Abstract
In flowering plant plastids and mitochondria, multiple organellar RNA editing factor (MORF/RIP) proteins are required at most sites for efficient C to U RNA editing catalyzed by the RNA editosome. MORF proteins harbor a conserved stretch of residues (MORF-box), form homo- and heteromers and interact with selected PPR (pentatricopeptide repeat) proteins, which recognize each editing site. The molecular function of the MORF-box remains elusive since it shares no sequence similarity with known domains. We determined structures of the A. thaliana mitochondrial MORF1 and chloroplast MORF9 MORF-boxes which both adopt a novel globular fold (MORF domain). Our structures state a paradigmatic model for MORF domains and their specific dimerization via a hydrophobic interface. We cross-validate the interface by yeast two-hybrid studies and pulldown assays employing structure-based mutants. We find a structural similarity of the MORF domain to an N-terminal ferredoxin-like domain (NFLD), which confers RNA substrate positioning in bacterial 4-thio-uracil tRNA synthetases, implying direct RNA contacts of MORF proteins during RNA editing. With the MORF1 and MORF9 structures we elucidate a yet unknown fold, corroborate MORF interaction studies, validate the mechanism of MORF multimerization by structure-based mutants and pave the way towards a complete structural characterization of the plant RNA editosome.
INTRODUCTION
In almost all land plants, RNA editing alters the DNA-encoded information in mitochondria and plastids on the transcript level. As part of the maturation of primary transcripts, specific cytosines are selectively altered to uridines. Requirement of several proteins of different classes for the C to U RNA editing in plant organelles has been elucidated. The target nucleotide is identified by an individual pentatricopeptide repeat (PPR) protein which recognizes and binds to the unique RNA motif upstream of the cytosine to be edited (1,2). The PPR proteins involved in RNA editing belong to the E or DYW subclasses. The E subclass PPR proteins are characterized by a canonical C-terminal extension, the E domain (3–7). The DYW subclass PPR proteins contain not only an E domain but also an additional C-terminal extension, the DYW domain, which harbors amino acid patterns conserved in cytidine deaminases, binds zinc atoms and may thus provide the C to U deaminase activity (8–10). Since E subclass PPR proteins have no such putative catalytic domains, it is hypothesized that all E subclass PPR proteins form a complex with at least one DYW subclass protein. For example, two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids (11). All RNA editing sites, about 500 in mitochondria and about 40 in plastids, are considered to be addressed by the ∼200 E or DYW subclass PPR proteins in flowering plants.
Besides the E domain containing PPR proteins, several proteins of different classes appear to influence RNA editing, usually identified as lowered or lost RNA editing in mutants of the respective gene. Thus, several RRM (RNA recognition motif) domain containing proteins are involved in RNA editing (12–16). Of these proteins, Cp31 and ORRM1 proteins are required for full processing of several RNA editing sites in plastids (12,13), while ORRM2, ORRM3 and ORRM4 are involved in editing of several sites in mitochondria (15,16). Likewise, a zinc-binding domain containing protein OZ1 is also necessary for full RNA editing at several sites in plastids, but how this protein is involved in editing is unclear (17).
Required for most C to U RNA editing events in both plant mitochondria and plastids are members of the small family of MORF (RIP) proteins of which nine full-length members are encoded in Arabidopsis thaliana (18,19). In A. thaliana, the T-DNA insertion line morf1-2 is homozygous lethal, underlining an essential function of MORF1 and also other MORF proteins (19). The MORF proteins appear to fall into two categories, those that are required for many RNA editing events in either or both mitochondria and plastids and those that show loss of editing at few if any sites when mutated (19,20). Of the first group, MORF2 and MORF9 are involved in most of the editing sites in plastid mRNAs, and MORF1 and MORF3 are each required for full editing at ∼150 sites in mitochondria. The dual targeted MORF8/RIP1 protein is required for editing at >400 RNA editing sites in mitochondria and several sites in plastids (20). MORF proteins show interaction with PPR type RNA editing factors or RRM domain containing proteins, implying the formation of RNA editosomes with these proteins (16,18,19). Interestingly, most editing sites requiring a MORF protein need one or two other MORF proteins for efficient editing (20,21). A recent analysis of MORF–MORF protein–protein interactions confirmed that most MORF proteins can interact to form homomers and also heteromers. It was found that MORF proteins required for many sites, such as MORF1, can interact with most other MORF proteins, whereas those with few or no assigned target sites interact with only few members of the MORF protein family (22). These observations suggest that specific MORF–MORF interactions in plant organelles are important for the assembly of RNA editosomes with other editing factors.
To better understand the function of MORF proteins in the RNA editing process in mitochondria and plastids of flowering plants, we have determined structures of the MORF domains of the mitochondrial MORF1 and the plastid MORF9, that are required for many editing sites and bind to most MORF protein family members. We delineate the structural basis of MORF multimer formation, validate MORF dimerization by structure-based site-directed mutagenesis coupled to yeast two-hybrid or pulldown assays and shed light on a potential role of MORF proteins within the plant RNA editosome as suggested by structural similarities to bacterial RNA binding proteins.
MATERIALS AND METHODS
Cloning, expression and protein purification
A DNA fragment encoding the A. thaliana MORF1 MORF-box (amino acid residues 79–190) was cloned into pETM-11 to yield a protein (MORF179–190) with a TEV-cleavable N-terminal His6-tag (His6-MORF179–190). A DNA fragment encoding the A. thaliana MORF9 MORF-box (amino acid residues 86–186) was cloned into pETM-11 to yield a protein (MORF986–186) with a TEV-cleavable N-terminal His6-tag (His6-MORF986–186). For protein production, Escherichia coli Rosetta2 (DE3) cells were transformed with the respective plasmid, grown in terrific broth to an OD600 of 0.6 at 37°C, cooled to 20°C, induced with 0.5 mM IPTG and cultivated at 20°C over night. Cells were harvested by centrifugation and stored at −80°C. Cell pellets from expression cultures were resuspended in lysis buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl) supplemented with 0,01% (w/v) CHAPS and 1 mM DTT in the presence of a protease inhibitor cocktail (Roche). Cells were lysed using a Sonoplus sonifier (Bandelin) and cell debris was removed by centrifugation.
For purification of MORF179–190 and MORF986–186, the soluble fraction was passed over a Ni2+-NTA gravity flow column, pre-equilibrated with lysis buffer. The beads were washed with lysis buffer, containing 30 mM imidazole. His6-MORF179–190 or His6-MORF986–186 was eluted with lysis buffer supplemented with 300 mM imidazole. The eluate of His6-MORF179–190 was treated with a 1:40 protein mass ratio of tobacco etch virus (TEV) protease over night to remove the N-terminal His6-tag. The eluate of His6-MORF986–186 was incubated over night at room temperature for cleavage accordingly. Cleaved proteins were further purified via Superdex 200 gel filtration chromatography (GE Healthcare) in lysis buffer. Peak fractions of the monomers were pooled, passed over an equilibrated Ni2+-NTA gravity column and concentrated to 56 mg/ml (MORF179–190) or 40 mg/ml (MORF986–186), flash frozen in liquid nitrogen and stored at −80°C.
Multi angle light scattering (MALS) and analytical gel filtration
MALS experiments were performed at 18°C. Samples were passed over a Superdex 200 increase 10/300 (His6-MORF179–190) or a Superdex 75 10/300 gel filtration column (MORF179–190) coupled to a miniDAWN TREOS three-angle light scattering detector (Wyatt Technology) and a RefractoMax520 refractive index detector (ERC). Detectors were aligned, corrected for band broadening, and photodiodes were normalized with BSA as a reference. For calculation of the molecular mass, protein concentrations were determined from the differential refractive index with a specific refractive index increment (dn/dc) of 0.185 ml/g. Data were analyzed with ASTRA 6.1.4.25 (Wyatt Technology). For analytical gel filtration, MORF179–190 or MORF186–186 were passed over a Superdex 75 10/300 gel filtration column and UV absorbance at 280 nm was recorded.
