Abstract
Objectives:
Hispanics represent an understudied inflammatory bowel disease (IBD) population. Prior studies examining genetic predisposition to IBD in Hispanics are limited. In this study, we examined whether European-derived IBD variants confer risk in Hispanics and their influence on IBD phenotype in Hispanics compared to non-Hispanic whites (NHW).
Methods:
Self-identified Hispanics and NHWs with IBD were included. Hispanic controls were included for our genetic analyses. We performed single-variant testing at previously identified Crohn's disease (CD) and ulcerative colitis (UC) IBD variants in Hispanic cases and controls. These risk variants were used to compute individual genetic risk scores. Genetic risk scores and phenotype associations were compared between Hispanic and NHW.
Results:
A total of 1,115 participants were included: 698 controls and 417 IBD patients (230 Hispanics). We found evidence of association within our Hispanic cohort at 22 IBD risk loci, with ~76% of the risk loci demonstrating over-representation of the European risk allele; these included loci corresponding to IL23R and NOD2 genes. CD genetic risk score for Hispanics (199.67) was similar to the score for NHW (200.33), P=0.51; the same was true in UC. Genetic risk scores did not predict IBD phenotype or complications in Hispanics or NHW except for a younger age of CD onset in Hispanics (P=0.04).
Conclusions:
This study highlights the fundamental importance of these loci in IBD pathogenesis including in our diverse Hispanic population. Future studies looking at non-genetic mechanisms of disease are needed to explain differences in age of presentation and phenotype between Hispanics and NHW.
Introduction
Despite the rising Hispanic population with inflammatory bowel disease (IBD), only a few studies have looked at disease characteristics in this group.1, 2, 3, 4, 5 In our prior study, we found that differences in age of presentation and IBD phenotype exist within Hispanics (US vs. foreign-born) and between Hispanics and non-Hispanic whites (NHW).3 The exact mechanism explaining differences within Hispanics remains unexplored. Studies examining immigrants arriving into IBD-prevalent countries suggest that not all migrants have the same predisposing risk to IBD onset.6 This alludes to genetic influence on IBD expression in spite of a similar “IBD-promoting” environment.
The first large-scale trans-ancestry association study of IBD examining East Asian, Indian and Iranian populations found genetic heterogeneity among populations at European-established risk loci (including NOD2 and IL23R);7 highlighting the need to study diverse populations to understand differing mechanisms of IBD pathogenesis. Thus far, genetic studies of IBD in Hispanics lag behind GWAS studies investigating North Americas, European Caucasians, Asians and middle Easterners.7, 8, 9, 10, 11, 12 In a recent study examining Puerto Rican IBD patients, risk allele frequencies for NOD2 and IL23R were higher in the IBD Puerto Rican population compared to Puerto Rican controls.13 This study suggests that at least a handful of established European-derived IBD risk loci may also confer risk to IBD in Hispanics. In addition, given the relatively superficial nature of genome-wide association studies in examining genotype–phenotype associations, studies using a composite genetic risk score across various autoimmune diseases, including IBD, have shown stronger correlations with IBD phenotype than individual single-nucleotide polymorphism (SNPs).14, 15, 16, 17, 18
In this study, we have generated specific genotyping data to investigate the association of previously identified IBD risk variants with IBD in our Hispanic sample. We examined genetic risk score in the context of age of presentation and IBD phenotype, including extent of disease and surgeries. We proposed that differences between Hispanics and non-Hispanics exist in the frequency of established IBD risk alleles and in their global prediction (as a composite genetic risk score) of disease phenotype. Our study is the first to explore the genetic profile of a diverse sample of Hispanics with IBD and to incorporate the most recently described additional IBD risk alleles into a composite genetic risk score.
Methods
Study design
Data were collected with the approval of the Institutional Review Boards at the University of Miami Miller School of Medicine and Jackson Memorial Hospital (Miami, FL, USA). Information was collected prospectively from adult Gastroenterology Clinics. Patients provided detailed demographic and medical information. Foreign-born patients were asked year of immigration US immigration. Blood samples were collected and stored within the Biorepository facility at the John P. Hussman Institute for Human Genomics (HIHG) for genetic analyses. Self-identified white Hispanics and NHWs were included. IBD patient samples were genotyped on the ImmunoChip array (a custom designed Illumina iSelect panel).19, 20 Control samples were self-identified Hispanics who were participating in a large genetic study of cardiovascular disease at our institution. General health questions are asked of controls in order to identify and exclude any moderate to severe cases of ulcerative colitis (UC) or Crohn's disease (CD) from this sample set. These unrelated controls were also genotyped within the HIHG using a custom Illumina genotyping array.
Phenotypic outcomes
IBD diagnosis and phenotype classification was confirmed by review of source documents and/or the clinical, endoscopic, radiologic, and pathologic findings available to the attending physician in clinic. Phenotyping for Crohn's disease and ulcerative colitis was performed according to the Montreal classification. We recorded information on surgical outcomes including abdominal surgeries (small bowel intestinal resection and subtotal or total proctocolectomies) and perianal surgeries (seton placement, mucosal advancement flaps, etc). Abdominal surgeries for CD were analyzed as a sum of outcomes and separately from perianal CD surgeries. UC surgeries (total proctocolectomies) were analyzed separately from surgeries done for CD. A history of IBD-related hospitalizations occurring from symptom onset to the time of the questionnaire were recorded.
