Abstract
R.SwaI, a Type IIP restriction endonuclease, recognizes a palindromic eight base pair (bp) symmetric sequence, 5΄-ATTTAAAT-3΄, and cleaves that target at its center to generate blunt-ended DNA fragments. Here, we report three crystal structures of SwaI: unbound enzyme, a DNA-bound complex with calcium ions; and a DNA-bound, fully cleaved complex with magnesium ions. We compare these structures to two structurally similar ‘PD-D/ExK’ restriction endonucleases (EcoRV and HincII) that also generate blunt-ended products, and to a structurally distinct enzyme (the HNH endonuclease PacI) that also recognizes an 8-bp target site consisting solely of A:T base pairs. Binding by SwaI induces an extreme bend in the target sequence accompanied by un-pairing and re-ordering of its central A:T base pairs. This result is reminiscent of a more dramatic target deformation previously described for PacI, implying that long A:T-rich target sites might display structural or dynamic behaviors that play a significant role in endonuclease recognition and cleavage.
Restriction endonucleases are components of microbial restriction-modification (R-M) systems that act as a pre-programmed or ‘innate’ form of immunity against infectious genetic elements such as viruses. These enzymes bind to double-stranded DNA molecules at specific base pair sequences, and hydrolyze the two DNA strands either within or nearby that sequence. Hydrolysis fragments the DNA, disrupting its genetic content and halting its further propagation. Thousands of restriction enzymes recognizing an equally diverse array of different DNA sequences have been characterized since their initial discovery in the early 1970s (reviewed in (1)). Together with the recently discovered CRISPR-Cas nucleases that act as a programmable, or ‘adaptive’ form of microbial immunity, these enzymes have revolutionized the fields of molecular biology, biochemistry, and medical genetics, and contributed enormously to our understanding of living processes (2).
Restriction enzymes and the systems to which they belong vary greatly with respect to amino acid sequence, substrate sequence, catalytic mechanism, domain and subunit composition, and oligomeric organization and size. Different combinations of these properties are the basis for their classification into four major groups, or ‘Types’, each with multiple sub-classes (defined in (3) and organized into the restriction endonuclease database (‘REBASE’) as described in (4)). Types I and III R-M enzymes (5,6) are multi-subunit assemblages that combine cleavage and DNA-modification together into large, unified molecular machines. Type II systems (7) are generally simpler, and for the most part comprise separate endonuclease and methyltransferase enzymes, each with all the elements needed for independent sequence-recognition and catalysis. Despite their simplicity, Type II endonucleases are highly diverse and built around several different folds and catalytic motifs that employ distinct DNA-hydrolysis mechanisms (8). They display a wide variety of structural organizations and are often embellished with additional structural domains. They assemble into several different quaternary arrangements that can lead to complex cooperative and allosteric behaviors (9).
In this study, we describe crystallographic analyses of the R.SwaI restriction endonuclease (hereafter called ‘SwaI’), which is encoded in Staphylococcus warneri (10). The enzyme recognizes and cleaves a long (8-base pair) palindromic sequence corresponding to 5΄-ATTT|AAAT-3΄ and produces blunt product ends. We describe the relationship between the structure of SwaI both to its closest known structural relatives, EcoRV (11) and HincII (12), which recognize shorter (6-bp) palindromic DNA targets and also generate blunt-ended product ends. We then compare the DNA-bound structures of SwaI to a structurally different restriction enzyme (the HNH endonuclease PacI) that also cleaves a palindromic 8-base pair target site consisting solely of A:T base pairs (5΄-TTAAT|TAA-3΄) (13). As was previously observed for PacI, recognition by SwaI of its long A:T-rich target site is accompanied by unusual and dramatic disruption of the target site duplex.
MATERIALS AND METHODS
Endonuclease cloning, expression and purification
The genes for the SwaI restriction endonuclease and modification methyltransferase were cloned into Escherichia coli ER2566 and characterized as described (14). The endonuclease gene was inserted into the chloramphenicol-resistant, low copy plasmid, pHKT7, and expressed from an inducible T7 promoter. The methyltransferase gene was inserted into the ampicillin-resistant, high copy plasmid, pHKUV5, and expressed from a constitutive lac UV5 promoter.
Recombinant E. coli ER2566 cells containing the SwaI genes were plated from a glycerol stock onto LB containing 100 μg/ml ampicillin, 25 μg/ml chloramphenicol and incubated at 22°C. A single colony was picked and inoculated into 1 liter of rich broth containing the same antibiotics, and incubated overnight at 30°C without shaking. This culture was diluted into 100 l LB (Amresco DF204) containing antifoam. After 2.6 h at 37°C, 200 rpm agitation and 50 l/l·min aeration, at a cell density of 160 Klett units, 7 g of IPTG was added to a final concentration of 0.3 mM in order to induce endonuclease synthesis. The culture was cooled and harvested 2 h later by continuous-flow centrifugation. The resulting wet cell mass, of ∼500 g, was stored at −80°C until needed.
Two hundred fify grams of frozen cell pellet was thawed and re-suspended in 800 ml of lysis buffer (40 mM KPO4 pH 6.8, 5% glycerol, 0.25 mM EDTA, 7 mM beta-mercaptoethanol (BME)) containing 50 mM NaCl. One gram of chicken egg white lysozyme was added. The suspension was stirred for 60 min until viscous, and then sonicated three times for 6 min, with 20 min intervals for cooling. The lysate was clarified by centrifugation for 30 min at 14 000 rpm. The pellet was discarded, and the supernatant was centrifuged twice more until no further debris could be removed. The clear supernatant was loaded onto a 280-ml Heparin HyperD column (Pall), washed with the same buffer until the A280 returned to background level, and then chromatographed by HPLC (AKTA, GE) with lysis buffer containing a linear NaCl gradient spanning 50–600 mM NaCl. SwaI activity eluted around 350 mM. Active fractions were pooled (300 ml) and applied directly to an 80-ml ceramic hydroxyapatite column (Bio-Rad) equilibrated with Lysis Buffer. A gradient of KPO4 at pH 6.8 from 40 mM to 1 M was applied; SwaI eluted around 270 mM. Pooled fractions were dialyzed overnight against Column Buffer (20 mM Tris pH 8.0, 5% glycerol, 0.25 mM EDTA, 7 mM BME) containing 50 mM NaCl. The dialysate (60 ml) was pumped through a 19-ml Source15Q column (GE Healthcare) to remove nucleic acids, and then loaded onto a 15-ml Heparin TSK column (Tosoh Bioscience) for concentration. This was chromatographed with Column Buffer containing a gradient of NaCl from 50 mM to 1 M. Active fractions were pooled, dialyzed into ‘Diluent B’ (New England Biolabs: 10 mM Tris pH 7.4, 300 mM NaCl, 50% glycerol, 0.1 mM EDTA, 1 mM DTT), loaded onto a Superdex-75 size-exclusion column, and eluted with Column Buffer containing 300 mM NaCl. Fractions containing homogeneous SwaI were pooled (45 ml) and stored frozen at −80°C. The specific activity of purified SwaI was calculated to be 2.8 × 106 units/mg of protein.