Crystallographic analyses
MORF179–190 crystallized by sitting drop vapor diffusion (1 μl protein plus 1 μl reservoir) at 4°C with a reservoir containing 0.1 M Tris, pH 8.0 and 2.4 M ammonium sulfate (space group P1) or 0,1 M Bicine, pH 9.0 and 2.4 M ammonium sulfate (space group P21). Crystals were cryo-protected by a mixture of 90% (v/v) lithium sulfate and 10% of the respective reservoir solution. For derivatization experiments, crystals were incubated for 2 min in the respective cryo-protectant solution supplemented with 0.5 M NaBr and flash frozen in liquid nitrogen.
MORF986–186 crystallized by sitting drop vapor diffusion (0.1 μl protein plus 0.1 μl reservoir plus 30 nl additive) at 4°C with a reservoir containing 0.08 M sodium cacodylate, pH 6.5, 14.4% (w/v) PEG 8000, 20% (v/v) glycerol, 0.16 M calcium acetate and 0.3 M NaCl as an additive. Crystals were cryo-protected in reservoir solution and flash frozen in liquid nitrogen. Diffraction data were collected at 100 K at beamline 14.1 of the BESSY II storage ring, Berlin, Germany (23). All diffraction data were processed with XDS (24).
The structure of MORF179–190 was solved by single-wavelength anomalous dispersion (SAD) with 26 Br-sites in space group P1 employing SHARP/AUTOSHARP (25). Initial phases were improved by density modification with SOLOMON (25). The initial experimental electron density was of good quality and allowed the building of all four MORF179–190 molecules in the asymmetric unit with PHENIX.AUTOBUILD (26,27). The second MORF179–190 crystals grown at pH 9.0 were processed in space group P21 and identified as pseudo-merohedrally twinned by PHENIX.XTRIAGE (26). The structure of the latter crystals was solved by molecular replacement with PHASER employing the coordinates of MORF179–190. The structure of MORF986–186 was solved by molecular replacement with PHASER employing the coordinates of a MORF179–190 monomer lacking the N-terminal β-strand. Structural models were completed through alternating rounds of automated refinement using PHENIX.REFINE (28) and manual model building using COOT (29). The pseudo-merohedrally twinned data were refined with PHENIX.REFINE in space group P21 with a twin fraction of 0.14 and the twin operator h, −k, l.
Pulldown assays
MORF1 cDNA fragments corresponding to amino acid residues from M1 to Y406 and from F61 to G241 were amplified by PCR and cloned into pMAL-TEV-MCS41 and pET41-GFP vectors by the In-Fusion® HD Cloning kit (TAKARA), respectively. The pMAL-TEV-MCS41 vector was created as follows: pET-41-a (Novagen) multiple cloning site between SacI to XhoI was amplified by PCR and cloned into SacI–HindIII sites into the pMAL-TEV vector by the In-Fusion® HD Cloning kit (TAKARA). The pET41-GFP vector was created as follows: GFP CDS from the pSM-GFP (GenBank: U70495.1) was amplified and cloned into the pET-41-a (Novagen) NotI site by the In-Fusion® HD Cloning kit. For mutated MORF1 proteins, the inverted PCR products with primers containing respective point mutations are circularized by the In-Fusion® HD Cloning kit. MORF1 protein fragments (residues 1–406) were expressed as fusion proteins with maltose-binding protein (MBP) label at their N termini on the one hand and MORF1 protein fragments (residues 61–241) with N-terminal GST and His tags and GFP labels at the C termini on the other hand. GST-His-GFP-tagged proteins were purified from E. coli via Ni-NTA-agarose (Qiagen) followed by a purification step with glutathione agarose (Sigma) and dialysis against 1× PBS. MBP-tagged MORFs were bound to amylose resin (New England Biolabs). To investigate interactions between MORF1 and MORF1 mutants, 20 μl of amylose resin saturated with MBP-MORF1 was incubated for 16 h at 4°C with 20 μg of MORF1 or the respective mutated MORF1 with GFP label. Resins were boiled for 5 min in SDS loading dye after five washing steps and the proteins were separated by SDS-PAGE on two gels in parallel. After blotting, proteins bound to the resin were detected by anti-MBP antibody (New England Biolabs) and the respective pulled down GFP-labeled MORF proteins were identified with an anti-GFP antibody (Roche Applied Science).
Yeast two-hybrid analysis
The coding sequences without target peptide, according to prediction by TargetP (30), of MORF1 (amino acid residues 61–406) and its respective mutants were cloned into the bait (pGBKT7) and / or the prey (pGADT7) vector of the Matchmaker GAL4 Two-Hybrid System 3 (Clontech Laboratories) for expression in yeast cells (PJ69-4A) according to the protocol and other publications (22,31,32) via the In-Fusion HD cloning system (Clontech Laboratories). Successfully cotransfected yeast cells were selected on medium lacking leucine and tryptophan. Growth was assayed on synthetic dropout (SD) medium lacking adenine, histidine, leucine and tryptophan and for rigorous probing also containing 2.5, 5 and 7.5 mM 3-amino-1,2,4-triazole (3AT). In each assay, 5 μl of an overnight liquid culture adjusted to A600 nm of 0.3 was dripped on the agar medium plate. Pictures were taken after 6 days of growth.
RESULTS
Purification and structure determination of the MORF1 MORF domain
One hallmark which unifies plant editosomal MORF proteins is a conserved stretch of around 100 amino acids termed MORF-box which in the light of our results here we now address as MORF domain (19). The fold of this region could not be predicted as sequence similarity to known structures is too low. However, it was possible to subdivide and classify the MORF domain as a sequence of four motifs (13,18).
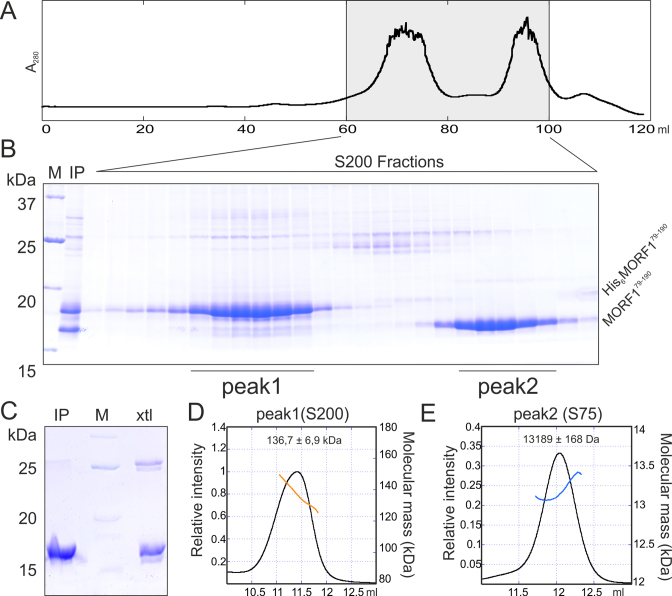
To gain first structural and functional insight into this key domain of MORF proteins, we set out to determine the structure of the MORF domain from A. thaliana MORF1. The corresponding protein fragment (MORF179–190) was cloned, expressed in fusion with an N-terminal His6-tag and purified via Ni-NTA sepharose and gel filtration. MORF proteins are known to form multimers (22). Surprisingly, the oligomerization state of MORF179–190 changed from multimeric to a lower molecular weight upon cleavage of the N-terminal His6-tag as shown by gel filtration analysis (Figure 1A and B). To determine the oligomerization state of the uncleaved and cleaved protein fractions, we performed multi-angle light scattering coupled to size exclusion chromatography (SEC-MALS). While the cleaved MORF179–190 fraction with a calculated molecular weight of 13 kDa was unambiguously a monomer, the oligomerization state of His6-MORF179–190 ranged between 120 and 150 kDa with an average of 136 kDa (Figure 1D and E). Apparently, the N-terminal His6-tag mediated an artificial MORF multimerization while the MORF-box alone is monomeric in solution.
Figure 1.
