Abstract
The most common variant causing Pompe disease is c.-32-13T>G (IVS1) in the acid α-glucosidase (GAA) gene, which weakens the splice acceptor of GAA exon 2 and induces partial and complete exon 2 skipping. It also allows a low level of leaky wild-type splicing, leading to a childhood/adult phenotype. We hypothesized that cis-acting splicing motifs may exist that could be blocked using antisense oligonucleotides (AONs) to promote exon inclusion. To test this, a screen was performed in patient-derived primary fibroblasts using a tiling array of U7 small nuclear RNA (snRNA)-based AONs. This resulted in the identification of a splicing regulatory element in GAA intron 1. We designed phosphorodiamidate morpholino oligomer-based AONs to this element, and these promoted exon 2 inclusion and enhanced GAA enzyme activity to levels above the disease threshold. These results indicate that the common IVS1 GAA splicing variant in Pompe disease is subject to negative regulation, and inhibition of a splicing regulatory element using AONs is able to restore canonical GAA splicing and endogenous GAA enzyme activity.
Keywords: Pompe disease, U7 snRNA screen, antisense oligonucleotides, exon inclusion, pre-mRNA splicing, metabolic disorder, lysosomal storage disorder, IVS1, lysosomal storage disease, acid α-glucosidase, GAA, skeletal muscle
Introduction
Pre-mRNA splicing is a highly regulated process, the outcome of which is critical for homeostasis and disease. The diversity of splicing variants positively correlates with evolutionary complexity, as it allows for an expansion of the possible protein isoforms that are derived from the same genetic information.1 The regulation of splicing is still relatively poorly understood, which is caused by the many factors that can affect splicing outcome. At the level of RNA sequences, splicing regulatory elements are loosely defined. The splice site junctions, as well as the polypyrimidine (pY) tract and branch point, are relatively easy to identify. Splice regulatory elements exist, including the exonic and intronic splice enhancer and silencer elements (ESEs, ESSs, ISEs, and ISSs, respectively), but these are more difficult to predict.2 The expression levels of splicing regulatory proteins can also play an important role in splicing outcome.3 Furthermore, the speed of RNA polymerase II transcription is thought to affect splicing efficiency, because splicing mainly occurs co-transcriptionally, and the available splice sites that are present in the protruding pre-mRNA within a certain time frame compete for the splicing machinery.4, 5 Additional levels of regulation are chromatin modifications and the composition of the UTRs.6, 7 All together, these diverse levels of regulation highlight the requirement for experimental testing of alternative splicing to validate predictions.
Disruption of splicing has been documented in a large number of human disorders. Approximately 9% of pathogenic variants annotated in the Human Gene Mutation Database are linked to splicing defects (http://www.hgmd.cf.ac.uk/). In this database, variants that affect splicing predominantly concern those that are located at splice site junctions.8 This is caused by the inherent detection bias for these variants, which are mostly identified using splice prediction algorithms. However, variants can also alter splicing by the generation of new splice sites9, 10 or by disturbing or generating other splice regulatory elements.11, 12 In addition, variants in the coding region can have more than one effect (i.e., a missense variant can also have an effect on splicing).9 Furthermore, indirect effects on target genes are known (e.g., in response to mutations in master splicing factors such as the muscleblind family, SF3B1, and U2AF35).13, 14 The consequences of such events can be diverse and may include (partial) exon skipping and/or (partial) intron retention. The final mRNA may be in frame, which in certain cases may result in a protein product that retains some biological activity. A reading frameshift usually results in mRNA decay and absence of a protein product, although the extent at which decay occurs may vary. It is likely that many more pathogenic splicing variants exist that are located outside the region of canonical splice sites, and that the percentage of identified variants that affect splicing will increase in the future.
Antisense oligonucleotides (AONs) can modulate splicing by binding to the pre-mRNA and blocking splicing regulatory sequences. One type of AON has been modified from the naturally occurring U7 small nuclear RNA (snRNA), which is normally utilized in histone pre-mRNA processing but has been modified to target the pre-mRNA of choice.15 U7 snRNA-based AONs are stabilized by a stem loop and also contain an antisense sequence that is used for targeting.16 Another strategy is the use of chemically modified AONs that are capable to bind to RNA via Watson-Crick base pairing but are insensitive to RNase-mediated degradation due to their modified backbone chemistry. Various backbones with different properties are known, which include phosphorodiamidate morpholino oligomer (PMO), 2′-O-methyl phosphorothioate, 2-O-methoxyethyl phosphorothioate, and tricyclo-DNA modifications.17
A straightforward approach to modulate splicing using AONs is to block a splice junction. This will result in exon skipping, which can be advantageous, for example, to bypass a mutation hotspot and to restore the reading frame. This will only work when the resulting truncated protein is at least partially active, which is the case for the dystrophin protein, providing a potential therapy for Duchenne muscular dystrophy.18, 19 This approach has been tested in multiple clinical trials and has moved forward toward conditional approval by the US Food and Drug Administration (FDA).20 Many other examples have been published, in which a cryptic splice site is blocked with an AON to restore canonical splicing (reviewed in Havens and Hastings17). We recently showed that this is possible in the case of GAA variants that cause Pompe disease.9 Restoration of splicing becomes more challenging in cases where a splicing variant causes exon skipping and requires promotion of exon inclusion. When there is no clear cryptic splice site involved that can be blocked by an AON, such cases require the identification of ISS or ESS elements that are critical for exon inclusion. Blocking of these splice silencers with an AON may in turn promote exon inclusion. Few examples for promotion of exon inclusion exist; notably, for spinal muscular atrophy, caused by variants in the SMN1 gene, AONs have been designed that reactivate the normally silent sister gene SMN2. SMN2 contains an exonic variant that causes exon skipping, and AONs have been identified that block ESS or ISS sequences and promote exon inclusion.21, 22 One of these, which targets the ISS, is currently being tested in clinical trials.23
Because AONs are sequence specific, they are usually only suitable for a subgroup of patients with similar gene variants. In Pompe disease, a common splicing variant occurs in the majority of Caucasian patients with the childhood/adult form. This well-described variant, c.-32-13T>G (also known as IVS1, or r.-32-13u>g at the RNA level), is located in the pY tract of exon 2 of the acid α-glucosidase (GAA) pre-mRNA and causes weakening of the splice acceptor site, resulting in complete or partial skipping of exon 2.24, 25, 26, 27 This exon contains the translation start codon, and its skipping results in mRNA degradation and a subsequent reduction of GAA protein production. Approximately 10%–15% of transcripts are spliced normally and produce a low level of wild-type GAA protein, which explains why these patients develop the childhood/adult form of Pompe disease. A minimum GAA activity of 20% of the average healthy control values is required to prevent the disease.28 The restoration of splicing from the IVS1 variant using AONs would potentially provide a therapeutic option for the majority of Caucasian patients with Pompe disease. This may be beneficial in addition to or instead of the current enzyme replacement therapy, which has a partial and heterogeneous response and is extremely expensive. However, splicing correction from the IVS1 variant requires promotion of exon inclusion, and no negatively acting splicing regulatory sequences in the GAA gene that are amenable to inhibition using AONs are currently known.
