Abstract
Genomic DNA methylation maps (methylomes) encode genetic and environmental effects as stable chemical modifications of DNA. Variations in DNA methylation, especially in regulatory regions such as promoters and enhancers, are known to affect numerous downstream processes. In contrast, most transcription units (gene bodies) in the human genome are thought to be heavily methylated. However, epigenetic reprogramming in cancer often involves gene body hypomethylation with consequences on gene expression. In this study, we focus on the relatively unexplored phenomenon that some gene bodies are devoid of DNA methylation under normal conditions. Utilizing nucleotide-resolution methylomes of diverse samples, we show that nearly 2000 human genes are commonly hypomethylated. Remarkably, these genes occupy highly specialized genomic, epigenomic, evolutionary and functional niches in our genomes. For example, hypomethylated genes tend to be short yet encode significantly more transcripts than expected based upon their lengths, include many genes involved in nucleosome and chromatin formation, and are extensively and significantly enriched for histone-tail modifications and transcription factor binding with particular relevance for cis-regulation. Furthermore, they are significantly more prone to cancer-associated hypomethylation and mutation. Consequently, gene body hypomethylation represents an additional layer of epigenetic regulatory complexity, with implications on cancer-associated epigenetic reprogramming.
INTRODUCTION
DNA methylation is a stable epigenetic marker deeply implicated in the regulation of gene expression, development and disease. Numerous studies on DNA methylation have established that the majority of Cytosine-phosphate-guanine (CpGs) in the human genome are heavily methylated ((1,2) and others). Short stretches of CpGs that are devoid of DNA methylation are concentrated in regulatory regions, such as enhancers and promoters (3–5). Because most CpGs are heavily methylated, and hypomethylation of specific CpGs are frequently implicated in regulatory processes, DNA methylation studies often address the mechanisms and consequences of hypomethylation at specific CpGs.
In this paper, we focus on DNA methylation of transcription units (‘gene bodies’). In the human genome, gene bodies are typically extensively methylated (4,6). Interestingly, methylated gene bodies are not necessarily associated with repressive chromatin states (6–10). In contrast to the inhibitory role of DNA methylation near transcription start sites (TSS), methylation of gene bodies often align with active transcription (6–10), a phenomenon referred to as the ‘DNA methylation paradox’ (11). In general, the functional roles of gene body DNA methylation are still being debated (4,6,12,13). There is some evidence that gene body DNA methylation affects splicing, and hence methylation of specific positions within gene bodies may directly confer splicing signals (14,15). Analyses of empirical data support the notion that gene body DNA methylation may also suppress cryptic promoters encoded within transcription units (12,16). Other data suggest that gene body DNA methylation is a mechanistic consequence of chromatin accessibility levels of DNA to methylation enzymes (6).
Interestingly, it has been observed that many gene bodies ‘lose’ DNA methylation and become hypomethylated in cancer (17–19). Cancer-associated hypomethylation of gene bodies was shown to be associated with reduced transcription compared to normal cells (18). In some cases, gene body hypomethylation has been directly and casually linked to the alterations of gene expression in cancer (20). Furthermore, some regions appear to be consistently hypomethylated in different cancer methylomes, indicating that there may exist potentially common underlying mechanisms for cancer-associated hypomethylation (18). Thus, understanding how gene body hypomethylation is regulated will contribute to our knowledge of the epigenetic reprogramming in cancer.
In this paper, we focus on the intriguing yet little understood phenomenon that some genes exhibit hypomethylation in normal cells. Singer et al. (21) analyzed DNA methylation data of a human fibroblast cell line and primary B-cells and demonstrated that a number of exons were hypomethylated in these cells. Keller et al. (22) have shown that a substantial number of gene bodies in the human genome were hypomethylated in the muscle tissue and that hypomethylated gene bodies were present in many vertebrate genomes. In addition, hypomethylated exons were enriched in various histone modification signatures (21). These observations indicate that hypomethylation of gene bodies may represent yet another layer of epigenetic regulatory complexity that is currently underappreciated.
As a first step toward elucidating mechanisms of gene body hypomethylation, we examined nucleotide-resolution whole genome DNA methylation maps (methylomes) of diverse human tissues. We show that a substantial number of gene bodies are hypomethylated in these normal tissues. Remarkably, these body-hypomethylated genes exhibit extremely unique genomic, functional and evolutionary features compared to the rest of genes in the human genome. Moreover, hypomethylated genes in normal human tissues are significantly over-represented in those that undergo cancer-associated hypomethylation. These results indicate that body-hypomethylated genes occupy a unique epigenetic niche within the human genome and that their regulation may share pathways involved in cancer-associated hypomethylation.
MATERIALS AND METHODS
Identification of body-hypomethylated genes
We analyzed the University of California Santa Cruz (UCSC) hg38 known transcript table using the R (http://www.r-project.org/) Bioconductor TxDb.Hsapiens.UCSC.hg38.knownGene annotation package. We identified the longest transcript for each autosomal gene and discarded overlapping transcripts to avoid the inclusion of promoter regions of genes overlapping with gene bodies of other genes. The whole genome bisulfite sequencing (WGBS) data are from ovary, sperm, placenta, embryonic stem cell, colon, liver, adrenal gland, B-cell, neuron and hair follicle (10,23–29). To avoid the inclusion of regulatory regions, which are typically hypomethylated, we excluded 100 bp downstream the TSS of each gene. In addition, we only considered genes with at least five mapped CpGs in the whole genome bisulfite data set, used by Mendizabal and Yi (5). The mean sequencing depth ranged between 14 and 63, with more than 81% of total CpGs in the genome are analyzed in all samples (Supplementary Table S1). From the whole genome methylome data, CpGs whose fractional methylation levels were below 0.2 (i.e. up to 20% of reads indicated methylation) were defined as ‘hypomethylated’. These sites are similar to those previously classified as unmethylated or hypomethylated in (26,30) or sparsely methylated in (5). We consequently defined genes whose average methylation level is below 0.2 as hypomethylated gene bodies. Out of the 17 423 genes included in the analyses, we identified 1799 genes with a hypomethylated gene body in at least one tissue, and 469 genes with consistent hypomethylation in all tissues analyzed.
