Abstract
Although studies of the differentiation from mouse embryonic stem (ES) cells to vascular endothelial cells (ECs) provide an excellent model for investigating the molecular mechanisms underlying vascular development, temporal dynamics of gene expression and chromatin modifications have not been well studied. Herein, using transcriptomic and epigenomic analyses based on H3K4me3 and H3K27me3 modifications at a genome-wide scale, we analysed the EC differentiation steps from ES cells and crucial epigenetic modifications unique to ECs. We determined that Gata2, Fli1, Sox7 and Sox18 are master regulators of EC that are induced following expression of the haemangioblast commitment pioneer factor, Etv2. These master regulator gene loci were repressed by H3K27me3 throughout the mesoderm period but rapidly transitioned to histone modification switching from H3K27me3 to H3K4me3 after treatment with vascular endothelial growth factor. SiRNA knockdown experiments indicated that these regulators are indispensable not only for proper EC differentiation but also for blocking the commitment to other closely aligned lineages. Collectively, our detailed epigenetic analysis may provide an advanced model for understanding temporal regulation of chromatin signatures and resulting gene expression profiles during EC commitment. These studies may inform the future development of methods to stimulate the vascular endothelium for regenerative medicine.
INTRODUCTION
To date, many studies on vascular development have consisted of gene knockout and knockdown experiments using mice and zebrafish (1–6). Although these studies resulted in new discoveries related to vascular development in vertebrates, they could not identify the precise molecular mechanisms underlying vascular endothelial cell (EC) differentiation. Recent studies indicated that embryonic stem (ES) cell differentiation recapitulates endogenous developmental processes, including vascular development (7). Therefore, the detailed investigation of EC differentiation from ES cells may provide valuable insights into EC development following mature vascularization in vivo. Because the vascular network is indispensable for homeostasis in the human body, the most effective EC differentiation system is required for future regenerative medicine.
It was recently proposed that some transcription factor networks determine cell fate (8,9). For example, four transcription factors, i.e. Klf4, Oct4, Sox2 and c-Myc are sufficient to reprogram somatic cells to pluripotent cells (10). In relation to vascular ECs, transient ETV2 expression and suppression of TGFβ with constitutive expression of FLI1 and ERG can induce direct conversion from amniotic cells to mature ECs (11). It was recently reported that the unique Ets family transcription factor Etv2 directly converts human fibroblasts into ECs (12). Several combinations of transcription factors induce or directly mediate the conversion to not only ECs but also cardiomyocytes; thus, transcription factors are able to maintain the specificity of terminally differentiated cells. However, the mechanism by which the epigenetic modifications are erased and re-written during EC fate determination and conversion is largely unknown.
Recent advances in next-generation sequencing have clarified that not only the expression of transcription factors but also epigenetic modifications including DNA methylation and histone modification influence gene expression, resulting in differences between cell types (13). Therefore, the machinery underlying gene expression and differentiation to ECs might be determined by lineage-specific transcription factors in concert with the specific epigenetic modulators. We previously reported that the transcription factor GATA2 regulates EC-specific gene expression through epigenetic modifications, in particular with histone modifications and chromatin conformation changes (14). We demonstrated that GATA2 binds to and upregulates EC-specific genes, and also represses cell transition factors such as TGFβ, Snail and Slug (14). However, the epigenetic mechanisms of the EC-specific differentiation system mediated by GATA2 and other transcription factors remain to be elucidated.
The Sox transcription factor family consists of 20 members in mice, classified into eight groups (15). Among these, SoxF factors, including Sox7, Sox17 and Sox18, specifically bind to the consensus sequence 5΄-(A/T)(A/T)CAA(A/T)G-3΄ (16,17). Many studies have indicated that Sox factors play pivotal roles in embryonic development. In particular, SoxF factors are related to cardiovascular development and EC differentiation (1,18). In fact, although SoxF factor-null mice exhibit severe vascular defects, a comprehensive analysis of each SoxF gene target has not been performed.
In this study, we conducted sequential microarray and chromatin immunoprecipitation-sequencing (ChIP-seq) analyses on our previously established EC differentiation system (19). Our results indicate that four transcription factors, i.e. Gata2, Fli1, Sox7 and Sox18, were induced after expression of Etv2 mediated via vascular endothelial growth factor (VEGF) stimulation. In addition, these four transcription factors had switched histone modifications, from H3K27me3 to H3K4me3, after VEGF stimulation. Knockdown of these factors enabled us to comprehensively identify their downstream target genes. Moreover, knockdown of all four transcription factors almost completely abrogated EC differentiation. Comprehensive gene expression analysis indicated that these transcription factors not only upregulated EC-specific genes but also downregulated other lineage commitment genes. By using bioinformatics approaches, we uncovered the essential combinations of transcription factors essential for EC commitment and the epigenetic landscape during EC differentiation. These results might contribute to the development of an efficient EC differentiation method, and to the evaluation of high-quality ECs for regenerative medicine.
MATERIALS AND METHODS
Cell culture
Mouse ES cells were maintained as previously described (20). Induction of differentiation was performed using differentiation medium (DM; α minimal essential medium (MEM; Gibco, Carlsbad, CA, USA) supplemented with 10% foetal calf serum (Japan Bioserum Co Ltd, Hiroshima, Japan) and 5 × 10−5 M 2-mercaptoethanol (Gibco)) as previously described (7). In brief, undifferentiated ES cells were cultured without leukaemia inhibitory factor on collagen-coated dishes (Becton Dickson, Bedford, MA, USA) at a cell density of 0.75 to 1 × 103 cells/cm2 for 96 h. Cells were harvested and purified by magnetic cell sorting (MACS) with an anti-Flk-1 antibody. Purified Flk-1-positive cells were seeded onto collagen-coated dishes at a density of 0.75 to 1 × 104 cells/cm2 in DM.
RNA collection and DNA microarray
Total RNA from mouse ES cells at 0, 6, 12, 24 and 48 h after stimulation with or without VEGF was collected and purified using the RNeasy Micro kit (QIAGEN, Hilden, Germany). Preparation of the cDNA and hybridization of the probe arrays were performed according to the manufacturer's instructions (Affymetrix, Santa Clara, CA, USA). Affymetrix GeneChip Mouse Genome 430 2.0 arrays containing over 39 000 transcripts were applied. Data were analysed according to the minimum information about a microarray experiment rule. The raw intensity data for each experiment were first normalized by Robust Multi-array Average algorithm (21). After removal of batch effects with ComBat (22), 44 754 probes were selected and used to create the heat maps. Additional RNA reverse transcription and quantitative polymerase chain reaction (qPCR) protocols are described in Supplementary Methods.
Creating heat maps by microarray data
From among 44 754 probes in Affymetrix GeneChip arrays, gene selections were performed following the pre-set criteria as described below. Specifically, gene sets correlating to EC differentiation were chosen according to average differences in gene expression score following VEGF stimulation of over 300, and exhibiting a fold change of >3.0 compared to the non-stimulated cells and ES cells. These thresholds minimized random noise fluctuations to the greatest possible degree. Alternatively, gene sets correlating to siRNA-mediated inhibition of EC differentiation were chosen based on the average differences in gene expression score of differentiated EC cells being over 100.0, and with a Z-score of the fold change compared to single-gene knockdown of more than 2.0. Selected genes were classified into clusters corresponding to the specific up- or downregulated patterns of gene expression yielded by each siRNA in the Gata2/Sox7/Sox18/Fli1 knockdown condition, using a hierarchal clustering algorithm, HOPACH (http://docpollard.org/) with the default parameter setting. Clustered patterns of gene expression are shown as heat maps. Additional details are provided in the Supplementary Methods.