Here, we used a screen to modulate endogenous GAA splicing using U7 snRNA-based AONs expressed via lentiviral transduction in patient-derived primary fibroblasts. This resulted in the identification of cis-acting splicing regulatory elements that were subsequently tested using PMO-based AONs. Two overlapping AONs were identified that promoted inclusion of exon 2 in cells from patients carrying the IVS1 variant. In cells from healthy controls, the AONs did not change GAA splicing or expression, confirming that they acted on splicing rather than elevating total GAA transcript levels. The AONs increased GAA enzymatic activity in patient-derived cells to above the disease threshold of 20%. These findings demonstrate the feasibility to correct aberrant splicing from the common IVS1 GAA variant by blocking a negative splicing element to promote exon inclusion.
Results
Aberrant Splicing of GAA Pre-mRNA in the Presence of the IVS1 Variant
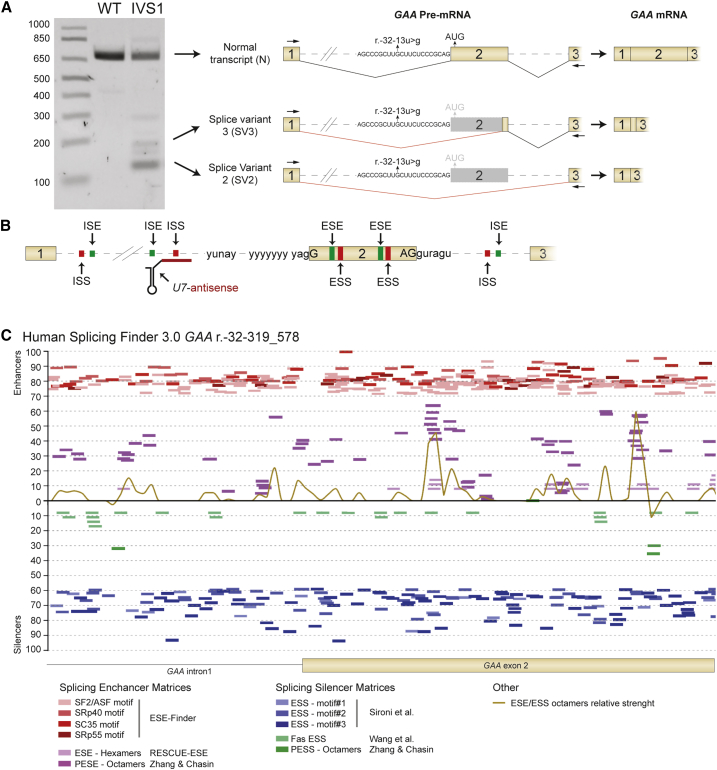
Previous work resulted in the characterization of aberrant splicing caused by the IVS1 GAA variant.24, 25, 26, 27 Splicing products include wild-type GAA mRNA (N) caused by leaky normal splicing, partial skipping of exon 2 caused by utilization of a cryptic splice site in exon 2 (SV3), and full skipping of exon 2 (SV2). These products can be identified by flanking exon RT-PCR in primary fibroblasts using primers that anneal to exons 1 and 3 (Figure 1A). Splicing is subject to regulation by additional cis-acting RNA sequences besides the sequences surrounding the canonical splice sites. These include ESSs, ISSs, ESEs, and ISEs and can be located distant from canonical splice sites (hypothetical example given in Figure 1B). Approximately 10%–15% of the GAA pre-mRNA is correctly spliced, indicating that the pathogenic effect of the IVS1 variant is not fully detrimental to splicing of GAA exon 2. We hypothesized that it may be possible to promote exon 2 inclusion by inhibition of a splicing negatively acting splicing regulatory element using an AON. To test this, in silico prediction was performed using several algorithms, which resulted in a plethora of splicing regulatory elements (Figure 1C). However, the results were algorithm dependent and many predicted enhancer and silencer sequences overlapped without indicating an obvious candidate silencer element that could be amenable to inhibition by an AON.
Figure 1.
In Silico Prediction to Identify Splicing Regulatory Elements Surrounding the IVS1 Variant
(A) Outline of the three major splicing products of the GAA pre-mRNA caused by the IVS1 variant in a patient-derived primary fibroblast. The gel illustrates the results of flanking exon RT-PCR analysis of GAA exon 2 using primers that anneal to exon 1 and exon 3. Loading from left to right is as follows: DNA size markers (in basepairs); WT indicates control fibroblasts and IVS1 denotes fibroblasts from patient 1. Cartoons of pre-mRNAs illustrate splicing events as described.24, 25, 26, 27 The location of the r.-32-13u>g (IVS1) variant in the pY tract is indicated. Spliced mRNA cartoons are shown on the far right. (B) Cartoon showing hypothetical splicing regulatory elements that may be subject to modulation (e.g., by a U7 snRNA). (C) In silico prediction in Human Splicing Finder 3.0 (http://www.umd.be/HSF3/) of exonic and intronic splicing silencers surrounding the GAA IVS1 variant. Algorithms used are indicated below the graph. PESE, putative exonic splicing enhancers; PESS, putative exonic splicing silencers.
