Abstract
Pompe disease is a metabolic myopathy caused by deficiency of the acid α-glucosidase (GAA) enzyme and results in progressive wasting of skeletal muscle cells. The c.-32-13T>G (IVS1) GAA variant promotes exon 2 skipping during pre-mRNA splicing and is the most common variant for the childhood/adult disease form. We previously identified antisense oligonucleotides (AONs) that promoted GAA exon 2 inclusion in patient-derived fibroblasts. It was unknown how these AONs would affect GAA splicing in skeletal muscle cells. To test this, we expanded induced pluripotent stem cell (iPSC)-derived myogenic progenitors and differentiated these to multinucleated myotubes. AONs restored splicing in myotubes to a similar extent as in fibroblasts, suggesting that they act by modulating the action of shared splicing regulators. AONs targeted the putative polypyrimidine tract of a cryptic splice acceptor site that was part of a pseudo exon in GAA intron 1. Blocking of the cryptic splice donor of the pseudo exon with AONs likewise promoted GAA exon 2 inclusion. The simultaneous blocking of the cryptic acceptor and cryptic donor sites restored the majority of canonical splicing and alleviated GAA enzyme deficiency. These results highlight the relevance of cryptic splicing in human disease and its potential as therapeutic target for splicing modulation using AONs.
Keywords: myogenic progenitors, induced pluripotent stem cell, antisense oligonucleotide, Pompe disease, IVS1 mutation, skeletal muscle differentiation, pre-mRNA splicing, pseudo exon, exon inclusion, lysosomal storage disorder
Introduction
Cell type-specific alternative pre-mRNA splicing is evolutionary conserved and plays a critical role in cell type-specific functions.1, 2 The involved mechanisms include the differential expression of splicing regulatory proteins, guided by cell type-specific master splicing factors.3 In human disease, aberrant pre-mRNA splicing plays a causal role in at least 9% of known cases (http://www.hgmd.cf.ac.uk/), and this likely represents an underestimation.4 Aberrant splicing is amenable to modulation with drugs that either target splicing regulatory proteins or that block cis-acting splicing regulatory elements such as canonical or cryptic splice sites or exonic or intronic splicing silencer/enhancer sequences. To test these drugs for possible further clinical development, in vitro and in vivo models are required that closely mimic splicing events that occur in the relevant cell type of human patients. In particular when targeting cis-acting splicing regulatory elements with sequence-specific antisense oligonucleotides (AONs), the availability of human cells is important as evolutionary conservation at the RNA level is generally poor.
We recently identified AONs that correct aberrant splicing in fibroblasts from patients with childhood/adult onset Pompe disease caused by the common variant c.-32-13T>G (IVS1) in the gene encoding the lysosomal enzyme acid alpha glucosidase (GAA).5 Pompe disease is a monogenic autosomal recessive disorder characterized by progressive muscle wasting. Classic infantile patients are affected at birth by hypertrophic cardiomyopathy and skeletal muscle wasting.6 These patients carry GAA mutations that result in a residual GAA enzyme activity of <1% of the average activity in healthy controls. Patients with childhood/adult Pompe disease have a later disease onset ranging from 3 to ∼60 years of age and have progressive skeletal muscle wasting, but no cardiac phenotype.7 Residual GAA enzyme activity in these patients is between 1% and 20% of average healthy control values. Enzyme replacement therapy (ERT) for Pompe disease is available in which recombinant human GAA is administered (bi)weekly intravenously.6, 8, 9 Although ERT is effective, not all patients respond well, its long-term efficacy is insufficient, and it is very expensive.10, 11, 12 For these reasons, an alternative treatment option is required.
The IVS1 GAA variant occurs in 90% of adults and 50% of children in the Netherlands with Pompe disease and in similarly high percentages of Caucasian Pompe patients in other countries.13, 14 Therefore, it forms an attractive target for a potential novel therapy. The IVS1 variant is located in the polypyrimidine tract (pY-tract) of GAA exon 2 and weakens the recognition of the splice acceptor site of this exon.13, 15, 16 AONs that restored splicing were identified in a screen for cis-acting splicing silencer elements, and these targeted an element in intron 1, 175 nucleotides upstream of the affected exon 2.5 It was unclear from this study whether the AON-mediated inhibition of a putative splicing silencer element would be operational in differentiated skeletal muscle cells, which represent the cell type relevant for Pompe disease, and what the underlying mechanism was. To this end, we developed an in vitro model using patient-derived skeletal muscle cells. We generated induced pluripotent stem cells (iPSCs) from patients with childhood/adult Pompe disease caused by the IVS1 GAA variant. iPSCs were differentiated into myogenic progenitors using a modified transgene-free protocol,17 and myogenic progenitors were expanded and differentiated into multinucleated myotubes to test the efficacy and mechanism of AONs that correct the IVS1 GAA variant. This showed that the IVS1 variant promoted utilization of a pseudo exon, and that the simultaneous blocking of the 3′ and 5′ cryptic splice sites of this pseudo exon with AONs resulted in restoration of the majority of canonical splicing and GAA enzyme activity. The results highlight the feasibility to correct the IVS1 variant in skeletal muscle cells from Pompe patients using AONs, and they underscore the potential of modulating cryptic splicing to correct human disease.
Results
Generation and Characterization of iPSCs from Pompe Patients Carrying the IVS1 GAA Variant
To generate iPSCs from patients with childhood/adult Pompe disease caused by the IVS1 GAA variant, primary fibroblasts were reprogrammed using a polycistronic lentiviral vector of Oct4, Sox2, Klf4, and c-Myc.18 Two patients were modeled. Patient 1 carried the IVS1 variant on one allele and the c.525delT variant on the other allele. The c.525delT variant causes a reading frameshift, and the mRNA expressed from this allele is degraded, facilitating detection of splicing forms from the IVS1 allele.19 Patient 2 carried the IVS1 variant on one allele and the c.923A > C missense variant on the second allele.20 This missense variant allows mRNA expression without degradation to at least 50% of normal levels. Fibroblasts from a healthy control were also reprogrammed; a second, independent healthy control has been described previously.21 The three lines generated here were characterized using standard procedures. Immunofluorescent analysis indicated that all three lines expressed pluripotency markers including NANOG, OCT4, SSEA4, TRA-1-80 and TRA-1-61 (Figure S1A). In vitro differentiation showed that all three lines were capable of differentiating into cell-types of the three germ layers monitored by expression of α-fetoprotein (AFP), smooth muscle actin (SMA), and class III β-tubulin (TUJI) (Figure S1B). Microarray analysis showed expression of essential pluripotent genes comparable to human embryonic stem cell (hESC) lines H1 and H9 and reduction of fibroblast-associated genes (Figure S1C). All iPSC lines contained normal karyotypes (Figure S1D).
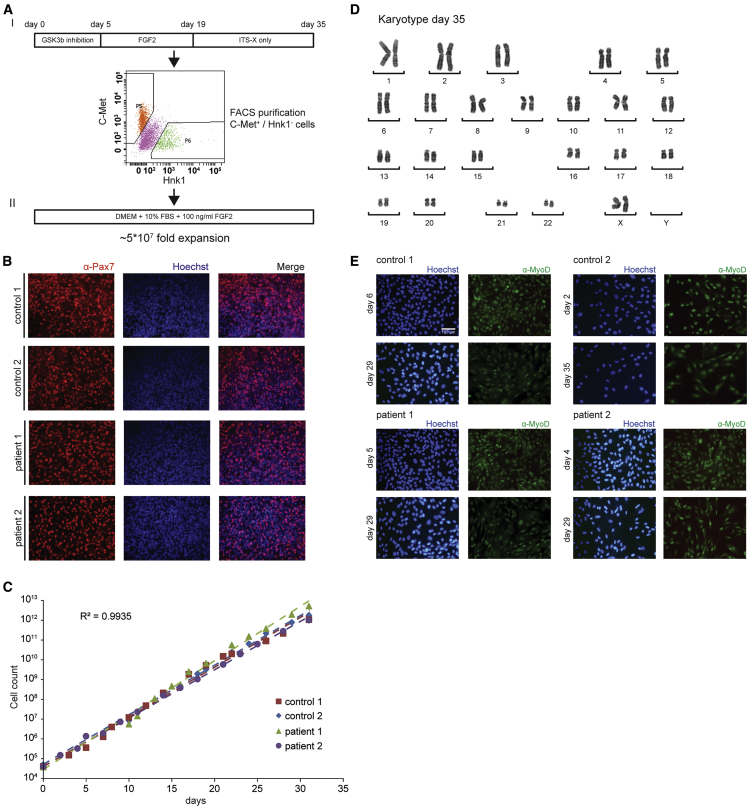
Differentiation of iPSCs into Myotubes via Expandable Myogenic Progenitors
To differentiate iPSCs into skeletal muscle cells, we used a recently published transgene-free protocol.17 This protocol consists of activation of WNT signaling using GSK3β inhibition, followed by an expansion phase mediated by FGF2 (Figure 1AI). Initial experiments resulted in Pax7+ cells after a 35-day period (Figure 1B), suggesting that muscle progenitors had formed. These cells were present in patches among other cell types. All experiments yielded Pax7+ cells, but percentages of obtained Pax7+ cells depended on concentration and duration of GSK3β inhibitor CHIR99021. Treatment with a higher concentration of CHIR99021 (3.5 μM) for a longer duration (5–6 days) was well tolerated without signs of toxicity and resulted in the generation of higher yields of Pax7+ cells. The generation of Pax7+ cells using this protocol was highly reproducible and was observed in all >30 independent differentiation experiments, although yields still varied between experiments (data not shown). Pax7+ cells were purified by fluorescence-activated cell sorting (FACS) using C-Met+/Hnk1− cells. These could be further differentiated into multinucleated myotubes within 5–9 days (Figure S2A). Typical yields of purified cells were between 50,000 and 250,000 myogenic progenitors derived from 1 × 106 iPSCs after a 35-day protocol. Cells purified at lower yields showed decreased capacity to form multinucleated myotubes (data not shown).