Gene ontology and gene family enrichment analyses
Gene ontology (GO) enrichment analyses were performed with the GOstats R package using P < 0.05 with a hypergeometric test and correcting for multiple testing using a false discovery rate (FDR) of 5%. Following the classification of human genes on 1011 gene families from The HUGO Gene Nomenclature Committee (HGNC) (http://www.genenames.org/cgi-bin/genefamilies/), we tested overrepresentation of hypomethylated genes at these gene categories. In particular, the frequency of hypomethylated genes in each family was compared with the frequency of hypomethylated genes in the whole human gene set. We tested overrepresentation using one-tailed hypergeometric test (31), and corrected for multiple testing via FDR at 0.05 level. We only show gene families with at least five observations at the hypomethylated gene list and at least one observation at constitutively hypomethylated gene list.
TSS, expression, chromatin modification and transcription factor binding sites analyses
We analyzed cap analysis of gene expression (CAGE) experiment data for the B-cell line sample (RPMI1788) from the FANTOM5 consortium data set (32) using the FANTOM5humanSamples R Bioconductor package. Expression data were obtained from RNAseq Atlas (http://medicalgenomics.org/rna_seq_atlas/download) for the following tissue types: adipose, colon, heart, hypothalamus, kidney, liver, lung, ovary, skeletal muscle, spleen and testes.
We analyzed ChIP-Seq experiments of six chromatin marks (H3K4me1, H3K4me3, H3K9me3, H3K27me3, H3K36me3 and H3K27ac) from 97 samples from Roadmap Epigenomics Consortia (from http://egg2.wustl.edu/roadmap/data/byFileType/alignments/consolidated/). For each gene, we computed the proportion of the gene body length occupied by peaks corresponding to each chromatin mark ranging from 0 to 1 (0 meaning 0 bp out of the total gene body length was occupied by peaks, and 1 means 100% of gene length overlapped with histone modification peaks). Then, we subtracted the values of hypomethylated genes to those of other genes, obtaining values ranging from −1 to 1 (−1 meaning the mark is present in 100% of body gene length in other genes and 0% in hypomethylated genes, and 1 meaning the mark is present in 100% of gene length at hypomethylated genes and 0% in other genes).
Transcription factor binding data from ChIP-seq experiments was obtained from the ENCODE project (33), including 161 transcription factor (TFs) and 91 cell types (wgEncodeRegTfbsClusteredV3 table in UCSC). For each gene we computed the gene body per base transcription factor binding site as the number of base pairs occupied by peaks at ChiP-seq experiments divided by the total gene body length. We used this value to examine the relationship between gene body methylation and gene body transcription factor binding. To take into account potential confounding effects of other variables, we used a linear model incorporating eight additional factors (protein connectivity, gene expression levels, gene expression tissue-specificity, gene UTR length, gene intron length, intron number, solvent accessibility and disorder content) obtained from (34).
Cancer hypomethylation analysis
We examined gene body methylation patterns in cancer using the MethHC database (35) (http://methhc.mbc.nctu.edu.tw/php/index.php) that systematically integrates methylation data from The Cancer Genome Atlas. Specifically, we examined 18 different cancer types: bladder urothelial carcinoma (blca), breast invasive carcinoma (brca), cervical squamous cell carcinoma and endocervical adenocarcinoma (cesc), colon adenocarcinoma (coad), head and neck squamous cell carcinoma (hnsc), kidney renal clear cell carcinoma (kirc), kidney renal papillary cell carcinoma (kirp), liver hepatocellular carcinoma (lihc), lung carcinoma (luad), lung squamous cell carcinoma (lusc), pancreatic adenocarcinoma (paad), prostate adenocarcinoma (prad), rectum adenocarcinoma (read), sarcoma (sarc), skin cutaneous melanoma (skcm), stomac adenocarcinoma (stad), thyroid carcinoma (thca) and uterine corpus endometrial carcinoma (ucec). For each of these cancer types, we analyzed the top 250 hypomethylated genes, 250 hypermethylated genes and 250 of the most differentially methylated genes as identified in the database.
We downloaded the Catalogue of Somatic Mutations in Cancer (COSMIC) v79 database from http://cancer.sanger.ac.uk/cosmic. We only considered 1 983 856 mutations identified in whole-genome and whole-exome sequencing projects that are also predicted to be pathogenic according to FATHMM prediction (36).
Evolutionary analyses
The rate of human-mouse synonymous versus non-synonymous substitutions (dN/dS) per were obtained from Ensembl using biomaRt R/Bioconductor package (37). To study the evolutionary age of hypomethylated genes we studied the assigned phylostrata for each gene by Domazet-Loso and Tautz (38). We studied each phylostratum individually and by grouping them in five categories by joining the following phylogenetic levels together: 1–3, categories representing time from before Holozoa split; 4–6, before Bilateria split; 7–10, before Vertebrata split; 11–14, before Mammalia split; and 15–19 thereafter. We reported how the frequency of the hypomethylated genes in every phylostratum compared to the expected frequencies as log-odds ratios following Domazet-Loso and Tautz (38). Specifically, we used a two-tail hypergeometric test (31) and corrected for multiple testing using a FDR at a 0.05 level. The patterns using 5 grouped categories were mostly consistent between the analyses using 19 ages, with the unique exception of an underrepresentation of hypomethylated genes in age 4 around the Holozoa origins, which was only observed in the original 19 category analyses (Supplementary Figure S7).
