Abstract
Ribosome biogenesis in Saccharomyces cerevisiae involves a regulon of >200 genes (Ribi genes) coordinately regulated in response to nutrient availability and cellular growth rate. Two cis-acting elements called PAC and RRPE are known to mediate Ribi gene repression in response to nutritional downshift. Here, we show that most Ribi gene promoters also contain binding sites for one or more General Regulatory Factors (GRFs), most frequently Abf1 and Reb1, and that these factors are enriched in vivo at Ribi promoters. Abf1/Reb1/Tbf1 promoter association was required for full Ribi gene expression in rich medium and for its modulation in response to glucose starvation, characterized by a rapid drop followed by slow recovery. Such a response did not entail changes in Abf1 occupancy, but it was paralleled by a quick increase, followed by slow decrease, in Rpd3L histone deacetylase occupancy. Remarkably, Abf1 site disruption also abolished Rpd3L complex recruitment in response to starvation. Extensive mutational analysis of the DBP7 promoter revealed a complex interplay of Tbf1 sites, PAC and RRPE in the transcriptional regulation of this Ribi gene. Our observations point to GRFs as new multifaceted players in Ribi gene regulation both during exponential growth and under repressive conditions.
INTRODUCTION
In eukaryotic cells, ribosome biogenesis is a large scale, highly complex process that requires the activity of hundreds of genes transcribed by all three nuclear RNA polymerases (1–3). In rapidly dividing cells, four ribosomal RNAs (rRNAs) and ∼80 ribosomal proteins (RPs) are required as building blocks for the production of thousands of ribosomes every minute (3). Moreover, building the cytoplasmic 60S and 40S ribosomal particles involves the sequential, orchestrated action of small nucleolar RNAs (snoRNAs), one of the most important families of nuclear non-protein-coding (nc) RNAs (4), and over 200 proteins involved in rRNA maturation, ribosomal particle assembly and export from the nucleus (2).
At the transcriptional level, ribosome biogenesis has been best characterized in Saccharomyces cerevisiae, where it involves at least 750 individual transcription units. These include both homogeneous and well-defined families, such as the ∼150 active copies of large rRNA and 5S rRNA genes, the 138 genes for cytoplasmic RPs and the 77 snoRNA genes, and the more composite group of protein-coding genes whose products contribute to ribosome production without being a structural part of it (1). Genes of the latter group are most commonly referred to as Ribi genes or, collectively, as the Ribi regulon, to indicate that their expression is tightly co-regulated in response to environmental or genetic perturbations (5,6). Many (∼150) Ribi genes code for rRNA modification and ribosome assembly factors, but the regulon also includes RNA polymerase (Pol) I and Pol III subunits, nucleotide metabolism enzymes, aminoacyl-tRNA synthetases, as well as translation and tRNA metabolism factors (6). A different term, ‘RRB regulon’, was initially used to define a set of 65 transcriptionally coregulated genes required for ribosome and rRNA biosynthesis. Later, the RRB regulon was expanded to include other yeast genes based on their conformity to the features of the first defined set. This was done through a comprehensive system-level analysis integrating transcriptional response profiles, functional gene annotations and computational analysis of transcriptional control regions (7,8). The RRB and Ribi regulons largely overlap, even though it is often difficult to distinguish unambiguously between RRB/Ribi genes and other yeast genes (8).
In terms of gene structure, the most distinctive feature of a Ribi gene is the presence in its promoter region of two regulatory elements, called PAC and RRPE, generally located within the first 200 base pairs (bp) upstream of the transcription start site (TSS) (1,6,8,9). The two Myb-like, helix-turn-helix transcription factors Dot6 and Tod6 have been identified as the cognate factors of the PAC element (10–12), while the RRPE is recognized by Stb3 (13). Motif recognition by these factors mediates transcriptional repression through recruitment of Rpd3L histone deacetylase, in a process antagonized by signals from TORC1, Sch9 and protein kinase A (PKA), with the Sch9 protein kinase directly phosphorylating both Dot6/Tod6 and Stb3 (5,14,15). Both RRPE and PAC thus seem to mediate transcriptional repression, even though the RRPE has previously been associated with the activation of Ribi gene transcription mediated by the zinc finger protein Sfp1 (16).
Based on current knowledge, Ribi gene promoters are generally perceived as differing from typical yeast promoters (17), in particular from those of RP genes, in that they have been best characterized with respect to repressive regulatory elements, while the presence of common activator-binding regulatory elements (UAS) has not been emphasized nor systematically addressed. Sparse previous observations suggested that the promoter regions of some genes involved in ribosome biogenesis are associated with transcription factors (TF) like Tbf1 and Abf1, known to bind large sets of promoters (1,18–21). With the aim of understanding how general is the presence of activator-binding sites in Ribi gene control regions, we carried out a systematic analysis of evolutionarily conserved motifs in Ribi promoter of Saccharomyces using phylogenetic footprinting (22). We found that the vast majority of Ribi gene promoters contain, at positions typical of upstream activating sequences (UASs), binding sites for Abf1, Reb1 and, less frequently, Rap1 and Tbf1 proteins, collectively referred to as General Regulatory Factors (GRFs) because of their widespread presence as multipurpose DNA binding proteins in yeast (23,24). These TFs were found to associate with and to be required for full expression of target Ribi gene promoters under rich medium growth conditions. The transcriptional response to glucose starvation was also profoundly altered at Ribi genes unable to bind the cognate GRFs, and in the case of the DBP7 gene it was found to rely on the simultaneous presence of GRF, PAC and RRPE recognition elements. Taken together our data support the idea that GRFs are key players in the regulation of Ribi genes as previously observed also for snoRNA and RP genes.
MATERIALS AND METHODS
Computational analysis
The list of 236 Ribi genes analyzed was obtained from Supplementary Table S1 of (6). Coding sequences were recovered from the Saccharomyces Genome Database (SGD). Orthologous sequences from Saccharomyces paradoxus, S. mikatae, S. bayanus and S. kudriavzevii were identified through a BLASTN search at SGD (http://seq.yeastgenome.org/cgi-bin/blast-fungal.pl). Upstream sequences of each Ribi gene (600 bp upstream of ATG) in the different genomes were obtained from NCBI. Upstream sequences of orthologous genes were aligned with ClustalX, and the alignments visualized with GeneDoc (available upon request). The identification of potential binding sites for transcription factors (TFs) was based on visualization of conserved sequence blocks and integration with information reported in the SGD and in the Yeast Promoter Atlas (YPA) (25). The promoter characterization was then completed with the help of the Jaspar database (26), MatInspector (http://www.genomatix.de) and literature to identify and label known cis-regulatory sequences.
Strain construction and cultures
Saccharomyces cerevisiae strains carrying mutations in the promoter regions of selected Ribi genes were obtained with two subsequent steps of homologous recombination in order to introduce a URA3 selection marker at the Ribi gene locus, and then replace URA3 with mutated versions of the gene. URA3 cassettes were PCR-synthesized as previously described (18), in two PCR rounds to increase the length of the flanking homology regions, using the oligonucleotide primers listed in Supplementary Table S1. Each deletion cassette (1–2 μg) was then used to transform ABF1, REB1 or TBF1 TAP-tagged strains from the Yeast TAP-fusion library (Open Biosystems). Second step replacements with mutated gene cassettes were carried out as described (18). Mutant gene replacement cassettes were constructed through mutagenic PCR using the oligonucleotide primers listed in Supplementary Table S1. The SDS3-13xMyc tagged strains were created by a single step of transformation with a PCR-synthesized fragment. In this case, PCR cassette was produced by a one-step PCR with Phusion DNA Polymerase on the pFA6a-13Myc-KanMX6 vector (27) using specific primers listed in Supplementary Table S1. A list of S. cerevisiae strains used throughout this work is reported in Supplementary Table S2. All strains were grown in YPD medium (1% yeast extract, 2% peptone, 2% dextrose). For ChIP and expression analysis overnight cell cultures pre-grown in YPD medium were diluted into fresh medium to a cell density of 4 × 106 cells/ml and grown at 30°C until they reached a density of 1.5–2 × 107 cells/ml before being collected for further analysis.