Size exclusion chromatography demonstrated that the enzyme in solution elutes as a single sharp peak corresponding to a stable protein dimer of 53 kDa, in close agreement with the calculated mass based on the amino acid sequence of 2 × 26.8 kDa (data not shown).
Mutagenesis and crude-extract assays
Site-directed mutagenesis of SwaI residues Asp76, Asp93 and Lys95 was performed by PCR using Deep Vent DNA Polymerase (New England Biolabs). Double-stranded plasmid DNA containing the swaIR gene was mixed with complementary, 51-nt mutagenic oligonucleotides (IDT) containing the triplet GCG, to code for alanine in place of the target amino acid. The primers were extended by temperature cycling to form linear, full-length plasmids with duplicated ends incorporating the mutation. These were gel-purified (Zymo Research) and transformed into E. coli ER2566 containing the SwaI methyltransferase. Transformants that had resolved the terminal duplication in vivo to regenerate circular plasmids were selected by plating onto LB containing chloramphenicol and ampicillin, and incubated overnight at 37°C. Individual colonies were picked, inoculated into 5 ml LB containing chloramphenicol and ampicillin, grown overnight at 37°C, and the plasmids recovered by mini-prep spin-column purification. The complete nucleotide sequence of the swaIR gene within these plasmids was determined to verify that only the desired mutation was present.
One isolate of each sequence-verified mutant was grown at 37°C overnight in 10 ml LB containing chloramphenicol and ampicillin, alongside a control culture of ER2566 expressing wild-type SwaI. The cultures were harvested by centrifugation, re-suspended in 2 ml of sonication buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT), and lysozyme was added to a final concentration of 1 mg/ml. The suspensions were stored on ice for 1 h, and then disrupted by sonication and clarified by micro-centrifugation. The clarified extracts were assayed for SwaI endonuclease activity by incubation with purified phage T7 DNA or supercoiled plasmid pXba DNA (each containing one SwaI site) in NEBuffer 3.1 at 25°C for 1 h. Assays were performed as 2-fold titrations by adding 1 μl of clarified extract to 1 μg of DNA in 50 μl of reaction buffer, and successively transferring 25 μl aliquots to four additional tubes each containing 0.5 μg of DNA in 25 μl reaction buffer.
Crystallization, Data Collection and structure determination
The purified enzyme was dialyzed into a final buffer consisting of 25 mM Tris–HCl (pH 7.5) and 150 mM NaCl and concentrated to ∼20 mg/ml in that same buffer. Crystals of the unbound (‘apo’) enzyme were grown by equilibration against a reservoir solution containing 18% PEG1000 (v/v), 100 mM Tris–HCl (pH 8.5), 40 mM Ca(OAc)2, and 150 mM NaBr. Crystals of the uncleaved DNA-bound complexes were grown by mixing one microliter drops of the protein (± a 1.2–1.5 molar excess of double stranded DNA) with an equal volume of a crystallization reservoir solution consisting of 24–28% PEG1000 (w/v), 100 mM Tris–HCl, pH8.5, 5 mM CaCl2, 10 mM DTT and 5% iso-propanol, and then allowing the drop to equilibrate via vapor phase diffusion against 500 μl of the same reservoir solution. The sequence of the DNA strands that yielded crystals of the bound complex used for the structure determination corresponded to 5΄-GGGCGGAGGCATTTAAATGCCGCGCGG- 3΄ and its complement 5΄-CCCGCGCGGCATTTAAATGCCTCCGCC-3΄. Crystals of the cleaved enzyme-DNA complex were grown under similar conditions as above, and then washed extensively and incubated in the same reservoir solution, with 10 mM MgCl2 replacing CaCl2. The space group and unit cell dimensions of the crystals are listed in Table 1.
Table 1. SwaI Data collection and refinement statistics.
| Data set Id | Se-Peak | Se-inflection | Cleaved | Unbound |
|---|---|---|---|---|
| Wavelength (Å) | 0.9794 | 0.9796 | 1.5418 | 0.9202 |
| PDB ID CODE | 5TGX | 5TH3 | 5TGQ | |
| Data collection: | ||||
| Space group | P21 | P21 | P21 | P22121 |
| a (Å) | 109.86 | 109.86 | 109.75 | 48.39 |
| b (Å) | 57.06 | 57.06 | 57.07 | 65.23 |
| c (Å) | 112.79 | 112.79 | 113.44 | 67.57 |
| β (°) | 107.06 | |||
| Resolution (Å) | 50–2.3 | 50–2.8 | 50–2.33 | 50–1.98 |
| Unique reflections | 53972 | 32759 | 56787 | 17667 |
| Redundancy* | 6.4 (4.1) | 7.5 (7.5) | 7.1 (4.7) | 12.5 (6.4) |
| Completeness (%)* | 96.1 (79.9) | 100 (100) | 98.2 (85.9) | 96.3 (71.0) |
| I/σ(I) | 12.5 (1.1) | 16.3 ( 6.3) | 17.8 (1.35) | 31.6 (1.13) |
| Rmergea (%)* | 12.7 (85.1) | 10.6 (44.9) | 9.5 (98.4) | 6.5 (104.8) |
| B(iso) (Å2) | 30.46 | 33.9 | 39.5 | |
| Refinement statistics: | ||||
| Protein atoms# | 7582 | 7674 | 1905 | |
| DNA atoms# | 2316 | 2130 | —– | |
| Heavy atoms | 16 Se- | 15 Se | —– | |
| Catalytic metal ions | 4 Ca2+ | 4 Mg2+ | 2 Ca2+ | |
| Solvent molecules | 243 | 51 | 89 | |
| R-factorb (%)* | 19.8 | 19.85 | 19.8 | |
| R-freeb (%)* | 24.2 | 23.89 | 27.9 | |
| Rmsd | ||||
| Bond length (Å) | 0.017 | 0.0178 | 0.021 | |
| Angles (o) | 1.928 | 2.127 | 2.185 | |
| Ramachandran distribution (%) | ||||
| Core region | 96.70 | 96.52 | 95.48 | |
| Allowed region | 2.85 | 2.90 | 3.62 | |
| Outliers | 0.46 | 0.58 | 0.90 | |
*Highest resolution shell values in parenthesis.
#Crystals containing SeMet and Iodide.
aRmerge = Σ|Ihi – <Ih>|/ΣIh, where Ihi is the ith measurement of reflection h, and <Ih> is the average measured intensity of reflection h.
b R-factor/R-free = Σh|Fh(o) – Fh(c)|/Σh|Fh(o)|, where R-free was calculated with 5% of the data excluded from refinement.
Diffraction data were first collected on crystals of the uncleaved enzyme–DNA–calcium complex, containing selenomethionyl (SeMet) derivatized protein, at beamline 5.0.2 of the Advanced Light Source (ALS) synchrotron X-ray facility (Lawrence Berkeley National Laboratory). The crystal used for data collection was treated for 5 min with 1% H2O2 (v/v) in the same mother liquor prior to flash-cooling in liquid nitrogen. Data sets were collected with incident X-rays at two wavelengths, corresponding to the selenium fluorescence signal peak (0.9794 Å) and inflection (0.9796 Å), allowing the structure to be solved via multiple anomalous difference (MAD) phasing.