RESULTS
Many gene bodies are hypomethylated in normal human tissues
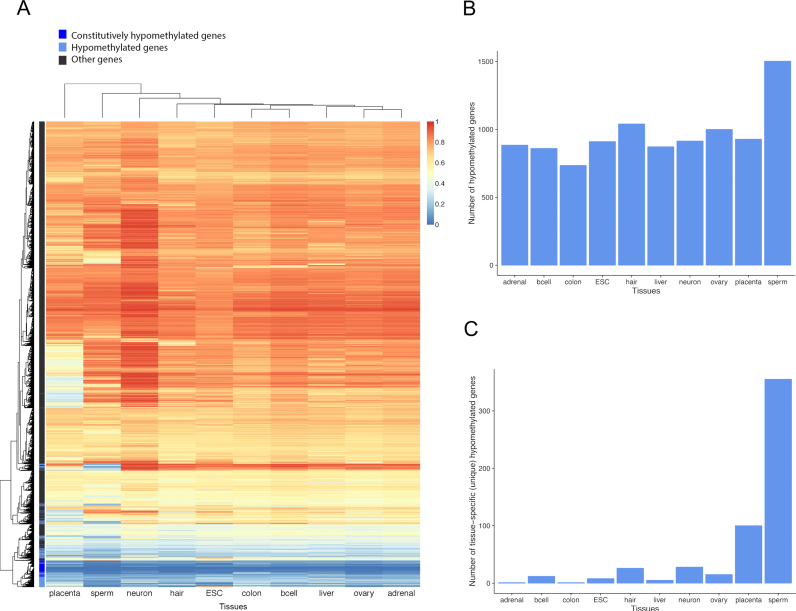
We examined deep-coverage nucleotide-resolution whole genome methylomes of 10 diverse normal human tissues. These tissues were selected to represent different developmental stages (embryonic stem cells, germ cells, fetal and adult somatic tissues) from all three germ layers, as used in (5) (Supplementary Table S1). We excluded the first 100 bp downstream of the TSS of each gene and only used gene bodies harboring at least five mapped CpGs to avoid bias due to a small number of CpGs and/or those that reside in promoter CpG islands. Among the 17 423 gene bodies satisfying these criteria, 1799 were nearly devoid of DNA methylation (defined as the mean fractional methylation level < 0.2) in at least one of the 10 tissues (Figures 1A and B). We refer to these genes as ‘hypomethylated genes’ or ‘body-hypomethylated genes’ henceforth. The placenta and sperm contain particularly high numbers of unique hypomethylated genes (Figure 1C), similar to what has been observed for CpG islands (5), and also in agreement with earlier studies demonstrating overall hypomethylation of these tissues (39–41). Remarkably, we found 469 genes that are hypomethylated in all 10 tissues examined (Figures 1A and B). For convenience, these genes are referred to as ‘constitutively hypomethylated genes’ henceforth. These genes show similar low-methylation across exons and introns (Figure 2A), indicating that hypomethylation is a general feature of these genes. The rest of human genes are referred to as ‘other genes’ henceforth. Consistent with previous studies (8,34,42), methylation levels of promoters are not necessarily correlated with those of gene bodies, and hypomethylated genes show high heterogeneity of promoter methylation levels (Supplementary Figure S1).
Figure 1.
(A) Gene-body DNA methylation levels of all 17 423 genes included in this study. Color bar on the right represents the mean fractional methylation levels. (B) The numbers of body-hypomethylated genes in the 10 tissues. (C) The numbers of tissue-specific hypomethylated genes.
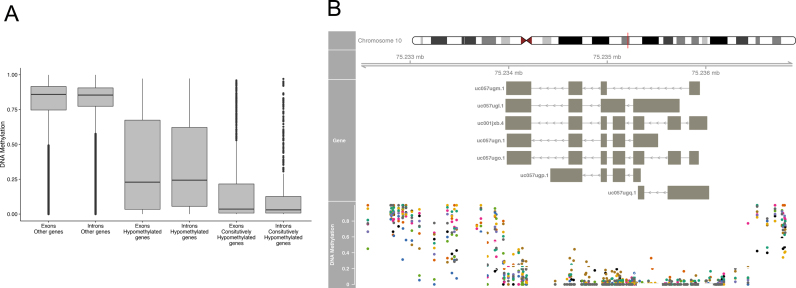
Figure 2.
(A) Most human genes show pervasive DNA methylation at exons and introns, whereas hypomethylated genes show lack of DNA methylation in both exons and introns.(B) Example of a constitutively body-hypomethylated gene. DNA methylation is remarkably low along the exons (indicated by boxes) and introns along the COMTD1 gene in all 10 tissues analyzed (colored dots: adrenal in green, B-cell in orange, colon in purple, embryonic stem cell (ESC) in pink, hair follicle in green, liver in yellow, neuron in brown, ovary in gray, placenta in blue and sperm in black).