Chromatin immunoprecipitation (ChIP) and ChIP-seq assay
ChIP and ChIP-seq assays were performed as described previously (23). In brief, cells were collected and crosslinked with 1% formaldehyde for 10 min. After neutralization by using 0.2 M glycine for 5 min, cells were collected, re-suspended in sodium dodecyl phate (SDS) lysis buffer (10 mM Tris–HCl, 150 mM NaCl, 1% SDS, 1 mM ethylenediaminetetraacetic acid (EDTA); pH 8.0, protease inhibitor cocktail) and fragmented by sonication (Sonifier 250, Branson; 10 min, 60% duty, output level 4). The sonicated solution was diluted in ChIP dilution buffer (20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) to a volume of 10.3 ml, and 10 ml was used for IP, whereas 300 μl was used as INPUT. Specific antibodies were bound with Dynabeads Magnetic beads (Life Technologies, Madison, WI, USA) and applied to the diluted sonicated solution for IP. Antibodies against H3K4me3 (kindly gifted by Dr Kimura (Tokyo Institute of Technology, Japan)) and H3K27me3 (07–449, Millipore, Billerica, MA, USA) were used (24,25). The prepared DNA was quantified using Q-bit (Life Technologies) and more than 10 ng of DNA was processed for the ChIP-seq assay. ChIP-DNA was prepared for sequencing according to a modified version of the genomic DNA protocol (Illumina, San Diego, CA, USA). Additional detailed procedures are provided in Supplementary Methods.
ChIP-seq data analysis
The sequence reads of the DNA fragments obtained by chromatin ChIP for H3K4me3, H3K27me3 and INPUT control were mapped onto a reference mouse genome, mm9, using the Illumina alignment program ELAND (included in the CASAVA 1.8.2 platform). The read enrichment (i.e. the normalized numbers of the sequence reads mapped onto the particular genomic sites) on the promoter of each gene was calculated within 1000 bp upstream/downstream of the transcription start site (TSS). Based on the total of 10 values from the read enrichments for each 5 samples of H3K4me3 and H3K27me3, the genes selected by the gene expression profiles were classified into 4 classes. Details of the data processing are available upon request and additional information on the analysis related to the reproducibility testing with duplicate ChIP-seq experiments is provided in the Supplementary Methods.
Histone modification heat map
H3K4me3 and H3K27me3 modification levels were analysed by using the Integrated Genome Viewer with ChIP-seq around the TSSs (upstream/downstream 1000 bp from TSS) of the identified genes in VEGF-stimulated cells. The modification levels were defined as the number of sequence reads mapped within 1000 bp upstream/downstream of the TSSs, and were normalized by the number of total reads and the size of the target promoter regions (i.e. 2000 bp).
Identification of highly expressed genes in ECs
To identify genes that exhibit an EC-specific pattern, we utilized the expression profiles generated using Affymetrix HT Human Genome U133A and U133B Arrays in primary cultured ECs (coronary artery ECs (HCAECs), dermal microvascular ECs (HMVECs), human umbilical vein ECs (HUVECs), intestinal ECs (IEns) and retinal ECs (REs)) and non-ECs (bronchial smooth muscle cells (BSMCs), coronary artery smooth muscle cells (CASMCs), bronchial/tracheal epithelial cells (NHBEs), epidermal keratinocytes-adult cells (NHEK-Ads), epidermal keratinocytes-neonate cells (NHEK-neos), mesangial cells (NHMCs), normal human prostate epithelial cells (PrECs), skeletal muscle cells (SkMCs), umbilical artery smooth muscle cells (UASMCs), uterine smooth muscle cells (UtSMCs), astrocytes (ASTs), pro-adipocytes (HN_dAs), hepatocytes (Heps), intestinal epithelial cells (IEs), lung fibroblasts (HN_LFs), neuronal precursors (HN_NPs), osteoblasts (HN_Os), small airway epithelial cells (HN_SAEs), macrophages (Mphages), monocytes (Mocytes), smooth muscle cells (Sm_mscl), small airway epithelial cell (SAECs) and fibroblasts (Fibros)). These data are available from our RefExA database (http://157.82.78.238/refexa/). Additionally, the expression profiles of 10 ECs and 6 non-ECs were downloaded from the NCBI Gene Expression Omnibus (GEO accession number: GSE 21212). The raw data of the total 15 ECs and 29 non-ECs were normalized using RMA normalization (21) and corrected for batch effect using ComBat (22). To identify differentially and highly expressed genes in ECs compared to non-ECs, a one-sided Welch's t-test was applied to the data from each gene. Genes were considered highly expressed if they had a false discovery rate (FDR)-adjusted P-value bellow 0.001. This EC-specific gene set was further ranked on the basis of the FDR-adjusted P-value and used for further analysis.
Visualization and comparison of mouse cells during EC differentiation and isolated from defined normal tissues
To investigate the transition state of the cells during EC development and compare these cells with those of various types of normal mouse tissues, the expression profiles of 61 tissue types from native male C57BL6 mice were downloaded from the NCBI Gene Expression Omnibus (GEO accession number: GSE10246). The raw data were normalized using the MAS5 algorithm (Median score of gene expression: 500), log2-transformed and corrected for batch effect using ComBat (22). We focused on the top 100 ranked highly expressed genes in human ECs that mapped to their 79 mouse orthologues based on Ingenuity Knowledge Base (http://www.ingenuity.com/science/knowledge-base). To visualize the cells in the presence or absence of VEGF as well as the mouse normal tissues in three-dimensional (3D) space, classical multi-dimensional scaling (MDS) was performed on the expression profiles of the 79 EC-specific genes.
Immunostaining
Immunostaining for cultured cells was performed as described previously (20). Briefly, 4% paraformaldehyde-fixed cells were blocked with 1% skim milk (BD Biosciences, Franklin Lakes, NJ, USA) and incubated overnight with primary antibodies at 4°C. For immunofluorescence staining, anti-mouse or -rabbit IgG antibodies conjugated with Alexa488 or Alexa546 (Invitrogen, Carlsbad, CA, USA) were used as secondary antibodies. Nuclei were visualized with 4΄,6 diamidino-2-phenylindole (DAPI) (Invitrogen). Pecam1 and α-Smooth muscle actin (SMA) positive cells were visualized using a phycoerythrin-conjugated anti-Pecam1 antibody (1:300; Mec13.3, BD Pharmingen, San Diego, CA, USA) or anti-α-SMA antibody (1:300; 1A4, Sigma-Aldrich, St Louis, MO, USA). Stained cells were photographed using an inverted fluorescence microscope (Eclipse TE2000-U; Nikon, Tokyo, Japan) and an AxioCam HRc digital camera system, and the images were processed using AxioVision software (Carl Zeiss, Oberkochen, Germany).