Characterization of MORF179–190 purification. (A) Preparative gel filtration chromatogram of purified and TEV cleaved MORF179–190. Absorption was counted at 280 nm wavelength. (B) SDS-PAGE analysis of the gel filtration run shown in (A), First two lanes show the protein standard (M) and the gel filtration input (IP). Molecular mass of the standard proteins in kDa are shown on the left, protein names on the right. (C) SDS-PAGE analysis of dissolved MORF179–190 crystals. IP- purified sample used for crystallization, xtl – dissolved crystals, M – protein standard. (D) SEC-MALS analysis of His6- MORF179–190 black curve – refractive index, orange curve – molecular mass at the corresponding elution volume. Input sample and respective column as indicated. The averaged molecular masses and standard deviations across the main peaks are indicated. (E) SEC-MALS analysis of MORF179–190 as in (D), blue curve – molecular mass at the corresponding elution volume.
The monomeric MORF179–190 fraction was further purified and crystallized at high ammonium sulfate concentrations. Analysis of the dissolved crystals by SDS-PAGE confirmed the presence of MORF179–190 and traces of an apparent MORF179–190 dimer at twice the molecular weight. The dimer may have been formed during crystallization by a disulfide bridge which was not completely reduced by DTT in the SDS loading dye (Figure 1C). Complete datasets for sulfur SAD were collected up to 1.94 Å resolution and the space group was determined to be P21 with pseudo-merohedral twinning. As sulfur SAD and molecular replacement strategies failed possibly due to the crystal twinning and unsuitable homology models, respectively, we screened for other untwinned space groups and conducted heavy atom derivative soaking experiments in parallel. Finally, we solved the structure by the SAD method employing NaBr as a derivative. The space group of the derivative dataset was determined to be P1 and datasets were collected up to a resolution of 1.5 Å with an anomalous signal sufficient for structure determination. Notably, likely owing to the low isoelectric point of MORF179–190, most of the 26 Br sites were located near hydrophobic residues and coordinated by anion π-stacking (Figure S1A). The resulting solvent-flattened electron density map was of a high quality and allowed model building of the entire structure which was refined to low R/Rfree factors of 0.1573/0.1923 while maintaining good stereochemistry (Table S1, Figure S1A and B).
The MORF domain adopts a globular fold
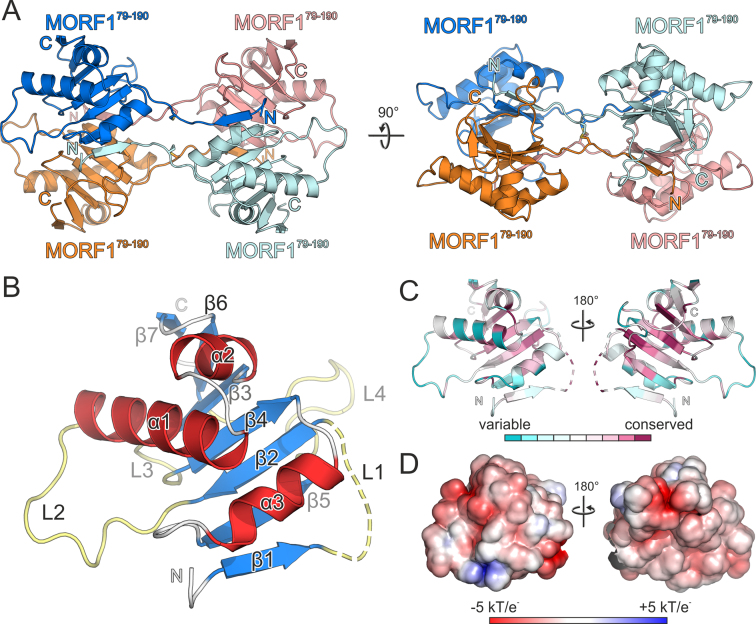
In our experimentally determined structure derived from the P1 crystals, the asymmetric unit comprises four globular molecules which are not related by crystallographic symmetry operators. MORF proteins are known to multimerize via the MORF domain and thus, we set out to differentiate between crystal packing contacts and physiologically relevant interfaces. The initial assignment of the interfaces was confirmed to be the most likely physiologigal assembly by the PISA interaction server (Figure 2A) (33). To distinguish between physiological molecular interactions and crystal packing contacts, we analyzed P21 twinned crystals of MORF179–190 for cross-validation as a native dataset. The P21 structure was solved by molecular replacement with four molecules of the MORF domain in the asymmetric unit and refined with the twin operators h, −k, l to low R/Rfree factors (0.163 / 0.192) while maintaining good stereochemistry (Table S1). Both structures share the prominent tetrameric assembly initially assigned to the P1 crystals (Figures 2A, S2A) (33,34). Two molecules each form two tight hydrophobic and symmetrical head-to-head interfaces with their globular moieties, each burying an average of 400 Å2 surface area. The interaction is exclusively mediated by a β-sheet surface while three α-helices form the outer shell of the dimer (Figure 2A). The extended N-termini of two neighboring MORF domains contact the globular parts of two distant molecules head to tail. This way, each terminus tethers both globular domains with an average buried surface area (BSA) of around 610 Å2, this way extending the hydrophobic dimerization interface of the distant molecules to 937 Å2. All N-termini of the MORF domains interact at the center of the tetrameric assembly, mainly via hydrophobic contacts and two disulfide bonds burying 200 Å2 of molecular surface area (Figure S2B). The overall P1 and P21 structures are in good agreement with an root mean square deviation (RMSD) of 0.64–0.91 Å. The highest degree of structural flexibility was observed in the center of the tetrameric interface formed by the N-termini (Figure S2A).
Figure 2.
Crystallization and structural characterization of MORF179–190. (A) Ribbon plot of the crystallized MORF179–190 tetramer, orthogonal view on the right. Two adjacent molecules (marine blue, orange) form a tight dimer and contact a distant equivalent dimer (cyan, salmon) with their N-termini and vice versa. Disulfide bridges are shown as sticks and colored by atom type. (B) Close-up view and domain architecture of a single MORF domain with β-sheet 1 folded back onto the globular moiety. Major loops L1–L4 (light yellow), β-sheets β1–β7 (marine blue) and α-helices α1-α3 (red) labeled accordingly. Minor loops and turns are shown in gray. (C) Conservation scores including 124 non-redundant MORF-related sequences obtained by CONSURF (37) plotted on the structure of MORF179–190. (D) Electrostatic surface potential as indicated obtained by APBS and plotted on the surface of MORF179–190. Dashed lines in the ribbon plots represent modeled residues G84-D86 of MORF179–190. Rotation symbols indicate the views relative to (A).
The extended N-termini, contacting two distant globular domains with β-strands and forming a relatively large central interface appear rather unusual. They most likely represent a domain swap where the strain of folding back the N-terminal β-strand is released by formation of distant intramolecular contacts during crystallization. Notably, the hinge region (loop 1) bridging the globular domains and the N-terminal sheet has different conformations in the P1 and P21 MORF179–190 crystal structures. We interpret this as a conformational flexibility which is commonly observed for hinge regions involved in domain swaps (Figure S2A) (35,36). In our MORF domain construct, the N-terminal His6-tag may have hindered the first β-strand to fold back onto the same molecule and instead generated unspecific multimers via intermolecular interactions (Figure 1A, B, D and E). After His6-tag cleavage most likely key anchoring residues at the N-terminus of β-strand 1 are missing which alleviates the strain of folding it back. As a consequence, the dimerization interface spanning 937 Å2 is reduced to 400 Å2 in solution and dimerization is thus impeded (Figure 1D and E). Taken together, MORF-tetramerization via the N-termini displays typical features of a domain swap. Consequently, we built a working model of a physiological MORF1-dimer burying a total surface area of almost 1000 Å2, where both N-terminal β-strands are folded back and join the respective globular moieties (Figures 2B and 4B). Modeling of the loop demanded repositioning of three residues (G84–D86) which were suspended from structural interpretation and are either shown as dashed lines in ribbon plots or entirely omitted in figures.
