Abstract
Clinical diagnosis of esophageal cancer (EC) at early stage is rather difficult. This study aimed to profile the molecules in serum and tissue and identify potential biomarkers in patients with EC. A total of 64 volunteers were recruited, and 83 samples (24 EC serum samples, 21 serum controls, 19 paired EC tissues, and corresponding tumor-adjacent tissues) were analyzed. The gas chromatography time-of-flight mass spectrometry (GC/TOF-MS) was employed, and principal component analysis was used to reveal the discriminatory metabolites and identify the candidate markers of EC. A total of 41 in serum and 36 identified compounds in tissues were relevant to the malignant prognosis. A marked metabolic reprogramming of EC was observed, including enhanced anaerobic glycolysis and glutaminolysis, inhibited tricarboxylic acid (TCA) cycle, and altered lipid metabolism and amino acid turnover. Based on the potential markers of glucose, glutamic acid, lactic acid, and cholesterol, the receiver operating characteristic (ROC) curves indicated good diagnosis and prognosis of EC. EC patients showed distinct reprogrammed metabolism involved in glycolysis, TCA cycle, glutaminolysis, and fatty acid metabolism. The pivotal molecules in the metabolic pathways were suggested as the potential markers to facilitate the early diagnosis of human EC.
1. Introduction
Esophageal cancer (EC) is the eighth most common malignancy globally and the fourth leading cause of cancer mortality in China [1]. Although surgical resection remains the first choice of curative treatment for localized cancer, treatment outcomes and 5-year survival rates are far from satisfactory, because the early symptoms of EC are insidious and most patients present with incurable disease [2, 3]. Therefore, more accurate and robust biomarkers are in great demand for early screening and diagnosis of EC.
Metabolomic analysis is a systemic tool focusing on endogenous low molecular weight compounds to quantitatively assess metabolic features and has been shown to be effective for elucidation of biomarkers, metabolic pathways, and disease diagnosis [4–7]. Metabolic reprogramming has been widely observed in various tumors [8–11]. Metabolomics increases the possibility of validation of candidate biomarkers in the prospective studies through an accurate screening process for marker identification [12–14]. This approach also enhances the ability of researchers to analyze metabolomic data of specific biomarkers to gain insight into disease biology [15]. Previous studies have applied nuclear magnetic resonance (NMR) spectrometry and gas chromatography/mass spectrometry (GC/MS) [16, 17] to profile metabolites in serum, urine, and tissue of EC separately and respectively. Jin et al. [18] identified metabolomic signatures in serum of patients with lymph node metastasis based on GC/MS, while Davis et al. [19] used NMR spectroscopy to profile metabolic phenotypes of urine in EC and Barrett's esophagus. Although these studies identified panels of discriminant metabolites and suggested potential markers, the metabolites were case dependent and not correlated to the perturbed metabolites in EC tissues.
Hence, the current study was designed to profile metabolites in both serum and tissue samples from clinical patients with primary EC based on gas chromatography time-of-flight mass spectrometry (GC/TOF-MS). We aimed to screen potential markers that were characterized in serum and EC tissues and evaluate the sensitivity and specificity of potential markers for EC diagnosis.
2. Materials and Methods
The study protocol was approved by the Ethics Committee of Nanjing Medical University, and informed consent was obtained from all patients and volunteers. The inclusion and exclusion criterions in this study were assessed based on the entire body. Inclusion criterions were (i) age > 18 years and no prior chemotherapy or radiotherapy before enrollment and (ii) definite pathological diagnosis. Exclusion criterions were (i) a comorbidity of a metabolic disease, such as diabetes mellitus, gout, hyperlipidemia, or hemopathy; (ii) pregnancy or lactation; (iii) any symptom of massive stress or acute disease in the previous 2 weeks, such as large area of burns, psychic trauma, fever, cough, vomiting, and diarrhea; and (iv) use of specific drugs during the last 2 weeks, such as antibiotics, hormones, or nonsteroid anti-inflammatory drugs.
Serum samples were collected from 21 volunteers and 24 EC patients under fasting conditions. Tissue samples were collected from an additional 19 EC patients. Primary EC tissue specimens were excised from the central no-necrotic zone to ensure harvesting of cancer cells. Tumor-adjacent tissues were excised 2–3 cm from the tumor margin. All tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until assayed. Serum samples were collected in ethylenediaminetetraacetic sodium anticoagulated tubes and then centrifuged at 4000g for 10 min. At least 500 μL of serum was collected and frozen at −80°C until assayed. Detailed information regarding reagents, sample preparation, and GC/TOF-MS analysis is provided in the Supplementary Materials available online at https://doi.org/10.1155/2017/5469597.