For glucose starvation experiments, overnight cell cultures in YPD were diluted into fresh medium to a cell density of 4 × 106 cells/ml and grown at 30°C to a density of 1.5 × 107 cells/ml. Samples were collected, representing the 0 min time point; the remaining cultures were then centrifuged at 3000 rpm for 3΄ at room temperature and suspended in YP medium (1% yeast extract, 2% peptone) pre-warmed at 30°C. Cells were collected at various time points (generally 10, 20 and 30 min) after the shift.
Gene expression analysis
Total RNA was extracted from yeast cells using the RNeasy mini kit (Qiagen) with DNase I treatment according to the manufacturer's suggestions. Total RNA (500 ng per sample) was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories) with the hexamer random priming protocol. The cDNA samples were diluted 1:5 and used as templates in quantitative (q) PCR reactions with Applied Biosystems 7300 Real-Time PCR System and SYBR® Green Master Mix (Life Technologies). The oligonucleotide primers we used are listed in Supplementary Table S1. The concentration of each primer pair was optimized as described for ChIP-qPCR (see below). In all experiments, expression levels of the analyzed genes were normalized to those of HHT2 (used as an internal standard). For the analyses in Figure 3, the expression levels upon promoter mutation were then expressed as 2−ΔΔCT (with ΔΔCT = ΔCT mutant strain – ΔCT wt strain). In Figures 4 and 6 the expression levels were calculated as 2–ΔΔCT with ΔΔCT = ΔCT mutant strain – ΔCT wt strain at t0.
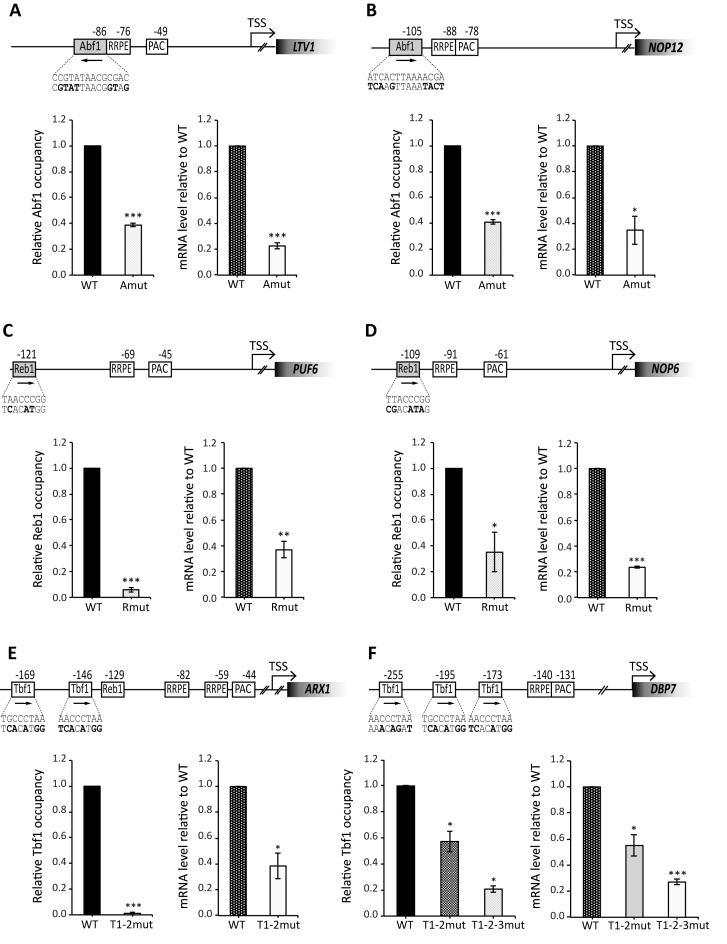
Figure 3.
GRF requirements for Ribi gene expression. Functional analysis of six different Ribi gene promoters displaying binding sites respectively for Abf1 (LTV1, panel A; NOP12, panel B), Reb1 (PUF6, panel C; NOP6; panel D) and Tbf1 (ARX1, panel E; DBP7, panel F). Strains carrying mutations in the GRF binding site and a TAP-tagged allele of either ABF1 (LTV1 and NOP12), REB1 (PUF6, NOP6) or TBF1 (ARX1, DBP7) were analyzed for GRF occupancy at promoter and mRNA expression. For each gene, a schematic representation of the promoter region and of its alteration in the mutant construct is shown. Distances reported are with respect to TSS and mutated sequence in the GRF binding site is indicated under the corresponding wt sequence (mutated bases are in bold). Left graph: GRF occupancy in wild type (WT; black bars) and mutated strain (gray–white bars) assessed by ChIP-qPCR and calculated as fold-enrichment relative to HHT2. GRF enrichment at each promoter is reported as relative to the level of enrichment measured in the wt strain. Right graph: Ribi gene expression levels in wild type (WT, gray dotted bars) and mutant strains (white bars). mRNA levels were measured by RT-qPCR analysis of total RNA extracted from exponentially growing cells and expressed as relative to the expression level in the corresponding wt strain. Data are represented as mean ± SEM calculated in three independent experiments. An unpaired Student's t-test was used to compare the means of measurements. *P < 0.05; **P < 0.01; ***P < 0.001.
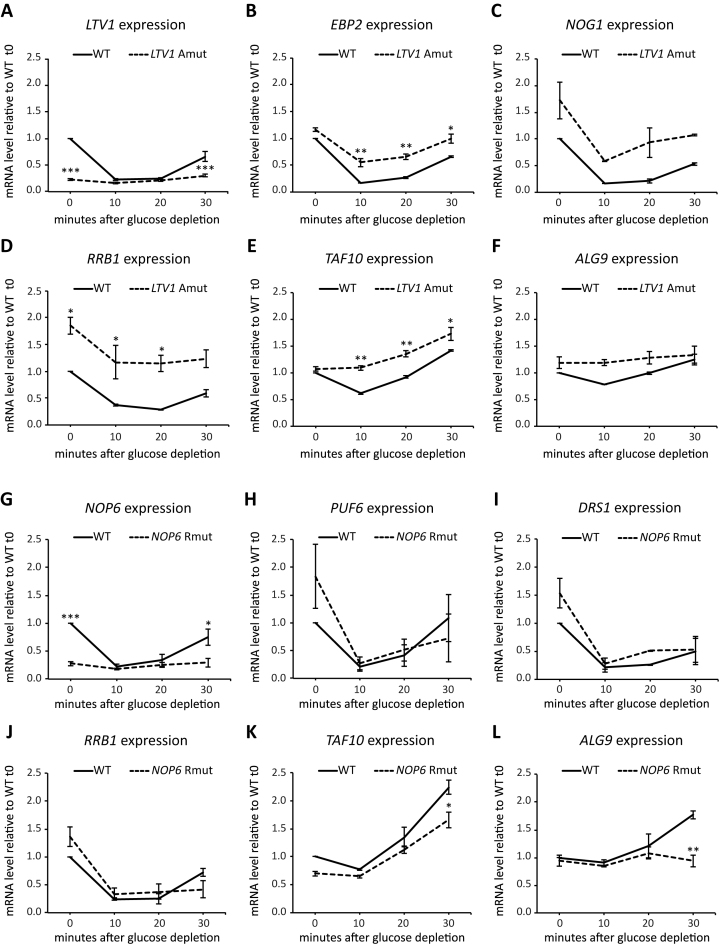
Figure 4.
Nutrient-dependent regulation of expression from wt and mutant Ribi gene promoters. (A–F) Exponentially growing yeast strains carrying either wild type (WT) or Abf1 site-mutated LTV1 gene (LTV1 Amut) were shifted to YP medium without glucose. Samples were collected before (0) and 10, 20 or 30 min after the shift for total RNA extraction and RT-qPCR analysis. Expression levels of LTV1 (A), EBP2 (B), NOG1 (C), RRB1 (D), TAF10 (E) and ALG9 (F) in wt and mutant strains at each time point are reported as relative to mRNA level measured in wt cells before shift (0). (G–L) Exponentially growing yeast strains carrying either wild type (WT) or Reb1 site-mutated NOP6 gene (NOP6 Rmut) were shifted to YP medium without glucose. Samples were collected before and 10, 20 or 30 min after the shift for total RNA extraction and RT-qPCR analysis. Expression levels of NOP6 (G), PUF6 (H), DRS1 (I), RRB1 (J), TAF10 (K) and ALG9 (L) in wt and mutant strains at each time point are reported as relative to mRNA level measured in wt cells before shift (0). In all panels, data are reported as mean ± SEM of three independent replicates. A one-way ANOVA test was used to compare the means of measurements for the mutant with respect to the corresponding wt strain at each time point. *P < 0.05; **P < 0.01; ***P < 0.001 using a Tukey post-hoc test.