Identification of Repressors of GAA Exon 2 Inclusion
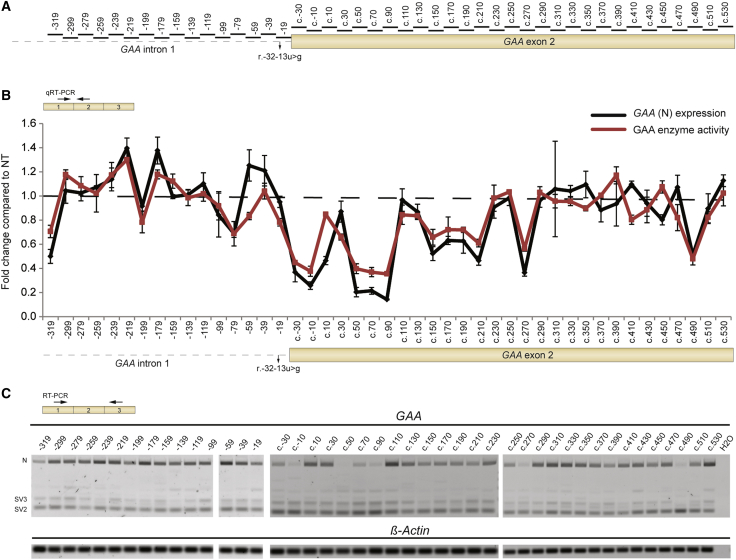
To identify splicing regulatory elements experimentally, an unbiased screen was performed using a non-overlapping tiling array of AONs that cover part of intron 1 and the complete exon 2 of the GAA pre-mRNA (Figure 2A). Because of the high costs involved when testing a large series of chemically modified AONs, AONs were expressed as U7 snRNAs using a lentiviral vector. The original U7 snRNA vector was adapted to enable one-step cloning of an AON and intermediate throughput screening (Figure S1A).29 To validate the U7 snRNA vector, a control experiment was performed on primary fibroblasts derived from a patient with adult Pompe disease (patient 1), who carried the IVS1 variant on one allele and the c.525delT on the second allele. A U7 snRNA-based AON was targeted to the splice sites of exon 4 of cyclophilin A (CypA) pre-mRNA (Figure S1B). This AON was capable of inducing skipping of exon 4, as shown by flanking exon RT-PCR (Figure S1C) and exon internal qRT-PCR analysis (Figure S1D). This confirmed previous reports29 and demonstrated that the modified U7 snRNA construct can be used to modulate splicing. Subsequently, the U7 snRNA screen using the AONs indicated in Figure 2A was performed on fibroblasts from patient 1. The screen resulted in the identification of a number of U7 snRNA-based AONs that modulated inclusion of exon 2 in the GAA mRNA, as shown with qRT-PCR analysis (Figure 2B, black line). Importantly, only mRNA from the allele carrying the IVS1 variant was detected due to the frameshift induced by the c.525delT variant on the second allele, which results in mRNA degradation. Notably, U7 snRNAs targeting two regions in intron 1, at c.-32-179 and c.-32-219, promoted inclusion of GAA exon 2. Exclusion of exon 2 was promoted by U7 snRNAs that targeted regions in the 5′ part of exon 2. Similar results were obtained using flanking exon RT-PCR analysis (Figure 2C). Promotion and inhibition of exon 2 inclusion resulted in increased and decreased GAA enzymatic activity, respectively (Figure 2B, compare black and red lines).
Figure 2.
U7 snRNA Screen to Identify Splicing Regulatory Elements Involved in GAA Exon 2 Splicing
(A) Locations of U7 snRNA-based AONs generated for the screen in (B). (B) Screen to identify splicing regulatory elements that negatively regulate splicing of GAA exon 2. Primary fibroblasts from patient 1 (IVS1, c.525delT) were transduced with U7 snRNA-expressing lentiviruses (200 ng p24 protein as determined by ELISA). The effects on GAA exon 2 expression were measured using qRT-PCR (black line; GAA [N] expression; primers are indicated in the upper left cartoon). Effects on GAA enzymatic activity are indicated by the red line. The cartoon of GAA pre-mRNA below the graph indicates the positions of the AONs tested. Data are expressed relative to non-transduced (NT) fibroblasts and represent means ± SD of three biological replicates. Samples were normalized for β-Actin expression. (C) The experiment in (B) was also analyzed by flanking exon RT-PCR of GAA exon 2. β-Actin mRNA was used as loading control. Primers are indicated in the upper left cartoon. (n = 3).
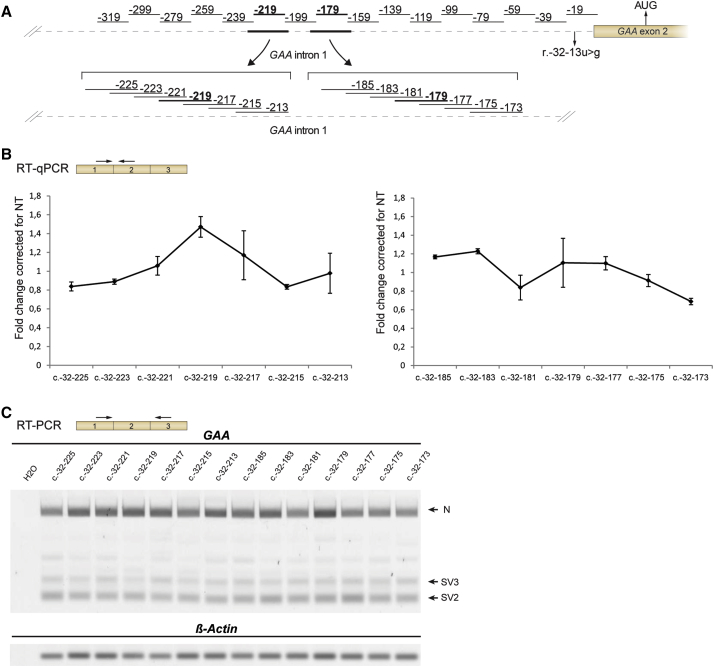
Next, the effect of the lentiviral amount was tested using p24 ELISA; 200 ng yielded an optimal ratio between biological effect and nonspecific reduction of cell viability and GAA expression seen at high viral amounts (Figure S2). To fine-tune the optimal location of U7 snRNAs targeting c.-32-179 and c.-32-219, a microwalk was performed around these regions with U7 snRNAs that shifted 2 nt (Figure 3A). This showed that the locations identified using the initial U7 snRNA screen were peak values. In addition, the microwalk revealed two nearby locations at c.-32-183 and c.-32-185 whose inhibition promoted exon 2 inclusion (Figures 3B and 3C). Taken together, the U7-based snRNA screen of intron 1 and exon 2 identified regions potentially involved in repression of GAA exon 2 inclusion.