Figure 1.
Expansion of Purified iPSC-Derived Myogenic Progenitors
(A) I, Scheme for differentiation of iPSCs into myogenic progenitors and FACS purification; II, Expansion of purified myogenic progenitors. The expansion medium is indicated. The fold expansion is indicated. (B) Immunofluorescent staining for Pax7 (in red) in selected areas from a culture dish derived from non-purified myogenic progenitors following the 35-day differentiation protocol. The nuclei were stained with Hoechst (blue). (C) Linear proliferation curves for all four iPSC-derived myogenic progenitor lines during expansion. The average R2 shown was calculated for all data points of the four lines and indicates high concordance between the four lines. (D) Karyotype analysis after expansion of purified myogenic progenitors at day 35 (a representative example of ten nuclei). (E) Immunofluorescent analysis of MyoD in FACS purified and expanded myogenic progenitors. Myogenic progenitors were expanded in proliferation medium and stained at the start of expansion and after expansion to 1012 cells. Representative pictures are shown in the image.
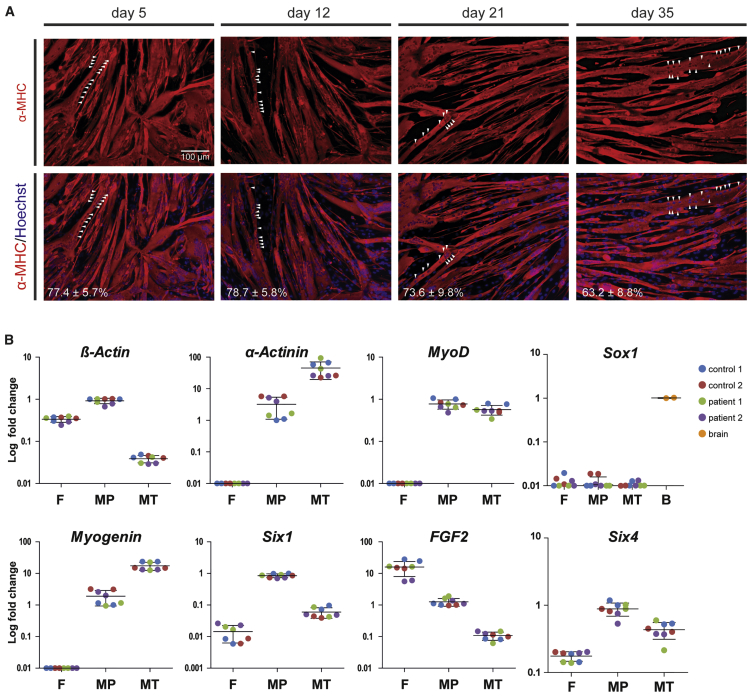
To improve the yields of myogenic progenitors and the robustness of myogenic differentiation, expansion of muscle cells after FACS was tested. FGF2 was included in the expansion medium because it is known to induce proliferation and to inhibit differentiation of myogenic cells.22 We tried five conditions (Table S1) and only basic DMEM supplemented with 10% FBS and 100 ng/mL FGF2 was supportive of myogenic progenitor proliferation and resulted in an expansion of at least 5 × 107-fold in 15 passages (Figures 1AII and 1C). Upon expansion, the karyotype remained normal (Figure 1D). MyoD was expressed in virtually all cells during the expansion period (Figure 1E). Cryopreservation of the expanded culture was possible at any moment and yielded high viabilities upon thawing. Expanded cells showed efficient and homogeneous differentiation into multinucleated myotubes with similair fusion index that expressed myosin heavy chain (MHC) (Figures 2A, S2B, and S2C). Proliferating myogenic progenitors were characterized by high expression of the myogenic markers MyoD, Myogenin, Six1, and Six4, moderately high expression of the myogenic differentiation marker α-actinin and of FGF2, while the neural crest marker Sox1 was not expressed (Figure 2B). In addition, at any stage of expansion, cells could be differentiated into multinucleated myotubes with a high fusion index (tested in >500 differentiations performed to date) (Figures S2B and S2C). Multinucleated myotubes showed high expression of the myogenic differentiation marker α-actinin, while β-actin expression was reduced relative to myogenic progenitors (Figure 2B). We conclude that expansion of myogenic progenitors provided sufficient amounts of cells to allow the testing of AONs on their potential to correct aberrant splicing from the IVS1 allele in myotubes.
Figure 2.
Differentiation of iPSC-Derived Myogenic Progenitors into Multinucleated Myotubes
(A) Myogenic progenitors retain their capacity to differentiate into multinucleated myotubes during expansion. Myogenic progenitors were expanded, and at several time points during expansion a subculture was differentiated for 4 days and stained for expression of the myogenic differentiation marker MHC (MF-20 antibody; red). The nuclei were stained with Hoechst (blue). The white arrowheads point to examples of aligned nuclei present in a single myotube. The fusion index of each differentiation is shown at the bottom. The data are means ± SDs of five technical replicates. (B) mRNA expression of iPSC-derived myogenic progenitors (purified and expanded) and myotubes thereof. Equal amounts of total RNA were isolated from fibroblasts (F), myogenic progenitors (MP), and myotubes (MT), generated after 4 days of differentiation, and mRNA expression of the indicated genes was determined by qRT-PCR analysis. Log fold change was calculated relative to control 1 sample 1. The lines represent means. Biological duplicates are shown in the image.
Splicing Modulation in iPSC-Derived Myotubes
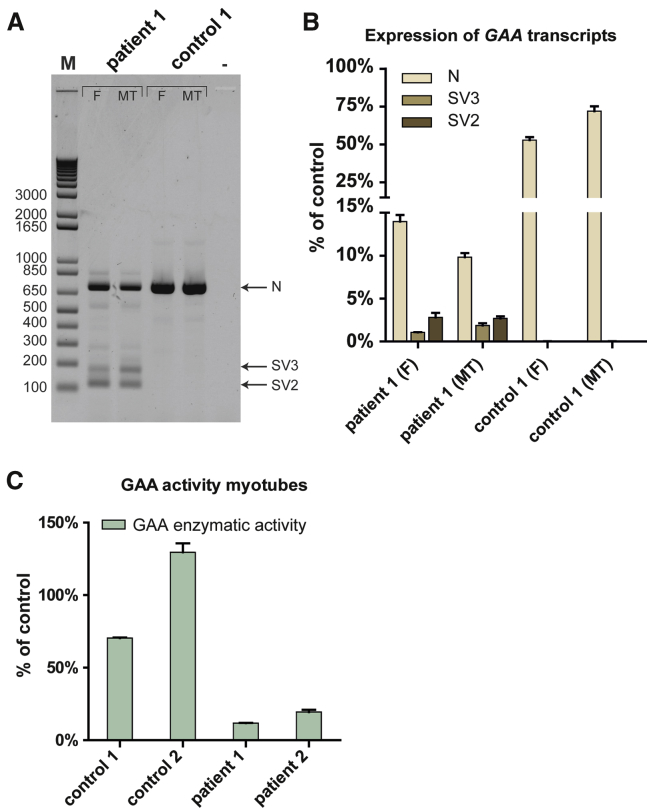
Since splicing regulation can be cell type-specific,2 it was a priori possible that the IVS1 variant caused qualitative or quantitative splicing differences in skeletal muscle cells compared to fibroblasts. It was also not clear to what extent the results on splicing modulation by AONs in fibroblasts could be extended to skeletal muscle cells. To test this, patient-derived iPSCs were differentiated into myotubes, and the effect of the IVS1 variant on splicing was compared to the effect observed in fibroblasts. Figure 3A shows the results of the flanking exon RT-PCR analysis. The effect of the IVS1 variant was similar in fibroblasts compared to skeletal muscle myotubes. Products from myotubes at the position of the splicing products N, SV2, and SV3 were sequenced and found to be identical to their counterparts in fibroblasts (data not shown). Control myotubes did not show obvious aberrant GAA pre-mRNA splicing, similar to fibroblasts. Quantitative analysis of individual splicing products using qRT-PCR showed that the wild-type splicing product N was expressed to slightly higher levels in control myotubes, but to slightly lower levels in Pompe myotubes compared to fibroblasts (Figure 3B). The SV3 product was slightly higher in Pompe myotubes compared to Pompe fibroblasts. Myotubes from both Pompe patients showed partial residual GAA enzyme activity consistent with residual leaky wild-type splicing (Figure 3C). This indicates that the IVS1 variant causes similar aberrant splicing in fibroblasts and skeletal muscle cells.
Figure 3.
GAA Splicing and Enzyme Activity in Fibroblasts and Myotubes from Pompe Patients
(A) Equal amounts of total RNA from primary fibroblasts (F) and their corresponding iPSC-derived myotubes (MT), derived from patient 1 or a healthy control, were analyzed by flanking exons 1–3 RT-PCR of GAA exon 2. N: WT product, SV2: splice variant 2, SV3: splice variant 3. (B) As (A), but now as analyzed by qRT-PCR of individual splicing products. To facilitate comparison between different cell types, no normalization was used and all products were compared to the value of average control fibroblast product N levels using the delta-Ct method. (C) GAA enzyme activity in iPSC-derived multinucleated myotubes. Myogenic progenitors from the cells indicated were differentiated for 4 days, and the GAA enzyme activity was determined. The average activity present in the two healthy controls paralleled those present in fibroblasts (data not shown) and was used to calculate the percentage of residual activity in myotubes from the two patients.
