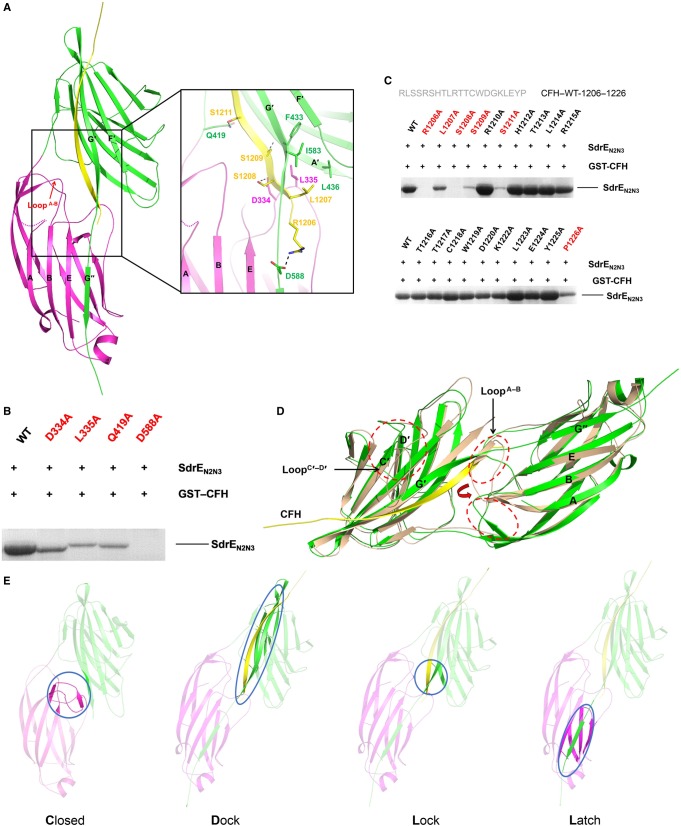
Figure 3. Crystal structure of the SdrEN2N3–CFH1206–1226 complex.
(A) Cartoon representation of the SdrEN2N3–CFH1206–1226 complex. N2 and N3 subdomains from the SdrE and the CFH peptide are coloured purple, green and yellow, respectively. LoopA–B is indicated by a red arrow. Key residues involved in the side-chain interactions between SdrE and CFH are shown as sticks in the zoom-in view. Hydrogen bonds involved in the SdrE–CFH interaction are shown as black dashes. (B) GST pull-down assays of GST–CFH1206–1226 with SdrEN2N3 and its mutants. (C) Identification of the key residues on CFH–CCP20 for SdrE binding. Key residues are coloured red. (D) Structural comparison of apo-SdrEN2N3 (wheat) and the SdrEN2N3–CFH1206–1226 complex (green). LoopA–B and LoopC′–D′ are circled by red dashes and labelled in black. The change of the two conformations of LoopA–B is indicated by a red arrow. (E) A CDLL model for the SdrE–CFH interaction is shown as a cartoon.
Isothermal titration calorimetry
The interaction between SdrEN2N3 protein and GST–CFH1206–1226 was analysed using isothermal titration calorimetry (ITC) with a MicroCal iTC200 instrument (GE Healthcare) at 20°C in 50 mM Tris, pH 7.8, 400 mM NaCl and 0.5 mM tris(2-carboxyl)phosphine (TCEP). Experimental data were fitted to a single binding-site model and analysed using the ITC data analysis module of Origin 7.0 (MicroCal) provided by the manufacturer (Figure 1E).
Results and discussion
The C-terminal tail of CFH–CCP20 binds to SdrEN2N3 with high affinity
The CCP19–20 region of CFH was reported to be capable of discriminating between host and complement-activating cells [27–31]. It is essential for binding the C3d part of C3b, and for the self cell markers such as GAGs and sialic acids. Some pathogen-secreted proteins, including the PspC [32] protein from Streptococcus pneumoniae and OspE [13] from B. burgdorferi, were also reported to recruit CFH through their interaction with the CCP19–20 units. SdrE is a newly identified CFH-binding protein that belongs to MSCRAMM family, which normally uses the extracellular domains N2 and N3 to bind ligands. Therefore, we hypothesized that SdrEN2N3 recruits CFH via interactions with CFH–CCP19–20 (Figure 1A).
