Abstract
The enzyme Dicer is best known for its role as a riboendonuclease in the small RNA pathway. In this canonical role, Dicer is a critical regulator of the biogenesis of microRNA and small interfering RNA, as well as a growing number of additional small RNAs derived from various sources. Emerging evidence demonstrates that Dicer's endonuclease role extends beyond the generation of small RNAs; it is also involved in processing additional endogenous and exogenous substrates, and is becoming increasingly implicated in regulating a variety of other cellular processes, outside of its endonuclease function. This review will describe the canonical and newly identified functions of Dicer.
Keywords: endoribonuclease Dicer, microRNA, small interfering RNA, viral small RNA
Introduction
The Dicer enzyme is a member of the ribonuclease (RNase) III family. It is most well known as the endonuclease that functions in the RNA interference (RNAi) pathway to cleave long double-stranded RNA (dsRNA) molecules into short dsRNA molecules, known as small RNAs, including microRNA (miRNA) and small interfering RNA (siRNA). Indeed, Dicer is considered a key factor in the biogenesis of most small regulatory RNAs, and the majority of Dicer studies have focused on this role. However, increasing evidence shows that Dicer also has functions outside of the small RNA pathway. Dicer's endonuclease function is not only involved in small RNA biogenesis, but also in the processing of other endogenous and exogenous substrates. Furthermore, its function is not limited to cleavage, but may regulate other cellular processes. This review will describe the canonical and newly identified functions of Dicer.
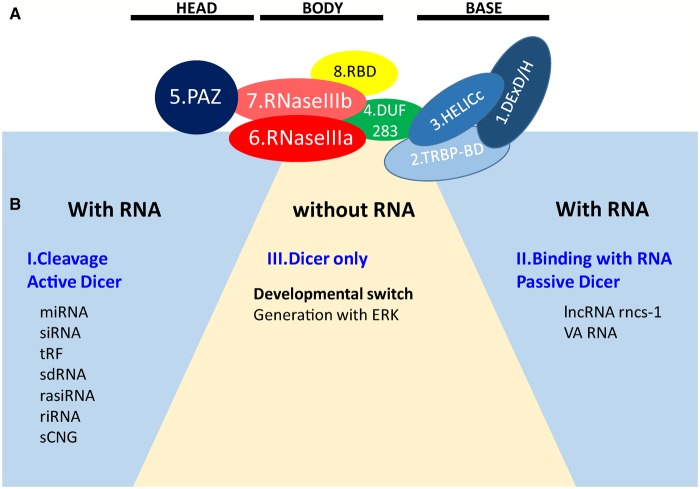
Dicer structure and domains
Although a full-length mammalian Dicer has not been crystallized, many studies have predicted its structure and the positions of its domains and interacting partners [1–5]. The main functional domains of Dicer are ordered from the N- to the C-terminus as follows: helicase domain included with DExD/H, TRBP-BD and HELICc, DUF283, PAZ (Piwi/Argonaut/Zwille) domains, RNase IIIa and IIIb domains, and dsRNA-binding domain (RBD) (Figure 1). Cryo-electron microscopy and crystallography showed that Dicer resembles the shape of the letter L, with a head, a body, and a base [6,7]. At the head is the PAZ domain, which contains binding pockets for the 3′ overhang of a dsRNA substrate. The PAZ domain of Dicer is unique in that it has an extra loop enriched in basic amino acids, which changes the electrostatic potential and molecular surface of the pocket. These differences may affect Dicer RNA binding and handing off the substrate to other protein complexes [8]. The PAZ domain also has a phosphate-binding pocket that recognizes the phosphorylated 5′ end of small RNAs [3]. On the lower half of the Dicer body are the RNase IIIa and IIIb domains, which form the catalytic core of Dicer; each domain is thought to be responsible for the cleavage of one strand of the dsRNA substrate [9]. The DExD/H domain is located in the base of the L and forms a clamp near the RNase III domain active site [1,10]. A recent study showed that the DUF283 domain of Dicer is capable of binding single-stranded nucleic acid [11].
Figure 1. Dicer structure and functions.
(A) The structure of Dicer. The number one indicates the amino-terminal. The amino-terminal helicase domain forms a clamp-like structure in the base of the L shape and is thought to reorganize and wrap around dsRNA. DExD/H, DExD/H box helicase domain; TRBP-BD, trans-activation response RNA-binding protein-binding domain; HELICc, helicase conserved carboxy-terminal domain; DUF283, domain of unknown function; PAZ, Piwi/Argonaute/Zwille domain; two RNase III domains; RBD, dsRNA-binding domain. (B) Multifaceted Dicer function. I. Active Dicer recognizes many types of RNA and can cleave small RNAs. II. Passive Dicer can be stably bound to RNA without endonuclease activity. Dicer does not efficiently process RNA if no free ends are available [33,177] which explains the resistance of the long non-coding RNA rncs-1 against dicing [178]. III. Dicer alone can function as a binding protein. For example, Ras/Erk signaling and Dicer regulate oogenesis, and Dicer was identified as a putative ERK substrate [166].
Evolutionary relationships among Dicer homologs
Various numbers of Dicer family proteins can be found in different organisms. Dicer probably arose from an early eukaryotic origin, as it is absent from archaebacteria, but can be found in many eukaryotic organisms, including plants, fungi, and metazoans (Figure 2A) [12,13]. The ancestral RNase III domains are found in eubacteria [14]; but these capabilities were not conserved in eukaryotic Dicer [15]. Dicer has been lost from many parasitic protozoa [16,17] as well as from fungi lacking RNAi, including the model budding yeast Sccharomyces cerevisiae [18]. The evolutionary phylogenetic tree of animal Dicers shows that an ancient duplication gave rise to Dicer1 and Dicer2 genes very early in metazoan evolution [19]. Both the miRNA and siRNA pathways rely on a single Dicer protein in vertebrates, and in the Nematoda phylum of invertebrates [9,20]. However, other invertebrates, including the fly Drosophila melanogaster and the prawn Litopenaeus vannamei of the Arthropoda phylum, and the fluke Clonorchis sinensis and the planarian Schmidtea mediterranea of the Platyhelminthes phylum retain both the Dicer1 and Dicer2 genes [21]. In Drosophila, Dicer1 is essential in the miRNA pathway, while Dicer2 facilitates the siRNA pathway [22]. Consistent with a role in immune defense, Dicer2, the siRNA-dedicated Dicer in Drosophila, is more closely related to the common ancestral Dicer protein than the miRNA pathway-dedicated Dicer1 [23]. This evidence suggests one common Dicer design evolved during metazoan evolution, from a universal factor for the miRNA and siRNA pathways, into a factor specifically adapted for either pathway.
Figure 2. Phylogenetic tree of Dicer and domain analysis.
(A) Phylogenetic diversity of eukaryotic Dicer proteins. We inferred the Dicer family phylogeny using the maximum likelihood method. The number has been indicated in bootstrap value. (B) Complex Dicer protein domain composition. DCL, Dicer-like.
Selective Dicer-mediated cleavage of various RNAs is driven by the evolutionarily conserved helicase domain (Figure 2B) [21,24,25]. Definitive functions for eukaryotic Dicer can be ascribed to different activities within the helicase domain, which is thought to facilitate movement of the protein along long dsRNA molecules [26]. This domain is analogous to the retinoic acid-inducible gene I-like family in the helicase superfamily 2 [27,28]. In Drosophila, the helicase domain of Dicer1, which cannot hydrolyze ATP, selectively interacts with the loops of precursor miRNAs (pre-miRNAs) and inhibits cleavage of long dsRNAs [25,29]. In contrast, the helicase domain of Dicer2 requires ATP and processes long dsRNAs to produce siRNAs. In Caenorhabditis elegans, Dicer also needs ATP and the helicase domain to process long dsRNAs with blunt or 5′ overhanging termini; mutation of this domain results in weak production of endogenous siRNAs but maintains the ability to dice pre-miRNAs [26,30]. In Arabidopsis thaliana, DCL1 mainly facilitates the biogenesis of imperfect stem-loop RNAs into 21 nt miRNAs [31] and also requires ATP [32]. However, although the helicase domains of mammalian Dicer proteins are strongly conserved with Drosophila Dicer2 [25], human Dicer does not require ATP [33,34], and cleaves both pre-miRNAs and long dsRNAs [35,36].
Canonical endonuclease function of Dicer
In the canonical pathway, the biogenesis of most small RNA classes, including miRNAs and many siRNAs, occurs through a stepwise process in the cytoplasm. This process involves association with Dicer and specific members of the large family of Argonaute (AGO) proteins, and assembly into various effector complexes, including the RNA-induced silencing complex (RISC). First, RNAs form a pre-RISC with Dicer [37], aided by the Hsc70/Hsp90 chaperone machinery [38–40]. The RNA is then cleaved by Dicer, in concert with two different dsRNA-binding proteins, the TRBP (HIV trans-activating response RNA-binding protein) and PACT (protein activator of PKR) [41]. After the double-stranded small RNA is captured, one of the two strands is selected as the guide, or active strand, and incorporated into an AGO protein to form the RISC. The duplex RNA is unwound and AGO2 degrades the other strand, known as the passenger strand. In general, the thermodynamic asymmetry of the end of the small RNA duplex is a major factor in determining which strand will be the guide strand; the guide strand is typically the strand with the less stable 5′ end [42,43]. Notably, the guide strand is already determined by the polarity of small RNA duplexes upon loading into the RISC, before unwinding occurs [44–48]. The activated RISC then recognizes a specific target site by complementary intermolecular base pairing throughout the single-stranded guide RNA molecule [49]. The resulting RISC-bound miRNAs or siRNAs either guide the sequence-specific degradation of complementary RNAs or inhibit the translation of partly complementary target messenger RNAs by post-transcriptional gene silencing in the cytoplasm, depending in part on the nature and degree of guide/target complementarity [50,51].