Genotyping and genetic risk scores
There are >200 IBD susceptibility variants described in a largely European-derived ancestry population.7, 8 The ImmunoChip array produced raw genotype data on >192,000 SNPs, including 232 previously published IBD risk variants. These raw data were loaded into the Genome Studio software suite for genotype calling, using the PLINK21 software package and R (Version 3.0.2)22 for further statistical analysis. Standard genotyping quality control checks were performed to identify genotyping or sample errors. SNPs were examined for genotyping efficiency (SNPs dropped if call rate <95%) and deviations from Hardy Weinberg equilibrium (SNPs dropped if P<0.001). Controls were genotyped using the Affymetrix SNP array 6.0, with raw genotype data having undergone similar standard quality control.23 In controls 80/232 SNPs were genotyped, with an additional 139 SNPs being successfully imputed. Imputation to 1000 Genomes was performed using IMPUTE2 (ref. 24) as described previously.23 All the imputed SNPs included in the analyses were of good quality (INFO>0.45). The genotyping arrays utilized to produce the genetic data for both the cases and controls are designed and run using the same chemistry and the same platform (i.e., the Illumina Infinium platform). For the analyses presented in the manuscript, we simply extracted the available genetic data for the variants (or SNPs) demonstrating previous associations. Following these standard measures of quality control, we were left with 219 of 232 SNPs with high quality genotyping and/or imputation data available for both our single variant and genetic risk score analyses.
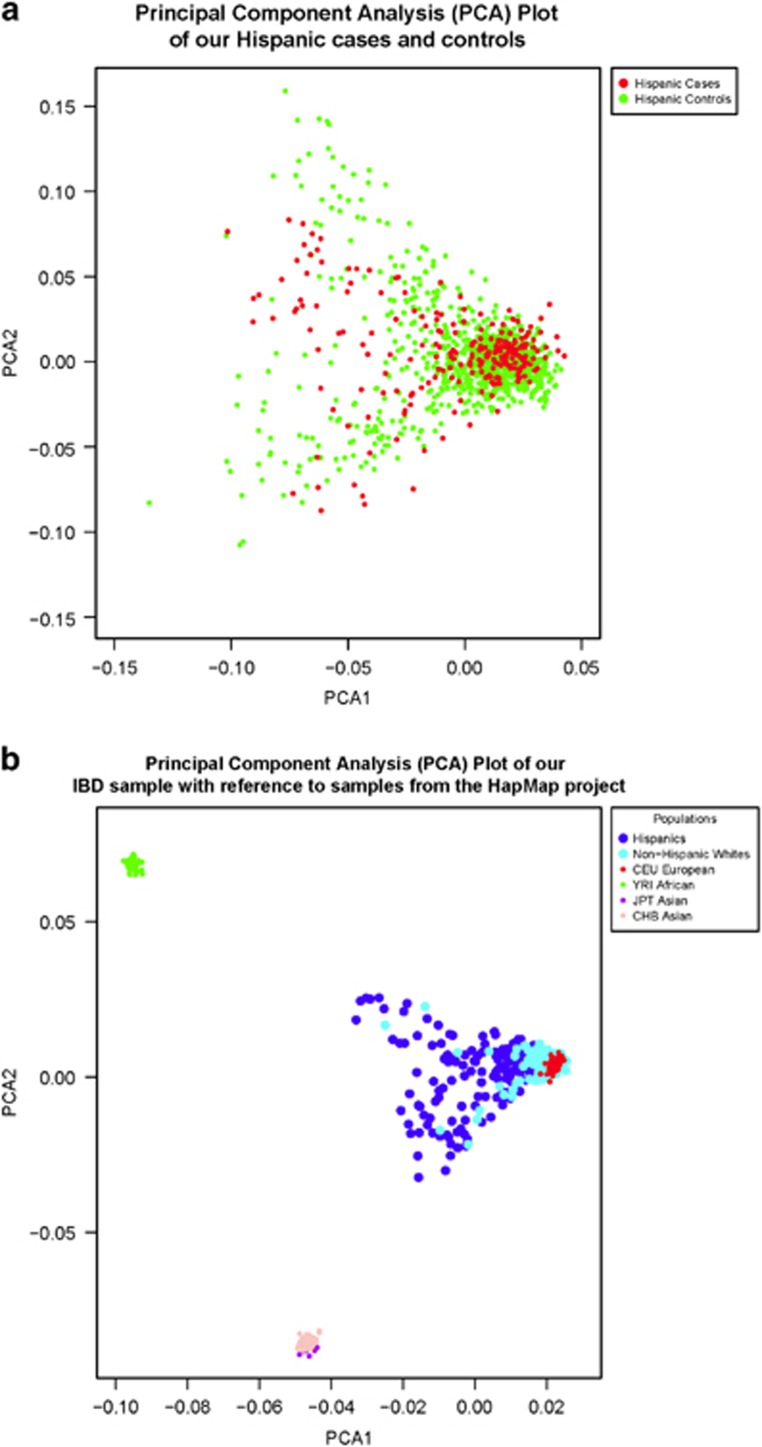
We calculated principal components (PCs) using Eigenstrat25 to both remove significant population sample outliers and to use as covariates in logistic regression. To calculate the PCs, we utilized 7,900 SNPs pruned for linkage disequilibrium (pairwise r2<0.1) from 22,635 total SNPs found in common between the genotyped cases and controls (no imputed SNPs were used to calculate PCs), and excluding the known associated IBD SNPs and those across the HLA region. Eight control samples were excluded as they were more than six s.d.'s from the mean on any of the first 10 PCs. In Figure 1a we plot the first two PCs for our Hispanic cases and controls and illustrate the population substructure of our Hispanic and non-Hispanic samples relative to those of the HapMap project (Figure 1b). These figures show that Hispanic cases and controls are genetically similar.
Figure 1.