Additional data sets were collected on the unbound apo-enzyme at the ALS, and on the cleaved product complex (in the presence of magnesium) using a Rigaku rotating anode generator and and RAXIS-IV++ phosphor imaging plate area detector. Those latter structures were solved via molecular replacement, using the coordinates of a DNA-bound SwaI monomer as a molecular phasing search model. All data were processed and scaled using the DENZO/SCALEPACK (HKL2000) program package (15). The program PHENIX (16) was used for initial phase determination of the protein-DNA-calcium complex, and for generation of initial electron density map. The Refmac5 algorithm (17) and CCP4i graphical interface (18), in the CCP4 program suite (19) were used for refinements. The graphic package COOT (20) was used for model building. Figures were generated with PYMOL (21). Refinement statistics for all three crystal structures described in this study are provided in Table 1.
RESULTS
Overall structure and fold of SwaI
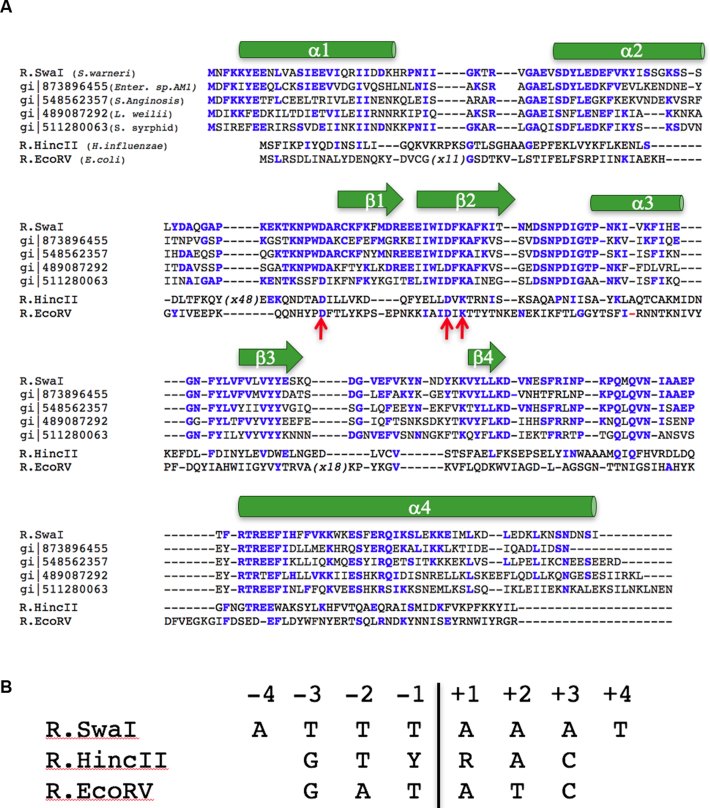
The SwaI restriction endonuclease is a homodimer containing 226 amino acids per subunit, corresponding to a mass of 26.8 kD and an estimated pI of 6.9. The sequence of the enzyme contains five methionine residues per protein chain, that were used for the initial phasing and structure determination of the DNA-bound enzyme complex. A search for sequence homologues using NCBI BLASTP (22) produces only five significant hits, all corresponding to hypothetical proteins of bacterial origin, with overall sequence identities ranging from 57% to 47%, corresponding to E-values of 2 × 10−89 to 7 × 10−63. A total of 56 residues (∼24%) are fully conserved across all six protein sequences (Figure 1A).
Figure 1.
(A) Alignment of SwaI against four of its closest sequence homologues identified within the NCBI protein database (39), and against its two nearest structural homologues (HincII and EcoRV) identified within the RCSB protein structure database (40). One additional significant sequence homologue (WP_066163638; REBASE Bsp13219ORF4205P) is not shown for clarity. Conserved residues are shown as colored bold residues. The overall identity between SwaI and its nearest sequence relatives is 46–57% identity, while the identity between SwaI and HincII or EcoRV is 14% and 11%, respectively (corresponding to 32 and 25 conserved residues, respectively). The position and boundaries of secondary structural elements in SwaI are shown above the aligned sequences, and the position of the conserved active site residues are indicated with red arrows. The ‘X48’ notation in the HincII sequence correspond to a large unique insertion of additional residues, that are not shown, relative to all the other aligned sequences. (B) Sequence and numbering of the DNA target sites recognized and cleaved by SwaI, HincII and EcoRV. ‘Y’ and ‘R’ refer to any pyrimidine and purine base, respectively, which corresponds to promiscuous recognition at positions ±1 by HincII.
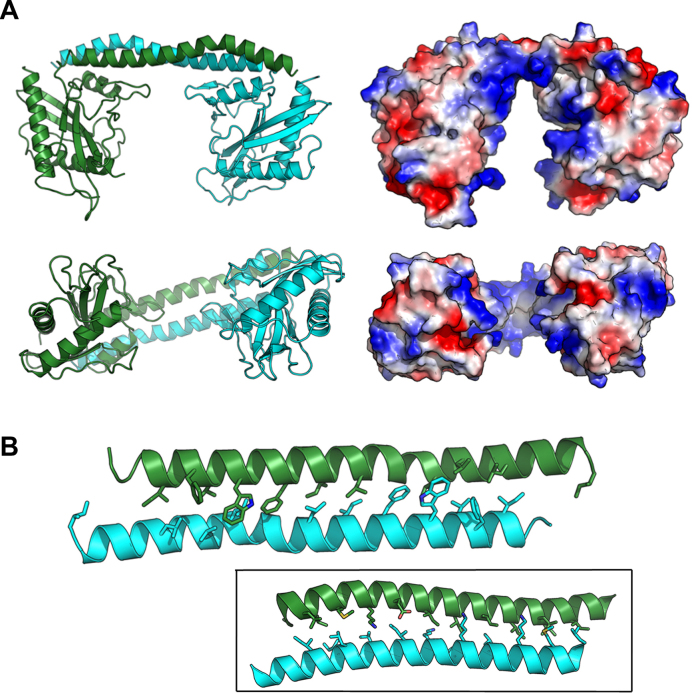
The crystal of the unbound SwaI enzyme contains one subunit per asymmetric unit, which together with its symmetry mate make up a functional dimer. The visible amino acids in the structure of unbound SwaI (Figure 2A) correspond to the entire 226-residue peptide chain of each subunit, with the exception of two surface loops (corresponding to residues 31–35 and 134–139) that are disordered.
Figure 2.