Additional endonuclease functions of Dicer
In addition, Dicer controls the fate of many other RNA species (Table 1). Dicer-derived small RNAs share several characteristic features. Small RNAs are typically ∼20–30 nucleotides (nt) long; have a characteristic and highly specific structure (i.e. 2-nt 3′ overhangs, and 5′ phosphate and 3′ hydroxyl groups); and contain both passenger and guide strands [24]. This section will describe the role of Dicer in processing a variety of RNA substrates into small RNAs, which together may form a very complex regulatory network.
Table 1. Types of endogenous small RNAs generated by Dicer.
| Origin of small RNA | Small RNA | Mechanism of action | |
|---|---|---|---|
 |
Pre-miRNA | miR320a-3p miR484-3p miR3615-3p miR7706-3p |
miRNA biogenesis [176] |
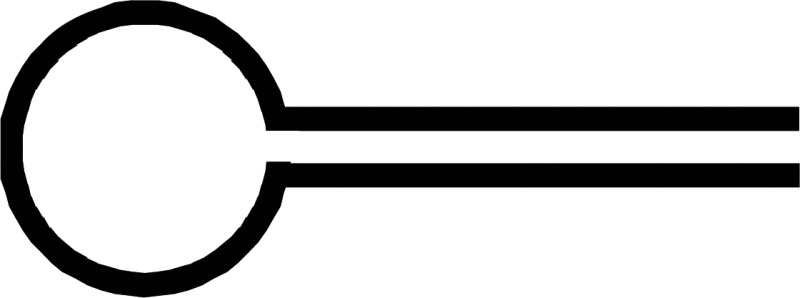 |
Pre-miRNA | Let-7b,d,g, miR-98, etc. | miRNA biogenesis [176] |
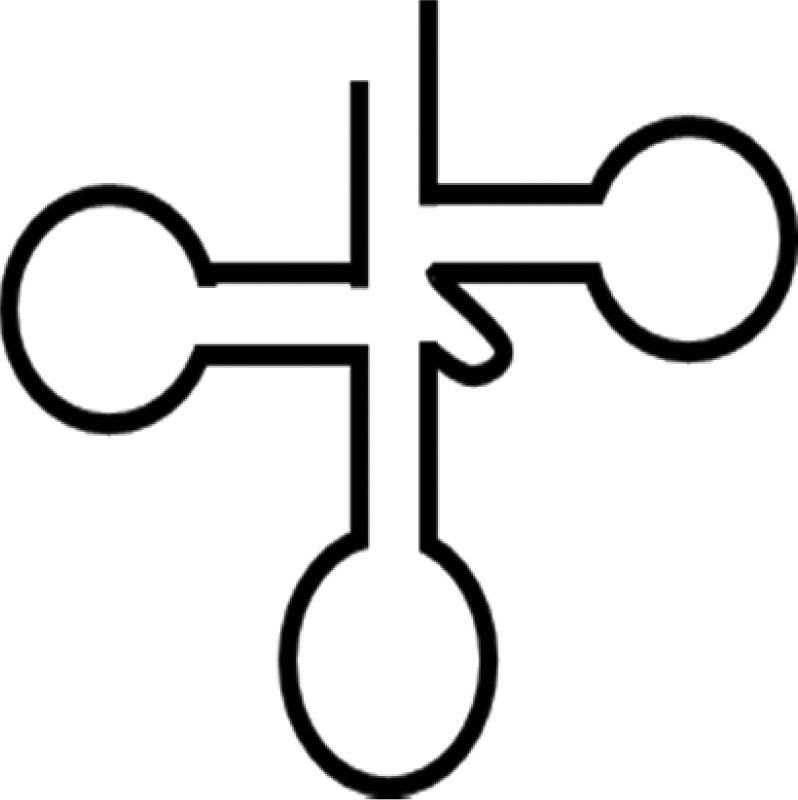 |
tRNA | 5′ tRFs 3′ CCA tRFs |
RNAi regulation Translation regulation [53,55–59] |
| tRNA | 3′ U tRFs | RNAi RNA metabolism [52,54] |
|
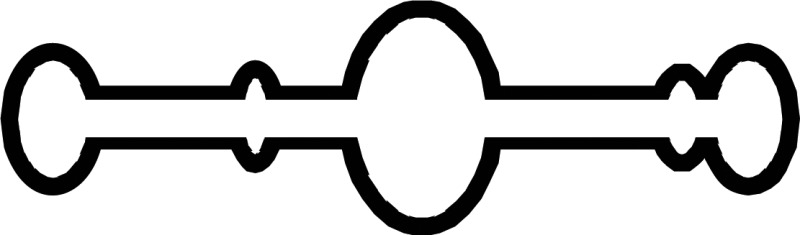 |
snoRNA | sdRNA | sdRNA RNA metabolism [61–69] |
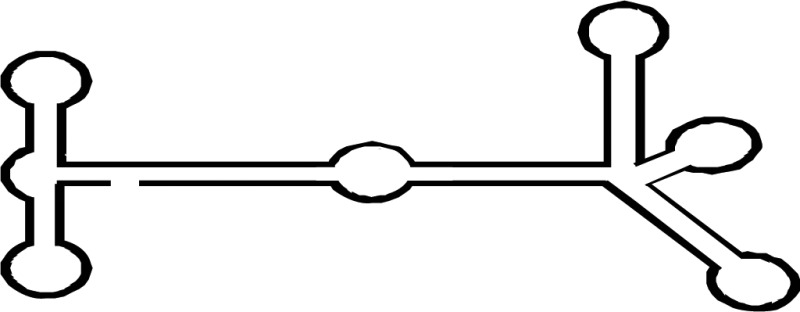 |
LINE1 SINEs |
rasiRNA riRNA |
Degradation/repeat-induced small RNA [72–77] |
 |
Triplet repeat RNA | sCNG CAG sRNA |
Degradation /siRNA [78–81] |
Red dot: m7 G cap.
Dicer-derived small RNAs from tRNA
Dicer is important for the production of transfer RNA (tRNA)-related fragments (tRFs), a heterogeneous class of small RNAs. tRFs are generated from tRNAs, and high-throughput sequencing and analysis show that they map to known tRNA genes [52–54]. The most abundant forms of tRFs can be classified into several groups. Three types of tRFs are derived from mature tRNA or pre-tRNA: 3′ U tRFs (tRF-1), 5′ tRFs (tRF-5), and 3′ CCA tRFs (tRF-3). 3′ U tRFs are generated by tRNA 3′-endonuclease ELAC2 (RNaseZ) [52]. 5′ tRFs are produced from the 5′ end of the tRNA generated at any point of tRNA processing, provided the 5′ leader sequence is removed by RNaseP, and are formed by a cleavage in the D loop. 3′ CCA tRFs are derived from the 3′ ends of mature tRNA by cleavage at the T loop and carry the trinucleotide CCA at the acceptor stem. Both 5′ tRFs and 3′ CCA tRFs have 5′ phosphate and 3′ hydroxyl ends, similar to miRNAs, and can be cleaved by Dicer protein complexes [53,55–58].
3′ CCA tRFs are processed in a Dicer-dependent manner [53]. Both in vitro and in vivo studies show that tRFs derived from human tRNA(Gln) are dependent on Dicer [53,55]. The tRNA(Gly)-derived tRF has been suggested to have miRNA-like functions. Dicer is implicated in the generation of CU1276, a 22-nt tRNA(Gly)-derived small RNA expressed in mature B cells, which is physically associated with AGO and represses an endogenous target mRNA. It inhibits the mRNA of the single-stranded DNA-binding protein RPA1, which suppresses cell proliferation and modulates the DNA damage response [58]. The miRNA biogenesis pathway utilizes tRNA processing enzymes and is found in specific herpesviruses. Murine γ-herpesvirus 68 miRNA encodes a viral tRNA-like sequence transcribed by RNA polymerase III to generate tRNA-like primary miRNA precursors (pri-miRNAs). Pri-miRNAs carry a 5′ tRNA moiety and are cleaved by a cellular tRNaseZ. Dicer processes the 3′ tRNA moiety to generate mature viral small RNAs [59].
The lupus autoantigen La is an RNA chaperone associated with the miRNA pathway. La functions as a gatekeeper that ensures correct tRNA maturation and protects the miRNA pathway from tRF. In the absence of La (La knockdown cells), various pre-tRNAs bind to Dicer, and the tRFs derived from the 3′ ends of tRNAs are bound to AGO [60].
Dicer-derived small RNAs from non-coding RNA
Dicer plays a role in processing several types of non-coding RNAs, which has implications for various regulatory mechanisms.
Dicer and snoRNA
Processing of non-coding small nucleolar RNA (snoRNA) to shorter, more stable snoRNA-derived RNAs (sdRNAs) is widespread, with sdRNAs identified in vertebrates, including humans [61–68], plants [63], fission yeast [63], and protozoa [69]. A high-throughput sequencing study of human small RNAs demonstrated that specific processing led to accumulation of small RNAs emerging from well-characterized non-coding RNAs, including snoRNAs [61]. sdRNAs are associated with AGO proteins and show miRNA capabilities, which may have implications for control of mRNA processing and translation [62,69]. Association with Dicer varies by type; many sdRNAs derived from box H/ACA snoRNAs are processed by Dicer, while processing of box C/D snoRNAs can be Dicer-independent [57,62]. In contrast, knockdown of Dicer and AGO in Giardia lamblia (protozoa) led to identification of small fragments with miRNA characteristics and showed that Dicer is required for miR2 production from box C/D snoRNA [69]. The identification of small fragments derives from the scaRNA of ACA45 (SCARNA15) by deep sequencing of human small RNAs associated with AGO proteins. The scaRNAs are a specific class of snoRNA that localize to Cajal bodies and guide the modification of RNA polymerase II transcribed spliceosomal small nuclear RNAs (snRNAs) U1, U2, U4, and U5 [70]. ACA45 is a 108-nt double-hairpin RNA that can be processed by Dicer to generate 20- to 22-nt ACA45 small RNA products [62].