These PC analysis (PCA) plots illustrate the clustering of samples according to their genetic background, utilizing genotype data from 7,900 linkage disequilibrium pruned single-nucleotide polymorphism (SNPs) overlapping our Hispanic cases/controls and our non-Hispanic cases. (a) Hispanic cases and controls. Using PC1 and PC2, we highlight that our Hispanic cases and controls share a common global ancestry structure. (b) Ancestral background distribution of our data set relative to the HapMap populations. In addition to samples in our data set (“Hispanics” and “non-Hispanics”), data from the International HapMap Project populations (CEU, YRI, JPT, and CHB) are included for illustration purposes. Asian (JPT and CHB) and African (YRI) population samples cluster in distinct groups, with the vast majority of our non-Hispanic whites samples clustering with the CEU population. As anticipated, our Hispanic samples demonstrate evidence of admixture as they spread from the cluster representing European ancestry.
We calculated separate genetic risk scores for both CD and UC within our cases. These risk scores are comprised of the weighted sum of the 219 SNPs demonstrating association with either CD or UC.7 Scores were calculated by multiplying the number of risk alleles for each individual SNP by the natural log of the reported odds ratio for the risk allele, and then summing across all SNPs (i.e., all SNPs associated with either IBD or CD were used to compute a genetic risk score for CD cases and all SNPs associated with either IBD or UC were used to compute a score for UC cases).7, 14 This genetic risk score was calculated for each affected individual based on his or her specific diagnosis and used as a proxy for genetic risk burden in each individual.
Statistical analysis
Demographics, IBD phenotype and mean genetic risk scores were compared between Hispanics and NHWs using χ2 and analysis of variance. A case/control dosage-based test of association for 219 previously reported IBD SNPs7, 8 was performed in PLINK21 using a logistic model and adjusting for the first three PCs. For this analysis, the genotype data for the 219 SNPs genotyped in cases and the 80 SNPs genotyped in controls was converted to dosage using GTOOL (Version 0.7.5) and merged with the imputed dosage data for the 139 SNPs that were imputed in controls. To reduce type one error and to account for multiple testing, permutation testing was performed using the max(T) permutation procedure implemented in PLINK,21 specifying 10,000 permutations. Both uncorrected and permuted P values for these single variant analyses are provided in Table 2 (Supplementary Table 1).
Genotype–phenotype associations were performed using R (Version 3.0.2).22 Genetic risk score, corresponding to an individual's specific diagnosis (either CD or UC), was analyzed as a continuous variable and incorporated into general linear models. We performed multivariable regression analysis of genetic risk scores with multiple outcomes (e.g., number of surgeries, etc) and adjusted for population stratification using the first three PCs and utilized the max(T) permutation procedure (permuting the phenotypes 10,000 times across the samples) to report permuted non-parametric P values (Table 3). The assumptions of linearity, independence of errors, homoscedasticity, unusual points and normality of residuals were met in our multivariable regression model.
Results
Population demographics
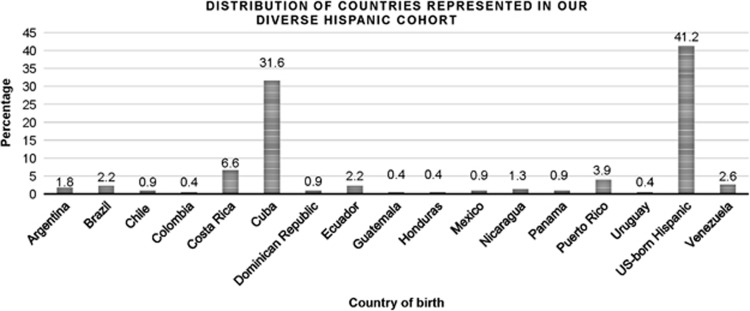
Our research study included 1,115 participants from the culturally and ethnic diverse population present in South Florida: 698 Hispanic controls, 230 Hispanics with IBD and 187 non-Hispanics with IBD. Most Hispanics with IBD were foreign-born (58.8%) with Cubans representing the largest proportion of foreign-born Hispanics (72 patients, 53.3%). In all, foreign-born Hispanics originated from 16 different countries (Figure 2). This patient cohort is distinct from the one described in an earlier publication on the phenotype of Hispanic patients.3 US-born Hispanics were mostly first generation (83.3%). We found that our Hispanic cases and controls are ancestrally well matched and are related as expected with ancestral populations (Figure 1).
Figure 2.
This bar graph displays the distribution of countries represented in our sample of Hispanics. US-born Hispanics represent the majority followed by Cubans.
IBD patient characteristics
In our IBD sample of Hispanics and NHWs, all groups had mostly CD (~63% Table 1). There were 261 CD patients and 156 UC patients including both Hispanic and NHWs. US-born Hispanics were diagnosed younger than foreign-born Hispanics, similar to what we found in our prior cohort.3 All demographic characteristics can be seen in Table 1. Clinical characteristics including location of luminal disease, CD-behavior (e.g. fistulizing and perforating) and surgeries were similar between Hispanics and NHWs with UC and CD (data not shown).
Table 1. Patient demographics according to ethnicity.