Cell sorting and flow cytometry
After induction of Flk-1 positive cells, cultured cells were harvested and stained with an allophycocyanin (APC)-conjugated anti-Flk1 antibody (AVAS12). Flk-1 positive cells were sorted by auto MACS (Miltenyi Biotec, Bergisch Gladbach, Germany) using anti-APC MicroBeads (Miltenyi Biotec). At day 2 after Flk-1 sorting, cultured cells were harvested and stained with monoclonal antibodies for phycoerythrin-conjugated Pecam1 (Mec13.3, BD Pharmingen) or VE-cadherin (11D4.1, BD Pharmingen), and then subjected to analysis by FACS Aria (Becton Dickinson, Monona, WI, USA).
Data access
The array and sequence data can be accessed through the Gene Expression Omnibus (GEO) under NCBI accession number GSE94829.
Statistical analysis
Data are represented as mean ± standard deviations of at least three independent experiments. Unless otherwise stated, statistical significance between multiple samples was determined by ANOVA to ensure comparable variance, then individual comparisons performed by Bonferroni post-hoc test (KaleidaGraph 4.1 Synergy Software, Inc. Japan). Statistical significance between two groups was determined by standard two-tailed Student's t-test. Comparison of gene ontology sets was assessed using the EASE Score test, a modified Fisher Exact P-value (https://david.ncifcrf.gov/). P < 0.05 was considered significant. An FDR < 1% was used for ChIP-seq experiments to differentially regulated genes.
RESULTS
Identification of highly expressed genes in ECs
First, to characterize the endothelial-specific gene regulation, we identified genes exclusively expressed in ECs. The expression profiles of the 15 EC and 29 non-EC lines studies were downloaded from our RefExA database and the NCBI GEO database (GSE21212). After normalization and preprocessing of the data, we generated a list of 200 genes that were significantly differentially and highly expressed in ECs compared to non-ECs with –log10(q-value) >3 (Supplementary Table S1), as well as a heat map of the expression profiles for the 200 EC-genes (Supplementary Figure S1). Consistent with our previous arrays in ECs, Flk-1, Endomucin (EMCN), VE-cadherin and Pecam1 were selected as EC-specific expressed genes (14). Moreover, ECs and non-ECs were completely separated based on the comparative unbiased gene expression levels even if the cells that were spatially closely aligned to ECs, such as blood cells (lymphocytes) and smooth muscle cells (SMCs), were separately defined as a completely independent cluster (Supplementary Figure S1).
Model of EC differentiation from mouse embryonic stem cells
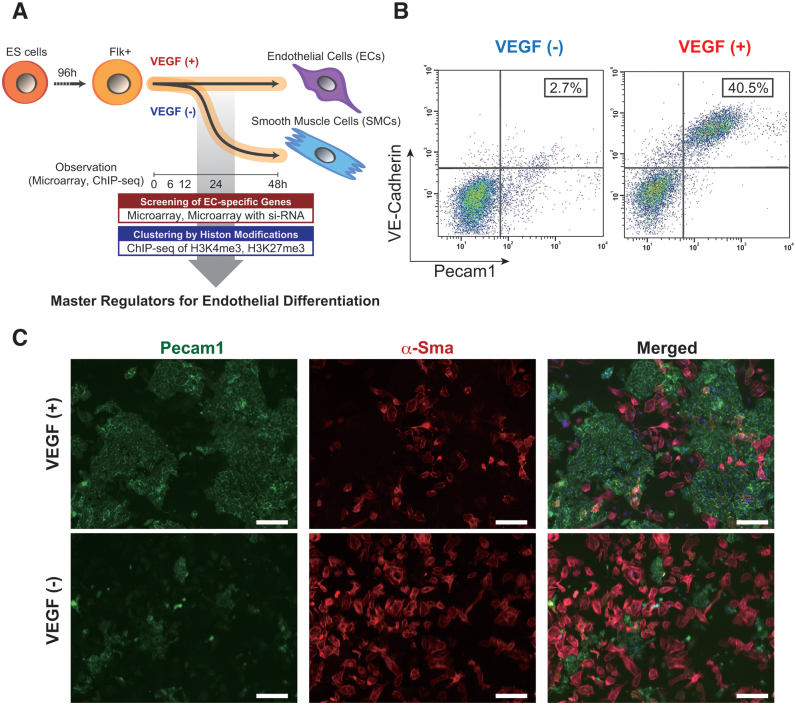
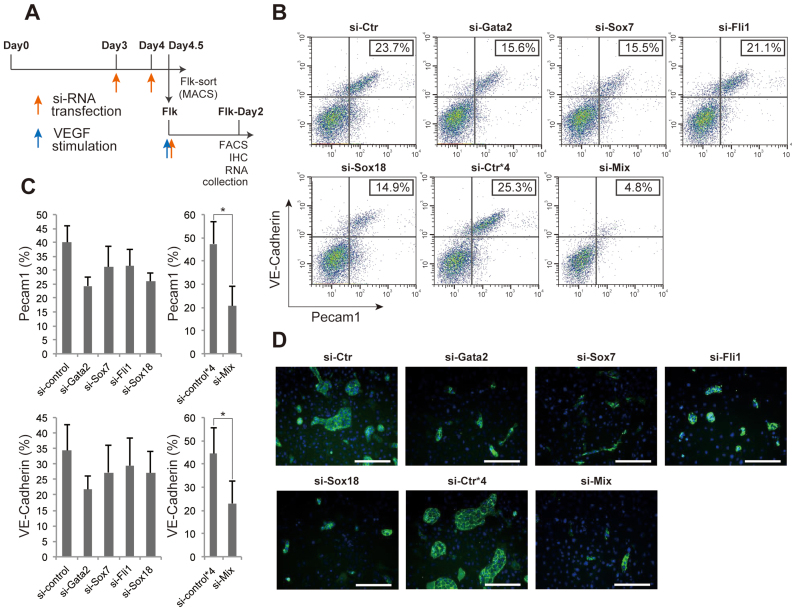
Next, we wished to investigate how comprehensive gene expression patterns change during EC differentiation. To that end, we used our previously reported EC differentiation system (19) for generation of ECs from mouse ES cells. In this system, Flk-1-positive cells differentiated into ECs in the presence of VEGF, whereas cells differentiated into SMCs in the absence of VEGF (Figure 1A). Consistent with previous report, only these two cell types (ECs and SMCs) are produced in this in vitro culture system (7). We confirmed EC differentiation using the selected EC-specific markers, Pecam1 and VE-cadherin, from Supplementary Table S1 (6). In a FACS analysis, 40.5% of the cells were Pecam1 and VE-Cadherin double positive, which was considered as representing the EC fraction, 48 h after VEGF stimulation (Figure 1B). In contrast, only a small proportion (2.7%) differentiated into ECs in the absence of VEGF (Figure 1B). Immunocytochemistry indicated that Pecam1-positive cells were obviously induced under VEGF stimulation. In contrast, most cells differentiated into α-SMA-positive-SMC populations in the absence of VEGF (Figure 1C). These data demonstrate that this differentiation system is suitable for further surveying the comprehensive analysis of gene expression status and epigenetic modifications during EC differentiation.
Figure 1.
In vitro EC differentiation system from ES cells. (A) Schematic view of the strategy to comprehensive data analysis with dynamic EC differentiation. (B) Flow cytometry data representation. X and Y-axis indicate Pecam1 and VE-cadherin positive cell counts, respectively. The rate of both Pecam1 and VE-cadherin positive ECs derived from total Flk-1 cell-derived cells were indicated into the panel (%). Cells were treated in the absence (left) or presence (right panel) of 50 ng/ml VEGF. (C) Immuno-staining data with antibodies to Pecam1 (green, left) and α-SMA (red, middle panel). Merged images were shown in right. Cells were treated with (upper) or without 50 ng/ml VEGF (lower panel). Scale bar: 50 μm.