Figure 6.
Rpd3L histone deacetylase recruitment at LTV1 promoter. Indicated strains expressing the Sds3-13xMyc-tagged protein were grown exponentially in YPD at 30°C and shifted to YP medium without glucose. Samples were collected before shift and after 10, 20 or 30 min for ChIP-qPCR analysis of Sds3-13xMyc association with the LTV1 promoter. Fold-enrichment relative to the control ADH4 promoter is represented in either wild type (WT, black bars) and promoter-mutated LTV1 gene at Abf1 binding site (LTV1 Amut, white dotted bars). Data are presented as mean ± SEM of four independent replicates. For wild type, a one-way ANOVA test was used to compare the means of measurements at each time point respect to WT t0; for the enrichment in the mutant a one-way ANOVA test was used to compare the means of measurements of mutants with respect to WT at each time point. *P < 0.05, **P < 0.01, ***P < 0.001 using a Tukey post-hoc test.
Chromatin immunoprecipitation assays
Yeast strains used for ChIP either were from the yeast TAP-tagged collection or they were generated in this study starting from TAP-tagged strains. ChIP of TAP-tagged strains was performed essentially as reported (28). Input and immunoprecipitated (IP) DNA samples were analyzed by qPCR using Applied Biosystems 7300 Real-Time PCR System and SYBR® Green Master Mix (Life Technologies). Oligonucleotides for qPCR (listed in Supplementary Table S1) were designed with Primer Express or Primer3 software, in order to produce 100–150 bp amplicons. Reaction conditions, in particular the concentration of each primer pair, were optimized in order to satisfy the requirements for quantification according to the Livak method (29). ΔCT was first calculated as the difference between IP and input CT values (after correcting for input sample dilution), for both the target gene and the internal standard gene (HHT2 in the ChIP experiments shown in Figures 3 and 5; ADH4 in the experiments of Figure 6). The enrichment of the target gene in IP DNA was then calculated as ΔΔCT = ΔCT target – ΔCT standard. The resulting fold-enrichment values were normalized either to the -fold enrichment of the WT strain with which promoter mutants were compared (for the ChIP experiments reported in Figure 3) or to the -fold enrichment of the t0 sample (for the experiments reported in Figure 5).
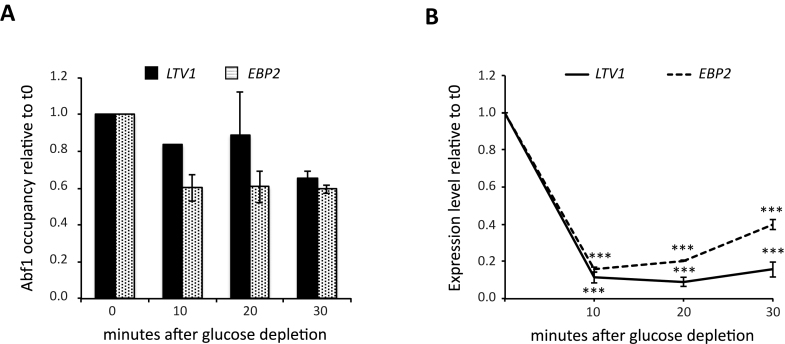
Figure 5.
Effects of glucose starvation on Abf1 association with Ribi gene promoters. An exponentially growing Abf1 TAP-tagged yeast culture was shifted to YP medium without glucose. Samples were collected before (0) and 10, 20 or 30 min after shift for ChIP-qPCR analysis of Abf1 enrichment and RT-qPCR analysis of LTV1 and EBP2 gene expression. (A) Abf1 enrichment for LTV1 (black bars) and EBP2 (white dotted bars); GRF occupancy level at each time point is reported as relative to the level measured in cells before shift in YP (0). (B) Expression level of LTV1 (black line) and EBP2 (black dotted line) reported as relative to the mRNA level measured in cells collected before shift in YP (0). Data are reported as mean ± SEM of two independent replicates. For eacg gene, a one-way ANOVA test was used to compare the means of measurements for each time point with respect to t0. ***P < 0.001 using a Tukey post-hoc test.
ChIP-seq analysis
To map Abf1 and Reb1 genome-wide, ChIP on Abf1 and Reb1 TAP-tagged strains was carried out as described above. Then 10 ng of immunoprecipitated dsDNA were used for library preparation with the Ovation® SP Ultralow Library Systems (NuGEN). Libraries were sequenced with Illumina HIseq-2500 to obtain single end 50 bp-long reads. Analysis was carried out as in (30) with some variation. Reads were mapped to the S. cerevisiae (SacCer3) genome using Bowtie software. Only reads that aligned to a unique position in the genome with no more than two sequence mismatches were retained for further analysis. Duplicate reads that mapped to the same exact location in the genome were counted only once to reduce clonal amplification effects. The genome was tiled into 10-bp windows. Each read was extended by 150 bases (we refer to tags as the extend read counts within a bin) and was counted as one read to each window to which it partially or fully matched. The total counts of the input and ChIP samples were normalized to each other. The input sample was used to estimate the expected counts in a window; the average value for all windows was assigned to windows with zero counts. Finally, we used the Poisson distribution to estimate the probability of observing the ChIP counts within a window given the expected counts in the input sample window. We considered all windows with P-values less than 1.0E–8 to have significant peaks. P-value ≤1.0E–8 was chosen to give False Discovery Rate (FDR) <1%. Normalization of IP tags was done by combining the total number of tags of the two inputs of the two different strains. ChIP-seq analysis was performed in a single experiment for each TAP-tagged strain.
Accession numbers
The Abf1 and Reb1 ChIP-seq data have been deposited at the Gene Expression Omnibus database (record GSE81112).
RESULTS
Phylogenetic footprinting analysis reveals that Ribi gene promoters are demarcated by Abf1, Reb1 and other GRF binding sites upstream of PAC and RRPE
The PAC and RRPE control elements have long been known to be landmarks of the Ribi gene promoters (9,31), and are now generally considered as cis-acting repressor elements (5,14). In contrast with the general notion that all protein-coding genes contain one or more Upstream Activating Sequences (UASs), which require binding of the associated TFs to induce expression (17), no UAS associated with the Ribi regulon have been identified to date. To identify potential UAS elements in Ribi gene promoter regions we implemented a bioinformatic approach based on the detection of conserved sequence motifs in the aligned orthologous Ribi promoter sequences of five Saccharomyces species: S. cerevisiae, S. paradoxus, S. bayanus, S. mikatae and S. kudriavzevii (32). The analysis was conducted on a set of 236 protein-coding genes which, based on stringent annotation and expression criteria, were considered to compose the Ribi regulon in a previous study (6). Starting from the S. cerevisiae sequence of each gene, the orthologs in the other four hemiascomycetes were identified (Supplementary Table S3). The upstream sequences from the different genomes were then aligned. Out of 236 promoter regions, 231 could be successfully aligned. For the majority of Ribi genes, the alignments revealed upstream sequence tracts conserved among the orthologs, recurring at similar positions upstream of several different genes (Supplementary Table S4). As expected based on previous studies, no conserved TATA box was detected in most cases. In contrast, the PAC and RRPE motifs were found in 67% and 79% of Ribi gene promoters, respectively, with ∼56% of all promoters containing both elements. These results were expected on the basis of previous studies. By comparing our results with those reported in (6), focusing on the same set of promoters, we noticed that we identified a higher number of PAC- and/or RRPE-containing promoters. This discrepancy can be explained by our use of alignments among orthologs, instead of a simple motif search in S. cerevisiae upstream regions, allowing us to recognize also non-canonical PAC and RRPE as conserved sequence blocks. As shown in Figure 1, the positions of these two motifs with respect to the ATG were consistent with previous studies (8). The RRPE was mainly found 100–150 bp upstream of the ATG (roughly corresponding to positions –75 and –125 from the transcription start site, TSS), while the PAC tends to be located slightly more downstream, at positions ranging from 75 to 125 bp upstream of ATG (corresponding to positions –50 to –100 upstream of TSS).