|
Hispanics with IBD
N=230 |
Controls N=698 | NHW with IBD N=187 | |||||
|---|---|---|---|---|---|---|---|
| US born N=94 | Foreign-born N=135 | Combined N=229a | Hispanics vs. NHW P value | US vs. foreign-born Hispanic P value | |||
| Crohn's disease n (%) | 58 (62) | 84 (62) | 142 (63) | NA | 118.0 (63) | 1.00 | 1.00 |
| Female n (%) | 49 (52) | 63 (47) | 112(49) | 226 (32) | 99.0 (53) | 0.43 | 0.42 |
| Age of diagnosis mean (years, s.d.) | 22.7 (8) | 36.6 (14) | 30.9 (20) | NA | 29.2 (16) | 0.22 | <1.00E-03 |
| Age of symptom onset mean (years, s.d.) | 22.0 (9) | 36.2 (21) | 30.3 (15) | NA | 27.3 (18) | 0.03 | <1.00E-03 |
| Disease duration from symptom onset mean (years, s.d.) | 8.8 (10) | 8.6 (10) | 8.7 (10) | NA | 13.5 (17) | <1.00E-03 | 0.86 |
| Family history, n (%)b | 10.0 (11) | 20.0 (15) | 30.0 (13) | – | 39.0 (21) | 0.02 | 0.04 |
| Active smoking at diagnosisb | 10.0 (11) | 16.0 (15) | 26.0 (13) | NA | 32.0 (20) | 0.11 | 0.5 |
| CD, n (%) | 8.0 (15) | 12.0 (17) | 20.0 (16) | – | 24.0 (23) | 0.22 | 0.53 |
| UC, n (%) | 2.0 (6) | 4.0 (11) | 6.0 (8) | – | 8.0 (15) | 0.43 | 0.39 |
IBD, inflammatory bowel disease; NHW, non-hispanic whites; s.d., standard deviation; UC, ulcerative colitis.
One patient was missing country of birth.
Data was incomplete for some UC patients
The bold highlights the significant P-values.
Associations with previously identified IBD risk loci in Hispanic patients
We tested 219 previously identified IBD susceptibility variants for association in our data set. We found nominally significant (P<0.05) evidence of association for 22 variants (6 IBD, 10 CD, and 6 UC; Table 2). Results for the remaining 197 variants tested can be found in Supplementary Table 1. After correcting for multiple testing, our most significant single-variant associations for CD and IBD were both found within the IL23R gene (encodes a subunit of the receptor for the pro-inflammatory cytokine interleukin-23) locus on chromosome one (rs7517847 and rs80174646 respectively), with our top UC association found on chromosome 16 nearest the KIR2DL1/2 locus. Our top two most significant variants are also the most significant variants seen by Liu et al. for all known associations in Europeans.7 A total of 166 out of 219 (76%) variants tested have over-representation of the European risk allele in IBD cases (Table 2; Supplementary Table 1). We did however also find statistically significant evidence for two SNPs (rs11739663 for UC and rs7438704 for CD) demonstrating effects in the opposite direction (i.e., protective for our cohort) compared to previous reports.7, 8
Table 2. Association results of established IBD risk variants in Hispanics.
| CHR | SNP | Position (bp) | Trait | Candidate genes | RA (EUR) | OR (EUR) | RAF (hisp cases) | RAF (hisp controls) | RAF (EUR) | RAF (AMR) | RAF (AFR) | OR | SE | P valuea |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs3806308 | 20,142,866 | UC | G | 1.19 | 0.70 | 0.61 | 0.63 | 0.56 | 0.71 | 1.57 | 0.179 | 9.50E-03 | |
| 1 | rs7517847 | 67,681,669 | CD | IL23R,IL12RB2 | A | 1.40 | 0.67 | 0.55 | 0.58 | 0.43 | 0.84 | 1.66 | 0.140 | <0.0001b |
| 1 | rs80174646 | 67,708,155 | IBD | IL23R,IL12RB2 | C | 1.88 | 0.96 | 0.92 | 0.93 | 0.94 | 0.96 | 2.31 | 0.262 | 8.00E-04 |
| 1 | rs2651244 | 70,995,562 | UC | G | 1.06 | 0.77 | 0.66 | 0.60 | 0.78 | 0.91 | 1.83 | 0.204 | 2.60E-03 | |
| 1 | rs4845604 | 151,801,680 | UC | RORC | G | 1.18 | 0.90 | 0.85 | 0.86 | 0.87 | 0.71 | 2.27 | 0.374 | 2.72E-02 |
| 2 | rs925255 | 28,614,794 | CD | FOSL2,BRE | G | 1.11 | 0.66 | 0.60 | 0.54 | 0.72 | 0.75 | 1.37 | 0.143 | 2.77E-02 |
| 2 | rs7608910 | 61,204,856 | IBD | REL,C2orf74 | G | 1.13 | 0.45 | 0.37 | 0.36 | 0.27 | 0.38 | 1.35 | 0.112 | 7.60E-03 |
| KIAA1841,AHSA2 | ||||||||||||||
| 2 | rs17229285 | 199,523,122 | UC | G | 1.10 | 0.62 | 0.54 | 0.50 | 0.58 | 0.91 | 1.41 | 0.171 | 4.63E-02 | |
| 4 | rs7438704 | 48,363,245 | CD | TXK,TEC,SLC10A4 | G | 1.09 | 0.53 | 0.60 | 0.64 | 0.54 | 0.46 | 0.73 | 0.132 | 1.66E-02 |
| 4 | rs13126505 | 102,865,304 | CD | A | 1.20 | 0.09 | 0.05 | 0.06 | 0.03 | 0.00 | 1.88 | 0.245 | 8.70E-03 | |
| 5 | rs11739663 | 594,083 | UC | SLC9A3 | A | 1.08 | 0.70 | 0.79 | 0.77 | 0.82 | 0.52 | 0.63 | 0.177 | 6.80E-03 |
| 5 | rs11742570 | 40,410,584 | CD | PTGER4 | G | 1.28 | 0.65 | 0.57 | 0.59 | 0.44 | 0.61 | 1.37 | 0.137 | 2.25E-02 |
| 5 | rs10051722 | 130,104,076 | CD | A | 1.09 | 0.73 | 0.66 | 0.68 | 0.55 | 0.64 | 1.34 | 0.150 | 4.74E-02 | |