Genome-wide transcriptome analyses from the EC differentiation
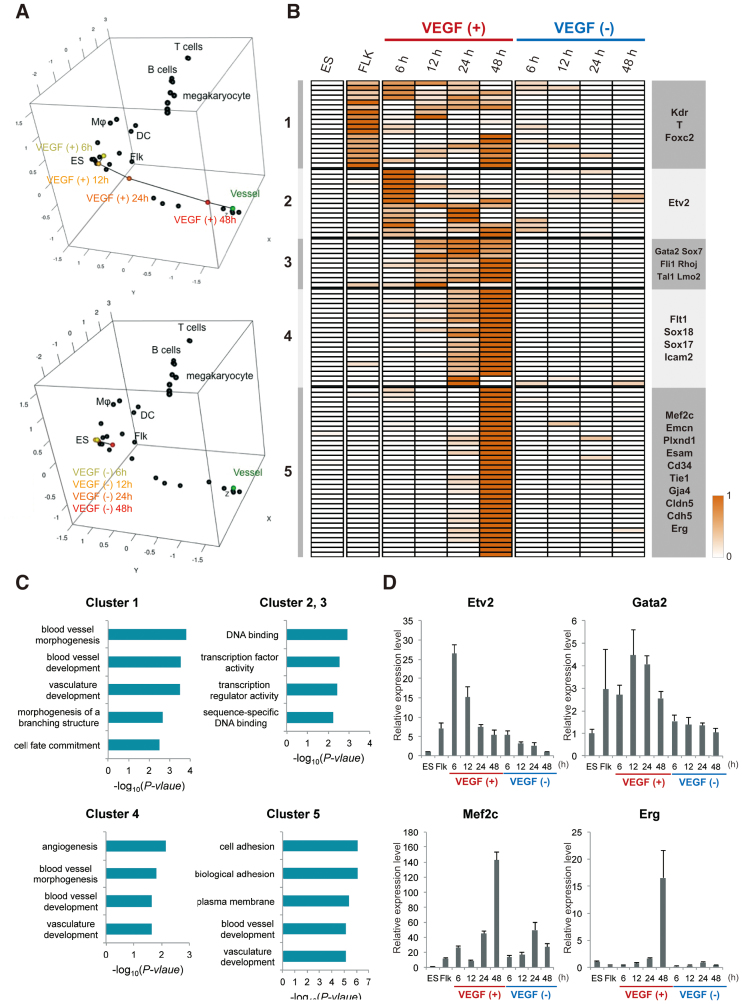
To analyse gene expression profiles, we performed microarray analyses at 0, 6, 12, 24 and 48 h after treatment of the cells in the presence or absence of VEGF. To investigate the transition state of the cells during endothelial development and compare the cells with the 61 mouse normal tissues in the GNF Mouse Gene Atlas (26), we focused on the top 100 EC-specific genes in humans that had been mapped to their 79 mouse orthologues. The expression profiles and heat maps of the 79 EC-specific genes for the cells undergoing EC differentiation and the 61 normal mouse tissues are described in Supplementary Table S2 and Figure S2. We further performed classical MDS based on the expression profiles of the 79 EC-specific genes in mouse to map the cells in the presence or absence of VEGF along with the 61 mouse normal tissues to the 3D space (Figure 2A). As shown in Figure 2A, FLK-1-positive cells gradually differentiated into ECs in the presence of VEGF on the 3D sub-space of MDS.
Figure 2.
Global and dynamic transcriptome analysis on the EC differentiation. (A) Three-dimensional (3D) MDS plots. The cells in the presence or absence of VEGF and the normal mouse tissues were mapped to the 3D space based on the expression profiles of the 79 EC-specific genes by using classical MDS. Black dots represent each cell line. Connected black line indicates gradual differentiation to mature ECs. Upper and lower 3D MDS indicate in the presence or absence of VEGF, respectively. DC and Flk into the box represent dendritic cells and Flk-sorted mesoderm cells, respectively. (B) Heat map representation of the VEGF responsive genes. Genes were classified based on the VEGF treated time points using the algorithm ‘HOPACH’. (C) Functional annotations for each cluster are shown. The P-values of each category analysed from DAVID are shown in the bar graphs. (D) Validation of the representative genes in (B) was conducted by the qPCR. The graph value indicates the expression levels relative to in ES cells. Data are shown as the mean and the standard deviation from triplicates.
Next, we searched the genes upregulated via VEGF during EC differentiation. Microarray data obtained from cells cultured in the following conditions were analysed. ES and Flk-1-positive mesoderm cells were sequentially analysed for 0, 6, 12, 24 and 48 h in the presence or absence of VEGF (Figure 2B). In total, we could identify 97 genes as an EC differentiation related genes, which were then classified into five clusters depending on the time-course oriented expression pattern (Figure 2B and Supplementary Table S3). Moreover, to verify how Pecam1 and Sma positive cells develop in this culture on the time course, after treatment with or without VEGF, we carried out qPCR analysis with Pecam1 and Sma specific primers at the indicated time points. Significant induction of Pecam1 started after 48 h for VEGF treatment. However, Pecam1 was barely expressed in the absence VEGF. In contrast, Sma expression was markedly and gradually induced without VEGF treatment (Supplementary Figure S3). Even though only a small but significant induction of Sma mRNA was observed in the presence of VEGF, the levels were much weaker than in the absence of VEGF (Supplementary Figure S3). These data suggested that VEGF inhibited non-EC lineage differentiation from the Flk-1-positive multi-potent cells.
To further investigate the relationship between VEGF-responsive genes and gene function, we performed functional clustering using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/). This functional heat map illustrated that genes were specifically induced at each time point with clusters 1–5. Cluster 1 included Kdr, Hey1, T and Foxc2, which are involved in function, vascular development and cell fate commitment based on Gene Ontology (GO) terms (Figure 2C). Clusters 2, 3 and 4 include genes expressed during EC differentiation such as Gata2, Sox7, Sox17, Sox18, Fli1 and Etv2. These genes are related to ‘sequence-specific DNA binding’ (clusters 2 and 3, Figure 2C), ‘angiogenesis’ and ‘vasculogenesis’ (cluster 4, Figure 2C) based on GO terms. Finally, cluster 5 included many endothelial specific marker genes including VE-cadherin, Emcn, Esam, Tie1 and Claudin5. These genes were termed as ‘cell adhesion’ and ‘biological adhesion’ genes based on GO terms (Figure 2C). We validated the upregulation of representative genes involved in each cluster by using qPCR with independent samples (Figure 2D). Although some of the EC-inducible genes were expressed at levels lower than those determined by microarray analysis, it was clear that their expression in the cells cultured in the absence of VEGF was much lower than that in the VEGF-stimulated cells. Taken together, these data indicate that lineage-specific transcription factors as shown in clusters 2, 3 and 4 were primarily induced, followed by the upregulation of cell-surface or cell type-specific genes as shown in cluster 5 during the later stage of EC differentiation.