Distinct genomic features and functional enrichment of body-hypomethylated genes
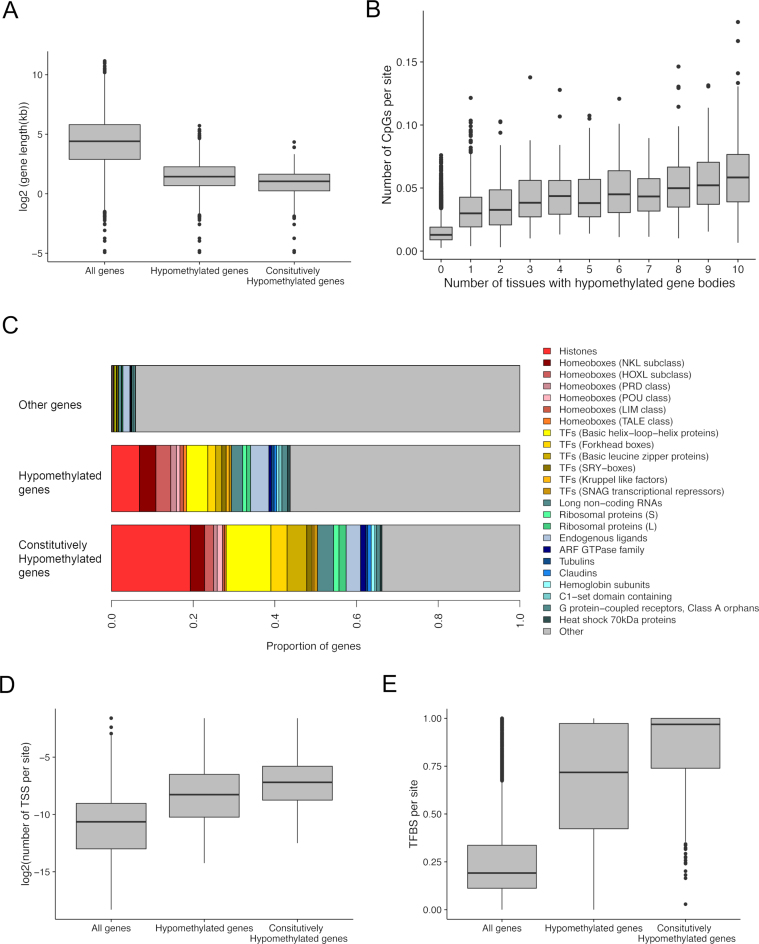
Hypomethylated genes are distinct in several genomic aspects compared to other genes. First, they are in average over one order of magnitude shorter compared to the genomic mean gene length, 4.0 kb (±105 bps [SE]) versus 54.3 kb (±780 bps [SE]), respectively (P < 10−15 by Mann–Whitney test, Figure 3A). Constitutively hypomethylated genes are even shorter (mean = 2.45 kb ± 96 bps [SE]). Second, hypomethylated genes have fewer exons compared to the genomic background. For example, only 4% of all genes are single-exon genes, yet 44% of constitutively hypomethylated genes harbor only a single exon. Interestingly, the mean exon length of the hypomethylated genes is longer than that of other genes (Supplementary Figures S2A and B). In addition, body-hypomethylated genes tend to have fewer isoforms (Supplementary Figure S2C).
Figure 3.
(A) Hypomethylated genes are shorter compared to the rest of the human genes. (B) The number of CpG sites normalized by gene body length increase with the hypomethylation breadth of genes (Spearman's rho = 0.43, P < 10−15). Constitutively body-hypomethylated genes have the highest density of CpG sites per length. (C) Significantly overrepresented gene families (HGNC classification) at hypomethylated genes. (D) Hypomethylated genes harbor greater numbers of TSS per site than other genes. (E) Hypomethylated genes harbor greater numbers of transcription factor binding sites (TFBS) per site than other genes.
However, we also find long genes with multiple exons and alternative transcripts among the list of hypomethylated genes (Supplementary Table S2). For example, the gene COMTD1 harbors 7 exons and 7 splice variants, yet it is constitutively hypomethylated in all 10 tissues examined (Figure 2B). Third, hypomethylated genes tend to be CpG-rich compared to other genes when normalized by gene length (Figure 3B). For example, compared to the mean density of CpGs in all genes (1.3 CpG/100 bps), CpG density in constitutively hypomethylated genes is 4.7 CpG/100 bps (P < 10−15 by Wilcoxon test). Intriguingly, the number of tissues where a gene is hypomethylated linearly increases with CpG density (Spearman's rho = 0.43, P < 10−15, Figure 3B). However, not all CpG-dense genes are hypomethylated. For example, among genes with more than 3.5 CpGs/100 bps (corresponding to roughly the top 5% of genes with respect to the relative CpG proportion), 53% of genes are methylated in at least one tissue. Thus, CpG density is not the determining factor of gene body hypomethylation.
Strikingly, a large number of transcription factors associated with developmental regulation are found in the constitutively hypomethylated set of genes. For example, many homeobox genes, as well as genes harboring DNA-binding motifs, such as those with helix-loop-helix DNA binding domains, are found in this list (Figure 3C, Supplementary Table S2). In addition, many histone genes are also hypomethylated (Figure 3C, Supplementary Table S2), indicating an intriguing relationship between DNA hypomethylation and DNA packaging processes. Functional GO analyses supported the association of nucleosome related functions among gene-body hypomethylated genes, including the enrichment of transcriptional regulation (i.e. GO:0001067, GO:0001077) and sequence-specific DNA-binding (i.e. GO:0043565, GO:0001159) processes (Supplementary Table S3).
Hypomethylated genes are enriched in non-polyadenylated genes
It is well known that many histone gene transcripts lack poly(A) tails because they are transcribed during the S-phase of cell cycle and not subject to polyadenylation (43). Since body-hypomethylated genes include a disproportionately large number of histone genes, we investigated if they are associated with a specific cell cycle and lack polyadenylation (similar to histone genes). We used deep sequencing data of adenylated and non-adenylated transcripts from H9 human ESC and HeLa cells (44) for this purpose. Indeed, non-adenylated genes are strongly enriched among hypomethylated genes (Supplementary Figure S3). More than half of genes without poly(A) tails are found in body-hypomethylated genes, which is a highly significant enrichment considering that non-adenylated genes account for less than 1% of all genes. In constitutively methylated genes, over 10% (48 genes out of 469) are non-polyadenylated (P < 10−15 by hypergeometric test). However, most of these non-adenylated constitutively hypomethylated genes are histone genes (e.g. 53 out of 72 non-adenylated genes). Therefore, even though non-adenylated genes are highly enriched in hypomethylated genes, most hypomethylated genes harbor poly(A) tails.
Hypomethylated genes have more transcripts than genomic background when normalized by length
One functional consequence of gene body DNA methylation is the regulation of alternative splicing (14,15,45). Specifically, DNA methylation may ‘mark’ splicing boundaries to generate alternative transcripts. It is thus possible that hypomethylated genes are those genes that are not alternatively transcribed. On the other hand, according to the idea that gene body DNA methylation suppresses the initiation of spurious transcripts (12,46), hypomethylated genes may generate more transcripts than other genes because those intragenic transcripts are not silenced. Thus, the predictions of these two hypotheses can be examined in the context of hypomethylated genes.