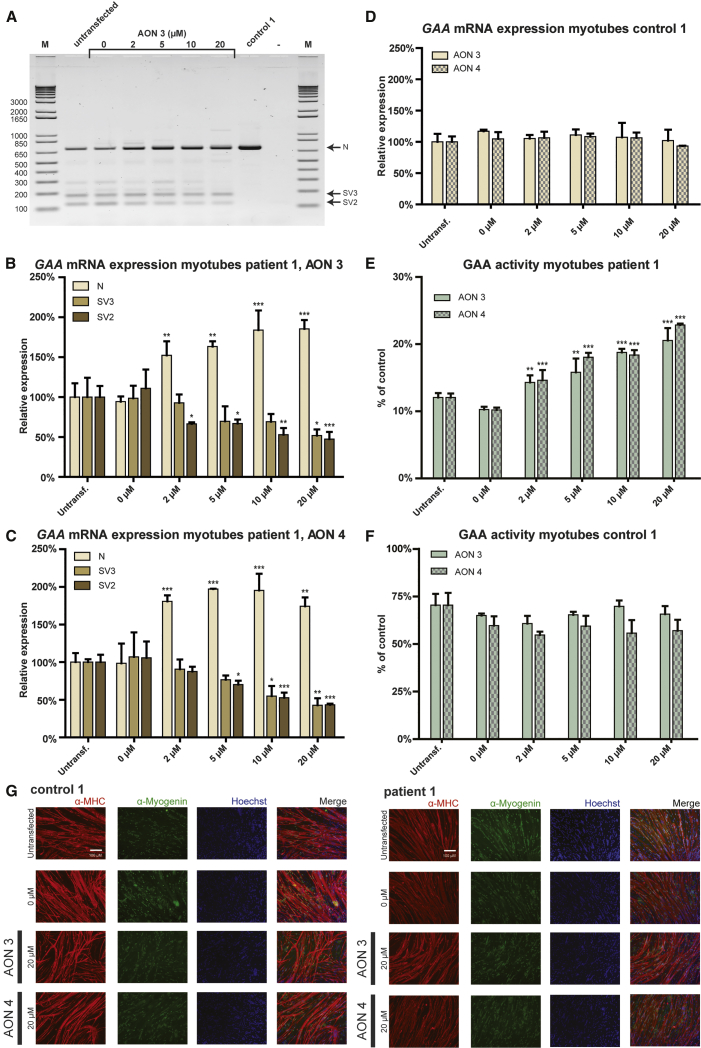
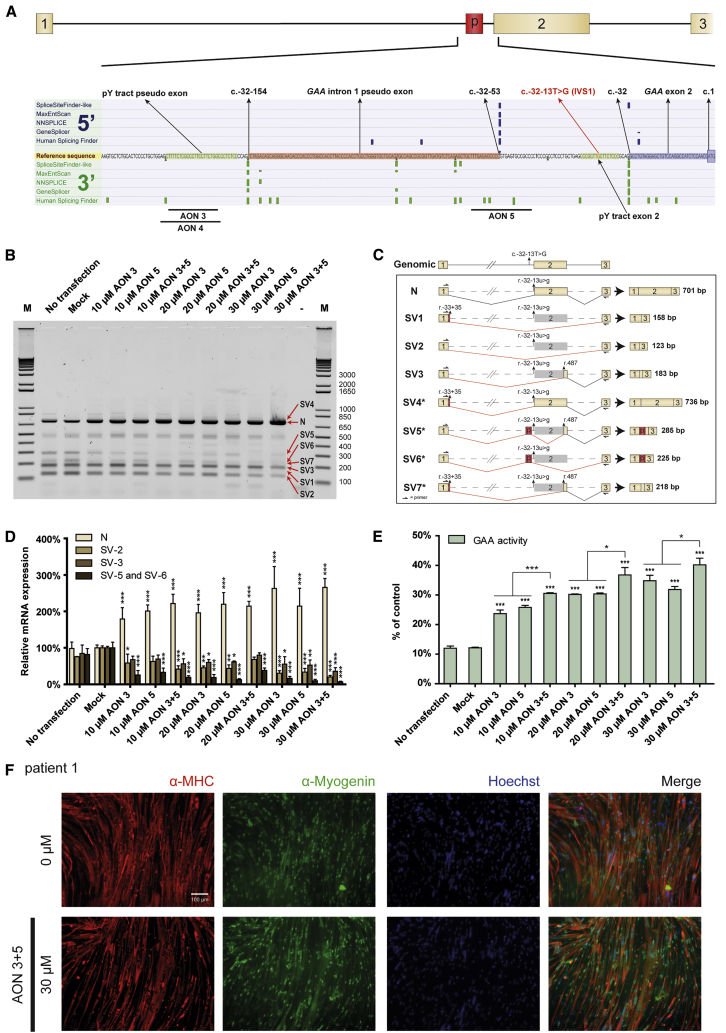
Next, we tested the effect of AONs 3 and 4, which we identified recently to promote exon inclusion in fibroblasts5 on exon 2 inclusion in myotubes. Figure 4A shows the effect of AON 3 on GAA exon 2 splicing in myotubes from patient 1, as analyzed by RT-PCR using primers annealing to exon 1 and 3. AON 3 caused a concentration-dependent increase in exon 2 inclusion, as judged from the increase in the amounts of the wild-type variant (N) and a concomitant decrease in the amounts of the partial (SV3) and full exon 2 skip (SV2) variants. This was confirmed by quantitative analysis using qRT-PCR with primers specific for individual splicing variants in myotubes from patient 1 treated with AON 3 (Figure 4B) and AON 4 (Figure 4C) and in myotubes from patient 2 treated wth AON 3 (Figure S3A) and AON 4 (Figure S3B). No effect of AONs 3 and 4 on GAA expression and exon 2 splicing was observed in myotubes from control iPSCs (Figures 4D and S3C). These results indicate that AONs 3 and 4 corrected aberrant exon 2 splicing caused by the IVS1 variant in patient-derived myotubes by promoting exon 2 inclusion during splicing. AON 3 and 4 enhanced GAA enzymatic activity in myotubes derived from patient 1 (Figure 4E) and patient 2 (Figure S3D) up to 2-fold and was ineffective in myotubes from control 1 (Figure 4F) and control 2 (Figure S3E). These efficacies were similar to those observed in fibroblasts.5 AON treatment was well tolerated. Stainings with anti-MHC and anti-myogenin antibodies showed no effects of AON treatment of the transfection itself, AON 3 or 4 (Figures 4G, S3G, and S3H) on differentiation. In addition, qRT-PCR analysis showed no consistent changes in expression of MyoD, Myogenin, LAMP1, or LAMP2 (Figure S3F). Taken together, the AONs 3 and 4 corrected aberrant GAA exon 2 splicing in patient-derived myotubes, and they elevated GAA enzyme activity with similar efficacy as in fibroblasts.
Figure 4.
Quantitative Analysis of GAA Exon 2 Splicing in Expanded iPSC-Derived Myotubes
(A) Flanking exon RT-PCR analysis of the effect of AON 3 on GAA exon 2 splicing in myotubes from patient 1 and control 1. (B) Effect of AON 3 on GAA exon 2 splicing in myotubes from patient 1 as analyzed with qRT-PCR analysis of individual splicing products. The data were normalized against expression of four genes that showed no consistent changes in expression: MyoD, Myogenin, LAMP1, and LAMP2 (see Figure S3F). (C) As (B), but now for AON 4. (D) Effect of AONs 3 and 4 on GAA exon 2 splicing in myotubes from control 1 as analyzed with qRT-PCR analysis of splice product N. The control cells have undetectable levels of aberrant splice products SV2 and SV3. The normalization was as in (B). (E) Effects of AON 3 and 4 on GAA enzymatic activity in myotubes from patient 1. (F) As (E), but now in myotubes from control 1. (G) AON treatment does not affect myogenic differentiation. Immunofluorescent stainings of myotubes after treatment with AONs 3 and 4 are shown. Red: MHC (anti-MF-20); green: Myogenin; blue: nuclei (Hoechst). 0 μM: mock transfection. Representative pictures are shown in the image. The quantitative data are means ± SDs of three biological replicates. *p < 0.05, **p < 0.01, and ***p < 0.001.
Identification of a Pseudo Exon that Competes with Exon 2 Splicing
As it was unclear how AONs 3 and 4 restored exon 2 inclusion, we were interested to investigate their mechanism of action. We noted that the target sequence of these AONs showed similarity to a pY-tract, which is usually present between 5–40 nt upstream of a splice acceptor. We then performed in silico analysis of splice sites using Alamut, and this predicted a strong natural cryptic splice acceptor site 12–13 nt downstream of the binding site for AONs 3 and 4 (Figure 5A). At 102 nt further downstream, a strong natural cryptic splice donor was predicted. These predictions were found for the wild-type GAA gene and were unchanged by the IVS1 variant, suggesting that these cryptic splice sites defined a hypothetical natural pseudo exon. Usage of the natural pseudo exon was observed in a minigene construct harboring the IVS1 variant. Mutation of the natural cryptic splice sites of the natural pseudo exon abolished its inclusion in the context of the minigene harboring the IVS1 variant (Figures S4A–S4C). This suggested the possibility that AONs 3 and 4 may act by inhibiting usage of a natural pseudo exon rather than by repressing a putative intronic splice silencer.
