Abstract
m-miR-184 (mutant miR-184, r.57c > u) appears in familial hereditary ocular diseases, including keratoconus, cataracts, EDICT (endothelial dystrophy, iris hypoplasia, congenital cataract, and stromal thinning) syndrome, severe keratoconus, and non-ectatic corneal thinning. The biological function of m-miR-184 in these ocular diseases remains unclear. With the emergence of high-throughput sequencing, it is now possible to discover many different biological components simultaneously. Using two different RNA libraries, we sequenced the complete transcriptome of HLE cells treated with miR-184, m-miR-184, and a negative control. Data were integrated in an effort to identify any novel gene affected by m-miR-184. Notably, we concluded that ALDH5A1 and GABRA3 were disordered by m-miR-184, which might lead to ocular disease. Moreover, circRNA (circular RNA) expression was highy random across miR-184, m-miR-184, and negative control treatment groups. The sequences of the circRNAs did reveal a particularly high level of ALU sequences. In summary, we provide a new avenue for understanding the role of m-miR-184 in ocular diseases.
Keywords: miR-184, circRNA, human lens epithelial, mutation, ALU
Introduction
miRNAs (microRNAs) are endogenous non-coding RNA (ncRNA) molecules of ∼22 nt, which inhibit target gene expression by either inducing mRNA degradation or by blocking protein translation.1 Some miRNAs have important cellular functions, including cellular differentiation, proliferation, and apoptosis.2, 3 The seed region is present from bases two to seven of the mature miRNA, and it is especially important for recognition of target mRNA and proper regulation of protein expression. miRNAs have a different expression pattern in ocular tissues.
The most abundantly expressed miRNA in the cornea and lens is miR-184.4, 5 miR-184 effects its biological function through the competitive inhibition of miR-205 when binding to the mRNA of INPPL1 (inositol polyphosphate-like 1) and ITGB4 (integrin, beta 4).6 Takahashi et al.7 reported that decreased levels of miR-184 are responsible for the aberrant activation of Wnt signaling in ischemia-induced retinal neovascularization. In addition, an earlier study reported that miR-184 is implicated in ocular neovascularization in the OIR (oxygen-induced retinopathy) model and the non-ischemia retinopathy (laser-induced choroidal NV [neovascularization]) model.8 Further, it is reported that miR-184 is involved in a competitive RNA network in control of mouse secondary cataract disease. An miR-184 inhibitor can attenuate secondary cataract expansion, migration of lens epithelial cells, and signs of epithelial-to-mesenchymal transition, such as α-smooth muscle actin expression.9 Recently, studies have reported a heterozygous c-to-u transition (r.57c > u) in the seed region of miR-184 in a three-generation Northern Ireland family with keratoconus and cataracts, a family with EDICT (endothelial dystrophy, iris hypoplasia, congenital cataract, and stromal thinning) syndrome, and a five-generation family with cataracts and varying corneal abnormalities, including severe keratoconus and non-ectatic corneal thinning, from Galicia, Spain.6, 10, 11, 12 However, no mutation was detected within the genomic region of miR-184 in keratoconus patients from Saudi Arabia.13 The biological function of the m-miR-184 (mutant miR-184, r.57c > u) is still not clear in ocular disease.
With the wide-scale adoption of high-throughput sequencing techniques, circRNAs (circular RNAs) were recently described as a novel type of endogenous ncRNA, and they represent a potential research hotspot in the field of RNA. circRNAs derived from the exon and intron of genes have been identified in all domains of life, including eukaryotes, archaea, bacteria, and viruses.14, 15 Functional studies have found that circRNAs can act as a sponge for certain miRNAs; an example of this is ciRS-7, which harbors more than 70 conventional miR-7 binding sites.16 This new circRNA/miRNA regulatory interaction has already generated an interesting new field of study.
However, no study has addressed whether mature miRNA or its mutant counterparts can influence the expression of circRNA. In an effort to enrich our understanding of the m-miR-184-mediated progression of ocular disease, we explored a variety of biological entities produced only in the presence of m-miR-184. We sequenced the complete RNA panel after depletion of rRNA. Furthermore, since circRNAs have been shown to have a sponge role in interaction with miRNA in cells, we also sequenced libraries of circRNA obtained from the same biological samples.
In this study, we attempted to describe the expression of previously identified genes and circRNAs and identify those that may be differentially regulated by m-miR-184 in ocular disease. Thus, the miR-184, m-miR-184, or a NC (negative control, a random sequence miRNA mimic molecule that has been extensively tested in human cell lines and tissues and validated not to produce identifiable effects on known miRNA function) was transfected into HLE (human lens epithelial) cells, and the effects on circRNA and the cellular transcriptome were investigated.
Results
RNA Expression Levels in miR-184, m-miR-184, and NC Treatment Cells
A total of six samples (n = 2 miR-184/m-miR-184/NC) was sequenced after rRNA depletion. In total from all 6 samples, 214.6 million raw reads were produced, in pair end, with an average of 35 million reads per sample. After quality trimming, 211.6 million high-quality reads remained (Data S1). We mapped the clean reads to the Ensembl human genome, release_77. The proportion of total reads in the six miRNA treatment transcriptome libraries that mapped to the genome ranged from 90.51% to 91.81%.