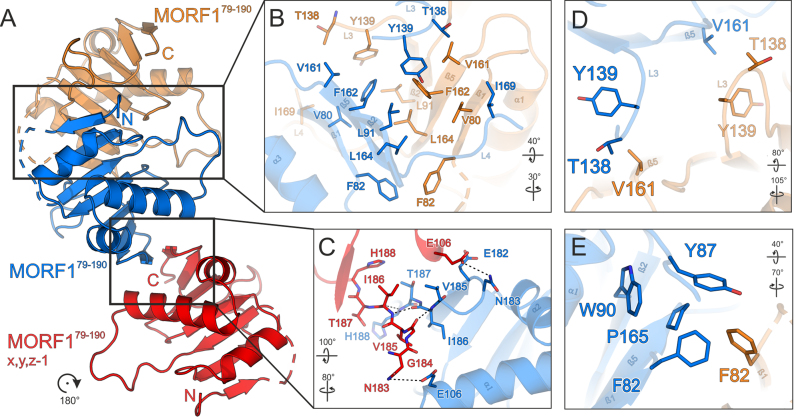
Figure 4.
Structural basis for dimerization and higher order multimerization of the MORF domain. (A) Overview of MORF179–190 interaction modes. A central MORF179–190 molecule (marine blue) forms a tight symmetrical dimer via a hydrophobic interaction surface with a second MORF179–190 molecule (orange). A third MORF179–190 molecule (red) may interact via the C-terminal β-sheet 7. (B) Close-up view of the main hydrophobic protein-protein interface of MORF179–190 shown in (A). (C) Close-up view of the secondary protein–protein interface of MORF179–190 shown in (A). (D) Close-up view of residues interacting with MORF179–190 V161 illustrating the symmetry of the hydrophobic interface. (E) Close-up view of the region around P165. All coloring as in (A). Interacting residues are shown as sticks and colored by atom type. Carbon – as for the respective molecule; nitrogen – blue; oxygen – red; sulfur – yellow. Dashed lines represent hydrogen bonds. Dashed lines in the ribbon plots represent modeled residues G84-D86 of MORF179–190. Rotation symbols indicate views relative to Figure 2A.
Further cross-validation of both MORF179–190 crystal structures with respect to crystal packing left one additional potential physiological interface which—in each crystal lattice—was formed by only two out of the four molecules in the asymmetric unit (Figure 4A and C). This second interface buries an average BSA of ∼400 Å2 and has a lower PISA score than the main hydrophobic interface. Nevertheless, since the interface was observed in two independent crystal lattices, it may play a role in higher order MORF1 multimerization, especially in an extended context of larger MORF termini, MORF heteromers and PPR-proteins (21).
Structure of the MORF domain
MORF179–190 folds to a globular domain with seven β-strands and three α-helices interspersed by four major loops (Figure 2B). The secondary structure elements but also loops 1 and 4 are highly conserved among MORF family members in A. thaliana and argue for a paradigmatic fold of the MORF domains (Figure 3). To further assess evolutionary aspects, we calculated conservation scores with CONSURF supplying 124 unique sequences with identities in the range of 35–90% to MORF1 (37). To this end, mapping the conservation scores of MORF-related proteins from all organisms onto the MORF179–190 structure shows a higher degree of conservation for the β-sheets and the hydrophobic core than for the molecule's surface. In contrast to loops 2–4, the first loop is highly conserved which may highlight its importance to stabilize the MORF fold and multimerization (Figure 2C). The less conserved surface of the MORF domain including loops 2–4 possibly reflects the functional diversity among MORF family members regarding editing site specificity and multimer formation (13,19,21,22). In agreement with other MORF proteins, the MORF179–190 domain surface has an overall negative charge distribution mirrored by the low isoelectric point of 4.15 (Figure 2D).
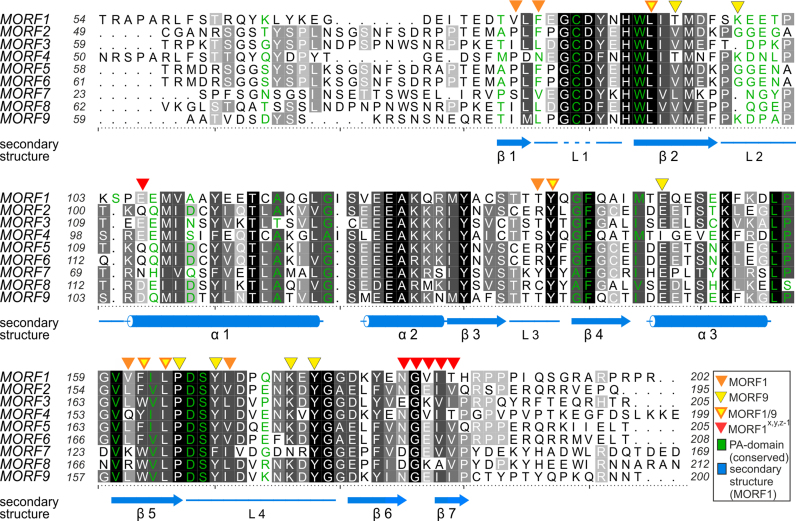
Figure 3.
Alignment of MORF domains of Arabidopsis thaliana. The alignment was prepared by Chimera employing Clustal Omega (40) and shaded with ALSCRIPT (47). Proteins are identified on the left of the aligned sequences. Higher conservation is indicated by a darker background. Numbering refers to the respective proteins. Below the alignment, secondary structure elements (α – α-helix, β – β-sheet, L – loop) of MORF179–190 are shown in marine blue and numbered, a dashed line indicates the modeled loop of residues G84-D86. Orange triangles above the alignment indicate residues of MORF179–190, which are forming the main hydrophobic dimerization interface. Yellow triangles indicate the MORF986–186 dimerization interface. Yellow triangles with orange frame mark positions of residues which both contribute to the dimerization interface of MORF179–190 and MORF986–186. Red triangles above the alignment mark residues of MORF179–190 that form a secondary interface as observed in two different crystal lattices. Green - Residue identities between MORF179–190 and the PA domain of human proprotein convertase subtilisin/kexin type 9 (sequence of PDB ID: 2W2N).
Structural basis for MORF domain dimerization
MORF1 dimerization observed in our structure states a paradigmatic interaction likely to be present in other MORF proteins. The main interface conferring head-to-head symmetric MORF1 dimerization is formed exclusively by hydrophobic interactions (Figures 3 and 4B). Due to the symmetry of the interface, each contact is bipartite at least in a MORF homomer, i.e. is mirrored by the other molecule. F82 makes an interaction with the equivalent residue F82 and sustains a minor hydrophobic contact to P165 of the adjacent molecule (Figures 4B and E). V80 contacts I169, tethering β-sheet 1 and loop 4. V80 and I169 display a medium to low degree of conservation within the MORF family, respectively, and may be important determinants for heteromer specificity (Figure S2C). As the only significant residue of β-strand 2 which is involved in dimer formation, L92 contacts a key residue at the center of the interface, F162. Loop 2 significantly contributes to MORF dimerization with two residues, T138 contacting V161 and Y138 sustaining a hydrophobic interaction with V161 and F162. The most significant structural element in the dimer interface is β-strand 5 which defines the center of the hydrophobic interactions and harbors V161, F162 and L164. Leucine 164 is highly conserved and contacts L164 as well as F162 in the neighboring molecule. F82, V161 and F162 as central parts of the interface, are not conserved within the MORF domain family and may confer selectivity for homo- and heteromer formation (Figures 4B and S2C).
The second interaction surface within the MORF179–190 crystal structures is noteworthy since it independently occurs in two different crystal lattices (Figures 3 and 4C). This interaction relies on residues 185–188 and residues 184–187 (β-strands 7) of the first and second molecule, respectively. Residues V185, I186 and T187 maintain hydrophobic side-chain contacts through the respective β-strands. In addition, hydrogen bonds are formed by T187 and H188 of the adjacent molecules. Lastly, in both molecules N183 maintains a hydrogen bond with E106 in α-helix 1 (Figure 4C). None of the residues involved in the second interface shows a significant degree of conservation among MORF related proteins (not shown).
