Abstract
Chronic infection with hepatitis B virus (HBV) remains a problem of global significance and improving available treatment is important to prevent life-threatening complications arising in persistently infected individuals. HBV is susceptible to silencing by exogenous artificial intermediates of the RNA interference (RNAi) pathway. However, toxicity of Pol III cassettes and short duration of silencing by effectors of the RNAi pathway may limit anti-HBV therapeutic utility. To advance RNAi-based HBV gene silencing, mono- and trimeric artificial primary microRNAs (pri-miRs) derived from pri-miR-31 were placed under control of the liver-specific modified murine transthyretin promoter. The sequences, which target the X sequence of HBV, were incorporated into recombinant hepatotropic self-complementary adeno-associated viruses (scAAVs). Systemic intravenous injection of the vectors into HBV transgenic mice at a dose of 1 × 1011 per animal effected significant suppression of markers of HBV replication for at least 32 weeks. The pri-miRs were processed according to the intended design, and intrahepatic antiviral guide sequences were detectable for 40 weeks after the injection. There was no evidence of toxicity, and innate immunostimulation was not detectable following the injections. This efficacy is an improvement on previously reported RNAi-based inhibition of HBV replication and is important to clinical translation of the technology.
Keywords: HBV, RNAi, Pol II, scAAV8, transgenic mice, interferon response, proinflammatory cytokines
Introduction
Despite availability of a vaccine to prevent hepatitis B virus (HBV) transmission, complications arising from chronic infection with the virus remain an important global health problem.1 About 700,000 people die each year as a result of HBV-related cirrhosis and hepatocellular carcinoma (HCC).2 Currently, licensed treatments rarely eliminate HBV and development of durable and effective anti-HBV therapy is a priority to limit mortality in the world’s 240 million chronic carriers.3, 4, 5, 6
The genome of HBV has a remarkably compact arrangement. Open reading frames (ORFs) overlap with each other and encompass the entire genome (Figure 1A). There are four ORFs: surface (S), polymerase (P), core (C), and X. Cis-elements that regulate transcription and viral replication are embedded in the protein coding sequences. Following hepatocyte infection, encapsidated relaxed circular DNA (rcDNA) is transported to the nucleus where it is released and “repaired” to form covalently closed circular DNA (cccDNA). This stable replication intermediate7, 8 is the template for transcription of pregenomic RNA and protein coding mRNA. Persistence of cccDNA has been an obstacle to curing HBV carriers from the infection and durable inhibition of HBV replication is desirable to eliminate the virus.
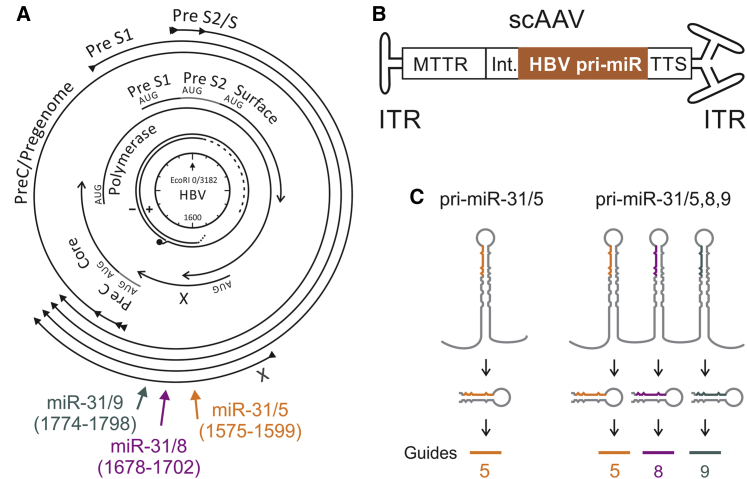
Figure 1.
Schematic Illustration of HBV Targets, Structure of Recombinant scAAVs, and Processing of apri-miR Sequences
(A) Relaxed circular DNA found in the virion, with + and − strands indicated, is depicted at the center of the diagram. The nucleotide coordinates are indicated in a clockwise direction from the unique EcoRI site. Core, polymerase, surface, and X, the four viral open reading frames, are shown as overlapping arrows immediately surrounding the genome. The preC/pregenomic, Pre S1, Pre S2/S, and X transcripts are indicated as the outermost arrows. The regions targeted by the three artificial pri-miRs, with cognate nucleotide coordinates given in parentheses, are indicted as axial arrows. (B) Structure of the scAAVs used to deliver anti-HBV expression cassettes. Inverted terminal repeats (ITRs) flank the cassettes, which comprise a liver-specific modified MTTR promoter, an intron (Int.), HBV-targeting apri-miR-31, and Pol II transcription termination signal (TTS). (C) Intended mechanism of processing of artificial anti-HBV mono- and tricistronic pri-miR transcripts that generate guide 5 alone or a combination of guides 5, 8, and 9. Adapted from Ivacik et al.21
The highly economical use of its genetic material limits sequence plasticity of HBV. This feature has been exploited to advance therapy that is based on nucleic acid hybridization. Several studies have shown that HBV is susceptible to RNA interference (RNAi)-mediated silencing (reviewed in Ivacik et al.9). This is despite some evidence that the viral X protein is capable of countering the cellular silencing mechanism.10 Both synthetic and expressed RNAi activators are capable of reprogramming the RNAi pathway to silence HBV replication in vivo. A combinatorial approach has previously been shown to knock down HIV-1 replication and prevent emergence of escape mutants.11, 12, 13, 14 The compact arrangement of the HBV genome restricts plasticity, and the number of RNAi effectors necessary to prevent escape is likely to be fewer than the four that are required for HIV-1. Use of synthetic RNAi activators against HBV has now reached a stage of evaluation in clinical trials.15 The formulation termed ARC-520, which includes cholesterol-conjugated siRNA and a hepatocyte-targeting peptide, showed promise in early clinical trials (https://www.clinicaltrials.gov/ct2/show/NCT02065336). However, the candidate drug has been placed on hold by the United States Food and Drug Administration after concerns relating to evidence of non-clinical toxicology were raised. A drawback of using synthetic siRNAs as therapy for chronic HBV infection is that efficacy against the virus is usually short lived and repeat administrations are required. Lasting renewable transcription of expressed artificial microRNAs (amiRs), produced from stable DNA templates within transduced cells, is thus attractive. Expression cassettes encoding the RNAi activators also have the advantage of compatibility with highly efficient recombinant viral vectors.