We analyzed the number of transcripts by counting the number of TSS as detected by the CAGE method in the FANTOM 5 data set (32). When all genes are analyzed together, heavily DNA methylated genes indeed harbor a large number of TSS, which could indicate that DNA methylation enables alternative transcription. Hypomethylated genes, on average, have fewer transcripts per gene compared to the genomic background. However, this observation is largely confounded by the fact that hypomethylated genes are short and have fewer exons. In fact, when normalized by gene lengths, hypomethylated genes, on average, encode a greater number of TSS compared to other genes (Figure 3D). This result supports the idea that gene body DNA methylation suppresses spurious intragenic transcripts (12).
Enrichment of distinctive histone modifications and transcription factor binding in hypomethylated genes
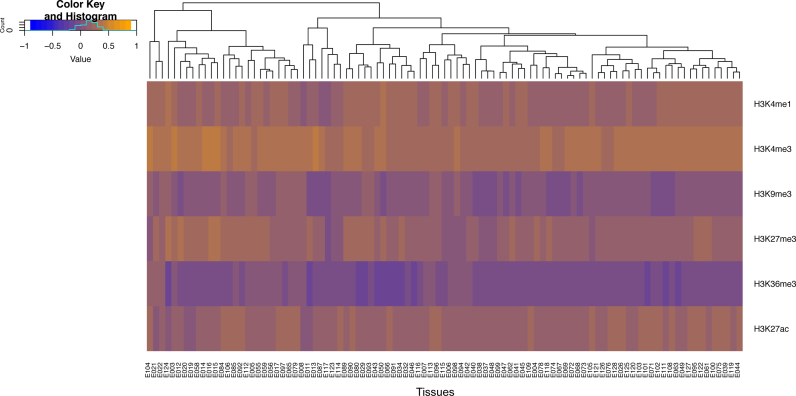
We examined ChIP-seq experiments of 6 chromatin marks (H3K4me1, H3K4me3, H3K9me3, H3K27me3, H3K36me3 and H3K27ac) at 97 tissue samples from the Roadmap Epigenomics Consortium (10). We computed the proportion of each gene (normalized by length) occupied by different chromatin marks. We then compared these proportions between hypomethylated genes and the rest of the human genes (Materials and Methods). The results are shown in Figure 4. Most human genes (‘other genes’) harbor histone modifications associated with regions of active transcription, such as H3K36me3 and H3K9me3, in agreement with previous studies (47). In contrast, we found extensive occupancy of several histone modifications, namely, H3K4me3, H3K4me1 and H3K27ac, on the body-hypomethylated genes (Figure 4).
Figure 4.
Distinctive occupancies of six histone modifications along the gene bodies of hypomethylated versus other genes. The occupancy values range between 1 and −1, where 1 (orange) indicates the specific modification is present in 100% of the gene body length in hypomethylated genes but 0% along the body of other genes. Conversely, the value −1 (blue) indicates the specific modification covers 100% of other genes but is absent (0%) in hypomethylated genes.
Gene body methylation and transcription factor binding at gene promoters show a positive genome-wide association (34). In contrast, the direct relationship between DNA methylation and transcription factor binding in gene bodies themselves is not well understood yet. We found a very strong negative correlation between gene body methylation and experimentally measured transcription factor binding per base at gene bodies at all 10 tissues analyzed (Supplementary Table S4). This correlation held after controlling for other known potential confounders (34) (Supplementary Table S5). This result indicates a high level of transcription factor binding activity within hypomethylated gene bodies. Indeed, transcription factor binding sites occupy an extensive proportion of the transcriptional units of hypomethylated gene bodies (median 72% compared to 18% of other genes, Figure 3E). This result is in line with the active TSS and chromatin mark enrichments found at hypomethylated gene bodies, supporting a regulatory role of those transcriptional units.
Gene bodies hypomethylated in normal tissues are prone to hypomethylation and hypermutation in cancer regardless of tissue origin
Previous studies have documented the cancer-associated hypomethylation of gene bodies occurring in different tumor types (17). Therefore, we asked whether genes that are body-hypomethylated in normal tissues are also hypomethylated in cancer samples, and whether these genes are implicated in malignant neoplasms. We analyzed a recently compiled extensive data set of DNA methylation from 18 different cancers from The Cancer Genome Atlas as integrated at the MethHC database (35). Analyses of these data reveal the following intriguing findings. First, hypomethylated genes in normal tissues also appear as the most hypomethylated genes in the genome for all cancer types studied (overrepresentation at the top 250 most hypomethylated genes, enrichment > 1.76, P < 0.00014, χ2-test, Supplementary Table S6). Second, hypomethylated genes are significantly more prone to be differentially methylated between tumor samples versus tissue-matched non-cancerous samples, which was found in 12 out of the 18 cancers analyzed (average enrichment of 3.65, P-value < 0.001 based on 1000 bootstraps, Supplementary Table S7). Of note, the majority of the tumor types analyzed have no equivalent tissue in the samples we used to define body-hypomethylated gene in normal conditions, suggesting that the association between gene body hypomethylation and cancer could be even stronger in tissue-matched data sets.
To further explore the relationship between gene body hypomethylation and human cancer, we studied the presence of pathogenic somatic mutations in transcriptional units genome-wide. We analyzed around two million somatic mutations considered pathogenic in cancer and identified in whole-genome and whole-exome sequencing projects and deposited in the COSMIC (47). Hypomethylated genes harbor significantly higher number of cancerous mutations per base than other genes (P < 10−15, Wilcoxon test, Supplementary Figure S4A). For example, the gene with the highest number of per base pair mutations is a histone gene HIST1H3B, which is constitutively hypomethylated at its gene body, harboring 69 mutations total. Interestingly, these genes accumulate mutations in a lower rate compared to other genes during evolutionary time, as shown by lower synonymous versus non-synonymous substitutions (dN/dS) values (Supplementary Figure S4B). Therefore, gene body regions of hypomethylated genes seem to evolve under stronger selective constraints than other genes but they tend to accumulate higher number of pathogenic mutations than other genes during somatic cancerous processes.