3. Results
3.1. Study Population
The clinical characteristics of the patients and volunteers are summarized (Table 1). There were no significant differences in gender, age, and body weight between the EC patients and healthy controls (P > 0.05). The vast majority (90.70%) of patients had ESCC of histological grades I–III. Most patients (95.35%) had cancer in the middle or lower thoracic or esophagogastric junction, and almost half (48.84%) had lymph node metastasis. Of all patients, 36 consented to computer tomography (CT) examinations, yet three of the 36 CT examinations revealed no lesions. Besides, most patients had normal hepatic and renal function.
Table 1.
Patients' characteristics in the study of esophageal cancer.
| Characteristics | Patients supplying serum | P 1 | Patients supplying tissue | P 2 | Healthy control |
|---|---|---|---|---|---|
| Gender (n, male/female) | 19/5 | 1.000 | 15/4 | 1.000 | 17/4 |
| Age [mean (range)] | 60.2 (48–86) | 0.960 | 65.4 (44–87) | 0.122 | 60.0 (45–86) |
| Weight [mean (range)] | 61.6 (42–78) | 0.597 | 62.4 (42–79) | 0.895 | 62.8 (47–75) |
| Histology (n) | |||||
| ESCC | 22 | — | 17 | — | — |
| Adenocarcinoma | 2 | — | 2 | — | — |
| Histologic grade (n) | |||||
| I | 6 | — | 2 | — | — |
| II | 8 | — | 10 | — | — |
| III | 10 | — | 7 | — | — |
| Location (n) | |||||
| Cervical | 0 | — | 0 | — | — |
| Upper thoracic | 1 | — | 1 | — | — |
| Middle thoracic | 11 | — | 7 | — | — |
| Lower thoracic/EGJ | 12 | — | 11 | — | — |
| Lymph node metastasis (n) | |||||
| Yes | 13 | — | 8 | — | — |
| No | 11 | — | 11 | — | — |
| CT (n/n) | 17/20 | — | 16/16 | — | — |
| ALT [mean ± SD] | 24.87 ± 35.99 | — | 17.63 ± 6.18 | — | — |
| Creatininea [mean ± SD] | 78.21 ± 24.34 | — | 79.79 ± 27.50 | — | — |
aSerum creatinine; EGJ: esophagogastric junction; CT: computer tomography; ALT: alanine transaminase; SD: standard deviation; P1: P value of the EC patients supplying serum compared with the healthy control; P2: P value of the EC patients supplying tissue compared with the healthy control.