Short hairpin (sh)RNA-encoding sequences have commonly been used as artificial mimics of precursor microRNA (pre-miR). Typically, these sequences are transcribed from constitutively active Pol III promoters, such as U6 or H1.16, 17 A drawback of Pol III shRNA expression cassettes, elucidated while investigating use of expressed RNAi activators to silence HBV gene expression, is that overexpression of the pre-miR mimics from a U6 promoter may disrupt natural miR processing to result in lethal toxicity.12, 18 Research aimed at advancing therapeutic expressed sequences has thus been focused on improving regulation of transcription of RNAi activators in transduced cells. Pol II transcriptional regulatory elements have been favored as they are capable of precise and tissue-specific control of expression of RNAi effecters in targeted cells. To enhance compatibility of Pol II promoters with RNAi activators, primary miR (pri-miR) mimics rather than shRNAs have been incorporated into artificial expression cassettes.13, 19, 20 The design takes advantage of the natural transcription of pri-miRs from Pol II promoters. The polycistronic nature of natural pri-miR expression also enables incorporation of multiple antiviral sequences into an expression cassette to improve efficacy. Simultaneous targeting of different viral sequences is also desirable to suppress emergence of viral escape mutants.
Using natural human pri-miR-31 as a scaffold, efficient anti-HBV artificial mono- and trimeric pri-miR expression cassettes have been generated in our laboratory.14, 20 Recombinant lentivirus-mediated delivery of artificial pri-miRs (apri-miRs) to neonatal HBV transgenic mice resulted in durable suppression of markers of HBV replication.21 Transcription of the anti-HBV apri-miRs from a liver-specific murine transthyretin receptor (MTTR) promoter,22 was sustained over a 3 month period of analysis without evidence of toxicity.21 A recent study demonstrated that a monomeric apri-miR, expressed from a liver-specific promoter, was capable of silencing HBV replication in transgenic mice over a period of 84 days.23 To evaluate the utility of anti-HBV mono- and trimeric apri-miRs over a longer time period under conditions that would resemble treatment in a clinical setting, we have generated self-complementary adeno-associated viral vectors (scAAVs) that encode the anti-HBV expression cassettes. These vectors were administered to HBV transgenic mice that stably replicate the virus. The safety and versatility of AAVs makes them attractive for use in experimental and clinical settings.24 Methods for preparing AAVs are now well established and features such as hepatotropism may be conferred by pseudotyping with the serotype 8 capsid to generate scAAV8 vectors. Moreover, an interesting feature that is relevant to use of the vectors to treat HBV infection is that HBV replication in hepatocytes renders the cells more susceptible to transduction by AAVs.25 We show that systemic administration of preparations of scAAV8 encoding apri-miRs achieve sustained knockdown over a period of 32 weeks. Inhibition of viral replication was more efficient in animals treated with vectors encoding the trimeric cassette. This durable silencing with a safe vector that has potential clinical utility is an important feature.
Results
Expressed amiRs Targeting HBV
apri-miR expression cassettes were based on previously characterized HBV-targeting sequences.14, 20, 21 The natural pri-miR-31 sequence was adapted to serve as a scaffold for monocistronic (miR-31/5) and tricistronic (miR-31/5,8,9) antiviral sequences that target sites within the X ORF (Figure 1A). The X sequence is multifunctional and conserved in HBV isolates,26, 27 which makes it a good target for RNAi-mediated silencing. Moreover, X is contained within all HBV transcripts, enabling simultaneous cleavage of all the viral RNAs. To control expression in a liver-specific manner, the MTTR Pol II promoter was used to initiate transcription of the amiR sequences. Expression cassettes, comprising the promoter, an intron, miR-31/5 or miR-31/5,8,9, and a transcription termination signal were incorporated into scAAVs (Figure 1B). According to the intended design, primary transcripts of miR-31/5 and miR-31/5,8,9 are processed using the canonical miR pathway to generate pre-miR intermediates and guide RNAs of approximately 21 nt in length (Figure 1C). Although repetitive sequences, derived from the miR-31 scaffold, were present in the vectors, rearrangement was not observed (data not shown). This was in accordance with our similar previous findings when using recombinant adenoviral28 or lentiviral vectors.21
Processing of scAAV-Delivered Anti-HBV amiRs in Transduced Liver-Derived Cells
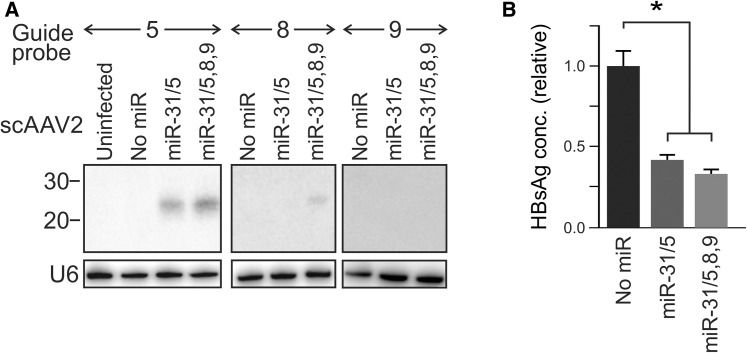
Liver-derived Huh7 cells were transduced with scAAVs pseudotyped with serotype 2 capsid (scAAV2), which encoded miR-31/5, miR-31/5,8,9, or the enhanced green fluorescent protein (eGFP). After infecting cells with the eGFP reporter-encoding vectors at a multiplicity of infection (MOI) of 100,000, fluorescence imaging showed that all cells were transduced (data not shown). To confirm processing of the apri-miRs, low molecular weight northern blot analysis was carried out. Putative guide sequences of 20–22 nt were detected after hybridization to probes complementary to anticipated mature miR-31/5 and miR-31/8 sequences (Figure 2A). Guide 5 was only detectable in RNA extracts of cells treated with scAAV2 miR-31/5 and scAAV2 miR-31/5,8,9. A comparison of bands corresponding to guides 5, 8, and 9 showed variation in quantities of the individual mature miRs. The guide 5 sequence was present in high amount in transduced cells, followed by guide 8, while the guide 9 sequence was least abundant and usually undetectable. Despite the guide 9 sequence being undetectable on northern blot, functional analysis in transfected cultured cells and in mice subjected to hydrodynamic injection indicated that the guide 9 sequence was indeed active (Figures S1 and S2). These data also demonstrated that the trimeric expression cassette is effective against a target with a mutation in the cognate of guide 5, which suggests that potential for viral escape would be limited. We have made similar observations following transient transfection of Huh7 cells20 or infection of these cells with miR-31/5,8,9-expressing lentiviral vectors.21 Variability in processing of the components of the tricistronic apri-miRs is likely to be the cause of differences in the concentrations of guide RNAs. Detection of the mature miRs did not appear to be influenced by the melting temperature of the guide and probe hybrids and the concentration of each mature guide sequence correlated with silencing efficacy.
Figure 2.
Processing and Antiviral Efficacy of HBV-Targeting Artificial pri-miR-31 Derivatives in Cultured Cells
(A) Northern blot analysis showing small RNA sequences detected following transduction of Huh7 cells with scAAVs expressing pri-miR-31/5, pri-miR-31/5,8,9, or no miR sequences. After stripping the blots, hybridization to a probe complementary to the U6 small nuclear RNA was used to confirm equal loading of the lanes. (B) Assessment of knock down efficacy of AAVs containing monomeric or trimeric HBV-targeting pri-miRs. The control cells received the scAAV construct that did not contain a miR sequence. The data are represented as the means, and the error bars indicate the standard error of the mean. The statistically significant differences are indicated by asterisks (*p ≤ 0.05).