Figure 5.
Blocking of a Natural Pseudo Exon Restores GAA Exon 2 Splicing
(A) The splicing silencer in intron 1 is predicted to be the pY-tract of a natural pseudo exon. Alamut was used to predict splice sites around the splicing silencer identified previously.5 Note that predictions were independent of the IVS1 variant. A strong 3′ splice site was predicted at c.-32-154, and a strong 5′ splice site at c.-32-53, which suggested the presence of a natural pseudo exon, indicated by “p” in the cartoon. The canonical 3′ splice site of exon 2 at c.-32 showed strong prediction and is also indicated. (B) Blocking of natural pseudo exon splicing restores GAA exon 2 splicing. AON 5 was designed to block the predicted 5′ splice site, and AONs 3 and 5 were tested alone or in combination in myotubes from patient 1. Flanking RT-PCR analysis of GAA exon 2 was performed. (C) Cartoons of splicing products that were identified by TOPO cloning (see Table 1) of products obtained from the flanking exon RT-PCR in (B). (D) Analysis of the experiment in (B) by qRT-PCR of individual splicing products. Splicing to the natural pseudo exon is represented by SV5 and SV6, and these products were quantified using unique PCR primers. The data were normalized against expression of four genes that showed no consistent changes in expression: MyoD, Myogenin, LAMP1, and LAMP2 (see Figure S4E). (E) Analysis of the experiment in (B) on GAA enzyme activity. (F) Combined treatment with AONs 3 and 5 does not interfere with myogenic differentiation to myotubes. Immunofluorescent staining results are shown for treatment of iPSC-derived myotubes obtained from patient 1. Red, MHC (anti-MF-20); green, myogenin; blue, nuclei (Hoechst); 0 μM, mock transfection. Representative pictures are shown in the image. The quantitative data are means ± SDs of three biological replicates. *p < 0.05, **p < 0.01, and ***p < 0.001.
To test this, we first analyzed whether splice products comprising the putative natural pseudo exon exist in cells from Pompe patients. To this end, mRNA isolated from patient-derived myotubes was analyzed by flanking exon RT-PCR of exon 2, and PCR products were cloned into a TOPO vector. There were 96 clones that were analyzed by Sanger sequencing, and this resulted in the identification of eight splice variants (Figures 5B and 5C; Table 1 [column: colony count mock transfection] and S4D). The predicted natural pseudo exon was indeed detected in two splice products in which exon 2 was fully (SV6) or partially (SV5) skipped, but at a low abundance. It is likely that both products are subject to mRNA degradation due to the lack of the translation start codon. Nevertheless, these could also be identified on agarose gels following flanking exon PCR of exon 2 (Figure 5B). Other low abundant splice products (SV1, SV4, and SV7) utilized a previously described cryptic splice donor nearby exon 1.15, 16 However, these never contained the natural pseudo exon. These data suggest that the predicted natural pseudo exon exists in vivo in splice products in which exon 2 is partially or fully skipped due to the IVS1 variant.
Table 1.
Analysis of Splice Variants by TOPO Cloning
| Splice Variant | Colony Count Mock Transfection | Colony Count Transfection of 15 μM AON 3 and 5 |
|---|---|---|
| Total | 96 | 96 |
| Readable | 93 | 90 |
| N | 14 | 45 |
| SV1 | 3 | 3 |
| SV2 | 44 | 16 |
| SV3 | 24 | 23 |
| SV4 | 0 | 2 |
| SV5 | 4 | 1 |
| SV6 | 2 | 0 |
| SV7 | 2 | 0 |
Short introns are unfavorable for successful splicing and have a typical minimum length of 70–80 nt.23 The length of the intron between the natural pseudo exon and exon 2 is 52 nt, which violates this rule. This suggested the possibility that inclusion of the natural pseudo exon competes with exon 2 inclusion, which would be in agreement with the mutually exclusive inclusion of the natural pseudo exon or exon 2 in splice products. Such scenario would explain why AONs 3 and 4 promote exon 2 inclusion, namely by repression of inclusion of the natural pseudo exon via interfering with the pY-tract of the natural cryptic splice acceptor site. We hypothesized that repression of the natural cryptic splice donor would likewise promote exon 2 inclusion. To test this, AON 5 was designed to target the natural cryptic splice donor site of the natural pseudo exon (Figure 5A). In patient-derived myotubes, AON 5 promoted exon 2 inclusion (product N) and repressed inclusion of the natural pseudo exon (products SV5 and SV6), as shown by flanking exon RT-PCR and splicing product-specific qRT-PCR (Figures 5B, 5D, and S4E). AON 5 was equally effective in splicing correction compared to AON 3, suggesting that both AONs prevent utilization of the natural pseudo exon. GAA enzyme activity was enhanced by AON 5 to similar levels compared to AON 3 (Figure 5E) and myotube differentiation was not altered by AON treatment (Figure 5F). These results are in agreement with the idea that the natural pseudo exon competes with exon 2 splicing and that natural pseudo exon skipping by AONs promotes exon 2 inclusion.
To test whether the simultaneous targeting of the cryptic splice acceptor and donor sites of the pseudo exon would further promote exon inclusion, a combination of AON 3 plus AON 5 was tested in patient-derived myotubes. At the same total AON concentrations, combinations of AON 3 plus AON 5 were equally effective as single AON treatments in promoting exon 2 inclusion and repressing aberrant exon 2 splicing (Figures 5B and 5D). We used TOPO cloning as above to analyze all products that arise from treatment with AON 3 plus AON 5 (Table 1). No additional products besides the eight known splicing products were identified. Compared to mock treated cells, cells treated with AON 3 plus 5 showed an increase in the number of clones with a wild-type exon 2 insert from 14 to 45 (3.2- fold), while the number of clones that contained the natural pseudo exon was reduced 6-fold from 6 to 1 (Table 1). When tested on GAA enzyme activity, combinations of AONs 3 plus 5 were slightly more efficient compared to single AONs and caused enhancement of up to 4-fold (Figure 5E). In addition, 76 kDa active GAA protein levels were quantified and demonstrated similar levels of active GAA protein as shown by the GAA enzyme activity assay (Figure S5B and S5C). It is unclear why the mRNA data do not fully match the GAA enzyme activity data. We note that the error bars are smaller in the GAA enzyme assay, as this assay is very sensitive and accurate, suggesting a technical reason. The restoration of canonical splicing was efficient, but incomplete, as small amounts of aberrant splice products were still detected. This was in particular the case for SV2 and SV3. This suggests that prevention of inclusion of the pseudo exon was not sufficient to fully restore aberrant splicing. The maximal possible enhancement of GAA enzyme activity using this approach is ∼3.5–5-fold: patients with the IVS1 allele have on average 10%–15% leaky wild-type splicing and full restoration will amount to a maximum of 50% (derived from one allele) of the average value of healthy controls. AONs 3 plus 5 promoted GAA exon 2 inclusion and GAA activity in myotubes with ∼3.3-fold, indicating that these corrected 66%–99% of exon 2 splicing. We conclude that the simultaneous inhibition of the cryptic splice donor and acceptor sites of the natural pseudo exon restore the majority of GAA splicing and GAA enzymatic activity in patient-derived skeletal muscle cells.
Discussion
We evaluated the effect of the common IVS1 GAA variant on splicing in skeletal muscle cells, the relevant cell type for childhood/adult Pompe disease, in vitro. In addition, we tested whether previously identified AONs that promoted exon 2 inclusion in fibroblasts carrying the IVS1 variant were also capable of doing so in skeletal muscle cells. By reprogramming patient-derived fibroblasts to iPSCs, we generated myogenic progenitors using a transgene-free protocol and expanded purified myogenic cells to allow quantitative analysis in multinucleated myotubes. This showed that the IVS1 variant caused exon skipping, and that AONs promoted exon inclusion and enhanced GAA enzymatic activity in myotubes similar as in fibroblasts. We found that the AONs targeted the pY-tract of a cryptic splice acceptor site that was part of a pseudo exon. The simultaneous blocking of 5′ and 3′ splice sites of the pseudo exon resulted in promotion of exon inclusion and rescue of the majority of GAA enzyme activity. The results suggest that the splicing machinery utilizes alternative splice sites when the canonical splice acceptor site is weakened by the IVS1 variant, and that blocking of such cryptic splice sites using AONs is capable to restore canonical exon inclusion.