To test our hypothesis, we constructed four CFH fragments (CFH1107–1138, CFH1139–1168, CFH1166–1196 and CFH1195–1231) within CCP19–20 fused to a GST tag at the N-terminus. In vitro GST pull-down assays confirm that the C-terminal 37 amino acids of CFH–CCP20 (CFH1195–1231) are capable of interacting with SdrEN2N3 (Figure 1B). To identify the shortest region in CFH–CCP20 responsible for SdrEN2N3 binding, shorter CFH fragments were used for GST pull-down assays. As shown in Figure 1(C and D), a minimal segment containing 21 amino acid residues (CFH1206–1226) is sufficient for SdrEN2N3 interaction.
Kajander et al. [33] reported that the CCP19–20 region binds the C3d fragment primarily through the CCP20 site, but binds C3b through the CCP19 site, and proposed a comprehensive molecular mechanism for target discrimination mediated by CFH in the alternative pathway. On host-cell surfaces, CFH binds to the C3b site and cell surface markers through CCP19 and CCP20 sites, respectively. This causes the rapid down-regulation of the alternative pathway and converts C3b to C3d on host-cell surfaces, resulting in additional CFH proteins recruited to the surface. While localized on the surfaces of pathogens lacking host-cell markers, CCP19–20 binds to C3b with lower affinity, subsequently activating the alternative pathway and initiating pathogen elimination [33,34]. To evade complement-mediated destruction, some pathogens use surface proteins to sequester CFH to their cell surface. After confirming that the CFH–CCP20 domain is recognized by SdrEN2N3, we determined the binding affinity of CCP20 to SdrEN2N3, revealing that SdrEN2N3 strongly interacts with CFH1206–1226, with a dissociation constant (Kd) value of ∼0.4 µΜ (Figure 1E), which is as high as that measured for the CCP20–C3d interaction [33]. These findings indicate that the relatively strong binding of SdrE to CCP20 promotes efficient recruitment of CFH to the cell surface of S. aureus for complement disguise.
A novel ‘close' state of apo-SdrEN2N3 with the ligand-binding groove occupied by LoopA–B
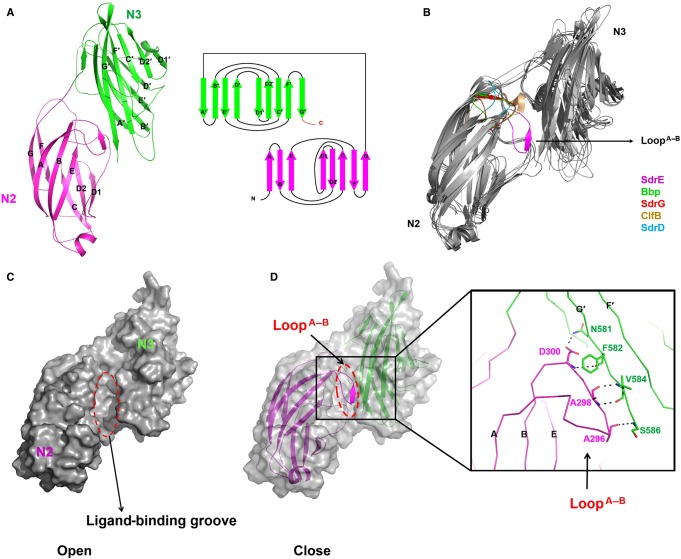
To investigate the molecular basis for CFH recognition by SdrE, we solved the crystal structures of apo-SdrEN2N3 and SdrEN2N3 in complex with CFH1206–1226. SdrEN2N3 exists as a monomer in both solution and its crystal form (see Supplementary Figure S1). Four apo-SdrEN2N3 monomers were found in one asymmetrical unit. Each subunit consists of two distinct domains: N2 (residues 270–419) and N3 (residues 420–586) (Figure 2A), packing against each other and separated by a short linker region. The N2 and N3 domains contain eight (A↑B↓C↑D1↑D2↓E↑F↓G↑) and nine (A′↓B′↑C′↓D′↑D1′↓D2′↑E′↓F′↑G′↓) antiparallel β-strands, respectively. The topologies of N2 and N3 domains are structurally similar, with both resembling the IgG-like fold, except that N2 lacks the D-strand (Figure 2A). Structural comparison with other reported MSCRAMM proteins reveals similar overall structures between SdrEN2N3 and Bbp (PDB: 5CF3, RMSD of 2.7 Å over 304 Cα atoms), SdrG (PDB: 1R19, RMSD of 2.6 Å over 276 Cα atoms), ClfB (PDB: 4F24, RMSD of 3.4 Å over 237 Cα atoms) and SdrD (PDB: 4JE0, RMSD of 4.7 Å over 229 Cα atoms), but a novel ‘close' state with the ligand-binding groove occupied by LoopA–B (Figure 2B). The LoopA–B in SdrE homologues either turns around to interact with the N2–N3 linker and G-strand in Bbp, SdrG and ClfB, or locates by the side of the groove as a loop in SdrD, to leave the groove in an ‘open' state for ligand binding. However, the LoopA–B in SdrE extends from the N2 domain to form a pair of antiparallel β-sheets with the G′-strand in the N3 domain. The β-sheet is stabilized by four pairs of main-chain hydrogen bonds, resulting in the ligand-binding groove of SdrEN2N3 being locked in a ‘close' state (Figure 2C and D).