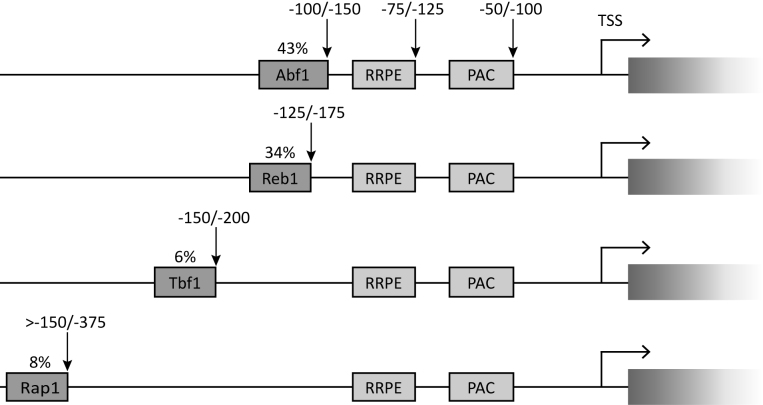
Figure 1.
Conserved sequence elements in Saccharomyces Ribi gene promoters. Graphical representations of promoter architecture of Ribi genes as derived from in silico analysis. The ∼20% of Ribi genes which did not display any conserved GRF binding site is not represented in the Figure. Indicated above each GRF box is the percentage of Ribi gene promoters in which the motif occurs. Vertical arrows pointing to the downstream end of the boxes are to indicate their range of distances (in base pairs, above the arrows) with respect to the TSS. The consensus sequences considered for the different elements reported are: PAC, GCGATGAGMT; RRPE, TGAAAAWTTTY; Abf1, RTCAYNNNN(N)ACGR; Reb1, RTTACCCK; Tbf1, ARCCCTAA; Rap1, WACAYCCRTACATY (M, A or C; W, A or T; R, A or G; Y, C or T; K, G or T; N, any nucleotide). These consensus sequences are based on the following references: PAC and RRPE (1); Abf1 (10,12,55); Reb1 (10,55); Tbf1 (10,12,18); Rap1 (56).
Intriguingly, the majority of Ribi gene promoter region alignments clearly displayed well-conserved sequence blocks upstream of the RRPE that corresponded to motifs recognized by the GRFs Abf1 (98 Ribi promoters), Reb1 (78 Ribi promoters), Rap1 and Tbf1 (19 and 15 Ribi gene promoters, respectively) (Figure 1; Supplementary Table S4). Globally, a large majority (close to 80%) of the 231 successfully aligned Ribi promoter regions was found to contain at least one binding site for one of the four GRFs. Most of the promoters with binding sites for either Abf1 or Reb1 had a single binding site for the cognate TF (double Abf1 binding sites were observed in only six promoters; only a few promoters contained more than one Reb1- or Rap1-binding sites). In contrast, Tbf1-marked promoters tend to display multiple binding sites for this GRF, a previously noted general trend for Tbf1-bound non-snoRNA promoters (18). Notably, 31 Ribi gene promoters had binding sites for more than one GRF protein. In a few promoters, binding sites for three different GRFs were also observed (e.g. YPL126W, which displays two Tbf1 binding sites together with an Abf1 and a Reb1 site within a ∼60 bp region; see Supplementary Table S4). Noteworthy, of the 134 promoters with both RRPE and PAC, 107 (80%) also had a GRF binding motif. These observations suggest that Ribi gene transcription might be regulated through a complex interplay of positively and negatively acting cis elements.
As schematically illustrated in Figure 1, both Abf1 and Reb1 sites are predominantly located at positions ranging from 150 to 100 bp upstream of the TSS (typically ∼25 bp upstream of the RRPE, which in turn tends to be located ∼25 bp upstream of the PAC element). Rap1 and Tbf1 recognition motifs are found at more variable distances from the TSS, with a tendency towards more upstream placements with respect to Abf1 and Reb1. There is no significant orientation preference of the evolutionarily conserved Abf1-binding nor of the Reb1 binding motifs found in Ribi promoter regions. Rap1 and Tbf1 binding sites are also equally represented in the two possible orientations, in agreement with the notion that UAS function is generally orientation-independent (17). The relative position of genes on the chromosome (e.g. adjacent genes with divergent, convergent or tandemly oriented promoter regions) has been recognized as an additional level of transcriptional control influencing coregulation of genes belonging to the same regulon (33). For simplicity, however, in our study divergent/convergent Ribi gene promoter pairs were not considered differently from tandemly oriented pairs, because previous analyses have revealed that gene pair co- or differential regulation can be achieved through a diverse range of mechanisms (34,35).
Inspection of the alignments also revealed potential binding sites for other TFs in addition to GRFs. One of them, Sfp1, is reported to recognize a sequence motif closely resembling the RRPE (1); it is thus not surprising to find it among the candidate TFs binding to Ribi gene promoters. However, even though Sfp1 has been strongly implicated in Ribi gene regulation, its physical interaction with their promoters could not be demonstrated, and it was proposed to act indirectly (16,36).
Abf1, Reb1 and other GRFs bind to the majority of Ribi gene upstream regions
To test whether the predicted GRF binding sites in Ribi gene promoter regions are actually bound by the cognate transcription factors in yeast cells, we performed ChIP experiments with strains carrying a TAP-tagged allele of either ABF1 or REB1, and identified precipitated DNA by high-throughput sequencing (ChIP-seq). This analysis was conducted on exponentially growing yeast cultures in rich medium, a condition under which Ribi genes are expected to be fully active (6). Approximately, a thousand binding sites for both Abf1 and Reb1 were identified (of which ∼270 were associated with both proteins). Such sites were spread throughout the entire genome, with most of them located within intergenic regions.
For both Abf1 and Reb1, association profiles averaged across promoters and plotted with respect to the transcription start site displayed a maximum occupancy between positions –100 and –200, in rough agreement with the predicted location of the corresponding binding sites (Figure 2A and B). Examples of occupancy profiles of individual Ribi genes are reported in Figure 2C and D. The comparison of the average occupancy profile of the 236 Ribi genes with that of all genes revealed a modest increase of enrichment around the expected position for both Abf1 and Reb1. When the average profiles of Abf1 and Reb1 enrichment were restricted to the subsets of Ribi genes whose in silico analysis revealed the corresponding binding motifs, enrichment of both factors to cognate Ribi promoters was much increased (Figure 2A and B). Accordingly, the majority of Ribi genes containing Abf1 or/and Reb1 binding motifs in their promoters were found to be significantly enriched in the cognate factors by ChIP-seq (Supplementary Table S4). Overall, ChIP-seq analyses showed that, under rich medium growth conditions, Abf1 and Reb1 together occupy 103 of the 236 Ribi gene promoters (Supplementary Table S4). Compared to previously published genome-wide location data for Abf1, ∼70% of Ribi gene promoters found to be Abf1-associated by us were also found associated with Abf1 by both ChIP-chip (20) and ChIP-seq (37) analyses, while ∼90% of Reb1-bound Ribi promoters according to our analysis were shared with those previously found by ChIP-chip (38), ChIP-seq (37) and ChIP-exo (39) experiments (Supplementary Table S5).
Figure 2.
