Abstract
Spinal muscular atrophy (SMA) is a recessive disease caused by mutations in the SMN1 gene, which encodes the protein survival motor neuron (SMN), whose absence dramatically affects the survival of motor neurons. In humans, the severity of the disease is lessened by the presence of a gene copy, SMN2. SMN2 differs from SMN1 by a C-to-T transition in exon 7, which modifies pre-mRNA splicing and prevents successful SMN synthesis. Splice-switching approaches using antisense oligonucleotides (AONs) have already been shown to correct this SMN2 gene transition, providing a therapeutic avenue for SMA. However, AON administration to the CNS presents additional hurdles. In this study, we show that systemic delivery of tricyclo-DNA (tcDNA) AONs in a type III SMA mouse augments retention of exon 7 in SMN2 mRNA both in peripheral organs and the CNS. Mild type III SMA mice were selected as opposed to the severe type I model in order to test tcDNA efficacy and their ability to enter the CNS after maturation of the blood brain barrier (BBB). Furthermore, subcutaneous treatment significantly improved the necrosis phenotype and respiratory function. In summary, our data support that tcDNA oligomers effectively cross the blood-brain barrier and offer a promising systemic alternative for treating SMA.
Keywords: splice switching, spinal muscular atrophy, antisense oligonucleotide, tricyclo-DNA
Introduction
Spinal muscular atrophy (SMA) is an autosomal-recessive inherited disease characterized by the degeneration of α-motor neurons of the anterior horn of spinal cord, which leads to progressive muscle atrophy. It is caused by mutations in the gene SMN1 encoding the survival motor neuron protein (SMN), and it has an incidence of ∼1 in 6,000 live births.1 SMN is a 37-kDa protein located in both the cytoplasm and the nucleus, where it is concentrated in several intense foci referred to as gemini of Cajal bodies (gems).2 This ubiquitous protein plays an essential role in uridine-rich small nuclear ribonucleoprotein (snRNP) assembly in the cytoplasm3 and also participates in snRNP importation into the nucleus. Once in the nucleus, SMN relocates to gems, while released snRNPs accumulate in Cajal bodies to form part of the splicing machinery. One might expect that reduced SMN levels should broadly affect the processing of pre-mRNAs in all cell types, though this clearly does not occur in SMA patients. The neuron-specific consequences of SMA are still widely debated, though have been partially attributed to SMN’s role in the axonal transport of particular mRNAs, which suggests a rationale for the prominent effect of SMN depletion on spinal motor neuron survival.4 The clinical consequences of SMA range from severe to mild (types 1–4), tending to affect proximal muscle groups primarily, and with deleterious effects on respiratory function.
In humans, the severity of disease is lessened by the presence of SMN2 (a centromeric copy of the gene).5 SMN2 differs from SMN1 (a telomeric copy) by only one nucleotide in the coding sequence: a translationally silent C to T at position +6 (C6T) altering the splicing of exon 7. Consequently, 10% of SMN2 pre-mRNAs are correctly spliced, but 90% lack exon 7 (SMN(Δ7)).6, 7 Translation of respective mRNAs gives rise to a very limited amount of fully functional full-length SMN protein in addition to a truncated SMN(Δ7) protein, which is rapidly degraded.8, 9 As one might expect, the severity of SMA is inversely proportional to the SMN2 copy number, which varies from 1 to 8 in humans,10 suggesting the potential of SMN2 correction as a valid therapeutic strategy.
The splicing of SMN2 exon 7 has been well characterized and requires many different elements, including a suboptimal intron 6 branch point,11 an extended inhibitory context,12, 13 a conserved tract domain and an inhibitory 3′-cluster,12 an intronic silencer element in intron 7 (ISS-N1),14 and a terminal stem-loop (TSL) structure on the 5′ splice site of exon 715 (Figure S1). The particular genomic organization of SMN genes has enabled novel therapeutic approaches aiming at re-including SMN2 exon 7 to produce fully functional SMN protein in the absence of the SMN1 gene. This has already been achieved using antisense oligonucleotide (AON) strategies aimed at blocking the exonic splice silencer generated by the C6T transition, annealing the intronic splice silencer downstream of exon 7, or decreasing the strength of the acceptor splice site of exon 8.16
Various classes of AONs have been developed to modulate mRNA splicing. Oligonucleotide studies have shown efficient re-inclusion of SMN2 exon 7, in particular those using 2′-O-methoxyethyl (2′MOE) with 17-mer AONs targeting ISS-N1. This particular AON demonstrated phenotypic rescue (ear and tail necrosis) of type III SMA mice (a mild phenotype) after intracerebroventricular (i.c.v.) injection17 and increased survival of type I SMA mice (a severe model) by subcutaneous injections at postnatal day 0 (P0) and P3.18 Studies have also been conducted using phosphorodiamidate morpholino oligomer (PMO) AONs on different mice models using ICV injection that result in a significantly extended lifespan.19, 20 Interestingly, a cumulative benefit was obtained when ICV injections were coupled with intraperitoneal (i.p.) injections, further reinforcing the peripheral function of SMN in SMA mice, which was previously demonstrated by Hua and colleagues.18, 21 This peripheral role of SMN strongly suggests that the optimal biodistribution of AONs across tissue types is important for therapeutic efficacy (at least in SMA mice models).
Notwithstanding these advances in chemistry and design, the uptake of AONs is still limited in many tissues, including the heart and the CNS, a crucial target in SMA. This is due to the inability of most AONs to cross the blood-brain barrier (BBB). Accordingly, in the majority of SMA animal studies and in trials for the recently US Food and Drug Administration (FDA)-approved nusinersen (an AON targeting ISS-N1), treatment is administered via the intrathecal or i.c.v. route. Despite encouraging data, these routes of administration present obvious clinical challenges and neglect the issue of delivery of AONs to the periphery.
In this paper, we explore the therapeutic potential of tricyclo-DNA (tcDNA) AONs, a class of conformationally constrained DNA analogs that display enhanced binding properties to DNA and RNA,22 allowing them to be designed as shorter sequences compared with chemistries such as 2′MOE or PMO. Moreover, tcDNA was recently shown to cross the BBB,23 a key property that allows us to effectively evaluate the central and peripheral restoration of SMN following systemic delivery.