Figure 3.
U7 snRNA-Based Miniscreen of Targets Identified with the Large Screen
(A) Two hits from the screen shown in Figures 2B and 2C (in bold) were further tested in a microwalk using the U7 snRNA system. Primer locations are indicated in the cartoon. (B) Results of the microwalk, as analyzed by qRT-PCR. (C) As in (B), using RT-PCR analysis. Results are expressed relative to non-transduced fibroblasts and represent means ± SD of three biological replicates.
Blockage of Splicing Repressor Sequences Using Antisense Oligonucleotides
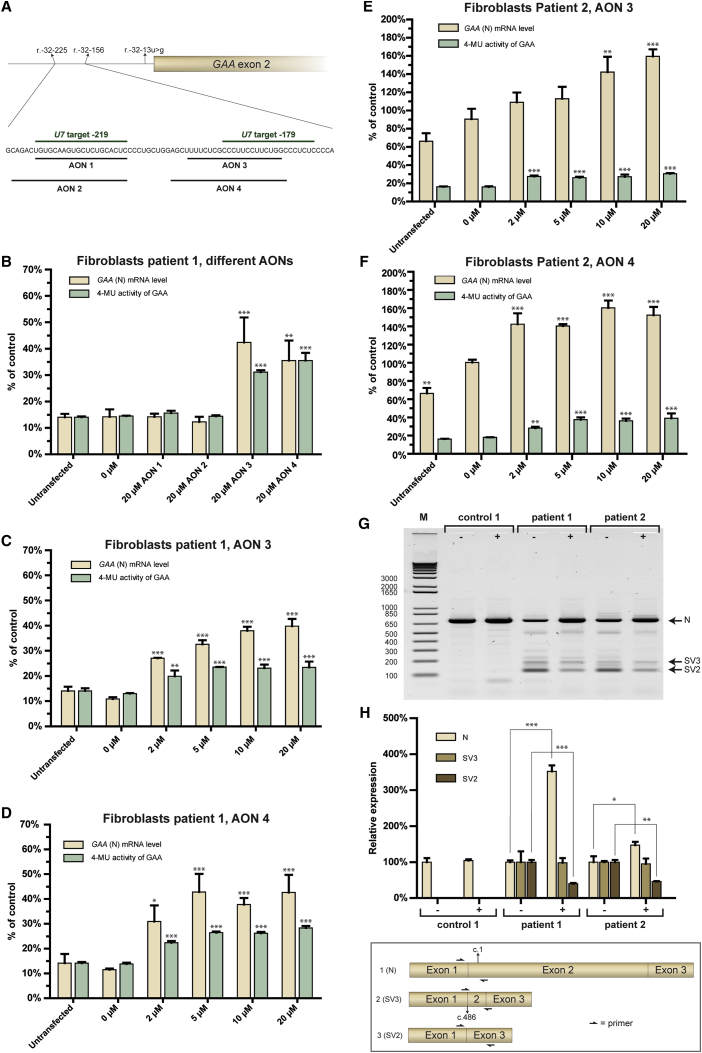
To test whether morpholino-based AONs can modulate aberrant splicing caused by the IVS1 variant, AONs were designed based on the results from the U7 snRNA screen shown in Figure 3. Specific requirements of low G content on chemical synthesis posed constrains to the regions that could be targeted but it was possible to design AONs that targeted the two major putative repressor sequences at c.-32-219 (AONs 1 and 2) and c.-32-179 (AONs 3 and 4) (Figures 4A and S3A). To test whether primary fibroblasts were amenable to splicing modulation using morpholino-based AONs, AONs targeting CypA were used as positive control. Two PMO-based AONs that target the 3′ donor splice site of exon 4 in the CypA pre-mRNA (Figures S3A and S3B) were able to promote skipping of exon 4 and exons 3 and 4 after transfection into primary fibroblasts, as shown by flanking exon RT-PCR and qRT-PCR analysis (Figures S3C and S3D), which is in agreement with our previous report.9 Next, AONs 1–4 targeting the identified regions in the GAA pre-mRNA were tested by transfecting primary fibroblasts from patient 1. qRT-PCR analysis of exon 2 inclusion showed that AONs 3 and 4 promoted exon 2 inclusion more than 2-fold, whereas AONs 1 and 2 were ineffective (Figure 4B). Concomitant effects on GAA enzymatic activity were found, with more than 2-fold enhancement by AONs 3 and 4, while AONs 1 and 2 had no effect. This showed that PMO-based AONs that targeted the region of c.-32-179 promoted exon 2 inclusion, and that other PMO-based AONs tested were ineffective.
Figure 4.
Splicing Correction of GAA Exon 2 Using PMO-Based AONs
(A) Positions in the GAA pre-mRNA to which PMO-based AONs1–4 anneal. (B) Effect of AONs1–4 in fibroblasts from patient 1. GAA exon 2 inclusion in the mRNA was measured using qRT-PCR analysis (GAA [N] mRNA level) and GAA enzymatic activity using 4-methylumbelliferylα-D-glucopyranoside (4-MU) as substrate. Data are expressed relative to the average levels in healthy control fibroblasts and were corrected for β-Actin expression. (C) As in (B) but now using a concentration range of AON 3. (D) As in (B) but now using a concentration range of AON 4. (E) As in (C) but now with the treatment of patient 2 fibroblasts. (F) As in (D) but now with the treatment of patient 2 fibroblasts. (G) Flanking exon RT-PCR analysis of the effect of AON 4 on GAA exon 2 inclusion in fibroblasts from patient 1 and 2. −, 0 μM AON; +, 20 μM AON. (H) qRT-PCR analysis of individual splicing products of GAA exon 2 splicing. The N, SV2, and SV3 products were quantified using primers as outlined in the cartoon, and the effect of AON 4 on GAA exon 2 splicing was determined in fibroblasts from patients 1 and 2 and control 1. Data are corrected for β-Actin expression and normalized per splicing variant for expression in untreated cells to visualize the effect per variant. Note that patient 2 carried a pathogenic missense GAA variant on the second allele. GAA mRNA is normally expressed from this allele, which partially masks the effect on mRNA expression from the IVS1 allele. Patient 1 has no GAA mRNA expression from the second allele due to the presence of a variant that causes a frame shift and subsequent mRNA degradation. Data are means ± SDs of three biological replicates. *p < 0.05; **p < 0.01; ***p < 0.001.