Several in vitro models for Pompe disease have been published. Most of these concern the classic infantile form, and in these reports iPSCs were differentiated into cardiomyocytes that showed hallmarks of Pompe disease including GAA deficiency, glycogen accumulation, and newly identified glycan processing abnormalities.24, 25, 26, 27
Classic infantile patients are characterized by left ventricle hypertrophic cardiomyopathy, severe skeletal muscle wasting resulting in hypotonia and respiratory failure, and CNS abnormalities. In childhood and adult forms of Pompe disease, skeletal muscle wasting is prominent, but cardiac abnormalities are more rare, and there is no evidence for CNS abnormalities.28, 29, 30 A recent report described the generation of myocytes, using MyoD overexpression, from iPSCs derived from adult Pompe patients who did not carry the IVS1 variant.26 These cells showed increased levels of total glycogen. To our knowledge, our report presents the first iPSC models for childhood/adult Pompe disease that carry the IVS1 variant. Following a 4-day differentiation protocol into myotubes, we did not observe major differences in size and number of lysosomes using LAMP1 immunofluorescent analysis (data not shown), despite the GAA enzyme deficiency in these cells. We anticipate that the development of lysosomal abnormalities in childhood/adult cells in vitro may take time, as it does in patients. Disease onset is heterogeneous and may occur between the age of 3 and 60 years. This may be explained by modifying factors that affect Pompe disease onset and progression.14, 31 We are currently optimizing culture conditions to allow long-term analysis of differentiated myogenic cells to test this. We note that lysosomal pathology and glycogen accumulation in vitro in fibroblasts are only detectable by immunofluorescent LAMP1 staining or by biochemical analysis of glycogen content, respectively, following a period of 3 weeks of confluent, non-proliferating culture and only in cells from classic infantile patients, but never in cells from childhood/adult patients.32 This suggests that lysosomal glycogen only accumulates under certain conditions and can be prevented by active cell proliferation. In childhood/adult Pompe patients, lysosomal pathology in skeletal muscle biopsies is very heterogeneous. Typically, virtually normal myofibers lay adjacent to strongly affected fibers.6 It is poorly understood why certain fibers escape pathology, while others do not. Future research is required to identify critical determinants of lysosomal glycogen accumulation.
Transgene-free differentiation of human iPSCs into myogenic cells has been achieved by a number of laboratories.17, 33, 34, 35, 36, 37, 38 The basic protocol consists of activation of the WNT signaling pathway, followed by an expansion period using FGF2. BMP inhibition has been used to prevent formation of neural crest.34 Alternatively, neural crest and other non-myogenic cells have been removed by FACS, similar as performed here.17, 33 In our hands, recoveries and differentiation capacities were heterogeneous directly following FACS. We showed that it is feasible to expand the purified cells using medium containing FGF2. This resulted in a stable condition that allowed for rapid expansion in proliferation medium and viable freezing or differentiation at any desired time point up to the point of 5 × 107-fold expansion. These conditions allowed the reproducible testing of drugs such as AONs. This may present a general strategy for drug testing in iPSC-derived models of other neuromuscular disorders. Other laboratories also have shown expansion of myogenic progenitors.33, 35, 36, 39 Hosoyama and colleagues35 demonstrated expansion in a sphere based culture. Shelton et al.39 used a pre-plating protocol to expand myogenic progenitors in the original plate used to induce myogenic differentiation. Choi and coworkers33 reported expansion of purified iPSC-derived myogenic progenitors of at least 1 × 104-fold. These cells were generated using a modified protocol that used DAPT instead of FGF2 followed by FACS with NCAM+/HNK− markers for purification. Caron et al.36 pre-plated cells 10 days after starting the differentiation protocol. It will be interesting to compare the different protocols for generating and expanding myogenic progenitors in more detail to test whether they yield myogenic progenitors with similar or different properties. This should elucidate whether myogenic cells generated in vitro relate to different types of myogenic progenitors that have been identified in vivo, such as fetal or adult muscle stem cells from different anatomical locations and in different states of activation.
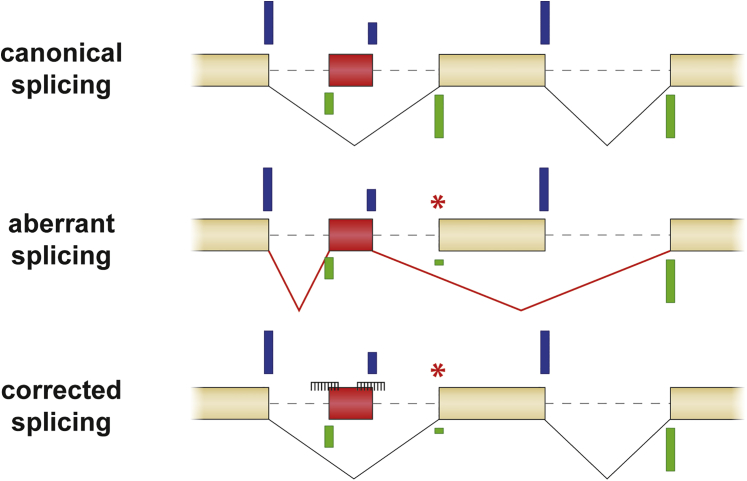
Master splicing factors have been identified that regulate tissue-specific splicing. In skeletal muscle, these include Rbfox1, RBM38, and MBNL3.3 This highlights the need to evaluate splicing in the cell type of interest, in particular, when testing potential drugs that modulate splicing such as AONs. In this case, GAA splicing abnormalities caused by the IVS1 variant and the effects of AONs on GAA splicing appeared similar in fibroblasts and myotubes. This suggests that splicing of exon 2 is not subject to regulation by the aforementioned master splicing factors and may be regulated by more general splicing factors that are also expressed in fibroblasts. We hypothesize that the IVS1 variant weakens the canonical splice acceptor of exon 2 such that the general splicing machinery prefers to utilize the nearby cryptic splice site of the pseudo exon (Figure 6). In healthy cells, this cryptic splice site is overruled by the strength of the canonical splice acceptor. Blocking of the cryptic splice acceptor, but also of the cryptic splice donor of the pseudo exon, prevents the splicing machinery from utilizing these sites, and when this occurs efficiently, the best available option is the weakened, but functional canonical splice acceptor of exon 2. While we originally anticipated that exon 2 splicing may be regulated by classical splice silencer motifs, we were surprised to find that this it is in fact regulated by cryptic splicing. In this respect, the IVS1 variant resembles other cases of cryptic splicing in Pompe disease that we analyzed previously and that could be restored using AONs.40 This finding highlights that the possibility of a shift toward cryptic splicing in human disease should be considered when designing AON-based therapies (Figure 6).
Figure 6.
Model for Promotion of Exon Inclusion in Cells with the IVS1 GAA Variant
Top: natural cryptic splice sites define a pseudo exon that is silent in healthy individuals. Middle: a pathogenic variant weakens a canonical splice acceptor site, resulting in inclusion of the pseudo exon and skipping of the canonical exon. Bottom: simultaneous blocking of the natural cryptic splice acceptor and splice donor sites of the pseudo exon prevents inclusion of the pseudo exon and promotes inclusion of the canonical exon. Splice site strength is indicated by the length of the blue (donor sites) and green (acceptor sites) bars. The natural pseudo exon is indicated in red. AONs are indicated as combs. The boxes represent exons, and the dashed lines introns.
Further work is required to translate these findings to a clinical setting. A combination of two AONs that have a larger effect than individual AONs at the same total AON dose may offer the possibility to lower the dose, which should be confirmed in animal and clinical studies. Encouraging progress toward clinical implementation has been reported for other neuromuscular disorders including SMA41, 42 and DMD43, 44 (reviewed by Havens et al.45). We anticipate that it will be important to promote cellular uptake of AONs in skeletal muscle cells applied via the circulation. Ongoing efforts focus on various strategies including cell penetrating peptides, exosomes, nanoparticles, and stimulation of ATP-dependent transport using hexose.46, 47, 48, 49, 50 Implementation of enhancers of cellular uptake may form the next generation of AONs that will be tested in vivo as potential targeted therapies of human disease.
Materials and Methods
Generation of iPSCs
Primary fibroblasts from control 1, patient 1 (c.-32-13T>G/c.525delT), and patient 2 (c.-32-13T>G/c.923A>C) (see van der Wal et al.5) were reprogrammed into iPSCs using a polycistronic lentiviral vector of Oct4, Sox2, Klf4, and c-Myc as described.18 The Erasmus Medical Center (MC) institutional review board approved the study protocol. iPSC line for control 2 was a gift from Christian Freund and Christine Mummery and has been characterized previously.21 iPSCs were cultured on γ-irradiated mouse embryonic feeder (MEF) cells. The iPSC culture medium consisted of DMEM/F12 medium (Invitrogen), 20% knockout serum replacement (Invitrogen), 1% non-essential amino acids (Gibco), 1% penicillin/streptomycin/L-glutamine (100×, Gibco), 2 mM β-mercaptoethanol (Invitrogen), and 20 ng/mL basic fibroblast growth factor (PeproTech). Human ES lines H1 and H9 were obtained from WiCell Research Institute, Madison, WI, USA. The identity of cell lines used in this study was confirmed by DNA sequence and microarray analyses. All cell lines were routinely tested for mycoplasma infection using the MycoAlert Mycoplasma Detection Kit (Lonza) and were found negative.
Immunofluorescence
Cells were cultured on normal tissue culture dishes and fixed with 4% paraformaldehyde (Merck) in PBS for 10 min at room temperature, washed with PBS, and permeabilized for 5 min with 0.1% Triton X-100 (AppliChem) in PBS. Blocking was performed for 30 min at room temperature with blocking solution containing PBS-T (0.1% Tween, Sigma-Aldrich) with 3% BSA (Sigma-Aldrich). Primary antibodies (Table S2) were diluted into 0.1% BSA in PBS-T and incubated 1 hr at room temperature. After incubation cells were washed three times for 5 min with PBS-T and incubated with the secondary antibodies (1:500, Alexa Fluor-594-α-goat, Alexa Fluor-488-α-mouse, Alexa Fluor-594-α-rabbit, Alexa Fluor-488-α-rabbit, Invitrogen or horse anti-mouse biotin, Vector Laboratories) in PBS-T for 30 min at room temperature. For incubations with secondary biotinylated antibodies, cells were washed three times for 5 min with PBS-T and incubated with Streptavidine 594 (1:500, Invitrogen). The cells were subsequently washed two times for 5 min with PBS and incubated for 15 min with Hoechst (1:15,000, Thermo Scientific). Cells were imaged in PBS.