MORF multimerization has been investigated by yeast two-hybrid (Y2H) studies of the MORF1 domain and fragments thereof. These data can be harnessed for cross-validation with the MORF1 domain interfaces of our structures (Figures 3, 4A and B). It was shown that C-terminal residues of the MORF1 domain (156 – 241) are mandatory for homodimer formation while N-terminal fragments covering residues 61–155 do not support MORF1 multimers in the Y2H assays (22). Strikingly, β-strand 5 with key interacting hydrophobic residues V161, F162 and L164 is missing from these N-terminal fragments underlining the dimer interface in our MORF179–190 structure. In addition, β-strand 5 stabilizes the very N-terminal β-strand 1 which extends the main dimer interface from 400 to almost 1000 Å2 of BSA. Apparently, the sequence of β-strand 1 in isolation does not support MORF1 multimers. Conclusively, the Y2H data support β-strand 1 as part of the intramolecular globular MORF domain and to be folded back as modeled (Figure 2B). If β-strand 1 was able to contact a remote globular domain independently, interaction of the N-terminal fragments with the globular domain would have been observed in the Y2H screen (22). In turn, all fragments containing β-strand 5 do confer MORF1 multimerization. Apparently irrespective of β-strand formation - as surrounding strands are not present - this hydrophobic stretch of residues confers MORF1-MORF1 dimerization.
Interestingly, with one very C-terminal fragment (178–241) a weak MORF1 multimerization is detected in Y2H assays (22). This can only be explained by the secondary MORF1 dimerization interface which almost exclusively spans the C-terminal residues 178–190 in the structure (Figure 4C). The observation that this fragment binds more weakly than the other C-terminal moieties and in turn cannot be explained by the larger hydrophobic interface supports a physiological relevance of the secondary interface observed in the MORF179–190 crystal structures. Prior studies indicate the possibility of more than two MORF proteins to be involved in one RNA editing event (21). The hydrophobic MORF dimerization interface is saturated and its mode of interaction leaves no possibility for higher order MORF multimers. Taken together, dimerization via the C-terminus of the MORF domain may play a key role for higher order MORF assemblies and ultimately editing site specificity in concert with PPR proteins (21).
Implications for MORF heterodimerization
MORF heteromerization has been investigated by extensive binding studies like Y2H, pull-down or immuno-co-localization which altogether draw a complex picture of the MORF interaction network (22). Some of the interactions consistent with all binding studies can be interpreted with our structure. According to our structural model for MORF dimerization, all MORF homomers have a central hydrophobic interface which is highly symmetric. Hence, one interaction conferred by the first molecule has an exact counterpart formed by the second molecule (Figure 4 and 4D). Since most residues of the dimer interface are not conserved within MORF family members and some are variable among A. thaliana MORF proteins, heterodimer formation gives rise to asymmetric interfaces (Figures 3 and S2C).
In Y2H screens, MORF1 multimerization depends on the C-terminal moiety of the MORF domain (residues 156–241) but a larger C-terminal MORF1 fragment (residues 123–241) is required to bind to MORF3 (22). In agreement with our structural data, differences in the C-terminal moiety of the MORF3 MORF domain may lead to a weaker interaction with MORF1. F162 lies at the center of the MORF1 hydrophobic interface. MORF3 in turn has a bulkier tryptophan (W166) at this position (Figure 4B). The extended dimensions of MORF3 W166 and introduction of one charge into the hydrophobic pocket of MORF1 may result in a sterical clash or repulsion which weakens the binding of MORF1 C-terminal fragments. Our structure explains how this specific attenuation of the central interface may be compensated by MORF3 residues contacting the N-terminal moiety of MORF1. In particular, MORF1 V161 is a leucine (L165) in MORF3. The exchange of V161 to a larger hydrophobic residue in MORF3 may reinforce the interaction with the highly conserved MORF1 residues T138 and Y139 only present in the larger MORF1 C-terminal fragment (Figures 3, 4D).
Interestingly, a P165S mutation in MORF1 weakens the interaction with MORF3 but does not change MORF1 homomer formation in one direction of Y2H analysis (22). In general, we expect this mutation to weaken a hydrophobic interaction network between Y87 (Y95 in MORF3), F82 (L89 in MORF3) and W90 (W97 in MORF3) which contributes to MORF dimer formation (Figure 4E). This weakening effect may have a stronger impact on a heteromeric MORF3-MORF1 interaction since this dimer is already impaired by the exchange of F162 to W166 in MORF3. Hence, the sum of both attenuating effects (P165S in MORF1 and MORF1 F162 vs. W166 in MORF3) may not be compensated by the consolidated interaction of L165 (MORF3), T138 and Y139 (MORF1). In general, the replacement of F162 in MORF1 by a tryptophan in other MORFs may weaken the central hydrophobic interface as for example observed for MORF7, yet this effect may be alleviated by the interface formed by MORF1 residues V161, T138 and Y139 and its varying equivalents in other MORF proteins. On the other hand, MORF1 residues V161 and T139 as well as adjacent residues (e.g. D95) are not conserved in other MORF proteins and may also introduce sterical clashes which cannot be compensated by a phenylalanine at the position of F162 in MORF1 (Figures 3 and 4D). Taken together, our structure provides a consistent model for MORF multimerization but further structures of other MORF domains as homodimers and ideally heterodimers are required to fully elucidate the molecular interactions underlying MORF multimerization.
The structure of chloroplast MORF986–186 confirms the MORF domain fold and its hydrophobic dimerization interface
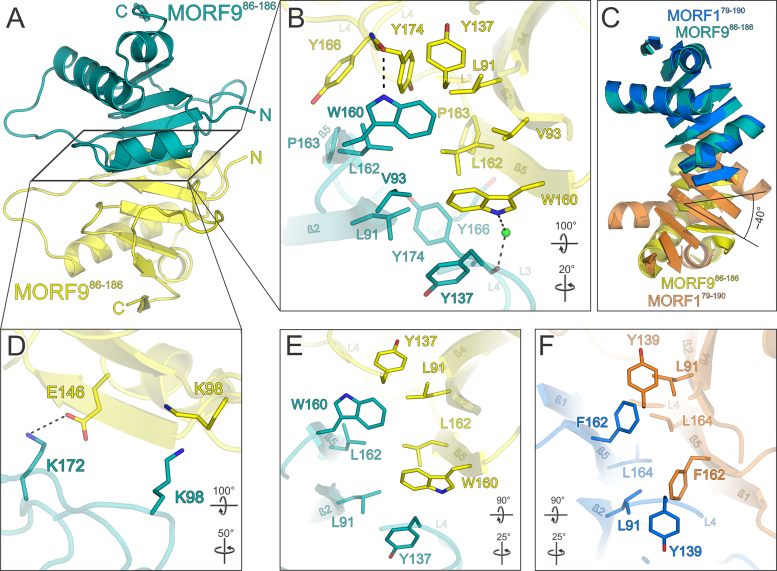
The structure of MORF179–190 allowed us to devise homology models of other MORF proteins and consequently employ these to predict domain borders for suitable crystallization constructs. In plant cells, MORF1 is localized to mitochondria, so we set out to obtain the structure of MORF9 which is only found in chloroplasts (19). To obtain a suitable construct for crystallization, MORF9 without its predicted N-terminal β-strand 1 (MORF986–186) was purified where we did not observe any signs of higher molecular weight aggregates as observed for MORF179–190 (Figure 1A and B). We compared purified MORF986–186 to the monomeric MORF179–190 protein by analytical size exclusion chromatography. MORF986–186 had exactly the same elution behavior as MORF179–190 monomer and is thus considered monomeric in solution. (Figures 1E, S3A, S3B). MORF986–186 was the only variant of the protein which yielded crystals of suitable quality of space group P6522. Complete datasets of MORF986–186 crystals were collected to 2.25 Å resolution and the structure was solved by molecular replacement employing the structure coordinates of a MORF179–190 monomer. The structure contains two MORF986–186 molecules in the asymmetric unit and was refined to low R/Rfree factors (0.195 / 0.238) while maintaining good stereochemistry (Table S1). The MORF domains of MORF1 and MORF9 share nearly 60% of identical residues which is also mirrored by a high degree of structural similarity with an RMSD of 2.56 Å for Cα atoms (Figures 3, S3C). Strikingly, MORF986–186 dimerizes in a similar fashion like MORF1 forming a nearly symmetric head to head interface (Figure 5A). Like F162 in MORF1, W160 is a central point of the MORF9 dimerization interface, making hydrophobic contacts to L91, V93, Y137, L162 and Y174 in both molecules (Figure 5B). Notably, unlike MORF1, the MORF9 dimer interface has a slight asymmetry, since W160 in one molecule directly contacts the backbone carbonyl of Y174 and in the other encages a water molecule to contact the carbonyl with hydrogen bonds. A second key residue for MORF dimerization is L162 in MORF9 (L164 in MORF1), which establishes hydrophobic contacts to its counterpart L162 as well as L91, W160 and Y174. Lastly, MORF9 P163 makes hydrophobic contacts to Y166 and Y174 in one molecule and to only Y166 in the other while the homologous MORF1 P165 has only a minor contribution to dimerization (Figures 5B, 4E). In the periphery of the central interface symmetrical contacts between E146 and K172 via a hydrogen bond are observed while K98 sustains a hydrophobic contact to its counterpart across the two molecules (Figure 5D). In comparison to the MORF1 dimer interface, MORF9 molecules are rotated by about 40° in plane with the β-sheet with respect to each other while the translational vector is negligible (Figure 5C). The center of rotation matches to the area holding two key interface residues, W160 (F162 in MORF1) and L162 (L164 in MORF1) which are as a consequence not as severely displaced as residues in the periphery. A possible explanation for this sliding rotation of the two β-sheet faces may be the missing N-terminal β-strand 1 in MORF986–186, which in MORF1 contributes to the dimerization interface with several residues (Figures 3, 4B) and consolidates the interaction. A comparison of all interface residue positions shared by MORF1 and MORF9 clearly shows that both proteins employ the same mode of dimerization (Figure 5E and F) and that these molecular contacts are paradigmatic for MORF dimer formation.