Figure 2. Crystal structure of apo-SdrEN2N3.
(A) Cartoon representation of the structure of apo-SdrEN2N3 and a schematic representation of the topology of the SdrEN2N3 fold. The N2 and N3 domains are shown in purple and green, respectively. (B) Structural comparison of apo-SdrEN2N3 with its homologous structures; LoopA–B is coloured purple for SdrE, green for Bbp, red for SdrG, wheat for ClfB and blue for SdrD. (C) Surface representation of the ‘open' state of apo-SdrEN2N3 with the LoopA–B deleted from the structure. (D) Cartoon representation of the ‘close' state of apo-SdrEN2N3. The N2 and N3 domains are shown in purple and green, respectively. The ligand-binding groove is highlighted by a dashed red ellipse. Key residues that stabilize the ‘close' state are shown as sticks.
It is not the first time that the ‘close' conformation of MSCRAMM proteins has been mentioned. A ‘dock, lock and latch' (DLL) model was proposed for the SdrG based on its apo- and Fg-peptide-bound structures [16]. In this model, the Fg peptide docks into the ‘open'-form groove first. Then the C-terminal tail of N3 extends forward to ‘lock' the peptide in place and an additional G′′-strand is formed to ‘latch' on to the peptide–SdrG complex by interacting with the N2 domain. A ‘close' conformation of SdrG, which was generated by introducing a disulfide bond between the latch and the N2 domain, cannot bind Fg, demonstrating that the ‘open' state of the ligand-binding groove of SdrG is required for the initial docking of the ligand [16]. However, a ClfA mutant, exhibiting an artificially closed binding groove through the introduction of a disulfide bond in a similar manner, retains the ability to bind Fg with lower affinity compared with that observed in wild-type proteins. Therefore, a different ligand-binding mechanism, the ‘latch, dock' (LD) model, was proposed for ClfA [18]. Although this ‘close' conformation was proposed, up until now no structural or other evidence was reported to clearly confirm this state of MSCRAMM proteins. The ‘close' state observed in the SdrE structure, in contrast to the previously proposed ‘close' state artificially formed by the ‘latch' of the N3 domain, is formed by LoopA–B protruding into the N2 domain to occupy the ligand-binding groove. This novel ‘close' state suggests a new ligand-binding mechanism for SdrE.
CFH1206–1226 is recognized by SdrEN2N3 via a CDLL mechanism
For further investigation, we solved the crystal structure of the SdrEN2N3–CFH1206–1226 complex. One SdrEN2N3 molecule and one CFH1206–1226 molecule form a heterodimer. Four SdrEN2N3–CFH1206–1226 heterodimers, named heterodimer A–D, are found in one asymmetrical unit (see Supplementary Figure S2). Two SdrEN2N3–CFH1206–1226 heterodimers form a tetramer via interaction of the C-terminal of four amino acids with the two CFH peptides (see Supplementary Figure S2). Gel filtration and PISA (proteins, interfaces, structures and assemblies) analysis clarify that the SdrEN2N3–CFH1206–1226 complex exists as a heterodimer in both solution and crystal form, respectively (see Supplementary Figure S2). Therefore, the tetrameric conformation associated with the SdrEN2N3–CFH1206–1226 structure was caused by crystal packing. The overall structures of the four SdrEN2N3 molecules are almost identical, with an average RMSD of 0.3 Å over 330 aligned Cα atoms. To simplify the description, we discuss only heterodimer A. As shown in Figure 3A, the CFH peptide forms a long β-strand and threads the ligand-binding groove between the N2 and N3 subdomains. Apart from four C-terminal residues (1223–1226), a well-defined electron density map is observed for the main chain of all other residues (1206–1222) and the side chain of some residues (see Supplementary Figure S3). Similar to Fg-bound Bbp, SdrG, ClfB and ClfA structures [16–19] (see Supplementary Figure S4), the CFH peptide forms an antiparallel β-sheet with G′-strand in N3, whereas the C-terminal G′-strand undergoes a rotation to surround the N-terminus of the CFH peptide. The C-terminal residues (587–598) missing in the apo-SdrEN2N3 structure are ordered as a G″-strand on CFH binding in the complex structure, and extend to the N2 domain to compose complete β-sheets together with strands A, B and E (Figure 3A). The buried surface between SdrEN2N3 and CFH is considerably larger, approximately 1100 Å2. A total of 13 pairs of antiparallel, main-chain hydrogen bonds are formed by SdrEN2N3 and the CFH peptide (see Supplementary Figure S5). All residues in SdrEN2N3 involved in the CFH binding belong to the G′-strand (572–586), demonstrating a critical role of the G′-strand in the SdrEN2N3–CFH interaction. In addition, side-chain interactions between SdrEN2N3 and CFH include four hydrogen bonds and one hydrophobic interaction. The carbonyl oxygen of Asp334 and Asp588 from SdrEN2N3 form hydrogen bonds with Ser1208 and Arg1206 from CFH, respectively. The main-chain nitrogen of SdrEN2N3F433 forms the hydrogen bond with the side-chain oxygen of CFHS1209. The side-chain nitrogen of SdrEN2N3Q419 contributes another hydrogen bond via interaction with the hydroxyl group of CFHS1211. A hydrophobic pocket created by Leu335, Leu436 and Ile583 from SdrEN2N3 mediates the hydrophobic interaction with the side chain of CFHL1207 (Figure 3A).