In our study, we first compare the efficacy of two different tcDNA sequences to induce exon 7 re-inclusion in vitro. We further investigate the in vivo efficacy of tcDNA in a mild SMA mouse model (type III SMA mouse) to induce exon re-inclusion in all tissues following systemic administration. We have selected the type III SMA mouse model in order to test tcDNA-AONs efficacy and their ability to enter the CNS after maturation of the BBB.24 This would not have been feasible with severe type I mice, which require injections around P0–P2 because of their very short lifespan (around P10).17, 18
After subcutaneous tcDNA treatment, we show efficient exon re-inclusion in all tissues, including the CNS, which leads to phenotypic improvement in type III SMA. Furthermore, for the first time in SMA mice, we describe rescue of respiratory function after treatment with tcDNA using whole-body plethysmography.
Results
tcDNA AON Effectively Restores Exon 7 Re-inclusion in SMA Type 1 Fibroblasts
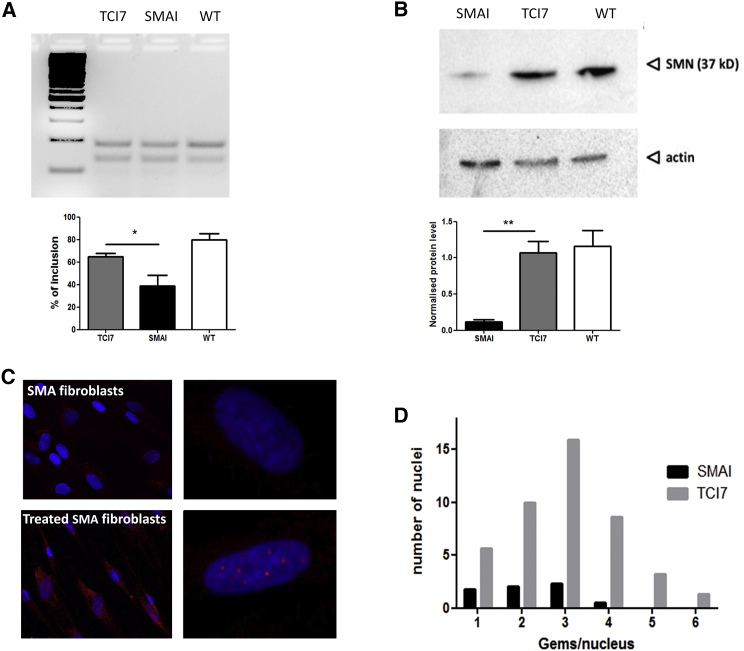
To evaluate the efficacy of tcDNA AONs, SMA type 1 fibroblast cells (GM03813, Coriell) were transfected with 30 μg oligonucleotide targeting the downstream ISS-N1 site in intron 7, thereby improving the strength of the 5′ donor site (15-mer tcDNA-AON I7; henceforth referred to as TCI7). Total RNA from transfected fibroblasts was analyzed by RT-PCR 48 hr after transfection to establish the inclusion levels of SMN2 exon 7. In TCI7-treated SMA fibroblasts, the level of exon 7 inclusion was augmented to 64% from a baseline of 40% in untreated SMA fibroblasts (Figure 1A).
Figure 1.
Evaluation of In Vitro Efficiency of tcDNA TCI7
(A) Detection of the inclusion of exon 7 of SMN2 mRNA by RT-PCR (primers on exons 6 and 8) in patient SMA I fibroblasts. PCR products were visualized on 3% agarose gel (top) and quantified by ImageJ (bottom). TCI7, fibroblasts treated with tcDNA AONs targeting the ISS-N1; SMAI, untreated SMAI fibroblasts (GM03813 Coriell); and WT, healthy fibroblasts of the mother (GM03814 Coriell). (B) Detection of the SMN protein by western blot in fibroblasts and normalization with actin. Lane 1, untreated SMAI fibroblasts (GM03813 Coriell); lane 2, SMA fibroblasts treated with TCI7; lane 3, healthy fibroblasts of the mother (GM03814 Coriell). Error bars represent SD. *p < 0.05 and **p < 0.01. (C) Detection of the SMN protein by immunostaining in untreated SMA fibroblasts (top) and SMA fibroblasts transfected with TCI7 (bottom). Cells were immunostained with the polyclonal antibody H-SMN 195 (Santa Cruz). The red staining shows the expression of SMN protein in gems. Nuclei are labeled in blue (DAPI). Left: original magnification ×20; right: original magnification ×63. For magnification ×63, the apotome is used to see the gems in the nucleus. (D) Counting the number of gems stained positively for SMN in each nucleus (n = 100 nuclei). The black bars correspond to untreated SMA type I, and gray bars correspond to cells treated with TCI7.
Despite the fact that SMA type 1 fibroblasts have no functional SMN1 gene and only two copies of the SMN2 gene, they still produce SMN protein generated from the two existing SMN2 gene copies; however, the truncated protein of the SMN2 gene is unstable and rapidly degraded, hampering normal cellular function. Western blot was conducted, and protein levels were recorded as a percentage of total actin expressed by respective fibroblast cell lines (GM03814, Coriell) (Figure 1B, WT). Untreated SMA type 1 fibroblasts expressed 10% of SMN protein at 37 kDa (Figure 1B, SMAI), whereas in TCI7-treated type 1 SMA fibroblasts induced a significant increase of total SMN protein, reaching mean levels of 90% as a proportion of total wild-type (WT) levels (Figure 1B, TCI7).
In addition, another tcDNA oligonucleotide sequence (17-mer tcDNA-AON TSL; henceforth referred to as TCTSL), which anneals to the TSL structure (Figure S1), was investigated, and analogous 2′-O-methyl compounds were looked at to compare their efficacies (Figure S2). RT-PCR and western blot results indicate that tcDNA AONs induce higher levels of exon 7 inclusion and SMN protein restoration than 2′OMe AONs (although this increase is not statistically significant). Regarding the targeted region, ISS7-N1-manipulation with AONs resulted in more inclusion of exon 7 and SMN protein expression, which prompted us to continue with this target for the remaining experiments.
To assess proper localization of SMN protein in gems, immunofluorescence was used. Compared with fibroblasts of healthy individuals, which normally display between 52 and 113 gems per 100 nuclei,25 most SMA type I patient fibroblasts have very limited amounts of SMN-containing nuclear gems (Figures 1C and 1D). To determine if TCI7 treatment increased levels of full-length SMN protein in its correct localization, we counted the number of gem-positive nuclei in treated and untreated SMA type 1 fibroblasts. As shown in Figure 1C, the restored SMN protein is correctly localized in nuclear gems in TCI7-treated fibroblasts, and the number of gem-positive nuclei is restored to the level found in fibroblasts of healthy individuals (Figure 1D).