3.2. Metabolic Phenotypes of EC
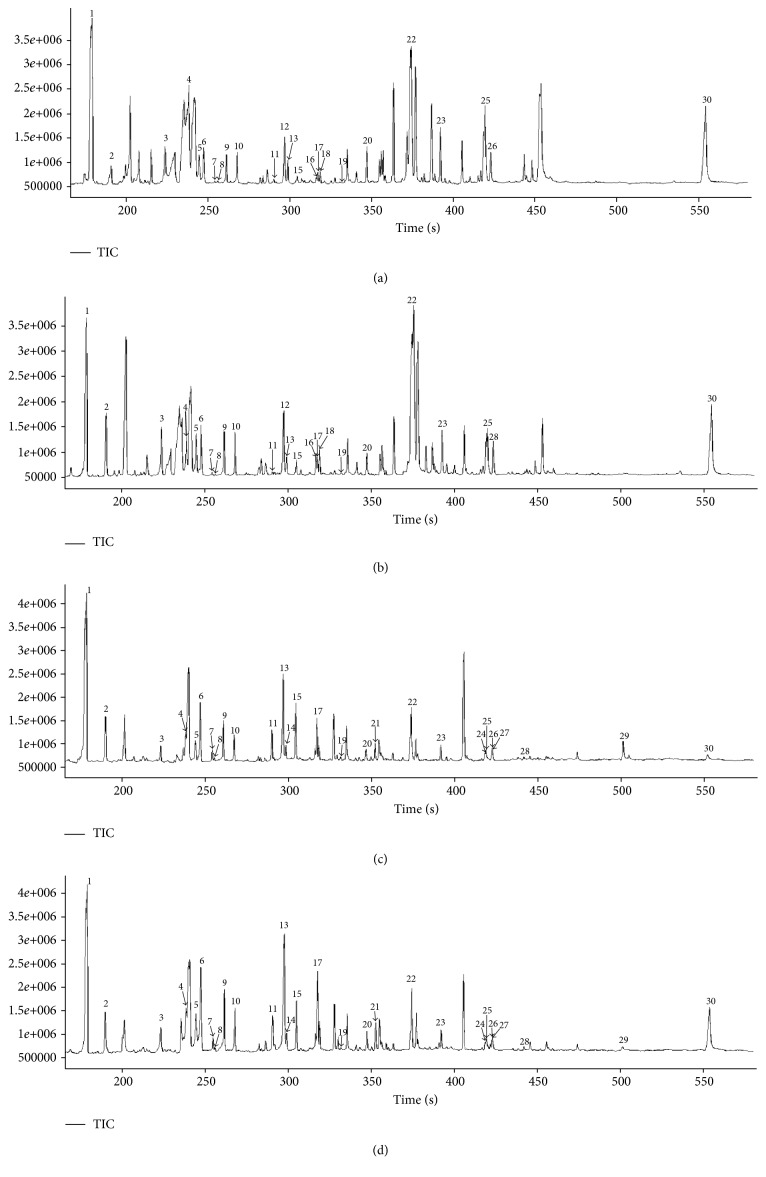
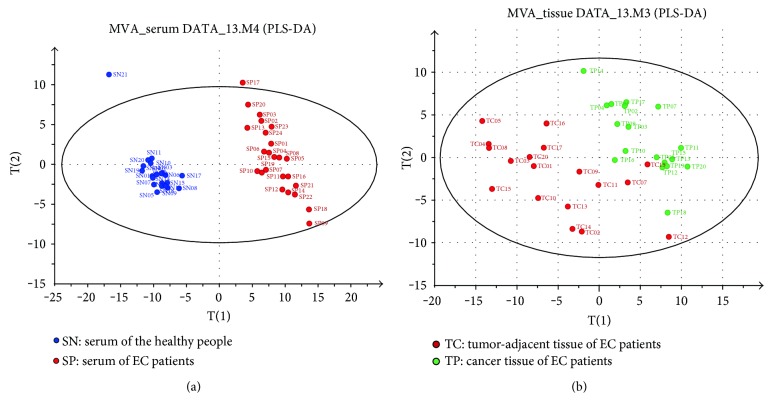
Representative total ion current chromatograms are presented in Figure 1. A total of 61 metabolites in serum and 58 in tissues were identified, which included carbohydrates, amino acids, organic acids, and fatty acids. Multivariate statistical analysis was performed using PLS-DA, and each dot in the score plot represented a sample. The scattering of samples in the score plots was exclusively dependent on the composition and concentration of the molecules in each sample. As shown in Figures 2(a) and 2(b), serum samples and cancer tissues were well separated with the controls located in two different regions. The higher explicative and predictive capacities of serum (R2Y = 98%, Q2Y = 95%) suggested differences in most of the detected variable molecules in serum between the EC patients and controls. The relatively lower R2Y (76.4%) and Q2Y (46.0%) values of tissues indicated that the differences in molecular compositions between the EC tissues and the tumor-adjacent tissues were not as great as those in serum, although still significant.
Figure 1.
Representative GC/TOF-MS total ion current (TIC) chromatograms of serums and tissues: (a) patient serum, (b) normal serum, (c) cancer tissue, and (d) tumor-adjacent tissue. Parts of the peaks were identified as 1, lactic acid; 2, alanine; 3, valine; 4, leucine; 5, proline; 6, glycine; 7, uracil; 8, fumaric acid; 9, serine; 10, threonine; 11, malic acid; 12, pyroglutamic acid; 13, asparagine; 14, hydroxyproline; 15, creatinine; 16, ornithine; 17, glutamic acid; 18, phenylalanine; 19, ribose; 20, glutamine; 21, hypoxanthine; 22, glucose; 23, hexadecanoic acid; 24, linoleic acid; 25, oleic acid; 26, tryptophan; 27, stearic acid; 28, glucose-6-phosphate; 29, maltose; and 30, cholesterol.
Figure 2.
