Abstract
The alternative proliferating-cell nuclear antigen (PCNA)-loader CTF18-RFC forms a stable complex with DNA polymerase ε (Polε). We observed that, under near-physiological conditions, CTF18-RFC alone loaded PCNA inefficiently, but loaded it efficiently when complexed with Polε. During efficient PCNA loading, CTF18-RFC and Polε assembled at a 3΄ primer–template junction cooperatively, and directed PCNA to the loading site. Site-specific photo-crosslinking of directly interacting proteins at the primer–template junction showed similar cooperative binding, in which the catalytic N-terminal portion of Polε acted as the major docking protein. In the PCNA-loading intermediate with ATPγS, binding of CTF18 to the DNA structures increased, suggesting transient access of CTF18-RFC to the primer terminus. Polε placed in DNA synthesis mode using a substrate DNA with a deoxidised 3΄ primer end did not stimulate PCNA loading, suggesting that DNA synthesis and PCNA loading are mutually exclusive at the 3΄ primer–template junction. Furthermore, PCNA and CTF18-RFC–Polε complex engaged in stable trimeric assembly on the template DNA and synthesised DNA efficiently. Thus, CTF18-RFC appears to be involved in leading-strand DNA synthesis through its interaction with Polε, and can load PCNA onto DNA when Polε is not in DNA synthesis mode to restore DNA synthesis.
INTRODUCTION
The eukaryotic DNA clamp proliferating-cell nuclear antigen (PCNA) is loaded onto the 3΄ end of a primer–template junction by replication factor C (RFC) and functions as a platform during DNA synthesis for loading of DNA polymerases and various factors involved in Okazaki-fragment processing, DNA-damage repair, chromatin assembly and sister-chromatid cohesion (1–4). RFC consists of one large subunit (RFC1) and four small subunits (RFC2–5), all of which belong to the AAA+ ATPase family (1,2,4). RFC hydrolyses ATP during PCNA loading. Three RFC1 paralogues, Ctf18, Rad17 and Elg1, exist in eukaryotes and associate with RFC2–5 to form alternative clamp-loader complexes, Ctf18-RFC, Rad17-RFC and Elg1-RFC. Ctf18-RFC and Elg1-RFC target PCNA (5,6), whereas the checkpoint loader Rad17-RFC targets the checkpoint clamp Rad9–Hus1–Rad1 (9-1-1 complex) (7,8). Elg1-RFC functions in the unloading of PCNA from chromatin (9–11).
Ctf18-RFC has two additional subunits, Dcc1 and Ctf8 (12,13), and forms a heptameric complex. Ctf18, Dcc1 and Ctf8 are required for the establishment of sister-chromatid cohesion, S-phase checkpoint response and proper telomere maintenance in Saccharomyces cerevisiae (12–19). In DNA replication, Ctf18 localises at the replication fork and recruits PCNA in the presence of the replication inhibitor hydroxyurea (20). Human Ctf18 (CTF18) is also enriched at the replication fork (21,22), and is required for normal replication-fork progression, DNA-damage response and proper cohesion establishment (23).
Biochemical studies have demonstrated that CTF18-RFC loads PCNA onto DNA and stimulates DNA polymerase δ (Polδ) (5,24). On the other hand, replication protein A (RPA)-directed unloading of PCNA by S. cerevisiae Ctf18-RFC has been reported with purified proteins (25), although the physiological relevance of this unloading has not been fully addressed. Direct interaction of CTF18-RFC with DNA polymerase ε (Polε), which mainly replicates leading-strand DNA (26), has been reported. The interaction occurs via a trimeric assembly consisting of CTF18, DCC1 and CTF8 (27). Interaction of these proteins is conserved in S. cerevisiae, and is involved in activation of the Rad53-dependent checkpoint pathway (28) and maintenance of genome stability (29). However, the actual biochemical activity resulting from this interaction remains to be elucidated.
Here, we described the functional significance of this protein interaction and demonstrated that Polε is required for PCNA loading by CTF18-RFC to a 3΄ primer–template junction under physiological conditions through their cooperative binding to DNA. These results suggest that CTF18-RFC is involved in the leading-strand DNA polymerase complex and accesses the 3΄ primer end transiently for PCNA loading if Polε loses its association to the primer end as a DNA polymerase.
MATERIALS AND METHODS
Purification of proteins
All recombinant human proteins were expressed in the High Five insect cell line via recombinant baculoviruses, except for PCNA and RPA, which were expressed using Escherichia coli expression systems. RFC, CTF18-RFC, CTF18-RFC(5), PCNA, RPA, the four-subunit complex of DNA polymerase δ consisting of p125, p66, His-p50 and p12 and the DNA polymerase ε complex consisting of His-p261, FLAG-p59, p17 and p12 (hereafter referred to as Polδ and Polε, respectively, unless noted otherwise) were purified as described previously (27,30,31).
p261N (the amino acid 1–1342 N-terminal fragment of p261, the 261 kDa catalytic subunit of Polε) was expressed with baculovirus harbouring p261 cDNA with a stop codon inserted at the corresponding position, and purified as previously described (27), except that a heparin-sepharose step was inserted between the second Ni-sepharose and final glycerol-gradient sedimentation steps. Exonuclease-deficient variants of Polε and p261N (Polεexo– and p261Nexo–) were purified by the same methods used for their wild-type counterparts, following expression from baculoviruses prepared by site-directed mutagenesis of p261 cDNA to substitute Asp275 to alanine and the insertion of a stop codon for p261Nexo– as above.
Preparation of gapped-DNA beads
Cfr10I-digested ends of 30 μg of 2.7 kb pUC19GAP1 (32) were biotinylated by incubation with 33 μM biotin-dCTP (PromoKine), 33 μM dGTP and 2 units of Klenow fragment (Clontech) in a 75 μl Klenow reaction mixture [10 mM Tris-HCl (pH 7.5), 7 mM MgCl2, 0.1 mM DTT] at 37°C for 1 h. This DNA, which harbours two Nt.BbvCI nicking endonuclease sites separated by 38 nt, was treated with 10 units of Nt.BbvCI (NEB), then heated at 80°C for 1 min followed by sepharose CL-4B chromatography (1.6 ml; GE Healthcare) in Tris-EDTA (TE) buffer [10 mM Tris-HCl (pH 8.0), 1 mM EDTA] containing 0.1 M NaCl, for separation of the short single-stranded (ss)DNA from the gapped-linear-plasmid DNA. The resultant gapped DNA (3 μg) was mixed with 500 μg of Dynabeads M-280 streptavidin (Life Technologies) in 100 μl of BW buffer [10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 M NaCl and 0.1% (v/v) Tween 20] to produce gapped-DNA beads, bound with ∼2 μg of DNA (Figure 1A).
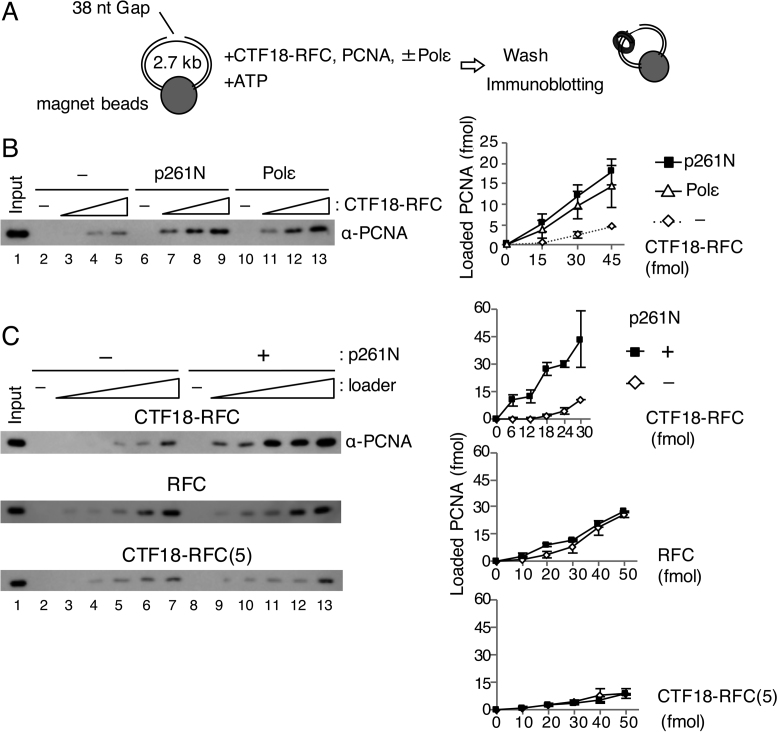
Figure 1.