Structure of the unbound (apo-enzyme) form of SwaI. (A) On the left are ribbon diagrams of the SwaI homodimer (shown in two orientations), with the two protein subunits colored green and cyan, respectively. The primary interface between the protein subunits is a long domain-swapped C-terminal helix that forms an antiparallel bundled structure similar to a coiled-coil. The folded domains that extend below those two helices correspond to the α/β fold that comprises the core nuclease domain from the PD-(D/E)xK family. On the right are electrostatic surface charge representations of the enzyme homodimer, shown in the same orientation as the ribbon diagram. Positively charged (i.e. basic) regions are blue, while negatively charged (i.e. acidic) regions are red. Note the large basic cleft that separates the two folded nuclease domains. (B) Close-up of the domain-swapped C-terminal helical bundle, showing the array of aliphatic and aromatic residues that form the packed interface between the helical protein backbones. Inset: Similar domain-swapped helices from a specificity (S) subunit from a type I R/M system in Methanocaldoccus jannaschii (PDB ID 1YF2) that dictate the distance between target recognition domains (TRDs) in type I enzyme assemblages.
The first 180 residues of each subunit of the R.SwaI homodimer display a single folded domain, corresponding to a mixed α/β topology in an α−α−β−β−α−β−β arrangement that typifies nucleases containing a PD-(D/E)xK catalytic motif. A fourth and final C-terminal helix, extending from residue 184 to the C-terminus, forms a long domain-swapped helix that is significantly kinked at residues 211–213 and is packed against its symmetry mate, forming an amphipathic two-helix bundle structure and inter-subunit contacts that closely resemble an antiparallel coiled-coil peptide fold (23) (Figure 2B). The two folded domains of the enzyme homodimer are each ∼35–40 Å in width, and are physically separated by a gap of ∼30 Å between the opposing protein surfaces (which is spanned by the domain-swapped C-terminal helices extending from the end of each protein subunit). The residues lining the interior surfaces of the gap between protein domains (including multiple surface loops on each folded α/β protein domain and on the underside of the C-terminal helices) are predominantly basic.
The domain-swapped, interdomain two-helix bundle that bridges the two catalytic domains of SwaI (Figure 2B) is similar in length and shape to the coiled-coil structure bridging the specificity domains found in the specificity (S) subunits of Type I restriction endonucleases (which, in turn, controls the recognition distance between the target recognition domains TRD1 and TRD2 in those enzymes) (5). However, the amino acid composition of the helices in the type I S-subunits, including the interface between them (Figure 2B, inset), is rather dissimilar to that found in the SwaI enzyme; they lack multiple aromatic residues that are found in the helical interface in SwaI and instead containing a large number of basic residues that form several intersubunit contacts.
DNA recognition and binding
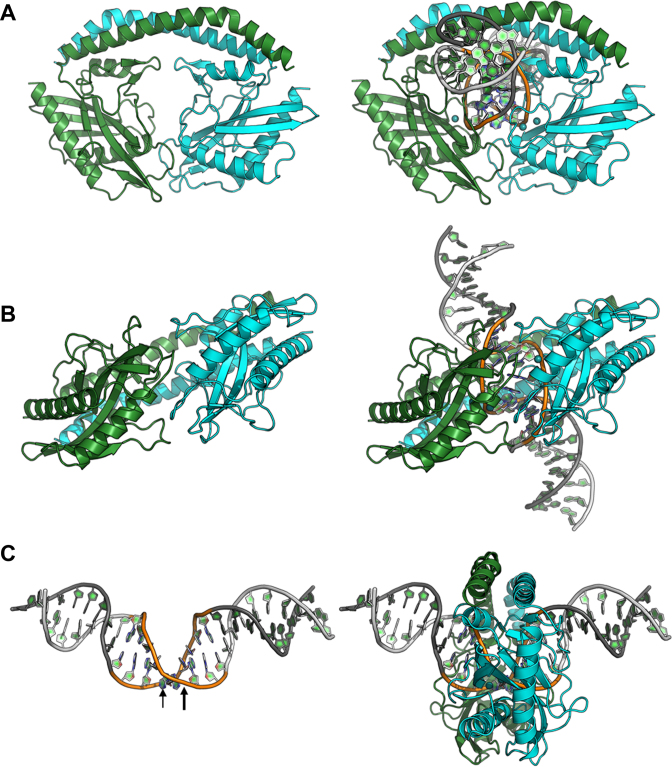
The structure of SwaI bound to its DNA target in the presence of calcium ions (as well as an additional structure of the complex solved in the presence of magnesium ions, described further below) demonstrate that the enzyme homodimer displays a significant conformational change corresponding to closure of the protein around the perimeter of the DNA duplex, and the formation of direct contacts to the DNA bases and backbone at various positions in both the major and minor groove (Figure 3A and B). The protein-bound DNA exhibits a sharp bend at the center of the DNA target, corresponding to a narrowed major groove across the central four base pairs and simultaneous widening of the minor groove and further separation of the target phosphates (Figure 3C).
Figure 3.
Structure of the DNA-bound form of SwaI in the presence of calcium ions. the structure of the same complex in the presence of magnesium ions (not shown) is virtually indistinguishable, with the exception of fully cleaved DNA ends in the two enzyme active sites (Figure 7). (A and B) Ribbon diagrams of the enzyme homodimer (colored as in Figure 2) with and without the bound DNA are shown for clarity. The enzyme undergoes a significant conformational closure around the DNA target site, that is augmented by smaller movements and ordering of surface loops along the nuclease domain (that present side chains to the DNA backbone and its nucleotide bases). (C) Bending of the DNA target (the 8 bases of the actual target are colored, and the location of the scissile phosphates are indicated with arrows) and corresponding position of the enzyme ring around the target site.
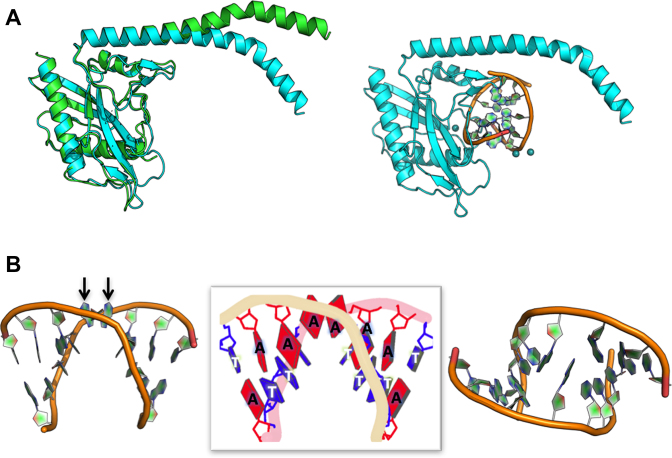
The conformational change exhibited by the protein is largely realized through bending of the domain-swapped C-terminal helices in each subunit (Figure 4A). Superposition of the nuclease core domain from individual protein subunits in the unbound and DNA-bound structures produces a relatively small backbone rmsd value, calculated over 285 superimposed residues, of ∼2 Å. In that superposition, the C-terminal tail residues from each subunit differ in position by over 20 Å. One of the two disordered loops in the DNA-free structure (residues 24–35) becomes ordered in the DNA-bound structures and contributes (i) a residue (Arg35) that is intimately involved in base contacts in the minor groove of the DNA and (ii) at least six possible H-bonds between the two protein subunits. Additional smaller conformational changes exhibited by several surface loops across the nuclease domain, that are located in the protein–DNA interface, result in additional contacts to the atoms of the DNA target site.
Figure 4.