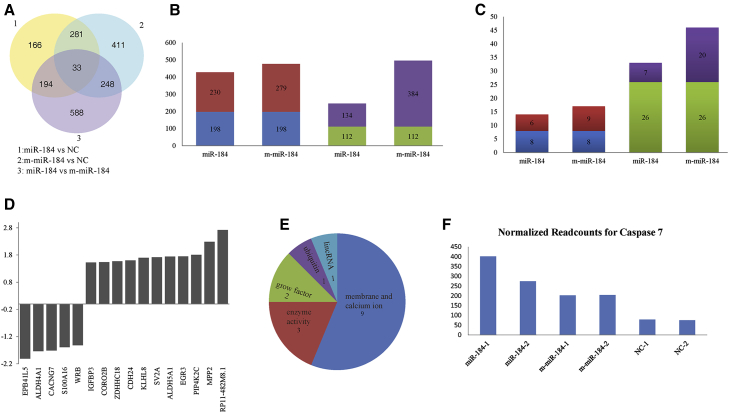
The distribution of the numbers of DEGs (differentially expressed genes) across the six miRNA treatment cells is shown in Figure 1A. We found 1,063 DEGs between HLE-miR-184 and HLE-m-miR-184 cells. However, there was no significantly enriched GO (gene ontology) term identified. Moreover, there were 674 DEGs identified when comparing HLE-miR-184 and HLE-NC cells. A GO-based enrichment analysis of these genes indicated that protein phosphorylation (GO: 0006468) was the most significantly enriched term (p = 2.75E−06). Furthermore, 973 DEGs were identified when comparing HLE-m-miR-184 and HLE-NC cells, and the biological process macromolecule modification (GO: 0043412) was shown to be the most significantly enriched GO term (p = 3.79E−06). Only 33 genes showed common differential expressions in comparisons of miR-184 versus NC, m-miR-184 versus NC, and miR-184 versus m-miR-184, indicating that HLE cells have a different response to miR-184, m-miR-184, and NC treatment (Data S2).
Figure 1.
Profiles of DEGs in miR-184-, m-miR-184-, and NC-Treated Cells
(A) Venn diagram of the DEGs in the HLE-miR-184 versus HLE-NC, HLE-m-miR-184 versus HLE-NC, and HLE-miR-184 versus HLE-m-miR-184. (B) The overlapping DEGs and unique DEGs between HLE-miR-184 versus HLE-NC and HLE-m-miR-184 versus HLE-NC. (C) The overlapping DEGs between HLE-miR-184 versus HLE-NC and HLE-m-miR-184 versus HLE-NC according to absolute fold change ≥ 1.5 and a corrected p value ≤ 0.05. In (B) and (C): blue column, overlapping upregulated DEGs; red column, unique upregulated DEGs; green column, overlapping downregulated DEGs; purple column, unique downregulated DEGs. (D) DEGs between miR-184- and m-miR-184-treated cells according to absolute fold change ≥ 1.5 and a corrected p value ≤ 0.05. (E) Pie chart of functional characteristics of DEGs in (D). (F) Caspase 7 expression.
Expression of Different Genes in HLE-miR-184 or HLE-m-miR-184 versus HLE-NC Cells
We aimed to perform a global analysis of the DEGs between the HLE-miR-184 and HLE-m-miR-184 and the HLE-NC cells. As shown in Figure 1B, there were 496 downregulated and 477 upregulated genes in HLE-m-miR-184 cells. However, there were only 246 downregulated and 428 upregulated genes in HLE-miR-184 cells. The number of overlapping downregulated and upregulated genes between HLE-miR-184 and HLE-m-miR-184 were 112 and 198, respectively, leaving 384 unique downregulated DEGs in HLE-m-miR-184 cells and 134 unique downregulated DEGs in HLE-miR-184 cells. Expression was normalized against NC in all cases.
Further, a minimum count after normalization of ten reads, absolute fold change ≥1.5, and a corrected p value ≤ 0.05 was set as the threshold for significant differential expression. In addition to the 26 overlapping downregulated genes, the numbers of the unique downregulated genes in HLE-m-miR-184 and HLE-miR-184 are 20 and 7 separately. The much higher proportion of downregulated genes in the HLE-m-miR-184 (Figure 1C) implies that m-miR-184 has a broader spectrum of targets when compared to miR-184.
Moreover, the most significantly downregulated gene, PLBD2 (phospholipase B domain containing 2, log2 fold change −2.3769, Q value 5.65E−53), has one perfect seed match with m-miR-184 and the most upregulated gene was INA, associated with morphogenesis of neurons (internexin neuronal intermediate filament protein, alpha, log2 fold change 2.1782, Q value 0.019968). Furthermore, these were different from the most downregulated gene, EPB41L5 (erythrocyte membrane protein band 4.1 like 5, log2 fold change −1.9299, Q value 1.11E−16) and the most upregulated gene FOXP4 (forkhead box P4, log2 fold change 1.7735, Q value 5.61E−26) in the HLE-miR-184 cells.
Expression of Different Genes between miR-184- and m-miR-184-Treated Cells
There were 16 significantly DEGs between miR-184- and m-miR-184-treated cells. Among these, nine were membrane and calcium ion association genes, including EPB41L5 (erythrocyte membrane protein band 4.1 like 5), CACNG7 (calcium channel, voltage-dependent, gamma subunit 7), S100A16 (S100 calcium-binding protein A16), CORO2B (coronin, actin-binding protein, 2B), ZDHHC18 (zinc finger, DHHC-type containing 18), CDH24 (cadherin 24, type 2), SV2A (synaptic vesicle glycoprotein 2A), WRB (tryptophan-rich basic protein), and MPP2 (membrane protein, palmitoylated 2); three are enzymatic genes, including ALDH4A1 (aldehyde dehydrogenase 4 family, member A1), ALDH5A1 (aldehyde dehydrogenase 5 family, member A1), and PIP4K2C (phosphatidylinositol-5-phosphate 4-kinase, type II, gamma) kinase. The expression level of ALDH4A1 was 1.74-fold higher and ALDH5A1 was 1.74-fold lower in m-miR-184- than miR-184-treated cells, and a KEGG (Kyoto Encyclopedia of Genes and Genomes)-based pathway analysis of ALDH4A1 and ALDH5A1 returned to the same alanine, aspartate, and glutamate metabolism pathway, implying an interesting relationship between these two genes. One was an ubiquitin protein gene KLHL8 (kelch-like family member 8), two were growth factor genes IGFBP3 (insulin-like growth factor-binding protein 3) and EGR3 (early growth response 3), and one was lincRNA (large intergenic ncRNA) RP11-482M8.1 (Figure 1D; Data S3). Among the 16 genes, 56% were membrane- and calcium ion-relevant genes (Figure 1E), indicating that the m-miR-184 mainly disorders membrane and calcium ion functions.