Dynamically changing chromatin states during EC differentiation
The chromatin signature is crucial for transcriptional regulation (13). However, its role during EC differentiation is largely unknown. To gain insight into histone modification patterns during EC differentiation, we performed biologically duplicate ChIP-seq for H3K4me3 and H3K27me3 in Flk-1 sorted cells with or without VEGF treatment for 0, 6 and 48 h. Histone code mapping (using biologic duplicates) in the presence or absence of VEGF gave quite similar patterns in the genome-wide scale (R2 > 0.88) (Supplementary Figure S4). H3K4me3 and H3K27me3 marks on the genomic regions are considered as representing transcriptionally active and silent genes, respectively (27). Across all conditions, H3K4me3 modification patterns with respect to TSS showed narrow peaks around the TSS such as in the β-actin (Actb) and Gapdh loci (Supplementary Figure S5C and D). In contrast, H3K27me3 modification patterns covered whole gene bodies such as the Hox cluster genes located in chromosome 6 (Supplementary Figure S5E).
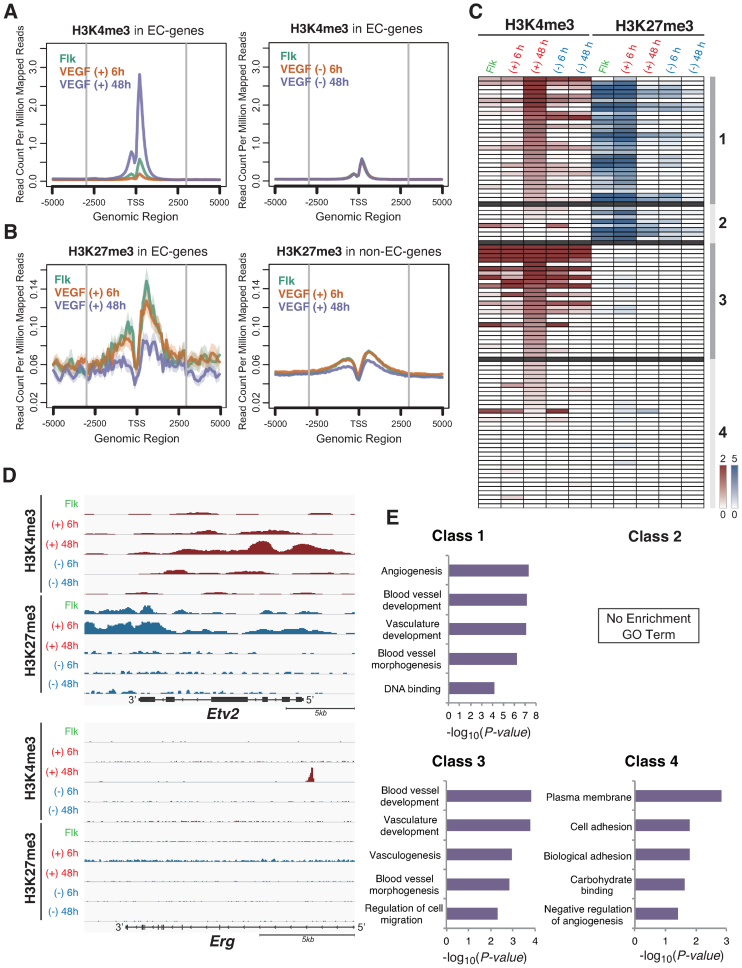
Among the 200 EC-specific genes as selected in Supplementary Figure S1, our data indicated that H3K4me3 marks were only elevated in EC differentiated cells via VEGF, compared with the enrichment levels around promoter regions (Figure 3A). Moreover, H3K27me3 marks were abundantly detected during the Flk-1 sorted mesenchymal stage, but gradually decreased under VEGF stimulation (Figure 3B, left). In contrast, the H3K27me3 enrichment levels in non-EC-specific genes were relatively low and did not change around promoter regions under the VEGF stimulation (Figure 3B, right). These results suggest that our initial global epigenetic profiles indicated during EC differentiation corresponded with the mRNA expression patterns at a genome-wide level.
Figure 3.
Global and dynamic histone code analysis on the EC differentiation. (A) Enrichment of the H3K4me3 modification around the promoters from 200 EC specific genes in Flk-1 positive mesoderm cells for 0 (green line), 6 (yellow line) and 48 h (blue line). VEGF treatment (left) or no treatment (right panel). The average histone code profiles are shown with −5 to +5 kbp from the transcription start sites (TSS). (B) Enrichment of the H3K27me3 modification around the promoters from 200 EC specific genes (left) and non-EC specific genes (right panel) for 0 (green line), 6 (yellow line) and 48 h (blue line) VEGF treatment. The average histone code profiles are shown with −5 to +5 kbp from the TSS. (C) Heat map representation of the VEGF responsive genes. Genes were classified based on the histone modification patterns using the algorithm ‘HOPACH’. Colour intensity: red (H3K4me3 mark) and blue (H3K27me3 mark) indicate higher and white indicate lower, shown relative to median. (D) UCSC mm9 genome browser view around the Etv2 and Erg genes. ChIP-seq tags were calculated by MACS and shown in Integrative Genomics Viewer (IGV). H3K4me3 peaks were shown in red, and H3K27me3 were in blue. (E) Functional annotations for each class are shown. The enrichment scores of each group from DAVID are shown in the bar graphs.
Next, we examined how chromatin patterns were dynamically changed during the EC differentiation steps in VEGF-stimulated cells. Thus, described four classes, based on the modification of H3K4me3 and H3K27me3 levels at 6 and 48 h after the VEGF stimulation (Figure 3C and Supplementary Table S4). Notably, we found that genes with high levels of H3K4me3 at 48 h after the VEGF stimulation presented two separate H3K27me3 modification patterns during the stages treated with VEGF for 0 and 6 h. Genes in Class 1 included active transcription factors such as Gata2 and Fli1 that had high levels of H3K27me3 in the stages after 0 and 6 h of VEGF treatment, indicating that these genes were repressed during the undifferentiated stage but that histone modification switching occurred with VEGF treatment. Moreover, the EC pioneer factor, Etv2, presented unique H3K27me3 modification patterns in the mesenchymal cell stage (Figure 3D). The H3K27me3 marks that were highly enriched on the Etv2 3΄-end region quickly disappeared after the EC differentiation stimuli. Instead of the repressive mark, transcriptionally permissive H3K4me3 levels gradually increased at the Etv2 5΄-proximal promoter region (Figure 3D and Supplementary Figure S5A). In contrast, the class 3 genes including Cdh5 (VE-cadherin), Egfl7, Rhoj and Aplnr, showed a lack of H3K27me3 enrichment of the regulatory regions but exhibited H3K4me3 marks after the EC fate fixation stage at 48 h after the VEGF treatment, which was considered to regulate their constitutive expression in ECs (Supplementary Figure S5B, F, G and H).
Subsequently, to examine the relationship between histone modification pattern and gene function, we performed functional annotation using DAVID. As shown in Figure 3E, class 1 genes included Gata2, Tal1, Fli1, Lmo2, Foxc2, Sox7, Sox17 and Sox18 (the histone codes are shown in Supplementary Figure S5I–O), which are transcription factors and associated with angiogenesis, blood vessel development and vasculature development. Taken together, these results suggest that EC differentiation-correlated transcriptional regulators acquire specific histone modification patterns and that such epigenetic changes direct the accurate EC differentiation via a transcription factor expression cascade.