PCNA loading by CTF18-RFC in the presence of Polε. (A) Schematic diagram of the PCNA loading assay with a gapped DNA attached to magnetic beads. A linearised 2.7 kb plasmid DNA harbouring a 38 nt gap was used. After incubation of the DNA beads with purified proteins and ATP, the loaded PCNA was recovered from the bead-bound fraction. (B) PCNA loading by 15–45 fmol of CTF18-RFC with either 100 fmol of p261N (lanes 6–9) or 100 fmol of Polε (lanes 10–13), or with neither (lanes 2–5). The 10 μl reaction mixture contained 30 mM NaCl, 40 mM creatine phosphate and 25 ng/μl creatine-phosphate kinase. Input control (17 fmol of trimeric PCNA) (lane 1) and 50% of bound samples (lanes 2–13) were analysed by immunoblotting with anti-PCNA antibody. The loaded PCNA was quantified and graphed with mean ± S.E. of two experimental replicates (right). (C) PCNA loading by 6–30 fmol of CTF18-RFC (top), 10–50 fmol of RFC (middle) or 10–50 fmol of CTF18-RFC(5) (bottom) in the same reaction mixture with (+) or without (−) 200 fmol of p261N. Input (lane 1 of each) used 17 fmol (top and middle) or 12 fmol (bottom) PCNA. The bound fractions (50%) were analysed for loaded PCNA (lanes 2–13), which was quantified and graphed as indicated at the right with mean ± S.E. of two experimental replicates.
ddAMP-labelling of the 3΄ end at the gap (Figure 6C) was achieved by incubation of 3.6 μg of gapped DNA with 140 μM each of TTP, dCTP and ddATP (GE Healthcare) and 4 units of Klenow fragment in a 70 μl Klenow reaction mixture on ice for 2 h, followed by addition of 5 μl of 0.5 M EDTA and conjugation with Dynabeads M-280 streptavidin, as described above.
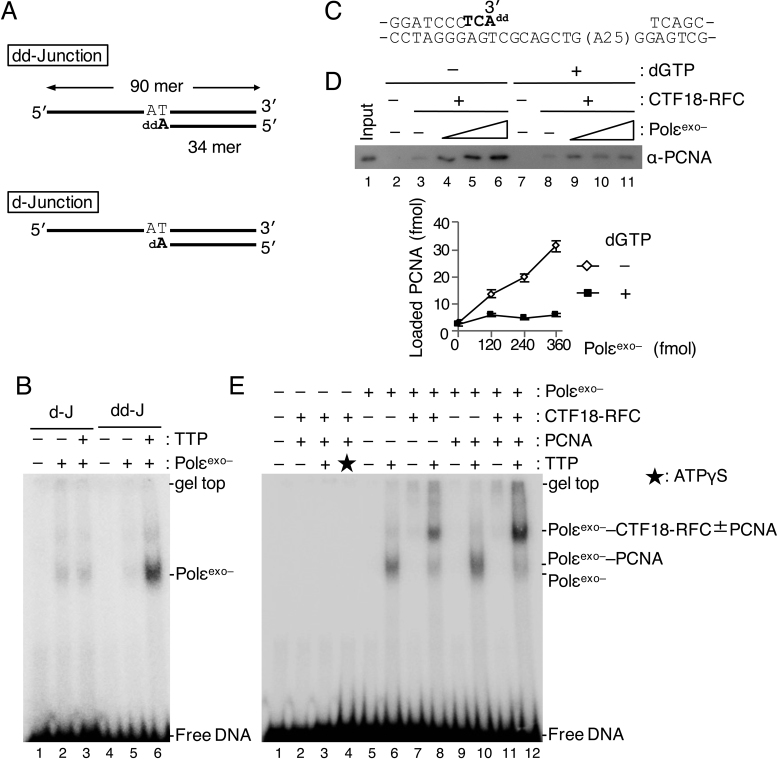
Figure 6.
Analyses of PCNA loading in the presence of Polε in synthesis mode. (A) Two 3΄ primer–template junction substrates with ddAMP (“dd-Junction”) or dAMP (“d-Junction”) at their 3΄ primer ends. (B) 60 fmol of Polεexo– was mixed with 25 fmol of d-Junction (“d-J”; lanes 2, 3) or dd-Junction (“dd-J”; lanes 5, 6) at 60 mM NaCl in the presence (+) or absence (−) of 100 μM TTP, and binding was analysed by EMSA after glutaraldehyde fixation. Lanes 1 and 4 were controls without Polεexo–. Bands produced by binding of Polεexo– to DNA are indicated. (C) To study PCNA loading in the presence of Polεexo– in synthesis mode, a gapped DNA with ddAMP at the 3΄ primer end was prepared. The sequence shows a 51 bp region with the 35 nt gap on the substrate DNA. The nucleotides shown in bold represent sequence extension to prepare the 3΄ primer end with ddAMP. (D) Comparison of PCNA loading with the gapped-DNA beads with Polεexo– in non-synthesising (–dGTP) and synthesising (+dGTP) modes. The indicated DNA beads (15 ng) were incubated with 100 fmol of CTF18-RFC and 6.2 pmol of PCNA in the presence of 0, 120, 240 and 360 fmol of Polεexo– in a 10 μl reaction mixture at 60 mM NaCl. dGTP (100 μM) was added in lanes 7–11. Input control of 12 fmol of PCNA (lane 1) and 100% bound fractions (lanes 2–11) were applied to immunoblotting with anti-PCNA antibody. Lanes 2 and 7 were the negative controls without CTF18-RFC and Polεexo–. Bound PCNA was quantified and graphed below with mean ± S.E. of two experimental replicates. (E) Assembly of 60 fmol each of CTF18-RFC and Polεexo– and 500 fmol of PCNA to 25 fmol of dd-Junction substrate was analysed by EMSA as above (B) using indicated combinations of proteins at 60 mM NaCl. Additions of 100 μM TTP and 250 μM ATPγS are indicated by ‘+’ and an asterisk (lane 3), respectively. DNA bands shifted at positions by added proteins are indicated at the right.
Preparation of oligo-DNA beads
Synthesised biotinylated 90-mer oligonucleotides were obtained from Sigma Genosys. BTN3 was annealed to BTN30 or BTN28 at a 1:2 ratio as described previously (33), resulting in 3΄ and 5΄ recessed (3΄/5΄) or 3΄ recessed (3΄) primer–template DNAs. Similarly, BTN5 was annealed with BTN32, resulting in 5΄ recessed primer–template DNA (5΄). Oligo-DNA beads were obtained by binding 100 pmol of biotinylated ssDNA (ss) or primer–template DNAs with 50 μl of streptavidin agarose ultra-performance beads (Solulink) in BW buffer (Figure 3B).
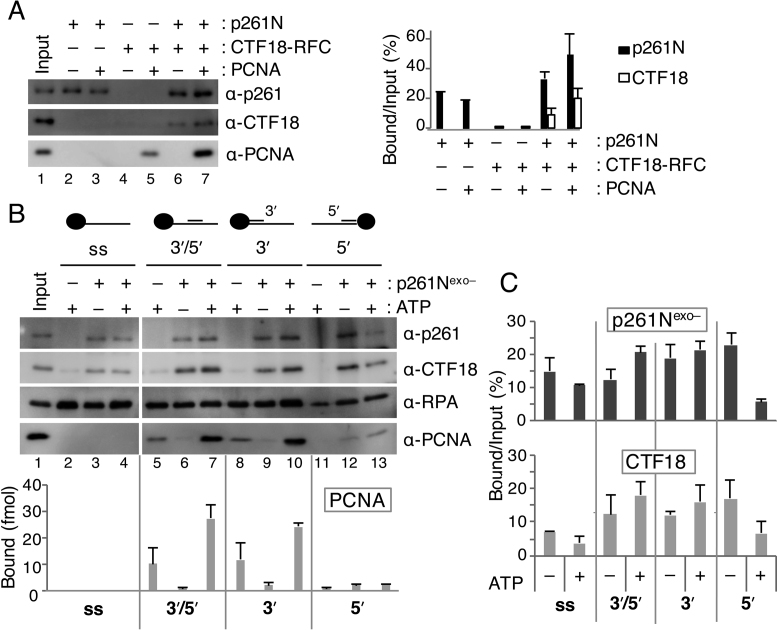
Figure 3.
Analyses of loaded PCNA and bound CTF18-RFC and p261N on various structures of DNA. (A) Pull-down assay with 30 ng of gapped-DNA beads at 100 mM NaCl using 100 fmol of p261N, 100 fmol of CTF18-RFC and 6.2 pmol of PCNA, as indicated. Input (10%; p261N, CTF18-RFC) and 12 fmol of PCNA (lane 1), and 50% bound fractions (lanes 2–7), were analyzed by immunoblotting with the indicated antibodies (left). Bound p261N and CTF18 from two experimental replicates are graphed with mean ± S.E. (right). (B) PCNA-loading assay with oligo-DNA beads containing conjugated ssDNA (ss), a 3΄ and 5΄ recessed primer–template DNA (3΄/5΄), a 3΄ recessed primer–template DNA (3΄), or a 5΄ recessed primer–template DNA (5΄). Reactions were performed at 120 mM NaCl with 6.2 pmol of PCNA, 4.2 pmol of RPA, 200 fmol of CTF18-RFC and the indicated combination of 2 mM ATP and 1 pmol of p261Nexo–. Input bands (lane 1) indicate 60 fmol of p261Nexo–, 10 fmol of CTF18-RFC, 340 fmol of RPA and 12 fmol of PCNA. Bound proteins were analysed with 50% samples (lanes 2–13). Band intensities of PCNA were quantified and graphed with the mean ± S.E. from two experimental replicates (bottom). (C) The amounts of p261Nexo– and CTF18 that bound to four different DNA bead substrates were compared with or without ATP by quantification of their bound % versus input as indicated by the graph, with mean ± S.E. of two experimental replicates.