Next, we computed all possible interactions of these 16 DEGs with miR-184 and m-miR-184 seed sequences using miRanda. All genes that were downregulated by m-miR-184 presented a perfect seed match with m-miR-184 seed sequence (7-mer-m8, ccatcc). Moreover, the genes that were downregulated by miR-184 also contained a perfect seed match with the miR-184 seed sequence (7-mer-m8, ccgtcc).
Expression Profile of Apoptosis Genes
Various stress conditions can induce apoptosis of the lens epithelial cells, leading to eventual cataract formation.17, 18, 19 We investigated the changes in apoptosis genes across the six miRNA treatment groups. The distribution of the expression of different apoptosis genes are shown in Data S4. Among the 96 genes known to be associated with apoptosis (Taqman Low Density Array configuration, Life Technologies), there were no different apoptosis-associated genes across miR-184, m-miR-184, and NC treatment cells. Further, we found one apoptosis gene, CASP7, upregulated in HLE-miR-184 and HLE-m-miR-184 when compared to HLE-NC cells (Figure 1F). It appears that the m-miR-184 does not affect the fate of HLE cells by disordering the apoptotic pathway. Further, the apoptosis rate of HLE cells transfected with miR-184, m-miR-184, or NC was determined by flow cytometry. Results indicated that there was no difference in the rate of apoptosis across these three samples (Figure 2F).
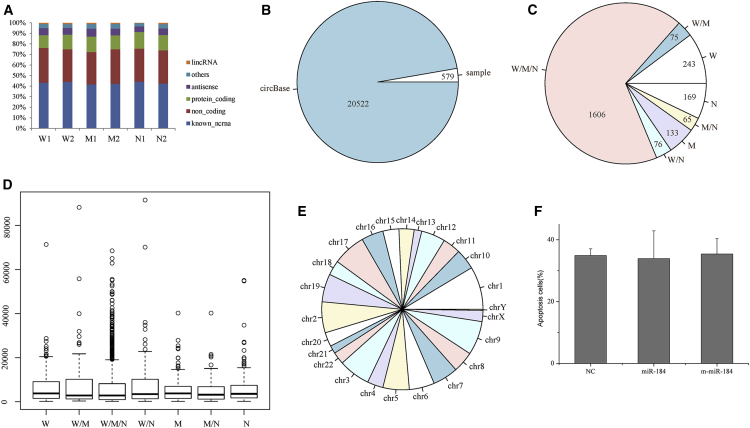
Figure 2.
Profiles of circRNAs Identified in Samples
(A) Functional characteristics of genes encompassing circular transcripts identified in this study. (B) Pie chart of circRNA annotation in circBase. (C) Numbers of circRNA distribution in samples. (D) Length distribution of circRNAs in samples. (E) Distribution of circRNAs in chromosome. (F) Apoptosis rate of HLE cells transfected with miR-184, m-miR-184, or NC. Results are displayed as mean ± SEM. W, miR-184-treated cells; M, m-miR-184-treated cells; N, negative control-treated cells.
circRNA Prediction
To identify putative miRNA-inducible circRNAs, we isolated RNA from HLE cells treated for 24 hr with miR-184, m-miR-184, or NC, and we performed two rounds of RNA sequencing using previously described procedures.14 Our data revealed that circRNAs are abundant in HLE cells. We identified a total of 20,522 circRNA candidates in the six miRNA-treated cells using RNA sequencing. Notably, only 2,367 circRNAs could be identified in both biological replicates. As shown in Figure 2A, apart from a few antisense, lincRNA candidates and others, most putative circRNAs overlapped known ncRNA, protein-coding, and antisense transcripts. In addition, of the 20,522 candidates, only 579 are listed in the circBase database20 (Figure 2B). Based on the circRNAs identified in both biological replicates, there were 2,000 distinct circRNA candidates in HLE-miR-184 cells, 1,879 circRNA candidates in HLE-m-miR-184 cells, and 1,916 circRNA candidates in HLE-NC cells. Overall, a total of 2,367 circRNAs was identified after removing the duplicate circRNAs between samples (Data S5). The detailed distribution and the length of the identified circRNAs are shown in Figures 2C and 2D. Further, no significant difference in the numbers of circRNAs originating from each chromosome was found (Figure 2E).
circRNA Quantification
The quantitation of circRNA raw counts was normalized using TPM (transcripts per million) normalization. Most circRNAs are low-abundance transcripts. Several circRNAs appeared differentially expressed but none of them displayed a corrected p value ≤ 0.05. Taken together, the individual circRNA expression seems to be highly random across the miR-184, m-miR-184, and NC treatment cells.
Seed-Matching Analyses
Next, we quantified the number of canonical miRNA seed sequences in each circRNA as follows: 7-mer-perfect match to the 6-nt miRNA seed augmented; 7-mer-A1-perfect match to the 6-nt miRNA seed augmented by an A at target position 1; and 7-mer-m8-perfect match to the 6-nt miRNA seed with an additional match to nucleotide 8 of the miRNA.21 We counted a total of three seed pairings for miR-184 or m-miR-184 against all circRNA sequences identified. Of 2,367 circRNAs, there were 149 circRNAs that had a miR-184 binding site, and the distribution of these circRNAs in the three different treatment groups was random. Of the 149 circRNAs, 111 had binding sites with m-miR-184. But the numbers of binding sites in circRNA that matched with m-miR-184 were less than those for miR-184. This indicated that the miR-184 and m-miR-184 binding site can exist in the same circRNA. In summary, the interaction between miR-184/m-miR-184 and circRNA is compatible and not exclusive, which indicates that the mechanism of interaction between miRNA and circRNA may be complex.