Figure 5.
Structure of the MORF986–186 MORF domain and its dimerization in relation to MORF1. (A) Overview of the MORF986–186 structure. A central MORF986–186 molecule (teal) forms a tight nearly symmetrical head-to-head dimer via a hydrophobic interaction surface with a second MORF986–186 molecule (yellow). (B) Close-up view of the main hydrophobic protein–protein interface of MORF986–186 shown in (A). (C) Superimposition of the MORF179–190 (marine blue and orange) and MORF986–186 (teal and yellow) dimers. Loops are omitted for clarity. The second MORF986–186 molecule (yellow) is rotated by an angle of ∼40° with respect to the respective MORF179–190 moiety (orange) as indicated for β-strand 4. (D) Close-up view of a peripheral MORF986–186 interface of the dimer shown in (A). (E and F) Comparison of the hydrophobic dimerization interfaces of MORF179–190 and MORF986–186. The selection of interacting residues is limited to positional or sequence identities shared by MORF1 and MORF9. Interacting residues are shown as sticks and colored by atom type. Carbon – as for the respective molecule; nitrogen – blue; oxygen – red; sulfur – yellow. Water oxygens are shown as green spheres. Dashed lines between atoms represent hydrogen bonds. Rotation symbols indicate views relative to Figure 2A.
Validation of MORF dimerization by yeast two-hybrid studies and pulldown assays employing structure-based MORF1 mutants
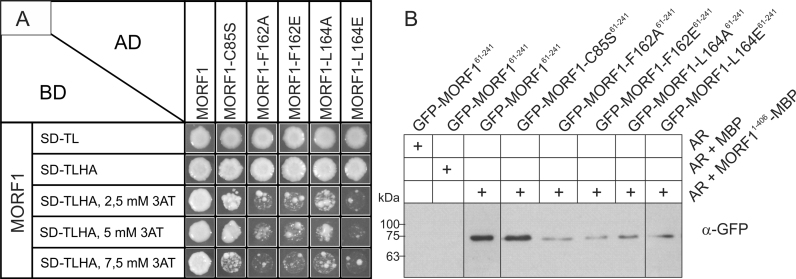
To validate the key interface residues of the MORF1-MORF1 interaction by structure-based point mutations, we mutated the most prominent interface residues F162 and L164. Here, mutations to alanine represent a conservative approach to probe the contribution of each, F162 and L164, to the interface and not impairing other residues while mutation to glutamate should severely distort the interface by introducing a polar residue (Figure 4B). In addition, C85 was mutated to a serine to rule out any significant contribution to MORF1 dimerization and thus identify β-strand 1 as part of a domain swap in the MORF179–190 crystal structure (Figure S2A and B). Dimerization was analyzed by Yeast-2-hybrid and pull-down assays. In Yeast-2-hybrid assays, all mutated MORF1 proteins, F162A, F162E, L164A, L164E, C85S and control show interaction with wild type MORF1 on the selection plate without 3AT (Figure 6A). However, the MORF1 interactions with the mutants at residues F162 and L164 are already strongly inhibited by low concentration (2.5 mM) of 3AT (Figure 6A), while neither wild type protein nor the C85S mutant were affected. These results suggest the crucial role of F162 and L164 in MORF1-MORF1 interaction and no significant contribution of C85 in multimerization which supports our remodeling of the domain-swapped β-strand 1 as part of the globular MORF domain. Pull-down assays with recombinant proteins mirror the results of Y2H assays. All MORF1 mutant proteins with altered amino acids of residues 162 or 164 bind only weakly to the wild type MORF1 protein while the C85S mutant and wild type MORF1 proteins show similar affinities to the wild type MORF1 protein (Figure 6B). These independent protein-protein interaction assays strongly support our interpretation of the two MORF1 and also the MORF9 crystal structures regarding the integrity of the MORF domain and also its mode of dimerization.
Figure 6.
Yeast two-hybrid and pulldown protein-protein interaction analyses with MORF1 and structure-based mutants. (A) Yeast colony growth of the MORF–MORF protein combinations after 6 days on control medium (SD-TL) and selection medium (SD-TLHA) containing different concentrations of 3AT. The C85S mutant shows interaction similar to the wildtype up to 5 mM 3AT while all other mutants are showing reduced growth at all 3AT concentrations, indicating weaker interaction. (B) Pull-down of GFP tagged MORF161-241 and its respective mutants with MBP tagged MORF1 bound to amylose resin (AR) as a bait. Shown are the respective MORF-GFP GFP antibody chemiluminescence signals in the respective gel lanes. Without any protein and with MBP as a bait no GFP antibody signal is detected. The C85S mutant shows a signal similar to the MORF161-241 wild type while all other mutated MORFs show reduced signal intensity.
The MORF domain shows structural similarities to a subtilisin protease associated domain and an NFLD domain
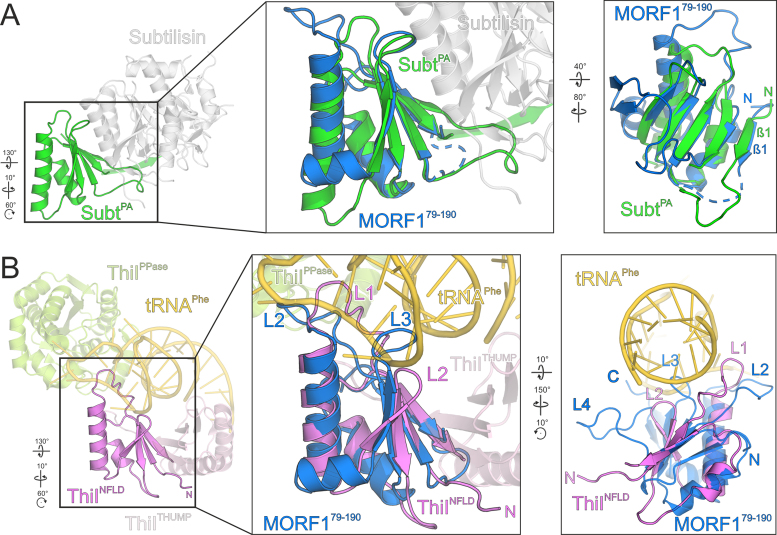
To compare the fold of the MORF domain to known structures, we carried out a DALI search with the MORF1 domain (38). The highest structural similarity was found to the subtilisin protease associated (PA) domain which was shown to assist folding of the subtilisin catalytic domain (39). In a sequence alignment, 24% of the residues of the PA domain align to MORF1 by sequence identity (Figure 3). In a sequence-based structural alignment, 68/87 aligned Cα atoms have an RMSD of 1.77 Å (Figure 7A) (40). Notably, some of the hits among the subtilisin prodomain family have the very N-terminus - which is extruded in the MORF1 domain crystals - folded back onto the globular moiety and forming a β-strand (Figure 7A). Although we cannot entirely rule out the tetrameric assembly as observed in the MORF179–190 crystal lattice, the MORF1 N-terminus is modeled in agreement with the members of the subtilisin pro domain family. Although the PA domain and MORF1 overall fold similarly, there are significant differences. For example, the transition of α-helix 1 and β-strand 3 of the PA domain is interspersed by a second α-helix in the MORF domain instead of a loop. Also, β-strands 6 and 7 as part of the MORF fold are missing in the PA domain. A functional relation between the PA domain and the MORF fold is rather unlikely.