PCNA loading
Gapped-plasmid-DNA beads containing the equivalent of 15 ng of DNA were suspended in 10 μl of reaction mixture [10 mM HEPES-NaOH (pH 7.8), 0.05% (v/v) Tween 20, 10 mM MgCl2, 0.2 mM EDTA, 0.01% (w/v) BSA and 0.5 mM DTT] containing 2 mM ATP, 540 ng (6.2 pmol) of PCNA and additional components as indicated. Assays shown in Figure 1 included 40 mM creatine phosphate and 250 ng of creatine-phosphate kinase. The DNA beads were incubated at 32°C for 30 min after addition of the indicated components and washed four times with 100 μl of HBS [10 mM HEPES-NaOH (pH 7.8), 3.2 mM EDTA, 0.05% (v/v) Tween 20 and 0.15 M NaCl] (Figure 1A). The assay with oligo-DNA beads was carried out similarly, except for inclusion of DNA beads with 500 fmol of oligonucleotides in the presence of 4.2 pmol of RPA and five washes of 500 μl of HBS after the reaction.
The proteins that bound to the DNA beads were eluted in 10 μl of sodium dodecyl sulfate (SDS) loading buffer [50 mM Tris-HCl (pH 6.8), 100 mM DTT, 2% (w/v) SDS, 0.05% (w/v) bromophenol blue and 10% (v/v) glycerol] and detected by immunoblotting after SDS-polyacrylamide-gel electrophoresis (SDS-PAGE), with anti-Polε p261 monoclonal antibody (ATCC, CRL-2284), anti-CTF18 monoclonal antibody (H00063922-M01; Abnova), anti-Polδ p125 goat polyclonal antibody (sc-8796; Santa Cruz), anti-PCNA rabbit polyclonal antiserum (in-house preparation) and anti-RPA polyclonal antibody (in-house preparation). The secondary antibodies were goat anti-mouse IgG antibody conjugated with HRP (Bio-Rad), goat anti-rabbit IgG antibody conjugated with HRP (Bio-Rad) and rabbit anti-goat IgG antibody conjugated with HRP (Zymed Laboratories).
Site-specific DNA photo-crosslinking
To prepare 3΄ end-labelled oligonucleotide substrates for photo-crosslinking, 45 pmol of RF-30 primer was annealed with 30 pmol of TEMP90-R to create APB-Junction or with 30 pmol of TEMP90-Rneo to create APB-End (see Table 1 for primer sequences), and incubated with 3.1 μM [α-32P] TTP (PerkinElmer Life Sciences), 20 μM α-S-dCTP (ChemCyte) and 4 units of Klenow fragment at 10°C for 30 min in a 20 μl Klenow reaction mixture, followed by a chase with 50 μM TTP at 10°C for 30 min. After phenol–chloroform (1:1) extraction, the product DNA was incubated with 2.1 nmol of azidophenacyl bromide (APB; Sigma) in a 50 μl volume for 3 h at room temperature in the dark. After removal of unreacted reagent by ethanol precipitation with ethachinmate (NIPPON GENE), the product DNA was dissolved in TE at 50 nM (calculated from the estimated recovery from incorporated TMP), and stored at 4°C in the dark. DNA substrates shown in Figure 6A had additional dAMP or ddAMP at their 3΄ ends.
Table 1. Oligonucleotides used in this study.
| Name | Sequence (5΄–3΄) |
|---|---|
| BTN5 | Biotin-tgaggttcagcaaggtgatgctttagatttttcatttgctgctggctctcagcgtggca-ctgttgcaggcggtgttaatactgaccgcct |
| BTN3 | tgaggttcagcaaggtgatgctttagatttttcatttgctgctggctctcagcgtggcactgttgc-aggcggtgttaatactgaccgcct-Biotin |
| BTN32 | aaaaatctaaagcatcaccttgctgaacctca |
| BTN30 | cagtgccacgctgagagccagcagcaaatg |
| BTN28 | aggcggtcagtattaacaccgcctgcaa |
| TEMP90-R | aggcggtcagtattaacaccgcctgcaacagtgccacgctgagagccagcagcaaatgaaaaatct-aaagcatcaccttgctgaacctca |
| TEMP90-R neo | gaaaaatctaaagcatcaccttgctgaacctcaaggcggtcagtattaacaccgcctgcaacagtg-ccacgctgagagccagcagcaaat |
| RF64 | aggcggtcagtattaacaccgcctgcaacagtgccacgctgagagccagcagcaaatgaaaaat |
| RF30 | tgaggttcagcaaggtgatgctttagattt |
| RF46 | tgaggttcagcaaggtgatgctttagatttttcatttgctgctggc |
| RF41 | tgaggttcagcaaggtgatgctttagatttttcatttgctg |
| RF36 | tgaggttcagcaaggtgatgctttagatttttcatt |
| RF31 | tgaggttcagcaaggtgatgctttagatttt |
| RF26 | tgaggttcagcaaggtgatgctttag |
| RF21 | tgaggttcagcaaggtgatgc |
BTN5, BTN3 and their annealing primers (BTN32, 30 and 28) have been described previously (33).
Photo-crosslinking oligonucleotide substrate labelled at a fixed position on the template strand was prepared by extension of RF64 on BTN3; 300 pmol of RF64 primer was annealed with 150 pmol of biotinylated BTN3 90-mer and bound to 5 μl of streptavidin agarose ultra-performance beads. The DNA beads were incubated in a 25 μl of Klenow reaction mixture with 4 μM [α-32P] TTP, 24 μM α-S-dCTP and 2 units of Klenow fragment at 10°C for 30 min. After three washes with BW buffer and one wash with TE, the attached RF64 was completely elongated with 250 μM dNTP and 2 units of Klenow fragment at 10°C for 30 min in a 20 μl Klenow reaction mixture. Free nucleotides and proteins were removed by two washes with BW buffer; then the elongated RF64 strand was eluted twice with 50 μl of 0.1 M NaOH. The eluted sample was neutralized by addition of 10 μl of 1 M Tris-HCl (pH 8.0) and 10 μl of 1 M HCl and reacted with 2.1 nmol of APB in a 130 μl reaction mixture, as above. Aliquots of the product were annealed 1:1.5 with RF21–RF46 oligonucleotides (Table 1) in TE containing 0.1 M NaCl, and stored at 4°C in the dark.
A 10 μl reaction mixture in a 1.5 ml tube containing 25 fmol of labeled oligonucleotide substrate, 60 mM NaCl and additional components as indicated was incubated at room temperature for 10 min and irradiated with 8 W ultraviolet (UV) C for 5 min at a 10 cm distance from the light source in a UVC 500 Ultraviolet crosslinker (GE Healthcare). The crosslinked DNA–protein complexes were further treated with 5 units of TurboNuclease (Accelagen) for 1 h at room temperature, and proteins that were conjugated with labeled nucleotides were separated by SDS-PAGE and visualised with a FLA-9500 phosphorimager (GE Healthcare) after drying of the gel.
Electrophoretic-mobility-shift assay (EMSA) after glutaraldehyde fixation
For the assay substrates, APB-Junction DNA obtained as described above was further extended with ddAMP or dAMP, and a dd-Junction and a d-Junction were obtained, respectively (Figure 6A). The substrate (25 fmol) was incubated in 5 μl of reaction mixture containing 60 mM NaCl and proteins as indicated at room temperature for 10 min and then for 5 min with addition of 0.5 μl of 10% (v/v) glutaraldehyde. The crosslinked sample was supplemented with 0.5 μl of electrophoretic-mobility-shift assay (EMSA) loading solution [100 mM Tris-HCl (pH 8.0), 20% (w/v) sucrose, 1 mg/ml bromophenol blue] and separated on a 5% polyacrylamide gel in TAE buffer [20 mM Tris-acetate (pH 7.8), 1.25 mM EDTA]. The shifted DNA was visualised by autoradiography with a FLA-9500 phosphorimager after fixing of the gel with 40% (v/v) methanol, 20% (v/v) acetic acid for 5 min followed by washing with water for 5 min and gel-drying under vacuum.
Holoenzyme assay with Polδ and Polε
A singly-primed template DNA was prepared by annealing M13mp18 ssDNA (TaKaRa) to TEMP90-R. The template DNA (30 ng) was then incubated in a 10 μl reaction mixture containing 25 mM HEPES-NaOH (pH 7.8), 10 mM MgOAc, 60 mM NaCl, 2 mM ATP, 0.01% (w/v) BSA, 0.5 mM DTT, 10 μM dNTP, [α-32P] TTP and indicated amounts of proteins at 32°C for 30 min. The product DNA was precipitated with ethanol, dissolved in 5 μl alkaline electrophoresis solution [0.3 M NaOH, 2 mM EDTA, 5% Ficoll] followed by electrophoresis in a 0.8% alkaline agarose gel at 40 V for 5 h, and the products were visualised by autoradiography as described above.
RESULTS
Polε stimulates loading of PCNA by CTF18-RFC under physiological conditions
To understand the functional significance of the interaction between CTF18-RFC and Polε, we examined the effects of Polε on PCNA loading by CTF18-RFC using a quantitative PCNA-loading assay with gapped-DNA beads (Figure 1A, 32). CTF18-RFC loaded PCNA onto DNA in a dose-dependent manner at 30 mM NaCl (Figure 1B). Addition of Polε resulted in an approximately 3-fold increase in PCNA loading (Figure 1B, lanes 11–13). The N-terminal half of Polε p261 (p261N), carrying the region necessary for interaction with CTF18-RFC (27), showed similar stimulation (Figure 1B, lanes 7–9). PCNA loading was also examined with two other loader complexes, the replicative PCNA loader RFC and CTF18-RFC(5), the pentameric derivative of CTF18-RFC lacking DCC1 and CTF8. Neither RFC nor CTF18-RFC(5) interact stably with Polε (27). In the absence of p261N, PCNA loading was similar with RFC and CTF18-RFC and slightly lower with CTF18-RFC(5) (Figure 1C, lanes 2–7). In the presence of p261N, only CTF18-RFC exhibited enhanced PCNA loading (Figure 1C, lanes 8–13), indicating that the specific interaction of CTF18-RFC with Polε is responsible for the enhanced PCNA loading.