TCI7 Induces Exon 7 Inclusion in SMA Type III Mice
Our in vivo studies were conducted using SMA type III transgenic mice, which exhibit a mild SMA phenotype. Type III SMA mice are knocked out for murine SMN and possess two transgenes comprising two copies of the human SMN2 gene, leaving a total of four copies of SMN2 genes (Smn−/−; SMN2+/+).24 Type III SMA mice were injected subcutaneously every week for a period of 4 or 12 weeks with a dose of 200 mg/kg TCI7 (or PBS as a control in the untreated mice) and sacrificed 2 weeks post-treatment. Since an element of our enquiry is to establish trans-BBB efficacy, the injection of mice commenced at P7, because at this stage of murine development the BBB already has very limited permeability.
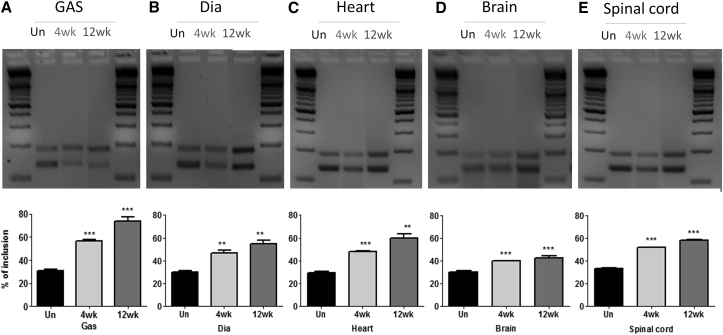
Mice were analyzed for exon 7 inclusion by RT-PCR in different tissues, including the gastrocnemius, diaphragm, heart, brain, and spinal cord (Figure 2). In untreated mice, the percentage of exon 7 inclusion in different tissues was ∼30%. TCI7 treatment significantly increased the inclusion of exon 7 in all tissues. This increase was particularly significant in the gastrocnemius muscle, where 55% of exon 7 inclusion is detected after 4 weeks of treatment and 75% is detected after 12 weeks of treatment. The degree of improvement was similar in diaphragm, heart, and spinal cord, with close to 50% inclusion after 4 weeks of treatment and up to 60% inclusion after 12 weeks. In brain, the level of exon 7 inclusion reached 40% after 12 weeks of treatment.
Figure 2.
Evaluation of tcDNA TCI7 Efficiency in SMA Mice
Detection of the inclusion of exon 7 of SMN2 mRNA by RT-PCR in different tissues (A, gastrocnemius [GAS and Gas]; B, diaphragm; C, heart; D, brain; E, spinal cord) of SMA mice type III and quantification using ImageJ (bottom). Un, untreated mice (n = 3); 4wk, mice treated for 4 weeks with 200 mg/kg per week by subcutaneous injection and sacrificed 2 weeks after treatment (n = 3); 12wk, mice treated for 12 weeks with 200 mg/kg per week by subcutaneous injection and sacrificed 2 weeks after treatment (n = 4). Error bars represent SD. *p < 0.05 and **p < 0.01 compared with saline controls.
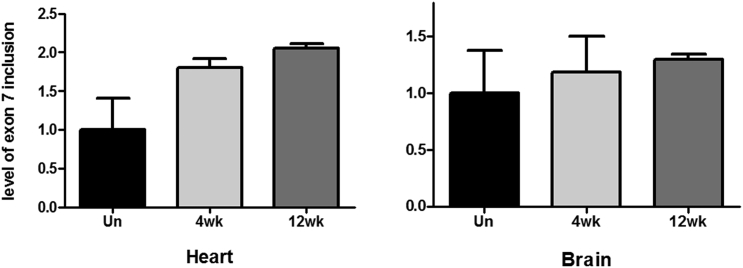
We also quantified by real-time qPCR the level of inclusion in heart and brain tissue (Figure 3). In cardiac muscle, real-time qPCR revealed that exon 7 inclusion was increased 1.8-fold after 4 weeks of treatment and over 2-fold after 12 weeks of treatment, confirming our previous semiquantitative results. In the brain, the inclusion was increased 1.3-fold after 12 weeks of treatment, verifying the ability of TCI7 to induce exon 7 inclusion in this tissue type.
Figure 3.
Quantitative Evaluation of Exon 7 Inclusion after tcDNA TCI7 Treatment in SMA Mice
Detection of the inclusion of exon 7 of SMN2 mRNA by qPCR in heart and brain of SMA type III mice. The primers used for amplification are located on exon 6 and 7 and normalized with GAPDH. Un, untreated mice; 4wk, mice treated for 4 weeks with 200 mg/kg per week by subcutaneous injection of TCI7; 12wk, mice treated for 12 weeks with 200 mg/kg per week by subcutaneous injection of TCI7; n = 3 per group.
TCI7 Treatment Is Consistently Sustained Long-Term
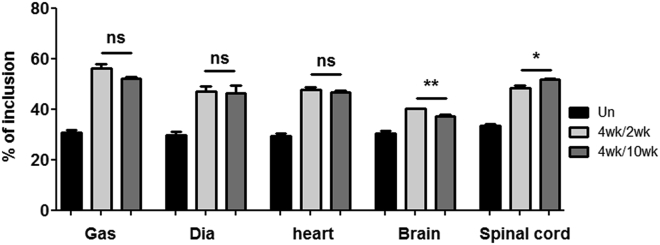
To evaluate the long-term effect of tcDNA treatment, we also analyzed exon 7 inclusion by RT-PCR 10 weeks after the end of a 4-week course of tcDNA treatment, comparing it with samples obtained 2 weeks after the end of the treatment. The level of exon 7 inclusion appears to be similar for the two treated groups of mice at 2 weeks or 10 weeks after the end of the treatment across analyzed tissues, suggesting that TCI7 or its effects persist in tissues for at least 10 weeks and are still inducing exon inclusion (Figure 4).
Figure 4.
Sustained Effect of tcDNA TCI7 Treatment in SMA Mice
Detection of the inclusion of exon 7 of SMN2 mRNA by RT-PCR in different tissues (gastrocnemius [Gas], diaphragm, heart, brain, and spinal cord) of SMA mice type III and quantification using ImageJ to evaluate the long-term effect of tcDNA treatment. Black bars represent untreated mice, gray bars represent mice treated for 4 weeks with 200 mg/kg per week by subcutaneous injection and sacrificed 2 weeks after treatment (called 4wk/2wk), and dark gray bars represent mice treated for 4 weeks with 200 mg/kg per week by subcutaneous injection and sacrificed 10 weeks after treatment (called 4wk/10wk). n = 3 per group; error bars represent SD. *p < 0.05 and **p < 0.01. ns, not significant.