Microarray Analysis
RNA samples to be analyzed by microarrays were prepared using RNeasy columns with on-column DNA digestion (QIAGEN). There were 300 ng of total RNA per sample that were used as input into a linear amplification protocol (Ambion), which involved synthesis of T7-linked double-stranded cDNA and 12 hr of in vitro transcription incorporating biotin-labeled nucleotides. Purified and labeled cRNA was then hybridized for 18 hr onto HumanHT-12 v4 expression BeadChips (Illumina) following the manufacturer’s instructions. After recommended washing, chips were stained with streptavidin-Cy3 (GE Healthcare) and scanned using the iScan reader (Illumina) and accompanying software. Samples were exclusively hybridized as biological replicates. The bead intensities were mapped to gene information using BeadStudio 3.2 (Illumina). Background correction was performed using the affymetrix robust multi-array analysis (RMA) background correction model.51 Variance stabilization was performed using the log2 scaling and gene expression normalization was calculated with the method implemented in the lumi package of R-Bioconductor. Data post-processing and graphics were performed with in-house developed functions in MATLAB. Hierarchical clustering of genes and samples was performed with one minus correlation metric and the unweighted average distance (UPGMA) (also known as group average) linkage method. The microarray data have been made available online at Gene Expression Omnibus: GSE75713.
Differentiation of iPSCs to Derivatives of the Three Germ Layers
iPSC colonies were washed once with PBS and treated for 45 min with 1 mg/mL collagenases IV (Invitrogen) at 37°C, scraped, and centrifuged for 15 s at 800 rpm. The pellet was slowly resuspended in EB medium (iPSC culture medium without FGF2) containing 10 μM Y-27632 dihydrochloride (Ascent Scientific) and plated on low binding plates (Cytoone). For endodermal differentiation, 10 μM SB 431542 (Ascent Scientific) was added to the EB medium. 6 days later, EBs were plated in 12 wells coated with 0.1% gelatin (Sigma-Aldrich) for endodermal and mesodermal differentiation or with matrigel-coated plates for ectodermal differentiation in endoderm/mesoderm/ectoderm medium (Table S3). Cells were fixed after 14 days of differentiation with 4% paraformaldehyde (Merck) in PBS for 5 min at room temperature and processed for immunofluorescence.
Karyotype Analysis
iPSCs or myogenic progenitors were detached with TrypLe (Gibco) for 5 min at 37°C. The pellet was incubated with 10 μg/mL colcemid (Gibco) for 30 min at room temperature. Cells were then centrifuged for 10 min at 1,100 rpm, resuspended in pre-warmed 0.075 M KCl, and incubated for 10 min at 37°C. After incubation cells were washed five times with fixation solution (3:1 methanol:acetic acid) and spread onto glass slides. Hoechst staining was performed as described above. There were 15 slides that were analyzed per cell line.
Differentiation of iPSCs to Myogenic Progenitors
Differentiation of iPSCs to myogenic progenitors cells was modified from Borchin et al.17 Briefly, 0.6 mm large iPSC colonies cultured in 10 cm dishes on MEF feeders were treated for 5 days with 3.5 μM CHIR99021 (Axon Medchem) in myogenic differentiation medium (DMEM/F12, 1× ITS-X and 1× Penicillin/Streptomycin/Glutamine, all Gibco). CHIR99021 was removed and cells were cultured in myogenic differentiation medium containing 20 ng/mL FGF2 (Prepotech) for 14 days and were then cultured for an additional 16 days in myogenic differentiation medium only. Medium was refreshed daily.
Purification of Myogenic Progenitors Using FACS
Following the 35-day protocol for differentiating iPSCs into a mixture of cells including myogenic progenitors as outlined above, cells were harvested and purified by FACS. To this end, cells were washed once with PBS, incubated for 5 min with TrypLe (Gibco) at 37°C, and gently detached with a pipetboy. The cell suspension was filtered through a 40 μM FACS strainer (Falcon) to remove cell aggregates. Cells were centrifuged for 4 min at 1,000 rpm and incubated with anti-HNK-1-FITC (1:100, Aviva Systems Biology) and anti-C-MET-APC (1:50, R&D Systems) antibodies for 30 min on ice in myogenic differentiation medium. Cells were washed three times with ice-cold 1% BSA in PBS before FACS sorting. Hoechst (33258, Life Technologies) was used as viability marker. Hoechst/C-MET-positive cells were sorted with a 100 μm nozzle and collected in ice-cold iPSC-myogenic progenitor proliferation medium (iPSC-MPC-pro medium) containing DMEM high glucose (Gibco) supplemented with 100 U/mL Penicillin/Streptomycin/Glutamine (Life Technologies), 10% fetal bovine serum (Hyclone, Thermo Scientific), and 100 ng/mL FGF2 (PeproTech). To reduce cell death, medium was supplemented with 1× RevitaCell Supplement (Gibco) during collection and the first 24 hr of cell culture. Sorting time was limited to 20 min per well. Plates/well were coated for 30 min at room temperature with ECM (E6909-5 mL, 1:200 in iPSC-MPC-pro medium, Sigma Aldrich). Sorted cells were plated either at 40,000 cells in one well of a 48-well plate or at 80,000 cells in one well of a 24-well plate, depending on the amount of cells.
Expansion of Myogenic Progenitors
At 1 day after plating FACS sorted myogenic progenitors, the medium was refreshed with iPSC-MPC-pro medium. When cells reached 90% confluence, cells were passaged using 2× diluted TrypLe in PBS and plated on ECM-coated plastic. Myogenic progenitors could be frozen at any time during expansion without loss of proliferation and differentiation capacity, using iPSC-MPC-pro medium supplemented with 10% DMSO.
Differentiation of Myogenic Progenitors into Multinucleated Myotubes
For differentiation to multinucleated myotubes, myogenic progenitors were grown to 90% confluence, and the medium was then replaced with myogenic differentiation medium (DMEM/F12, 1× ITS-X and penicillin/streptomycin/glutamine, all Gibco). After 4 days, myotubes were harvested.
Production of LV-OSKM Lentivirus
The LV-OSKM lentivirus was produced in HEK293T cells cultured in DMEM high glucose (Gibco) with 10% fetal bovine serum (Thermo Scientific) supplemented with 100 U/mL Penicillin/Streptomycin/Glutamine (Gibco). There were 80% confluent HEK293T cells that were transfected in 10 cm culture dishes with 3 μg LV-OSKM reprogramming vector,18 2 μg psPAX2, and 1 μg pVSV packaging vectors per dish using Fugene 6 transfection reagent according to the manufacturer’s protocol (Promega). For concentration, 72 hr after transfection, medium was filtered with 0.45 μm PVDF filters (Millipore) and centrifuged for 2 hr at 20,000 rpm with a Beckman Coulter Ultracentrifuge with SW 32 Ti Rotor. The pellet was dissolved in 25 μL DMEM low glucose (Gibco) per 10 cm dish and stored at −80°C.
AON Transfections
At day −1 or day 0 prior to differentiation, myogenic progenitors were transfected with Morpholino AONs (Table S4) using 4.5 μL endoporter reagent (Gene Tools) per milliliter of medium. After 4 days of differentiation myotubes were harvested. In our previous study, a control AON targeting CypA showed no effect on GAA mRNA expression and no toxic effects were observed.5 We tested in addition the effect of control AON1 (which targets GAA c.-32-219_-200) and the standard control from Gene Tools (which targets HBB c.316-162_138) in iPSC-derived skeletal muscle and found no effects on GAA protein expression and no toxicity (Figure S5).
RNA Isolation and cDNA Synthesis
RNeasy Mini Kit with DNase treatment (QIAGEN) was used to extract RNA. RNA was eluted in RNase-free water and stored at −80°C. There were 500 ng of RNA that were reverse transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad).
qRT-PCR
The qRT-PCR reaction consisted of a 15 μL mix with 7.5 μL iTaq Universal SYBR Green Supermix (Bio-Rad), 10 pmol/μL forward and reverse primers (Table S5), and cDNA (5×, 10×, or 20× diluted) measured with a CFX96 real-time system (Bio-Rad). For each plate, a standard curve was included with 4–6 dilutions. Quantification of expression was calculated relative to expression of four markers (Myog, MyoD, LAMP1, and LAMP2) in experiments where myotubes were used and to RNA input in experiments were multiple tissues (fibroblasts, myogenic progenitors, and myotubes) were compared.
Flanking Exon RT-PCR
RT-PCR was performed with the Advantage GC 2 PCR Kit (Clontech) with a GC-melt concentration of 0.5 M according to manufactory instructions with primers from Table S4 and ten times diluted cDNA. The RT-PCR reaction was analyzed on a 1.5% agarose gel containing 0.5 μg/mL ethidium bromide (Sigma-Aldrich).
GAA Enzyme Activity Assay
Cells were lysed for 10 min on ice with ice cold lysis buffer containing: 50 mM Tris (pH 7.5), 100 mM NaCl, 50 mM NaF, 1% Triton X-100, and one tablet Protease Inhibitor Cocktail cOmplete, with EDTA, (Roche). The substrate 4-methylumbelliferyl α-D-glucopyranoside (4-MU; Sigma-Aldrich) was used to measure GAA enzyme activity as described previously.52 A BCA protein assay kit (Pierce, Thermo Scientific) was used to determine total protein concentration.