Evaluation of silencing efficacy of the scAAV2-delivered miRs was also carried out in cultured Huh7 cells. HBV surface Ag (HBsAg) was measured in the culture supernatant of Huh7 cells after transfection with an HBV replication-competent plasmid29 and transduction with the scAAV2s containing monocistronic and tricistronic pri-miR cassettes (Figure 2B). When compared to the cells treated with the scAAV2s lacking miR cassettes, HBsAg was significantly reduced in cells infected with the vectors containing miR-31/5 or miR-31/5,8,9.
Antiviral Effects of AAVs Expressing miR-31/5 or miR-31/5,8,9 following Systemic Administration of the Vectors to HBV Transgenic Mice
The HBV transgenic mice used to assess efficacy of scAAVs contain an integrated replication-competent greater-than-genome-length viral sequence.30 Long term replication of HBV occurs in hepatocytes of these animals, but formation of cccDNA and infection of the murine liver cells does not occur. Nevertheless, the animals are useful as the long term replication of HBV in the transgenic mice simulates chronic HBV infection of humans.30, 31
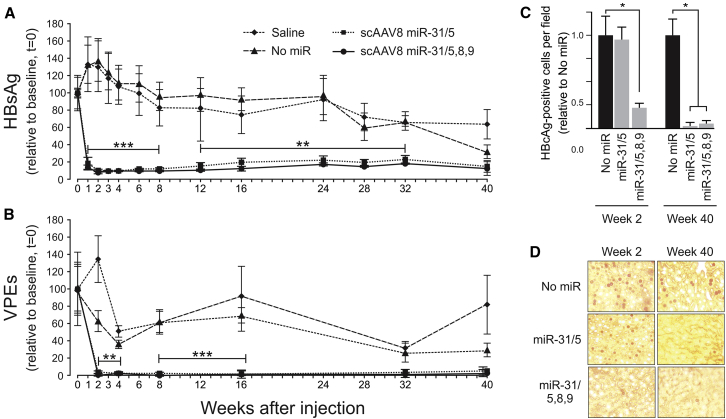
Following injection of 1 × 1011 scAAV8 particles into a tail vein of each animal, HBsAg and circulating viral particle equivalents (VPEs) were measured as indicators of viral replication (Figures 3A and 3B). Mice receiving saline or scAAVs lacking miR were used as controls. scAAV8 miR-31/5 and scAAV8 miR-31/5,8,9 both caused rapid and significant reduction in HBsAg and circulating VPEs by 2 weeks after the injection. Values for HBsAg decreased to approximately 5% of the controls, while the circulating VPEs diminished to 1%–2% of the values obtained for animals treated with saline or the control scAAV8. These low levels of HBsAg and circulating VPEs were sustained over 32 of the 40 weeks of the analysis. Declining efficacy against markers of replication correlated with the concentrations of AAV genome equivalents that were detectable in hepatocytes (Figure S3). By 40 weeks, the average HBsAg concentrations and circulating VPEs in the serum of control animals also had diminished. In our hands, steady decline and variability in markers of HBV replication over a period of approximately 40 weeks is a feature of the transgenic mice used here. Normalizing the data to values obtained from animals treated with the vector lacking miR revealed similar efficacy and the inhibitory effect had diminished by 40 weeks following injection of the scAAVs (Figures S4A and S4B). This analysis enabled better discrimination between efficacy of scAAV8 miR-31/5,8,9 and scAAV8 miR-31/5. Average values for HBsAg and VPEs, although not statistically significant, were lower in animals treated with scAAV8 miR-31/5,8,9 than they were for mice receiving scAAV8 miR-31/5. Intrahepatic HBV core antigen (HBcAg), detected using immunohistochemistry, was carried out on mice that were killed at 2 weeks and 40 weeks after injection with scAAV miR-31/5 and scAAV8 miR-31/5,8,9 (Figures 3C and 3D). This analysis revealed decreased numbers of positive cells in sections obtained from animals that had received the scAAV8 miR-31/5,8,9 vector for both time points. A decreased number of HBcAg-positive cells from livers of animals treated with the scAAV miR-31/5 monomeric cassette was only observed at the later time point of 40 weeks, but not at 2 weeks. This better efficacy of the trimeric cassette correlates with our previous observations.20, 21 Although HBV core particles in the hepatocytes of HBV transgenic mice are stable,32 the diminished number of cells that were positive for the viral antigen at 40 weeks suggests that in this model, HBcAg may be a more sensitive indicator of HBV replication than are HBsAg and VPEs.
Figure 3.
Evaluation of Long-Term Effects In Vivo of HBV-Targeting scAAVs on Concentrations of HBV Surface Ag, Circulating Hepatitis B VPEs, and Intrahepatic HBcAg
(A) HBV transgenic mice were injected intravenously with saline (diamond), scAAV8 with no artificial miR (triangle), scAAV8 pri-miR-31/5 (square), or scAAV8 pri-miR-31/5,8,9 (circle). Each animal received a dose of 1 × 1011 scAAV8 particles. HBsAg in serum samples was measured thereafter for a 40 week period and values presented relative to those obtained immediately prior to scAAV injection. (B) Circulating VPEs were measured using real-time quantitative PCR, and the data are also given relative to time 0. (C) HBcAg-immunopositive hepatocytes were detected per high power field from sections of animals that were treated with scAAV containing no artificial miR or scAAV8 pri-miR-31/5,8,9. Analysis was carried out on mice that were killed at week 2 and week 40 after injection with scAAVs. The data are presented relative to the number of nuclei detected in fields from the mice that received scAAV8 without the artificial miR. (D) Representative fields of liver sections from animals analyzed in (C). The means are given and standard error of the mean is indicated by the error bars. The statistically significant differences between the control and experimental samples, determined using the Student’s two-tailed paired t test, are indicated by asterisks (*p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.005).