ALU Element Enrichment in circRNA
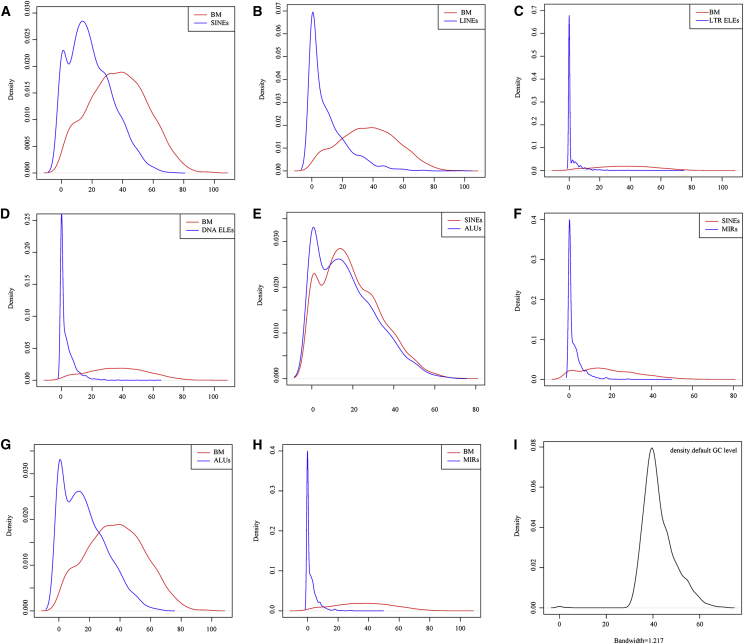
As ALUs are the most successful active TEs (transposable elements) in the human genome, in terms of copy number, and they occupy two-thirds of the SINEs (short interspersed nuclear elements), we tested whether ALUs contributed to the sequence of circRNAs. To investigate this, we divided SINEs into two subgroups, ALUs and MIRs (miniature inverted repeats), and we investigated them separately. After running the sequences in the RepeatMasker program, no repeat sequences were found in 587 circRNAs, but the vast majority (1,780 circRNAs) contained at least one repeat sequence. The four major types (LINE [long interspersed nuclear element], SINE, LTR element, and DNA element) of TEs are shown in Data S6. Results indicated that SINEs accounted for 87.33% of TEs in circRNAs and 80.41% of all of the SINEs contained at least one ALU sequence. Figure 3 shows the kernel density distribution of these four different TE types in circRNAs derived from Data S6. The percentage was converted to a scale from zero to one and plotted with the kernel density function using R statistical software. Figures 3A and 3G suggest that masked bases were mainly composed of SINEs and ALUs. Figure 3E shows the trend that the SINEs identified mainly consisted of ALUs.
Figure 3.
Kernel Density Distribution of Four Different TE Types in circRNAs
Kernel density plot of (A) BM/SINEs, (B) BM/LINEs, (C) BM/LTR ELEs, (D) BM/DNA ELEs, (E) SINEs/ALUs, (F) SINES/MIRs, (G) BM/ALUs, and (H) BM/MIRs. (I) Kernel density plot of GC level. BMs, bases masked; LTR ELEs, long terminal repeat elements; DNA ELEs, DNA elements; SINEs, short interspersed nuclear elements; LINEs, long interspersed nuclear elements; MIR, miniature inverted repeat.
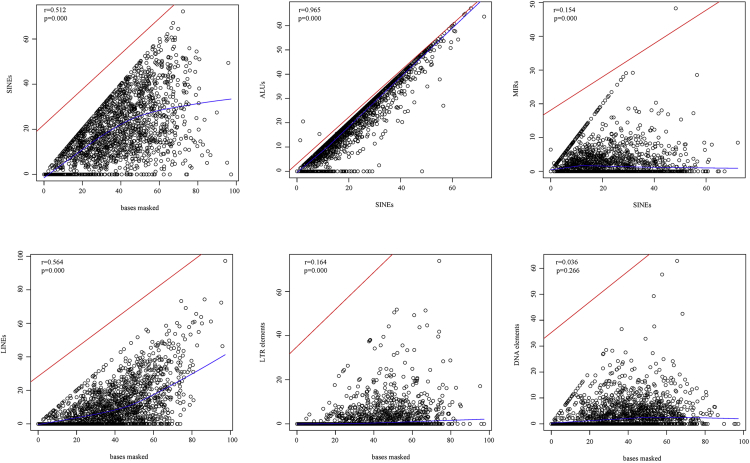
The relationships of different TEs were then used in a Pearson correlation coefficient analysis. As shown in Figure 4, the masked bases of circRNAs were correlated with SINEs and LINEs (r = 0.512 and r = 0.564, respectively). Moreover, SINEs highly correlated with ALU sequence (r = 0.965). There was low correlation between masked bases and LTR elements (r = 0.164, p < 0.05), but there was no correlation between masked bases and DNA elements (r = 0.036, p > 0.05).
Figure 4.
The Relationships of Different TEs Were Analyzed with the Pearson Correlation
Red line is the regression line and blue line is the fitting trend line of points.
Case Study: m-miR-184 Downregulated Genes
Since m-miR-184 (r.57c > u) appeared in familial hereditary ocular diseases, including keratoconus and cataracts, we decided to focus our data integration analysis on this mutant miRNA. Of the 19 significantly downregulated DEGs in HLE-m-miR-184 when compared to HLE-NC cells, 12 possessed at least one perfect seed match in their 3′ UTR with m-miR-184. A GO-based enrichment analysis of these genes returned the molecular function binding (GO: 0005488) to be the most significantly enriched (p = 0.032). Thus, the genes PLBD2, SV2A, RAB11FIP4 (RAB11 family-interacting protein 4 [class II]), ALDH5A1, KLHL8, PIP4K2C, EGR3, DUSP2 (dual-specificity phosphatase 2), MPP2, EPHB3 (EPH receptor B3), FKBP4 (FK506-binding protein 4, 59 kDa), and PRDM1 (PR domain-containing 1, with ZNF domain) all demonstrated a perfect seed match with m-miR-184 in their 3′ UTR, indicating that their expression may be inhibited by m-miR-184 directly (Data S7).