A lot of data support the hypothesis that Dicer has direct roles in post-transcriptional gene-silencing activity of many miRNA-like sdRNAs, but the mechanism underlying their biogenesis remains unknown. It has been shown that processing of box C/D RNAs into sdRNAs can be Dicer-independent, while production of box H/ACA-derived sdRNAs requires Dicer [57,62]. Dicer is able to process dsRNA from snoRNAs [54,62,63,71]. Thus, snoRNAs can serve as a source of short regulatory RNA species generated by Dicer, which may be involved in the control of processing and translation of various mRNAs.
Dicer and long interspersed nuclear element-1
Another example of non-coding RNA processing by Dicer involves LINE-1 (long interspersed nuclear element-1), a retrotransposon. The LINE-1 promoter has a bidirectional orientation, containing sense and antisense transcripts that form double-stranded long hairpin RNAs, which are then processed by Dicer into repeat-associated siRNAs (rasiRNAs). rasiRNAs are loaded into AGO proteins to silence LINE-1 by RNA-directed DNA methylation. Inhibition of biogenesis of rasiRNAs has been suggested to increase the processing of LINE-1 transcription via Dicer inhibition [72]. Other experiments using engineered LINE-1 reporters have shown that mutation of Dicer increases LINE-1 transcripts. The rasiRNAs generated by Dicer negatively regulate LINE-1 transcription at the post-transcriptional level [73,74].
Dicer and short interspersed element
Geographic atrophy (GA), an age-related macular degeneration disease of the retinal pigment epithelium, is associated with reduced expression of Dicer. Inhibition of Dicer leads to accumulation of Alu repeat RNAs of ∼300-nt derived from short interspersed elements (SINEs). This accumulation is cytotoxic and triggers interferon-mediated, caspase 8-dependent apoptosis. Injection of Alu repeat RNA into mouse eyes generates GA [75]. This condition is rescued by Dicer cleavage of Alu repeat RNAs, suggesting that Dicer-dependent degradation of the RNAs is critical for detoxification [75]. Dicer processing of Alu repeat RNA prevents activation of the host inflammasome [76]. The DR2 Alu repeat RNAs are processed into 28–65-nt repeat-induced small RNAs (riRNAs) under DCIER-dependent conditions. These riRNAs are stabilized by binding with AGO3 and recruit mRNA-decapping complexes, which block translation and degrade key stem cell transcripts like Nanog and Tdgf1 mRNAs [77].
Dicer-derived small RNAs from triplet repeat RNA
Dicer has direct and indirect roles in curtailing expression of toxic triplet repeat RNAs. Triplet repeat expansion disorders include myotonic dystrophy type 1 (DM-1), fragile X-associated tremor ataxia syndrome, Huntington's disease (HD), and spinocerebellar ataxia (SCA), which frequently encode RNAs with long internal triplet repeat structures [78]. In vitro studies showed that single-stranded CGG-RNA is cleaved by Dicer to generate short CGG-RNAs [79]. Dicer recognizes and processes the hairpin structure of expanded CNG repeats, producing small CNG-repeated RNAs (sCNG) [78,80]. The Dicer-dependence of sCNG biogenesis has been demonstrated in fibroblasts of patients with DM-1 (sCUG), HD (sCAG), and SCA (sCAG) [78]. The relevance of sCNG has been reported in HD, which is caused by an abnormal CAG expansion within the first exon of the Huntingtin (HTT) gene. The sCAGs from HTT RNA stimulate neurotoxic activity [81]. Importantly, the toxic activity of sCAG is dependent on Dicer.
Dicer-derived small RNAs from exogenous RNA
In addition to its role in processing endogenous RNA, Dicer is also important in processing exogenous RNA, including viral-associated (VA) RNA (Table 2). Viral infection can produce virus-derived small RNAs. For example, VA RNA I and II are 160-nt long non-coding RNAs from the adenovirus genome, which are processed similarly to miRNAs, resulting in the production of 22-nt VA-RNA-derived miRNAs (mivaRNAs). These mivaRNAs are processed by Dicer [82–84].
Table 2. Types of exogenous small RNAs generated by Dicer.
RNA genome viruses do not express small RNA in infected cells; this phenomenon is believed to be a mechanism of host immune evasion [85,86]. However, the bovine leukemia virus, a retrovirus with an RNA genome, encodes a conserved cluster of small RNAs that are produced as shorter RNA polymerase III-transcribed hairpins that are directly cleaved by Dicer [87].
Recent studies show that RNA viruses also process miRNA-like RNAs [88,89]. A small RNA within the Nef (negative regulatory factor) region of the human immunodeficiency virus type 1 (HIV-1) genome is proposed to play a role in inhibition of viral transcription [90]. A 19-nt small RNA is derived from hairpin structures, including TAR (trans-activation response element) RNA [91]. Dicer processes these structures into functional small RNAs that target the HIV-1 genome. The Nef-derived miRNA (miR-N367) reduces Nef expression and suppresses replication by reducing HIV-promoter activity.
Additional functions of Dicer
In addition to the critical role of Dicer in small RNA biogenesis, it also has important roles in genome regulation and surveillance, unconnected to small RNA production. For example, although its RNAi role occurs in the cytoplasm, Dicer can also localize to the nucleus [92,93] and interact with nuclear pore components [94]. Dicer is present in autophagy and is required for autophagosome formation [95–97]. Dicer is found in the exosomes that are tiny vesicles with RNAs and proteins that are secreted from cells [98,99]. Dicer can also bind to ‘passive' RNA-binding sites without cleavage or small RNA generation [100]. Such interactions act as a buffering system to stabilize passive-site RNAs and control the catalytic activity of the enzyme by sequestering it from other targets. Dicer can also process viral sources of dsRNAs to produce viral siRNAs (viral small interfering RNAs) involved in various antiviral silencing responses [101].
Dicer in the nucleus
Several lines of evidence suggest that Dicer plays important roles in the nucleus. The C-terminal dsRNA-binding domain of Dicer contains a nuclear localization signal, which allows it to shuttle between the nucleus and the cytoplasm [93]. Human Dicer is associated with a nuclear pore complex component, nucleoporin 153, which assists Dicer in transport and localization to the nucleus [94]. Recent studies have shown that mammalian Dicer can also function in the nucleus [102]. Dicer inhibition leads to altered nucleolar structure; a significant increase in nucleolar size was observed in cells depleted of Dicer [103]. Some studies show that Dicer is required for cleavage and processing of pre-ribosomal RNA (rRNA) [104,105], which occurs mainly in the nucleolus [106], suggesting a connection between nuclear Dicer, rRNA synthesis, and ribosome biogenesis. However, another study shows that Dicer is associated with rDNA, but did not process pre-rRNA [7]. This suggests Dicer may affect pre-rRNA processing in different ways, such as transcriptional regulation.
Nuclear siRNAs corresponding to heterochromatic loci are associated with silencing; Dicer and AGO1 mutations destroy the production of these endo-siRNAs and the assembly of heterochromatin in the fission yeast Schizosaccharomyces pombe [107,108]. These siRNAs produce sequence-dependent silencing by recruiting histone-modifying enzymes that initiate heterochromatin formation and correlate with AGO1 and other silencing components [107,109]. Such siRNA-mediated transcriptional silencing has been reported in Arabidopsis [110,111], Drosophila [112], and mammalian cells [113]. In mammalian cells, nuclear siRNA-mediated transcriptional gene silencing was induced by histone deacetylases and DNA methyltransferases [113]. Another study showed heterochromatin defects at centromeres in Dicer-inhibited cells [114], suggesting that the Dicer-related RNAi machinery is implicated in the formation of the heterochromatin structure. Recent studies show that mammalian Dicer can be located in the nucleus [102]. The nuclear localization and function of mammalian Dicer remains one of the least understood aspects of Dicer biology.
Dicer in exosomes
Exosomes are small extracellular vesicles with an average size of 30–100 nm, which were simply considered cell debris for many years. Today, exosomes are known to be rich in lipids, proteins, and RNAs, including protein-coding transcripts (i.e. mRNAs) as well as many types of non-coding RNAs (e.g. miRNAs, long non-coding RNAs, circular RNAs, snoRNAs, snRNAs, tRNAs, rRNAs, and piwi-interacting RNAs) [115]. Exosomes allow these RNAs to be transferred from parent cells to recipient cells, where they can regulate or serve as templates for protein production [116,117].
In addition to recognition of a novel form of cell–cell communication, the discovery of miRNAs in exosomes has suggested new applications for exosomes, such as potentially easy-access biomarkers, or, given that exosomes are non-immunogenic, possibly as novel therapeutics [118–124]. miRNAs transported via exosomes could function as gene expression regulators in recipient cells, and growing evidence from exosomes in cancer and other processes has expanded the known subsets of exosome functions [121,125–128]. The discovery of miRNAs in exosomes also suggests a mechanism through which dysregulation of endogenous miRNAs may affect disease pathogenesis.