Conformational changes that accompany protein-DNA binding. (A) The protein closure shown in Figure 3 is largely facilitated by a change in the bending of the C-terminal helix in each protein subunit. Other regions of the protein (i.e. the nuclease domain in each subunit) display much more limited motions that allow formation of atomic contacts to individual atoms in the DNA. (B) The DNA is bent by ∼50°, resulting in widening of the minor groove around the position of the two scissile phosphates. Of the 8 bp in the enzyme's target site, the central two A:T base pairs (at positions ±1) are disrupted. The two thymidine bases are left behind in the same relative position as if they were still engaged in base pairs, while the corresponding adenine bases (indicated by arrows in the left panel) have shifted towards the surface of the DNA duplex, where they form a reversed base stack that is flanked by two arginine residues and more distal phosphoribosyl groups of the flanking DNA backbone (shown in more detail in Figure 5).
At the same time that the protein undergoes the conformational rearrangement described above, bending of the DNA produces an unusual and significant disruption of the central two A:T base pairs at the middle of the target site. While the unpaired thymidine bases at each position (±1) remain in contact with their immediate 3΄ neighbor in the bent DNA duplex, their adenine counterparts display a dramatic movement: the two bases ‘leapfrog’ one another and form a reversed stack of two consecutive purine bases near the surface of the major groove (Figure 4B).
In contrast, as described below, very similar bending of palindromic DNA target sites by HincII and EcoRV (which also produce blunt-ended DNA targets, but recognize different target sequences and lengths) do not result in disruption of the Watson–Crick DNA base pairing arrangement at any position within their bound targets (11,12). The extent and nature of the overall DNA bending in all three structures is very similar, making it unlikely that the observed bend in their bound DNA substrates is inherent to the composition or sequence of their individual target sequences.
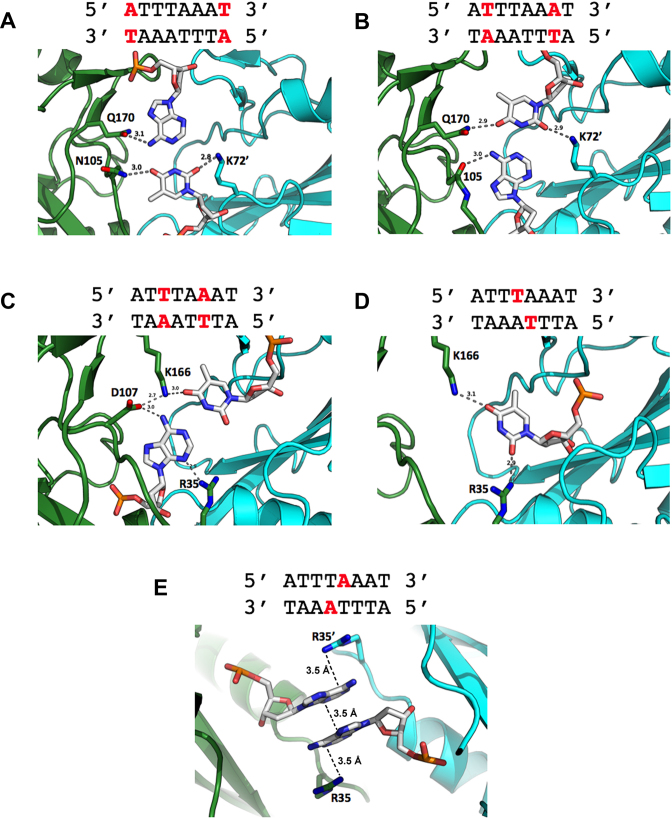
The observable contacts made by the protein to individual nucleotide bases in the target site are notable for their economical use of a small number of protein residues (illustrated and further described in Figure 5). Within each DNA half-site, seven out of eight bases are engaged in at least nine hydrogen-bond contacts in the major DNA groove and four additional contacts in the minor groove. The sum of these interactions involve just six amino acids: K72, N105 and Q170 interact with the first two base pairs of each half site (Figure 5, top panels), and R35, D107 and K166 interact with remaining two, albeit in a convoluted manner (Figure 5, lower panels). Four of these six protein side chains participate in contacts to multiple, neighboring bases.
Figure 5.
Atomic contacts made between SwaI and individual bases in its DNA target site. The individual panels show contacts to bases starting at the outermost base pairs (A–C) and progressively working towards the middle of the target site (D and E). For clarity, only contacts to bases in one half-site are shown (except for the adenine bases at the exact center of the target site); the contacts are identical in the two DNA half-sites. The eight bases in each DNA half-site are directly contacted by six amino acid side chains (five from one protein subunit, and one from the opposing subunit). Of those six protein residues, four appear to form bridging contacts to bases at immediately neighboring positions. Two contacts between protein backbone nitrogen atoms (from residues 105 and 107) and atoms on DNA bases (adenine N7 in panels 2 and 3, respectively) are not shown for clarity. Panel e illustrates the position and reversed base-stacking interactions formed between the adenine bases extracted from positions ±1 in the protein-DNA complex. That pair of bases is flanked by the side chains of arginine 35 and 35΄, which respectively form cation–π interactions with each base (while at the same time, also forming contacts to two additional bases). Additional non-specific contacts made between protein side chains and the DNA backbone are also not shown in the figure for clarity.
The unpaired thymidine of each of the central base pairs (at positions ±1) engages in two H-bonds—with K166 in the major groove, and R35 in the minor groove—while the partner adenine is removed from its original position in the DNA duplex, and forms a reversed stacking interaction with the corresponding adenine from the opposite strand. These two stacked adenine bases are flanked by a pair of symmetry-related arginine residues (Arg 35 and 35΄), which form cation-π interactions with each base (Figure 5, bottom panel). The same arginine residues also make apparent H-bond contacts both to adenine ±2 and to thymine ±1. Therefore, Arg 35 participates in multiple interactions spanning three separate paired and unpaired base positions in the DNA target.
Structural homologues
A search for structural homologues against SwaI using the DALI (24) and FATCAT (25) servers indicates that the two closest related molecules currently found in the RCSB PDB database are the R.HincII (‘HincII’) and R.EcoRV (‘EcoRV’) restriction endonucleases (aligned sequences and target sites shown in Figure 1A and B, respectively). Both of those enzymes are also homodimeric, Type IIP restriction endonucleases that recognize and cleave palindromic DNA target sites and also produced products with blunt ends. The more closely related of these two proteins, HincII, displays approximately a 3 Å backbone rmsd over 251 aligned residues, when using PDB ID 2gih (26) for comparison. Those two enzymes share 33 amino acids in common, corresponding to 14% sequence identity. EcoRV is more distantly related, displaying only 9% sequence identity and a 4 Å backbone rmsd versus SwaI across the same region of aligned residues.