The projection to latent structures and discriminant analysis score plot. (a) SN, normal serum; SP, patient serum. The two groups of serum were completely scattered into two different regions. There were two samples on the outside of the oval, which were far away from the others. It indicated that the metabolic profilings of the two people were significantly different from others, and we had eliminated the two samples when performing two sample t-tests. (b) TC, tumor-adjacent tissue; TP, cancer tissue.
3.3. Distinct Metabolites of EC Patients
Statistical analysis revealed that 42 compounds in serum and 37 in tissues were the discriminant molecules between the EC patients and controls (Table 2). In general, the concentrations of molecules in serum and tissues of the EC patients were diverse. The pivotal metabolites in glycolysis, pyruvic acid and lactic acid, showed a reverse tendency, while the level of glucose was low in both serum and tissues of the EC group. The majority of intermediates in the tricarboxylic acid (TCA) cycle (citric acid, α-ketoglutarate, fumaric acid, and malic acid) were elevated in both serum and tissue samples from the EC patients. Most concentrations of amino acids were lower in serum from the EC patients as compared with those from the controls, but at greater levels in cancer tissues than normal tumor-adjacent tissues. Interestingly, we found that the concentrations of most fatty acids were lower in serum collected from the EC patients but showed a reverse tendency in tissues. The levels of cholesterol, myo-inositol-1-phosphate, uracil, and hypoxanthine were higher in both serum and tissues of the EC patients than those of the controls.
Table 2.
The variation trend of metabolites in serum or tissue compared to the control.
| Metabolites | The regulatory tendency and statistical analysis relative to the controls | |||
|---|---|---|---|---|
| Groups | Serum | Tissue | ||
| Glycolysis | ||||
| Glucose | ↓↓ | ∗ | ↓ | / |
| Pyruvic acid, lactic acid | ↑↑ | ∗ | ↓ | ∗ |
| Glucose-6-phosphate | ± | / | ↑↑ | ∗ |
| Ribose | ↓↓ | ∗ | ↑↑ | ∗ |
| TCA cycle | ||||
| Citric acid, α-ketoglutarate, and malic acid | ↑↑ | ∗ | ↑↑ | ∗ |
| cis-Aconitic acid | ↑ | ∗ | ± | / |
| Succinic acid | ↓ | ∗ | ↑↑ | / |
| Fumaric acid | ↑ | ∗ | ↑ | / |
| Amino acid metabolism | ||||
| Arginine | ↓↓ | ∗ | ↑ | ∗ |
| Glutamic acid, glycine, and phenylalanine | ↓↓ | ∗ | ↑↑ | ∗ |
| Tryptophan, methionine | ↓ | ∗ | ↑ | ∗ |
| Serine, isoleucine, leucine, valine, proline, threonine, hydroxyproline, and ornithine | ↓ | ∗ | ↑↑ | ∗ |
| Tyrosine, kynurenic acid, asparagine, and histidine | ± | / | ↑↑ | ∗ |
| Alanine | ↓ | ∗ | ↑ | / |
| Cystine | ↓↓ | ∗ | ↑↑ | / |
| Taurine | ↑↑ | ∗ | ± | / |
| Pyroglutamic acid, creatinine | ↓ | ∗ | ↓ | ∗ |
| Urea | ± | / | ↓↓ | ∗ |
| Urate | ± | / | ↑ | ∗ |
| Lysine | ↑ | ∗ | ↑ | ∗ |
| Cysteine | ↑ | ∗ | ↑ | / |
| Glutamine | ↑↑ | ∗ | ↓ | ∗ |
| Fatty acid metabolism | ||||
| Monostearin, pipecolic acid | ↓↓ | ∗ | ± | / |
| Tetradecanoic acid, glyceric acid, docosahexaenoic acid, hexadecanoic acid, and 2-hydroxyvaleric acid | ↓ | ∗ | ± | / |
| Stearic acid | ± | / | ↑ | ∗ |
| Oleic acid, aminomalonic acid | ± | / | ↑↑ | ∗ |
| Cholesterol, myo-inositol-1-phosphate | ↑ | ∗ | ↑ | / |
| Linoleic acid | ↓ | ∗ | ↑ | ∗ |
| Nucleotide metabolism | ||||
| Uracil | ± | / | ↑↑ | ∗ |
| Hypoxanthine | ↑↑ | / | ↑ | ∗ |
↓, ↓↓ show downregulation by at least 20% or 50%, respectively.