To verify our complex structure, SdrEN2N3 residues Asp334, Leu335, Gln419 and Asp588, which interact with CFH through side chains, were mutated to alanine, and their interaction with CFH1206–1226 was evaluated by GST pull-down assays. As shown in Figure 3B, the mutations D334A, L335A and Q419A significantly weaken the interaction between SdrEN2N3 and CFH, whereas the mutation D588A absolutely abolishes the interaction. Consistent with the structural analysis, our mutagenesis studies confirm the important roles of Asp334, Leu335, Gln419 and Asp588 from SdrE in the SdrEN2N3–CFH interaction. To identify residues of CFH1206–1226 that are important for binding SdrEN2N3, each residue within CFH1206–1226 (except for alanine and glycine) was sequentially substituted by alanine (Figure 3C). The binding ability of each mutant for SdrEN2N3 was tested in vitro by GST pull-down assays. Remarkably, the mutations R1206A and S1208A absolutely abolish the interaction of CFH with SdrEN2N3. Interaction between CFH and SdrEN2N3 is significantly weakened by the mutations S1209A and S1211A, whereas mutation L1207A results in the loss of half the binding capacity of CFH with SdrEN2N3. Therefore, Arg1206, Leu1207, Ser1208, Ser1209 and Ser1211 are the key residues of CFH for SdrEN2N3 binding.
To investigate the ligand-binding mechanism associated with the SdrEN2N3–CFH interaction, we compared the apo-SdrEN2N3 structure with that of the CFH1206–1226-bound form. The overall structure of SdrEN2N3 in either apo- or CFH1206–1226-bound form is similar, with an RMSD of 2.8 Å over 293 aligned Cα atoms. Several significant conformational changes caused by the interaction of the CFH peptide are observed (Figure 3D). First, LoopA–B rotates ∼180° away from the ligand-binding groove to interact with the N2–N3 linker and the G-strand, which converts the ‘close’ groove to the ‘open' conformation for CFH binding. Second, residues from Val468 to Leu476 from LoopC′–D′ become disordered after the ‘dock' procedure with the CFH peptide. This region of LoopC′–D′ localizes in close proximity to the CFH-binding site in the apo-SdrEN2N3 structure; however, after CFH binding, this region could be dislocated and become disordered. The third and most significant difference is the formation of the ‘lock' and ‘latch' structures in the SdrEN2N3–CFH complex. The C-terminus of the G′-strand bends around the N-terminal region of the CFH peptide to ‘lock’ the ligand in place, and extends further to form an additional G′′-strand to create β-sheets with β-strands in the N2 domain, thereby functioning as a ‘latch'. The novel ‘close' state of apo-SdrEN2N3 and the structural comparison of apo-SdrEN2N3 and CFH1206–1226-bound SdrEN2N3 indicate a novel ligand-binding mechanism as a ‘CDLL' model among MSCRAMM proteins (Figure 3E). In addition, the ‘close' conformation implicates potential regulatory mechanisms associated with SdrE by unknown cellular components.