| 6 | rs116392568 | 31,274,380 | CD | HLA-C,PSORS1C1 | G | 1.16 | 0.45 | 0.35 | 0.36 | 0.30 | 0.27 | 1.48 | 0.138 | 3.90E-03 |
| NFKBIL1,MICB | ||||||||||||||
| 8 | rs13277237 | 130,604,563 | IBD | G | 1.06 | 0.51 | 0.44 | 0.43 | 0.44 | 0.56 | 1.33 | 0.111 | 9.60E-03 | |
| 9 | rs10781499 | 139,266,405 | IBD | CARD9,PMPCA, | A | 1.17 | 0.47 | 0.40 | 0.40 | 0.57 | 0.25 | 1.31 | 0.110 | 1.53E-02 |
| SDCCAG3,INPP5E | ||||||||||||||
| 13 | rs9525625 | 43,018,030 | CD | AKAP1, TFSF11 | A | 1.08 | 0.61 | 0.53 | 0.46 | 0.54 | 0.71 | 1.47 | 0.141 | 6.50E-03 |
| 16 | rs2066844 | 50,745,926 | CD | NOD2,ADCY7 | A | 2.13 | 0.07 | 0.05 | 0.05 | 0.02 | 0.00 | 2.01 | 0.311 | 2.26E-02 |
| 18 | rs7240004 | 46,395,022 | IBD | SMAD7 | A | 1.07 | 0.63 | 0.58 | 0.63 | 0.58 | 0.42 | 1.33 | 0.128 | 2.46E-02 |
| 19 | rs516246 | 49,206,172 | CD | DBP,SPHK2 | A | 1.12 | 0.53 | 0.44 | 0.44 | 0.34 | 0.49 | 1.46 | 0.139 | 5.60E-03 |
| IZUMO1,FUT2 | ||||||||||||||
| 19 | rs17771967 | 55,380,214 | UC | NLRP7,NLRP2 | G | 1.07 | 0.55 | 0.43 | 0.44 | 0.41 | 0.37 | 1.73 | 0.178 | 1.40E-03 |
| KIR2DL1,LILRB4 | ||||||||||||||
| 20 | rs6087990 | 31,349,908 | IBD | DNMT3B | G | 1.05 | 0.47 | 0.42 | 0.37 | 0.63 | 0.76 | 1.30 | 0.115 | 2.04E-02 |
AFR, African; AMR, Ad Mixed American; CD, Crohn's disease; CHR, chromosome; EUR, European; IBD, inflammatory bowel disease; RA, risk allele; RAF, risk allele frequency; SNP, single-nucleotide polymorphism; OR, odds ratio; UC, ulcerative colitis.
Permuted P value.
This result was the most significant after 10,000 permutations.
The bold indicates those effects that are opposite in direction for our Hispanic cohort. Position is based on human genome 19 and dbSNP 137. Trait=the phenotype with the largest MANTRA Bayes factor from Liu et al.,7 Supplementary Table 1. Candidate gene(s) as previously defined (Jostins et al.;8 Liu et al.7) RA (Based on Liu et al.7) Risk allele frequencies (RAFs) for EUR, AMR, and AFR are phase 3 allele frequencies from 1000 Genomes (www.1000genomes.org).
Permuted P value was calculated using max (T) permutation as implemented in PLINK (using 10,000 permutations).
As one would expect, the reported 1000 Genomes super population allele frequencies for each of the variants we tested differs between the ancestral populations (AFR, AMR, and EUR) that comprise the diversely admixed Hispanic population.26 Most interestingly, our two variants (rs11739663 and rs7438704) demonstrating protective effects have a much lower frequency for the risk allele within the African super population compared to either the European or admixed American population frequencies. These data suggest that our Hispanic cohort may have more African derived haplotypes across these loci that potentially confer protective effects. Within our control population, we find that the risk allele frequencies of all but two variants (rs9525625 and rs7240004 nearest SMAD7) track most closely to the reported European allele frequencies. Notably, the frequency of these two variants within controls most closely resemble the AMR super population and may be a reflection of local ancestry differences and admixture proportions found within the Hispanic population of largely Caribbean origin within South Florida.
Genetic risk score and onset of IBD in Hispanics and non-Hispanic whites
The mean genetic risk score for Hispanics with CD 199.67 (range 50.57; s.d. 9.06) was similar to the mean genetic risk score for NHWs with CD 200.33 (range 95.10; s.d. 10.29), P=0.51. The same was true for UC genetic risk scores (Hispanics 161.58 (range 39.228 s.d. 8.39) vs. non-Hispanics 162.23 (range 36.58 s.d. 7.95), P=0.50). We found no difference in mean genetic risk scores between US and foreign-born Hispanics (data not shown). In addition, because our cohort is mainly Cuban, we compared genetic risk score between Cubans and non-Cuban Hispanics and found that there was no difference in mean scores (data not shown). Multivariable regression analysis demonstrated that a higher genetic risk score was associated with a younger age of IBD symptom onset in Hispanic patients with CD (P=0.04) but not Hispanics with UC (P=0.76); see Table 3. Among NHWs, a higher CD genetic risk score did not predict a younger age of onset (P value=0.14). We did not see a correlation between genetic risk score and age of diagnosis in either ethnicity with UC (Table 3).