TCI7 Treatment Significantly Improves Respiratory Function in SMA Mice
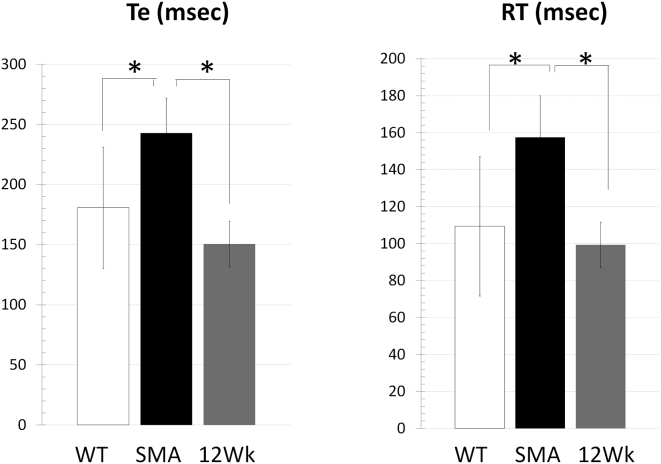
SMA type III patients display respiratory dysfunction, such as thoraco-abdominal asynchrony, which is often caused by paralysis of the rib cage muscles in the presence of normal diaphragmatic activity.26 Notwithstanding, to our knowledge, respiratory function has never been studied in SMA type III mice. We investigated respiratory function in SMA type III mice by whole-body plethysmography, a non-invasive technique that measures various respiratory parameters (displayed in Table 1). Compared with age-matched WT control FVB mice, Te (expiration time), RT (relaxation time), and Penh are significantly increased in SMA type III mice (Table 1). Some parameters are not altered in SMA type III mice, such as Ti (inspiration time), f (breathing frequency), TV (tidal volume), and MV (minute ventilation), consistent with what is observed in patients compared to healthy individuals26 (Table 1). TCI7 treatment in SMA type III mice restored RT and Te back to WT levels, suggesting that an increase of exon 7 inclusion to 60% in the diaphragm is correlated with an improvement in respiratory function (Figure 5).
Table 1.
Respiratory Function Evaluation in WT FVB Mice and SMA Type III Mice
| Ti (ms) | Te (ms) | PIF (mL/s) | PEF (mL/s) | TV (mL) | EV (mL) | RT (ms) | MV (mL) | f (bpm) | EIP (ms) | EEP (ms) | Penh | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FVB | ||||||||||||
| Mean | 129.6 | 180.8 | 2.9 | 1.9 | 0.2 | 0.2 | 109.5 | 44.4 | 205.5 | 1.2 | 19.9 | 0.444 |
| SEM | 7.18 | 50.45 | 0.30 | 0.33 | 0.03 | 0.03 | 37.71 | 8.68 | 20.47 | 0.17 | 4.37 | 0.061 |
| SMA | ||||||||||||
| Mean | 130.1 | 242.8 | 2.4 | 1.3 | 0.2 | 0.2 | 157.5 | 33.3 | 179.0 | 1.2 | 23.4 | 0.353 |
| SEM | 8.81 | 29.20 | 0.35 | 0.23 | 0.01 | 0.01 | 22.48 | 5.23 | 15.41 | 0.08 | 2.40 | 0.026 |
| p Value | ||||||||||||
| 0.5130 | 0.0472 | 0.4490 | 0.0747 | 0.1816 | 0.2039 | 0.0423 | 0.1400 | 0.1136 | 0.8545 | 0.1899 | 0.0097 | |
| = | + | = | = | = | = | + | − | − | = | + | − | |
Te, expiratory time; EEP, end-expiratory pause; EIP, end-inspiratory pause; PIF, peak inspiratory flow; PEF, peak expiratory flow.
Figure 5.
Respiratory Function Evaluation in SMA Mice after tcDNA TCI7 Treatment
Respiratory function in SMA mice treated with 200 mg/kg tcDNA for 12 weeks compared to SMA control mice and WT mice (n = 4 per group). Ti, Te, end-inspiratory pause (EIP), end-expiratory pause (EEP), RT, f, and Penh are shown. Error bars represent mean ± SD. *p < 0.05 compared with the saline controls.
TCI7 Treatment Halts Tissue Necrosis in SMA III Mice
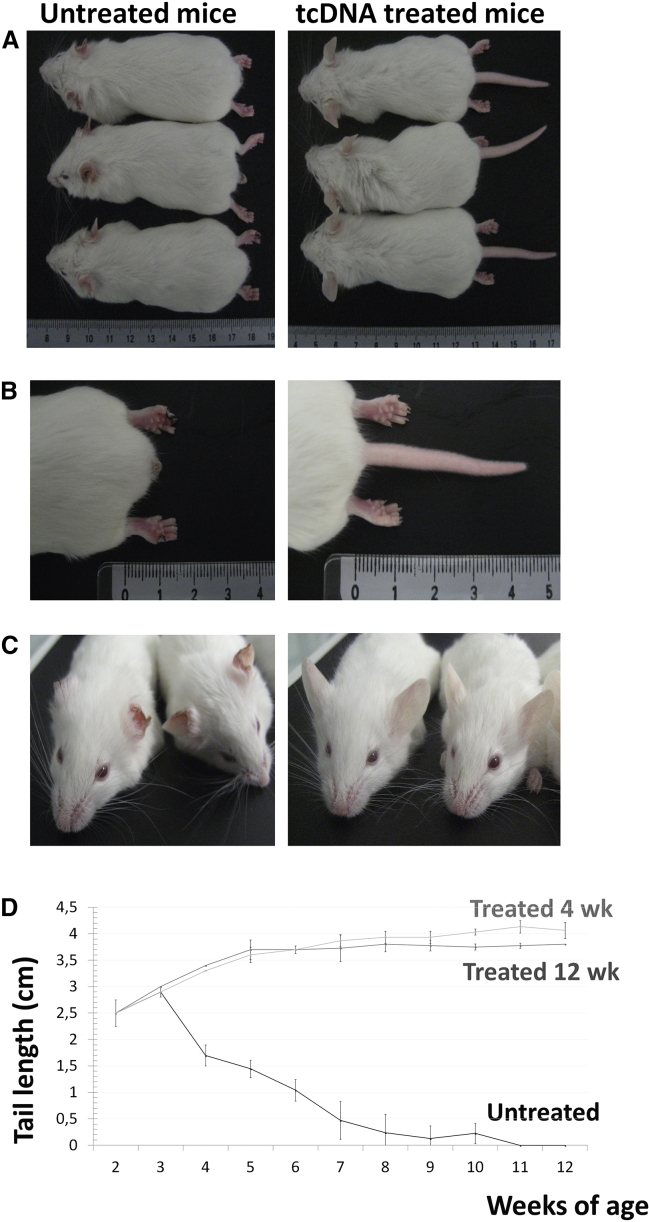
Finally, we investigated the effect of TCI7 treatment on the SMA III phenotype. SMA type III mice typically display progressive necrosis of tails, ears, and toes.24 Tail lengths in all mice were measured weekly throughout the course of treatment (12 weeks). Untreated mice at week 12 post-treatment underwent complete tail necrosis, while treated SMA mice were unaffected (Figures 6A, 6B, and 6D). Interestingly, treatment with 200 mg/kg TCI7 for 4 weeks also resulted in tail lengths comparable to those of mice treated for 12 weeks (Figure 6D), suggesting that this phenotypic benefit is sustained long-term. Moreover, like SMA controls, none of the treated mice presented necrosis of toes and ears at 12 weeks of age (Figures 6B and 6C). A dosing regimen of 100 mg/kg was also evaluated, and this treatment resulted in slightly shorter tails compared with mice treated with 200 mg/kg, suggesting that the effect of TCI7 is dose dependent (data not shown).