Intrahepatic Processing of miR-31/5 and miR-31/5,8,9
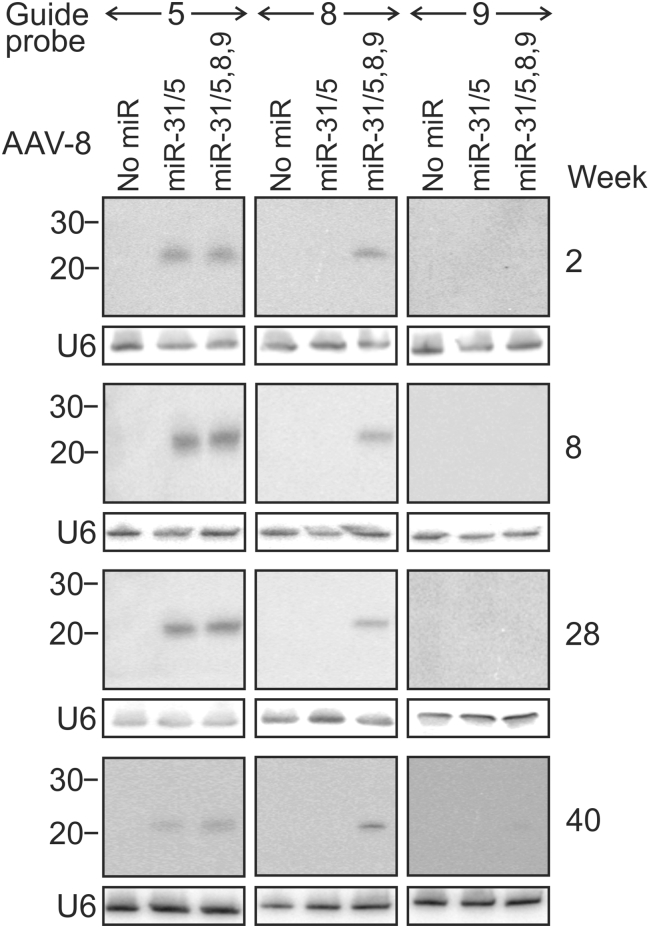
Northern blot hybridization was carried out to evaluate processing of apri-miRs in the livers of HBV transgenic mice. Animals were killed at 2, 8, 28, and 40 weeks after they had received the scAAVs. Probes complementary to guides 5, 8, and 9 were again used to detect the matured amiRs (Figure 4). As with the data obtained from analysis of cultured cells, the guide 5 sequence was most abundant and the RNA was present for the entire 40 weeks of the analysis. Guide 8 RNA was also detectable for 40 weeks in hepatic extracts from animals that had received scAAV8 miR-31/5,8,9 vectors. As expected, this sequence was not found in RNA isolated from mice treated with scAAV8 miR-31/5. Undetectable guide 9 sequences in extracts from mice treated with scAAV8 miR-31/5,8,9 correlates with data from previous studies20, 21 and observations in cultured cells (Figure 2A). Steady-state levels of guide 5 appeared to correlate with the intrahepatic copies of scAAV (Figure S3). However, guide 8 has a more constant concentration, despite the decrease in vector copy number over time. This apparent anomaly may result from greater sensitivity of the guide 8 probe for the mature miR. RNA species that are larger than the guides were absent in the blots that encompassed larger areas of the northern hybridization (Figure S5). This observation indicates that processing of the artificial cassettes is efficient and intermediates of the mature miRs comprising apri-miRs and apre-miRs exist transiently.
Figure 4.
Processing In Vivo of pri-miR-31/5 and pri-miR-31/5,8,9 Sequences Delivered with scAAV8
Northern blot hybridization was carried out on total RNA extracted from livers of mice that were treated with scAAV lacking artificial miR (No miR), scAAV8 miR-31/5, or scAAV8 miR-31/5,8,9. The blots were initially hybridized to probes complementary to the sequences of guides 5, 8, or 9. Thereafter, the blots were stripped and rehybridized with probes complementary to the U6 small nuclear RNA, which was used to confirm equal loading of the lanes. The animals were killed at weeks 2, 8, 28, or 40 after injection with the recombinant viral vectors. The positions of bands from the molecular weight markers corresponding to RNA fragments of 20 and 30 nt are indicated on the left.
Assessment of Toxic and Immunostimulatory Effects following Administration of scAAV8 miR-31/5,8,9 and scAAV8 miR-31/5 to HBV Transgenic Mice
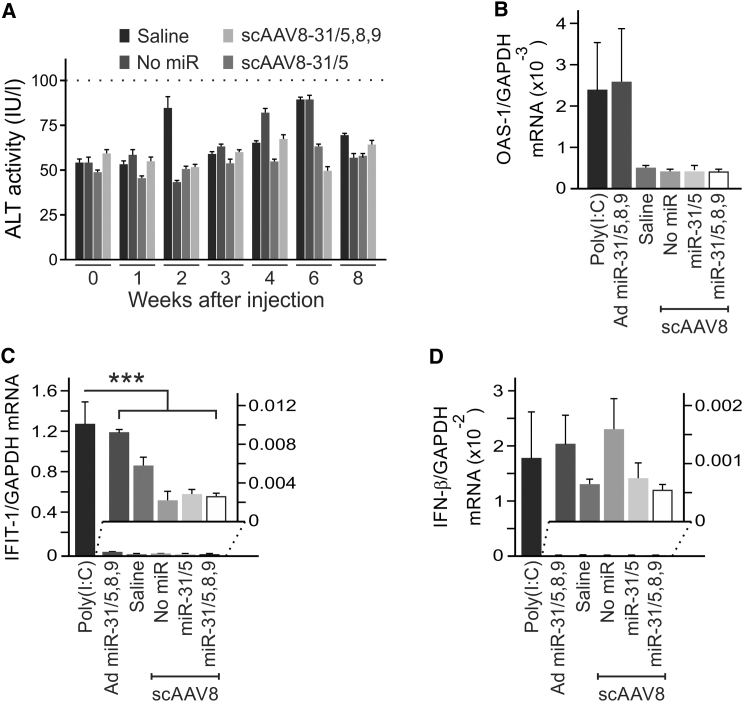
Toxicity caused by overexpression of RNAi activators may be significant and is potentially lethal.12, 18 Several analyses were therefore carried out to exclude toxicity of scAAV8 miR-31/5,8,9 and scAAV8 miR-31/5. The average mass of all groups of mice steadily increased for the duration of the 40 weeks of the study (Figure S6). No differences in weight gain between animals from each of the control and miR-treated groups were observed. A histological evaluation was also carried out to exclude evidence for microscopically detectable pathology. Sections stained with hematoxylin and eosin or Sirius red were within normal limits and there were no differences between groups (Figure S7). Measurement of alanine transaminase (ALT) activity in the serum also indicated that the scAAVs containing the MTTR miR-31-derived cassettes did not cause hepatotoxicity (Figure 5A). All groups of animals had average ALT activities that were below the upper limit of normal (<100 U/L) for a period of 8 weeks after receiving the vectors.
Figure 5.
Assessment of Hepatotoxicity and Induction of IFN Response Genes following Intravenous Administration of scAAVs
(A) HBV transgenic mice were injected with saline or the indicated scAAVs. Blood samples were collected prior to injection and at weeks 1, 2, 3, 4, 6, and 8 after administration. The threshold of the accepted normal range, <100 IU/L, is indicated. The means (±SEM) were calculated from replicate injections of four different mice. (C and D) Hepatic mRNA was prepared 6 hr after intravenous injection of mice with poly (I:C), saline, an adenoviral vector expressing miR-31/5,8,9 (Ad miR-31/5,8,9), scAAV with no artificial miR, scAAV8 pri-miR-31/5, or scAAV8 pri-miR-31/5,8,9. After reverse transcription, concentrations of mRNA from (B) OAS-1, (C) IFIT-1, and (D) IFN-β and GAPDH genes were determined using quantitative PCR. The data are presented as a ratio of the concentrations of mRNA from the IFN response genes relative to the concentration of the GAPDH mRNA. The values represent the means and standard error of the means following injection of four different animals. The statistically significant differences relative to the saline control, determined using the Student’s two-tailed paired t test, are indicated by asterisks (***p ≤ 0.005).