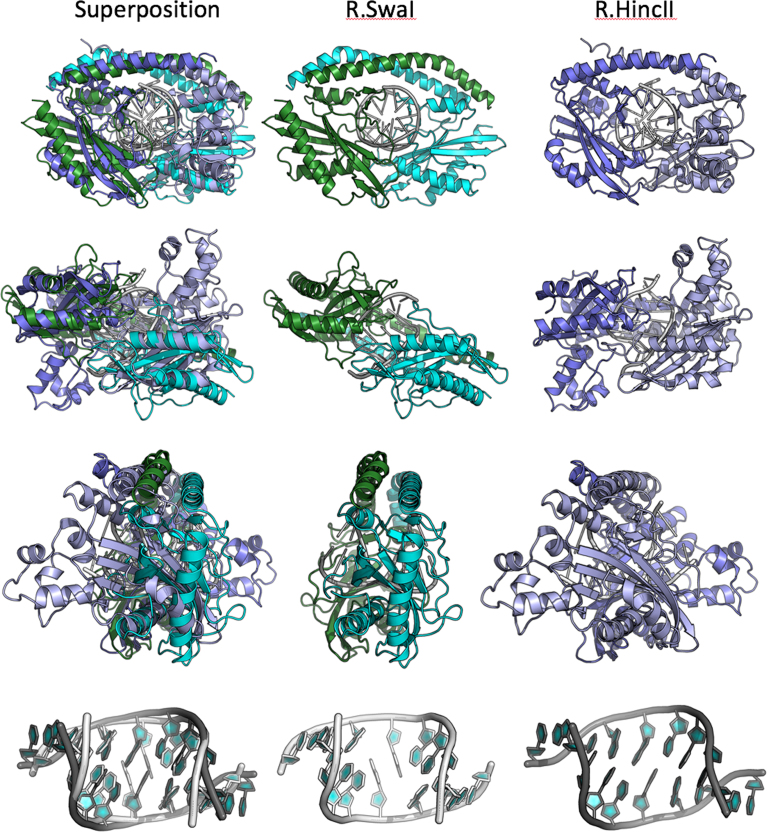
Superposition of the DNA-bound complexes of SwaI and HincII (Figure 6) indicates that both enzymes display similar overall tertiary structures and homodimeric organizations, and both also encircle their DNA targets. In both cases, the complex places the nuclease core domain and catalytic residues in similar positions near the scissile phosphates that produce blunt-ended products upon cleavage. Although the overall architectures displayed by the enzyme-DNA complexes are similar, the angle at which each enzyme's DNA target site penetrates the protein ring differs by ∼10–15°, and each enzyme possesses unique elaborations on their core folds.
Figure 6.
Superposition and comparison of the DNA-bound structures of SwaI and HincII. The left panels in each row show the superposition of both structures; the middle and right panels show the same elements in their individual molecular complexes. Each superposition is calculated based on the core nuclease residues of the helix and 3 β-strands that comprise the core elements of the PD-(D/E)xK nuclease motif (see Figure 7). Note that while the nuclease domains superimpose relatively closely, that the corresponding orientations of additional protein structural elements (such as the domain-swapped helices) and the bound DNA differ by at least 10° from one another. The bottom row shows the superposition of the two bound DNA targets for SwaI and HincII. While the overall bend angle and positions of the flanking bases (at positions 2–4 in each half-site) are similar, the effect of DNA binding and bending on the central two base pairs are radically different. In SwaI the central A:T base pairs are completely disrupted, whereas in HincII (and in EcoRV, not shown) they remain in a bent (but still base-paired) conformation.
Although the length (8 bp versus 6 bp), the sequence (ATTT|AAAT versus GTY|RAC), and the intermolecular angles of the bound DNA duplex engaged by SwaI and HincII differ as described above, the overall conformation and bend of the DNA substrates are similar, with the exception of the central two base pairs in each complex (Figure 6, bottom panels). Whereas the central two base pairs in the SwaI–DNA complex are disrupted, resulting in an unusual set of interactions between opposing adenine bases and the protein, in the HincII structure the same base pairs retain their Watson–Crick base pairing.
Active site architecture and mechanism of DNA cleavage
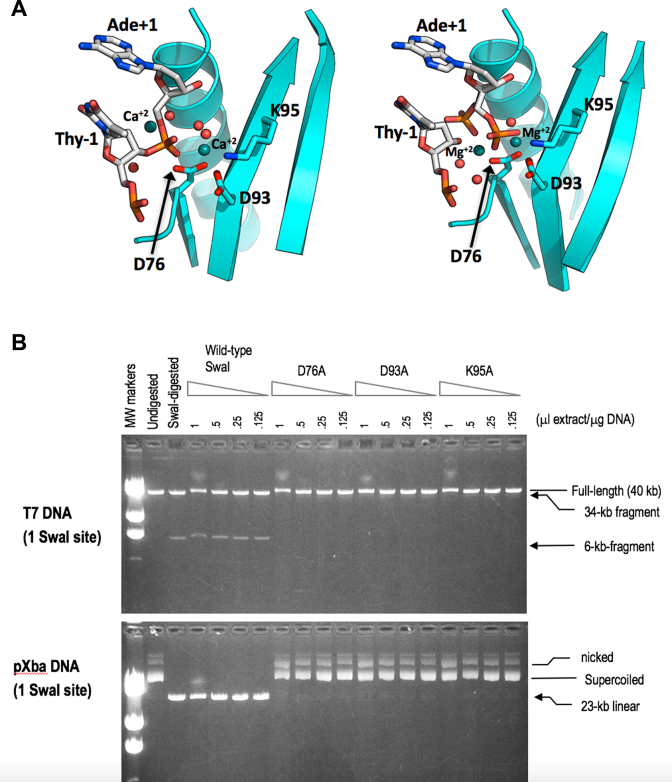
Examination of the structures of the SwaI–DNA complex in the presence of calcium ions (which results in a largely uncleaved complex) and in the presence of magnesium (resulting in a fully cleaved product complex) indicates that SwaI displays an active site structure and metal-dependent mechanism of phosphoryl hydrolysis typical of PD-(D/E)xK restriction endonucleases (27) (Figure 7A). In both structures, the target phosphate is flanked by two bound metal ions, one of which is in contact with a non-bridging phosphate oxygen and two conserved aspartates (D76 and D93), while the other is in contact with the 3΄ oxygen leaving group (as well as a 5΄ phosphate in the cleaved complex). A conserved lysine (K95) is positioned appropriately to act as a general base in the reaction and assist in activation of a metal-bound water that participates in hydrolysis.
Figure 7.
(A) The active site of SwaI in the presence of calcium ions (left panel) and in the presence of magnesium ions (right panel). In both structures, a pair of bound metal ions flank each scissile phosphate, near the location of an incoming nucleophilic water (and corresponding non-bridging oxygens) and near the 3΄ oxygen leaving group, respectively. The former metal ion is coordinated by six oxygen moieties: two acidic side-chain oxygens from conserved aspartate residues D76 and D93; the backbone carbonyl of F94; a non-bridging phosphate oxygen; and two water molecules. The conserved active site lysine (K95) is positioned appropriately to participate in activation of one of the metal-bound water molecules that can then serve an incoming nucleophile for in-line hydrolytic displacement. In the structure solved in the presence of magnesium, the DNA appears to be fully cleaved, and the free 5΄ phosphates in each DNA strand are observed in two distinct positions, as indicated in the figure and corresponding refined model. (B) DNA-cleavage assays of wild-type SwaI and catalytic-site mutants. Cell-extracts of wild type SwaI, and of alanine-substitutions of the three key residues of the presumptive catalytic site (D76, D93, and K95) were prepared and assayed by 2-fold serial dilution on linear phage T7 DNA (upper panel) and circular plasmid pXba DNA (lower panel). Wild type SwaI cleaved both substrates, converting T7 into 34-kb and 6-kb fragments, and pXba into the 23-kb linear form. The mutated enzymes, in contrast, displayed a trace of DNA-nicking activity at the highest enzyme concentration, but no DNA-cleavage activity, supporting the idea that these residues are essential for catalysis.