GAA Protein Detection
iPSC-derived myogenic progenitors from control 2 and patient 1 were differentiated for 4 days into myotubes, treated with AONs as described above, and cells were then harvested with ice-cold RIPA buffer (150 mM NaCl, 0.1% SDS, 50 mM Tris, pH 7.4, 2 mM EDTA, and 1% Triton X-100) supplemented with Protease Inhibitor Cocktail cOmplete, with EDTA (Roche). Total protein concentrations and GAA activities were measured as described above. For immunoblot analysis, 10 μg of total protein was heat denaturated for 5 min at 95°C with sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromophenol blue, and 5% β-mercaptoethanol) and loaded on a Criterion TGX 4%–15% gel (Bio-Rad). Proteins were transferred onto 0.45 μm nitrocellulose blotting membranes (GE Healthcare). The membranes were blocked for 1 hr in Odyssey blocking buffer (LI-COR Biosciences) (4× diluted in PBS) and stained for 1 hr with rabbit-α-GAA (Abcam 137068 1:1,000) and mouse-α-GAPDH (Millipore MAB374 1:1,000) for 1 hr in Li-Cor blocking buffer. The blot was washed for three times with PBS-T (0.1% Tween, Sigma-Aldrich), incubated for 30 min with IRDye 700/800 secondary antibodies (LI-COR Biosciences) in PBS-T, and washed three times with PBS-T. Immunoblots were scanned with the Odyssey Infrared Imaging system (LI-COR Biosciences). Bands were quantified with Odyssey software (LI-COR Biosciences). Standard curves were generated for each antibody (GAA and GAPDH) using 2-fold dilution series. The formulas generated from the linear range were used to quantify the active form of GAA (76 kD) and GAPDH. Values were corrected for background, which was obtained from an empty position on the blot.
Statistical Analysis
All data represent mean ± SD, and p values refer to two-sided t tests. Bonferroni multiple testing correction was applied where necessary. A p value < 0.05 was considered to be significant. Data showed normal variance. There was no power calculation in any of the experiments. No randomization method was used. No samples were excluded from the analyses. Experiments on expansion of iPS-derived muscle progenitors, differentiation into myotubes, and AON treatment have been performed at least two times. Investigators were not blinded to the identity of the samples.
Author Contributions
E.v.d.W., A.J.B., A.T.v.d.P., and W.W.M.P.P. conceived and designed the study and drafted the manuscript. E.v.d.W., A.J.B., T.J.M.v.G., and S.L.M.i.G. performed the experiments. M.J.A.-B. performed the bioinformatics analysis. W.W.M.P.P., H.Z., and H.R.S. supervised the study. E.v.d.W., A.J.B., T.J.M.v.G., S.L.M.i.G, H.Z., M.J.A.-B., H.R.S., A.T.v.d.P., and W.W.M.P.P. were involved in data interpretation and approved the final manuscript.
Conflicts of Interest
A.T.v.d.P. has provided consulting services for various industries in the field of Pompe disease under an agreement between these industries and Erasmus MC. All the other authors declare no conflict of interest.
Acknowledgments
We thank Martina Radstaak and Hannie Douben for technical assistance, Dr. Schambach for the OSKM-lentiviral vector, Drs. Christian Freund and Christine Mummery for providing a control fibroblast and iPSC line, Michelle Minneboo for brain cDNA, Philip Lijnzaad for advice on statistical analysis, and Dr. Arnold Reuser for discussion. This work was funded by the Sophia Children’s Hospital Foundation (SSWO) (grant S-687), the Prinses Beatrix Spierfonds/Stichting Spieren voor Spieren (grant W.OR13-21), and Tex net.
Footnotes
Supplemental Information includes five figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.03.002.
Supplemental Information
References
- 1.Shapiro M.B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lei Q., Li C., Zuo Z., Huang C., Cheng H., Zhou R. Evolutionary insights into RNA trans-splicing in vertebrates. Genome Biol. Evol. 2016;8:562–577. doi: 10.1093/gbe/evw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jangi M., Sharp P.A. Building robust transcriptomes with master splicing factors. Cell. 2014;159:487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van der Wal E., Bergsma A.J., Pijnenburg J.M., van der Ploeg A.T., Pijnappel W.W.M.P. Antisense oligonucleotides promote exon inclusion and correct the common c.-32-13T > G (IVS1) GAA splicing variant in Pompe disease. Mol. Ther. Nucleic Acids. 2017;7:90–100. doi: 10.1016/j.omtn.2017.03.001. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Ploeg A.T., Reuser A.J. Pompe’s disease. Lancet. 2008;372:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 7.van der Beek N.A., de Vries J.M., Hagemans M.L., Hop W.C., Kroos M.A., Wokke J.H., de Visser M., van Engelen B.G., Kuks J.B., van der Kooi A.J. Clinical features and predictors for disease natural progression in adults with Pompe disease: a nationwide prospective observational study. Orphanet J. Rare Dis. 2012;7:88. doi: 10.1186/1750-1172-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kishnani P.S., Beckemeyer A.A. New therapeutic approaches for Pompe disease: enzyme replacement therapy and beyond. Pediatr. Endocrinol. Rev. 2014;12(Suppl 1):114–124. [PubMed] [Google Scholar]
- 9.Schoser B., Stewart A., Kanters S., Hamed A., Jansen J., Chan K., Karamouzian M., Toscano A. Survival and long-term outcomes in late-onset Pompe disease following alglucosidase alfa treatment: a systematic review and meta-analysis. J. Neurol. 2016 doi: 10.1007/s00415-016-8219-8. Published online July 2, 2016. [DOI] [PubMed] [Google Scholar]
- 10.Anderson L.J., Henley W., Wyatt K.M., Nikolaou V., Waldek S., Hughes D.A., Lachmann R.H., Logan S. Effectiveness of enzyme replacement therapy in adults with late-onset Pompe disease: results from the NCS-LSD cohort study. J. Inherit. Metab. Dis. 2014;37:945–952. doi: 10.1007/s10545-014-9728-1. [DOI] [PubMed] [Google Scholar]
- 11.Regnery C., Kornblum C., Hanisch F., Vielhaber S., Strigl-Pill N., Grunert B., Müller-Felber W., Glocker F.X., Spranger M., Deschauer M. 36 months observational clinical study of 38 adult Pompe disease patients under alglucosidase alfa enzyme replacement therapy. J. Inherit. Metab. Dis. 2012;35:837–845. doi: 10.1007/s10545-012-9451-8. [DOI] [PubMed] [Google Scholar]
- 12.de Vries J.M., van der Beek N.A., Hop W.C., Karstens F.P., Wokke J.H., de Visser M., van Engelen B.G., Kuks J.B., van der Kooi A.J., Notermans N.C. Effect of enzyme therapy and prognostic factors in 69 adults with Pompe disease: an open-label single-center study. Orphanet J. Rare Dis. 2012;7:73. doi: 10.1186/1750-1172-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huie M.L., Chen A.S., Tsujino S., Shanske S., DiMauro S., Engel A.G., Hirschhorn R. Aberrant splicing in adult onset glycogen storage disease type II (GSDII): molecular identification of an IVS1 (-13T-->G) mutation in a majority of patients and a novel IVS10 (+1GT-->CT) mutation. Hum. Mol. Genet. 1994;3:2231–2236. doi: 10.1093/hmg/3.12.2231. [DOI] [PubMed] [Google Scholar]
- 14.Kroos M.A., Pomponio R.J., Hagemans M.L., Keulemans J.L., Phipps M., DeRiso M., Palmer R.E., Ausems M.G., Van der Beek N.A., Van Diggelen O.P. Broad spectrum of Pompe disease in patients with the same c.-32-13T->G haplotype. Neurology. 2007;68:110–115. doi: 10.1212/01.wnl.0000252798.25690.76. [DOI] [PubMed] [Google Scholar]
- 15.Dardis A., Zanin I., Zampieri S., Stuani C., Pianta A., Romanello M., Baralle F.E., Bembi B., Buratti E. Functional characterization of the common c.-32-13T>G mutation of GAA gene: identification of potential therapeutic agents. Nucleic Acids Res. 2014;42:1291–1302. doi: 10.1093/nar/gkt987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boerkoel C.F., Exelbert R., Nicastri C., Nichols R.C., Miller F.W., Plotz P.H., Raben N. Leaky splicing mutation in the acid maltase gene is associated with delayed onset of glycogenosis type II. Am. J. Hum. Genet. 1995;56:887–897. [PMC free article] [PubMed] [Google Scholar]
- 17.Borchin B., Chen J., Barberi T. Derivation and FACS-mediated purification of PAX3+/PAX7+ skeletal muscle precursors from human pluripotent stem cells. Stem Cell Reports. 2013;1:620–631. doi: 10.1016/j.stemcr.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warlich E., Kuehle J., Cantz T., Brugman M.H., Maetzig T., Galla M., Filipczyk A.A., Halle S., Klump H., Schöler H.R. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol. Ther. 2011;19:782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermans M.M., De Graaff E., Kroos M.A., Mohkamsing S., Eussen B.J., Joosse M., Willemsen R., Kleijer W.J., Oostra B.A., Reuser A.J. The effect of a single base pair deletion (delta T525) and a C1634T missense mutation (pro545leu) on the expression of lysosomal alpha-glucosidase in patients with glycogen storage disease type II. Hum. Mol. Genet. 1994;3:2213–2218. doi: 10.1093/hmg/3.12.2213. [DOI] [PubMed] [Google Scholar]