Dicer, AGO2, and TRBP proteins are detected in exosomes derived from breast cancer cells, but not from non-tumorigenic breast cells [98]. Exosomes derived from cancer cells, but not normal cells, were enriched in mature miRNAs, suggesting that pre-miRNAs can convert into mature miRNA in exosomes from cancer but not from non-tumorigenic breast cells. Tumor-derived exosomes isolated from cells or fluids of patients with breast cancer can be transferred to non-cancer cells in a Dicer-dependent manner. Mice form tumors when injected with cancer-derived exosomes, except in the presence of Dicer inhibition. Thus, Dicer is important for the transformation of normal cells to cancer cells, following exposure to exosomes. Exosomes carrying Dicer-derived miRNAs can affect cell–cell communication. Dicer-sufficient macrophage-derived exosomes carry miRNAs into Dicer-deficient endothelial cells to measurably reduce targeted sequences [99]. Thus, exosomes released by cancer cells may bioengineer miRNAs with the assistance of Dicer, resulting in tumor boosters.
Exosomes derived from HIV-1-infected cells contain TAR RNA and Dicer, as well as components of the host miRNA machinery [129]. The inclusion of TAR RNA and these components requires CRM1 transport receptor-mediated export from the nucleus; after packaging, the viral exosomes are released into the extracellular space. In recipient cells, exosomal TAR RNA down-regulates apoptosis by reducing expression of pro-apoptotic protein Bim and transcriptional regulator cyclin-dependent kinase 9. Thus, viral exosomes may provide a mechanism for intercellular viral spread in HIV-infected hosts.
Dicer DNase function
Dicer also plays a role in the recognition and processing of DNA in apoptosis [130]. The caspase CED-3 cleaves the two RNase III domains of Dicer in C. elegans, leaving a truncated though catalytically active protein consisting only of the C-terminal RNase III and dsRNA-binding domains. This truncated Dicer inactivates its RNase function, gains DNase function, and initiates breaks on chromosomal DNA. The DNA fragments are metabolized to complete apoptotic DNA degradation.
Two faces of Dicer viral interactions
Dicer antiviral function
Antiviral RNAi is a powerful mechanism to protect against viral infections. Specific features of viral RNA are sensed as foreign, and Dicer cleaves the double-stranded viral RNA into viral siRNA [10,131–134]. As described above, VA RNAs are processed by Dicer into small RNAs (mivaRNAs). These are incorporated into the RISC and may play an important role in suppressing RNAi or miRNA regulation during viral infection. VA RNA I is quickly generated after infection of host cells and amasses high copy numbers to support adenovirus survival [135–138]. VA RNA I has RNA-silencing properties, in addition to its well-established role in blocking dsRNA-dependent protein kinase PKR activation [139–141]. Activated PKR is designed to stop translation in virus-infected cells by phosphorylating a translation initiation factor [84]. VA RNA is a highly structured RNA that is bound by the nuclear export protein Exportin 5. It is rapidly transcribed after infection and produces very high copy numbers. VA RNA I saturates the Exportin 5 pathway, thereby inhibiting the nuclear export of pre-miRNAs. Additionally, due to its pre-miRNA-like secondary structure, VA RNA I suppresses Dicer mRNA movement to the cytoplasm by saturating the Exportin 5-dependent export pathway [139,142]. On the other hand, VA RNA I plays an important role in blocking antiviral function, thereby inhibiting Dicer mRNA. This suggests that Dicer has roles in antiviral function.
Although VA RNA is a poor substrate for Dicer cleavage, a few VA RNAs are cleaved to ∼22-nt mivaRNAs. mivaRNA association with the RISC is crucial for adenovirus replication [143]. A recent study showed that mivaRNAs down-regulate adenovirus replication, and knockdown of Dicer significantly promotes adenovirus replication by inhibiting production of mivaRNAs [82]. This suggests that Dicer performs its antiviral function against adenovirus via cleavage of VA RNAs.
Vaccinia virus (VACV) infection shows that inhibition of Dicer protein is associated with reduction in pre-miRNA processing [144]. Flaviviruses produce a small pathogenic RNA called subgenomic flavivirus RNA (sfRNA), which suppresses RNA silencing in insect and mammalian cells. The sfRNAs are associated with Dicer in infected cells and compete with endogenous small RNAs, resulting in decreased Dicer activity [145–148]. The human herpesvirus Epstein–Barr virus (EBV)-derived miRNA miR-BART6-5p targets human Dicer mRNA [149]. This suggests that viral miRNA influences Dicer transcription via a more subtle mechanism. This phenomenon shows the competition between Dicer of host cells and viral miRNAs of exogenous RNA.
EBV miRNA can be edited by adenosine deaminase acting on RNA to convert adenosine into inosine. A-to-I-edited miR-BART6-3p decreases the efficiency with which the miRNA encoded on the opposite strand, miR-BART6-5p, is loaded into the RISC in human cells. miR-BART6-5p targets human Dicer though four binding sites in its 3′UTR. Therefore, editing of miR-BART6-3p has a positive effect on Dicer expression. In turn, Dicer levels affect the expression levels of multiple genes that regulate the infectious and lytic states of EBV. Thus it is postulated that editing of miR-BART6-3p could be an indirect way to modulate miRNA biogenesis and thereby the viral life cycle [149].
Viruses encode proteins that suppress RNAi in mammalian cells, e.g. E3L (VACV), B2 (Nodamura virus), NSs (La Crosse virus), Tat (HIV), and VP30, VP35, and VP40 (Ebola virus) [142,150–154]. In particular, B2 of the Nodamura virus binds exogenous short hairpin RNAs and endogenous pre-let-7d and inhibits Dicer small RNA biogenesis [152]. The helicase domain of Dicer interacts with the lysine 51 residue of HIV Tat. The interaction between Tat and Dicer is dependent on the presence of small RNA, as RNase treatment inhibits this interaction. Tat interferes with Dicer processing of miRNA biogenesis [91]. VP30 and VP35 can function through direct interaction with Dicer or Dicer-associated factors TRBP and PACT, and inhibit the production of functional small RNAs [153,154]. These results demonstrate that viral RNA-binding proteins have the potential to interfere with miRNA biogenesis through RNA–Dicer interactions.
Dicer proviral function
Virus-derived small RNAs that direct specific antiviral defenses through biogenesis of a small RNA may be dependent on the cleavage activity of Dicer. However, it is still under debate whether infection by virus induces or suppresses antiviral small RNA.
A. thaliana encodes multiple Dicer proteins (DCL 1–4) involved in distinct endogenous RNA-silencing pathways and antiviral defenses, which show functional redundancy and specificity [155,156]. Inactivation of DCL4 but not DCL2 or DCL3 induces a high level of viral replication, indicating that DCL4 is essential for intracellular antiviral silencing. While DCL2 can produce abundant 22-nt viral siRNAs when DCL4 is absent, these siRNAs are less efficient in mediating an antiviral defense [157,158]. As described above, Dicer has two types in Drosophila. Dicer1 produces mature miRNAs in an ATP-dependent manner. Dicer2 cleaves cell-derived dsRNA precursors to produce endogenous siRNAs [159] and exogenous siRNAs [160,161], and mediates host defenses against viruses by activating Toll immune signaling [160]. The specific type of Dicer that processes virus-derived dsRNA into small RNAs in A. thaliana and D. melanogaster suggests an antiviral strategy, but other Dicer types do not produce antiviral small RNAs from viruses. In contrast, humans have one type of Dicer that processes both miRNAs and siRNAs. These various roles of Dicer led us to hypothesize that Dicer might regulate both antiviral and proviral mechanisms.
Infection of human cells with a wide range of viruses, including yellow fever virus, Sindbis virus, Venezuelan equine encephalitis virus, measles virus, influenza A virus, reovirus, vesicular stomatitis virus, HIV-1, or herpes simplex virus 1, failed to reveal any enhancement in viral replication in Dicer-inhibited human cells (human cells with the absence of Dicer) [162]. miRNAs derived from the Dengue virus genome include six different miRNA-like molecules (viral small RNA: vsRNA 1–6) [163]. Inhibition of vsRNA-5 leads to significant increases in viral replication. However, inhibition of the other vsRNAs has no direct effect on viral replication. Importantly, viral replication proceeded more slowly in human Dicer-deficient cells. KUN-miR-1 is derived from the terminal 3′ stem loop of West Nile virus (WNV) [164], and its production reduces inhibition of Dicer, with a significant reduction in WNV replication.
Dicer as a developmental switch
The Dicer protein can command specific interactions with other proteins. For example, Ras/ERK signaling and Dicer regulate C. elegans oogenesis; Dicer is a putative ERK substrate [165]. Phosphorylation of Dicer by ERK localizes it to the nucleus; it remains present during most of oogenesis but is rapidly lost from the terminal oocyte shortly before fertilization [166]. An ERK-mediated switch in Dicer activity could regulate genes during oogenesis and in the oocyte–embryo transition.
Dicer and cancer
In addition to other diseases described throughout this review, many groups have reported a correlation between Dicer expression and cancer. For example, Dicer expression shows significant changes in different stages of lung adenocarcinoma [167]. Reduced expression of Dicer is associated with poor prognosis in some types of lung, breast, skin, endometrial, and ovarian cancer [168–172]. In contrast, Dicer is overexpressed in metastatic lesions of prostate cancer [173], and is increased in Burkitt lymphoma [174]. There is no clear correlation between Dicer expression, cancer type, and disease progression; whether these inconsistencies can be attributed to technical artifacts, to tissue specificity, or to some other biological process remains unknown. Thus, whether Dicer acts as a tumor suppressor or an oncogene remains controversial [175]. However, given Dicer's dual roles in other biological processes, one can speculate that Dicer may function as both a tumor suppressor and an oncogene.