Identification of the transcriptional network for vascular development
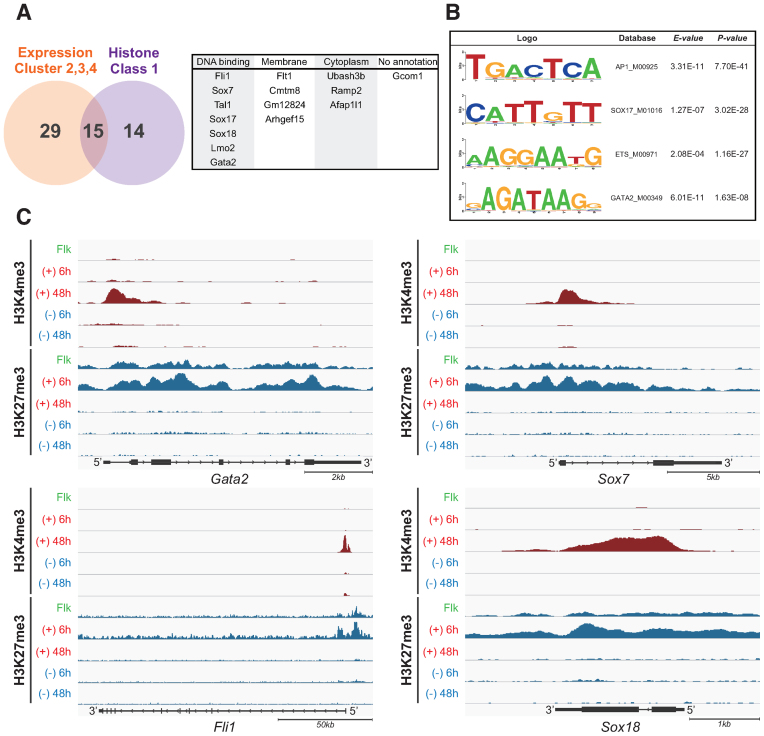
We next sought to determine the critical transcription factors for EC differentiation from the combination of global expression data and the histone dynamics. From the microarray data of VEGF-treated time points, we selected 44 potential EC regulators, which were in clusters 2, 3 and 4 in Figure 2B (a detailed gene list is shown in Supplementary Table S3). In comparison, dynamic histone modification analysis suggested that 29 genes displayed a histone switching pattern from H3K27me3 to H3K4me3 (Histone class 1; detailed gene lists are shown in Supplementary Table S4); these were also considered as EC regulators. Thus, we merged both clusters, resulting in the identification of 15 overlapping genes (Figure 4A). Among these, half were DNA binding transcription factors, four genes encoded membrane proteins and three genes were classified as encoding cytoplasm expressed proteins. However, the remaining gene, gcom1, was annotated as being of unknown function (Figure 4A). Considering the relationship between chromatin dynamics and expression profiling, we considered that the listed seven transcription factors likely included the critical determinant factors for EC definition.
Figure 4.
Identification of the master transcription factor for EC differentiation. (A) Venn diagram depicting the overlap of expression cluster 2,3 and 4 in Figure 2B and histone class 1 in Figure 3C. The numbers indicate the overlapped or unique genes count. The common 15 genes are classified by GO term (right panel). (B) Determination of enriched transcription factor motifs calculated from the enhancer regions of HUVECs database. The MODIC method (32) was used for identification of enriched sequences and the size of the character reflects the degree of enrichment. E-value and P-value indicate the probability that de novo enriched sequences obtained from FAIRE-seq are matched to the shown ‘Web logo’ and known consensus motifs by chance, respectively. (C) UCSC mm9 genome browser view around Gata2, Fli1, Sox7 and Sox18 genes. ChIP-seq tags were calculated by MACS and are shown in IGV. H3K4me3 peaks were shown in red, and H3K27me3 were in blue.
Recently, many studies have demonstrated that enhancer regions are important to determine cell fate and specificity, in addition to promoter regions (28–30); therefore, we considered using motif analysis in the enhancer regions. We believe that not only gene expression and histone dynamics data but also enhancer analysis would be useful to determine the master transcription factors in EC differentiation. However, to date no information is available regarding defined ECs in mouse and it is very difficult to obtain the necessary enhancer information from ES cell-derived ECs owing to the limited available cell volume in this differentiation system. Therefore, we chose to utilize HUVECs as a well-defined primary culture mature EC system and carried out formaldehyde-assisted isolation of regulatory elements (FAIRE)-seq to determine open chromatin regions (31) and obtain de novo motif identification analysis within the enhancer data from ECs. As shown in Figure 4B, the MODIC method mediated (32) predicted sequence motifs from FAIRE-seq that corresponded to binding motifs were for AP-1, GATA, ETS and the SOX family. Notably, our chosen list in Figure 4A corresponded to each family; i.e. GATA2, FLI1 and SOX 7/17/18 transcription factors. Moreover, all the identified transcription factors had been revealed as exhibiting ‘histone switch’ from H3K27me3 to H3K4me3 during EC differentiation (Figure 4C and Supplementary Figure S5J). Taken together, the results of these genomic and epigenomic approaches suggested that Gata2, Fli1 and Sox 7/17/18 are candidates as master regulators for EC differentiation.
Next, to verity the importance of these transcription factors at the functional level, in addition to the expression level and histone code profiling, we performed knockdown experiments using each siRNA following the indicated time points during EC differentiation (Figure 5A). In these knockdown experiments, we confirmed the knockdown efficiency by qPCR at day 2 after the VEGF stimulation (Supplementary Figure S6A). Compared with the si-control, only the siRNA against Sox17 failed to show any defects in differentiation to ECs, as determined by the EC-specific expression of Pecam1 and VE-cadherin (Supplementary Figure S6B and C). However, among the reminder of the gene lists, siRNA against either Gata2, Sox7, Fli1 or Sox18 slightly blocked EC differentiation whereas it was almost abrogated by the combined siRNAs against all four factors (Figure 5B and C). Immunocytochemistry with antibodies against Pecam1 also showed that each siRNA and the combined siRNAs treatment significantly and completely arrested the EC differentiation, respectively (Figure 5D). In addition, as a negative control for Ets and the cluster shown in Figure 4A, we utilized siRNAs against Ets1 and Ubash3b, respectively. These siRNA treatments failed to interfere with EC differentiation following VEGF treatment (Supplementary Figure S6D). Together, these data demonstrate that four transcription factors with Gata2, Fli1, Sox 7 and Sox18 are indispensable for mature EC fate definition. Even if the expression of one of these transcription factors is knocked down, terminal EC differentiation was significantly impaired.
Figure 5.
Functional validation of the master transcription factor for EC differentiation. (A) Time points for siRNA transfection during EC differentiation. During the first 4.5 days of cells culture, siRNA was twice transfected at the days 3 and 4, and then sorted Flk-1 positive cells were subjected to siRNA transfection, again. Red and blue arrows represent the timing of siRNA transfection and VEGF stimulation, respectively. (B) Flow cytometry data representation. X and Y-axis indicate Pecam1 and VE-cadherin positive cell counts, respectively. The rate of both Pecam1 and VE-cadherin positive ECs derived from total Flk-1 cell-derived cells were indicated into the panel (%). (C) Quantitative evaluation of the effect of each or combined siRNA transfection. The results show the mean and standard deviations of Pecam1 (upper) or VE-cadherin (lower) positive EC induction rate (%) from Flk-1 cells by FACS, obtained triplicate. n = 3; *P < 0.05 versus si-control. (D) Immuno-staining data with antibody to Pecam1 (green) and DAPI (blue). Si-Ctr*4 indicates that 4-fold higher amount of si-control was added to the cells compared to si-Ctr. Si-Mix means that si-Gata2, si-Sox7, si-Fli1 and si-Sox18 were added to the cells. Scale bar: 50 μm.