- 20.Hermans M.M., van Leenen D., Kroos M.A., Beesley C.E., Van Der Ploeg A.T., Sakuraba H., Wevers R., Kleijer W., Michelakakis H., Kirk E.P. Twenty-two novel mutations in the lysosomal alpha-glucosidase gene (GAA) underscore the genotype-phenotype correlation in glycogen storage disease type II. Hum. Mutat. 2004;23:47–56. doi: 10.1002/humu.10286. [DOI] [PubMed] [Google Scholar]
- 21.Dambrot C., van de Pas S., van Zijl L., Brändl B., Wang J.W., Schalij M.J., Hoeben R.C., Atsma D.E., Mikkers H.M., Mummery C.L., Freund C. Polycistronic lentivirus induced pluripotent stem cells from skin biopsies after long term storage, blood outgrowth endothelial cells and cells from milk teeth. Differentiation. 2013;85:101–109. doi: 10.1016/j.diff.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Clegg C.H., Linkhart T.A., Olwin B.B., Hauschka S.D. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 1987;105:949–956. doi: 10.1083/jcb.105.2.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubé F., Francastel C. Mammalian introns: when the junk generates molecular diversity. Int. J. Mol. Sci. 2015;16:4429–4452. doi: 10.3390/ijms16034429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi T., Kawagoe S., Otsu M., Shimada Y., Kobayashi H., Hirayama R., Eto K., Ida H., Ohashi T., Nakauchi H., Eto Y. The generation of induced pluripotent stem cells (iPSCs) from patients with infantile and late-onset types of Pompe disease and the effects of treatment with acid-α-glucosidase in Pompe’s iPSCs. Mol. Genet. Metab. 2014;112:44–48. doi: 10.1016/j.ymgme.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Raval K.K., Tao R., White B.E., De Lange W.J., Koonce C.H., Yu J., Kishnani P.S., Thomson J.A., Mosher D.F., Ralphe J.C., Kamp T.J. Pompe disease results in a Golgi-based glycosylation deficit in human induced pluripotent stem cell-derived cardiomyocytes. J. Biol. Chem. 2015;290:3121–3136. doi: 10.1074/jbc.M114.628628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato Y., Kobayashi H., Higuchi T., Shimada Y., Ida H., Ohashi T. Metabolomic profiling of pompe disease-induced pluripotent stem cell-derived cardiomyocytes reveals that oxidative stress is associated with cardiac and skeletal muscle pathology. Stem Cells Transl. Med. 2016 doi: 10.5966/sctm.2015-0409. Published online August 18, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato Y., Kobayashi H., Higuchi T., Shimada Y., Ida H., Ohashi T. TFEB overexpression promotes glycogen clearance of Pompe disease iPSC-derived skeletal muscle. Mol. Ther. Methods Clin. Dev. 2016;3:16054. doi: 10.1038/mtm.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wens S.C., Schaaf G.J., Michels M., Kruijshaar M.E., van Gestel T.J., In ‘t Groen S., Pijnenburg J., Dekkers D.H., Demmers J.A., Verdijk L.B. Elevated plasma cardiac troponin T levels caused by skeletal muscle damage in Pompe disease. Circ. Cardiovasc. Genet. 2016;9:6–13. doi: 10.1161/CIRCGENETICS.115.001322. [DOI] [PubMed] [Google Scholar]
- 29.van der Beek N.A., Soliman O.I., van Capelle C.I., Geleijnse M.L., Vletter W.B., Kroos M.A., Reuser A.J., Frohn-Mulder I.M., van Doorn P.A., van der Ploeg A.T. Cardiac evaluation in children and adults with Pompe disease sharing the common c.-32-13T>G genotype rarely reveals abnormalities. J. Neurol. Sci. 2008;275:46–50. doi: 10.1016/j.jns.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Ebbink B.J., Poelman E., Plug I., Lequin M.H., van Doorn P.A., Aarsen F.K., van der Ploeg A.T., van den Hout J.M. Cognitive decline in classic infantile Pompe disease: An underacknowledged challenge. Neurology. 2016;86:1260–1261. doi: 10.1212/WNL.0000000000002523. [DOI] [PubMed] [Google Scholar]
- 31.Wens S.C., van Gelder C.M., Kruijshaar M.E., de Vries J.M., van der Beek N.A., Reuser A.J., van Doorn P.A., van der Ploeg A.T., Brusse E. Phenotypical variation within 22 families with Pompe disease. Orphanet J. Rare Dis. 2013;8:182. doi: 10.1186/1750-1172-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Ploeg A.T., Kroos M., van Dongen J.M., Visser W.J., Bolhuis P.A., Loonen M.C., Reuser A.J. Breakdown of lysosomal glycogen in cultured fibroblasts from glycogenosis type II patients after uptake of acid alpha-glucosidase. J. Neurol. Sci. 1987;79:327–336. doi: 10.1016/0022-510x(87)90239-5. [DOI] [PubMed] [Google Scholar]
- 33.Choi I.Y., Lim H., Estrellas K., Mula J., Cohen T.V., Zhang Y., Donnelly C.J., Richard J.P., Kim Y.J., Kim H. Concordant but varied phenotypes among Duchenne muscular dystrophy patient-specific myoblasts derived using a human iPSC-based model. Cell Rep. 2016;15:2301–2312. doi: 10.1016/j.celrep.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Chal J., Oginuma M., Al Tanoury Z., Gobert B., Sumara O., Hick A., Bousson F., Zidouni Y., Mursch C., Moncuquet P. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015;33:962–969. doi: 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- 35.Hosoyama T., McGivern J.V., Van Dyke J.M., Ebert A.D., Suzuki M. Derivation of myogenic progenitors directly from human pluripotent stem cells using a sphere-based culture. Stem Cells Transl. Med. 2014;3:564–574. doi: 10.5966/sctm.2013-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caron L., Kher D., Lee K.L., McKernan R., Dumevska B., Hidalgo A., Li J., Yang H., Main H., Ferri G. A human pluripotent stem cell model of facioscapulohumeral muscular dystrophy-affected skeletal muscles. Stem Cells Transl. Med. 2016;5:1145–1161. doi: 10.5966/sctm.2015-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shelton M., Metz J., Liu J., Carpenedo R.L., Demers S.P., Stanford W.L., Skerjanc I.S. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Reports. 2014;3:516–529. doi: 10.1016/j.stemcr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu C., Tabebordbar M., Iovino S., Ciarlo C., Liu J., Castiglioni A., Price E., Liu M., Barton E.R., Kahn C.R. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shelton M., Kocharyan A., Liu J., Skerjanc I.S., Stanford W.L. Robust generation and expansion of skeletal muscle progenitors and myocytes from human pluripotent stem cells. Methods. 2016;101:73–84. doi: 10.1016/j.ymeth.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 40.Bergsma A.J., In ‘t Groen S.L., Verheijen F.W., van der Ploeg A.T., Pijnappel W.P. From cryptic toward canonical pre-mRNA splicing in Pompe disease: a pipeline for the development of antisense oligonucleotides. Mol. Ther. Nucleic Acids. 2016;5:e361. doi: 10.1038/mtna.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond S.M., Hazell G., Shabanpoor F., Saleh A.F., Bowerman M., Sleigh J.N., Meijboom K.E., Zhou H., Muntoni F., Talbot K. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 2016;113:10962–10967. doi: 10.1073/pnas.1605731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aartsma-Rus A., Muntoni F. 194th ENMC international workshop. 3rd ENMC workshop on exon skipping: towards clinical application of antisense-mediated exon skipping for Duchenne muscular dystrophy 8-10 December 2012, Naarden, The Netherlands. Neuromuscul. Disord. 2013;23:934–944. doi: 10.1016/j.nmd.2013.06.369. [DOI] [PubMed] [Google Scholar]
- 44.Straub V., Balabanov P., Bushby K., Ensini M., Goemans N., De Luca A., Pereda A., Hemmings R., Campion G., Kaye E. Stakeholder cooperation to overcome challenges in orphan medicine development: the example of Duchenne muscular dystrophy. Lancet Neurol. 2016;15:882–890. doi: 10.1016/S1474-4422(16)30035-7. [DOI] [PubMed] [Google Scholar]
- 45.Havens M.A., Hastings M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han G., Gu B., Cao L., Gao X., Wang Q., Seow Y., Zhang N., Wood M.J., Yin H. Hexose enhances oligonucleotide delivery and exon skipping in dystrophin-deficient mdx mice. Nat. Commun. 2016;7:10981. doi: 10.1038/ncomms10981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehto T., Ezzat K., Wood M.J., El Andaloussi S. Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev. 2016;106(Pt A):172–182. doi: 10.1016/j.addr.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Abushahba M.F., Mohammad H., Thangamani S., Hussein A.A., Seleem M.N. Impact of different cell penetrating peptides on the efficacy of antisense therapeutics for targeting intracellular pathogens. Sci. Rep. 2016;6:20832. doi: 10.1038/srep20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boisguérin P., Deshayes S., Gait M.J., O’Donovan L., Godfrey C., Betts C.A., Wood M.J., Lebleu B. Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv. Drug Deliv. Rev. 2015;87:52–67. doi: 10.1016/j.addr.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu R., Liu J., Ji X., Liu Y. Synthetic nucleic acids delivered by exosomes: a potential therapeutic for generelated metabolic brain diseases. Metab. Brain Dis. 2013;28:551–562. doi: 10.1007/s11011-013-9434-y. [DOI] [PubMed] [Google Scholar]
- 51.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergsma A.J., Kroos M., Hoogeveen-Westerveld M., Halley D., van der Ploeg A.T., Pijnappel W.W. Identification and characterization of aberrant GAA pre-mRNA splicing in pompe disease using a generic approach. Hum. Mutat. 2015;36:57–68. doi: 10.1002/humu.22705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.