Table 3. Multivariable regression analysis of genetic risk scores with CD and UC outcomes.
| Phenotypic outcome | Ethnicity | Genetic risk score beta coefficient | R-squared | Permuted P value |
|---|---|---|---|---|
| Crohn's disease | ||||
| Age of diagnosis | Hispanics | −0.229 | 0.059 | 0.046 |
| NHW | −0.162 | 0.063 | 0.191 | |
| Age of symptom onset | Hispanics | −0.231 | 0.060 | 0.039 |
| NHW | −0.170 | 0.054 | 0.144 | |
| Location of disease | ||||
| Ileal disease | Hispanics | −0.026 | 0.047 | 0.184 |
| NHW | 0.031 | 0.035 | 0.126 | |
| Ileocolonic disease | Hispanics | −0.002 | 0.011 | 0.946 |
| NHW | −0.028 | 0.041 | 0.253 | |
| Colonic disease | Hispanics | 0.015 | 0.023 | 0.443 |
| NHW | −0.007 | 0.054 | 0.737 | |
| Isolated upper GI tract | Hispanics | 0.204 | 0.080 | 0.098 |
| NHW | −0.826 | 0.014 | 0.602 | |
| Crohn's behavior | ||||
| Stricturing | Hispanics | 0.023 | 0.022 | 0.233 |
| NHW | 0.022 | 0.017 | 0.274 | |
| Penetrating | Hispanics | 0.017 | 0.015 | 0.536 |
| NHW | −0.039 | 0.039 | 0.156 | |
| Perianal disease | Hispanics | 0.029 | 0.033 | 0.158 |
| NHW | −0.004 | 0.029 | 0.841 | |
| History of hospitalizations | Hispanics | 0.067 | 0.038 | 0.079 |
| NHW | −0.023 | 0.069 | 0.690 | |
| Abdominal surgeries | Hispanics | 0.026 | 0.036 | 0.050 |
| NHW | 0.011 | 0.013 | 0.317 | |
| Perianal surgeries | Hispanics | 0.049 | 0.026 | 0.082 |
| NHW | −0.007 | 0.016 | 0.786 | |
| Ulcerative colitis | ||||
| Age of diagnosis | Hispanics | 0.068 | 0.013 | 0.720 |
| NHW | 0.136 | 0.073 | 0.550 | |
| Age of symptom onset | Hispanics | 0.059 | 0.013 | 0.760 |
| NHW | 0.147 | 0.500 | 0.530 | |
| Location of disease | ||||
| Proctitis | Hispanics | 0.053 | 0.041 | 0.390 |
| NHW | −0.063 | 0.018 | 0.395 | |
| Left-sided colitis | Hispanics | 0.010 | 0.045 | 0.716 |
| NHW | −0.037 | 0.119 | 0.327 | |
| Pan-colitis | Hispanics | −0.014 | 0.046 | 0.592 |
| NHW | 0.051 | 0.132 | 0.175 | |
| History of hospitalizations | Hispanics | −0.077 | 0.020 | 0.270 |
| NHW | 0.108 | 0.066 | 0.150 | |
| Colectomy | Hispanics | −0.018 | 0.060 | 0.720 |
| NHW | 0.176 | 0.04 | 0.090 | |
CD, Crohn's disease; IBD, inflammatory bowel disease; NHW, non-hispanic whites; UC, ulcerative colitis.
Beta coefficient: the beta value is a measure of how strongly each predictor variable (in this case genetic risk score) influences the dependent variable (in this case IBD phenotype).
R-squared: is a number that indicates the proportion of the variance in the dependent variable (IBD phenotype) that is predictable from the independent variable (genetic risk score).
Bold means significant p-values (P<0.05).
The mean genetic risk score for the control group was 185.24 (range 59.64; s.d. 9.01), using the CD risk alleles and 157.28 (range 51.88; s.d. 8.06), using the UC risk alleles. When comparing the genetic risk scores between the Hispanic IBD cases and controls, the risk scores were significantly higher in the CD and UC cases compared to controls (P<0.0001).
Genetic risk score and phenotypic manifestations of disease in Hispanics and non-Hispanic whites
Regression analyses using genetic risk score as an independent predictor of CD disease location and behavior was not significant (Table 3). No association existed between UC genetic risk score and extent of disease in UC was observed (Table 3). A higher genetic risk score was weakly associated with an increased risk of abdominal surgeries in Hispanics with CD (P=0.05), but was not significant in non-Hispanics (Table 3). No relationship was seen between colectomies for UC patients or predictive of number of IBD-related hospitalizations (Table 3).
Discussion
To our knowledge, this is the first study to examine genetic burden in Hispanics and its relationship to age of disease onset, diagnosis and detailed IBD phenotype. It is also the first study to examine the genetic profile of 200 established IBD risk variants in a diverse sample of Hispanics. Although the prevalence of IBD is lower in Latin America, we found that the composite genetic risk score of established European risk loci was similar between Hispanics (US and foreign-born) and NHWs with IBD. These findings do not explain differences in age of IBD presentation observed between US and foreign-born Hispanics, and why US-born Hispanics have a similar age of presentation to their NHW counterparts; we have replicated these phenotypic findings in this new cohort.3 Our findings highlight the critical factor played by the environment and substantiate that IBD onset occurring after migration may be strongly influenced by exposure to an environmental trigger that cannot be circumvented by genetic burden as measured herein.
An important aspect of our study was to determine whether the IBD risk loci identified in large genome-wide association studies of European populations also confer risk in the diverse Hispanic population. We observed nominally significant associations for 22 (6 IBD, 10 CD, and 6 UC) previously established risk variants in our Hispanic population data set. For 20/22 variants, the direction of effect was consistent with previously meta-analyses.7 Within the 22 risk loci, we found significant associations across SNPs corresponding to IL23R, NOD2, and Smad7 genes. A recent study in the African American population found that many IBD risk variants, including those in IL23R and NOD2, also demonstrate evidence for association in African Americans.27 In addition, our findings parallel the results reported in the Puerto Rican IBD study, even though our sample is comprised of only 3.9% Puerto Ricans. In a recent study of Puerto Rican IBD patients, the haplotype structure for both the IL23R and NOD2 regions were found to be predominantly of European ancestry. We can extrapolate from their study that the same (European) haplotypic structure may be seen at these loci in our admixed Hispanic data set, composed largely of Cubans.