Although some of these genes are not really informative, we noticed that ALDH5A1, a mitochondrial NAD (+)-dependent succinic semi-aldehyde dehydrogenase, and GABRA3, a GABA-A receptor, were downregulated in HLE-m-miR-184 cells. Both ALDH5A1 and GABA (γ-aminobutyric acid) associate with the alanine, aspartate, and glutamate metabolism pathway. It is reported that the SSADH (succinic semi-aldehyde dehydrogenase) deficiency is a rare autosomal recessive disorder, and it affects the degradation of GABA, leading to intractable seizures and psychomotor retardation.22 Studies have also shown that inherited disorders of GABA metabolism, including SSADH and GABA-T (GABA transaminase) deficiencies, are associated with developmental delay, hypotonia, and epilepsy.23 Furthermore, because the 3′ UTR of ALDH5A1 has five perfect putative seed matches with m-miR-184, the downregulation of ALDH5A1 may be directly caused by m-miR-184. As there are no putative seed matches on the GABRA3 3′ UTR, the downregulation of GABRA3 may be regulated by m-miR-184 indirectly. Furthermore, we analyzed the nine upregulated genes in HLE-m-miR-184 (Data S7). We found that the GRIK5, glutamate receptor, was upregulated. L-glutamate is also an important intermediate in the alanine, aspartate, and glutamate metabolism pathway. It is notable that GRIK5, ALDH5A1, and GABRA3 all affect this alanine, aspartate, and glutamate metabolism pathway.
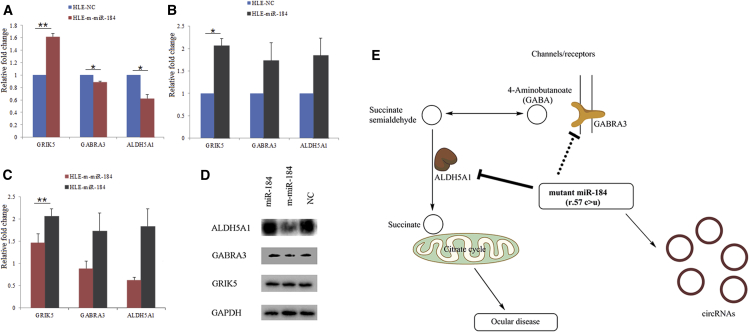
The specific expression changes of the GRIK5, ALDH5A1, and GABRA3 were also investigated by real-time PCR. The results indicated that mRNA levels of ALDH5A1 decreased 0.619 ± 0.068-fold (p < 0.05), GABRA3 decreased 0.883 ± 0.017-fold (p < 0.05), and GIRK5 increased 1.616 ± 0.054-fold (p < 0.01) in HLE-m-miR-184 cells (Figure 5A). The GIRK5 gene expression increased 2.056 ± 0.165-fold (p < 0.05), but ALDH5A1 and GABRA3 were unaffected in the HLE-miR-184 (Figure 5B). Expression was normalized against HLE-NC in all cases. Moreover, the mRNA expression of GIRK5 was greater in HLE-miR-184 compared with HLE-m-miR-184 (Figure 5C). Western blot analysis indicated that ALDH5A1 and GABRA3 expression was reduced and GIRK5 was unaffected when HLE was transfected with m-miR-184 (Figure 5D).
Figure 5.
GRIK5, ALDH5A1, and GABRA3 Expression Was Analyzed by Real-Time PCR/Western Blot and Hypothesis of an Ocular Disease Mechanism
(A) The mRNA expression levels of GRIK5, ALDH5A1, and GABRA3 in HLE-m-miR-184 versus HLE-NC. (B) The mRNA expression levels of GRIK5, ALDH5A1, and GABRA3 in HLE-miR-184 versus HLE-NC. (C) The mRNA expression levels of GRIK5, ALDH5A1, and GABRA3 in HLE-m-miR-184 versus HLE-miR-184. Expression of GRIK5, ALDH5A1, and GABRA3 was determined by real-time PCR after miR-184, m-miR-184, or NC transfecting HLE for 24 hr. The error bars represent the data in triplicates. Results are displayed as mean ± SEM (**p < 0.01 and *p < 0.05). (D) ALDH5A1, GABRA3, and DGRIK5 expression levels were determined by western blot after HLE cells were transfected with m-miR-184, miR-184, or NC for 48 hr. GAPDH was used as an internal control. (E) m-miR-184 exposure affects the alanine, aspartate, and glutamate metabolism pathway by the downregulation of ALDH5A1 and GABRA3, contributing to dysfunction of the citrate cycle.
Taken together, we are now able to formulate the following hypothesis: the overexpression of m-miR-184 in HLE cells directly inhibits ALDH5A1 expression and downregulates GABRA3, disordering the alanine, aspartate, and glutamate metabolism pathway (Figure 5E). In the absence of ALDH5A1, succinate semi-aldehyde cannot be fully dehydrogenated to succinate. Succinate is an important intermediate in the molecular citrate cycle. The insufficiency of succinate may cause disorder of the citrate cycle and ultimately lead to ocular disease.
Discussion
The miRNAs are small ncRNAs binding to complementary sequences in the 3′ UTR of target mRNA transcripts, inhibiting mRNA translation or promoting mRNA degradation.1 miR-184 located on the q arm of chromosome 15 is known to play a key role in neurological development and apoptosis and is highly expressed in mouse brain, mouse corneal epithelium, lens, and retinal pigment epithelium.24, 25, 26 Also, miR-184 can be oxidatively modified by ROS (reactive oxygen species) and mismatch with the 3′ UTRs of Bcl-xL and Bcl-w. The mismatch of oxidized miR-184 with Bcl-xL and Bcl-w is involved in the apoptosis of rat heart cell line H9c2 and mouse models.27 Furthermore, m-miR-184 is found in families with EDICT syndrome or cataracts and varying corneal abnormalities. How the m-miR-184 influences ocular disease is not clear. Recently, evidence has suggested that circRNA can act as a sponge for certain miRNAs.16 So we focused on describing the differential expression of mRNAs and circRNAs in cells expressing m-miR-184 in an effort to elucidate a possible mechanism for ocular disease.