The master transcription factors contribute to EC-lineage commitment via separate downstream target genes
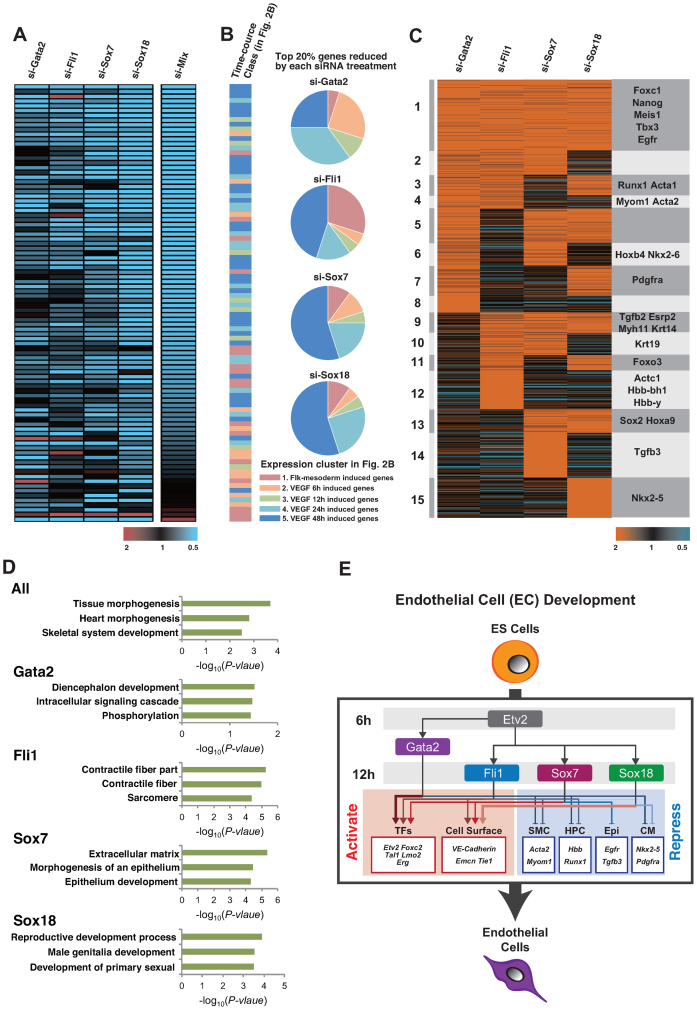
Because Gata2, Fli1, Sox7 and Sox18 are transcription factors that do not have well-recognized downstream target genes for the EC fate determination period, we performed a microarray analysis following each siRNA transfection during the EC differentiation step. We first identified a total of 97 genes that were induced at the EC differentiation step and showed reduction in the presence of siRNAs against the combined four master transcription factors. All selected genes were sorted in ascending order of their knockdown pattern (Figure 6A and Supplementary Table S5). Notably, each transcription factor-regulated downstream gene set did not completely overlap. For example, Gata2 knockdown reduced the expression of Etv2, Tal1 and Sox18, which are included in cluster 2, 3 and 4 gene expression profiles in Figure 2B. As shown in the pie-chart in Figure 6B, the gene sets categorized with orange, green and blue colours were predominantly reduced by si-Gata2 treatment. In contrast, Sox18 knockdown led to reduced expression of the EC marker genes Cdh5, Tie1 and Erg, which are included in the cluster 5 gene expression profile in Figure 2B. This demonstrated the lowest population of gene sets with orange and green colours, whereas the highest population included those having light blue and blue colours in the pie-chart (Figure 6B and Supplementary Table S6). In addition, si-Fli1 reduced the expression of Arhgap18, Foxc2 and Emcn, whereas si-Sox7 reduced the expression of a different gene set with Cdh5, Lmo2 and Tie1. Collectively, these findings suggest that among the four transcription factors, Gata2 and Sox18 upregulated genes in the early- and late-phase of EC differentiation, respectively, and Fli1 and Sox7 upregulated separate genes during EC differentiation (Supplementary Figure S7 and Table S7).
Figure 6.
Regulatory gene circuit of the master transcription factors for EC differentiation. (A) Heat map representation of downregulated gene groups by siRNA treatment. The colour intensity indicates the fold expression level. Red-higher and Blue-lower, shown relative to the median in black (left panel). Genes were classified based on expression patterns using the algorithm ‘HOPACH’. (B) Relationship between the master transcription factor-regulated genes and the induced time points via VEGF treatment in Figure 2B are shown. Colours in the bar graph and pie-chart are shown corresponding to the clusters in Figure 2B. Pink, orange, green, light blue and blue represent clusters 1, 2, 3, 4 and 5, respectively. (C) Heat map representation of upregulated genes grouped by the siRNA treatment. The colour intensity indicates the expression level; Red-higher and blue-lower, shown in relation to the median in black. Genes are classified based on expression patterns using the algorithm ‘HOPACH’. Representative genes in each cluster are displayed in the right column. (D) Functional annotations for each class are shown. The enrichment scores of each group from DAVID are shown in the bar graphs. All means cluster 1 in C. Gata2 means cluster 4, 6, 7 and 8, in C. Fli1; cluster 4, 10, 11 and 12, Sox7; cluster 6, 10, 13 and 14, and Sox18; means 7, 11, 13 and 15, in C. (E) Schematic representation of the genetic regulatory network linking four transcription factors and EC differentiation. TFs; transcription factors, SMC; smooth muscle cells, HPC; hematopoietic stem cells, Epi; epithelial cells, CM; cardiomyocytes. Representative activated (red) or repressed (blue) genes by each transcription factor are shown in the box.
Subsequently, we considered the upregulated gene set obtained by the silencing of these four transcription factors. Notably, the selected heat map revealed that the genes commonly induced via siRNAs against the four master regulators (cluster 1) was not dominant; rather, each transcription factor revealed a unique downstream gene set (clusters 2–15) (Figure 6C). Moreover, these induced gene groups involved non-EC expressed genes (Figure 6C and Supplementary Table S8). For example, SMC marker genes involving smooth muscle actin α, α-SMA (Acta2) and Myom1 (cluster 4) were upregulated in si-Gata2 and si-Fli1 transfected cells. Haemoglobin β (Hbb) in the si-Fli1-unique cluster 12 and Runx1 in cluster 3 belong to the haematopoietic cell lineage. Nkx2.5 is a cardiomyocyte marker gene, which was in the si-Sox18-unique cluster 15. In addition, each EC differentiation master transcription factor was divided according to functional cluster (Figure 6D). Representatively, si-Fli1 transfection upregulated Krt19 (cluster10), Actc1(cluster 12) and Acta2 (cluster 4), which are classified as contractile fibre or sarcomere genes by DAVID analysis (Figure 6D). Taken together, these findings suggest that the identified EC master transcriptional factors not only upregulate EC-specifically expressed genes but also downregulate expression of the other lineage commitment genes (Figure 6E).