Next, the concentration-dependent effects of AONs 3 and 4 were tested after transfection in fibroblasts from patient 1 (Figures 4C and 4D). AONs 3 and 4 induced a concentration-dependent increase in both the expression of full-length GAA transcript and GAA enzymatic activity. Effects were almost maximal at 2 μM AON and reached a maximum at 5–20 μM. These results were confirmed in a fibroblast cell line from patient 2 (Figures 4E and 4F). This patient (genotype IVS1/c.923A>C) contained a missense GAA variant on allele 2, which was expressed at the mRNA level and led to a high “background” GAA cDNA level in the RT-PCR assays. We conclude that AONs 3 and 4 show in vitro activities that are sufficient to elevate canonical GAA pre-mRNA splicing, and thus GAA enzymatic activity, to above the disease threshold of 20% of the average level present in healthy controls that has been established in the diagnostic department of our center.
To confirm the mechanism by which AON 3 and 4 enhanced GAA enzymatic activity (i.e., by modulating splicing rather than total gene expression), analysis of individual splicing products was performed. Of note, full-skip (SV2) and cryptic splicing transcripts (SV3) lack the translation start codon and are subject to mRNA degradation, hampering accurate relative quantification. The effect of AON 4 was tested in control fibroblasts and in fibroblast from patients 1 and 2. Semiquantitative RT-PCR analysis using primers annealing to exon 1 and exon 3 showed that in patients with IVS1, AON 4 treatment caused an increase in the amount of the full-length (N) transcript, while the amount of the full-skip (SV2) transcript was reduced (Figure 4G). The amount of cryptic splicing transcript SV3 remained unchanged. AON 4 had no effect on GAA expression in control cells. Quantitative analysis by qRT-PCR utilized splicing product-specific primers,27 and this confirmed the semiquantitative analysis (Figure 4H). These results suggest that AON 4 enhances expression of wild-type GAA mRNA by promoting exon 2 inclusion during aberrant splicing caused by the IVS1 variant, rather than acting on enhancing total GAA gene expression. This is further supported by the lack of effect on GAA expression in control cells.
Discussion
The purpose of this work was to test whether aberrant GAA splicing caused by the common IVS1 variant could be restored using AONs. To this end, we performed a lentiviral U7 snRNA-based screen with the aim to identify splicing regulatory sequences that could be blocked to promote exon inclusion. Hits from this screen were tested using PMO-based AONs. Two AONs were identified that promoted exon inclusion and GAA enzyme activity to above the disease threshold of 20% of average healthy control levels. These results identify splicing regulatory elements that negatively regulate the IVS1 variant, and they provide proof of concept for the restoration of canonical splicing in cells from patients with Pompe disease.
The IVS1 GAA variant represented a challenging case, because it is located in the pY tract of exon 2 and weakens the recognition of the splice acceptor site of exon 2. It was not obvious whether it was possible to enhance exon inclusion using AONs, as no negative splice regulatory elements in the GAA gene have been described that could be amenable to inhibition using an AON. This contrasts with other, more rare, GAA variants that affect splicing by causing a major shift from canonical toward cryptic splicing. In these cases, AONs that prevent cryptic splicing could restore canonical splicing.9 We were encouraged by the observation that the IVS1 variant also allows a low level of leaky wild-type splicing, indicating that canonical splicing was still possible and that the pY tract was still partially functional. Previous findings that small drugs could enhance exon 2 splicing strengthen this hypothesis.26 We identified a splicing regulatory element in intron 1 that was located approximately 280 nt upstream of the canonical splice acceptor of GAA exon 2. In an accompanying work,30 we show that this does not concern a classical ISS motif, but that it is the pY tract of a cryptic splice acceptor site that, together with a downstream splice donor, forms a pseudo exon.
Current prediction programs for the identification of cis-acting elements that modulate splicing outcome vary widely in outcome, as illustrated in Figure 1C. We therefore performed an unbiased screen using U7 snRNA-based AONs modified from Liu et al.16, 29 The lentiviral vector employed here was modified to enable one-step cloning with a >90% success rate. Lentiviral transduction showed near 100% infection of target cells, ensuring expression of the U7 snRNAs in all target cells (data not shown). An alternative approach would be to order all desired AONs and to transfect directly in target cells, but this is expensive and not feasible for most research laboratories. The U7 snRNA-based approach identified splicing regulatory motifs that both reduce (intron 1) and enhance (5′ part of exon 2) inclusion of GAA exon 2 (Figure 2B). We were unable to identify these sequences using currently available splicing prediction programs,31 highlighting the need for experimental work. This approach should be more generally applicable to unravel basic mechanisms of splicing and to identify therapeutic targets for splicing variants in human disease.
Metabolic disorders such as Pompe disease are good candidates for AON-mediated splicing correction. Often the enzyme deficiency only needs elevation to levels above a threshold to be fully functional.32, 33, 34 Hits from the U7 snRNA screen were tested using PMO-based AONs, and these promoted exon inclusion and GAA enzyme activity to above the disease threshold of 20%. This threshold was evident from the evaluation of fibroblast samples (using 4-MU as substrate) from ∼700 individuals in our center, ∼350 of which were diagnosed with Pompe disease based on clinical examination, muscle pathology, and biochemical and DNA analysis. While GAA enzyme activity levels in patients never exceeded 20%, the level in healthy individuals ranged from 40% to 180%. We note that the GAA enzyme activity assay is very sensitive to cell culture and assay conditions, and different patient and normal ranges (and therefore disease thresholds) may apply depending on the diagnostic center. Usually the IVS1 variant is present in combination with a variant on the second allele with no residual enzymatic activity, causing childhood/adult Pompe disease, although homozygosity of the IVS1 variant has been described in some patients with Pompe disease.35, 36 Full restoration of the IVS1 allele in compound heterozygous patients would provide a maximum of 50% GAA activity relative to the average levels in healthy controls, and this activity is within the range of healthy controls.