Figure 6.
Phenotype Rescue following tcDNA Treatment in SMA Mice
(A) Phenotypic rescue in type III SMA mice at 12 weeks of age after tcDNA treatment, compared to SMA control. (B) Toes and tails necrosis rescue in treated mice (right picture) compared to SMA control (left picture). (C) Ears rescue in treated mice (right picture) compared to SMA control (left picture). (D) Tails length of mice according to weeks. The darker gray curve corresponds to the size of the tails of type III SMA mice treated with TcI7 during 12 weeks (n = 4), the black curve is the size of the tails of type III SMA untreated mice (n = 11) and the lighter gray one corresponds to the tails length of type III SMA mice treated with TcI7 during 4 weeks only (n = 3).
Discussion
Splice switching to induce SMN re-inclusion using AON is a well-established strategy for the treatment of SMA. As previously mentioned, an AON of the 2′O-methoxyethyl subtype, nusinersen (marketed as Spinraza), has recently been approved by the FDA for the treatment of SMA. After positive phase 1 results with an improvement of the Hammersmith functional motor scale at 3 months post-treatment,27 interim analyses of phase 3 studies (ENDEAR and CHERISH) have demonstrated promising outcomes for patients. However, it is notable that nusinersen is administered intrathecally, a route of administration that is frequently less well tolerated and typically requires specialist training compared with alternative routes of administration (e.g., subcutaneous or intravenous). In a study analyzing the patient experience of nusinersen, 32% of lumbar punctures (LPs) resulted in adverse events.28 While the LP event rate in SMA patients is not higher than that usually reported in children, it is noteworthy that all patients in this trial underwent general anaesthesia, which is not without its own risks.
CNS delivery of most AONs does not occur with systemic (e.g., intravenous or subcutaneous) injection. In SMA, it is necessary for treatment to reach the CNS. To surmount the issue of AONs crossing the BBB, i.c.v. injections can be administered, or intrathecal pumps may be used clinically to deliver treatment to the spinal cord in patients. In the case of nusinersen, intrathecal injection is crucial, since it is unable to cross the BBB, though this neglects the issue of oligonucleotide penetration of peripheral tissues. It has been demonstrated several times (albeit in animal models) that peripheral rescue in SMA is important and that CNS treatment alone is not sufficient. While this phenomenon may not directly apply to human forms of the disease, it remains an unaddressed issue.
Consequent to these challenges, we believe that an unmet need remains for an AON treatment in SMA that not only deals with the issue of peripheral and central tissue penetration but also provides a simpler route of administration that is more easily performed and better tolerated. Previous research in our group using a Duchenne muscular dystrophy mouse model (named mdx) has demonstrated that tcDNA is able to cross the BBB.23 Using tcDNA administered intravenously, mdx mice benefitted from an improvement in cardio-respiratory function and furthermore, correction of behavioral features controlled by central mechanisms. Given tcDNA’s proven ability to cross the BBB while maintaining efficacy in the periphery when administered systemically, it is a strong candidate as a treatment for SMA.
To properly investigate this intrinsic property of tcDNA in SMA, we elected to use type III SMA mice rather than the more severe type I mouse model to allow sufficient time for maturation of the BBB. After successful in vitro proof-of-concept studies in SMA patient fibroblast cell lines, we have shown that subcutaneous administration of TCI7 in type III SMA mice (at P7) induces significantly increased levels of exon 7 re-inclusion in all examined tissues. Importantly TCI7 penetration includes the CNS, demonstrating the ability of tcDNA to cross the BBB.
TC17 splice-switching treatment improves the SMA phenotype by preventing tail, ear, and toe necrosis, further implying its therapeutic potential in SMA. It remains unclear why necrosis phenotypes are associated with SMA, a feature more prevalent in SMA mice than in human SMA phenotypes. The Hsieh-Li model used here presents with necrosis of the tail, ears, and toes, and many therapeutic studies conducted in severe forms of SMA mice report such events when lifespan is increased.19, 20, 29, 30 There are few documented cases of SMA type I patients with a single copy of SMN2 that have presented with finger necrosis, but the etiology is unclear.31, 32 Parson’s group has demonstrated that the dense capillary bed of skeletal muscle is dramatically decreased in severe SMA mice, potentially exacerbating the already-affected muscle. This mechanism of capillary reduction may also explain necrosis in mild SMA mice,33 but the precise role of SMN protein in necrosis is not established and may be due to a defect in innervations.34 Some oligonucleotide studies delay this necrosis phenotype via i.c.v. injection, and a recent study demonstrated a rescue of necrosis following subcutaneous injection between P23 and P31 after the onset of tail-tip necrosis.17, 35 These data demonstrate not only that the phenotype can be rescued by central administration but also that restoration of the periphery is achievable via systemic administration.36, 37
A further peripheral consequence of SMA is its respiratory complications, which are generally linked to intercostal muscle weakness and thoracic cage deformation.26 In some patients, atelectasis is reported,38, 39 and structural lung damage is observed after autopsy. In severe SMA mice, lung abnormalities are also found.40 For the first time, in this study, we have investigated respiratory function in type III SMA mice and also report some dysfunctions. Interestingly, TCI7 treatment restores some parameters of respiration, which could reflect the increased level of exon 7 re-inclusion in the diaphragm. Given TCI7’s efficacy in the brain and spinal cord, one cannot exclude its effect on restoring CNS control of respiration.