Innate immunostimulation following in vivo administration of exogenous RNAi activators is also potentially toxic. To evaluate activation of expression of genes that participate in the IFN response, RNA was extracted from mice 6 hr after they were injected with scAAV8 miR-31/5,8,9, scAAV8 miR-31/5, scAAV8 lacking miR, poly(I:C), adenoviral vectors expressing miR-31/5,8,9,33 or saline. Quantitative reverse transcription PCR was carried out to measure mRNA derived from IFN-β, IFN-induced protein with tetratricopeptide repeats (IFIT)-1, and oligoadenylate synthase-1 (OAS-1) genes relative to the amount of GAPDH mRNA (Figures 5B–5D). mRNA concentrations of the IFN response genes were similar in liver cells of mice that had received saline, the scAAV8 without miR, scAAV8 miR-31/5,8,9, and scAAV8 miR-31/5. These values were lower than those obtained from samples extracted from mice that had received poly (I:C), a positive control for stimulation of the innate immune response. Animals receiving recombinant adenoviruses (Ads) expressing miR-31/5,8,9 were also used as a control group.33 Although highly efficiently hepatotropic, Ads have innate immunostimulatory properties. Concentrations of transcripts from the IFN response genes in the mice receiving scAAV miR-31/5,8,9 and scAAV8 miR-31/5 were consistently lower than those detectable in mice receiving poly (I:C). The Ad miR-31/5,8,9 vector induced OAS-1 expression that was similar to that of the poly (I:C)-treated animals, but the stimulatory effect on IFIT-1 and IFN-β was minimal.
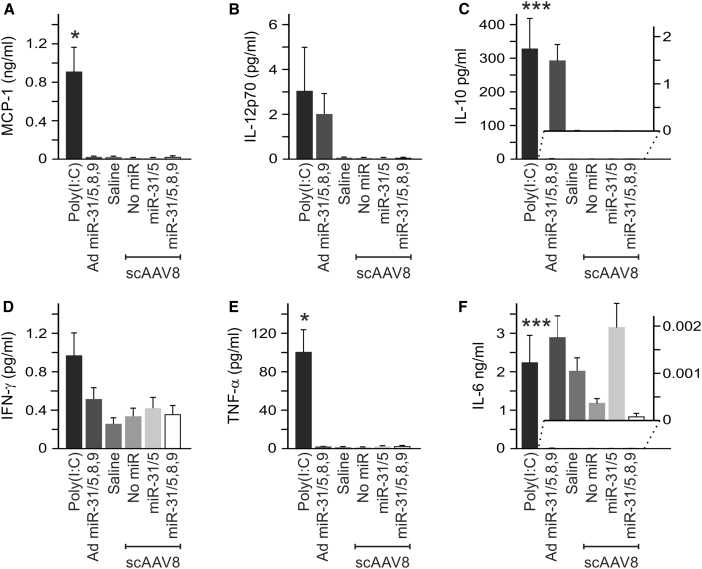
Effects of the scAAVs on release of pro-inflammatory cytokines were assessed as further assessment of innate immunostimulation. This was performed using a cytometric bead array (CBA) (Figures 6 and S8). Concentrations of interleukin 6 (IL-6), interleukin 10 (IL-10), monocyte chemoattractant protein-1 (MCP-1), interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin 12p70 (IL-12p70) were measured in the serum of mice 6 hr after they had received the scAAVs, poly (I:C), Ad miR-31/5,8,9, or saline. IL-6, IL10, MCP-1, and TNF-α were significantly elevated in the animals that received poly (I:C). In animals receiving saline and the scAAVs, serum concentrations of each of the cytokines were similar and low. Collectively, our findings indicate that the HBV-silencing effected by scAAV8 miR-31/5,8,9 and scAAV8 miR-31/5 mediates prolonged knockdown of markers of HBV replication without evidence for toxicity and activation of an immunostimulatory response.
Figure 6.
Evaluation of Cytokine Induction in HBV Transgenic Mice following Intravenous Administration of scAAVs Containing pri-miRs
(A–F) Concentrations of MCP-1 (A), IL-12p70 (B), IL-10 (C), IFN-γ (D), TNF-α (E), and IL-6 (F). Measurements were made using a cytometric bead array assay to quantify the cytokines at the time point of 6 hr after injection of the indicated scAAVs, adenoviral vector, or saline. Administration of poly (I:C) was used as the positive control. The means (±SEM) were calculated from replicate injections of four mice per group. The statistically significant differences relative to the saline control, determined using the Student’s two-tailed paired t test, are indicated by asterisks (*p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.005).
Discussion
Chronic infection with HBV continues to be an important global health problem. Available therapies have modest curative efficacy and consequently the risk of complicating liver cancer and cirrhosis remains high in carriers of the virus.34 To limit the life-threatening complications of the infection, there is a need to improve on current clinically useful anti-HBV agents. Availability of treatments that have a durable and safe effect against the virus is desirable to counter the stable replication intermediate comprising cccDNA. Several different approaches are being employed to eliminate HBV infection, and these include use of new nucleoside and nucleotide analogs, inhibitors of viral capsid assembly, immunomodulators, gene editing strategies, and silencing by harnessing the RNAi pathway. Harnessing gene therapy to treat HBV infection offers advantages, such as rational drug design, and the field is now advancing impressively. Advances with hepatotropic delivery of therapeutic nucleic acids have also been remarkable. Evidence now indicates that gene therapy-based treatment, alone or in combination with other therapeutics, is feasible and may lead to complete or functional cure from HBV infection.9, 35, 36
Renewable production of therapeutic gene silencers from stable DNA templates is useful to achieve sustained silencing. This feature is particularly important to inhibit gene expression of pathogens, such as HBV, that cause chronic infection. Pol III promoters, such as the U6 small nuclear (sn)RNA37 and RNase P H1,38 have been widely used to encode shRNAs that mimic pre-miRs. Small size and constitutive transcription of short sequences in most tissues are useful features of the Pol III promoters. Compatibility of DNA expression cassettes with viral vectors, such as AAVs, is valuable for delivery of therapeutic sequences. AAVs are well characterized, have a good safety profile, and use of the vectors has extensively been evaluated in clinical trials.39 The utility of AAVs was exploited here to generate hepatotropic self-complementary particles that delivered silencing sequences to the liver.