These results form a proof of concept for the development of a potential alternative treatment of a large percentage of Caucasian patients with Pompe disease. Current enzyme replacement therapy has limitations and is based on intravenous delivery of rhGAA that is taken up by target cells via mannose-6-phosphate receptor-mediated endocytosis. AON-mediated splicing correction presents a different strategy, as it enhances endogenous production of wild-type GAA enzyme. Further work is required to test the potential of the identified AONs for clinical implementation. The first step is to test AONs in the relevant cell type, skeletal muscle cells, as splicing can be cell-type specific.37 This is described in our accompanying work.30 Another aspect is cellular uptake by skeletal muscle in vivo following systemic delivery. AON treatments are well tolerated without serious adverse events, as shown in phase I and II clinical trials using 2′-O-methoxyethyl phosphorothioate and PMO chemistries for spinal muscular atrophy and Duchenne muscular dystrophy, respectively.23, 38 This may accelerate testing of AONs for the treatment of Pompe disease in a clinical setting. The ongoing development of improved methods for AON delivery, including tricyclo-DNA-based backbones39 and cell-penetrating peptides like Pip6A,40 is expected to be relevant for the future testing of AON-based drugs for the treatment of Pompe disease.
Materials and Methods
Obtaining Patient Fibroblasts
Dermal fibroblasts from one control (control 1) and two patients (patients 1 and 2) with Pompe disease were obtained via skin biopsy with informed consent. The Erasmus Medical Center (MC) institutional review board approved the study protocol. All patient and control primary cell lines were negative for HIV, hepatitis B, and hepatitis C as tested by qPCR analysis at the diagnostic Department of Virology of the Erasmus MC Rotterdam. Both patient cell lines contain the IVS1 mutation on one allele. The second allele was c.525delT for patient 1 and c.923A>C (his>pro) for patient 2, which both are established pathogenic GAA variants (www.pompecenter.nl).
Nomenclature
Indications to variants and/or locations on the cDNA or (pre-)mRNA conform to Human Genome Variation Society (HGVS) standards (http://www.hgvs.org/mutnomen/).41
Modification of the U7 snRNA Vector for Efficient One-Step Cloning of AON Sequences
The U7 snRNA gene and promoter were amplified by PCR from female mouse genomic DNA using Fw-ms-U7 snRNA-Pst1 and rv-ms-U7 snRNA-SalI primers, which included PstI and SalI overhang restriction sites. The PCR fragment (425 bp) was cloned into a pCRII-TOPO vector according to the manufacturer’s manual (Invitrogen). SMopt and NsiI sites were generated by site-directed mutagenesis according to an inner and outer primer design with Fw- and Rv-U7 snRNA-SMopt or Fw- and Rv-U7 snRNA-NsiI as inner primers and with Fw-M13 and Rv-M13 as outer primers (Table S1), and they were subcloned using the PstI and SalI sites upstream of the polypurine tract fragment of the lentiviral vector used for reprogramming, from which OSKM and the spleen focus-forming virus (SF) promoter were removed.
Cloning of AONs into the U7 snRNA Vector
AONs were inserted via PCR amplification using a forward primer that contained the desired antisense sequence and the unique NsiI restriction site and the reverse primer Rv-ms- U7 snRNA-SalI. The amplified PCR product was purified by agarose gel electrophorese, extracted using the gel extraction kit (QIAGEN), digested with NsiI and SalI, purified with a PCR purification kit from (QIAGEN), and cloned into the NsiI and SalI sites of the U7 snRNA vector. Clones were verified by sequencing with the Fw-ms-U7 snRNA-PstI (Table S1) and restriction enzyme digestion.
Cell Culture
Both HEK293T cells and human primary fibroblasts were cultured in high-glucose DMEM (Gibco) supplemented with 100 U/mL penicillin/streptomycin/glutamine (Gibco/Thermo Scientific) and 10% fetal bovine serum (Hyclone/Thermo Scientific). Cells were passaged after reaching 80%/90% confluence with TrypLE (Gibco/Thermo Scientific). All cell lines were routinely tested for mycoplasma infection using the MycoAlert Mycoplasma Detection Kit (Lonza) and were negative.
Virus Production
Lentiviruses were produced by co-transfecting HEK293T cells at 80% confluence in a 10-cm culture dish with the lentivirus transfer vector (3 μg SF-U7 snRNA vectors) and packaging plasmids (2 μg psPAX2 and 1 μg pVSV vectors) using Fugene 6 transfection according to the manufacturer’s protocol (Promega). Lentiviruses were harvested from the medium after 72 hr of transfection and were filtered using a 0.45-μm polyvinylidene fluoride (PVDF) filter (Millipore). After filtering, lentiviruses were concentrated by high-speed centrifugation for 2 hr at 20,000 rpm in a Beckman Coulter Ultracentrifuge with a SW32 Ti rotor at 4°C. The supernatant was removed and the pellet was resuspended in 25 μL low-glucose DMEM (Invitrogen) per plate. The virus was stored in aliquots at −80°C.
p24 ELISA
Viral titers were determined with the HIV-1 p24 antigen ELISA kit (Retrotek; Zeptomix) according to the manufacturer’s manual. Each virus was diluted 1:400,000 and 1:1,000,000 and the optical density at 450 nm (OD450) was measured with the Varioskan reader (Thermo Scientific).
Transduction of U7 snRNA Vectors
One day before infection, 6 × 104 cells per single well of a 12-well plate of primary fibroblasts derived from patient 1 were seeded. One day later, the cells were infected with 200 ng virus containing the SF-U7 snRNA constructs; after 24 hr, cells were washed three times with PBS before fresh medium was added. After 4 days, cells were washed with PBS and harvested with RLT buffer from the RNAeasy kit for RNA isolation (QIAGEN). For the GAA enzyme activity assay, cells were harvested after 12 days.
Morpholino Transfections
Fibroblasts were transfected with morpholino AONs using Endoporter reagent (Gene-Tools). Cells were grown to 90% confluence before transfection. Endoporter reagent was used at a concentration of 4.5 μL/mL medium. Morpholinos were dissolved in sterile water to a concentration of 1 mM and the appropriate volume was added to each culture well. Cells were harvested 3–5 days after AON addition.