SdrE functions as a ‘clamp' to capture the C-terminal tail of the CFH for complement evasion
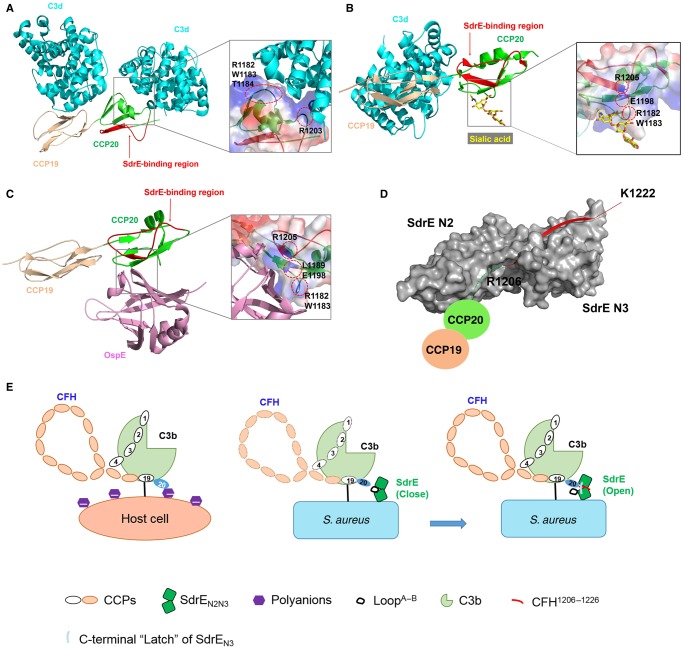
CFH functions as a host-recognition molecule and complement regulator in the alternative pathway. Normal recognition of host cells by the CFH requires the interaction of different CFH–CCP complexes with C3b and markers on the cell surfaces. CCP1–4 and CCP19–20 represent binding sites for C3b [4,35]. The crystal structure of the CCP19–20–C3d complex revealed that C3d binds to both CCP19 and -20 [33] (Figure 4A). CCP6 and CCP20 are capable of binding heparin and other polyanions, including GAGs [36,37], and CCP20 was also reported to be a sialic acid-binding site [38] (Figure 4B). Among all 20 CCP units, the C3b and host cell marker-binding sites on CCP19–20 play key roles in host recognition/discrimination and complement regulation. The dual interaction of CFH with C3b via the CCP19 site and with cell surface polyanions (heparin, GAGs, sialic acid) via the CCP20 site ensures optimal binding between the CFH and the host-cell surface. This results in rapid down-regulation of the alternative pathway and recruitment of additional CFH proteins to the surface to protect host cells. In contrast, microbes that lack such polyanions have far fewer CFH proteins located on their surfaces and are thereby susceptible to subsequent complement-mediated destruction. To escape elimination by the alternative pathway, some pathogens have developed novel strategies to recruit CFH to their surfaces, thus disguising themselves as normal host cells and evading immune attack. N. meningitidis subverts immune responses by using the surface protein fHbp to mimic host carbohydrates to recruit CFH [12]. Structural analysis combined with biochemical studies demonstrates that fHbp interacts with CFH–CCP6. The interaction interface between fHbp and CFH–CCP6 overlaps with the GAG-binding site on CFH–CCP6. Another outer surface protein, OspE of B. burgdorferi, was recently reported to bind to the same site as GAGs and sialic acid in CFH–CCP20 (Figure 4B and C) [13]. However, the binding site for SdrE on the CFH is unique (Figure 4D). In the present study, we identified Arg1206, Leu1207, Ser1208, Ser1209 and Ser1211 of the CFH as the key residues required for SdrEN2N3 binding (see Figure 3C). These differ from the residues in CFH–CCP20 identified for C3d binding (1182, 1183, 1184 and 1203) [33], binding of the cell markers heparin, GAGs and sialic acid (1182, 1183, 1186, 1188, 1189, 1198, 1215 and 1230) [38,39], and binding to the pathogen surface protein OspE (1182, 1183, 1189, 1198 and 1215) (Figure 4A–D) [13]. Although SdrE recruits CFH by interacting with CCP20, which represents a common binding site for surface proteins from different pathogens, our structural and biochemical analysis demonstrated that the interaction region in CCP20 and the CFH-binding mechanism associated with SdrE were unique (Figure 4A–D).
Figure 4. Structural comparison of the SdrEN2N3–CFH1206–1226 complex with C3d/sialic acid/OspE–CFH complexes.
(A–D) Cartoon representation of crystal structures of CFH–CCP19–20 complexed with (A) C3d (cyan), (B) sialic acid (yellow), (C) OspE (pink) and (D) SdrEN2N3 (grey). CCP19 and CCP20 are coloured wheat and green, respectively. The SdrE-binding region on CCP20 is coloured and labelled red. Binding sites on CCP20 for C3d, sialic acid and OspE are shown in zoom-in windows, with the key residues on CCP20 labelled in black and highlighted by dashed red circles. (E) Proposed model for the CFH–SdrE-mediated immune evasion mechanism.