Conclusions
Accumulating evidence continues to demonstrate the critical role of the Dicer enzyme in not only the biogenesis of miRNA and siRNA in the canonical RNAi pathway, but also in controlling the fate of many other small RNA species. Furthermore, the list of non-endonuclease roles of Dicer continues to grow. Future directions in Dicer research should include: expanding our understanding of Dicer in the biogenesis and regulation of a growing network of endogenous small RNAs, which have interrelated implications for the regulation of complex physiological processes, including cancer; delineating the regulation of Dicer itself, including the causes and consequences of Dicer subcellular localization; and determining the role of Dicer in response to exogenous RNAs, and how this connects to the immune system.
Acknowledgements
We thank S.T. Wilkinson at City of Hope for advise on scientific writing.
Abbreviations
- AGO
Argonaute
- CRM
chromosome region maintenance
- DCL
Dicer-like
- DExD/H
DEAD-like and helicase domain
- DM-1
myotonic dystrophy type 1
- dsRNA
double-stranded RNA
- EBV
Epstein–Barr virus
- ERK
extracellular signal-regulated kinase
- GA
geographic atrophy
- HD
Huntington's disease
- HIV-1
human immunodeficiency virus type 1
- HSV
herpes simplex virus
- HTT
Huntingtin
- LINE-1
long interspersed nuclear element-1
- miRNA
microRNA
- mivaRNAs
VA-RNA-derived miRNA
- Nef
negative regulatory factor
- PACT
protein activator of PKR
- PAZ
Piwi/Argonaut/Zwille
- PKR
RNA-dependent protein kinase
- pre-miRNAs
precursor miRNAs
- pri-miRNAs
primary miRNA precursors
- rasiRNA
repeat-associated siRNA
- RBD
dsRNA-binding domain
- riRNA
repeat-induced small RNA
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RNase
ribonuclease
- rRNA
ribosomal RNA
- SCA
spinocerebellar ataxia
- scaRNA
Cajal body- specific RNA
- sCNG
small CNG
- sdRNA
snoRNA-derived RNAs
- sfRNA
subgenomic flavivirus
- sfRNA
subgenomic flavivirus RNA
- SINE
short interspersed elements
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNAs
- TAR
trans-activation response element
- TRBP
HIV trans-activating response RNA-binding protein
- tRF
transfer RNA-related fragments
- tRNA
transfer RNA
- VA RNA
viral-associated RNA
- VACV
vaccinia virus
- viral siRNA
viral small interfering RNA
- vsRNA
viral small RNA
- WNV
West Nile virus.
Funding
This work was funded in part by the National Heart, Lung, and Blood Institutes under grants [R01AI029329] and [R01Hl074704].
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Lau P.-W., Guiley K.Z., De N., Potter C.S., Carragher B. and MacRae I.J. (2012) The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 19, 436–440 doi: 10.1038/nsmb.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson R.C., Tambe A., Kidwell M.A., Noland C.L., Schneider C.P. and Doudna J.A. (2015) Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 57, 397–407 doi: 10.1016/j.molcel.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian Y., Simanshu D.K., Ma J.-B., Park J.-E., Heo I., Kim V.N. et al. (2014) A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell 53, 606–616 doi: 10.1016/j.molcel.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor D.W., Ma E., Shigematsu H., Cianfrocco M.A., Noland C.L., Nagayama K. et al. (2013) Substrate-specific structural rearrangements of human Dicer. Nat. Struct. Mol. Biol. 20, 662–670 doi: 10.1038/nsmb.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H.-W., Noland C., Siridechadilok B., Taylor D.W., Ma E., Felderer K. et al. (2009) Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 16, 1148–1153 doi: 10.1038/nsmb.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svobodova E., Kubikova J. and Svoboda P. (2016) Production of small RNAs by mammalian Dicer. Pflugers Archiv 468, 1089–1102 doi: 10.1007/s00424-016-1817-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinkkonen L., Hugenschmidt T., Filipowicz W., Svoboda P. and Aramayo R. (2010) Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS ONE 5, e12175 doi: 10.1371/journal.pone.0012175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z. et al. (2006) Structural basis for double-stranded RNA processing by Dicer. Science 311, 195–198 doi: 10.1126/science.1121638 [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., Kolb F.A., Jaskiewicz L., Westhof E. and Filipowicz W. (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118, 57–68 doi: 10.1016/j.cell.2004.06.017 [DOI] [PubMed] [Google Scholar]
- 10.Flemr M., Malik R., Franke V., Nejepinska J., Sedlacek R., Vlahovicek K. et al. (2013) A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155, 807–816 doi: 10.1016/j.cell.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 11.Kurzynska-Kokorniak A., Pokornowska M., Koralewska N., Hoffmann W., Bienkowska-Szewczyk K. and Figlerowicz M. (2016) Revealing a new activity of the human Dicer DUF283 domain in vitro. Sci. Rep. 6, 23989 doi: 10.1038/srep23989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerutti H. and Casas-Mollano J.A. (2006) On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 50, 81–99 doi: 10.1007/s00294-006-0078-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shabalina S.A. and Koonin E.V. (2008) Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587 doi: 10.1016/j.tree.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamontagne B., Larose S., Boulanger J. and Elela S.A. (2001) The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Issues Mol. Biol. 3, 71–78 PMID: [PubMed] [Google Scholar]
- 15.Johanson T.M., Lew A.M. and Chong M.M.W. (2013) MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. 3, doi: 10.1098/rsob.130144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ullu E., Tschudi C. and Chakraborty T. (2004) RNA interference in protozoan parasites. Cell. Microbiol. 6, 509–519 doi: 10.1111/j.1462-5822.2004.00399.x [DOI] [PubMed] [Google Scholar]
- 17.Baum J., Papenfuss A.T., Mair G.R., Janse C.J., Vlachou D., Waters A.P. et al. (2009) Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 37, 3788–3798 doi: 10.1093/nar/gkp239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drinnenberg I.A., Weinberg D.E., Xie K.T., Mower J.P., Wolfe K.H., Fink G.R. et al. (2009) RNAi in budding yeast. Science 326, 544–550 doi: 10.1126/science.1176945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee K., Campos H. and Kolaczkowski B. (2013) Evolution of animal and plant Dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 30, 627–641 doi: 10.1093/molbev/mss263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J.K., Gabel H.W., Kamath R.S., Tewari M., Pasquinelli A., Rual J.-F. et al. (2005) Functional genomic analysis of RNA interference in C. elegans. Science 308, 1164–1167 doi: 10.1126/science.1109267 [DOI] [PubMed] [Google Scholar]
- 21.Gao Z., Wang M., Blair D., Zheng Y., Dou Y. and Rossi J. J. (2014) Phylogenetic analysis of the endoribonuclease Dicer family. PLOS ONE 9, e95350 doi: 10.1371/journal.pone.0095350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J. et al. (2004) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 doi: 10.1016/S0092-8674(04)00261-2 [DOI] [PubMed] [Google Scholar]
- 23.Murphy D., Dancis B. and Brown J.R. (2008) The evolution of core proteins involved in microRNA biogenesis. BMC Evolut. Biol. 8, 92 doi: 10.1186/1471-2148-8-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidwell M.A., Chan J.M. and Doudna J.A. (2014) Evolutionarily conserved roles of the Dicer helicase domain in regulating RNA interference processing. J. Biol. Chem. 289, 28352–28362 doi: 10.1074/jbc.M114.589051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsutsumi A., Kawamata T., Izumi N., Seitz H. and Tomari Y. (2011) Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat. Struct. Mol. Biol. 18, 1153–1158 doi: 10.1038/nsmb.2125 [DOI] [PubMed] [Google Scholar]
- 26.Welker N.C., Maity T.S., Ye X., Aruscavage P.J., Krauchuk A.A., Liu Q. et al. (2011) Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell. 41, 589–599 doi: 10.1016/j.molcel.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jankowsky E. (2011) RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 36, 19–29 doi: 10.1016/j.tibs.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairman-Williams M.E., Guenther U.-P. and Jankowsky E. (2010) SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 20, 313–324 doi: 10.1016/j.sbi.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma E., Zhou K., Kidwell M.A. and Doudna J.A. (2012) Coordinated activities of human dicer domains in regulatory RNA processing. J. Mol. Biol. 422, 466–476 doi: 10.1016/j.jmb.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welker N.C., Pavelec D.M., Nix D.A., Duchaine T.F., Kennedy S. and Bass B.L. (2010) Dicer's helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16, 893–903 doi: 10.1261/rna.2122010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurihara Y. and Watanabe Y. (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. U.S.A. 101, 12753–12758 doi: 10.1073/pnas.0403115101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Z., Han M.-H. and Fedoroff N. (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl Acad. Sci. U.S.A. 105, 9970–9975 doi: 10.1073/pnas.0803356105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Kolb F.A., Brondani V., Billy E. and Filipowicz W. (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21, 5875–5885 doi: 10.1093/emboj/cdf582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B. and Rådmark O. (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21, 5864–5874 doi: 10.1093/emboj/cdf578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma E., MacRae I.J., Kirsch J.F. and Doudna J.A. (2008) Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 380, 237–243 doi: 10.1016/j.jmb.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soifer H.S., Sano M., Sakurai K., Chomchan P., Saetrom P., Sherman M.A. et al. (2008) A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 36, 6511–6522 doi: 10.1093/nar/gkn687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betancur J.G. and Tomari Y. (2012) Dicer is dispensable for asymmetric RISC loading in mammals. RNA 18, 24–30 doi: 10.1261/rna.029785.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyoshi T., Takeuchi A., Siomi H. and Siomi M.C. (2010) A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat. Struct. Mol. Biol. 17, 1024–1026 doi: 10.1038/nsmb.1875 [DOI] [PubMed] [Google Scholar]