In vivo association of Abf1 and Reb1 to Ribi gene promoters. (A) Average binding profiles (normalized total counts) of Reb1 across the TSS regions of annotated yeast promoter regions for different set of gene lists as indicated in the inset (see text and Supplementary material). (B) Average binding profiles (normalized total counts) of Abf1 across the TSS regions of annotated yeast promoter regions for the same genes lists as in (A). (C) Genome browser plots showing normalized tags counts (and significant peaks in blue) of Abf1 and Reb1 binding for ribi genes (EFT2, LTV1 and EBP2) that showed significant enrichment for binding of Abf1 (but not Reb1). Bound gene is highlighted in green color. Relevant TF peaks and the corresponding Ribi genes are boxed. (D) Genome browser plots showing normalized tags counts (and significant peaks in blue) of Abf1 and Reb1 binding for Ribi genes (PUF6 and NOP6) that showed significant enrichment for Reb1 binding (but not Abf1) and NHP2, a Ribi gene whose promoter contains significant region of binding for both factors. Bound gene is highlighted in green color. Relevant TF peaks and the corresponding Ribi genes are boxed.
Moreover, considering previously published genome-wide location data for Tbf1 (18), we could confirm that 9 of the 15 Ribi genes computationally identified as Tbf1 targets are indeed Tbf1-associated according to ChIP-seq analysis. A similar comparison for Rap1 (39) showed that 8 of the 19 Ribi genes predicted as Rap1 targets were actually Rap1-associated. Overall, genome-wide location data indicate that at least 50% of a stringent set of 236 Ribi genes are associated to a GRF in vivo, suggesting that GRFs, in particular Abf1 and Reb1, are important players in Ribi gene transcriptional regulation.
GRF requirement for Ribi gene expression under rich media conditions
To verify whether Abf1 and the other GRFs are actually involved in transcription of the associated Ribi genes, we evaluated the effect of site-specific mutations introduced into the promoter regions of selected target genes in the native chromosomal context. To this end, six non-essential Ribi genes were selected: the Abf1 targets YKL143W (LTV1) and YOL041C (NOP12), the Reb1 targets YDR496C (PUF6) and YDL213C (NOP6), and the Tbf1 targets YKR024C (DBP7) and YDR101C (ARX1). These genes code for: a component of the GSE complex required for efficient nuclear export of the ribosomal small subunit (Ltv1) (40); proteins variously involved in the biogenesis of either 60S (Nop12, Puf6) or 40S (Nop6) ribosomal subunits (41–43); a putative RNA helicase (Dbp7) required for 60S ribosomal subunit assembly (44); a nuclear export factor for the ribosomal pre-60S subunit (Arx1) (45). All of these genes were found to be associated in vivo with the cognate GRF under rich medium conditions as estimated by genome-wide analyses, and display both PAC and RRPE motifs downstream of GRF binding sites. For each of these genes, a strain containing TAP-tagged GRFs was generated in which a specific mutation disrupting the GRF-binding site was introduced at the native locus. As shown in Figure 3 (A, B), disruption of Abf1 binding sites in both LTV1 and NOP12 promoter regions negatively affected Abf1 binding (more markedly in the case of LTV1) and significantly reduced gene expression levels. Similarly, Reb1 site mutations in PUF6 and NOP6 promoter regions led to a significant loss of Reb1 enrichment, accompanied by a significant reduction of both PUF6 and NOP6 mRNA levels (Figure 3C and D). The same approach was adopted to test the role of Tbf1-promoter interactions in the transcription of DBP7 and ARX1. The promoter of DBP7 contains three Tbf1 binding motifs within a region from ∼250 to ∼175 bp upstream of the ATG, followed by contiguous RRPE and PAC elements; the ARX1 promoter region has a similar organization, with two Tbf1-binding sites only. As shown in Figure 3E, mutation of both Tbf1 binding motifs in ARX1 promoter region resulted in loss of Tbf1 binding and a marked decrease in ARX1 expression. In the case of DBP7 (Figure 3F), additive effects of Tbf1 site mutations were observed on both Tbf1 enrichment at the promoter region and gene expression, with the triple Tbf1 site mutant displaying DBP7 mRNA levels corresponding to ∼25% of the wt. It must be noted that considerable levels of residual transcription were observed for all Ribi gene promoter mutants, ranging from ∼20% to ∼60% of wt indicating that Abf1, Reb1 and Tbf1 are not absolutely required, yet contribute to full expression of Ribi genes, at least in rich medium, as we previously observed for Tbf1-dependent expression of yeast snoRNA genes (18) and Abf1-dependent expression of RP gene promoters (46).
Abf1 and Reb1 are required for nutrient-dependent regulation of Ribi gene expression
To address more in depth the role of GRFs in Ribi gene expression, we asked whether GRF binding is required for Ribi gene down-regulation upon nutrient deprivation, a regulatory response known to involve PAC- and RRPE-binding proteins Dot6, Tod6 and Stb3 (14,47).
We chose to focus on two Ribi genes, LTV1 and NOP6, representative of Abf1- and Reb1-associated promoters, respectively. Promoter-mutated strains for each of these two genes (the same shown to be transcription-defective in the previous section) were compared to the corresponding isogenic non-mutated strains in their response to glucose starvation. Expression levels of either LTV1 or NOP6 were measured by RT-qPCR before depletion (t0, reflecting the situation of logarithmically growing cells in rich medium) and after 10, 20 and 30 min of glucose starvation. As shown in Figure 4 (panels A and G), a rapid and significant down-regulation followed by slower recovery of expression was observed for both genes upon glucose depletion. A similar behavior was observed for all the other Ribi genes whose expression was measured in this experiment (EBP2, NOG1, RRB1, PUF6 and DRS1, panels B, C, D, H, I, J). The time-dependent response of Ribi gene expression to glucose starvation resembles the one previously reported for the same genes to other types of stress like heat shock (7). Remarkably, this response was abolished by disruption of the GRF binding site in both LTV1 and NOP6 promoters. In this case, the same weak level of Ribi gene expression observed before depletion was maintained unchanged upon glucose starvation, indicating that nutrient-dependent transcriptional repression mediated by RRPE and PAC binding proteins can only be exerted on GRF-activated promoters. When the response to glucose starvation of two other non-mutated Ribi genes was analysed in the ltv1 promoter mutant strain, some alterations in absolute expression levels and/or in time-dependent responses were observed. In the case of EBP2 (Figure 4B), whose promoter is bound by Abf1, the overall response profile to glucose starvation was similar to the one observed in the wt, but the extent of starvation-dependent down-regulation was significantly lower (only ∼2-fold after 10 min in the ltv1 mutant strain, compared to 6-fold in the reference non-mutated strain). A similar behavior was displayed by NOG1, another Abf1-associated Ribi gene (Figure 4C). Conversely in the case of RRB1, a Ribi gene whose promoter is not associated with Abf1, we observed (Figure 4D) higher expression levels in the ltv1 promoter mutant with respect to wt at each time point of the experiment but, at variance with EBP2, the response amplitude upon starvation was similar in the two strains. A similar tendency to higher expression levels in the ltv1 mutant, but without significant variation in the response profiles, was shared by two non-Ribi genes, TAF10 and ALG9, which were used as controls and displayed a less than 2-fold down-regulation after 10 min of glucose starvation (Figure 4E and F). In the case of the nop6 promoter mutant, unable to bind Reb1, the response to glucose starvation of PUF6 and DRS1, two non-mutated Ribi genes with Reb1 bound to their promoter, did not differ significantly from their response in a reference wt strain (Figure 4H and I). A similar behaviour was observed for the other three genes analysed (the Ribi gene RRB1 and the unrelated TAF10 and ALG9, Figure 4J, K, L) except for a tendency to reduced recovery in the nop6 mutant strain after 30 min of glucose starvation.
It is clear from the above results that in rich medium the absence of bound GRF, in mutated LTV1 and NOP6 promoters, results in expression levels very similar to those obtained from the corresponding non-mutated promoters after 10 min of glucose starvation and that, in this context, glucose starvation produces no further reduction of transcription levels from mutated promoters. A possible mechanistic frame accommodating such observations is that, under starvation conditions, the GRF dissociates from promoter resulting in reduced transcription. We addressed this possibility by ChIP analysis of Abf1 association to wt LTV1 and EBP2 promoters in an ABF1 TAP-tagged strain subjected to glucose starvation protocol. As shown in Figure 5, for both genes the strong expression down-regulation in response to starvation was associated with no or modest decrease in Abf1 occupancy, which might contribute to some extent to the parallel decrease in gene expression.