↑, ↑↑ show upregulation by at least 20% or 50%, respectively.
± shows marginal regulation without statistical significance; ∗ shows P < 0.05; / shows P > 0.05.
3.4. Diagnostic Value of Potential Biomarkers of EC
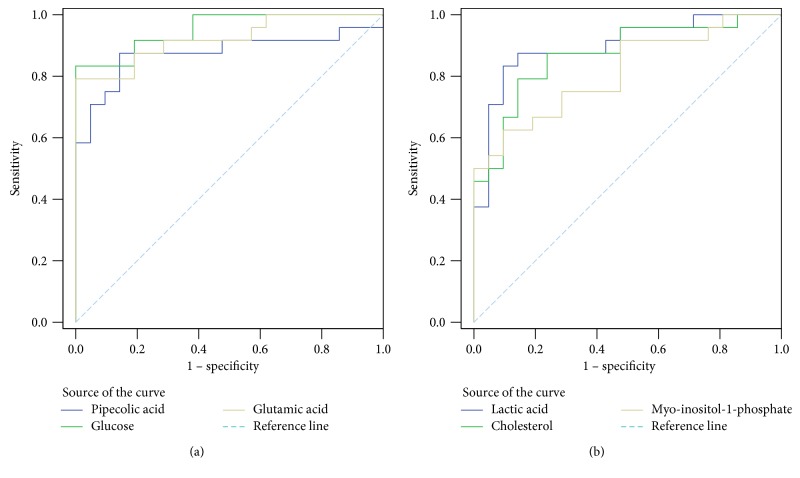
To characterize the EC patients from the healthy controls, receiver operating characteristic (ROC) curve analysis of potential biomarkers in serum was performed (Figures 3(a) and 3(b)), and cutoff values were calculated according to the Youden indices, sensitivities, specificities, and AUC values. For example, the cutoff value of glucose was 109.60, with a sensitivity of 83.3%, specificity of 100.0%, and AUC value of 0.952 (95% confidence interval = 0.897–1.000) (Tables 3 and 4). The ROC curves indicated that these markers had good diagnostic and prognostic values for EC.
Figure 3.
ROC curve analysis of potential serum biomarker levels. (a) Low levels of biomarkers in the EC group. (b) High levels of biomarkers in the EC group.
Table 3.
ROC curves of low-level biomarkers in the EC group.
| Marker | Cutoff valuea | Sensitivity | Specificity | AUC | P value | 95% CIb |
|---|---|---|---|---|---|---|
| Pipecolic acid | 8.50 | 87.5% | 85.7% | 0.875 | <0.001 | 0.762–0.988 |
| Glucose | 109.60 | 83.3% | 100.0% | 0.952 | <0.001 | 0.897–1.000 |
| Glutamic acid | 27.44 | 79.2% | 100.0% | 0.923 | <0.001 | 0.844–1.000 |
| Oleic acid | 14.62 | 83.3% | 66.7% | 0.794 | 0.001 | 0.663–0.924 |
aCutoff value (×105); b95% confidence interval of the difference.
Table 4.
ROC curves of high-level biomarkers in the EC group.
| Marker | Cutoff valuea | Sensitivity | Specificity | AUC | P value | 95% CIb |
|---|---|---|---|---|---|---|
| Lactic acid | 661.68 | 83.3% | 90.5% | 0.899 | <0.001 | 0.805–0.992 |
| Cholesterol | 695.62 | 79.2% | 85.7% | 0.869 | <0.001 | 0.764–0.974 |
| Myo-inositol-1-phosphate | 6.19 | 62.5% | 90.5% | 0.813 | <0.001 | 0.690–0.937 |
aCutoff value (×105); b95% confidence interval of the difference.