Admixture analysis of Cubans using ancestry informative markers illustrates that Cubans have largely a European contribution, estimated at 72%.28 Therefore, these data suggest that the contribution of NOD2 and IL23R to the development of CD in our Hispanics is likely similar to that of European patients. The IL23R locus demonstrates a complex set of association signals with IBD, with both risk and protective effects having been previously shown potentially resulting from the effects of the genetic variants on differential splicing.29 Moreover, our most significant variant (rs7517847) found in IL23R has not only previously demonstrated highly significant association with CD, but has also been found to be associated with plasma omega-6 polyunsaturated fatty acid levels (linoleic acid) in a recent genome-wide association study.30 Although many studies suggest that a high dietary ratio of ω6/ω3 polyunsaturated fatty acids is potentially risk-conferring for CD, evidence across studies is equivocal but suggests that the modern western diet is highly likely to play a role in chronic human disease.31, 32, 33 While our present results are modest, taken together these findings may begin to help explain our observations of increased numbers of CD Hispanic patients in our clinics compared to the expected prevalence of CD within this admixed population.
We also find evidence of association near the Smad7 (SMAD family member 7) gene, which encodes a protein that is overexpressed in IBD patients and is responsible for suppressing anti-inflammatory signaling and is associated with colon cancer34, 35 (Table 2). Our study suggests that this locus is relevant to IBD risk in the Hispanic population. Another interesting finding is the association to a gene involved in DNA methylation. CD Hispanics were 1.3x more likely to carry the risk allele for rs6087990 corresponding to the DNMT3B (DNA methyltransferase 3B) gene, which encodes for a DNA methyltransferase responsible for de novo methylation. A recent systematic review of cancer found a decreased risk between rs6087990 and colon cancer in the Asian population.36 Further studies are needed to better delineate the potential role this gene may play in colon cancer risk for Hispanic IBD patients and the downstream epigenetic effects altering gene expression.
In this study, we found that a high genetic risk score predicted a younger age of Crohn's disease onset in Hispanics. While we may not have replicated this finding in our non-Hispanic cohort,16 we were able to augment these findings with data on Hispanics. Predicting risk of complications in CD or UC remains imperfect. In this study, we observe that our composite genetic risk score does not correlate with IBD phenotypes or surgeries in either UC or CD. However, other studies differ from our findings.37, 38 The largest genotype–phenotype association study, using 193 SNPs and 23 HLA types to generate genetic risk scores, examined phenotypes across 34,819 patients of European ancestry and found that IBD “subphenotype” (i.e., ileal Crohn's, colonic Crohn's) may be determined in part by genetics.18 Our study findings may differ from the former study either as a result of a different population examined or because our sample size could not detect associations of this small effect size. Although our findings were not significant, the possibility of a genetic risk score as a prediction tool for disease phenotype remains enticing, particularly in phenotypes that warrant further classification, such as IBD- unclassified. The genetic risk score as a measure of overall genetic burden is a contemporary model used in cardiovascular and autoimmune diseases, including IBD that has shown stronger correlations with phenotype than individual SNPs.14, 16, 17, 39
One of the limitations of our study is the size of our data set and for this reason we chose to focus on the known genetic associations to ask a very specific question regarding where or not previously identified variants confer risk in Hispanic IBD. The smaller sample size in NHWs, may explain the negative associations noted between genetic risk score and age of onset and phenotype, including ileal disease–observed in prior recent studies.16, 18 However, even with a small sample size and using sound statistical methodology, including permutation testing, we were able to replicate previous findings for several European risk alleles in Hispanic cases and found that a composite weighted score of these established European risk alleles is still applicable to Hispanics with IBD. Last, since our focus was on Hispanics, we did not include a control population of NHWs. Genetic association studies comparing NHWs with IBD to NHW controls already exist and are larger than the cohort of non-Hispanics in our database, thereby abating the need for us to replicate these findings.8, 9, 10, 11
In summary, our results provide biological insight into the pathogenesis of IBD in Hispanics and a better understanding of the genetic architecture across this diverse population. Our findings highlight the potential applicability of a genetic risk score in the prediction of age of onset in the Hispanic IBD population. When immigrating, time in the US is necessary to manifest IBD even in the face of a similar genetic burden. These findings provide the framework for future studies to focus on gene-environment interactions and unidentified environmental exposures that may explain the rising incidence of IBD in US Hispanic populations.
Study Highlights

Footnotes
Guarantorof the article: Oriana Damas, MD.
Specific author contributions: Oriana M. Damas: study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, statistical analysis, and obtained funding; Lissette Gomez: analysis and interpretation; Evadnie Rampersaud: analysis and interpretation of data, statistical analysis, and study supervision; Maria A. Quintero: study concept and design, and acquisition of data; Susan Slifer: study concept and design, analysis and interpretation of data; Gary W. Beecham: shared Hispanic genotype control data for this project; David H. Kerman: critical revision of the manuscript for important intellectual content; Amar R. Deshpande: study concept and design, interpretation of data, and critical revision of the manuscript for important intellectual content; Daniel A. Sussman: study concept and design, interpretation of data, and critical revision of the manuscript for important intellectual content; Maria T. Abreu: study concept and design, analysis and interpretation of data, drafting of the manuscript, and obtained funding; Jacob L. McCauley: study concept and design, analysis and interpretation of data, drafting of the manuscript, statistical analysis, and supervision.
Financial support: This work was supported by the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant (R01DK104844 to M.T.A. and J.L.M.), UCB, IBD Working Group Research Award (O.M.D.), The Micky & Madeleine Arison Family Foundation Crohn's & Colitis Discovery Laboratory (to M.T.A.), and The Martin Kalser Chair in Gastroenterology (to M.T.A).
Potential competing interests: None.
Supplementary Material
References
- Ennis SR, Ríos-Vargas M, Albert NG. The Hispanic Population: 2010. 2010 Census 2011. Retrieved from: https://www.census.gov/prod/cen2010/briefs/c2010br-04.pdf.