As previously demonstrated in the mdx model, we show here that tcDNA has excellent uptake in cardiac muscle, with similar levels of exon re-inclusion to that of diaphragmatic tissue. While cardiac issues are not particularly prevalent in SMA patients or mice models (save for congenital structural defects),41 SMA I patients with artificially prolonged lifespans have been shown to exhibit autonomic dysfunction,42 which may benefit from peripheral restoration of SMN.
Peripheral rescue of SMA may not be properly resolved by CNS administration alone, and tcDNA’s uptake profile across all tissues is a potential solution to this issue. That being said, other promising candidates that offer a similar uptake profile using systemic administration, including vector- and AON-based strategies, are currently under investigation.
Many adeno-associated vectors (AAVs) have been shown to efficiently cross the BBB,43 and as an example, a group using self-complementary AAV serotype 9 (scAAV9) has demonstrated transduction efficacy throughout the spinal cord after a single systemic injection in newborn and adult mice and non-human primates, potentially surmounting the issue of peripheral versus central treatment of SMA.44, 45 A clinical trial aiming at replacing SMN1 directly, using these vectors, is currently underway (www.clinicaltrials.gov identifier NCT02122952).
However, the well-documented immunogenicity of the AAV capsid is likely to complicate repeated administration (if required) of such a therapy, which is not an issue with AON-based approaches. Combined treatments consisting of an initial AAV injection followed by sequential AON administration could be a viable treatment approach.
Recently, a peptide-conjugated PMO AON (Pip6a-PMO) has been administered intravenously in SMA mice at P0 and P2, achieving significant lifespan extension.46 More importantly, this group demonstrated corrected SMN2 transcripts in the CNS after tail vein administration at 7.5 weeks, suggesting that this AON may penetrate the BBB. However, this was demonstrated in an asymptomatic mouse model; thus, TCI7 is the only AON to demonstrate geno- and phenotypic correction of central and peripheral SMA pathology with systemic administration as late as P7. Furthermore, the peptide moiety of Pip6a-PMO may be more likely to induce immunological reactions upon systemic administration than the naked chemistry of tcDNA, rendering clinical translation a lengthier and more complex process.
It is still debated how SMN deficiency leads to peripheral pathology, but the major goal of a tcDNA-based therapeutic strategy is to achieve systemic efficacy to ensure the best resolution of disease in SMA patients. tcDNA’s proven efficacy in restoring SMN2 exon 7 re-inclusion, in addition to its unique uptake profile and naked chemistry, offers these properties following less invasive administration, without the need for patient anaesthesia. tcDNA could therefore represent a particularly attractive AON therapy for SMA requiring whole-body treatment.
Materials and Methods
Cell Culture and tcDNA Transfection
Human fibroblasts from a 3-year-old type I SMA patient (GM03813, Coriell Cell Repositories) and WT fibroblasts (GM03814, Coriell Cell Repositories) were grown in DMEM with 20% fetal bovine serum and 1% penicillin-streptomycin (100 U/mL). tcDNA AONs were synthetized by Synthena as previously described,47, 48 and 2′OMePS was obtained from Eurogentec. The two sequences used were SMN2 TSL (39;55) 5′-(pTTAATTTAAGGAATGTG)-3′ and SMN2 TCI7(10;24) 5′-(pCTTTCATAATGCTGG)-3′.
2′OMePS and tcDNA transfection was performed with oligofectamine (Invitrogen) and incubated for 48 hr without serum and antibiotics.
RT-PCR and Real-Time qPCR
Total RNA was extracted 48 hr post-transfection using TRIzol reagent (Invitrogen), and first-strand cDNA synthesis was performed using Super Script II (Invitrogen) and random hexamers. PCR reactions were carried out with Master Mix 2x Phusion GC (Finnzymes) in a total volume of 50 μL from 200 ng cDNA, with 10μM each of the SMN-Ex6-FW (5′- GCTGATGCTTTGGGAAGTATGTTA-3′) and SMN-Ex8-Re primers (5′- ATTCCAGATCTGTCTGATCG-3′). The PCR products were separated by electrophoresis on a 3% agarose gel.
Real-time qPCR reactions were run in Opticon2 (Bio-Rad). cDNA was used as a template with primers specific for SMN and GAPDH, a housekeeping gene used as control for variations in the amount of template loaded to each reaction. The SMN primers forward 5′-GCTGATGCTTTGGGAAGTATGTTA-3′ in exon 6 and reverse 5′-CCTTAATTTAAGGAATGTGAGCACC-3′ in exon 7 were used. 20 ng cDNA was included in a 20 μL mix containing iTaq Universal SYBR green supermix (Bio-Rad) and 0.2 μM of each specific primer. The run conditions were as follows: 15-min polymerase activation step at 95°C, followed by 50 cycles of two-step qPCR (15 s of 95°C denaturation and 1 min of 60°C combined annealing/extension).
Western Blot
Fibroblast protein extracts were obtained in lysis buffer (10 mmol/L HEPES [pH 7.9], 100 mmol/l KCl, 1 mmol/l EDTA, 1 mmol/l 1,4-dithiothreitol, 1× complete protease inhibitor cocktail [Roche], and 0.5% NP-40). Equal amounts of protein (determined by Bradford Protein Assay [Pierce]) were mixed with 2× loading buffer (125 mmol/l Tris [pH 6.8], 2% sodium dodecyl sulfate, 10% glycerol, 0.01% bromophenol blue, and 10% β-mercaptoethanol).25 10 μg protein from each sample was resolved by SDS-PAGE 4%–12% Bis-Tris Gels (Invitrogen) and transferred onto a nitrocellulose membrane. The membrane was blocked with 10% milk in PBS-Tween buffer. SMN immunoblot was performed overnight using rabbit polyclonal antibody SMN H-195 (dilution 1/500, Santa Cruz Biotechnology). A goat anti-rabbit secondary antibody conjugated with horseradish peroxidase was used to detect the protein SMN (dilution 1/50,000). Signals were detected with the SuperSignal West Pico Chemiluminescent kit (Thermo Scientific). The membrane was then washed, re-blocked, and probed with a mouse monoclonal anti-actin antibody (dilution 1/5,000, Sigma Aldrich) followed by a secondary sheep anti-mouse antibody conjugated with horseradish peroxidase (dilution 1/15,000). The signal was detected as described earlier. Membranes were converted to numerical pictures by scanning, and band intensities were analyzed using ImageJ 1.46r software (http://gate2.inist.fr/login?url=http://rsb.info.nih.gov/ij/) and normalized to actin protein.