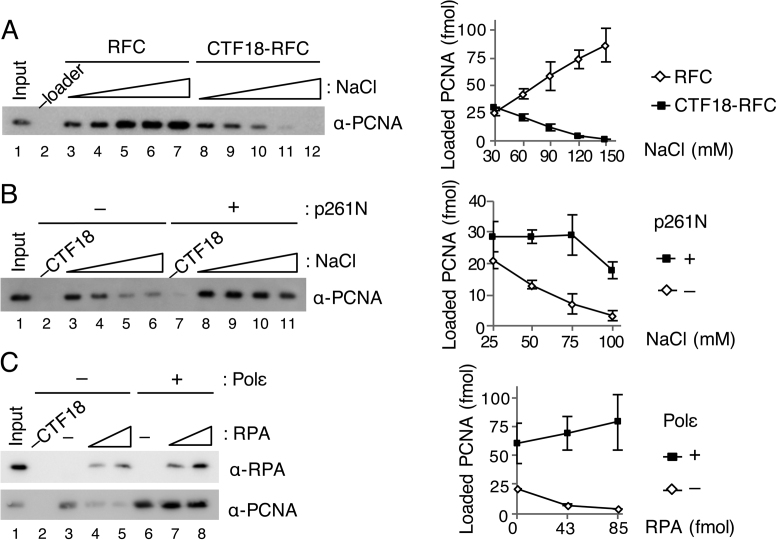
CTF18-RFC has been reported to be less active for PCNA loading than RFC, reflecting higher salt sensitivity of ssDNA-stimulated ATPase and 3΄ primer–template junction binding in CTF18-RFC than in RFC (5). The effects of salt concentration on PCNA loading were therefore examined. These experiments omitted creatine phosphate (which is added to maintain the ATP concentration during assays), to avoid its effect on salt concentration and enable an accurate study of the salt effect. RFC and CTF18-RFC loaded similar amounts of PCNA at 30 mM NaCl, but PCNA loading with RFC increased as the NaCl concentration increased, whereas PCNA loading with CTF18-RFC decreased with increasing NaCl, and was very low at near-physiological salt concentrations >0.1 M NaCl (Figure 2A). PCNA loading with CTF18-RFC alone decreased from 21 fmol to 3.2 fmol from 25 mM to 0.1 M NaCl. However, in the presence of p261N, 18 fmol of PCNA was loaded even at 0.1 M NaCl (Figure 2B). Active PCNA loading by CTF18–RFC at physiological salt concentrations seems to require interaction with Polε.
Figure 2.
Effect of salt concentrations and RPA on PCNA loading by CTF18-RFC. (A) Titration of NaCl concentration from 30 mM to 150 mM for PCNA loading by 50 fmol each of RFC (lanes 3–7) or CTF18-RFC (lanes 8–12). PCNA in 50% bound fractions (lanes 2–12) and input control (12 fmol) were detected by immunoblotting (left). The band intensities were quantified and graphed with mean ± S.E. of two experimental replicates (right). (B) Titration of NaCl (25–100 mM) for PCNA loading by 30 fmol of CTF18-RFC with (lanes 7–11) or without (lanes 2–6) 200 fmol of p261N. PCNA in 50% bound fractions (lanes 2–11) and input control (12 fmol) were detected by immunoblotting (left), and their band intensities were quantified and graphed with mean ± S.E. of two experimental replicates (right). (C) Effects of RPA (42, 85 fmol) on PCNA loading with 30 ng gapped-DNA beads at 60 mM NaCl using 100 fmol CTF18-RFC with (lanes 6–8) or without (lanes 3–5) 200 fmol Polε. The total bound fractions (lanes 2–8) and 42 fmol RPA and 12 fmol PCNA as the input control (lane 1) were analysed by immunoblotting with the indicated antibodies (left). Band intensities of PCNA were quantified and graphed with the mean ± S.E. of three experimental replicates (right).
It has been reported that RPA also influences the efficiency of PCNA loading by S. cerevisiae Ctf18-RFC (25). Indeed, the presence of RPA in a PCNA loading reaction makes the reaction more physiological. When we added increasing amounts of RPA to the reaction with CTF18-RFC alone at 60 mM NaCl, less PCNA was loaded than in the absence of RPA (Figure 2C, lanes 3–5). This result is consistent with a previous report showing that saturating amounts of RPA inhibit loading of PCNA by S. cerevisiae Ctf18-RFC (25). In the presence of Polε, however, PCNA loading by CTF18-RFC was maintained or slightly increased even in the presence of RPA (Figure 2C, lanes 6–8). Collectively, these data suggest that under physiological conditions. i.e., in the presence of RPA and a high salt concentration, CTF18-RFC can only load PCNA after it has formed a complex with Polε.
CTF18-RFC–p261N complex loads PCNA efficiently at 3΄ primer–template junction through cooperative binding at the site
S. cerevisiae Polε has greater affinity for various DNA structures than Polδ (34). This led us to hypothesize that CTF18-RFC might be recruited to its target site through its interaction with Polε. Protein binding to substrate DNA was therefore analysed during PCNA loading (Figure 3A). p261N bound to the gapped DNA in the absence of other proteins, and addition of PCNA did not affect this binding (Figure 3A, lanes 2, 3). Similar DNA binding was observed with Polε, but not Polδ, even with PCNA (Supplementary Figure S1A and B, lanes 2, 3). In the absence of other proteins, CTF18-RFC bound to DNA at a very low level, and a limited PCNA loading occurred (Figure 3A, lanes 4, 5 and Supplementary Figure S1A and B, lanes 4, 5). When p261N was present, >10% of the input CTF18-RFC was retained on the DNA with or without PCNA, and 3-fold more PCNA was loaded than in the absence of p261N (Figure 3A, lanes 6, 7). Similarly, increased CTF18-RFC retention and PCNA loading were observed with Polε, but not Polδ (Supplementary Figure S1A, B, lanes 6, 7). Notably, about 2-fold greater binding of p261N and Polε to DNA occurred in the presence of CTF18-RFC than in its absence (Figure 3A, Supplementary Figure S1A, lanes 2, 3, 6, 7). These results indicated that CTF18-RFC and Polε bound to DNA cooperatively. Thus, CTF18-RFC could access target DNA through cooperative binding, which would further lead to enhanced PCNA loading at near-physiological salt concentrations.
RFC targets 3΄ primer–template junctions to load PCNA (7). The mechanism of PCNA loading by CTF18-RFC was analyzed through binding to different target DNA structures, which were attached to agarose beads (Figure 3B). PCNA loading on the DNA was analyzed in the presence of RPA, which prevents PCNA from sliding off the DNA ends. In the absence of other proteins, CTF18-RFC loaded PCNA onto 3΄ and 5΄ recessed or 3΄ recessed primer–template DNAs (Figure 3B, lanes 5, 8), but not onto ssDNA or 5΄ recessed primer–template DNA (Figure 3B, lanes 2, 11). Thus, CTF18-RFC, like RFC, specifically loads PCNA at 3΄ primer–template junctions.
The DNA structural requirements for PCNA loading were also analysed in the presence of p261N. Because p261N retains strong 3΄–5΄ exonuclease activity (35), p261Nexo– was generated. In p261Nexo–, the highly conserved Asp275 of the exonuclease motif of B-family polymerases (26) is substituted with alanine. Purified p261Nexo– did not exhibit any detectable nuclease activity, and it augmented PCNA loading by CTF18-RFC slightly more effectively than p261N (Supplementary Figure S2). In the presence of p261Nexo– and ATP, increased PCNA loading was observed with 3΄ and 5΄ recessed or 3΄ recessed primer–template DNAs (Figure 3B, lanes 7, 10), but not with ssDNA (Figure 3B, lane 4). Small amounts of PCNA were detected with 5΄ recessed primer–template DNA (Figure 3B, lanes 12, 13) in the presence of p261Nexo– but in the absence of ATP. The small amounts of PCNA detected with this template could be due to p261Nexo–-mediated binding of PCNA to DNA not to direct loading of PCNA onto DNA. In the absence of ATP, both CTF18-RFC and p261Nexo– bound non-specifically to all the DNA structures that were tested (Figure 3B, lanes 3, 6, 9, 12). Notably, in the absence of other proteins, p261Nexo– showed dose-dependent binding to several DNA structures (Supplementary Figure S2D), implying that p261Nexo– could be the dominant cause of this non-specific DNA binding. In the presence of PCNA and ATP, CTF18-RFC and p261Nexo– binding to DNA with a 3΄ primer–template junction increased (Figure 3B, lanes 7, 10), whereas binding to other structures decreased (Figure 3B, lanes 4, 13 and Figure 3C), compared with the absence of ATP. Thus, the cooperative action of CTF18-RFC and p261Nexo– could increase specificity for 3΄ primer–template junctions during PCNA loading.