Demonstration that expression of HBV-targeting shRNAs from a U6 promoter may cause lethal toxicity in vivo was an important development.12, 18 The limited range of transcriptional control by Pol III promoters is a drawback for their therapeutic application. Pol II promoters have greater flexibility and ability to regulate transcription in a tissue-specific manner and respond to subtle regulatory signals. We harnessed this property to transcribe trimeric pri-miR mimics from the liver-specific MTTR Pol II promoter. Sustained expression from the MTTR Pol II promoter-containing cassettes enabled silencing of HBV replication over a prolonged period. This inhibitory action following single administration of the recombinant scAAVs has potential therapeutic utility and was an improvement over what has previously been described.40 The trimeric cassette has an important advantage of preventing emergence of HBV escape mutants. Michler and colleagues23 also demonstrated good silencing of HBV in transgenic mice following administration of 1 × 1011 scAAVs expressing a single apri-miR from a liver-specific promoter. This effect, which was observed after administration of a similar dose of scAAV to what we report here, accords with our observations. Chen and colleagues40 reported on inhibition of markers of HBV replication for 22 weeks following a single systemic administration of 1 × 1012 scAAVs to HBV transgenic mice, which is 10-fold higher than the number of vector particles that was injected in our study. Liver-specific expression of potent gene silencers that are more lastingly effective at a lower dose of vector particles are useful to limit side effects. Sequential administration of scAAVs pseudotyped with different capsids is useful to avoid a host’s immune response to the vectors and prolong anti-HBV efficacy.40 This feature is also applicable to scAAVs expressing miR-31/5,8,9.
Safety of exogenous RNAi activators is vitally important for their use in a therapeutic context. Unintended side effects may derive from immunostimulation by duplex RNA, silencing of host cellular sequences, disruption of natural RNAi mechanisms, and effects of the vectors. Our data have shown that scAAVs pseudotyped with capsid 8 to achieve hepatotropic delivery of apri-miRs do not have discernible toxic effects. Since the apri-miRs are transcribed within the nucleus, and unlike synthetic siRNAs delivered with non-viral vectors do not traverse the endosomal compartment, Toll-like receptor (TLR) activation is not expected. Although pattern recognition receptors may be activated in the cytoplasm, our observations show no evidence for stimulation of the IFN response by apri-miR-expressing scAAVs. Moreover, the scAAVs used here do not cause release of pro-inflammatory cytokines. Non-specific indicators of hepatotoxicity, including ALT and histological evaluation, were also not perturbed following scAAV administration. Markedly elevated ALT was a notable feature of the liver injury caused by expression of anti-HBV U6 shRNAs.12, 18 Collectively, our data indicate that scAAVs encoding mono- and trimeric HBV-targeting sequences safely achieve lasting silencing of the virus in vivo.
Developing new approaches to affect a cure from HBV infection has become a very active field of investigation (reviewed in Revill et al.34). Advances with improved understanding of the basic biology of HBV replication have facilitated identification of new targets that may be used to inactivate the virus. Improved insights into the deficiencies of innate, cell-mediated, and humoral immunity that occur in HBV chronic carriers have also led to new approaches to treatment. Demonstration that Toll-like receptor 7 stimulation41 and IFN-induced degradation of cccDNA by APOBEC3A and 3B,42 are active against the virus are interesting new advances. Use of gene editors to mutate HBV cccDNA specifically has also showed promise (reviewed in Ely et al.35). As with RNAi-based therapies, ensuring specificity of the therapeutic for the intended target is important. Overall, advancement of curative therapy for HBV infection is at an exciting stage. Ultimate successful treatment is likely to use combination strategies, and RNAi-based therapeutic approaches such as are described here may be a useful component of future therapies.
Materials and Methods
Plasmids Encoding Viral Targets and Anti-HBV Sequences
Anti-HBV sequences targeting X were extracted from previously described pTZ57R plasmid containing miR-31/5 or miR-31/5,8,9 sequences which were each linked to the MTTR promoter.21, 28 Both miR-31/5 and miR-31/5,8,9 MTTR cassettes were excised with Asc I, blunt ended, and then inserted into Eco RV sites of the scAAV-generating transgene plasmid (pBS-H1-RSV-GFP).43 To limit risk of recombination during scAAV propagation, this plasmid contains ITRs from serotype 2 (ITR2) and serotype 4 (ITR4) that flank the transgene. Insertion of MTTR miR-31/5 and MTTR miR-31/5,8,9 resulted in substitution of the antiviral sequences for the RSV-GFP cassette to create pAAVMTTR31/5 and pAAVMTTR31/5,8,9. To create the empty plasmid (pAAV), the GFP sequence was deleted from pBS-H1-RSV-GFP by digestion with Nhe I and Bam HI, blunt ending, and then self-ligation of the backbone. Plasmids encoding the individual apri-miR cassettes under control of the CMV promoter/enhancer, together with mutant and wild-type targets have been described previously.20 Hydrodynamic injection with the pCH-9/309129 replication competent sequence, plasmids containing antiviral cassettes, and targets was carried out as described.20
Production and Propagation of Anti-HBV AAVs
scAAV vectors produced in this study were pseudotyped with serotype 2 (scAAV2) or serotype 8 (scAAV8) capsids for use in cell culture or in vivo, respectively. Large scale production, amplification, and purification of scAAVs were carried out according to published methods with minor modifications.43, 44 Briefly, HEK293 cells were co-transfected with recombinant scAAV genome-containing plasmid (pAAVMTTR31/5 or pAAVMTTR31/5,8,9, or pAAV, or pBS-H1-RSV-GFP), the plasmid carrying AAV2 or AAV8 rep and cap genes, and plasmid encoding adenoviral helper genes, pXX-6 and pACG, respectively.43 After 72 hr of incubation at 37°C, cells were harvested and lysed using three cycles of freeze thawing. To digest the host DNA and RNA, lysates were incubated with 50 U/mL of benzonase at 37°C for 1 hr. The lysates were then cleared by centrifugation and the supernatants collected. The virus was purified by iodixanol gradient centrifugation. There were 50 μL aliquots of the pure virus that were stored at −80°C. Titers of viral particle preparations were determined using quantitative PCR (qPCR) with primer sets targeting the promoter regions of the vectors (MTTR Forward: 5′ GCACTGGGAGGATGTTGAGT 3′ and MTTR Reverse: 5′ CCCCTGTTCAAACATGTCCT3′ or RSV Forward: 5′TCTGAGGGGACTAGGGTGTG3′ and RSV Reverse: 5′GGACTCCTAACCGCGTACAA3′). Preparation of the adenoviral vectors used as controls for some of the investigations has been described previously.28
Detection of Markers of HBV Replication
To assess inhibition of HBV gene expression in Huh7 cells, cultures were initially transfected with an HBV replication competent plasmid containing a greater-than-genome length viral sequence (pCH-9/3091).29 After 5 hr, the transfected cells were infected with AAV at an MOI of 1,000 to 1,000,000. HBV replication inhibition in vivo was assessed by injecting six HBV transgenic mice per group with 1 × 1011 viral particles via a tail vein. HBsAg was measured using the Monolisa Ag HBs Plus Immunoassay Kit (Bio-Rad). Assays were carried out on culture supernatants at 48 and 72 hr post AAV infection or in serum of mice over a period of 40 weeks. Serum HBV particle equivalents were measured using real-time qPCR with FastStart SYBR Green Master Mix (Roche Diagnostics). Primers specific for the HBV surface ORF were used as previously reported.45 For HBV core antigen staining, AAV-infected mice were sacrificed 2 or 40 weeks after injection and livers fixed in 10% buffered formalin (Sigma). Fixed tissues were embedded in paraffin and sectioned according to standard procedures. Immunohistochemical staining of tissue sections was performed using anti-HBV core antigen primary antibody (HBcAg, Abcam) and an Ultra-Sensitive ABC Peroxidase Staining Kit (Pierce, Thermo Scientific). All procedures were carried out according to manufacturers’ instructions.