Our analysis reports 16 significant DEGs between m-miR-184 and miR-184 treatments, nine of which are membrane and calcium ion genes. Furthermore, all downregulated DEGs have perfect seed sequence matches with m-miR-184. Moreover, we report 47 significant DEGs between HLE-miR-184 and HLE-NC and 63 significant DEGs between HLE-m-miR-184 and HLE-NC cells. We found that the number of downregulated DEGs in HLE-m-miR-184 cells is almost three times higher than in HLE-miR-184 cells. Results indicated that m-miR-184 downregulates a broader spectrum of genes when compared to the wild-type miR-184. Also, the upregulation of GRIK5 and downregulation of ALDH5A1 and GABRA3 in HLE-m-miR-184 cells suggest that there is some differential regulation of the alanine, aspartate, and glutamate metabolism pathway, which may be linked to ocular disease.
The apoptosis of HLE cells is a common cause of non-congenital cataract development in humans and animals.28 Our data investigated a global change in the apoptosis genes across the six miRNA treatment cells. There are no significant differences in the expression of these genes in any of the groups. We only found CASP7 to be upregulated in HLE-miR-184 and HLE-m-miR-184 when compared to the control. Further, we have shown that there was no difference in apoptosis rate across the three groups. It appears that the m-miR-184 is not associated with the apoptotic pathway when compared to miR-184.
Although the sequencing data and real-time PCR results showed that GRIK5 was highly expressed in HLE-m-miR-184 cells, the protein expression was not confirmed by western blot. Both mRNA and protein levels of ALDH5A1 and GABRA3 were downregulated in HLE-m-miR-184 cells, but there was a greater degree of ALDH5A1 downregulation than that of GABRA3. Integration of these different data types leads us to the hypothesis that the overexpression of m-miR-184 in HLE cells directly inhibits ALDH5A1 expression, indirectly downregulating GABRA3. A KEGG enrichment analysis shows that these two genes are related to the alanine, aspartate, and glutamate metabolism pathway. Fonnum and Henke29 reported that there is a marked reduction of glutamate and aspartate in the optic nerve and the superficial tectal layers after retinal ablation. Also, there is a good correlation between the topographical distribution of glutamate carboxylase activity and the concentration of GABA in the tectum.29 There is evidence indicating that the dysregulation of ALDH5A1, GABA, and GRIK5 is associated with epilepsy, psychomotor retardation, developmental delay, and hypotonia.22, 23 Obviously, this hypothesis provides a new perspective for the association of m-miR-184 with ocular disease.
Moreover, our study reports the first prediction of circRNA expression patterns in the presence of m-miR-184 and miR-184. Our prediction reveals that, within a single cell type, circRNA expression patterns appear globally variable. Total circRNA expression and the number of chromosomal origins for the circRNA also appear to be random under our conditions. We further observed the interaction between miR-184/m-miR-184 and circRNA. We found that the miR-184 and m-miR-184 can bind the same circRNA, although the numbers of binding sites for the mutant form seem lower than for the wild-type miRNA. Our analysis suggests that the interaction between these miRNAs and the circRNA is compatible and not exclusive. Further, of the total 2,376 circRNAs, 1,780 are observed within TEs, indicating that this is a common phenomenon. Most of the TEs were composed of SINEs (87.33%), and ALUs accounted for 80.41% of these SINEs. These results suggest a tendency for masked bases in circRNA to be composed of ALUs. Further, SINEs show a high correlation with ALUs (r = 0.965) and masked bases show correlation with SINEs (r = 0.512) and LINEs (r = 0.564) after Pearson correlation coefficient analysis. Taken together, our study shows that circRNAs are abundant in ALU sequences and have miR-184 and m-miR-184 binding sites; but, we have not identified a specific biological function for these ncRNAs.
Our study presents an innovative data integration analysis in which mRNA, miR-184/m-miR-184, and circRNAs are studied. We provide a catalog of predicted circRNAs in HLE cell lines, which may be of relevance for other research groups.
The identification of the alanine, aspartate, and glutamate metabolic pathway as a target for m-miR-184 offers a new avenue for understanding ocular disease progression; but, further complete proteomic and metabolomic profiling is needed to elucidate how m-miR-184 influences ocular disease through disorder of ALDH5A1 and GABRA3 expression and the long-term effects of m-miR-184 on cells, especially regarding potential clinical application.
Materials and Methods
Reagents
Culture reagents and molecular biology products including Opti-MEM reduced serum medium were purchased from Gibco (Life Technologies). Lipofectamine RNAiMAX transfection reagent was acquired from Invitrogen (Life Technologies). The synthesized miRNA mimics (miR-184 and m-miR-184) and the negative control were from GenePharma.
Cell Culture and Treatment
HLE cells obtained from ATCC (CRL-11421) were used as a cell model. HLE cells were maintained as a monolayer in a humidified incubator containing 5% CO2 at 37°C. The cells were cultured in growth medium comprising DMEM/F-12 50/50 (DMEM/Han’s F-12 50/50 Mix; Corning) with L-glutamine, 15 mM HEPES, and 10% FBS (fetal bovine serum). Transfections were done using 30 nM miR-184, m-miR-184, and the NC in HLE cells at a cell density of 1 × 107 cells. Transfections were done using the Lipofectamine RNAiMAX transfection reagent protocol. This experiment was done in duplicate.
RNA Isolation
After the cells had been transfected with miRNAs for 24 hr, total RNA was extracted using Trizol reagent according to the manufacturer’s instructions (Ambion, Life Technologies). RNA degradation and contamination was monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN). RNA concentration was measured using Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies). RNA integrity was assessed by RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies).
Library Preparation for Transcriptome Sequencing
A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using Next Ultra RNA LibraryPrep Kit for Illumina (New England Biolabs), following the manufacturer’s recommendations. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina), according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq2500 platform and 125-bp paired-end reads were generated. The raw data from this paper have been submitted to the genebank database https://www.ncbi.nlm.nih.gov/sra/ under accession numbers: SRP077606, SRP077680, and SRP077681.