- 39.Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T. et al. (2010) Hsc70/Hsp90 chaperone machinery mediates ATP-Dependent RISC loading of small RNA duplexes. Mol. Cell 39, 292–299 doi: 10.1016/j.molcel.2010.05.015 [DOI] [PubMed] [Google Scholar]
- 40.Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I. et al. (2010) In vitro assembly of plant RNA-Induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39, 282–291 doi: 10.1016/j.molcel.2010.05.014 [DOI] [PubMed] [Google Scholar]
- 41.Fukunaga R., Han B.W., Hung J.-H., Xu J., Weng Z. and Zamore P.D. (2012) Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 151, 533–546 doi: 10.1016/j.cell.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khvorova A., Reynolds A. and Jayasena S.D. (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 doi: 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- 43.Schwarz D.S., Hutvágner G., Du T., Xu Z., Aronin N. and Zamore P.D. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 doi: 10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi K., Tsukumo H., Nagami T., Siomi H. and Siomi M.C. (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19, 2837–2848 doi: 10.1101/gad.1370605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matranga C., Tomari Y., Shin C., Bartel D.P. and Zamore P.D. (2005) Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 doi: 10.1016/j.cell.2005.08.044 [DOI] [PubMed] [Google Scholar]
- 46.Rand T.A., Petersen S., Du F. and Wang X. (2005) Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629 doi: 10.1016/j.cell.2005.10.020 [DOI] [PubMed] [Google Scholar]
- 47.Kawamata T., Seitz H. and Tomari Y. (2009) Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 16, 953–960 doi: 10.1038/nsmb.1630 [DOI] [PubMed] [Google Scholar]
- 48.Yoda M., Kawamata T., Paroo Z., Ye X., Iwasaki S., Liu Q. et al. (2010) ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 17, 17–23 doi: 10.1038/nsmb.1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolia N.H. and Joshua-Tor L. (2007) Slicer and the Argonautes. Nat. Chem. Biol. 3, 36–43 doi: 10.1038/nchembio848 [DOI] [PubMed] [Google Scholar]
- 50.Pillai R.S., Bhattacharyya S.N. and Filipowicz W. (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 17, 118–126 doi: 10.1016/j.tcb.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 51.Hock J., Weinmann L., Ender C., Rudel S., Kremmer E., Raabe M. et al. (2007) Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 8, 1052–1060 doi: 10.1038/sj.embor.7401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee Y.S., Shibata Y., Malhotra A. and Dutta A. (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649 doi: 10.1101/gad.1837609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole C., Sobala A., Lu C., Thatcher S.R., Bowman A., Brown J.W. S. et al. (2009) Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15, 2147–2160 doi: 10.1261/rna.1738409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z., Ender C., Meister G., Moore P.S., Chang Y. and John B. (2012) Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 40, 6787–6799 doi: 10.1093/nar/gks307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P. and Blelloch R. (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785 doi: 10.1101/gad.1705308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schopman N.C. T., Heynen S., Haasnoot J. and Berkhout B. (2010) A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 7, 573–576 doi: 10.4161/rna.7.5.13141 [DOI] [PubMed] [Google Scholar]
- 57.Langenberger D., Cakir M.V., Hoffmann S. and Stadler P.F. (2013) Dicer-processed small RNAs: rules and exceptions. J. Exp. Zool. B: Mol. Dev. Evol. 320, 35–46 doi: 10.1002/jez.b.22481 [DOI] [PubMed] [Google Scholar]
- 58.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K. et al. (2013) tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl Acad. Sci. U.S.A. 110, 1404–1409 doi: 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bogerd H.P., Karnowski H.W., Cai X., Shin J., Pohlers M. and Cullen B.R. (2010) A mammalian herpesvirus uses non-canonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell 37, 135–142 doi: 10.1016/j.molcel.2009.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hasler D., Lehmann G., Murakawa Y., Klironomos F., Jakob L., Grässer F.A. et al. (2016) The lupus autoantigen La prevents Mis-channeling of tRNA fragments into the human MicroRNA pathway. Mol. Cell 63, 110–124 doi: 10.1016/j.molcel.2016.05.026 [DOI] [PubMed] [Google Scholar]
- 61.Kawaji H., Nakamura M., Takahashi Y., Sandelin A., Katayama S., Fukuda S. et al. (2008) Hidden layers of human small RNAs. BMC Genomics 9, 1–57 doi: 10.1186/1471-2164-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W. et al. (2008) A human snoRNA with microRNA-like functions. Mol. Cell 32, 519–528 doi: 10.1016/j.molcel.2008.10.017 [DOI] [PubMed] [Google Scholar]
- 63.Taft R.J., Glazov E.A., Lassmann T., Hayashizaki Y., Carninci P. and Mattick J.S. (2009) Small RNAs derived from snoRNAs. RNA 15, 1233–1240 doi: 10.1261/rna.1528909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brameier M., Herwig A., Reinhardt R., Walter L. and Gruber J. (2011) Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 39, 675–686 doi: 10.1093/nar/gkq776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langenberger D., Bermudez-Santana C.I., Stadler P.F. and Hoffmann S. (2010) Identification and classification of small RNAs in transcriptome sequence data. Pac. Symp. Biocomput. 80–87 PMID: [DOI] [PubMed] [Google Scholar]
- 66.Ono M., Scott M.S., Yamada K., Avolio F., Barton G.J. and Lamond A.I. (2011) Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 39, 3879–3891 doi: 10.1093/nar/gkq1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott M.S., Avolio F., Ono M., Lamond A.I. and Barton G.J. (2009) Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 5, e1000507 doi: 10.1371/journal.pcbi.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Politz J.C., Hogan E.M. and Pederson T. (2009) MicroRNAs with a nucleolar location. RNA 15, 1705–1715 doi: 10.1261/rna.1470409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saraiya A.A. and Wang C.C. (2008) snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 4, e1000224 doi: 10.1371/journal.ppat.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darzacq X., Jády B.E., Verheggen C., Kiss A.M., Bertrand E. and Kiss T. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21, 2746–2756 doi: 10.1093/emboj/21.11.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taft R.J., Simons C., Nahkuri S., Oey H., Korbie D.J., Mercer T.R. et al. (2010) Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 17, 1030–1034 doi: 10.1038/nsmb.1841 [DOI] [PubMed] [Google Scholar]
- 72.Faulkner G.J. (2013) Retrotransposon silencing during embryogenesis: dicer cuts in LINE. PLoS Genet. 9, e10003944 doi: 10.1371/journal.pgen.1003944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heras S.R., Macias S., Plass M., Fernandez N., Cano D., Eyras E. et al. (2013) The microprocessor controls the activity of mammalian retrotransposons. Nat. Struct. Mol. Biol. 20, 1173–1181 doi: 10.1038/nsmb.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang N. and Kazazian H.H. Jr (2006) L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13, 763–771 doi: 10.1038/nsmb1141 [DOI] [PubMed] [Google Scholar]
- 75.Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G. et al. (2011) DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–330 doi: 10.1038/nature09830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tarallo V., Hirano Y., Gelfand B.D., Dridi S., Kerur N., Kim Y. et al. (2012) DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149, 847–859 doi: 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu Q.D., Tanasa B., Trabucchi M., Li W., Zhang J., Ohgi K.A. et al. (2012) DICER- and AGO3-dependent generation of retinoic acid–induced DR2 Alu RNAs regulates human stem cell proliferation. Nat. Struct. Mol. Biol. 19, 1168–1175 doi: 10.1038/nsmb.2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krol J., Fiszer A., Mykowska A., Sobczak K., de Mezer M. and Krzyzosiak W.J. (2007) Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell 25, 575–586 doi: 10.1016/j.molcel.2007.01.031 [DOI] [PubMed] [Google Scholar]
- 79.Handa V., Saha T. and Usdin K. (2003) The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 31, 6243–6248 doi: 10.1093/nar/gkg818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Galka-Marciniak P., Urbanek M.O. and Krzyzosiak W.J. (2012) Triplet repeats in transcripts: structural insights into RNA toxicity. Biol. Chem. 393, 1299–1315 doi: 10.1515/hsz-2012-0218 [DOI] [PubMed] [Google Scholar]
- 81.Bañez-Coronel M., Porta S., Kagerbauer B., Mateu-Huertas E., Pantano L., Ferrer I. et al. (2012) A pathogenic mechanism in Huntington's disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet. 8, e1002481 doi: 10.1371/journal.pgen.1002481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Machitani M., Sakurai F., Wakabayashi K., Tomita K., Tachibana M. and Mizuguchi H. (2016) Dicer functions as an antiviral system against human adenoviruses via cleavage of adenovirus-encoded noncoding RNA. Sci. Rep. 6, 27598 doi: 10.1038/srep27598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Machitani M., Yamaguchi T., Shimizu K., Sakurai F., Katayama K., Kawabata K. et al. (2011) Adenovirus Vector-Derived VA-RNA-Mediated innate immune responses. Pharmaceutics 3, 338–353 doi: 10.3390/pharmaceutics3030338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mathews M.B. and Shenk T. (1991) Adenovirus virus-associated RNA and translation control. J. Virol. 65, 5657–5662 PMCID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Houzet L. and Jeang K.-T. (2011) MicroRNAs and human retroviruses. Biochim. Biophys. Acta 1809, 686–693 doi: 10.1016/j.bbagrm.2011.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cullen B.R. (2010) Five questions about viruses and microRNAs. PLoS Pathog. 6, e1000787 doi: 10.1371/journal.ppat.1000787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kincaid R.P., Burke J.M. and Sullivan C.S. (2012) RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl Acad. Sci. U.S.A. 109, 3077–3082 doi: 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langlois R.A., Shapiro J.S., Pham A.M. and tenOever B.R. (2012) In vivo delivery of cytoplasmic RNA virus-derived miRNAs. Mol. Ther. 20, 367–375 doi: 10.1038/mt.2011.