RNA Isolation and cDNA Synthesis
RNA was extracted with the RNeasy Mini Kit with DNase treatment (QIAGEN) and was stored at −80°C in RNase-free water. cDNA was synthesized from 500 ng RNA using the iScript cDNA synthesis kit (Bio-Rad).
qRT-PCR
cDNA was diluted 5, 10, or 20 times and used with 7.5 μL iTaq Universal SYBR Green Supermix (Bio-Rad) with 10 pmol/μL forward and reverse primers (Table S1) in a CFX96 real-time system (Bio-Rad). Cycle threshold (Ct) values were related to amounts using standard curves of four to six dilutions. Quantification of expression was calculated relative to β-Actin expression.
Flanking Exon RT-PCR of GAA
cDNA (diluted 10 times) with GC GAA exon 1–3 fw and GC GAA exon 1–3 rv primers (Table S1) were used for RT-PCR with the Advantage GC 2 PCR kit (Clontech/Takara) and a GC-melt concentration of 0.5 M according to the manufacturer’s protocol. The whole GC-PCR reaction was analyzed on a 1.5% agarose gel containing 0.5 μg/mL ethidium bromide (Sigma-Aldrich).
GAA Enzyme Activity Assay
Cells were harvested with ice-cold lysis buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 50 mM NaF, 1% Triton X-100, and one tablet of Roche cOmplete Protease Inhibitor Cocktail, with EDTA) and incubated for 10 min on ice. Samples were centrifuged at 14,000 rpm for 10 min at 4°C. GAA enzyme activity was measured using 4-MU (Sigma-Aldrich) as substrate as described.27 Total protein concentration was determined using a BCA protein assay kit (Pierce/Thermo Scientific).
Because GAA enzyme activity data strongly depend on assay conditions and so we could evaluate enzyme activities relative to the disease threshold, we normalized the data based on two criteria: (1) the enzyme activity of patient cells measured in a diagnostic setting rather than a research setting, and (2) the average enzyme activity present in healthy control fibroblasts. These results, expressed as the percent of control, are presented in Figure 4. Enzyme data without normalization are shown in Figure S4.
Statistical Analysis
All data represent means ± SD, and p values refer to two-sided Student’s t tests. The Bonferroni multiple testing correction was applied where necessary. A p value < 0.05 was considered significant. Data showed normal variance. There was no power calculation in any of the experiments. No randomization method was used. No samples were excluded from the analyses. Investigators were not blinded to the identity of the samples.
Author Contributions
E.v.d.W., A.J.B., A.T.v.d.P., and W.W.M.P.P. conceived and designed the study and drafted the manuscript. E.v.d.W., A.J.B., and J.M.P. performed the experiments. W.W.M.P.P supervised the study. E.v.d.W., A.J.B., J.M.P., A.T.v.d.P., and W.W.M.P.P. were involved in data interpretation and approved the final manuscript.
Conflicts of Interest
A.T.v.d.P. has provided consulting services for various industries in the field of Pompe disease under an agreement between these industries and Erasmus MC. The other authors declare no conflicts of interest.
Acknowledgments
We thank Philip Lijnzaad for advice on statistical analysis and Dr. Arnold Reuser for discussion. This work was funded by grants from the Sophia Children’s Hospital Foundation (SSWO) (S-687) and the Prinses Beatrix Spierfonds/Stichting Spieren voor Spieren (W.OR13-21).
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.03.001.
Supplemental Information
References
- 1.Chen L., Bush S.J., Tovar-Corona J.M., Castillo-Morales A., Urrutia A.O. Correcting for differential transcript coverage reveals a strong relationship between alternative splicing and organism complexity. Mol. Biol. Evol. 2014;31:1402–1413. doi: 10.1093/molbev/msu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y., Rio D.C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jangi M., Sharp P.A. Building robust transcriptomes with master splicing factors. Cell. 2014;159:487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moehle E.A., Braberg H., Krogan N.J., Guthrie C. Adventures in time and space: splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014;11:313–319. doi: 10.4161/rna.28646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Movassat M., Crabb T.L., Busch A., Yao C., Reynolds D.J., Shi Y., Hertel K.J. Coupling between alternative polyadenylation and alternative splicing is limited to terminal introns. RNA Biol. 2016;13:646–655. doi: 10.1080/15476286.2016.1191727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naftelberg S., Schor I.E., Ast G., Kornblihtt A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 8.Krawczak M., Thomas N.S., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 9.Bergsma A.J., In ‘t Groen S.L., Verheijen F.W., van der Ploeg A.T., Pijnappel W.P. From cryptic toward canonical pre-mRNA splicing in Pompe disease: a pipeline for the development of antisense oligonucleotides. Mol. Ther. Nucleic Acids. 2016;5:e361. doi: 10.1038/mtna.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber A., Kreth J., Müller U. Intronic PRRT2 mutation generates novel splice acceptor site and causes paroxysmal kinesigenic dyskinesia with infantile convulsions (PKD/IC) in a three generation family. BMC Med. Genet. 2016;17:16. doi: 10.1186/s12881-016-0281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stucki M., Suormala T., Fowler B., Valle D., Baumgartner M.R. Cryptic exon activation by disruption of exon splice enhancer: novel mechanism causing 3-methylcrotonyl-CoA carboxylase deficiency. J. Biol. Chem. 2009;284:28953–28957. doi: 10.1074/jbc.M109.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 13.Wang E.T., Cody N.A., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 15.Soldati D., Schümperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol. 1988;8:1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorman L., Suter D., Emerick V., Schümperli D., Kole R. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc. Natl. Acad. Sci. USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havens M.A., Hastings M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzin C.H., Feng J., Yan J., Scaringe W., Liu Q., den Dunnen J., Mendell J.R., Sommer S.S. Mutation rates in the dystrophin gene: a hotspot of mutation at a CpG dinucleotide. Hum. Mutat. 2005;25:177–188. doi: 10.1002/humu.20132. [DOI] [PubMed] [Google Scholar]