RNA-Seq Analysis
RNA-seq (RNA sequencing) reads were trimmed to the first 80 bp and then aligned to the Ensembl human genome using Bowtie v2.2.330 using the default parameters. The paired-end clean reads were then aligned to the reference genome using TopHat v2.0.12.31
Library Preparation for circRNA Sequencing
A total of 5 μg RNA was treated with Epicenter Ribo-zero rRNA Removal Kit and RNase R (Epicenter). Sequencing libraries were generated using the Next Ultra Directional RNA LibraryPrep Kit for Illumina (NEB), following the manufacturer’s recommendations. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq PE cluster kit (Illumina). After cluster generation, the library preparations were sequenced on an Illumina HiSeq 4000 platform, and 150-bp paired-end reads were generated.
circRNA Prediction and Annotation from Sequencing Data
circRNA sequences were predicted using the protocol described by Memczak et al.14 Briefly, the RNA-seq reads mapping to the human genome were discarded, retaining only the spliced reads. Next, we extracted 20mers from both ends and aligned them independently to find unique anchor positions within spliced exons. Finally, the anchor alignments were extended to the original read sequence alignments, and the inferred breakpoint was flanked by the required GU/AG splice signals. Non-unique mappings and ambiguous breakpoints were discarded. Genome references used for all mapping and subsequent analyses were from the Ensembl human genome, release_77. All circRNAs identified from sequencing data were annotated in the circBase database.
Differential Expression Analysis
Following the primary analysis, each biological entity (mRNA and circRNA) was associated with a read count number. The mRNA raw counts were normalized using FPKM (fragments per kilobase of transcript sequence per million base pairs sequenced) normalization.32 Differential expression analysis of the two groups was performed using the DESeqR package (1.18.0),33 and p values were adjusted using the Benjamini and Hochberg approach for controlling the false discovery rate.34 Genes with an adjusted p value < 0.05 were assigned as differentially expressed. Further, we estimated significant differentially expressed entities between the two treatment groups using the following criteria: (1) a minimum count after normalization of ten reads, (2) absolute fold change ≥ 1.5, and (3) a corrected p value ≤ 0.05. Since we worked with two biological replicates per condition, we also removed the biological entities for which the two biological replicates were discordant (e.g., opposite fold change direction) or for which a single sample represented >75% of all the read counts. The quantitation of circRNA raw counts was normalized as TPM (transcript per million tags).35
miRNA Target Prediction
Putative interactions between the miR-184/m-miR-184 seed sequence and the predicted mRNA/circRNAs were evaluated using miRanda, investigating only perfect seed matching without gap or Wobble pairing. To this end, the 3′ UTR of all mRNAs expressed in our samples was downloaded using Ensembl Biomart, and the circRNA BED output file was converted into FASTA format DNA sequences.
The Analysis of circRNA Feature
Accumulating evidence demonstrates that widespread ALU elements are distributed throughout the human genome in a non-random manner and play an important role in genome and gene evolution and epigenetic and gene regulation.36, 37 As one of the most important and abundant TEs that is still active in the human genome, ALU, a SINE family TE, has demonstrated its indispensable regulatory function.38 There are three major subfamilies as follows: the oldest ALUJ, intermediately aged ALUS, and the youngest ALUY.39 ALUs regulate gene expression and the function of RNAs, including miRNAs, circRNAs, and lncRNAs (long ncRNAs).40
Moreover, recent findings indicate that inverted ALU repeats appear to promote the formation of circRNAs by facilitating back-splicing of pre-mRNAs.41 We analyzed circRNA sequence on RepeatMasker program and assigned all the TEs to four major types (LINE, SINE, LTR, and DNA). The kernel density of the four TE types was calculated, and the relationship of different TEs was then used in a Pearson correlation coefficient analysis. The GC content of the circRNA was calculated too.
Real-Time PCR and Western Blot Analysis
Differentially expressed ALDH5A1, GABRA3, and GIRK5 were validated by real-time PCR and western blot according to the previously published experimental methods used in our laboratory.42
Author Contributions
Y.L. and K.Y. conceived and designed the experiments. Y.L. and S.L. performed the experiments. Y.L. analyzed the data. Y.L. wrote and edited the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (2016FZA7009), the Zhejiang Provincial Natural Science Foundation of China (LY17H120001), the Key Program of Natural Science Foundation of China (81130018), the Program of National Natural Science Foundation of China (81371001), the Program of National Natural Science Foundation (81428005); and the Project of National Clinical Key Discipline of the Chinese Ministry of Health, Zhejiang ProvinceKey Research and Development Program.
Footnotes
Supplemental Information includes seven data sheets and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.02.008.