Since the repression of Ribi gene transcription has been related to the recruitment of the histone deacetylase complex Rpd3L (5,14), we also asked whether Abf1 binding to promoter could affect Rpd3L recruitment upon glucose starvation. To this end, we generated a 13xMyc-tagged version of the Rpd3L subunit Sds3 in the ltv1-Amut strain (whose LTV1 promoter is unable to bind Abf1) and in the corresponding wt strain. We then repeated the glucose depletion experiment monitoring by ChIP-qPCR analysis the binding of Sds3 to LTV1 promoter in the two strains. As shown in Figure 6, the wt LTV1 promoter displayed low, yet detectable occupancy by Sds3 at t0, corresponding to exponentially growing cells. After the shift to glucose-depleted medium, the Sds3 occupancy increased dramatically, with a maximum after 10 min followed by slight decrease at longer times. By contrast, in the ltv1-Amut strain Sds3 association was compromised at each time point of the experiment. This result suggests that, at least for this promoter, bound Abf1 is not only required for full expression under rich media conditions, but also for the recruitment of Rpd3L to ensure proper transcriptional repression under glucose starvation conditions.
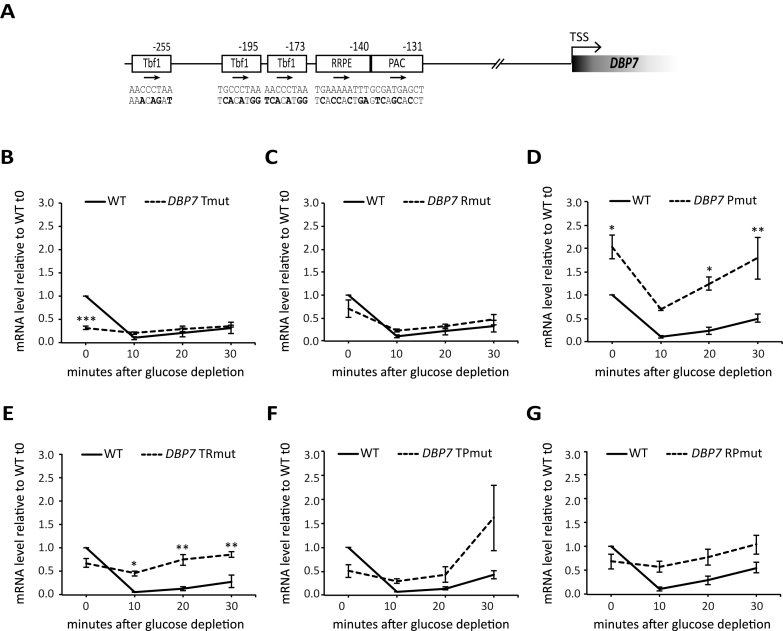
Complex interplay between UAS and other cis-acting regulatory elements in the control of DBP7 transcription
A more extensive promoter mutagenesis was carried out in the case of DBP7, one of the Tbf1 targets among Ribi genes (18). Six strains carrying different promoter-mutated alleles of DBP7 were constructed: Tmut, with all the three Tbf1 binding sites disrupted; Rmut and Pmut, lacking RRPE and PAC elements, respectively; TRmut and TPmut, combining the triple Tbf1 site disruption with mutated RRPE or PAC, respectively; and RPmut, combining RRPE and PAC disruption (Figure 7). The response of these strains to glucose depletion in terms of DBP7 mRNA expression levels was analyzed as described for Abf1-dependent and Reb1-dependent Ribi promoters in Figure 4. In the wt strain, DBP7 expression was already strongly down-regulated after 10 min of glucose starvation, with a conspicuous recovery after 30 min. The triple Tbf1 site disruption caused a marked decrease in DBP7 expression under rich medium conditions (t0 in Figure 7B; see also Figure 3F). The same weak level of DBP7 gene expression observed before depletion did not change significantly upon glucose starvation, indicating that nutrient-dependent transcriptional repression mediated by RRPE- and PAC-binding proteins can be exerted on DBP7 promoter only if it is activated by Tbf1. The combined mutation of RRPE and PAC, the two elements mediating negative regulation of Ribi genes, resulted in a complete loss of DBP7 response to glucose starvation (Figure 7G). When either RRPE or PAC were individually mutated, the regulatory response of DBP7 tended to be much less severely altered than in the case of the double mutant (Figure 7C and D). These observations corroborate previous data showing that the two motifs cooperate to produce growth- and nutrient-dependent gene regulation (14,48). We observed that in the dbp7-Pmut strain (panel D) DBP7 expression was considerably increased with respect to wt even in exponentially growing cells (t0 in Figure 7D), and it was maintained at higher-than-wt levels at all time points after glucose depletion (possibly suggesting a repressive role for PAC element under all conditions). Somewhat intriguing were the behaviors of mutants carrying combined disruption of Tbf1 sites and either RRPE or PAC (Figure 7E and F). In both cases, the expression drop caused by Tbf1 site disruption under rich medium conditions (t0) was mitigated by mutational inactivation of either negative cis-acting element, suggesting that negative modulation by these elements is operating to some extent even under pro-growth conditions. Moreover, a comparison of the behavior of single motif mutant in the RRPE (Rmut, Figure 7C) with the corresponding multiple motif mutant in which the Tbf1 UAS was also disrupted (TR mut, Figure 7E) is particularly informative, because the two mutants support very similar levels of transcription (∼70% of wt) in rich medium (t0), but the negative modulation upon glucose depletion, which is still observed at the Rmut promoter, is compromised in the double RRPE/UAS mutant, as if Tbf1 played an active role in repression in this context.
Figure 7.
Mutational analysis of DBP7 promoter. (A) Schematic representation of DBP7 promoter region with conserved cis-regulatory element. All positions reported are with respect to the TSS; the wt sequence and the mutated version of the regulatory element in the different mutants are indicated below the corresponding box (mutated bases, bold character). (B–G) Exponentially growing yeast strains carrying either wild type (WT, black line) or a promoter-mutated DBP7 gene (dashed line in each panel); (B) Tmut, mutant in the three Tbf1 binding sites; (C) Rmut, mutated in the RRPE; (D) Pmut, mutated in the PAC element; (E) TRmut, mutated in both RRPE and all Tbf1 sites; (F) TPmut, mutated in both PAC and all Tbf1 sites; (G) RPmut, mutated in both RRPE and PAC elements) were shifted from YPD to YP medium without glucose. Samples were collected before (0) and 10, 20 or 30 min after the shift for total RNA extraction and RT-qPCR analysis of DBP7 expression. The DBP7 mRNA levels for wt and mutant strains reported in each panel are expressed as relative to those observed in the corresponding wt cells before shift in YP. Data were collected from three independent replicates and are reported as mean ± SEM. A one-way ANOVA test was used to compare the means of measurements for the mutant with respect to wt at each time point. *P < 0.05, **P < 0.01, ***P < 0.001 using a Tukey post-hoc test. Reported data for the wt were the same for panels B and C, while they were derived from independent measurements in panels D–G.
DISCUSSION
This study extends our knowledge of the coordinated transcriptional regulation of ribosome biogenesis in budding yeast, by showing that the regulated expression of Ribi genes (i.e. genes whose products influence ribosome production without being a structural part of it) requires promoter-associated GRFs (Abf1, Reb1 or Tbf1). At Ribi promoters these TFs interact with cognate upstream activating sequences (UAS), are required for full transcription in exponentially growing cells and cooperate with the negative cis-acting elements PAC and RRPE in determining the overall transcriptional output in response to different growth conditions. In the case of Abf1, the role in repression might mechanistically involve histone deacetylase recruitment.
Until now, the commonly accepted model for Ribi gene regulation was focused mainly on the two negatively acting cis-regulatory elements as the major determinant of the regulatory properties of Ribi genes. Nevertheless, some evidence suggested that RRPE and PAC motifs could act in concert with more upstream located, constitutively activating element(s) to control transcription in response to environmental signals (48). Moreover, a possible role for the protein Sfp1 as a positive regulator was identified although its mode of action remains elusive (6,16,49).