Immunofluorescence
Transfected fibroblasts on slides were fixed with acetone/methanol (volume/volume). Fixed cells were blocked in PBS + 5% BSA for 1 hr. SMN immunostaining was performed with rabbit polyclonal antibody SMN h-195 (dilution 1/100 in PBS +1% BSA) for 1 hr. Cells were washed in PBS and incubated with secondary anti-rabbit Alexa 594. Then, cells were washed in PBS and incubated 5 min with DAPI (dilution 1/50,000). Slides were then fitted with coverslips using Fluoromount-G (Southern Biotech) and incubated overnight at 4°C.
Animal Experiments
All procedures were performed in accordance with national and European legislation. All mice experiments were carried out at the Centre D’Évaluation Fonctionnelle, Université Pierre et Marie Curie (Paris, France). The initial breeding SMA type III model mice were purchased from Jackson Laboratory after material transfer agreement (MTA) accordance and were originally developed by Hsieh-Li et al.24 We used strain FVB.Cg- Tg(SMN2)2HungSMN1tm1Hung/J, founder line 2, stock number 005058. TC17 oligonucleotides were administered weekly to SMA mice by subcutaneous injections at a dose of 200 mg/kg per week under general anesthesia using isoflurane, starting at 7 days of age. Treated mice were sacrificed at various time points as indicated in Results, and muscles and tissues were harvested and snap-frozen in liquid-nitrogen-cooled isopentane and stored at −80°C before further analysis.
Respiratory Function
Mouse respiratory function was evaluated by whole-body plethysmography using an EMKA Technologies plethysmograph as described by TREAT-NMD (http://www.treat-nmd.eu/downloads/file/sops/dmd/MDX/DMD_M.2.2.002.pdf). Briefly, unrestrained conscious mice were placed in calibrated chambers containing a pneumatograph that measured pressure differentials within the compartment by a difference in air flow. Mice were allowed to acclimate in chambers for 45 min at a stable temperature and humidity. Data were then collected every 5 s using iox software (EMKA technologies). The inspiration time, Ti, was defined as the start of inspiration to the end of inspiration, and the expiration time was defined as the start of expiration to the end of expiration. The relaxation time, RT, was defined as the time from the start of expiration to the time when 65% of the total expiratory pressure occurred. Pause and enhanced pause (Penh) were defined and calculated by the formulas pause = (Te − RT)/RT and Penh = (PEP/PIP) × pause, where Te is expiratory time, PEP is peak expiratory pressure, and PIP is peak inspiratory pressure. The value of each parameter was calculated from an average of 60 recordings of 5 s representing a total of 5 min. Inclusion criteria for each recording were more than eight respiration events by 5 s and >0% of success rate as measured by iox software.
Statistical Analysis
Data were analyzed by GraphPad Prism5 software and are shown as the means ± SEM. “n” refers to the number of mice per group. Comparisons of statistical significance were assessed by unpaired Student’s t tests. Significant levels were set at *p < 0.05, **p < 0.01, and ***p < 0.001.
Author Contributions
V.R., G.G., and A.G. designed and performed the laboratory experiments. V.R. analyzed the experiments. V.R., J.-P.L.C., C.J.L., L.G., and A.G. wrote the manuscript. V.R. and A.G. conceived the project, designed the experiments, and supervised the entire study.
Conflicts of Interest
Christian Leumann and Luis Garcia are co-funders of Synthena, which produces tricyclo-DNA oligomers.
Acknowledgments
This work was supported by the Agence Nationale de la Recherche (Chair of Excellence HandiMedEx), the Association Monegasque contre les Myopathies, the Duchenne Parent Project France, and the Institut National de la Santé et de la Recherche Médicale (INSERM).
Footnotes
Supplemental Information includes two figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.02.009.
Contributor Information
Valérie Robin, Email: valerie.robin@uvsq.fr.
Aurélie Goyenvalle, Email: aurelie.goyenvalle@uvsq.fr.