Supplemental Information
References
- 1.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Lynam-Lennon N., Maher S.G., Reynolds J.V. The roles of microRNA in cancer and apoptosis. Biol. Rev. Camb. Philos. Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Garcia I., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 4.Ryan D.G., Oliveira-Fernandes M., Lavker R.M. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 5.Karali M., Peluso I., Gennarino V.A., Bilio M., Verde R., Lago G., Dollé P., Banfi S. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes A.E., Bradley D.T., Campbell M., Lechner J., Dash D.P., Simpson D.A., Willoughby C.E. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am. J. Hum. Genet. 2011;89:628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi Y., Chen Q., Rajala R.V.S., Ma J.X. MicroRNA-184 modulates canonical Wnt signaling through the regulation of frizzled-7 expression in the retina with ischemia-induced neovascularization. FEBS Lett. 2015;589:1143–1149. doi: 10.1016/j.febslet.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J., Yang X., Xie B., Chen Y., Swaim M., Hackett S.F., Campochiaro P.A. MicroRNAs regulate ocular neovascularization. Mol. Ther. 2008;16:1208–1216. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann A., Huang Y., Suetsugu-Maki R., Ringelberg C.S., Tomlinson C.R., Del Rio-Tsonis K., Tsonis P.A. Implication of the miR-184 and miR-204 competitive RNA network in control of mouse secondary cataract. Mol. Med. 2012;18:528–538. doi: 10.2119/molmed.2011.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iliff B.W., Riazuddin S.A., Gottsch J.D. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest. Ophthalmol. Vis. Sci. 2012;53:348–353. doi: 10.1167/iovs.11-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bykhovskaya Y., Seldin M.F., Liu Y., Ransom M., Li X., Rabinowitz Y.S. Independent origin of c.57 C > T mutation in MIR184 associated with inherited corneal and lens abnormalities. Ophthalmic Genet. 2015;36:95–97. doi: 10.3109/13816810.2014.977491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bykhovskaya Y., Caiado Canedo A.L., Wright K.W., Rabinowitz Y.S. C.57 C > T Mutation in MIR 184 is Responsible for Congenital Cataracts and Corneal Abnormalities in a Five-generation Family from Galicia, Spain. Ophthalmic Genet. 2015;36:244–247. doi: 10.3109/13816810.2013.848908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abu-Amero K.K., Helwa I., Al-Muammar A., Strickland S., Hauser M.A., Allingham R.R., Liu Y. Screening of the Seed Region of MIR184 in Keratoconus Patients from Saudi Arabia. BioMed Res. Int. 2015;2015:604508. doi: 10.1155/2015/604508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Petkovic S., Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Yan Q., Liu J.P., Zou L.J., Liu J., Sun S., Deng M., Gong L., Ji W.K., Li D.W. Apoptosis: its functions and control in the ocular lens. Curr. Mol. Med. 2010;10:864–875. doi: 10.2174/156652410793937741. [DOI] [PubMed] [Google Scholar]
- 18.Mrugacz M., Zelazowska-Rutkowska B., Bakunowicz-Lazarczyk A., Wysocka J., Kaczmarski M. [The role of apoptosis in induction ocular changes in patients with cystic fibrosis] Klin. Oczna. 2007;109:22–24. [PubMed] [Google Scholar]
- 19.Nickells R.W., Zack D.J. Apoptosis in ocular disease: a molecular overview. Ophthalmic Genet. 1996;17:145–165. doi: 10.3109/13816819609057889. [DOI] [PubMed] [Google Scholar]
- 20.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Ding Y., Liu Y., Zhang Y., Song J., Wang Q., Li M., Qin Y., Huang S., Yang Y. Succinic semialdehyde dehydrogenase deficiency of four Chinese patients and prenatal diagnosis for three fetuses. Gene. 2015;574:41–47. doi: 10.1016/j.gene.2015.07.078. [DOI] [PubMed] [Google Scholar]
- 23.Pearl P.L., Parviz M., Vogel K., Schreiber J., Theodore W.H., Gibson K.M. Inherited disorders of gamma-aminobutyric acid metabolism and advances in ALDH5A1 mutation identification. Dev. Med. Child Neurol. 2015;57:611–617. doi: 10.1111/dmcn.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weitzel R.P., Lesniewski M.L., Greco N.J., Laughlin M.J. Reduced methyl-CpG protein binding contributing to miR-184 expression in umbilical cord blood CD4+ T-cells. Leukemia. 2011;25:169–172. doi: 10.1038/leu.2010.227. [DOI] [PubMed] [Google Scholar]
- 25.Shalom-Feuerstein R., Serror L., De La Forest Divonne S., Petit I., Aberdam E., Camargo L., Damour O., Vigouroux C., Solomon A., Gaggioli C. Pluripotent stem cell model reveals essential roles for miR-450b-5p and miR-184 in embryonic corneal lineage specification. Stem Cells. 2012;30:898–909. doi: 10.1002/stem.1068. [DOI] [PubMed] [Google Scholar]
- 26.Li P., Peng J., Hu J., Xu Z., Xie W., Yuan L. Localized expression pattern of miR-184 in Drosophila. Mol. Biol. Rep. 2011;38:355–358. doi: 10.1007/s11033-010-0115-1. [DOI] [PubMed] [Google Scholar]
- 27.Wang J.X., Gao J., Ding S.L., Wang K., Jiao J.Q., Wang Y., Sun T., Zhou L.Y., Long B., Zhang X.J. Oxidative Modification of miR-184 Enables It to Target Bcl-xL and Bcl-w. Mol. Cell. 2015;59:50–61. doi: 10.1016/j.molcel.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Li W.C., Kuszak J.R., Dunn K., Wang R.R., Ma W., Wang G.M., Spector A., Leib M., Cotliar A.M., Weiss M. Lens epithelial cell apoptosis appears to be a common cellular basis for non-congenital cataract development in humans and animals. J. Cell Biol. 1995;130:169–181. doi: 10.1083/jcb.130.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fonnum F., Henke H. The topographical distribution of alanine, aspartate, gamma-aminobutyric acid, glutamate, glutamine, and glycine in the pigeon optic tectum and the effect of retinal ablation. J. Neurochem. 1982;38:1130–1134. doi: 10.1111/j.1471-4159.1982.tb05358.x. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L., Chen J., Li Z., Li X., Hu X., Huang Y., Zhao X., Liang C., Wang Y., Sun L. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teng L., Firpi H.A., Tan K. Enhancers in embryonic stem cells are enriched for transposable elements and genetic variations associated with cancers. Nucleic Acids Res. 2011;39:7371–7379. doi: 10.1093/nar/gkr476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynch V.J., Leclerc R.D., May G., Wagner G.P. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 38.Kazazian H.H., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 39.Kapitonov V., Jurka J. The age of Alu subfamilies. J. Mol. Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- 40.Daniel C., Behm M., Öhman M. The role of Alu elements in the cis-regulation of RNA processing. Cell. Mol. Life Sci. 2015;72:4063–4076. doi: 10.1007/s00018-015-1990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Y., Wu X., Ling Z., Yuan L., Cheng Y., Chen J., Xiang C. microRNA133a targets Foxl2 and promotes differentiation of C2C12 into myogenic progenitor cells. DNA Cell Biol. 2015;34:29–36. doi: 10.1089/dna.2014.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.