A previous study, focusing on genes differentially affected in an abf1 conditional mutant strain, reported an enrichment of Abf1 in the promoter region of genes belonging to the functional category of ribosome biogenesis (21). We also previously reported that a small subset of Ribi genes display one or more Tbf1 binding sites in their promoter region (18). Starting from these observations, we have now extended the analysis to all the genes of the Ribi regulon and we found that a large fraction of Ribi gene promoters are characterized by a GRF binding site bound in vivo by the cognate factor. The present study thus provides the first comprehensive picture of Ribi gene promoter architecture in budding yeast that, together with functional analysis, indicates GRFs, and in particular Abf1, as pivotal TFs in Ribi gene expression and transcriptional regulation.
When S. cerevisiae cells are logarithmically growing in a rich medium, GRFs are required for full Ribi gene expression. Upon nutritional perturbation, exemplified in this study by glucose depletion from growth medium, promoters defective in GRF binding fail to respond appropriately, showing that GRF-sustained gene transcription is essential in order to produce expression levels allowing for full range of down-regulation. We also found that GRF–UAS interactions cooperate in a non–trivial manner with TF-promoter interactions involving the PAC and RRPE cis-acting elements. While PAC is known to be recognized by Dot6/Tod6 transcriptional repressors whose main action is exerted at Ribi promoters, the RRPE is thought to act as a binding site for Stb3, a protein with both activator and repressor roles, and whose nutrient-dependent regulatory action is mainly exerted at RP genes and, to a lesser extent, Ribi gene promoters (5,13,47). At the DBP7 promoter, whose UAS contains multiple Tbf1 binding sites, mutating individually each of the three cis-acting elements (Tbf1 UAS, RRPE, PAC) resulted in slight (for RRPE, PAC) or dramatic (UAS) alterations of the transcriptional response to glucose starvation. Combining mutations of two out of the three cis-acting elements generally resulted in more severely altered responses.
Mechanistically, the lack of starvation-induced Ribi gene repression at GRF-less promoters might be explained by assuming that the lack of GRF binding has the same molecular consequences as starvation-induced repression, ultimately involving recruitment of the Rpd3L histone deacetylase complex by dephosphorylated Dot6/Tod6/Stb3 (5). In this scenario, the lack of promoter-bound GRFs would favour the establishment, even under pro-growth conditions, of a closed chromatin state similar to the one actively induced by PAC/RRPE-bound repressors in response to starvation at wt promoters. Such a role was previously described for Abf1, Reb1, Rap1 and Tbf1 at many yeast promoters, where they contribute to the generation of nucleosome-free regions favourable to transcription (18,23,50). Alternatively, UAS-bound GRFs at Ribi gene promoters might be mainly involved in direct recruitment of the transcription machinery, as previously documented for Rap1 at RP gene promoters (51). According to this view, constitutively defective Pol II recruitment would render the affected promoter almost unresponsive to chromatin-mediated repression. We favour the first hypothesis because of data presented in Figure 7, which show that transcription levels can be dramatically increased at GRF-less promoters by the combined mutation of either RRPE or PAC, thus demonstrating that wt or even higher levels of transcription machinery recruitment can be attained at promoters lacking GRF binding sites.
Data presented in Figures 5 and 6 further contribute to define mechanistically the regulatory scenario at Ribi gene promoters. First, the results of the experiment in Figure 5 show that the drop in Ribi gene transcription upon glucose depletion is not paralleled by a significant loss of promoter-bound Abf1. This observation points to repressor-induced histone-deacetylation as a plausible mechanism of Ribi gene repression that can also take place in the presence of a bound activator. The results in Figure 6 strengthen and unexpectedly extend this conclusion, by showing that starvation-induced Rpd3L deacetylase recruitment can only occur at the LTV1 Ribi gene promoter in the presence of bound Abf1. This GRF could be involved in Rpd3L recruitment either directly, through protein-protein interaction, or indirectly, by allowing for the maintenance of a local chromatin milieu favorable to binding of repressor proteins to PAC and RRPE and/or to Rpd3L recruitment by bound repressors (5). It should be added, however, that the above scenario is most likely an incomplete one. Another transcriptional regulator, Sfp1, has been shown to participate in the regulation of Ribi gene expression. In particular it was observed that Sfp1 is rapidly expelled from the nucleus in response to glucose starvation and other types of stress and that this relocalization correlates with repression of RP and Ribi regulons (6,49). The lack of starvation-induced repression at Ribi promoters unable to bind Abf1 or Reb1 suggests the possibility that Sfp1 relocalization in response to environmental stress requires GRF-activated promoters in order to exert an effect on Ribi gene expression.
The constancy (or slight decrease) of Abf1 association to Ribi gene promoters during the transcriptional response to glucose depletion is in sharp contrast with the recently reported increase in Abf1 occupancy at Ribi (and RP) gene promoters in response to TORC1 inactivation by rapamycin, a phenomenon which may be related to the optimization of transcriptional rescue upon re-establishment of pro-growth signalling (46). Evidently, the regulatory strategies operating at Ribi genes in response to the two types of nutritional perturbations are not identical. This is not surprising, as the response of Ribi genes to glucose starvation is thought to be mediated through the cAMP-protein kinase A (PKA) pathway and the PAC-binding repressor Dot6, whereas the TORC1 pathway is mainly responsive to nitrogen availability and was shown to act on Ribi gene transcription preferentially through the PAC-binding repressor Tod6 (14,52). Such differences in signalling pathways and responsive TFs acting at promoters might account for the different behaviors of Abf1 at Ribi promoters in response to different types of nutritional stress. On the other hand, the phosphorylation state of Abf1 has also been reported to change according to growth medium composition (53), thus suggesting the possibility that different nutritional cues might differently affect Abf1 association with Ribi promoters by inducing condition-specific Abf1 phosphorylation states.
Our analysis of the transcriptional response of Ribi promoters to nutritional perturbations in the context of defective Ribi promoter mutants (Figure 4) led to a further potentially interesting observation: Abf1 site mutation of the LTV1 promoter also produced alterations in the regulatory response of another Abf1-bound Ribi gene, EBP2. The influence of altered expression of one Ribi gene on the expression of other members of the same regulon suggests some form of regulatory cross-talk, whereby suboptimal expression of one regulon member would be compensated by an increase in expression of other members accompanied by an attenuation of their response to nutritional perturbation. Somehow altered expression levels before and after glucose depletion in the ltv1 promoter mutant strain, however, were also observed for a Ribi-unrelated gene, and the same type of analysis in another Ribi promoter mutant, nop6-Rmut, revealed an altered regulatory response at the mutated promoter only, without any significant alteration in the response of other Ribi or non-Ribi genes. One possible explanation for the ensemble of these observations is that reduced Ltv1 protein expression in the ltv1-Amut strain, which results in a slow growth phenotype (data not shown), specifically produces a wide spectrum effect on the ability of cells to respond to nutritional perturbation.
In conclusion, the requirement of Abf1 and other GRFs for proper transcriptional regulation of Ribi genes, demonstrated by this study, reveals a composite interplay of positive and negative cis-acting elements in their promoters. Moreover, it provides a wider perspective on the regulation of yeast ribosome biogenesis in response to nutrient availability. Under glucose limitation conditions, the reduction in PKA activity results in more efficient association of Dot6 and Stb3 repressors to PAC and RRPE elements, respectively (54). Under such conditions, as revealed by our study, the recruitment of these repressors and/or the consequent repressor-dependent recruitment of Rpd3L histone deacetylase might generally require a UAS-bound GRF at Ribi genes. In the case of Ribi genes, however, the outcomes of glucose signalling by adenylyl cyclase are reinforced and complicated by the response to glucose availability of Sch9 protein kinase, acting on Dot6/Tod6 and Stb3 (5). In turn Sch9 activity is mainly regulated by its direct TORC1-dependent phosphorylation, which responds to nitrogen availability. In this complex scenario, the recently described property of Abf1 to be reversibly recruited to and released from Ribi (and RP) gene promoters depending on signalling through the TOR pathway (46), but not in response to glucose starvation (this study), appears as a remarkable feature whose further investigation could help to clarify the fine-tuning of ribosome biogenesis in the nutritional control of yeast growth.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Enrico Baruffini (University of Parma) for plasmids, Rodolfo Negri (Sapienza Università di Roma) and Barbara Montanini (University of Parma) for discussions, Simone Ottonello (University of Parma) for continuous support.