Structural comparison of CFH peptide bound to SdrEN2N3 with CFH–CCP20 complexed with C3d, sialic acid or OspE reveals a big conformational change of the C-terminal tail of CCP20 on SdrE binding (Figure 4A–D). CCP20 interacts with C3d and sialic acid mainly through electrostatic interactions, mediated by its positively charged surface regions. The B. burgdorferi surface protein OspE also uses a similar mechanism to bind the same region on CCP20 where sialic acid binds. Unique among these CFH ligands, SdrE's structural features make it suitable to function as a ‘clamp' for capturing CFH's C-terminal tail. As shown in the model in Figure 4(E), SdrE adopts a ‘close' conformation after integration on to the S. aureus surface during infection. When the host complement component C3b is deposited on the S. aureus surface, CFH binds to C3b through CCP1–4 and CCP19. Simultaneously, LoopA–B of SdrEN2 rotates ∼180° to convert the ‘close' conformation to an ‘open' one to accommodate the C-terminal tail of CFH–CCP20. By undergoing a big conformational change, CCP20's C-terminal tail protrudes from the CFH to dock into the ligand-binding groove of SdrEN2N3 by forming an antiparallel β-sheet with the G′-strand of SdrEN3. In addition, SdrEN2N3 functions as a ‘clamp' to strongly stabilize the CFH–SdrE complex by locking and latching the CFH tail in its ligand-binding groove (see Figures 3 and 4C). Given that SdrE binds to CFH with a high affinity and functions as a ‘clamp' to capture CFH's C-terminal tail, SdrE efficiently recruits CFH proteins to S. aureus surfaces, followed by S. aureus recruitment of factor I using CFH–CCP1–4 and its surface protein ClfA to cleave C3b into iC3b [40]. In addition, to decrease C3b deposition, CFH also accelerates the decay of already formed C3 convertase C3bBb [41], thereby limiting the amplification of the complement pathway. By using an SdrE–CFH-mediated immune evasion strategy, S. aureus is capable of successfully disguising itself as a host cell to evade host complement attack.
Conclusion
Many pathogens have developed immune-evasion strategies by recruiting CFH to their surfaces to disguise themselves as normal host cells, thus escaping elimination by the complement pathway. In the present study, we identified the minimal fragment of CFH responsible for interaction with the S. aureus surface protein SdrE and determined the crystal structures of apo-SdrEN2N3 and the SdrEN2N3–CFH1206–1226 complex. Structural analysis and biochemical studies demonstrate that SdrE binds to a unique region of CFH–CCP20 relative to the binding sites on CCP20 for other CFH ligands, including host-cell markers and pathogenic virulence factors. In contrast to reported molecular mechanisms associated with CFH-mediated immune evasion, SdrE recognizes CFH–CCP20 with a CDLL mechanism and functions as a ‘clamp' to capture CFH's C-terminal tail for complement evasion. Our results not only provide insights into the molecular mechanism of S. aureus’s complement disguise, but also shed light on the development of new therapeutics for the increasingly serious infections of S. aureus.
Acknowledgements
We thank the staff at beamline BL17U1 and BL18U1 of the Shanghai Synchrotron Radiation Facility for assistance with data collection.