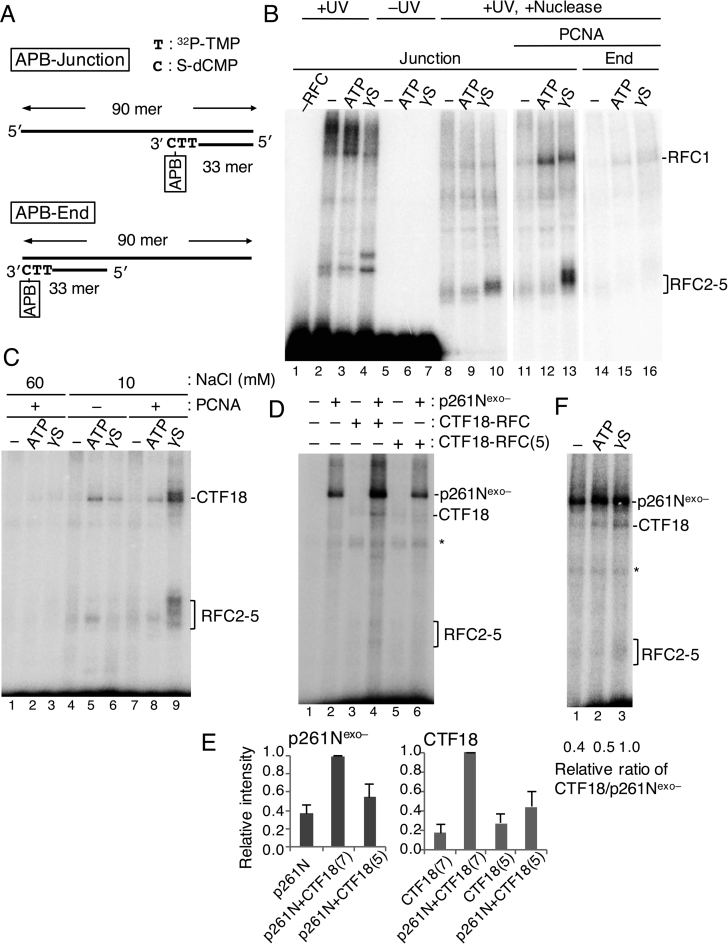
Analysis of the binding modes of CTF18-RFC and p261Nexo– to 3΄ primer ends by photo-crosslinking
Targeting of 3΄ primer ends by CTF18-RFC–Polε complexes during PCNA loading was addressed. Protein binding to DNA during the reaction was analysed via site-specific DNA photo-crosslinking with APB (36,37). This reagent couples with S-dNMP in a substrate DNA (Supplementary Figure S3). The substrate “APB-Junction” had APB coupled to a 3΄ primer–template junction (Figure 4A), so proteins that bound to the junction could be crosslinked via APB to the DNA substrate.
Figure 4.
Analyses of proteins directly bound to the 3΄ primer end at a primer–template junction during PCNA loading with CTF18-RFC–p261N complex by photo-crosslinking. (A) Substrate DNA “APB-Junction” for photo-crosslinking analyses has azidophenacyl bromide (APB) at the 3΄ primer end of the primer–template junction. Two 32P-TMP, and one S-dCMP, were incorporated at the 3΄ primer end of RF30 annealed to TEMP90-R, and a photoreactive crosslinker APB was conjugated to S-dCMP. The control DNA “APB-End” has APB at the blunted end. (B) Photo-crosslinking of 25 fmol APB-Junction (Junction; lanes 1–13) or APB-End (End; lanes 14–16) at 60 mM NaCl by 150 fmol of RFC (lanes 2–16) and 500 fmol of PCNA (lanes 11–16). A set of results without nucleotide (−) or with 2 mM ATP or 250 μM ATPγS (γS) is shown for each condition. Samples with (lanes 1–4, 8–16) or without (lanes 5–7) UV irradiation and with (lanes 8–16) or without (lanes 1–7) nuclease treatment were separated by SDS-PAGE and visualised. (C) Photo-crosslinking of 25 fmol of APB-Junction at 60 mM (lanes 1–3) or 10 mM (lanes 4–9) NaCl by 150 fmol of CTF18-RFC with or without 500 fmol of PCNA, as indicated. A set of results without nucleotides (−) or with 2 mM ATP or 250 μM ATPγS is shown for each condition. (D) Photo-crosslinking of 25 fmol of APB-Junction with combinations of 150 fmol of CTF18-RFC, 250 fmol of CTF18-RFC(5) and 150 fmol of p261Nexo–, as indicated, in the presence of 500 fmol of PCNA and 250 μM ATPγS. (E) Crosslinked bands in (D) corresponding to p261Nexo– and CTF18 were quantified, and their relative intensities were graphed on the right using the highest intensity bands (lane 4) as reference (1.0), with mean ± S.E. of three experimental replicates. “p261N”, “CTF18(7)” and “CTF18(5)” represent p261Nexo–, CTF18-RFC and CTF18-RFC(5), respectively. (F) Photo-crosslinking of the 25 fmol APB-Junction, with 150 fmol of p261Nexo–, 150 fmol of CTF18-RFC and 500 fmol of PCNA in the presence or absence of 2 mM ATP or 250 μM ATPγS. Crosslinked bands corresponding to p261Nexo– and CTF18 were quantified, and the relative values of the CTF18:p261Nexo– ratios are indicated below, with the ratio with ATPγS as 1.0.
When RFC was mixed with APB-Junction at 60 mM NaCl and irradiated with UV light, a high-molecular-mass smear and one or two bands migrating slower than RFC1 and RFC2–5 were detected after SDS-PAGE (Figure 4B, lanes 2–4). Detection of these bands depended on DNA–protein crosslinking, as no protein bands were detected without UV irradiation (Figure 4B, lanes 5–7). After nuclease digestion, some high-molecular-mass signals decreased, and bands corresponding to RFC2–5 were prominent in the presence of ATPγS (Figure 4B, lanes 8–10). In this analysis, labelled peptides will correspond to those most proximal to the APB-labelled nucleotide on individual DNA molecules during a defined period of UV-irradiation. In other words, a band of higher intensity will have a higher probability of being attached to the target site than a band of lower intensity. Thus, this result suggests that RFC2–5 is the major docking protein at the 3΄ primer end when ATP-bound RFC is bound to the 3΄ primer–template junction. Addition of PCNA to the reaction in the presence of ATPγS enhanced the RFC2–5 signal and the RFC1 signal (Figure 4B, lane 13). Addition of PCNA in the presence of ATP decreased the RFC2–5 signal, but enhanced the RFC1 signal, suggesting retention of RFC1 at the 3΄ primer end in the presence of loaded PCNA. These signals were specific for the 3΄ primer–template junction, because moving the target 3΄ primer end to a double-stranded end decreased most of the signals (“APB-End”; Figures 4A and B, lanes 14–16). The molecular mass of labelled protein could not be determined precisely with this assay, so the specific subunits of RFC2–5 that bound to 3΄ primer could not be distinguished, and possible PCNA binding to the site could not be detected.
CTF18-RFC had weak PCNA-loading activity at near-physiological salt concentrations compared with RFC (Figure 2), and binding of CTF18–RFC to the 3΄ primer end at 60 mM NaCl was hardly detectable even with PCNA (Figure 4C, lanes 1–3). At 10 mM NaCl in the presence of ATPγS and PCNA (lane 9), a similar binding profile was observed for CTF18 and RFC2–5 as was seen with RFC, but binding was limited with PCNA and ATP, and without PCNA (lanes 4–8). This result indicated that stable binding of CTF18-RFC to the 3΄ primer end only occurred in the intermediate state of PCNA loading, and that binding in the ATP-bound state without PCNA, or after PCNA loading and ATP hydrolysis, was less stable than the binding seen with RFC, demonstrating the weak intrinsic DNA binding of CTF18-RFC.
When p261Nexo– was added to the experiment, it showed a much greater level of binding to the 3΄ primer end than CTF18-RFC (Figure 4D, lanes 2, 3). As in the DNA pull-down assay (Figure 2), addition of p261Nexo– to CTF18-RFC increased binding of both p261Nexo– and CTF18 in the presence of ATPγS, demonstrating their cooperative binding (lane 4). About 3-fold more p261Nexo– and 5-fold more CTF18 bound to the 3΄ primer end than when they were included individually (Figure 4E). Again, this stimulation of the binding depended on their specific interaction, as CTF18-RFC(5) did not exhibit any significant increase (Figure 4D, lane 6). Both CTF18-RFC and p261Nexo– will make multiple contacts with the primer–template DNA, some of which will be competitive but not others. Thus, the presence of multiple non-competitive contacts will make the DNA binding of CTF18-RFC–p261Nexo– complex cooperative. In the case of the 3΄-end of the primer, CTF18-RFC and p261Nexo– will compete for this end. This competition will occur in the CTF18-RFC–p261Nexo– complex on the primer–template DNA, but with a predominance of p261N binding over that of CTF18, resulting in more crosslinking to p261N than to CTF18.
In contrast to the stabilisation of binding of RFC1 and RFC2–5 that was seen with RFC and ATPγS (Figure 4B), with CTF18-RFC, p261Nexo– and ATPγS, p261Nexo– was the major docking protein, and the signal of CTF18 binding was <5% of the p261Nexo– signal. Thus, in the complex of CTF18-RFC and p261N at the primer–template DNA, binding was cooperative, but with the 3΄ primer end mostly occupied by p261N. In the presence of ATP or the absence of nucleotides (Figure 4F), to minimise the level of the intermediate state of PCNA loading, the ratio of the binding signal of CTF18 to p261Nexo– was decreased compared with in the presence of ATPγS.