Figure 7.
The MORF domain resembles a subtilisin PA domain and an NFLD fold. (A) Left – human pro-protein convertase subtilisin/kexin type 9 (gray) with its protease associated (PA) domain (green), (PDB ID: 2W2N). Center panel – Close-up view of a superimposition of MORF179–190 (marine blue) and the PA domain (green). Right panel – side-view of the superimposition (center panel) illustrating the configuration of β-sheet 1 of the PA domain. Colors as in center panel. (B) Left – Thermothoga maritima 4-thio-uracil synthetase ThiI (PDB ID: 4KR6) with its THUMP (light pink), NFLD (pink) and PPase (light green) domains bound to a stretch of tRNA (gold). Center panel – close-up view of a superimposition of ThiI NFLD (pink) and MORF179–190 (marine blue). RNA binding protein loops of ThiI and respective loops of MORF179–190 are indicated. For clarity reasons, β-sheets 6 and 7 and L4 of MORF179–190 which do not align to NFLD are not shown. Right panel – side-view of the superimposition (center panel) illustrating the position of RNA binding loops of ThiI and MORF179–190 loops. Dashed lines in the ribbon plots represent modeled residues G84-D86 of MORF179–190. Rotation symbols indicate views relative to Figure 2A.
Strikingly, in the DALI search list the PA domain is followed by an N-terminal ferredoxin-like domain (NFLD). In conjunction with domains in thiouridine synthases, methylases and pseudouridine synthetases (THUMP), the NFLD binds bacterial and archaeal tRNAs and positions them for enzymatic modifications (41–43). An s4U8 tRNA modification plays a central role in bacterial UV protection as a sensor for near-UV radiation, the reaction mechanism elicited by ThiI remains elusive (43,44). The highest structural similarity of MORF1 to an NFLD was found for Pyrococcus horikoshii ThiI where 62/72 aligned Cα atoms superimpose with an RMSD of 2.0 Å. A sequence alignment of NFLD with MORF1 yields 22% identical residues, which is comparable to the degree conservation between MORF and PA domains. The MORF domain structurally differs from the NFLD and PA domain as it harbors an additional α-helix and two C-terminal β-sheets.
For superimposition with the MORF fold, we used the structure of Thermotoga maritima ThiI which has the same fold as the Pyrococcus horikoshii ortholog but was crystallized with a bound tRNA fragment (43). Strikingly, the NFLD domain, which positions the ThiI PPase domain for catalysis, superimposes well with the MORF domain (57/74 residues with an RMSD of 2.3 Å). The NFLD domain uses two loops to position the single stranded RNA loop tip while the THUMP domain coordinates the RNA stem. It is intriguing to think of a similar RNA binding mode for MORF1, employing L2, L3 and possibly also L4 to position the editing site for catalysis (Figure 7B). We tested MORF1 and MORF9 for specific RNA binding by band shift assays but were unable to detect any affinity (not shown). However, in the context of an editosomal reaction it cannot be excluded that MORF proteins, too, contact RNA.
DISCUSSION
Paradigmatic structures of two MORF domains shed light on MORF multimerization
The MORF1 tetrameric assembly observed in our crystal structure and the monomeric MORF domain in solution can be rationalized. During purification, unspecific His6-tagged multimers are observed (Figure 1A and B). We hypothesize that additional residues of the TEV cleavage site/His6-tag may have mediated these unspecific intramolecular contacts. Upon tag cleavage, MORF1 monomerizes and the first β-sheet is not sufficiently restrained to fold back onto the globular domain and foster dimer formation (Figure 1D). Under high protein concentrations during crystallization, intermolecular contacts are formed again in the crystal lattice facilitating crystallization with a relatively low turnover. Crystallization is a selective process and the low amounts of crystals in the droplets most likely reflect the low degree of tetramerization which would be unusual for a native tetramer but still favorable over the native dimer. Arguably, a physiological function in detaching the first β-strand from the globular MORF domain to establish intermolecular contacts of two distant molecules cannot be entirely ruled out, but does anyway not contradict the model for MORF dimerization. The MORF986–186 protein does not form unspecific multimers during purification, most likely due to the absence of the first β-strand which cannot contribute to the dimer interface at all (not shown). The striking structural similarity of MORF1 and MORF9 homodimer formation rationalizes prior studies and most likely applies to the entire MORF protein family. Based on the structures reported in this work, we will now aim at screening fragments to obtain a MORF dimer in solution and conduct further structure-based mutagenesis studies.
MORF proteins have been widely acknowledged as integral parts of flowering plant editosomes (13,18,19,21). The exact role of these proteins and the mode of multimerization have so far remained elusive due to missing structural information of this hitherto unknown fold. With our structures we provide the basis for the MORF domain, its dimerization and potentially higher order multimers which have been hypothesized (19,21). Symmetrical protein interfaces are frequently observed, which also applies to all MORF homomers dimerizing via the main hydrophobic interface (45). Since around half of the MORF interface residues are not conserved within the MORF protein family, all MORF heteromers fall into the category of asymmetric interfaces, which adds one level of complexity to MORF dimerization. Our structure allows an initial interpretation of prior MORF–MORF interaction studies, however, additional MORF homo- or heterodimeric structures are required to fully assess the molecular rules of MORF multimerization and ultimately assess their association with PPR proteins (22). To this end, our structure paves the way for consecutive structural studies of other MORF domains in isolation or as multimers.
MORF domain assemblies may assist specific RNA positioning for editosomal cytidine deamination
In this study we have discovered unexpected parallels between the NFLD of tRNA modifying enzymes and plant editosomal MORF proteins. The NFLD in prokaryotes is crucial for single stranded RNA recognition and substrate positioning, the same function may apply to the MORF domain. Regardless of a common evolutionary context, the contrast between the two systems lies in the mode of RNA recognition. While ThiI recognizes the stem and tip of the folded tRNA via NFLD loops and a THUMB domain, the MORF domain may bind to a linear RNA sequence. MORF dimers superimposed onto the NFLD domain are in agreement with a single stranded RNA binding mode where the protein loops of different MORF domains contact the RNA (Figure 7B). This way, heteromeric MORF assemblies with a defined composition may address RNA even sequence-specifically. PPR proteins are the prime entities in the plant editosome recognizing the RNA sequence. MORF proteins are known to bind to PPR proteins and may be responsible to bridge the gap between PPR tract and editing site and aid in substrate positioning consolidating earlier hypotheses (46). This may occur especially in cases where an additional level of selectivity is required or the enzymatic activity is added in trans. This implies not only a bridging function for substrate positioning but MORF proteins may add another level of complexity to the plant editosome since they can dimerize. Different combinations of MORF proteins may determine the fate of an editing site as observed at the eight MEF13 target sites which require all MORF3, MORF8 and MEF13 (21). Since MORF proteins are absent in some mosses and ferns they may have emerged as mediators of the next level of complexity, site specificity or regulation during evolution of the flowering plant editosome.