In the uncleaved complex, a water molecule is positioned in-line with the departing 3΄ oxygen, indicating that a standard nucleophilic displacement reaction mechanism, as has been well-established for many similar enzymes, is likely in place for SwaI. Mutational analysis of the putative active site residues D76, D93 and K95 confirmed that alanine substitutions at all three positions result in inactivation of the enzyme (Figure 7b).
Enzymatic digests comparing calcium ions in place of magnesium ions, indicate that in a 1 h incubation at 25°C, SwaI is completely inactive in 50 mM Tris–HCl pH 7.9, 100 mM NaCl, 0.1 mg/ml BSA, 10 mM CaCl2, but fully active in the same buffer containing 10 mM MgCl2 instead of 10 mM CaCl2 (Supplementary Data). While the crystal structure determined in the presence of calcium does indicate the presence of a minor (<50%) cleaved DNA species, the crystals used for that analysis were grown over a period of weeks at room temperature. Therefore, any cleavage products observed in that structure do not represent a physiologically relevant level of activity in the presence of calcium ions.
DISCUSSION
Even when Type II restriction endonucleases display primary sequences that differ significantly, they can share closely related tertiary folds, quaternary structures, catalytic mechanisms and similar cleavage products (7,8). A striking example of this principle is observed when comparing the enzymes SwaI, HincII and EcoRV. Pairwise alignments of any two of these restriction endonucleases indicates only 9–14% sequence identity (distributed rather uniformly across the entire length of their respective peptide chains) but these enzymes nonetheless display closely related tertiary structures (backbone rmsd values of ∼ 3–4 Å across ∼180 superimposable alpha carbons), as well as similar DNA-bound complexes. All three of these enzyme homodimers recognize and encircle palindromic base pair target sites, and they all cleave both DNA strands between the central nucleotides of their targets to create blunt ended DNA products.
Despite their similarities of form and function, the recognition properties of these enzymes differ significantly: EcoRV recognizes a single six base pair target site with high fidelity (5΄ GAT|ATC 3΄) (28), whereas HincII tolerates base pair alternatives at the central two positions of its target, displaying cleavage activity against the consensus sequence 5΄ GTY|RAC 3΄ (29) (where ‘Y’ corresponds to a pyrimidine and ‘R’ to a purine, and ‘|’ again indicates the site of cleavage). In contrast, SwaI recognizes an 8 nucleotide target site consisting solely of A:T base pairs (5΄ ATTT|AAAT 3΄) (10).
Crystallographic and biochemical analyses have previously demonstrated that EcoRV relies upon direct contacts between two threonine side chains and the extracyclic thymine methyl groups found at the central base pairs of its target, coupled with DNA bending that results in nearly complete unstacking between those base pairs to enforce recognition fidelity (30). Similar analyses of HincII have demonstrated that it exhibits ambiguous base pair discrimination at the same central base pairs (cleaving targets containing either of the two possible purine-pyrimidine steps with approximately equal efficiency). This is accomplished via a structural mechanism where the direct contacts to the central base pair positions described above for EcoRV are replaced with a single hydrogen bond to the N7 nitrogen of either purine base located at those same positions (12). That contact is complemented by DNA bending that is superficially similar to that displayed by EcoRV, but that instead results in a cross-strand stacking interaction between the same bases. Those interactions also appear to favor retaining the Py-Pu step at the center of the target site, rather than enforcing higher discrimination for a unique base pair step.
Despite its significantly different protein sequence relative to EcoRV and HincIII, SwaI displays considerable structural similarity to EcoRV and HincII and in the overall topology of its DNA bound complex, and also cleaves at the center of its target to create blunt product ends. Unlike both of those enzymes, however, SwaI recognizes and cleaves a single 8-base pair target site, containing only A:T base pairs (5΄-ATTT|AAAT-3΄). Unlike the mechanisms of specificity described above for EcoRV and HincII, particularly at the central base pairs of their target DNA palindromes, SwaI appears to rely upon a different mechanism to enforce fidelity at those same positions, that involves a dramatic disruption of the base pairing in the bound DNA duplex, and the formation of an unusual arrangement of unpaired nucleotide rings at the center of the bound DNA substrate.
The only previous described example involving a disruption and reorganization of multiple DNA base pairs within a restriction endonuclease–DNA complex is found in the structure the R.PacI restriction endonuclease, which (like SwaI) also recognizes and cleaves a long, palindromic DNA target comprising eight A:T base pairs (5΄-TTAAT|TAA-3΄) (13). Unlike SwaI, the PacI enzyme contains a ‘ββα-metal’ or ‘HNH’ nuclease-superfamily catalytic site and displays a completely unrelated tertiary structure. It also exhibits a completely different mode of DNA binding, and generates 5΄ overhangs rather than blunt ends. In the PacI complex, each and every base pair in the DNA target is removed and redistributed from its normal Watson-Crick base pairing arrangement (13). Because these two enzymes differ in almost every way in how they fold and function, it is tempting to speculate that their one remaining similarity (that they both recognize and cleave an 8 base pair target sites consisting solely of A:T base pairs) reflects an ability to exploit sequence-specific information that is inherent in (and perhaps unique to) such DNA target sites.
There are many known examples of A:T-rich DNA sequences being involved in key biological processes via their interaction with sequence-specific DNA-binding proteins, many of which clearly recognize such sequences via mechanisms that largely rely on shape recognition and complementarity, rather than formation of extensive networks of directional hydrogen-bonds within the protein-DNA interface. Classic examples include the interaction of the TATA binding protein with TATA box sequences in many eukaryotic promoters (reviewed in (31)) and the positioning of nucleosomes in a variety of AT-rich initiator elements (32,33). The ability of long tracts of AT-rich DNA sequences, often termed ‘A-tracts’, to form intrinsically curved DNA duplexes that can play a role in gene expression activity is also well documented (34,35). AT-rich repeat regions display a tendency to reversibly form hairpin and cruciform structures within the context of surrounding duplex DNA, due in part to their relatively low thermal stability in the duplex form combined with the inherent ability of palindromic sequences to form these structures (36). However, detailed structural and thermodynamic studies of such sequences (for example as described in (37)), have generally shown that when they are surrounded by sequences of higher GC content (similar to PacI and SwaI target sites found within the context of surrounding genomic DNA) they tend to maintain overall b-form duplex structure, while exhibiting localized fluctuation and bending that results in elevated variation of groove dimensions along that DNA sequence.