Analysis of the binding modes of CTF18-RFC and p261Nexo– on the template strand at template-primer junctions
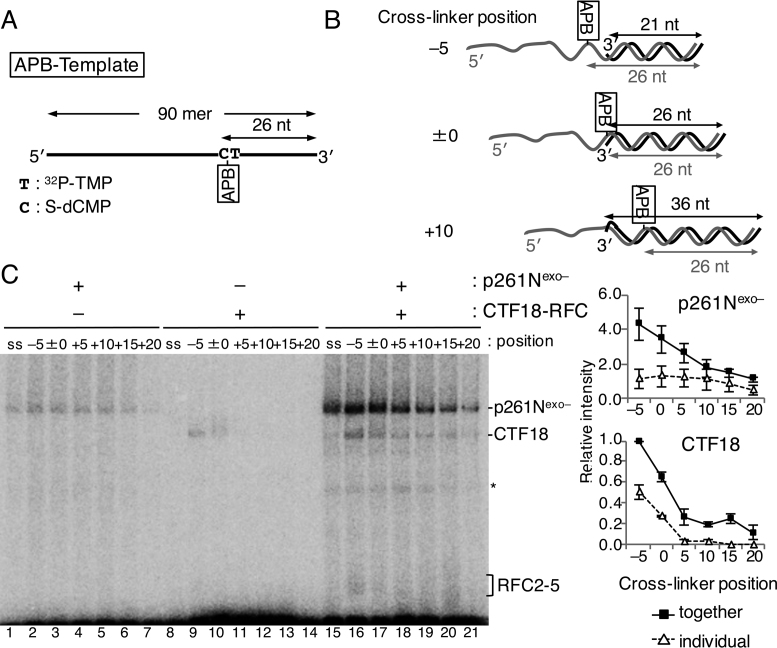
The binding modes of CTF18-RFC and p261Nexo– on the template strand at template-primer junctions were investigated with a 90-mer ssDNA with 32P-TMP and S-dCMP at positions 25 nt and 26 nt from the 3΄ end, respectively, enabling binding of APB (APB-Template, Figure 5A). Six primers, from 21 nt to 46 nt, were annealed at the 3΄ end of the 90-mer to make six constructs in which the position of the APB crosslinker relative to the 3΄ primer end (single–double-strand junction) varied from −5 (ssDNA region) to +20 (double-stranded-DNA region) (Figure 5B).
Figure 5.
Photo-crosslinking analyses of proteins directly bound to the template strand of a primer–template junction during PCNA loading by the CTF18-RFC–p261N complex. (A) The template DNA, “APB-Template”, had 32P-TMP and S-dCMP 25 nt and 26 nt from the 3΄ end on a 90-mer oligonucleotide. (B) Three representatives of the six primer–template junction substrates with differently positioned crosslinkers (azidophenacyl bromide; APB) relative to the 3΄ primer end are indicated. In ‘–5’, the APB is located in single-stranded DNA 5 nt from the junction, in ‘±0’ APB is at the junction and in ‘+10’ APB is in double-stranded DNA 10 nt from the junction. (C) Photo-crosslinked bands from 25 fmol of indicated substrate DNA and 150 fmol of p261Nexo– (lanes 1–7, 15–21), or 150 fmol of CTF18-RFC (lanes 8–21) as indicated in the presence of 500 fmol of PCNA and 250 μM ATPγS. Protein bands corresponding to p261Nexo–, CTF18 and RFC2–5 are indicated. Some degraded proteins are indicated with an asterisk. Results with APB-Template without primers are indicated as the controls (‘ss’; lanes 1, 8, 15). Relative band intensities of p261Nexo– and CTF18 using that of CTF18 in lane 16 as a reference (1.0) were measured and graphed on the right, with mean ± S.E. of three experimental replicates.
Binding profiles of p261Nexo– and CTF18-RFC in the presence of PCNA and ATPγS were studied with the six APB-Template substrates. p261Nexo– crosslinked to the APB obviously when it was in positions −5 to +15, whereas CTF18-RFC crosslinked to APB in positions −5 and ±0 (Figure 5C, lanes 2–7 and 9–14). In assays containing both p261Nexo– and CTF18-RFC (Figure 5C, lanes 16–21), increased crosslinking was observed, corresponding to cooperative binding. The binding of CTF18 at position −5 corresponded to ∼25% of the signal for binding of p261Nexo–, indicating that the association of CTF18-RFC with the template strand at this point (relative to that of p261Nexo–) was greater than the association with the 3΄ primer end (Figure 4C). The prominent binding of CTF18 at −5 and ±0 in the presence of ATPγS was not observed in the presence of ATP (Supplementary Figure S4), where the level of the intermediate state of PCNA loading should be low as described above. Thus, in the presence of ATPγS, the greater association of CTF18 with the region from the single–double-strand junction to the ssDNA template strand suggests a temporal association of CTF18-RFC to the region during PCNA loading.
PCNA loading by CTF18-RFC–Polε is blocked if Polε is in DNA-synthesis mode
All the preceding experiments were carried out without dNTPs, and therefore demonstrate the interactions of Polε in non-synthesizing mode. To investigate the interactions with Polε in DNA-synthesis mode, we prepared a primer–template junction substrate with dideoxynucleotide (ddNMP) at the 3΄ primer end (deoxidised 3΄ primer end, Figure 6A). With this substrate, in the presence of the next-incorporating dNTP, Polε is trapped in the act of extending DNA with deoxynucleotides (DNA-synthesis mode), as demonstrated by a structural study of yeast Polε (38). Indeed, discrete binding of Polα and Polδ to a similar substrate DNA was observed only in the presence of the incoming dNTP (39). To test the DNA-synthesis mode of Polε at the deoxidised 3΄ primer end, a 90 nt oligonucleotide was annealed with a 32P-labeled 34 nt primer terminating with ddAMP at its 3΄ end (‘dd-Junction’; Figure 6A). When this substrate was incubated with Polεexo– in the presence of TTP, the incoming nucleotide, stronger shifted bands were observed than without TTP (Figure 6B, E, lanes 5, 6) or with a substrate lacking the deoxidised 3΄ primer end (‘d-Junction’, Figure 6A, Figure 6B, lane 3). This result demonstrates a strategy to produce Polεexo– in synthesising mode on a substrate DNA.
On the basis of this result, gapped-DNA beads with ddAMP at the 3΄ primer end were prepared (Figure 6C), to examine the effect of dGTP, the incoming nucleotide, on PCNA loading by CTF18-RFC in the presence of Polεexo–. dGTP did not affect PCNA loading by CTF18-RFC alone in the presence of ATP (Figure 6D, lanes 3, 8), but it suppressed the stimulation of PCNA loading by the addition of Polεexo– (Figure 6D, lanes 4–6, 9–11).
Assembly of proteins at the 3΄ primer–template junction of the dd-Junction substrate was studied by EMSA after glutaraldehyde fixation. In the absence of Polεexo–, crosslinked CTF18-RFC was not detected, even with PCNA and ATPγS (Figure 6E, lanes 2–4). This result differed from the binding of RFC to a 3΄ primer–template junction (39), which was observed in the presence of PCNA and ATPγS, demonstrating again the relatively low affinity of CTF18-RFC for DNA. In the presence of Polεexo– and absence of CTF18-RFC, a band shift was observed only in the presence of TTP (Figure 6E, lane 6). This band was slightly supershifted by the addition of PCNA (Figure 6E, lane 10), representing the formation of a Polεexo––PCNA complex on the DNA without the active loading of PCNA. This novel shift by PCNA was more apparent with the smaller Polε variant, p261Nexo– (Supplementary Figure S5). Addition of CTF18-RFC to Polεexo– supershifted the Polεexo– band (Figure 6E, lane 8), indicating tethering of CTF18-RFC to Polε in synthesising mode at the 3΄ primer. Addition of PCNA produced a 2-fold to 3-fold increase in intensity of the supershifted band (Figure 6E, lane 12), compared with no PCNA, suggesting the presence of a stable assembly of Polεexo––CTF18-RFC including PCNA on the dd-Junction substrate. The supershift depended on TTP, indicating that the complex would be formed with Polεexo– in synthesising mode. This complex might represent a trimeric assembly that occurs with Polε in synthesising mode after PCNA association at the 3΄ primer–template junction.
DNA synthesis by CTF18-RFC–Polε–PCNA
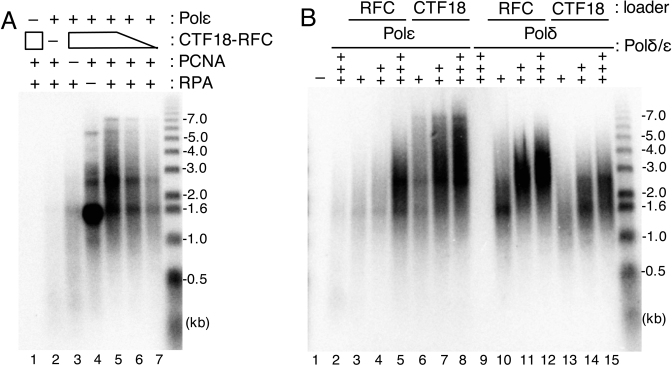
What is the functional significance of the trimeric assembly of CTF18-RFC–Polε–PCNA? To test the hypothesis that this novel complex functions as an active DNA polymerase complex, we performed a holoenzyme assay with CTF18-RFC, Polε, PCNA and RPA using a singly primed M13mp18 as template. Efficient DNA synthesis was observed with the four components (Figure 7A, lane 5). Omission of one of the components resulted in the severe or total loss DNA synthesis except for when RPA was omitted (lanes 1–4); in this case, DNA products with a size of about 1.5 kb accumulated. Thus, efficient initiation of DNA synthesis occurred with the trimeric complex, although RPA was further required for DNA elongation, probably via its ability to resolve secondary structures on the template DNA. A decrease in the amount of CTF18-RFC resulted in reduced DNA synthesis, but the mean-product-lengths were not affected significantly (lanes 5–7), suggesting that CTF18-RFC might be required for efficient initiation of DNA synthesis by Polε.