Abbreviations
- C3
complement component 3
- C5
complement component 5
- CCP
complement-control protein
- CFH
complement factor H
- Fg
fibrinogen
- GAG
glycosaminoglycan
- ITC
isothermal titration calorimetry
- LB
Luria–Bertani
- MSCRAMM
microbial surface component recognizing adhesive matrix molecule
- OspE
outer surface protein E
- SdrE
serine–aspartate repeat protein
Author Contribution
J. Zang, X. Zhang and M. Zhang provided the scientific direction and overall experimental design for the studies, Y. Zhang and T. Hang designed and performed the biochemical experiments, M. Wu was responsible for the crystal structure studies, and M. Wu and X. Zhang wrote the manuscript.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences [Grant No. XDB 08010101], the National Key Research and Development Program of China [Grant no. 2016YFA0400903] and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China [Grant no. 31621002]. This work was also supported by the National Natural Science Foundation of China [Grant Nos. U1532109, 31370756 and 31361163002], and the Scientific Research Grant of Hefei Science Centre of CAS [Grant Nos. 2015SRG-HSC043 and 2015HSC-UP019] to J. Zhang. This work was also supported by the Anhui Provincial Natural Science Foundation [Grant no. 1608085QC52] for X. Zhang and the National Natural Science Foundation of China [Grant no. 31400627] for M. Wu.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Janeway C.A. Jr and Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 doi: 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- 2.Zipfel P.F. (2009) Complement and immune defense: from innate immunity to human diseases. Immunol. Lett. 126, 1–7 doi: 10.1016/j.imlet.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 3.Carroll M.C. (1998) The role of complement and complement receptors in induction and regulation of immunity. Annu. Rev. Immunol. 16, 545–568 doi: 10.1146/annurev.immunol.16.1.545 [DOI] [PubMed] [Google Scholar]
- 4.Ferreira V.P., Pangburn M.K. and Cortés C. (2010) Complement control protein factor H: the good, the bad, and the inadequate. Mol. Immunol. 47, 2187–2197 doi: 10.1016/j.molimm.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricklin D., Hajishengallis G., Yang K. and Lambris J.D. (2010) Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11, 785–797 doi: 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harboe M. and Mollnes T.E. (2008) The alternative complement pathway revisited. J. Cell. Mol. Med. 12, 1074–1084 doi: 10.1111/j.1582-4934.2008.00350.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J., Wu Y.-Q., Ricklin D., Janssen B.J.C., Lambris J.D. and Gros P. (2009) Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat. Immunol. 10, 728–733 doi: 10.1038/ni.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kristensen T. and Tack B.F. (1986) Murine protein-H ss comprised of 20 repeating units, 61 amino-acids in length. Proc. Natl. Acad. Sci. U.S.A. 83, 3963–3967 doi: 10.1073/pnas.83.11.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esparza-Gordillo J., Soria J.M., Buil A., Almasy L., Blangero J., Fontcuberta J. et al. (2004) Genetic and environmental factors influencing the human factor H plasma levels. Immunogenetics 56, 77–82 doi: 10.1007/s00251-004-0660-7 [DOI] [PubMed] [Google Scholar]
- 10.de Córdoba S.R. and de Jorge E.G. (2008) Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin. Exp. Immunol. 151, 1–13 doi: 10.1111/j.1365-2249.2007.03552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zipfel P.F., Würzner R. and Skerka C. (2007) Complement evasion of pathogens: common strategies are shared by diverse organisms. Mol. Immunol. 44, 3850–3857 doi: 10.1016/j.molimm.2007.06.149 [DOI] [PubMed] [Google Scholar]
- 12.Schneider M.C., Prosser B.E., Caesar J.J.E., Kugelberg E., Li S., Zhang Q. et al. (2009) Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature 458, 890–893 doi: 10.1038/nature07769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharjee A., Oeemig J.S., Kolodziejczyk R., Meri T., Kajander T., Lehtinen M.J. et al. (2013) Structural basis for complement evasion by Lyme disease pathogen Borrelia burgdorferi. J. Biol. Chem. 288, 18685–18695 doi: 10.1074/jbc.M113.459040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp J.A., Echague C.G., Hair P.S., Ward M.D., Nyalwidhe J.O., Geoghegan J.A. et al. (2012) Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS ONE 7, e38407 doi: 10.1371/journal.pone.0038407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster T.J., Geoghegan J.A., Ganesh V.K. and Höök M. (2014) Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 doi: 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponnuraj K., Bowden M.G., Davis S., Gurusiddappa S., Moore D., Choe D. et al. (2003) A ‘dock, lock, and latch’ structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115, 217–228 doi: 10.1016/S0092-8674(03)00809-2 [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Wu M., Zhuo W., Gu J., Zhang S., Ge J. et al. (2015) Crystal structures of Bbp from Staphylococcus aureus reveal the ligand binding mechanism with fibrinogen α. Protein Cell 6, 757–766 doi: 10.1007/s13238-015-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesh V.K., Rivera J.J., Smeds E., Ko Y.