4. Discussion
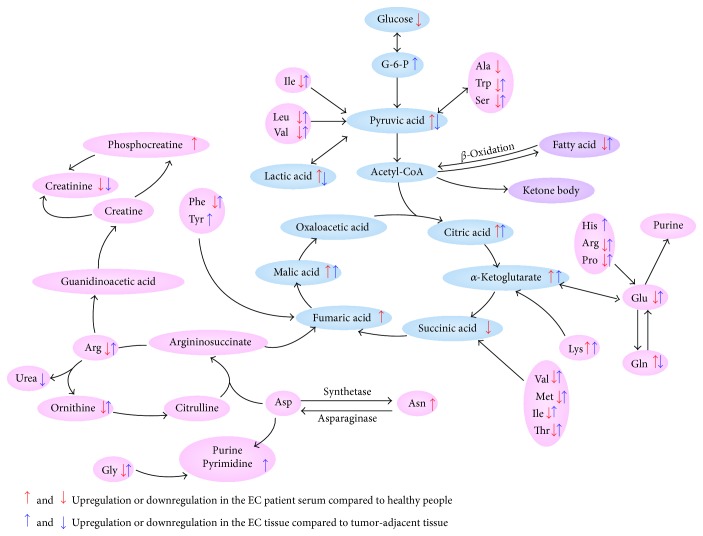
It is well documented that metabolism reprogramming occurs in various cancer cells [8, 20, 21]. According to the discriminant molecules in this study, most metabolic pathways were perturbed in the EC patients, at both systemic (peripheral blood) and local (EC tissues) levels (Figure 4). Measurement of these molecules has the potential to identify candidate markers suggestive of reprogrammed metabolism [22].
Figure 4.
Map illustrating the main metabolic pathways for the most relevant distinguishing metabolites. G-6-P, glucose-6-phosphate; Ala, alanine; Trp, tryptophan; Ser, serine; Ile, isoleucine; Leu, leucine; Val, valine; His, histidine; Arg, arginine; Pro, proline; Glu, glutamic acid; Gln, glutamine; Lys, lysine; Met, methionine; Thr, threonine; Asp, aspartic acid; Asn, asparagine; Phe, phenylalanine; Tyr, tyrosine; and Gly, glycine.
The Warburg effect is a process that cancer cells prefer anaerobic glycolysis rather than oxidative phosphorylation for energy production, even when provided with sufficient oxygen [23]. Apart from previous research, serum and tissues as a whole and ROC curve analysis in the present study identified glucose and lactic acid concentrations as potential biomarkers of EC. The low level of glucose in both EC serum and tissues suggested that cancer cells acquire large quantities of glucose, which accelerates lactic acid accumulation in serum via glycolysis. Lactic acid and pyruvate, which were increased in serum, but decreased in tissues of the EC patients, revealed enhanced gluconeogenesis of EC cells, consistent with gastric cancer [24]. Lactic acid in serum and cancer tissues may indicate poor prognosis in many types of cancers [25–27]. In this study, most intermediates of the TCA cycle accumulated both in EC serum and tissues. Most cancer cells were always under anoxic conditions due to accelerated proliferation, as compared with normal cells. So, the process of oxidative phosphorylation was limited and the metabolic pathway was impeded, which was a well-known barrier-like effect. This disruption in the TCA cycle further verified the presence of enhanced glycolysis.
Glutamine and glutamic acid play important roles in cancer cell proliferation. Glutamic acid is an important energy source via the TCA cycle after conversion to α-ketoglutarate [28]. Besides glutamic acid, glutamine contributes to de novo fatty acid synthesis and serves as a nitrogen source [29, 30]. In this study, glutamine in EC tissues was excessively consumed for the proliferation of cancer cells, resulting in decreased tissue concentrations. The intensive metabolism of glutamine suggested that EC cells require large amounts of glutamic acid from the systemic environment, resulting in decreased serum glutamic acid levels. The results of the present study demonstrated a significant increase in uracil content in EC tissues, which further confirmed that glutaminolysis was an indispensable metabolic pathway of de novo synthesis of purine and pyrimidine bases.
In this study, the level of most amino acids, such as tryptophan, serine, isoleucine, leucine, and valine, decreased in EC serum and increased in tissues. These amino acids participated in gluconeogenesis and energy production, which led to the large depletion of these amino acids from serum. In addition to energy production, serine, tryptophan, and glycine supply one carbon unit for synthesis of purines and pyrimidines. The branched-chain amino acids leucine, isoleucine, and valine are used as nitrogen sources by cancer cells [18]. Because of the rapid proliferation of cancer cells, copious amounts of asparagine were absorbed by the cancer tissues for production of hereditary material, which led to increased asparagine levels in EC tissues. In this study, we found decreased EC serum and increased tissue tryptophan levels, along with the elevated kynurenic acid concentrations in EC tissues, suggesting an enhanced cancer cell immune escape in EC [31].