Figure 7.
Analysis of DNA synthesis with CTF18-RFC–Polε–PCNA. Product DNA profiles after electrophoresis in alkaline agarose gels are indicated together with DNA size markers (right). (A) Holoenzyme assay was done with indicated combinations of 200 fmol Polε, 600 fmol CTF18-RFC, 2 pmol PCNA and 6 pmol RPA. Lower amounts of CTF18-RFC (400 or 200 fmol) were used in lanes 6 and 7. (B) Titration of Polε (+, ++, +++ for 147, 293, 440 fmol, respectively; lanes 2–8) or Polδ (+, ++, +++ for 53, 107, 160 fmol, respectively; lanes 9–15) in the holoenzyme assay with 600 fmols RFC (lanes 3–5, 10–12) or CTF18-RFC (lanes 6–8, 13–15) or without (lanes 2 and 9) in the presence of 6 pmol RPA and 2 pmol PCNA.
To determine whether efficient DNA synthesis with the four components is mediated by the specific assembly of CTF18-RFC–Polε and PCNA, we compared DNA syntheses with Polδ and Polε in the presence of RPA in reactions where PCNA was loaded by either RFC or CTF18-RFC. Similar to previously published results (5,24), Polδ synthesised DNA efficiently with PCNA loaded by RFC, but less efficiently with PCNA loaded by CTF18-RFC (Figure 7B, lanes 10–15). This difference may reflect the difference in the efficiency of PCNA loading by these two loaders. PCNA loaded by RFC also stimulated DNA synthesis by Polε via the previously reported interaction between Polε and PCNA (34) (lanes 3–5). DNA synthesis in the presence of CTF18-RFC was more efficient and produced longer DNA than DNA synthesis in the presence of RFC (lanes 6–8). Collectively, these results suggest that CTF18-RFC is more adapted as a PCNA loader for Polε than RFC and vice versa for Polδ. Furthermore, Polε in the CTF18-RFC–Polε–PCNA complex synthesised DNA more processively than Polε–PCNA, suggesting that it functions as a genuine functional DNA polymerase holoenzyme.
DISCUSSION
Our results showed that loading of PCNA by CTF18-RFC was stimulated by Polε through their specific interaction. The intrinsic loading of PCNA by CTF18-RFC was salt-sensitive and blocked by a saturating amount of RPA; thus, it would be almost inactive under near-physiological conditions. However, the activity was restored if CTF18-RFC was present in the complex with Polε (Figure 2), suggesting that CTF18-RFC functions as an active PCNA loader when it associates with Polε. Because Polε is the leading-strand DNA polymerase, CTF18-RFC can be expected to function as a major component in the replication-fork complex. Indeed, depletion of CTF18 in human cells impairs the normal progression of the replication fork (23), and CTF18 is enriched in chromatin in S phase (40) and localises at the replication fork (21,22). However, additional roles for CTF18-RFC–Polε in DNA synthesis for nucleotide-excision repair (41) and telomere maintenance (18,19) might also exist.
Cooperative binding of CTF18-RFC and Polε to DNA was observed in various assays. If both CTF18-RFC and p261N were present, they cooperatively bound to DNA in a fairly non-specific manner (Figure 3). However, they bound to DNA with a 3΄ primer–template junction more specifically than other structures under PCNA-loading conditions, indicating that their cooperative action in the presence of ATP and PCNA could drive efficient PCNA loading at physiological salt concentrations. DNA photo-crosslinking experiments demonstrated that, in the complex of CTF18-RFC–Polε, Polε associated predominantly with the junction and the association of CTF18-RFC was minor, although evidently sufficient for PCNA loading. PCNA loading by CTF18-RFC–Polε was suppressed when Polε was in DNA-synthesis mode, whereas efficient PCNA loading occurred with Polε in non-synthesising mode. Therefore, switching of DNA-synthesis modes of Polε could be an essential part of the mechanism for PCNA loading by CTF18-RFC.
DNA synthesis by the two polymerases was stimulated by PCNA, irrespective of the loader added to the assay. In the absence of a specific interaction between DNA polymerases and loaders, PCNA will stimulate both DNA polymerases according to the loading efficiency of PCNA onto DNA and the affinity between PCNA and the DNA polymerase. However, in the case of Polε and CTF18-RFC, the loaded PCNA was captured by the CTF18-RFC–Polε complex, whose assembly resulted in efficient and processive DNA synthesis by Polε. Thus, the assembly of the CTF18-RFC-Polε–PCNA complex is important for the functional status of the processive Polε holoenzyme at the replication fork.
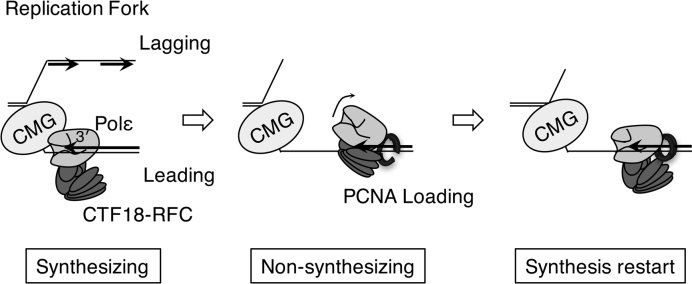
The model in Figure 8 summarises our observations. In the normal replication fork, CMG helicase and Polε form a processive DNA polymerase complex for leading-strand synthesis (42–44). CTF18-RFC would be involved in the complex by association with Polε. If the replication fork proceeded at a normal rate, CMG–Polε would be maintained and Polε would remain in DNA-synthesis mode. In this case, the associated CTF18-RFC would have little opportunity to load PCNA. Upon arrest of Polε by obstacles on the chromosome, Polε would decouple from CMG and shift to non-synthesising mode. CTF18-RFC would then be able to access the 3΄ primer end and load PCNA at the site. Participation of PCNA would stabilise the CTF18-RFC–Polε complex at the 3΄ primer end. As suggested above, the CTF18-RFC–Polε–PCNA complex would function as the secondary processive DNA polymerase complex, even without CMG, and would facilitate the restart of DNA synthesis. Because the arrest of Polε occurs frequently in replicating DNA at particular sequences and chromatin structures even in the absence of artificial factors that perturb DNA synthesis (Figure 7 and references 45,46), depletion of CTF18, which would impair the function of the backup system of DNA synthesis, would reduce the gross replication rate of the chromosome, as has been observed in human cells (23).
Figure 8.
A possible PCNA-loading mechanism by CTF18-RFC–Polε complex. Synthesising: In the normal replication fork, CMG helicase and Polε form a processive DNA polymerase complex for leading-strand synthesis. CTF18-RFC will associate with Polε in the complex, but PCNA loading will be minimised because Polε is in synthesising mode. Non-synthesising: Upon arrest of Polε by an obstacle on the chromosome, Polε is decoupled from CMG and shifts to non-synthesising mode. Under this condition, CTF18-RFC is able to access the 3΄ primer end and load PCNA at the site. Synthesis restart: Participation of PCNA in the CTF18-RFC–Polε complex at the 3΄ primer end will stabilise the complex and facilitate restart of the DNA synthesis by forming a secondary processive DNA polymerase complex lacking CMG.
Defects in Ctf18 cause a wide range of deficiencies in chromosomal-DNA metabolism (12–19,23,40,41), but its biochemical functions have been poorly understood. Our biochemical studies demonstrated that PCNA would be loaded onto the leading strand by CTF18-RFC during DNA replication. PCNA functions as a platform for various proteins that function in DNA replication (3). However, its distribution between leading and lagging strands will potentially be imbalanced by the asymmetric priming mechanisms on the two strands. Whether PCNA distributes on the leading and lagging DNA strands in a biased fashion, and whether PCNA-interacting factors function evenly on the two strands for chromatin assembly, chromosome cohesion and DNA repair when the distribution of PCNA is biased on the two strands is not known (47). Recent analyses in budding yeast (11) demonstrated that the ratio of PCNA on the lagging strand to that on the leading strand is only about two to one, which is smaller than the expected ratio on the basis of the number of priming events that occur on the two strands. Elg1-RFC has a role as an unloader of PCNA from the lagging strand in stalled replication forks (11). However, our data strongly suggest the presence of an active PCNA-loading mechanism on the leading strand, engaged by CTF18-RFC–Polε. Previous work suggests that this active loading will occur regularly during replication judging from the importance of CTF18-RFC in maintaining the normal replication rate (23). Thus, this mechanism might increase the dosage of PCNA on the leading strand, providing sufficient opportunities for PCNA-interacting factors to act to maintain the DNA structures.
Thus, PCNA loading by CTF18-RFC–Polε has two roles for DNA replication. One is to maintain leading-strand synthesis at template DNA structures that disrupt Polε progression, by supplying PCNA to restore DNA synthesis. The second role is active PCNA loading to the leading strand through the above mechanism, which balances PCNA dosage between the two DNA strands, enabling PCNA-binding proteins to function properly on both strands. To test these possibilities, it will be crucial to prepare Polε point mutants defective in their interaction with CTF18-RFC and determine whether they are defective in PCNA loading in vitro and in vivo.