Hypomethylated gene repertoire has been replenished throughout evolution by the addition of genes in specific functional categories
In distantly related animal genomes, such as those of invertebrates, a large number of genes are body-hypomethylated (22,48,49). Gene body hypomethylation has also been found in many plant genomes (50–52). We recently demonstrated that evolutionarily conserved genes are those that have maintained gene body DNA methylation for a long time, yet many vertebrate genomes still retain body-hypomethylated genes (22). It also has been shown that many cancer-related genes originated early during evolution (38). Therefore, we initially hypothesized that hypomethylated genes in the human genome may be those that have originated early in evolution and maintained hypomethylation. To test this hypothesis, we investigated the ‘evolutionary age’ of genes that records at which time point during evolution a gene has appeared based upon their phylogenetic distribution (38). In this classification, genes are classified into 19 phylogenetic ages, according to their presence/absence in sequenced genomes. We grouped these 19 phylogenetic stages into five categories to increase statistical power (Materials and Methods).
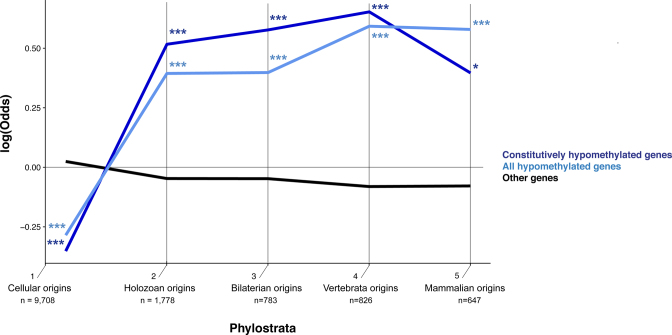
We found that hypomethylated genes do not primarily consist of those that have avoided DNA methylation during a long evolutionary timescale. Instead, they show a significantly different pattern of evolutionary origins compared to the rest of the human gene pool (Figure 5). Specifically, hypomethylated genes are under-represented in the very early evolutionary stages near the cellular and eukaryotic origins (expected 848, observed 645, Supplementary Table S8). On the other hand, hypomethylated genes are over-represented in the subsequent evolutionary stages, meaning that the current repertoire of hypomethylated human genes is shaped by an excess of genes added throughout evolution (Figure 5). We examined the functional enrichment of hypomethylated genes in each evolutionary stage compared to genes that originated in the same stage (results in Supplementary Table S9). This analysis revealed intriguing functional enrichment. We found that hypomethylated genes that originated in the early evolutionary stages (encompassing cellular origins, i.e. Holozoan and Bilaterian origins) are significantly overrepresented by GO categories involved in nucleosome and regulation of transcription. For example, the GO:0005667 (‘transcription factor complex’) is enriched for hypomethylated genes in all three evolutionary stages compared to other genes in those stages. Interestingly, hypomethylated genes that originated near the origin of vertebrates are enriched in immune-related functional categories. Finally, hypomethylated genes that have originated since the origin of mammals are enriched in various biosynthetic and metabolic processes. Notably, five out of six genes with the GO term ‘negative regulation of cell growth’ (GO:0030308) and that originated in this evolutionary stage are all hypomethylated, representing a highly significant enrichment (FDR-corrected Q-value < 0.05, Supplementary Table S9).
Figure 5.
Probabilities of enrichment or depletion of hypomethylated genes in the respective phylostrata (evolutionary origins). Log-odds ratios show the deviation of hypomethylated genes from the expected frequency based on whole set of human genes. (*P < 0.05; **P < 0.01; ***P < 0.001, two-tailed hypergeometric test corrected for multiple comparison by FDR). The number of total human genes at each evolutionary age category is also shown.
DISCUSSION
Methylation of gene bodies is one of the most phylogenetically widespread epigenetic mechanisms (48,53–56), yet its role(s) remain debated (4). In the human genome, most CpGs and gene bodies are heavily methylated, except those that reside in promoters and enhancers. In this paper, we show that in addition to these regulatory regions, gene bodies of many genes are also hypomethylated. The data show that nearly 2000 genes exhibit hypomethylation in at least 1 tissue examined, and 469 gene bodies are consistently hypomethylated. Note that we could only identify these genes using whole genome bisulfite sequencing data. Data generated from array-based methods (which interrogate a limited number of CpGs) or other reduced sampling methods do not offer a comprehensive view of whole genes.
A previous study examining distinctive methylation patterns of exons and introns in the human genome using methylomes from two cell types identified a number of exons classified as hypomethylated (21). In our study, we provide a whole gene perspective by examining hypomethylation profiles of whole gene bodies (exons and introns) and also across a large number of tissues with distinctive developmental origins. Hypomethylated genes identified in the current study show similarly low-methylation across exons and introns (Figure 2). Our study thus expands the complexity of the hypomethylation landscape of the human genome.
We show that body-hypomethylated genes have unique genomic, epigenomic and functional features. Hypomethylated genes tend to be short, harbor fewer exons than the rest of the genome, and they are guanine cytosine (GC)-rich. Some of these characteristics are reminiscent of CpG islands (3,57). However, the hypomethylated genes identified here do not typically qualify for CpG islands, which require the continuous presence or ‘clusters’ of hypomethylated CpGs. In addition, many CpG-rich and short genes are not hypomethylated (Results). Moreover, some hypomethylated genes are long and composed of multiple exons. Furthermore, to avoid any potential bias caused by the inclusion of short, single exon genes with CpG islands as hypomethylated gene bodies, we performed all the analyses again using only multi-exonic hypomethylated genes (n = 1225). Consistently same patterns were found from these analyses (Supplementary Figure S5 and Supplementary Tables S10–S13), indicating that our results are not biased by single exon genes. Moreover, genes harboring hypomethylated promoters are often highly expressed and/or broadly expressed genes (58,59). We thus tested if hypomethylated gene bodies exhibit similar expression characteristics using data from RNA-Seq Atlas (60). We do not find evidence that hypomethylated gene bodies encode highly or broadly expressed genes (Supplementary Figure S6). Together these results indicate that hypomethylated genes are not equivalent to CpG islands. Rather, they may constitute an additional class of epigenomic regulatory loci in the human genome.