Northern Blot Hybridization
Expression and processing of anti-HBV pri-miRs in vitro was assessed by infecting liver derived Huh7 cells with AAVs at a MOI of 100,000. Using TRI Reagent (Sigma), total RNA was extracted from cells 3 days after the infection. To assess pri-miR expression and processing in vivo, HBV transgenic mice were used according to a protocol approved by the University of the Witwatersrand Animal Ethics Screening Committee. A dose of 1 × 1011 viral particles/mouse was injected via a tail vein. Livers were then harvested from mice killed at 2, 8, 28, and 40 weeks after scAAV administration. Total RNA was isolated from liver homogenates using TRI Reagent (Sigma). Northern blot hybridization analysis of 30 μg of total liver RNA was performed according to previously described methods.45 Radioactively labeled probes with the following sequences were used to detect mature guides: guide 5 probe, 5′CCGTGTGCACTTCGCTTC 3′; guide 8 probe, 5′CAATGTCAACGACCGACC 3′; or guide 9 probe, 5′ TAGGAGGCTGTAGGCATA 3′. After stripping, blots were rehybridized to a U6 snRNA probe, with sequence 5′ TAGTATATGTGCTGCCGAAGCGAGCA 3′, to verify equal loading of RNA in each of the lanes.20
Measurement of Inflammatory Markers in Mice
To detect IFN-β, OAS-1, and IFIT-1 transcript concentrations, livers from mice were harvested 6 hr after AAV injection and RNA isolated using TRI Reagent (Sigma). 1 μg of RNA was used for reverse transcription using a QuantiTect Reverse Transcription Kit (QIAGEN). Resultant cDNA was then used for SYBR green-based real-time qPCR using FastStart SYBR Green Master Mix (Roche Diagnostics).
To assess induction of release of inflammatory cytokines by AAVs, serum concentrations of IL-6, IL-10, MCP-1, IFN-γ, TNF-α, and IL-12p70 were measured. Assays using a mouse inflammation kit (BD Biosciences) were carried out on serum taken immediately before and 6 hr after scAAV administration to groups of mice, which each comprised four animals. CBA reactions were run on a Fortessa LSR flow cytometer using the BD FACSDiva software. Data were analyzed using FCAP Array software 3.0 (BD Biosciences).
Analysis of Liver Toxicity
ALT activity was measured in serum samples collected at weeks 1, 2, 3, 4, 6, and 8 after scAAV injections. Assays were carried out using an Advia 1800 Chemistry System (Siemens) at the accredited facilities of the South African National Health Laboratory Service (NHLS, Johannesburg, South Africa). To assess for change in liver tissue morphology after AAV injection, livers were harvested from mice sacrificed at 2 or 40 weeks after injection with 1 × 1011 AAV viral particles and fixed in 10% buffered formalin (Sigma). The tissues were embedded in paraffin, sectioned, and then stained with hematoxylin and eosin (H&E) or Sirius red according to standard procedures (AMPATH laboratories, Auckland Park, South Africa).
Statistical Analysis
Statistical calculations were performed using the GraphPad Prism software (GraphPad Software). Data were expressed as the mean ± SEM. Differences were determined using the Student’s two-tailed t test and were considered significant when p ≤ 0.05.
Author Contributions
M.B.M., A.E., and P.A. conceptualized the study. M.B.M. carried out most of the experimental work with some assistance from A.E. W.G. provided interpretation of hepatic pathology associated with administration of recombinant AAVs. P.A. oversaw the research and wrote the manuscript with input from the other authors.
Acknowledgments
Financial assistance that was received from the South African National Research Foundation (NRF, GUNs 81768, 81692, 68339, 85981, and 77954), South African Medical Research Council, Johnson and Johnson Innovation, and the Poliomyelitis Research Foundation (PRF) is gratefully acknowledged. Help with tissue sectioning, carried out by Nerisha Phekoo, is appreciated. Plasmids required for preparation of scAAVs were kindly donated by Dr. Dirk Grimm, Heidelberg University.
Footnotes
Supplemental Information includes eight figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.04.007.
Supplemental Information
References
- 1.WHO. (2013). Hepatitis B virus Fact sheet No. 204 (Updated March 2015). http://www.who.int/mediacentre/factsheets/fs204_Jul2014/en/.