Based on the unusual details of DNA binding exhibited by PacI and SwaI towards similar DNA targets, one could ask (i) whether recognition of long symmetric sequences comprised solely of A:T nucleotide pairs might rely upon unique structural and dynamic properties of such sequences; and (ii) whether enzymes that act upon such sequences either recognize unique (and perhaps transiently populated) structural features displayed by such targets in the absence of bound protein, or instead induce such structural perturbations solely after DNA binding. Similar questions have been examined in the past for enzymes that act upon DNA substrates with flipped-out bases (for a recent review of experimental approaches and results addressing this question, see (38)). A variety of non-crystallographic methods for further studies of long A:T-rich target sequences of the types recognized by PacI and SwaI, including the use of fluorescent base analogues (as probes of DNA conformation during enzymatic action) and the use of rapid NMR relaxation techniques (to examine the dynamic behavior of such sequences prior to protein binding) may eventually provide important new insight into their properties and recognition mechanisms.
ACCESSION NUMBERS
Three macromolecular crystal structures: RCSB PDB 5TGX, RCSB PDB 5TH3 and RCSB PDB 5TGQ.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 GM105691]; Fred Hutchinson Cancer Research Center [Discretionary and Endowment Funds]. Funding for open access charge: U.S. Department of Health and Human Services; National Institutes of Health.
Conflict of interest statement. Three of the co-authors of this manuscript are employees of New England Biolabs; the enzyme described in this paper (R.SwaI restriction endonuclease) is a commercial product sold by that company. B.L.S. is a Senior Executive Editor for Nucleic Acids Research.
REFERENCES
- 1. Loenen W.A., Dryden D.T., Raleigh E.A., Wilson G.G., Murray N.E.. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 2014; 42:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roberts R.J. How restriction enzymes became the workhorses of molecular biology. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5905–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J., Blumenthal R.M., Degtyarev S., Dryden D.T., Dybvig K. et al. . A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003; 31:1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roberts R.J., Vincze T., Posfai J., Macelis D.. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015; 43:D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loenen W.A., Dryden D.T., Raleigh E.A., Wilson G.G.. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014; 42:20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rao D.N., Dryden D.T., Bheemanaik S.. Type III restriction-modification enzymes: a historical perspective. Nucleic Acids Res. 2014; 42:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pingoud A., Wilson G.G., Wende W.. Type II restriction endonucleases–a historical perspective and more. Nucleic Acids Res. 2014; 42:7489–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bujnicki J.M. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the “midnight zone" of homology. Curr. Protein Pept. Sci. 2003; 4:327–337. [DOI] [PubMed] [Google Scholar]
- 9. Gowers D.M., Bellamy S.R., Halford S.E.. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004; 32:3469–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lechner M., Frey B., Laue F., Ankenbauer W., Schmitz G.. SwaI, a unique restriction endonuclease from Staphylococcus warneri, which recognizes 5΄-ATTTAAAT-3΄. Fresenius Z. Anal. Chem. 1992; 343:123–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winkler F.K., Banner D.W., Oefner C., Tsernoglou D., Brown R.S., Heathman S.P., Bryan R.K., Martin P.D., Petratos K., Wilson K.S.. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993; 12:1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horton N.C., Dorner L.F., Perona J.J.. Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation. Nat. Struct. Biol. 2002; 9:42–47. [DOI] [PubMed] [Google Scholar]
- 13. Shen B.W., Heiter D.F., Chan S.-H., Wang H., Xu S.-Y., Morgan R.D., Wilson G.G., Stoddard B.L.. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure. 2010; 18:734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kong H., Higgins L.S., Dalton M.A.. Method for cloning and producing the SwaI restriction endonuclease. 2001; US Patent 6245545 B1.
- 15. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 16. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winn M.D., Murshudov G.N., Papiz M.Z.. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003; 374:300–321. [DOI] [PubMed] [Google Scholar]
- 18. Potterton E., Briggs P., Turkenburg M., Dodson E.. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003; 59:1131–1137. [DOI] [PubMed] [Google Scholar]
- 19. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. . Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 21. The PyMOL Molecular Graphics System. Version 1.8. Schrödinger, LLC. [Google Scholar]
- 22. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 23. Busch S.J., Sassone-Corsi P.. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990; 6:36–40. [DOI] [PubMed] [Google Scholar]
- 24. Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye Y., Godzik A.. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003; 19(Suppl. 2):ii246–ii255. [DOI] [PubMed] [Google Scholar]
- 26. Joshi H.K., Etzkorn C., Chatwell L., Bitinaite J., Horton N.C.. Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism. J. Biol. Chem. 2006; 281:23852–23869. [DOI] [PubMed] [Google Scholar]
- 27. Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski K.. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012; 40:7016–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schildkraut I., Banner C.D., Rhodes C.S., Parekh S.. The cleavage site for the restriction endonuclease EcoRV is 5΄-GAT/ATC-3΄. Gene. 1984; 27:327–329. [DOI] [PubMed] [Google Scholar]
- 29. Kelly T.J.J., O S.H.. A restriction enzyme from Haemophilus influenzae. J. Mol. Biol. 1970; 51:393–409. [DOI] [PubMed] [Google Scholar]
- 30. Horton N.C., Perona J.J.. Crystallographic snapshots along a protein-induced DNA-bending pathway. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5729–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998; 62:465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elmendorf H.G., Singer S.M., Pierce J., Cowan J., Nash T.E.. Initiator and upstream elements in the alpha2-tubulin promoter of Giardia lamblia. Mol. Biochem. Parasitol. 2001; 113:157–169. [DOI] [PubMed] [Google Scholar]
- 33. Iyer V., Struhl K.. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995; 14:2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carmona M., Magasanik B.. Activation of transcription at sigma 54-dependent promoters on linear templates requires intrinsic or induced bending of the DNA. J. Mol. Biol. 1996; 261:348–356. [DOI] [PubMed] [Google Scholar]
- 35. Olson W.K., Zhurkin V.B.. Biological Structure and Dynamics. 1996; Schenectady NY: Adenine Press; 341–370. [Google Scholar]
- 36. Benham C.J., Savitt A.G., Bauer W.R.. Extrusion of an imperfect palindrome to a cruciform in superhelical DNA: complete determination of energetics using a statistical mechanical model. J. Mol. Biol. 2002; 316:563–581. [DOI] [PubMed] [Google Scholar]
- 37. Ulyanov N.B., Bauer W.R., James T.L.. High-resolution NMR structure of an AT-rich DNA sequence. J. Biomol. NMR. 2002; 22:265–280. [DOI] [PubMed] [Google Scholar]
- 38. Jones A.C., Neely R.K.. 2-Aminopurine as a fluorescent probe of DNA conformation and the DNA-enzyme interface. Q. Rev. Biophys. 2015; 48:244–279. [DOI] [PubMed] [Google Scholar]
- 39. Gish W., Stated D.J.. Identification of protein coding regions by database similarity search. Nat. Genet. 1993; 3:266–272. [DOI] [PubMed] [Google Scholar]
- 40. Rose P.W., Prlic A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J. et al. . The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015; 43:D345–D356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.