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shapiro J.S., Varble A., Pham A.M. and Tenoever B.R. (2010) Noncanonical cytoplasmic processing of viral microRNAs. RNA 16, 2068–2074 doi: 10.1261/rna.2303610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Brisibe E.A. et al. (2004) HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1, 44 doi: 10.1186/1742-4690-1-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bennasser Y. and Jeang K.-T. (2006) HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology 3, 95 doi: 10.1186/1742-4690-3-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsui M., Li L., Janowski B.A. and Corey D.R. (2015) Reduced expression of Argonaute 1, Argonaute 2, and TRBP changes levels and intracellular distribution of RNAi factors. Sci. Rep. 5, 12855 doi: 10.1038/srep12855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doyle M., Badertscher L., Jaskiewicz L., Güttinger S., Jurado S., Hugenschmidt T. et al. (2013) The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA 19, 1238–1252 doi: 10.1261/rna.039255.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ando Y., Tomaru Y., Morinaga A., Burroughs A.M., Kawaji H., Kubosaki A. et al. (2011) Nuclear pore complex protein mediated nuclear localization of Dicer protein in human cells. PLoS ONE 6, e23385 doi: 10.1371/journal.pone.0023385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang P. and Zhang H. (2013) Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 14, 568–576 doi: 10.1038/embor.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kovaleva V., Mora R., Park Y.J., Plass C., Chiramel A.I., Bartenschlager R. et al. (2012) miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 72, 1763–1772 doi: 10.1158/0008-5472.CAN-11-3671 [DOI] [PubMed] [Google Scholar]
- 97.Gibbings D., Mostowy S., Jay F., Schwab Y., Cossart P. and Voinnet O. (2012) Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat. Cell Biol. 14, 1314–1321 doi: 10.1038/ncb2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A. et al. (2014) Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 26, 707–721 doi: 10.1016/j.ccell.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Squadrito M.L., Baer C., Burdet F., Maderna C., Gilfillan G.D., Lyle R. et al. (2014) Endogenous RNAs modulate MicroRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 8, 1432–1446 doi: 10.1016/j.celrep.2014.07.035 [DOI] [PubMed] [Google Scholar]
- 100.Rybak-Wolf A., Jens M., Murakawa Y., Herzog M., Landthaler M. and Rajewsky N. (2014) A variety of Dicer substrates in human and C. elegans. Cell 159, 1153–1167 doi: 10.1016/j.cell.2014.10.040 [DOI] [PubMed] [Google Scholar]
- 101.Ding S.-W. (2010) RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644 doi: 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- 102.Kalantari R., Chiang C.-M. and Corey D.R. (2016) Regulation of mammalian transcription and splicing by nuclear RNAi. Nucleic Acids Res. 44, 524–537 doi: 10.1093/nar/gkv1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liang X.-H. and Crooke S.T. (2011) Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 39, 4875–4889 doi: 10.1093/nar/gkr076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burger K., Muhl B., Rohrmoser M., Coordes B., Heidemann M., Kellner M. et al. (2013) Cyclin-dependent kinase 9 links RNA polymerase II transcription to processing of ribosomal RNA. J. Biol. Chem. 288, 21173–21183 doi: 10.1074/jbc.M113.483719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu H., Xu H., Miraglia L.J. and Crooke S.T. (2000) Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 275, 36957–36965 doi: 10.1074/jbc.M005494200 [DOI] [PubMed] [Google Scholar]
- 106.Lafontaine D.L.J. (2015) Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22, 11–19 doi: 10.1038/nsmb.2939 [DOI] [PubMed] [Google Scholar]
- 107.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I.S. and Martienssen R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 doi: 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 108.Hall I.M., Shankaranarayana G.D., Noma K.-i., Ayoub N., Cohen A. and Grewal S.I.S. (2002) Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 doi: 10.1126/science.1076466 [DOI] [PubMed] [Google Scholar]
- 109.Creamer K.M. and Partridge J.F. (2011) RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast. Wiley Interdiscip. Rev. RNA 2, 632–646 doi: 10.1002/wrna.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zilberman D., Cao X. and Jacobsen S.E. (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719 doi: 10.1126/science.1079695 [DOI] [PubMed] [Google Scholar]
- 111.Vaistij F.E., Jones L. and Baulcombe D.C. (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 14, 857–867 doi: 10.1105/tpc.010480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pal-Bhadra M., Bhadra U. and Birchler J.A. (2002) RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9, 315–327 doi: 10.1016/S1097-2765(02)00440-9 [DOI] [PubMed] [Google Scholar]
- 113.Morris K.V., Chan S.W.-L., Jacobsen S.E. and Looney D.J. (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305, 1289–1292 doi: 10.1126/science.1101372 [DOI] [PubMed] [Google Scholar]
- 114.Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y. et al. (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6, 784–791 doi: 10.1038/ncb1155 [DOI] [PubMed] [Google Scholar]
- 115.Kim K.M., Abdelmohsen K., Mustapic M., Kapogiannis D. and Gorospe M. (2017) RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA e1413–e1n/a doi: 10.1002/wrna.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hergenreider E., Heydt S., Treguer K., Boettger T., Horrevoets A.J.G., Zeiher A.M. et al. (2012) Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 14, 249–256 doi: 10.1038/ncb2441 [DOI] [PubMed] [Google Scholar]
- 117.Montecalvo A., Larregina A.T., Shufesky W.J., Stolz D.B., Sullivan M.L.G., Karlsson J.M. et al. (2012) Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766 doi: 10.1182/blood-2011-02-338004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Willms E., Johansson H.J., Mäger I., Lee Y., Blomberg K.E.M., Sadik M. et al. (2016) Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 6, 22519 doi: 10.1038/srep22519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Crescitelli R., Lässer C., Szabo T.G., Kittel A., Eldh M., Dianzani I. et al. (2013) Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2 doi: 10.3402/jev.v2i0.20677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gould S.J. and Raposo G. (2013) As we wait: coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles. 2 doi: 10.3402/jev.v2i0.20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iftikhar H. and Carney G.E. (2016) Evidence and potential in vivo functions for biofluid miRNAs: from expression profiling to functional testing: potential roles of extracellular miRNAs as indicators of physiological change and as agents of intercellular information exchange. BioEssays 38, 367–378 doi: 10.1002/bies.201500130 [DOI] [PubMed] [Google Scholar]
- 122.Bronisz A., Godlewski J. and Chiocca E.A. (2016) Extracellular vesicles and MicroRNAs: their role in tumorigenicity and therapy for brain tumors. Cell. Mol. Neurobiol. 36, 361–376 doi: 10.1007/s10571-015-0293-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheng L., Sharples R.A., Scicluna B.J. and Hill A.F. (2014) Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 3, 23743 doi: 10.3402/jev.v3.23743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hagiwara S., Kantharidis P. and Cooper M.E. (2014) MicroRNA as biomarkers and regulator of cardiovascular development and disease. Curr. Pharm. Des. 20, 2347–2370 doi: 10.2174/13816128113199990495 [DOI] [PubMed] [Google Scholar]
- 125.Bryniarski K., Ptak W., Martin E., Nazimek K., Szczepanik M., Sanak M. et al. (2015) Free extracellular miRNA functionally targets cells by transfecting exosomes from their companion cells. PLoS ONE 10, e0122991 doi: 10.1371/journal.pone.0122991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu J., Chen Q., Zen K., Zhang C. and Zhang Q. (2013) Synaptosomes secrete and uptake functionally active microRNAs via exocytosis and endocytosis pathways. J. Neurochem. 124, 15–25 doi: 10.1111/jnc.12057 [DOI] [PubMed] [Google Scholar]
- 127.Patton J.G., Franklin J.L., Weaver A.M., Vickers K., Zhang B., Coffey R.J. et al. (2015) Biogenesis, delivery, and function of extracellular RNA. J. Extracell. Vesicles 4, 27494 doi: 10.3402/jev.v4.27494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morel L., Regan M., Higashimori H., Ng S.K., Esau C., Vidensky S. et al. (2013) Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J. Biol. Chem. 288, 7105–7116 doi: 10.1074/jbc.M112.410944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Narayanan A., Iordanskiy S., Das R., Van Duyne R., Santos S., Jaworski E. et al. (2013) Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J. Biol. Chem. 288, 20014–20033 doi: 10.1074/jbc.M112.438895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakagawa A., Shi Y., Kage-Nakadai E., Mitani S. and Xue D. (2010) Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science 328, 327–334 doi: 10.1126/science.1182374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li Y., Lu J., Han Y., Fan X. and Ding S.-W. (2013) RNA interference functions as an antiviral immunity mechanism in mammals. Science 342, 231–234 doi: 10.1126/science.1241911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maillard P.V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S.W. et al. (2013) Antiviral RNA interference in mammalian cells. Science 342, 235–238 doi: 10.1126/science.1241930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Y., Basavappa M., Lu J., Dong S., Cronkite D.A., Prior J.T. et al. (2016) Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2, 16250 doi: 10.1038/nmicrobiol.2016.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kennedy E.M., Whisnant A.W., Kornepati A.V. R., Marshall J.B., Bogerd H.P. and Cullen B.R. (2015) Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc. Natl Acad. Sci. U.S.A. 112, E6945–E6954 doi: 10.1073/pnas.1513421112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kitajewski J., Schneider R.J., Safer B., Munemitsu S.M., Samuel C.E., Thimmappaya B. et al. (1986) Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2α kinase. Cell 45, 195–200 doi: 10.1016/0092-8674(86)90383-1 [DOI] [PubMed] [Google Scholar]