Inspired by the distinct functional enrichment and genomic features of hypomethylated genes, we sought to understand why these genes escape genomic DNA methylation. We tested the existing data to determine if hypomethylated genes tend to be predominantly non-polyadenylated genes and found that even though those genes are enriched in hypomethylated genes (mostly histone genes), the majority of hypomethylated genes are adenylated. Interestingly, we found ample evidence of transcription within the hypomethylated gene bodies, which contrasts with the hypothesis that DNA methylation encodes signals of alternative transcription. Rather, the abundance of transcripts in hypomethylated gene bodies is consistent with the idea that DNA methylation suppresses transcription initiation within gene bodies (12,46). Furthermore, we found strong enrichment of several histone modifications, namely, H3K4me1, H3K4me3 and H3K27ac, in hypomethylated gene bodies compared to the rest of the genome. Given that these modifications often associate with promoters and enhancers, these results indicate that hypomethylated genes may perform regulatory functions.
These observations are all in line with the currently unappreciated complex regulatory role of DNA methylation at gene bodies (e.g. 61). It is particularly interesting that hypomethylated genes are highly enriched in H3K27ac, which is a robust marker of enhancers (62). We thus propose that hypomethylated genes may function as global (in the case of constitutively hypomethylated genes) or tissue-specific regulators (in the case of tissue-specific hypomethylated genes).
Hypomethylated genes are found widely across a deep phylogeny spanning humans to archaea. However, human hypomethylated genes have not necessarily maintained hypomethylation during evolution. Instead, the current human hypomethylated gene repertoire has been continuously shaped by the addition of distinctive functional categories of genes throughout evolution. Furthermore, we show that genes hypomethylated in normal tissues are highly prone to cancer-associated hypomethylation and somatic mutations, irrespective of their tissue origins. Although the implications of cancer-associated hypomethylation of specific promoters (such as those of oncogenes) are well-recognized (4,17,63,64), the impact of cancer-associated gene body hypomethylation is less clear. Our results suggest that in addition to potential impact on expression, cancer hypomethylation of specific gene bodies may have far reaching consequences, as they disrupt unique epigenetic regulatory elements of the human genome. In conclusion, our results support gene body methylation as a plausible therapeutic target in cancer (20).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Yuhong Fan and JungKyoon Choi for discussion during the course of the research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Personnel Improvement Program by the Department of Education, Language Policy and Culture by the Basque Government [POS_2013_1_130 to IM]; the National Science Foundation [SBE-131719 to S.V.Y]; and the National Institutes of Health [1R01MH103517-01A1 to S.V.Y]. Funding for open access charge: National Institutes of Health [1R01MH103517-01A1 to S.V.Y].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al. . Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeng J., Konopka G., Hunt B.G., Preuss T.M., Geschwind D., Yi S.V.. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am. J. Hum. Genet. 2012; 91:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Illingworth R.S., Bird A.P.. CpG islands - ‘A rough guide’. FEBS Lett. 2009; 583:1713–1720. [DOI] [PubMed] [Google Scholar]
- 4. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 5. Mendizabal I., Yi S.V.. Whole-genome bisulfite sequencing maps from multiple human tissues reveal novel CpG islands associated with tissue-specific regulation. Hum. Mol. Genet. 2016; 25:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jjingo D., Conley A.B., Yi S.V., Lunyak V.V., Jordan I.K.. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012; 3:462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aran D., Toperoff G., Rosenberg M., Hellman A.. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2011; 20:670–680. [DOI] [PubMed] [Google Scholar]
- 8. Ball M.P., Li J.B., Gao Y., Lee J.H., LeProust E.M., Park I.H., Xie B., Daley G.Q., Church G.M.. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009; 27:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hellman A., Chess A.. Gene body-specific methylation on the active X chromosome. Science. 2007; 315:1141–1143. [DOI] [PubMed] [Google Scholar]
- 10. Roadmap Epigenomics Consortium Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. . Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones P.A. The DNA methylation paradox. Trends Genet. 1999; 15:34–37. [DOI] [PubMed] [Google Scholar]
- 12. Huh I., Zeng J., Park T., Yi S.V.. DNA methylation and transcriptional noise. Epigenetics Chromatin. 2013; 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lorincz M.C., Dickerson D.R., Schmitt M., Groudine M.. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004; 11:1068–1075. [DOI] [PubMed] [Google Scholar]
- 14. Maunakea A.K., Chepelev I., Cui K., Zhao K.. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013; 23:1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S.. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011; 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hunt B.G., Glastad K.M., Yi S.V., Goodisman M.A.. Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol. Evol. 2013; 5:591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009; 1:239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hon G.C., Hawkins R.D., Caballero O.L., Lo C., Lister R., Pelizzola M., Valsesia A., Ye Z., Kuan S., Edsall L.E. et al. . Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012; 22:246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kulis M., Heath S., Bibikova M., Queiros A.C., Navarro A., Clot G., Martinez-Trillos A., Castellano G., Brun-Heath I., Pinyol M. et al. . Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012; 44:1236–1242. [DOI] [PubMed] [Google Scholar]