This study opens for further studies directed at determining how, when and where CTF18-RFC and Polε interact in cells, and what is functional significance of the interaction in vivo. Future studies focusing on the cellular behaviours CTF18-2RFC and Polε during DNA replication will be necessary to elucidate the novel PCNA dynamics proposed by our biochemical studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr H. Kawakami (Kyushu University) for his kind advice on the photo-crosslink experiments and for providing α-S-dCTP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Scientific Research (KAKENHI) [25131714, 25440011, 26114714 and 16H04743]. Funding for open access charge: JSPS Grants-in-aid for Scientific Research (KAKENHI) [16H04743].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bloom L.B. Loading clamps for DNA replication and repair. DNA Repair. 2009; 8:570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hedglin M., Kumar R., Benkovic S.J.. Replication clamps and clamp loaders. Cold Spring Harb. Perspect. Biol. 2013; 5:a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moldovan G.L., Pfander B., Jentsch S.. PCNA, the maestro of the replication fork. Cell. 2007; 129:665–679. [DOI] [PubMed] [Google Scholar]
- 4. Kelch B.A. Review: The lord of the rings: Structure and mechanism of the sliding clamp loader. Biopolymers. 2016; 105:532–546. [DOI] [PubMed] [Google Scholar]
- 5. Bermudez V.P., Maniwa Y., Tappin I., Ozato K., Yokomori K., Hurwitz J.. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:10237–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanellis P., Agyei R., Durocher D.. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 2003; 13:1583–1595. [DOI] [PubMed] [Google Scholar]
- 7. Ellison V., Stillman B.. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5΄ recessed DNA. PLoS Biol. 2003; 1:231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Navadgi-Patil M.V., Burgers P.M.. A tale of two tails: Activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair. 2009; 8:996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kubota T., Nishimura K., Kanemaki M.T., Donaldson A.D.. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol. Cell. 2013; 50:273–280. [DOI] [PubMed] [Google Scholar]
- 10. Shiomi Y., Nishitani H.. Alternative replication factor C protein, Elg1, maintains chromosome stability by regulating PCNA levels on chromatin. Genes Cells. 2013; 18:946–959. [DOI] [PubMed] [Google Scholar]
- 11. Yu C., Gan H., Han J., Zhou Z., Jia S., Chabes A., Farrugia G., Ordog T., Zhang Z.. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell. 2014; 56:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mayer M.L., Gygi S.P., Aebersold R., Hieter P.. Identification of RFC (Ctf18p, Ctf8p, Dcc1p): An alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell. 2001; 7:959–970. [DOI] [PubMed] [Google Scholar]
- 13. Merkle C.J., Karnitz L.M., Henry-Sánchez J.T., Chen J.. Cloning and characterization of hCTF18, hCTF8, and hDCC1. Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J. Biol. Chem. 2003; 278:30051–30056. [DOI] [PubMed] [Google Scholar]
- 14. Hanna J.S., Kroll E.S., Lundblad V., Spencer F.A.. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 2001; 21:3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naiki T., Kondo T., Nakada D., Matsumoto K., Sugimoto K.. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 2001; 21:5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crabbé L., Thomas A., Pantesco V., De Vos J., Pasero P., Lengronne A.. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat. Struct. Mol. Biol. 2010; 17:1391–1398. [DOI] [PubMed] [Google Scholar]
- 17. Kubota T., Hiraga S., Yamada K., Lamond A.I., Donaldson A.D.. Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol. Cell. Proteomics. 2011; 10, M110.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiraga S., Robertson E.D., Donaldson A.D.. The Ctf18 RFC‐like complex positions yeast telomeres but does not specify their replication time. EMBO J. 2006; 25:1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao H., Moss D.L., Parke C., Tatum D., Lustig A.J.. The Ctf18RFC clamp loader is essential for telomere stability in telomerase-negative and mre11 mutant alleles. PLoS One. 2014; 9:e88633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K.P., Shirahige K., Uhlmann F.. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006; 23:787–799. [DOI] [PubMed] [Google Scholar]
- 21. Sirbu B.M., McDonald W.H., Dungrawala H., Badu-Nkansah A., Kavanaugh G.M., Chen Y., Tabb D.L., Cortez D.. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 2013; 288:31458–31467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A.. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014; 16:281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terret M.E., Sherwood R., Rahman S., Qin J., Jallepalli P.V.. Cohesin acetylation speeds the replication fork. Nature. 2009; 462:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shiomi Y., Shinozaki A., Sugimoto K., Usukura J., Obuse C., Tsurimoto T.. The reconstituted human Chl12-RFC complex functions as a second PCNA loader. Genes Cells. 2004; 9:279–290. [DOI] [PubMed] [Google Scholar]
- 25. Bylund G.O., Burgers P.M.. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol. Cell. Biol. 2005; 25:5445–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henninger E.E., Pursell Z.F.. DNA polymerase ε and its roles in genome stability. IUBMB Life. 2014; 66:339–351. [DOI] [PubMed] [Google Scholar]
- 27. Murakami T., Takano R., Takeo S., Taniguchi R., Ogawa K., Ohashi E., Tsurimoto T.. Stable interaction between the human proliferating cell nuclear antigen loader complex Ctf18-replication factor C (RFC) and DNA polymerase ε is mediated by the cohesion-specific subunits, Ctf18, Dcc1, and Ctf8. J. Biol. Chem. 2010; 285:34608–34615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. García-Rodríguez L.J., De Piccoli G., Marchesi V., Jones R.C., Edmondson R.D., Labib K.. A conserved Polε binding module in Ctf18-RFC is required for S-phase checkpoint activation downstream of Mec1. Nucleic Acids Res. 2015; 18:8830–8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okimoto H., Tanaka S., Araki H., Ohashi E., Tsurimoto T.. Conserved interaction of Ctf18-RFC with DNA polymerase ε is critical for maintenance of genome stability in Saccharomyces cerevisiae. Genes Cells. 2016; 21:482–491. [DOI] [PubMed] [Google Scholar]
- 30. Shiomi Y., Masutani C., Hanaoka F., Kimura H., Tsurimoto T.. A second proliferating cell nuclear antigen loader complex, Ctf18-Raplication Factor C, stimulates DNA polymerase η activity. J. Biol. Chem. 2007; 282:20906–20914. [DOI] [PubMed] [Google Scholar]
- 31. Narita T., Tsurimoto T., Yamamoto J., Nishihara K., Ogawa K., Ohashi E., Evans T., Iwai S., Takeda S., Hirota K.. Human replicative DNA polymerase δ can bypass T-T (6-4) ultraviolet photoproducts on template strands. Genes Cells. 2010; 15:1228–1223. [DOI] [PubMed] [Google Scholar]
- 32. Sun Q., Tsurimoto T., Juillard F., Li L., Li S., De León Vázquez E., Chen S., Kaye K.. Kaposi's sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11816–11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waga S., Stillman B.. Cyclin-dependent kinase inhibitor p21 modulates the DNA primer–template recognition complex. Mol. Cell. Biol. 1998; 18:4177–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chilkova O., Stenlund P., Isoz I., Stith C.M., Grabowski P., Lundström E.B., Burgers P.M., Johansson E.. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007; 35:6588–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Korona D.A., Lecompte K.G., Pursell Z.F.. The high fidelity and unique error signature of human DNA polymerase ε. Nucleic Acids Res. 2011; 39:1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang S.W., Nash H.A.. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:12183–12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lagrange T., Kim T.K., Orphanides G., Ebright Y.W., Ebright R.H., Reinberg D.. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hogg M., Osterman P., Bylund G.O., Ganai R.A., Lundström E.B., Sauer-Eriksson A.E., Johansson E.. Structural basis for processive DNA synthesis by yeast DNA polymerase ε. Nat. Struct. Mol. Biol. 2014; 21:49–55. [DOI] [PubMed] [Google Scholar]
- 39. Tsurimoto T., Stillman B.. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer–template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 1991; 266:1950–1960. [PubMed] [Google Scholar]
- 40. Shiomi Y., Hayashi A., Ishii T., Shinmyozu K., Nakayama J., Sugasawa K., Nishitani H.. Two different replication factor C proteins, Ctf18 and RFC1, separately control PCNA-CRL4Cdt2-mediated Cdt1 proteolysis during S phase and following UV irradiation. Mol. Cell. Biol. 2012; 32:2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G. et al. . Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010; 37:714–727. [DOI] [PubMed] [Google Scholar]
- 42. Georgescu R. E, Langston L., Yao N.Y., Yurieva O., Zhang D., Finkelstein J., Agarwal T., O’Donnell M.E.. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat. Struct. Mol. Biol. 2014; 21:664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Georgescu R.E., Schauer G.D., Yao N.Y., Langston L.D., Yurieva O., Zhang D., Finkelstein J., O’Donnell M.E.. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife. 2015; 4:e04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kang Y.H., Galal W.C., Farina A., Tappin I., Hurwitz J.. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6042–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Le H.P., Masuda Y., Tsurimoto T., Maki S., Katayama T., Furukohri A., Maki H.. Short CCG repeat in huntingtin gene is an obstacle for replicative DNA polymerases, potentially hampering progression of replication fork. Genes Cells. 2015; 20:817–833. [DOI] [PubMed] [Google Scholar]
- 46. Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., Nicolas A.. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011; 30:4033–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Georgescu R.E., Langston L.D., O’Donnell M.E.. A proposal: evolution of PCNA's role as a marker of newly replicated DNA. DNA Repair (Amst.). 2015; 29:4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.