-P., Bowden M.G., Wann E.R. et al. (2008) A structural model of the Staphylococcus aureus ClfA–fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 4, e1000226 doi: 10.1371/journal.ppat.1000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang H., Feng Y., Wang J., Liu B., Chen Y., Liu L. et al. (2012) Crystal structures reveal the multi-ligand binding mechanism of Staphylococcus aureus ClfB. PLoS Pathog. 8, e1002751 doi: 10.1371/journal.ppat.1002751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R. and Leslie A.G.W. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 doi: 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 doi: 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project, N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 doi: 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 23.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C. and Read R.J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 doi: 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov G.N., Vagin A.A. and Dodson E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 doi: 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 25.Emsley P. and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 26.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X. et al. (2007) Molprobity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 doi: 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jokiranta T.S., Cheng Z.-Z., Seeberger H., Jòzsi M., Heinen S., Noris M. et al. (2005) Binding of complement factor H to endothelial cells is mediated by the carboxy-terminal glycosaminoglycan binding site. Am. J. Pathol. 167, 1173–1181 doi: 10.1016/S0002-9440(10)61205-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jokiranta T.S., Hellwage J., Koistinen V., Zipfel P.F. and Meri S. (1998) Each of the three binding sites on factor H interacts with a distinct site on C3b. Mol. Immunol. 35, 360–360 doi: 10.1016/S0161-5890(98)90650-2 [DOI] [PubMed] [Google Scholar]
- 29.Jòzsi M., Oppermann M., Lambris J.D. and Zipfel P.F. (2007) The C-terminus of complement factor H is essential for host cell protection. Mol. Immunol. 44, 2697–2706 doi: 10.1016/j.molimm.2006.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferreira V.P., Herbert A.P., Hocking H.G., Barlow P.N. and Pangburn M.K. (2006) Critical role of the C-terminal domains of factor H in regulating complement activation at cell surfaces. J. Immunol. 177, 6308–6316 doi: 10.4049/jimmunol.177.9.6308 [DOI] [PubMed] [Google Scholar]
- 31.Ram S., Sharma A.K., Simpson S.D., Gulati S., McQuillen D.P., Pangburn M.K. et al. (1998) A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 187, 743–752 doi: 10.1084/jem.187.5.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammerschmidt S., Agarwal V., Kunert A., Haelbich S., Skerka C. and Zipfel P.F. (2007) The host immune regulator factor H interacts via two contact sites with the PspC protein of Streptococcus pneumoniae and mediates adhesion to host epithelial cells. J. Immunol. 178, 5848–5858 doi: 10.4049/jimmunol.178.9.5848 [DOI] [PubMed] [Google Scholar]
- 33.Kajander T., Lehtinen M.J., Hyvarinen S., Bhattacharjee A., Leung E., Isenman D.E. et al. (2011) Dual interaction of factor H with C3d and glycosaminoglycans in host-nonhost discrimination by complement. Proc. Natl. Acad. Sci. U.S.A. 108, 2897–2902 doi: 10.1073/pnas.1017087108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pangburn M.K., Ferreira V.P. and Cortes C. (2008) Discrimination between host and pathogens by the complement system. Vaccine 26, I15–I21 doi: 10.1016/j.vaccine.2008.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alsenz J., Lambris J.D., Schulz T.F. and Dierich M.P. (1984) Localization of the complement-component-C3b-binding site and the cofactor activity for factor I in the 38kDa tryptic fragment of factor H. Biochem. J. 224, 389–398 doi: 10.1042/bj2240389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt C.Q., Herbert A.P., Kavanagh D., Gandy C., Fenton C.J., Blaum B.S. et al. (2008) A new map of glycosaminoglycan and C3b binding sites on factor H. J. Immunol. 181, 2610–2619 doi: 10.4049/jimmunol.181.4.2610 [DOI] [PubMed] [Google Scholar]
- 37.Prosser B.E., Johnson S., Roversi P., Herbert A.P., Blaum B.S., Tyrrell J. et al. (2007) Structural basis for complement factor H-linked age-related macular degeneration. J. Exp. Med. 204, 2277–2283 doi: 10.1084/jem.20071069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaum B.S., Hannan J.P., Herbert A.P., Kavanagh D., Uhrin D. and Stehle T. (2015) Structural basis for sialic acid–mediated self-recognition by complement factor H. Nat. Chem. Biol. 11, 77–82 doi: 10.1038/nchembio.1696 [DOI] [PubMed] [Google Scholar]
- 39.Morgan H.P., Schmidt C.Q., Guariento M., Blaum B.S., Gillespie D., Herbert A.P. et al. (2011) Structural basis for engagement by complement factor H of C3b on a self surface. Nat. Struct. Mol. Biol. 18, 463–470 doi: 10.1038/nsmb.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hair P.S., Echague C.G., Sholl A.M., Watkins J.A., Geoghegan J.A., Foster T.J. et al. (2010) Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of Staphylococcus aureus and decreases complement-mediated phagocytosis. Infect. Immun. 78, 1717–1727 doi: 10.1128/IAI.01065-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunkelberger J.R. and Song W.-C. (2010) Complement and its role in innate and adaptive immune responses. Cell Res. 20, 34–50 doi: 10.1038/cr.2009.139 [DOI] [PubMed] [Google Scholar]