- 2.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montuclard C., Hamza S., Rollot F., Evrard P., Faivre J., Hillon P., Di Martino V., Minello A. Causes of death in people with chronic HBV infection: A population-based cohort study. J. Hepatol. 2015;62:1265–1271. doi: 10.1016/j.jhep.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Parvez M.K., Arbab A.H., Al-Dosari M.S., Al-Rehaily A.J. Antiviral natural products against chronic hepatitis B: recent developments. Curr. Pharm. Des. 2016;22:286–293. doi: 10.2174/1381612822666151112152733. [DOI] [PubMed] [Google Scholar]
- 5.Yang N., Bertoletti A. Advances in therapeutics for chronic hepatitis B. Hepatol. Int. 2016;10:277–285. doi: 10.1007/s12072-015-9661-x. [DOI] [PubMed] [Google Scholar]
- 6.Zeisel M.B., Lucifora J., Mason W.S., Sureau C., Beck J., Levrero M., Kann M., Knolle P.A., Benkirane M., Durantel D. Towards an HBV cure: state-of-the-art and unresolved questions--report of the ANRS workshop on HBV cure. Gut. 2015;64:1314–1326. doi: 10.1136/gutjnl-2014-308943. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Y., Yamamoto T., Cullen J., Saputelli J., Aldrich C.E., Miller D.S., Litwin S., Furman P.A., Jilbert A.R., Mason W.S. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 2001;75:311–322. doi: 10.1128/JVI.75.1.311-322.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addison W.R., Walters K.A., Wong W.W., Wilson J.S., Madej D., Jewell L.D., Tyrrell D.L. Half-life of the duck hepatitis B virus covalently closed circular DNA pool in vivo following inhibition of viral replication. J. Virol. 2002;76:6356–6363. doi: 10.1128/JVI.76.12.6356-6363.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivacik D., Ely A., Arbuthnot P. Countering hepatitis B virus infection using RNAi: how far are we from the clinic? Rev. Med. Virol. 2011;21:383–396. doi: 10.1002/rmv.705. [DOI] [PubMed] [Google Scholar]
- 10.Chinnappan M., Singh A.K., Kakumani P.K., Kumar G., Rooge S.B., Kumari A., Varshney A., Rastogi A., Singh A.K., Sarin S.K. Key elements of the RNAi pathway are regulated by hepatitis B virus replication and HBx acts as a viral suppressor of RNA silencing. Biochem. J. 2014;462:347–358. doi: 10.1042/BJ20140316. [DOI] [PubMed] [Google Scholar]
- 11.Song E., Lee S.K., Dykxhoorn D.M., Novina C., Zhang D., Crawford K., Cerny J., Sharp P.A., Lieberman J., Manjunath N., Shankar P. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 2003;77:7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm D., Wang L., Lee J.S., Schürmann N., Gu S., Börner K., Storm T.A., Kay M.A. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J. Clin. Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y.P., Haasnoot J., ter Brake O., Berkhout B., Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ely A., Naidoo T., Mufamadi S., Crowther C., Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol. Ther. 2008;16:1105–1112. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- 15.Gish R.G., Yuen M.F., Chan H.L., Given B.D., Lai C.L., Locarnini S.A., Lau J.Y., Wooddell C.I., Schluep T., Lewis D.L. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antiviral Res. 2015;121:97–108. doi: 10.1016/j.antiviral.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H., Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherer L.J., Frank R., Rossi J.J. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 19.Aagaard L.A., Zhang J., von Eije K.J., Li H., Saetrom P., Amarzguioui M., Rossi J.J. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ely A., Naidoo T., Arbuthnot P. Efficient silencing of gene expression with modular trimeric Pol II expression cassettes comprising microRNA shuttles. Nucleic Acids Res. 2009;37:e91. doi: 10.1093/nar/gkp446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivacik D., Ely A., Ferry N., Arbuthnot P. Sustained inhibition of hepatitis B virus replication in vivo using RNAi-activating lentiviruses. Gene Ther. 2015;22:163–171. doi: 10.1038/gt.2014.94. [DOI] [PubMed] [Google Scholar]
- 22.Costa R.H., Grayson D.R. Site-directed mutagenesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactions with its enhancer region. Nucleic Acids Res. 1991;19:4139–4145. doi: 10.1093/nar/19.15.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michler T., Große S., Mockenhaupt S., Röder N., Stückler F., Knapp B., Ko C., Heikenwalder M., Protzer U., Grimm D. Blocking sense-strand activity improves potency, safety and specificity of anti-hepatitis B virus short hairpin RNA. EMBO Mol. Med. 2016;8:1082–1098. doi: 10.15252/emmm.201506172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hösel M., Lucifora J., Michler T., Holz G., Gruffaz M., Stahnke S., Zoulim F., Durantel D., Heikenwalder M., Nierhoff D. Hepatitis B virus infection enhances susceptibility toward adeno-associated viral vector transduction in vitro and in vivo. Hepatology. 2014;59:2110–2120. doi: 10.1002/hep.26990. [DOI] [PubMed] [Google Scholar]
- 26.Koike K. Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer Lett. 2009;286:60–68. doi: 10.1016/j.canlet.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Zoulim F., Saputelli J., Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mowa M.B., Crowther C., Ely A., Arbuthnot P. Inhibition of hepatitis B virus replication by helper dependent adenoviral vectors expressing artificial anti-HBV pri-miRs from a liver-specific promoter. BioMed Res. Int. 2014;2014:718743. doi: 10.1155/2014/718743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marion P.L., Salazar F.H., Liittschwager K., Bordier B.B., Seeger C., Winters M.A., Cooper A.D., Cullen J.M. A transgenic mouse lineage useful for testing antivirals targeting hepatitis B virus. In: Schinazi R.F., editor. Frontiers in Viral Hepatitis. Elsevier; Amsterdam: 2003. pp. 197–210. [Google Scholar]
- 31.Guidotti L.G., Matzke B., Schaller H., Chisari F.V. High-level hepatitis B virus replication in transgenic mice. J. Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guidotti L.G., Matzke B., Pasquinelli C., Shoenberger J.M., Rogler C.E., Chisari F.V. The hepatitis B virus (HBV) precore protein inhibits HBV replication in transgenic mice. J. Virol. 1996;70:7056–7061. doi: 10.1128/jvi.70.10.7056-7061.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crowther C., Mowa M.B., Ely A., Arbuthnot P.B. Inhibition of HBV replication in vivo using helper-dependent adenovirus vectors to deliver antiviral RNA interference expression cassettes. Antivir. Ther. (Lond.) 2014;19:363–373. doi: 10.3851/IMP2713. [DOI] [PubMed] [Google Scholar]
- 34.Revill P., Testoni B., Locarnini S., Zoulim F. Global strategies are required to cure and eliminate HBV infection. Nat. Rev. Gastroenterol. Hepatol. 2016;13:239–248. doi: 10.1038/nrgastro.2016.7. [DOI] [PubMed] [Google Scholar]
- 35.Ely A., Moyo B., Arbuthnot P. Progress with developing use of gene editing to cure chronic infection with hepatitis B virus. Mol. Ther. 2016;24:671–677. doi: 10.1038/mt.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiffer J.T., Aubert M., Weber N.D., Mintzer E., Stone D., Jerome K.R. Targeted DNA mutagenesis for the cure of chronic viral infections. J. Virol. 2012;86:8920–8936. doi: 10.1128/JVI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertrand E., Castanotto D., Zhou C., Carbonnelle C., Lee N.S., Good P., Chatterjee S., Grange T., Pictet R., Kohn D. The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 38.Baer M., Nilsen T.W., Costigan C., Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mingozzi F., High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 40.Chen C.C., Sun C.P., Ma H.I., Fang C.C., Wu P.Y., Xiao X., Tao M.H. Comparative study of anti-hepatitis B virus RNA interference by double-stranded adeno-associated virus serotypes 7, 8, and 9. Mol. Ther. 2009;17:352–359. doi: 10.1038/mt.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanford R.E., Guerra B., Chavez D., Giavedoni L., Hodara V.L., Brasky K.M., Fosdick A., Frey C.R., Zheng J., Wolfgang G. GS-9620, an oral agonist of Toll-like receptor-7, induces prolonged suppression of hepatitis B virus in chronically infected chimpanzees. Gastroenterology. 2013;144:1508–1517. doi: 10.1053/j.gastro.2013.02.003. 1517 e1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucifora J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X., Sprinzl M.F., Koppensteiner H., Makowska Z., Volz T. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimm D., Pandey K., Nakai H., Storm T.A., Kay M.A. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarty D.M., Monahan P.E., Samulski R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 45.Crowther C., Mowa B., Arbuthnot P. Hepatic delivery of artificial micro RNAs using helper-dependent adenoviral vectors. In: Shum K., Rossi J., editors. Volume 1364. Springer New York; 2016. pp. 249–260. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.