In the present study, the EC patients had significantly higher cholesterol levels than the healthy controls. Excessive amounts of nonesterified free cholesterol are required to meet the metabolic requirements of rapidly proliferating cancer cells, which also tend to accelerate the release of free cholesterol from esterified cholesterol. The high cholesterol level in EC serum was in contrast to the findings of previous studies [19]. ROC curve analysis identified cholesterol as a potential biomarker of EC. Generally, concentrations of most free fatty acids (FFAs) are increased in EC serum but decreased in cancer tissues. We presume that the EC cells take in just enough FFAs to promote the release of fatty glyceride. In fact, the depressed 2-hydroxyvaleric levels in EC serum suggested inhibition of β-oxidation of FFAs.
The EC patients showed distinct reprogrammed metabolism involved in glycolysis, glutaminolysis, and TCA synthesis. Although the abundances of these discriminatory metabolites in serum and tissue were not always positively correlated, the metabolic phenotype of serum in the EC patients was closely associated with that of tissues. Future work involving larger populations of patients with EC should be performed to confirm our findings, and more studies are encouraged to investigate the influence of the lymph node metastasis on metabolic perturbation.
Supplementary Material
Supplementary Materials are detailed descriptions of reagents, sample preparation and GC/TOF-MS analysis.
Acknowledgments
This work was supported by grants from the Six Talent Peaks Project in Jiangsu Province (2012), the National Key Special Project of Science and Technology for Innovation Drugs of China (2013ZX09402202), and the Fundamental Research Funds for the Central Universities (YD2014SK0002).
Conflicts of Interest
All authors declared no conflict of interests.
Authors' Contributions
Lianzhen Yu and Jiye Aa are assigned to patient enrollment, EC diagnosis, experimental design, and manuscript revision. Xianlan Zhu and Kun Wang are assigned to patient enrollment, experimental design, data collection, and manuscript writing and revision. Gaoshuang Liu, Yuqing Wang, and Jin Xu handled the specimens for screening, data collection, and critical review of the manuscript. Linsheng Liu, Mengjie Li, and Jian Shi are assigned to data collection, statistical analysis, and manuscript preparation. Xianlan Zhu and Kun Wang contributed equally to this work.
References
- 1.Chen W., Zheng R., Zhang S., et al. Report of cancer incidence and mortality in China, 2010. Annals of Translational Medicine. 2014;2(7):p. 61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim E., Koroukian S., Thomas C. R., Jr Conditional survival of esophageal cancer: an analysis from the SEER registry (1988-2011) Journal of Thoracic Oncology. 2015;10(10):1490–1497. doi: 10.1097/JTO.0000000000000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen D., Zhang L., Wang X., et al. A 5.5-year surveillance of esophageal and gastric cardia precursors after a population-based screening in China. Journal of Gastroenterology and Hepatology. 2015;30(12):1720–1725. doi: 10.1111/jgh.13040. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson J. K., Lindon J. C. Systems biology: metabonomics. Nature. 2008;455(7216):1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 5.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson J. K., Connelly J., Lindon J. C., Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nature Reviews Drug Discovery. 2002;1(2):153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 7.Claudino W. M., Quattrone A., Biganzoli L., Pestrin M., Bertini I., Di Leo A. Metabolomics: available results, current research projects in breast cancer, and future applications. Journal of Clinical Oncology. 2007;25(19):2840–2846. doi: 10.1200/JCO.2006.09.7550. [DOI] [PubMed] [Google Scholar]
- 8.Yu L., Aa J., Xu J., et al. Metabolomic phenotype of gastric cancer and precancerous stages based on gas chromatography time-of-flight mass spectrometry. Journal of Gastroenterology and Hepatology. 2011;26(8):1290–1297. doi: 10.1111/j.1440-1746.2011.06724.x. [DOI] [PubMed] [Google Scholar]
- 9.Pasikanti K. K., Esuvaranathan K., Hong Y., et al. Urinary metabotyping of bladder cancer using two-dimensional gas chromatography time-of-flight mass spectrometry. Journal of Proteome Research. 2013;12(9):3865–3873. doi: 10.1021/pr4000448. [DOI] [PubMed] [Google Scholar]