- 19.Flanigan K.M., Dunn D.M., von Niederhausern A., Soltanzadeh P., Gappmaier E., Howard M.T., Sampson J.B., Mendell J.R., Wall C., King W.M., United Dystrophinopathy Project Consortium Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum. Mutat. 2009;30:1657–1666. doi: 10.1002/humu.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traynor K. Eteplirsen approved for Duchenne muscular dystrophy. Am. J. Health Syst. Pharm. 2016;73:1719. doi: 10.2146/news160063. [DOI] [PubMed] [Google Scholar]
- 21.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haché M., Swoboda K.J., Sethna N., Farrow-Gillespie A., Khandji A., Xia S., Bishop K.M. Intrathecal injections in children with spinal muscular atrophy: nusinersen clinical trial experience. J. Child Neurol. 2016;31:899–906. doi: 10.1177/0883073815627882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huie M.L., Chen A.S., Tsujino S., Shanske S., DiMauro S., Engel A.G., Hirschhorn R. Aberrant splicing in adult onset glycogen storage disease type II (GSDII): molecular identification of an IVS1 (-13T-->G) mutation in a majority of patients and a novel IVS10 (+1GT-->CT) mutation. Hum. Mol. Genet. 1994;3:2231–2236. doi: 10.1093/hmg/3.12.2231. [DOI] [PubMed] [Google Scholar]
- 25.Boerkoel C.F., Exelbert R., Nicastri C., Nichols R.C., Miller F.W., Plotz P.H., Raben N. Leaky splicing mutation in the acid maltase gene is associated with delayed onset of glycogenosis type II. Am. J. Hum. Genet. 1995;56:887–897. [PMC free article] [PubMed] [Google Scholar]
- 26.Dardis A., Zanin I., Zampieri S., Stuani C., Pianta A., Romanello M., Baralle F.E., Bembi B., Buratti E. Functional characterization of the common c.-32-13T>G mutation of GAA gene: identification of potential therapeutic agents. Nucleic Acids Res. 2014;42:1291–1302. doi: 10.1093/nar/gkt987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergsma A.J., Kroos M., Hoogeveen-Westerveld M., Halley D., van der Ploeg A.T., Pijnappel W.W. Identification and characterization of aberrant GAA pre-mRNA splicing in Pompe disease using a generic approach. Hum. Mutat. 2015;36:57–68. doi: 10.1002/humu.22705. [DOI] [PubMed] [Google Scholar]
- 28.Kroos M.A., Pomponio R.J., Hagemans M.L., Keulemans J.L., Phipps M., DeRiso M., Palmer R.E., Ausems M.G., Van der Beek N.A., Van Diggelen O.P. Broad spectrum of Pompe disease in patients with the same c.-32-13T->G haplotype. Neurology. 2007;68:110–115. doi: 10.1212/01.wnl.0000252798.25690.76. [DOI] [PubMed] [Google Scholar]
- 29.Liu S., Asparuhova M., Brondani V., Ziekau I., Klimkait T., Schümperli D. Inhibition of HIV-1 multiplication by antisense U7 snRNAs and siRNAs targeting cyclophilin A. Nucleic Acids Res. 2004;32:3752–3759. doi: 10.1093/nar/gkh715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van der Wal E., Bergsma A.J., Van Gestel T.J.G., In ‘t Groen S.L.M., Zaehres H., Araúzo-Bravo M.J., Scholer H.R., van der Ploeg A.T., Pijnappel W.W.M.P. GAA deficiency in Pompe disease is alleviated by exon inclusion in iPS-derived skeletal muscle cells. Mol. Ther. Nucleic Acids. 2017;7:101–115. doi: 10.1016/j.omtn.2017.03.002. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altarescu G.M., Goldfarb L.G., Park K.Y., Kaneski C., Jeffries N., Litvak S., Nagle J.W., Schiffmann R. Identification of fifteen novel mutations and genotype-phenotype relationship in Fabry disease. Clin. Genet. 2001;60:46–51. doi: 10.1034/j.1399-0004.2001.600107.x. [DOI] [PubMed] [Google Scholar]
- 33.Huemer M., Mulder-Bleile R., Burda P., Froese D.S., Suormala T., Zeev B.B., Chinnery P.F., Dionisi-Vici C., Dobbelaere D., Gökcay G. Clinical pattern, mutations and in vitro residual activity in 33 patients with severe 5, 10 methylenetetrahydrofolate reductase (MTHFR) deficiency. J. Inherit. Metab. Dis. 2016;39:115–124. doi: 10.1007/s10545-015-9860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M., Liu Y., Sun S., Zhang H., Wang W., Ning G., Li X. A prevalent and three novel mutations in CYP11B1 gene identified in Chinese patients with 11-beta hydroxylase deficiency. J. Steroid Biochem. Mol. Biol. 2013;133:25–29. doi: 10.1016/j.jsbmb.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 35.Musumeci O., Thieme A., Claeys K.G., Wenninger S., Kley R.A., Kuhn M., Lukacs Z., Deschauer M., Gaeta M., Toscano A. Homozygosity for the common GAA gene splice site mutation c.-32-13T>G in Pompe disease is associated with the classical adult phenotypical spectrum. Neuromuscul. Disord. 2015;25:719–724. doi: 10.1016/j.nmd.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Laforêt P., Nicolino M., Eymard P.B., Puech J.P., Caillaud C., Poenaru L., Fardeau M. Juvenile and adult-onset acid maltase deficiency in France: genotype-phenotype correlation. Neurology. 2000;55:1122–1128. doi: 10.1212/wnl.55.8.1122. [DOI] [PubMed] [Google Scholar]
- 37.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendell J.R., Goemans N., Lowes L.P., Alfano L.N., Berry K., Shao J., Kaye E.M., Mercuri E., Eteplirsen Study Group and Telethon Foundation DMD Italian Network Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann. Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goyenvalle A., Griffith G., Babbs A., El Andaloussi S., Ezzat K., Avril A., Dugovic B., Chaussenot R., Ferry A., Voit T. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 40.Godfrey C., Muses S., McClorey G., Wells K.E., Coursindel T., Terry R.L., Betts C., Hammond S., O’Donovan L., Hildyard J. How much dystrophin is enough: the physiological consequences of different levels of dystrophin in the mdx mouse. Hum. Mol. Genet. 2015;24:4225–4237. doi: 10.1093/hmg/ddv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E. HGVS recommendations for the description of sequence variants: 2016 update. Hum. Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.