Supplemental Information
References
- 1.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Liu Q., Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 3.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 4.Fallini C., Bassell G.J., Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldkötter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 8.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 9.Pellizzoni L., Charroux B., Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl. Acad. Sci. USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butchbach M.E. Copy number variations in the survival motor neuron genes: implications for spinal muscular atrophy and other neurodegenerative diseases. Front. Mol. Biosci. 2016;3:7. doi: 10.3389/fmolb.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholl R., Marquis J., Meyer K., Schümperli D. Spinal muscular atrophy: position and functional importance of the branch site preceding SMN exon 7. RNA Biol. 2007;4:34–37. doi: 10.4161/rna.4.1.4534. [DOI] [PubMed] [Google Scholar]
- 12.Singh N.N., Androphy E.J., Singh R.N. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh N.N., Androphy E.J., Singh R.N. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 14.Singh N.K., Singh N.N., Androphy E.J., Singh R.N. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh N.N., Singh R.N., Androphy E.J. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porensky P.N., Mitrpant C., McGovern V.L., Bevan A.K., Foust K.D., Kaspar B.K., Wilton S.D., Burghes A.H. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H., Janghra N., Mitrpant C., Dickinson R.L., Anthony K., Price L., Eperon I.C., Wilton S.D., Morgan J., Muntoni F. A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice. Hum. Gene Ther. 2013;24:331–342. doi: 10.1089/hum.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osman E.Y., Miller M.R., Robbins K.L., Lombardi A.M., Atkinson A.K., Brehm A.J., Lorson C.L. Morpholino antisense oligonucleotides targeting intronic repressor Element1 improve phenotype in SMA mouse models. Hum. Mol. Genet. 2014;23:4832–4845. doi: 10.1093/hmg/ddu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renneberg D., Bouliong E., Reber U., Schümperli D., Leumann C.J. Antisense properties of tricyclo-DNA. Nucleic Acids Res. 2002;30:2751–2757. doi: 10.1093/nar/gkf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goyenvalle A., Griffith G., Babbs A., El Andaloussi S., Ezzat K., Avril A., Dugovic B., Chaussenot R., Ferry A., Voit T. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 25.Marquis J., Meyer K., Angehrn L., Kämpfer S.S., Rothen-Rutishauser B., Schümperli D. Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol. Ther. 2007;15:1479–1486. doi: 10.1038/sj.mt.6300200. [DOI] [PubMed] [Google Scholar]
- 26.LoMauro A., Romei M., Priori R., Laviola M., D’Angelo M.G., Aliverti A. Alterations of thoraco-abdominal volumes and asynchronies in patients with spinal muscle atrophy type III. Respir. Physiol. Neurobiol. 2014;197:1–8. doi: 10.1016/j.resp.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haché M., Swoboda K.J., Sethna N., Farrow-Gillespie A., Khandji A., Xia S., Bishop K.M. Intrathecal injections in children with spinal muscular atrophy: Nusinersen Clinical Trial Experience. J. Child Neurol. 2016;31:899–906. doi: 10.1177/0883073815627882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passini M.A., Bu J., Roskelley E.M., Richards A.M., Sardi S.P., O’Riordan C.R., Klinger K.W., Shihabuddin L.S., Cheng S.H. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer K., Marquis J., Trüb J., Nlend Nlend R., Verp S., Ruepp M.D., Imboden H., Barde I., Trono D., Schümperli D. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum. Mol. Genet. 2009;18:546–555. doi: 10.1093/hmg/ddn382. [DOI] [PubMed] [Google Scholar]
- 31.Araujo Ap., Araujo M., Swoboda K.J. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J. Pediatr. 2009;155:292–294. doi: 10.1016/j.jpeds.2009.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudnik-Schöneborn S., Vogelgesang S., Armbrust S., Graul-Neumann L., Fusch C., Zerres K. Digital necroses and vascular thrombosis in severe spinal muscular atrophy. Muscle Nerve. 2010;42:144–147. doi: 10.1002/mus.21654. [DOI] [PubMed] [Google Scholar]
- 33.Somers E., Stencel Z., Wishart T.M., Gillingwater T.H., Parson S.H. Density, calibre and ramification of muscle capillaries are altered in a mouse model of severe spinal muscular atrophy. Neuromuscul. Disord. 2012;22:435–442. doi: 10.1016/j.nmd.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 34.Borisov A.B., Huang S.K., Carlson B.M. Remodeling of the vascular bed and progressive loss of capillaries in denervated skeletal muscle. Anat. Rec. 2000;258:292–304. doi: 10.1002/(SICI)1097-0185(20000301)258:3<292::AID-AR9>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 35.Hua Y., Liu Y.H., Sahashi K., Rigo F., Bennett C.F., Krainer A.R. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 2015;29:288–297. doi: 10.1101/gad.256644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez T.L., Kong L., Wang X., Osborne M.A., Crowder M.E., Van Meerbeke J.P., Xu X., Davis C., Wooley J., Goldhamer D.J. Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. J. Neurosci. 2012;32:8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahashi K., Ling K.K., Hua Y., Wilkinson J.E., Nomakuchi T., Rigo F., Hung G., Xu D., Jiang Y.P., Lin R.Z. Pathological impact of SMN2 mis-splicing in adult SMA mice. EMBO Mol. Med. 2013;5:1586–1601. doi: 10.1002/emmm.201302567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henrichsen T., Lindenskov P.H., Shaffer T.H., Loekke R.J., Fugelseth D., Lindemann R. Perfluorodecalin lavage of a longstanding lung atelectasis in a child with spinal muscle atrophy. Pediatr. Pulmonol. 2012;47:415–419. doi: 10.1002/ppul.21565. [DOI] [PubMed] [Google Scholar]
- 39.Leistikow E.A., Jones N.E., Josephson K.D., de Sierra T.M., Costakos D.T., Sprague D., Gorch D.H., Asonye U.O. Migrating atelectasis in Werdnig-Hoffmann disease: pulmonary manifestations in two cases of spinal muscular atrophy type 1. Pediatr. Pulmonol. 1999;28:149–153. doi: 10.1002/(sici)1099-0496(199908)28:2<149::aid-ppul12>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 40.Schreml J., Riessland M., Paterno M., Garbes L., Roßbach K., Ackermann B., Krämer J., Somers E., Parson S.H., Heller R. Severe SMA mice show organ impairment that cannot be rescued by therapy with the HDACi JNJ-26481585. Eur. J. Hum. Genet. 2013;21:643–652. doi: 10.1038/ejhg.2012.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudnik-Schöneborn S., Heller R., Berg C., Betzler C., Grimm T., Eggermann T., Eggermann K., Wirth R., Wirth B., Zerres K. Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J. Med. Genet. 2008;45:635–638. doi: 10.1136/jmg.2008.057950. [DOI] [PubMed] [Google Scholar]
- 42.Hachiya Y., Arai H., Hayashi M., Kumada S., Furushima W., Ohtsuka E., Ito Y., Uchiyama A., Kurata K. Autonomic dysfunction in cases of spinal muscular atrophy type 1 with long survival. Brain Dev. 2005;27:574–578. doi: 10.1016/j.braindev.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Hocquemiller M., Giersch L., Audrain M., Parker S., Cartier N. Adeno-associated virus-based gene therapy for CNS diseases. Hum. Gene Ther. 2016;27:478–496. doi: 10.1089/hum.2016.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duque S., Joussemet B., Riviere C., Marais T., Dubreil L., Douar A.M., Fyfe J., Moullier P., Colle M.A., Barkats M. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H., Kaspar B.K. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Hammond S.M., Hazell G., Shabanpoor F., Saleh A.F., Bowerman M., Sleigh J.N., Meijboom K.E., Zhou H., Muntoni F., Talbot K. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 2016;113:10962–10967. doi: 10.1073/pnas.1605731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner T., Pfleiderer W. Synthesis of 2′-deoxyribonucleoside 5′-phosphoramidites: New building blocks for the inverse (5′-3′)-oligonucleotide approach. Helv. Chim. Acta. 2000;83:2023–2035. [Google Scholar]
- 48.Renneberg D., Leumann C.J. Watson-Crick base-pairing properties of tricyclo-DNA. J. Am. Chem. Soc. 2002;124:5993–6002. doi: 10.1021/ja025569+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.