- Hou JK, El-Serag H, Thirumurthi S. Distribution and manifestations of inflammatory bowel disease in Asians, Hispanics, and African Americans: a systematic review. Am J Gastroenterol 2009; 104: 2100–2109. [DOI] [PubMed] [Google Scholar]
- Damas OM, Jahann DA, Reznik R et al. Phenotypic manifestations of inflammatory bowel disease differ between Hispanics and non-Hispanic whites: results of a large cohort study. Am J Gastroenterol 2013; 108: 231–239. [DOI] [PubMed] [Google Scholar]
- Nguyen GC, Torres EA, Regueiro M et al. Inflammatory bowel disease characteristics among African Americans, Hispanics, and non-Hispanic Whites: characterization of a large North American cohort. Am J Gastroenterol 2006; 101: 1012–1023. [DOI] [PubMed] [Google Scholar]
- Basu D, Lopez I, Kulkarni A et al. Impact of race and ethnicity on inflammatory bowel disease. Am J Gastroenterol 2005; 100: 2254–2261. [DOI] [PubMed] [Google Scholar]
- Benchimol EI, Mack DR, Guttmann A et al. Inflammatory bowel disease in immigrants to Canada and their children: a population-based cohort study. Am J Gastroenterol 2015; 110: 553–563. [DOI] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015; 47: 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011; 43: 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 2008; 40: 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 2010; 42: 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugathasan S, Loizides A, Babusukumar U et al. Comparative phenotypic and CARD15 mutational analysis among African American, Hispanic, and White children with Crohn's disease. Inflamm Bowel Dis 2005; 11: 631–638. [DOI] [PubMed] [Google Scholar]
- Ballester V, Guo X, Vendrell R et al. Association of NOD2 and IL23R with inflammatory bowel disease in Puerto Rico. PLoS One 2014; 9: e108204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Chibnik LB, Cui J et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol 2009; 8: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Colombel JF, Lemann M et al. Genotype/phenotype analyses for 53 Crohn's disease associated genetic polymorphisms. PLoS One 2012; 7: e52223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthakrishnan AN, Huang H, Nguyen DD et al. Differential effect of genetic burden on disease phenotypes in Crohn's disease and ulcerative colitis: analysis of a North American cohort. Am J Gastroenterol 2014; 109: 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibnik LB, Keenan BT, Cui J et al. Genetic risk score predicting risk of rheumatoid arthritis phenotypes and age of symptom onset. PLoS ONE 2011; 6: e24380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Boucher G, Jostins L et al. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016; 387: 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther 2011; 13: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium (IMSGC)International Multiple Sclerosis Genetics Consortium (IMSGC)Beecham AH International Multiple Sclerosis Genetics Consortium (IMSGC)Patsopoulos NA et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013; 45: 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core TeamR: A Language And Environment For Statistical Computing. R Foundation for Statistical Computing, ISBN 3-900051-07-0: Vienna, Austria, 2013. [Google Scholar]
- Beecham AH, Wang L, Vasudeva N et al. Utility of blood pressure genetic risk score in admixed Hispanic samples. J Hum Hypertens 2016; 30: 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium1000 Genomes Project ConsortiumAuton A 1000 Genomes Project ConsortiumBrooks LD et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Haritunians T, Okou DT et al. Characterization of genetic loci that affect susceptibility to inflammatory bowel diseases in African Americans. Gastroenterology 2015; 149: 1575–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheco-Teruel B, Parra EJ, Fuentes-Smith E et al. Cuba: exploring the history of admixture and the genetic basis of pigmentation using autosomal and uniparental markers. PLoS Genet 2014; 10: e1004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006; 314: 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W, Steffen BT, Lemaitre RN et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet 2014; 7: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costea I, Mack DR, Lemaitre RN et al. Interactions between the dietary polyunsaturated fatty acid ratio and genetic factors determine susceptibility to pediatric Crohn's disease. Gastroenterology 2014; 146: 929–931. [DOI] [PubMed] [Google Scholar]
- Calder PC. Polyunsaturated fatty acids, inflammatory processes and inflammatory bowel diseases. Mol Nutr Food Res 2008; 52: 885–897. [DOI] [PubMed] [Google Scholar]
- Chilton FH, Murphy RC, Wilson BA et al. Diet-gene interactions and PUFA metabolism: a potential contributor to health disparities and human diseases. Nutrients 2014; 6: 1993–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi C, De Simone V, Colantoni A et al. A functional role for Smad7 in sustaining colon cancer cell growth and survival. Cell Death Dis 2014; 5: e1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G, Neurath MF, Ardizzone S et al. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn's disease. N Engl J Med 2015; 372: 1104–1113. [DOI] [PubMed] [Google Scholar]
- Duan F, Cui S, Song C et al. Systematic evaluation of cancer risk associated with DNMT3B polymorphisms. J Cancer Res Clin Oncol 2015; 141: 1205–1220. [DOI] [PubMed] [Google Scholar]
- Weersma RK, Stokkers PC, van Bodegraven AA et al. Molecular prediction of disease risk and severity in a large Dutch Crohn's disease cohort. Gut 2009; 58: 388–395. [DOI] [PubMed] [Google Scholar]
- Cleynen I, Gonzalez JR, Figueroa C et al. Genetic factors conferring an increased susceptibility to develop Crohn's disease also influence disease phenotype: results from the IBDchip European Project. Gut 2013; 62: 1556–1565. [DOI] [PubMed] [Google Scholar]
- Thanassoulis G, Peloso GM, Pencina MJ et al. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium-The Framingham Heart Study. Circ Cardiovasc Genet 2012; 5: 113–121, CIRCGENETICS-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.