- 20. Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singer M., Kosti I., Pachter L., Mandel-Gutfreund Y.. A diverse epigenetic landscape at human exons with implication for expression. Nucleic Acids Res. 2015; 43:3498–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller T.E., Han P., Yi S.V.. Evolutionary transition of promoter and gene body DNA methylation across invertebrate-vertebrate boundary. Mol. Biol. Evol. 2016; 33:1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Court F., Tayama C., Romanelli V., Martin-Trujillo A., Iglesias-Platas I., Okamura K., Sugahara N., Simon C., Moore H., Harness J.V. et al. . Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014; 24:554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hodges E., Molaro A., Dos Santos C.O., Thekkat P., Song Q., Uren P.J., Park J., Butler J., Rafii S., McCombie W.R. et al. . Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol. Cell. 2011; 44:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kunde-Ramamoorthy G., Coarfa C., Laritsky E., Kessler N.J., Harris R.A., Xu M., Chen R., Shen L., Milosavljevic A., Waterland R.A.. Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing. Nucleic Acids Res. 2014; 42:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C.T., Low H.M., Kin Sung K.W., Rigoutsos I., Loring J. et al. . Dynamic changes in the human methylome during differentiation. Genome Res. 2010; 20:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. et al. . Global epigenomic reconfiguration during mammalian brain development. Science. 2013; 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Molaro A., Hodges E., Fang F., Song Q., McCombie W.R., Hannon G.J., Smith A.D.. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011; 146:1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ziller M.J., Gu H., Muller F., Donaghey J., Tsai L.T., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E. et al. . Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013; 500:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fernandez A.F., Assenov Y., Martin-Subero J.I., Balint B., Siebert R., Taniguchi H., Yamamoto H., Hidalgo M., Tan A.C., Galm O. et al. . A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012; 22:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivals I., Personnaz L., Taing L., Potier M.C.. Enrichment or depletion of a GO category within a class of genes: which test. Bioinformatics. 2007; 23:401–407. [DOI] [PubMed] [Google Scholar]
- 32. The Fantom Consortium and the RIKEN PMI and CLST (DGT) Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M. et al. . A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chuang T.J., Chiang T.W.. Impacts of pretranscriptional DNA methylation, transcriptional transcription factor, and posttranscriptional microRNA regulations on protein evolutionary rate. Genome Biol. Evol. 2014; 6:1530–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang W.Y., Hsu S.D., Huang H.Y., Sun Y.M., Chou C.H., Weng S.L., Huang H.D.. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015; 43:D856–D861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shihab H.A., Gough J., Cooper D.N., Day I.N., Gaunt T.R.. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013; 29:1504–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Durinck S., Spellman P.T., Birney E., Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Domazet-Loso T., Tautz D.. An ancient evolutionary origin of genes associated with human genetic diseases. Mol. Biol. Evol. 2008; 25:2699–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ehrlich M., Gama-Sosa M.A., Huang L.H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C.. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982; 10:2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monk M., Boubelik M., Lehnert S.. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987; 99:371–382. [DOI] [PubMed] [Google Scholar]
- 41. Schroeder D.I., Blair J.D., Lott P., Yu H.O., Hong D., Crary F., Ashwood P., Walker C., Korf I., Robinson W.P. et al. . The human placenta methylome. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lou S., Lee H.M., Qin H., Li J.W., Gao Z., Liu X., Chan L.L., Kl Lam V., So W.Y., Wang Y. et al. . Whole-genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biol. 2014; 15:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marzluff W.F., Wagner E.J., Duronio R.J.. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008; 9:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.L.. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011; 12:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y. et al. . Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010; 466:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bird A.P. Gene number, noise reduction and biological complexity. Trends Genet. 1995; 11:94–100. [DOI] [PubMed] [Google Scholar]
- 47. Forbes S.A., Beare D., Bindal N., Bamford S., Ward S., Cole C.G., Jia M., Kok C., Boutselakis H., De T.. COSMIC: high-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr. Protoc. Hum. Genet. 2016; 91, doi:10.1002/cphg.21. [DOI] [PubMed] [Google Scholar]
- 48. Sarda S., Zeng J., Hunt B.G., Yi S.V.. The evolution of invertebrate gene body methylation. Mol. Biol. Evol. 2012; 29:1907–1916. [DOI] [PubMed] [Google Scholar]
- 49. Zeng J., Yi S.V.. DNA methylation and genome evolution in honeybee: gene length, expression, functional enrichment covary with the evolutionary signature of DNA methylation. Genome Biol. Evol. 2010; 2:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niederhuth C.E., Bewick A.J., Ji L., Alabady M.S., Kim K.D., Li Q., Rohr N.A., Rambani A., Burke J.M., Udall J.A. et al. . Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016; 17:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmitz R.J., He Y., Valdes-Lopez O., Khan S.M., Joshi T., Urich M.A., Nery J.R., Diers B., Xu D., Stacey G. et al. . Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013; 23:1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takuno S., Ran J.H., Gaut B.S.. Evolutionary patterns of genic DNA methylation vary across land plants. Nat. Plants. 2016; 2:15222. [DOI] [PubMed] [Google Scholar]
- 53. Mendizabal I., Keller T.E., Zeng J., Yi S.V.. Epigenetics and evolution. Integr. Comp. Biol. 2014; 54:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Suzuki M.M., Bird A.. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008; 9:465–476. [DOI] [PubMed] [Google Scholar]
- 55. Takuno S., Gaut B.S.. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zemach A., McDaniel I.E., Silva P., Zilberman D.. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010; 328:916–919. [DOI] [PubMed] [Google Scholar]
- 57. Zeng J., Nagrajan H.K., Yi S.V.. Fundamental diversity of human CpG islands at multiple biological levels. Epigenetics. 2014; 9:483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Antequera F. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 2003; 60:1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Park J., Xu K., Park T., Yi S.V.. What are the determinants of gene expression levels and breadths in the human genome. Hum. Mol. Genet. 2012; 21:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Krupp M., Marquardt J.U., Sahin U., Galle P.R., Castle J., Teufel A.. RNA-Seq Atlas-a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics. 2012; 28:1184–1185. [DOI] [PubMed] [Google Scholar]
- 61. Chuang T.J., Chen F.C.. DNA methylation is associated with an increased level of conservation at nondegenerate nucleotides in mammals. Mol. Biol. Evol. 2014; 31:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Baylin S.B., Jones P.A.. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev Cancer. 2011; 11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Portela A., Esteller M.. Epigenetic modifications and human disease. Nat. Biotechnol. 2010; 28:1057–1068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.