- 10.Mondal S., Roy D., Camacho-Pereira J., et al. HSulf-1 deficiency dictates a metabolic reprograming of glycolysis and TCA cycle in ovarian cancer. Oncotarget. 2015;6(32):33705–33719. doi: 10.18632/oncotarget.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bro R., Kamstrup-Nielsen M. H., Engelsen S. B., et al. Forecasting individual breast cancer risk using plasma metabolomics and biocontours. Metabolomics. 2015;11(5):1376–1380. doi: 10.1007/s11306-015-0793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S., Liu H., Jin Y., Lin S., Cai Z., Jiang Y. Metabolomics study of alcohol-induced liver injury and hepatocellular carcinoma xenografts in mice. Journal of Chromatography B Analytical Technologies in the Biomedical and Life Sciences. 2011;879(24):2369–2375. doi: 10.1016/j.jchromb.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Wong V. W., Wong G. L., Chu W. C., et al. Hepatitis B virus infection and fatty liver in the general population. Journal of Hepatology. 2012;56(3):533–540. doi: 10.1016/j.jhep.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Zhang A., Sun H., Wang X. Power of metabolomics in biomarker discovery and mining mechanisms of obesity. Obesity Reviews. 2013;14(4):344–349. doi: 10.1111/obr.12011. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Ma Z., Min L., et al. Biomarker identification and pathway analysis by serum metabolomics of lung cancer. BioMed Research International. 2015;2015:p. 9. doi: 10.1155/2015/183624.183624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowda G. A., Zhang S., Gu H., Asiago V., Shanaiah N., Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Review of Molecular Diagnostics. 2008;8(5):617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Z., Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Analytical and Bioanalytical Chemistry. 2007;387(2):525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 18.Jin H., Qiao F., Chen L., Lu C., Xu L., Gao X. Serum metabolomic signatures of lymph node metastasis of esophageal squamous cell carcinoma. Journal of Proteome Research. 2014;13(9):4091–4103. doi: 10.1021/pr500483z. [DOI] [PubMed] [Google Scholar]
- 19.Davis V. W., Schiller D. E., Eurich D., Sawyer M. B. Urinary metabolomic signature of esophageal cancer and Barrett’s esophagus. World Journal of Surgical Oncology. 2012;10:p. 271. doi: 10.1186/1477-7819-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burch T. C., Isaac G., Booher C. L., et al. Comparative metabolomic and lipidomic analysis of phenotype stratified prostate cells. PLoS One. 2015;10(8):p. e0134206. doi: 10.1371/journal.pone.0134206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin X., Yun S. J., Jeong P., Kim I. Y., Kim W. J., Park S. Diagnosis of bladder cancer and prediction of survival by urinary metabolomics. Oncotarget. 2014;5(6):1635–1645. doi: 10.18632/oncotarget.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denkert C., Budczies J., Weichert W., et al. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Molecular Cancer. 2008;7:p. 72. doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 24.Aa J. Y., Yu L. Z., Sun M., et al. Metabolic features of the tumor microenvironment of gastric cancer and the link to the systemic macroenvironment. Metabolomics. 2012;8(1):164–173. doi: 10.1007/s11306-011-0297-0. [DOI] [Google Scholar]
- 25.Maria R. M., Altei W. F., Andricopulo A. D., et al. Characterization of metabolic profile of intact non-tumor and tumor breast cells by high-resolution magic angle spinning nuclear magnetic resonance spectroscopy. Analytical Biochemistry. 2015;488:14–18. doi: 10.1016/j.ab.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Halama A., Guerrouahen B. S., Pasquier J., et al. Metabolic signatures differentiate ovarian from colon cancer cell lines. Journal of Translational Medicine. 2015;13:p. 223. doi: 10.1186/s12967-015-0576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J., Liu L., Wei S., et al. Metabolomics study of esophageal adenocarcinoma. The Journal of Thoracic and Cardiovascular Surgery. 2011;141(2):469–475. doi: 10.1016/j.jtcvs.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 28.Wang L., Chen J., Chen L., et al. 1H-NMR based metabonomic profiling of human esophageal cancer tissue. Molecular Cancer. 2013;12:p. 25. doi: 10.1186/1476-4598-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metallo C. M., Gameiro P. A., Bell E. L., et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuneva M., Zamboni N., Oefner P., Sachidanandam R., Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. The Journal of Cell Biology. 2007;178(1):93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opitz C. A., Litzenburger U. M., Opitz U., et al. The indoleamine-2,3-dioxygenase (IDO) inhibitor 1-methyl-D-tryptophan upregulates IDO1 in human cancer cells. PLoS One. 2011;6(5):p. e19823. doi: 10.1371/journal.pone.0019823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials are detailed descriptions of reagents, sample preparation and GC/TOF-MS analysis.