Footnotes
Present addresses:
Maria Cristina Bosio, Dipartimento di Bioscienze, Università degli Studi di Milano, via Celoria 26, Milano, Italy.
Roberto Ferrari, Centre de Regulació Genómica (CRG), Barcelona, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Ministry of Education, University and Research [MIUR, PRIN 2009 to G.D.]; Italian Association for Cancer Research (AIRC, IG 16877). Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro (AIRC).
Conflict of interest statement. None declared.
REFERENCES
- 1. Bosio M.C., Negri R., Dieci G.. Promoter architectures in the yeast ribosomal expression program. Transcription. 2011; 2:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lempiainen H., Shore D.. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009; 21:855–863. [DOI] [PubMed] [Google Scholar]
- 3. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 4. Dieci G., Preti M., Montanini B.. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009; 94:83–88. [DOI] [PubMed] [Google Scholar]
- 5. Huber A., French S.L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M.P., Tyers M., Rougemont J., Beyer A.L., Loewith R.. Sch 9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011; 30:3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., Tyers M.. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004; 18:2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wade C., Shea K.A., Jensen R.V., McAlear M.A.. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 2001; 21:8638–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wade C.H., Umbarger M.A., McAlear M.A.. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast. 2006; 23:293–306. [DOI] [PubMed] [Google Scholar]
- 9. Hughes J.D., Estep P.W., Tavazoie S., Church G.M.. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000; 296:1205–1214. [DOI] [PubMed] [Google Scholar]
- 10. Badis G., Chan E.T., van Bakel H., Pena-Castillo L., Tillo D., Tsui K., Carlson C.D., Gossett A.J., Hasinoff M.J., Warren C.L. et al. . A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008; 32:878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freckleton G., Lippman S.I., Broach J.R., Tavazoie S.. Microarray profiling of phage-display selections for rapid mapping of transcription factor-DNA interactions. PLoS Genet. 2009; 5:e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu C., Byers K.J., McCord R.P., Shi Z., Berger M.F., Newburger D.E., Saulrieta K., Smith Z., Shah M.V., Radhakrishnan M. et al. . High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009; 19:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liko D., Slattery M.G., Heideman W.. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 2007; 282:26623–26628. [DOI] [PubMed] [Google Scholar]
- 14. Lippman S.I., Broach J.R.. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19928–19933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKnight J.N., Boerma J.W., Breeden L.L., Tsukiyama T.. Global promoter targeting of a conserved lysine deacetylase for transcriptional shutoff during quiescence entry. Mol. Cell. 2015; 59:732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fingerman I., Nagaraj V., Norris D., Vershon A.K.. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell. 2003; 2:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hahn S., Young E.T.. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011; 189:705–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Preti M., Ribeyre C., Pascali C., Bosio M.C., Cortelazzi B., Rougemont J., Guarnera E., Naef F., Shore D., Dieci G.. The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol. Cell. 2010; 38:614–620. [DOI] [PubMed] [Google Scholar]
- 19. Lavoie H., Hogues H., Mallick J., Sellam A., Nantel A., Whiteway M.. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010; 8:e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schlecht U., Erb I., Demougin P., Robine N., Borde V., van Nimwegen E., Nicolas A., Primig M.. Genome-wide expression profiling, in vivo DNA binding analysis, and probabilistic motif prediction reveal novel Abf1 target genes during fermentation, respiration, and sporulation in yeast. Mol. Biol. Cell. 2008; 19:2193–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yarragudi A., Parfrey L.W., Morse R.H.. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2007; 35:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M.. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003; 301:71–76. [DOI] [PubMed] [Google Scholar]
- 23. Ganapathi M., Palumbo M.J., Ansari S.A., He Q., Tsui K., Nislow C., Morse R.H.. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 2011; 39:2032–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fourel G., Miyake T., Defossez P.A., Li R., Gilson E.. General regulatory factors (GRFs) as genome partitioners. J. Biol. Chem. 2002; 277:41736–41743. [DOI] [PubMed] [Google Scholar]
- 25. Chang D.T., Huang C.Y., Wu C.Y., Wu W.S.. YPA: an integrated repository of promoter features in Saccharomyces cerevisiae. Nucleic Acids Res. 2011; 39:D647–D652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H. et al. . JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014; 42:D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Longtine M.S., McKenzie A. 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 28. Braglia P., Dugas S.L., Donze D., Dieci G.. Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007; 27:1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 30. Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A.J., Pellegrini M., Kurdistani S.K.. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012; 22:1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beer M.A., Tavazoie S.. Predicting gene expression from sequence. Cell. 2004; 117:185–198. [DOI] [PubMed] [Google Scholar]
- 32. Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S.. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003; 423:241–254. [DOI] [PubMed] [Google Scholar]
- 33. Arnone J.T., Robbins-Pianka A., Arace J.R., Kass-Gergi S., McAlear M.A.. The adjacent positioning of co-regulated gene pairs is widely conserved across eukaryotes. BMC Genomics. 2012; 13:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnone J.T., Arace J.R., Soorneedi A.R., Citino T.T., Kamitaki T.L., McAlear M.A.. Dissecting the cis and trans elements that regulate adjacent-gene coregulation in Saccharomyces cerevisiae. Eukaryot. Cell. 2014; 13:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yan C., Zhang D., Raygoza Garay J.A., Mwangi M.M., Bai L.. Decoupling of divergent gene regulation by sequence-specific DNA binding factors. Nucleic Acids Res. 2015; 43:7292–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gordan R., Hartemink A.J., Bulyk M.L.. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009; 19:2090–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kasinathan S., Orsi G.A., Zentner G.E., Ahmad K., Henikoff S.. High-resolution mapping of transcription factor binding sites on native chromatin. Nat. Methods. 2014; 11:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Venters B.J., Pugh B.F.. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009; 19:360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rhee H.S., Pugh B.F.. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011; 147:1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seiser R.M., Sundberg A.E., Wollam B.J., Zobel-Thropp P., Baldwin K., Spector M.D., Lycan D.E.. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 2006; 174:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu X., Kasper L.H., Mantcheva R.T., Mantchev G.T., Springett M.J., van Deursen J.M.. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Z., Lee I., Moradi E., Hung N.J., Johnson A.W., Marcotte E.M.. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009; 7:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia-Gomez J.J., Babiano R., Lebaron S., Froment C., Monsarrat B., Henry Y., de la Cruz J.. Nop6, a component of 90S pre-ribosomal particles, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. RNA Biol. 2011; 8:112–124. [DOI] [PubMed] [Google Scholar]
- 44. Daugeron M.C., Linder P.. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA. 1998; 4:566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hung N.J., Lo K.Y., Patel S.S., Helmke K., Johnson A.W.. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell. 2008; 19:735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fermi B., Bosio M.C., Dieci G.. Promoter architecture and transcriptional regulation of Abf1-dependent ribosomal protein genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2016; 44:6113–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liko D., Conway M.K., Grunwald D.S., Heideman W.. Stb3 plays a role in the glucose-induced transition from quiescence to growth in Saccharomyces cerevisiae. Genetics. 2010; 185:797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Slattery M.G., Heideman W.. Coordinated regulation of growth genes in Saccharomyces cerevisiae. Cell Cycle. 2007; 6:1210–1219. [DOI] [PubMed] [Google Scholar]
- 49. Marion R.M., Regev A., Segal E., Barash Y., Koller D., Friedman N., O'Shea E.K.. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14315–14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hartley P.D., Madhani H.D.. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009; 137:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Layer J.H., Miller S.G., Weil P.A.. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J. Biol. Chem. 2010; 285:15489–15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Conrad M., Schothorst J., Kankipati H.N., Van Zeebroeck G., Rubio-Texeira M., Thevelein J.M.. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014; 38:254–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Francesconi S.C., Eisenberg S.. The multifunctional protein OBF1 is phosphorylated at serine and threonine residues in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:4089–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Broach J.R. Nutritional control of growth and development in yeast. Genetics. 2012; 192:73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J. et al. . Transcriptional regulatory code of a eukaryotic genome. Nature. 2004; 431:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lascaris R.F., Mager W.H., Planta R.J.. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999; 15:267–277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.