ACCESSION NUMBERS
Structure coordinates and diffraction data were deposited with the Protein Data Bank (http://www.pdb.org) under accession codes 5MPW (MORF179–190, space group P1), 5MPX (MORF179–190, space group P21), 5MPY (MORF986–186).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dagmar Pruchner, Angelika Müller, and Bianca Wolf for excellent experimental help. We thank Markus Wahl for continuously supporting this project and providing infrastructure, Jan Wollenhaupt for sharing his MALS expertise, Claudia Alings for excellent help with protein crystallization, Britta Girbardt for excellent technical support and Agnieszka Pietrzyk for data collection. We acknowledge access to beamlines BL14.1/2/3 of the BESSY II storage ring (Berlin,Germany) via the Joint Berlin MX-Laboratory sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück Centrum, and the Leibniz-Institut für Molekulare Pharmakologie.
Authors Contributions: A.B., M.T. and G.W. designed research, S.H., M.T., M.S. and G.W. performed research, S.H., M.T. and G.W. analyzed data, S.H., M.T., A.B. and G.W. wrote the manuscript.
Footnotes
Present address: Gert Weber, Universität Greifswald, Institut für Biochemie, Molekulare Strukturbiologie, Felix-Hausdorff-Str. 4, 17487 Greifswald, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [TA642/10-1, TA 642/6-1, TA 642/3-1 to M.T.; BR726/11-2 to A.B.]; Heisenberg fellowship from Deutsche Forschungsgemeinschaft [TA642/4-2 to M.T.]. Funding for open access charge: Ulm University and Greifswald University budget.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kotera E., Tasaka M., Shikanai T.. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005; 433:326–330. [DOI] [PubMed] [Google Scholar]
- 2. Zehrmann A., Verbitskiy D., van der Merwe J.A., Brennicke A., Takenaka M.. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009; 21:558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andrés C., Lurin C., Small I.D.. The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 2007; 129:14–22. [Google Scholar]
- 4. Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B. et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004; 16:2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okuda K., Chateigner-Boutin A.-L., Nakamura T., Delannoy E., Sugita M., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T.. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009; 21:146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmitz-Linneweber C., Small I.. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008; 13:663–670. [DOI] [PubMed] [Google Scholar]
- 7. Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A.. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013; 47:335–352. [DOI] [PubMed] [Google Scholar]
- 8. Boussardon C., Avon A., Kindgren P., Bond C.S., Challenor M., Lurin C., Small I.. The cytidine deaminase signature HxE(x) n CxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phytol. 2014; 203:1090–1095. [DOI] [PubMed] [Google Scholar]
- 9. Hayes M.L., Giang K., Berhane B., Mulligan R.M.. Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. J. Biol. Chem. 2013; 288:36519–36529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salone V., Rüdinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., Small I., Knoop V., Lurin C.. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007; 581:4132–4138. [DOI] [PubMed] [Google Scholar]
- 11. Boussardon C., Salone V., Avon A., Berthomé R., Hammani K., Okuda K., Shikanai T., Small I., Lurin C.. Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell. 2012; 24:3684–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tillich M., Hardel S.L., Kupsch C., Armbruster U., Delannoy E., Gualberto J.M., Lehwark P., Leister D., Small I.D., Schmitz-Linneweber C.. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:6002–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun T., Germain A., Giloteaux L., Hammani K., Barkan A., Hanson M.R., Bentolila S.. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E1169–E1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vermel M., Guermann B., Delage L., Grienenberger J.-M., Maréchal-Drouard L., Gualberto J.M.. A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi X., Hanson M.R., Bentolila S.. Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res. 2015; 43:3814–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi X., Germain A., Hanson M.R., Bentolila S.. RNA recognition motif-containing protein ORRM4 broadly affects mitochondrial RNA editing and impacts plant development and flowering. Plant Physiol. 2016; 170:294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun T., Shi X., Friso G., Van Wijk K., Bentolila S., Hanson M.R.. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015; 11:e1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bentolila S., Heller W.P., Sun T., Babina A.M., Friso G., van Wijk K.J., Hanson M.R.. PNAS Plus: RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1453–E1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Härtel B., Brennicke A.. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:5104–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bentolila S., Oh J., Hanson M.R., Bukowski R.. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013; 9:e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glass F., Härtel B., Zehrmann A., Verbitskiy D., Takenaka M.. MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in Arabidopsis thaliana. Mol. Plant. 2015; 8:1466–1477. [DOI] [PubMed] [Google Scholar]
- 22. Zehrmann A., Härtel B., Glass F., Bayer-Császár E., Obata T., Meyer E., Brennicke A., Takenaka M.. Selective homo- and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana. J. Biol. Chem. 2015; 290:6445–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mueller U., Darowski N., Fuchs M.R., Förster R., Hellmig M., Paithankar K.S., Pühringer S., Steffien M., Zocher G., Weiss M.S.. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron Radiat. 2012; 19:442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vonrhein C., Blanc E., Roversi P., Bricogne G.. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007; 364:215–230. [DOI] [PubMed] [Google Scholar]
- 26. Adams P.D., Afonine P. V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P. V, Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D.. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. Biol. Crystallogr. 2008; 64:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Afonine P. V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emanuelsson O., Brunak S., von Heijne G., Nielsen H.. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007; 2:953–971. [DOI] [PubMed] [Google Scholar]
- 31. Braun P. Interactome mapping for analysis of complex phenotypes: Insights from benchmarking binary interaction assays. Proteomics. 2012; 12:1499–1518. [DOI] [PubMed] [Google Scholar]
- 32. Braun P., Aubourg S., Van Leene J., De Jaeger G., Lurin C.. Plant protein interactomes. Annu. Rev. Plant Biol. 2013; 64:161–187. [DOI] [PubMed] [Google Scholar]
- 33. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 34. Krissinel E. Crystal contacts as nature's docking solutions. J. Comput. Chem. 2010; 31:133–143. [DOI] [PubMed] [Google Scholar]
- 35. Dehouck Y., Biot C., Gilis D., Kwasigroch J.M., Rooman M.. Sequence-structure signals of 3D domain swapping in proteins. J. Mol. Biol. 2003; 330:1215–1225. [DOI] [PubMed] [Google Scholar]
- 36. Huang Y., Cao H., Liu Z.. Three-dimensional domain swapping in the protein structure space. Proteins Struct. Funct. Bioinforma. 2012; 80:1610–1619. [DOI] [PubMed] [Google Scholar]
- 37. Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., Ben-Tal N.. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013; 53:199–206. [Google Scholar]
- 38. Holm L., Rosenström P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gallagher T., Gilliland G., Wang L., Bryan P.. The prosegment-subtilisin BPN’ complex: crystal structure of a specific ‘foldase’. Structure. 1995; 3:907–914. [DOI] [PubMed] [Google Scholar]
- 40. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 41. Aravind L., Koonin E. V.. THUMP – a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001; 26:215–217. [DOI] [PubMed] [Google Scholar]
- 42. Waterman D.G., Ortiz-Lombardía M., Fogg M.J., Koonin E. V., Antson A.A.. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 2006; 356:97–110. [DOI] [PubMed] [Google Scholar]
- 43. Neumann P., Lakomek K., Naumann P.-T., Erwin W.M., Lauhon C.T., Ficner R.. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 2014; 42:6673–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Favre A., Michelson A.M., Yaniv M.. Photochemistry of 4-thiouridine in Escherichia coli transfer RNA1Val. J. Mol. Biol. 1971; 58:367–379. [DOI] [PubMed] [Google Scholar]
- 45. Plaxco K.W., Gross M.. Protein complexes: the evolution of symmetry. Curr. Biol. 2009; 19:R25–R26. [DOI] [PubMed] [Google Scholar]
- 46. Takenaka M. How complex are the editosomes in plant organelles. Mol. Plant. 2014; 7:582–585. [DOI] [PubMed] [Google Scholar]
- 47. Barton G.J. Alscript: a tool to format multiple sequence alignments. Protein Eng. Des. Sel. 1993; 6:37–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.