- 136.Bhat R.A. and Thimmappaya B. (1984) Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res. 12, 7377–7388 doi: 10.1093/nar/12.19.7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thimmappaya B., Weinberger C., Schneider R.J. and Shenk T. (1982) Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31, 543–551 doi: 10.1016/0092-8674(82)90310-5 [DOI] [PubMed] [Google Scholar]
- 138.Subramanian S., Bhat R.A., Rundell M.K. and Thimmappaya B. (1986) Suppression of the translation defect phenotype specific for a virus-associated RNA-deficient adenovirus mutant in monkey cells by simian virus 40. J. Virol. 60, 363–368 PMCID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu S. and Cullen B.R. (2004) Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 78, 12868–12876 doi: 10.1128/JVI.78.23.12868-12876.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Andersson M.G., Haasnoot P.C.J., Xu N., Berenjian S., Berkhout B. and Akusjarvi G. (2005) Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 79, 9556–9565 doi: 10.1128/JVI.79.15.9556-9565.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kamel W., Segerman B., Öberg D., Punga T. and Akusjärvi G. (2013) The adenovirus VA RNA-derived miRNAs are not essential for lytic virus growth in tissue culture cells. Nucleic Acids Res. 41, 4802–4812 doi: 10.1093/nar/gkt172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bennasser Y., Chable-Bessia C., Triboulet R., Gibbings D., Gwizdek C., Dargemont C. et al. (2011) Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat. Struct. Mol. Biol. 18, 323–327 doi: 10.1038/nsmb.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu N., Segerman B., Zhou X. and Akusjarvi G. (2007) Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the RNA-induced silencing complex and associate with polyribosomes. J. Virol. 81, 10540–10549 doi: 10.1128/JVI.00885-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Grinberg M., Gilad S., Meiri E., Levy A., Isakov O., Ronen R. et al. (2012) Vaccinia virus infection suppresses the cell microRNA machinery. Arch. Virol. 157, 1719–1727 doi: 10.1007/s00705-012-1366-z [DOI] [PubMed] [Google Scholar]
- 145.Schnettler E., Sterken M.G., Leung J.Y., Metz S.W., Geertsema C., Goldbach R.W. et al. (2012) Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J. Virol. 86, 13486–13500 doi: 10.1128/JVI.01104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pijlman G.P. (2014) Flavivirus RNAi suppression: decoding non-coding RNA. Curr. Opin Virol. 7, 55–60 doi: 10.1016/j.coviro.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 147.Kakumani P.K., Ponia S.S., S R.K., Sood V., Chinnappan M., Banerjea A.C. et al. (2013) Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J. Virol. 87, 8870–8883 doi: 10.1128/JVI.02774-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Moon S.L., Dodd B.J.T., Brackney D.E., Wilusz C.J., Ebel G.D. and Wilusz J. (2015) Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 485, 322–329 doi: 10.1016/j.virol.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iizasa H., Wulff B.-E., Alla N.R., Maragkakis M., Megraw M., Hatzigeorgiou A. et al. (2010) Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 285, 33358–33370 doi: 10.1074/jbc.M110.138362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li W.-X., Li H., Lu R., Li F., Dus M., Atkinson P. et al. (2004) Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl Acad. Sci. U.S.A. 101, 1350–1355 doi: 10.1073/pnas.0308308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Soldan S.S., Plassmeyer M.L., Matukonis M.K. and Gonzalez-Scarano F. (2005) La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J. Virol. 79, 234–244 doi: 10.1128/JVI.79.1.234-244.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sullivan C.S. and Ganem D. (2005) A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 79, 7371–7379 doi: 10.1128/JVI.79.12.7371-7379.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fabozzi G., Nabel C.S., Dolan M.A. and Sullivan N.J. (2011) Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 85, 2512–2523 doi: 10.1128/JVI.01160-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Haasnoot J., de Vries W., Geutjes E.-J., Prins M., de Haan P. and Berkhout B. (2007) The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 3, e86 doi: 10.1371/journal.ppat.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brodersen P. and Voinnet O. (2006) The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280 doi: 10.1016/j.tig.2006.03.003 [DOI] [PubMed] [Google Scholar]
- 156.Margis R., Fusaro A.F., Smith N.A., Curtin S.J., Watson J.M., Finnegan E.J. et al. (2006) The evolution and diversification of Dicers in plants. FEBS Lett. 580, 2442–2450 doi: 10.1016/j.febslet.2006.03.072 [DOI] [PubMed] [Google Scholar]
- 157.Wang X.-B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.X. et al. (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell. 23, 1625–1638 doi: 10.1105/tpc.110.082305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Andika I.B., Maruyama K., Sun L., Kondo H., Tamada T. and Suzuki N. (2015) Differential contributions of plant Dicer-like proteins to antiviral defences against potato virus X in leaves and roots. Plant J. 81, 781–793 doi: 10.1111/tpj.12770 [DOI] [PubMed] [Google Scholar]
- 159.Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M. et al. (2008) An endogenous small interfering RNA pathway in Drosophila. Nature 453, 798–802 doi: 10.1038/nature07007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Z., Wu D., Liu Y., Xia X., Gong W., Qiu Y. et al. (2015) DrosophilaDicer-2 has an RNA interference-independent function that modulates Toll immune signaling. Sci. Adv. 1, e1500228 doi: 10.1126/sciadv.1500228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sabin L.R., Zheng Q., Thekkat P., Yang J., Hannon G.J., Gregory B.D. et al. (2013) Dicer-2 processes diverse viral RNA species. PLoS ONE 8, e55458 doi: 10.1371/journal.pone.0055458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bogerd H.P., Skalsky R.L., Kennedy E.M., Furuse Y., Whisnant A.W., Flores O. et al. (2014) Replication of many human viruses Is refractory to inhibition by endogenous cellular MicroRNAs. J. Virol. 88, 8065–8076 doi: 10.1128/JVI.00985-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hussain M. and Asgari S. (2014) MicroRNA-like viral small RNA from dengue virus 2 autoregulates its replication in mosquito cells. Proc. Natl Acad. Sci. U.S.A. 111, 2746–2751 doi: 10.1073/pnas.1320123111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hussain M., Torres S., Schnettler E., Funk A., Grundhoff A., Pijlman G.P. et al. (2012) West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 40, 2210–2223 doi: 10.1093/nar/gkr848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Arur S., Ohmachi M., Nayak S., Hayes M., Miranda A., Hay A. et al. (2009) Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc. Natl Acad. Sci. U.S.A. 106, 4776–4781 doi: 10.1073/pnas.0812285106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Drake M., Furuta T., Suen K.M., Gonzalez G., Liu B., Kalia A. et al. (2014) A requirement for ERK-Dependent Dicer phosphorylation in coordinating oocyte-to-Embryo transition in C. elegans. Dev. Cell 31, 614–628 doi: 10.1016/j.devcel.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chiosea S., Jelezcova E., Chandran U., Luo J., Mantha G., Sobol R.W. et al. (2007) Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 67, 2345–2350 doi: 10.1158/0008-5472.CAN-06-3533 [DOI] [PubMed] [Google Scholar]
- 168.Karube Y., Tanaka H., Osada H., Tomida S., Tatematsu Y., Yanagisawa K. et al. (2005) Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 96, 111–115 doi: 10.1111/j.1349-7006.2005.00015.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R. et al. (2008) Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Eng. J Med. 359, 2641–2650 doi: 10.1056/NEJMoa0803785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pampalakis G., Diamandis E.P., Katsaros D. and Sotiropoulou G. (2010) Down-regulation of dicer expression in ovarian cancer tissues. Clin. Biochem. 43, 324–327 doi: 10.1016/j.clinbiochem.2009.09.014 [DOI] [PubMed] [Google Scholar]
- 171.Sand M., Gambichler T., Skrygan M., Sand D., Scola N., Altmeyer P. et al. (2010) Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Investig. 28, 649–653 doi: 10.3109/07357901003630918 [DOI] [PubMed] [Google Scholar]
- 172.Dedes K.J., Natrajan R., Lambros M.B., Geyer F.C., Lopez-Garcia M.A., Savage K. et al. (2011) Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur. J. Cancer 47, 138–150 doi: 10.1016/j.ejca.2010.08.007 [DOI] [PubMed] [Google Scholar]
- 173.Chiosea S., Jelezcova E., Chandran U., Acquafondata M., McHale T., Sobol R.W. et al. (2006) Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am. J. Pathol. 169, 1812–1820 doi: 10.2353/ajpath.2006.060480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kaul D. and Sikand K. (2004) Defective RNA-mediated c-myc gene silencing pathway in Burkitt's lymphoma. Biochem. Biophys. Res. Commun. 313, 552–554 doi: 10.1016/j.bbrc.2003.12.002 [DOI] [PubMed] [Google Scholar]
- 175.Klein S., Lee H., Ghahremani S., Kempert P., Ischander M., Teitell M.A. et al. (2014) Expanding the phenotype of mutations in DICER1: mosaic missense mutations in the RNase IIIb domain of DICER1 cause GLOW syndrome. J. Med. Gen. 51, 294–302 doi: 10.1136/jmedgenet-2013-101943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim Y.-K., Kim B. and Kim V.N. (2016) Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc. Natl Acad. Sci. U.S.A. 113, E1881–E1889 doi: 10.1073/pnas.1602532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fukunaga R., Colpan C., Han B.W. and Zamore P.D. (2014) Inorganic phosphate blocks binding of pre-miRNA to Dicer-2 via its PAZ domain. EMBO J. 33, 371–384 doi: 10.1002/embj.201387176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hellwig S. and Bass B.L. (2008) A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc. Natl Acad. Sci. U.S.A. 105, 12897–12902 doi: 10.1073/pnas.0805118105 [DOI] [PMC free article] [PubMed] [Google Scholar]