DISCUSSION
In this study, we characterized dynamic chromatin state transitions during murine EC differentiation from ES cells to provide insights into the coordinated regulation of histone modifications and mRNA expression. In ES cells, several studies have reported that important developmental genes are completely modified with both H3K4me3 and H3K27me3; these are termed ‘bivalent’ (33,34). However, our histone code dataset obtained with two separate time points after the Flk-1 positive mesoderm stage clearly indicated the increase of H3K4me3 marks following the reduction of H3K27me3 marks, rather than the bivalent switch. Such ‘histone switching’ was suggested to be extremely crucial for EC differentiation. We believe that our study provides the first global histone modification and gene expression profile database on ECs differentiated from common progenitors, though the complete epigenome databases were reported from other differentiation lineages (e.g. cardiomyocytes) (35).
H3K27 acetylation marks at genomic loci distal to the promoters provide a means to identify candidate enhancer motifs (36). Recent comprehensive analyses for genome-wide mapping of enhancer features resulted in a number of striking discoveries that enhancer activation is crucial for specific lineage differentiation (29). We determined that AP1, SOX17/18, ETS and GATA2 motifs are highly enriched using FAIRE-seq data in HUVECs compared with random promoters (Figure 5B). In addition, based on genetic and epigenetic data on EC differentiation, Fli1, Sox7, Tal1, Sox17, Sox18, Lmo2 and Gata2 are predicted to be indispensable transcription factors. Additionally, the target genes of each transcription factor differed according to our microarray data. From this analysis demonstrated that we could apply this integration method between histone switching genes and expression profiles to identify master regulators in various situations, including cell lineage differentiation.
Flk-1-positive mesoderm cells are known to give rise to not only ECs and blood cells, but also to other mesoderm lineage cells including vascular smooth muscle cells, cardiomyocytes and skeletal muscle cells (37). We found that the EC fate determinant transcription factors Gata2, Sox7, Sox18 and Fli1 functioned not only to differentiate to ECs but also to block differentiation to another lineage. Our comprehensive analyses with siRNA against the critical transcription factors indicate that Gata2 and Fli1 cooperatively inhibited the expression of blood cell or vascular smooth muscle cell marker genes. Similarly, Sox18 and Gata2 blocked cardiomyocyte commitment and Sox7 represses epithelial-mesenchymal transition-related genes. Moreover, all of these transcription factors could be inhibitory toward skeletal muscle cell differentiation. It is also interesting to note that Nanog and Sox2, as pluripotency markers (38), were identified in the induced gene group through the knockdown of these EC fate determination transcription factors (Supplementary Table S7). These results suggest that master transcription factors for specific lineages repress not only differentiation to closely aligned lineage cells but also repress potential commitment to far distant lineages through reprogramming.
Several studies have indicated that Etv2 is essential for development of the endothelial lineage in mice. Etv2 expression is detected in the earliest vessels (E8.25–E9.5) in embryos, followed by a sharp reduction beyond E11.5 (39,40). Consistent these in vivo findings, Etv2 was also transiently expressed and declined quickly in our in vitro experimental method (Figure 2B), which may suggest that a similar Etv2 activation system was functioning during the EC differentiation stage. Etv2 is widely known to be crucial for early vascular development; however, little is known regarding its downstream genes. Recently, the cooperative interaction of Etv2 and Gata2 was shown to be necessary for endothelial development using a co-immunoprecipitation assay (41). As shown in Figure 6A, si-Gata2 affected cluster 2 and 3 genes, including Etv2, as noted in Figure 2B, which are early-activated genes. In addition, we have previously reported that si-Etv2-reduced Gata2 mRNA expression (42). These results thus suggest that Gata2 regulates early-activated genes in cooperation with Etv2.
In addition to Etv2, it is generally accepted that transcription factors from the Ets family such as Ets1, Ets2, Fli1 and Erg are important for EC differentiation and maintenance (3,43,44). Furthermore, in the direct conversion from amniotic cells to functional ECs, enforced expression of Fli1 and ERG after transient Etv2 expression contributes to provide fully differentiated ECs (11). Consistent with the previous report of direct conversion, our findings present the first evidence for the expression of the ‘Ets switch’ from Etv2 to Fli1 and Erg (Figure 2B). Regarding histone modifications of Ets family protein genes, Etv2 and Fli1 presented repressive marks in the Flk-1 sorted cells and switched to active histone marks during differentiation. In sharp contrast, the gene for another Ets family protein, Erg, carried no repressive marks and presented the H3K4me3 mark after the EC fate determination, at 48 h after the VEGF stimulation (Figure 3D). These results suggest that during EC differentiation, the Etv2, Fli1 and Erg expression relay is epigenetically controlled and rigidly expressed following this order. Among the ETS family proteins, the detailed molecular mechanisms underlying their binding affinity on the DNA and the functional differences are largely unknown, although the Ets binding consensus sequence is observed in more than 200 genes involved in angiogenesis and haematopoiesis (45). ETS binding profiles therefore warrant further investigation.
Among 20 Sox transcription factors in mice, SoxF factors are reported to have functional and redundant roles in cardiovascular development in vivo (18,46). Our comprehensive expression and histone modification data clearly identified Sox7, 17 and 18 as candidates for master regulators of EC differentiation (Figure 4A). Consistent with this, Sox7 and 18 knockdown demonstrated abrogation of EC differentiation; however, Sox17 knockdown had little effect on EC fate definition in our in vitro EC differentiation model (Supplementary Figure S5B and C). The data that Sox17 may be independent of EC fate determination appears to differ from a previous report demonstrating that all SoxF family members mediate cardiovascular development in mice (4,18). Our data reflect the direct VEGF-mediated EC differentiation from ES cells. In contrast, cardiovascular development in vivo would reflect the complex final outcome, with spatially and temporally different microenvironments. Moreover, the SoxF family is believed to have redundant roles, especially between Sox17 and 18 (46). However, as the target genes regulated by Sox7 or 18 were different according to our microarray data (Figure 6A–C), the SoxF family members, or at least Sox7 and Sox17/18, might have different roles in our model. Further precise molecular analyses with each null mutation mouse and downstream target identifications in cells would be needed to clarify the distinct SoxF expression profiles and functions during EC development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Shogo Yamamoto and Akashi Izumi (RCAST, The University of Tokyo) for technical assistance with ChIP-seq and microarrays, and Dr Hiroshi Kimura (Tokyo Institute of Technology) for kindly providing histone modification antibodies. We also thank Editage (www.editage.jp) for English language editing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Young Scientists (B), Japan Society for the Promotion of Science [24710227 to Y.K.]; NOVARTIS Foundation (Japan) for the Promotion of Science (to Y.K.); Uehara Memorial Foundation (to Y.K.); Cooperative Research Program of the Institute for Frontier Medical Sciences, Kyoto University, Japan (in part) (to Y.K.); Leading-edge Research Promotion Fund from the Japan Society for the Promotion of Science [LS038 to T. M.]; Life Science promotion Fund (Daiichi-Sankyo) (to T.M.). Funding for open access charge: Grant-in-Aid, Japan Society for the Promotion of Science [15H01348 to T.M.]
Conflict of interest statement. None declared.
REFERENCES
- 1. Cermenati S., Moleri S., Cimbro S., Corti P., Del Giacco L., Amodeo R., Dejana E., Koopman P., Cotelli F., Beltrame M.. Sox18 and Sox7 play redundant roles in vascular development. Blood. 2008; 111:2657–2666. [DOI] [PubMed] [Google Scholar]
- 2. Francois M., Caprini A., Hosking B., Orsenigo F., Wilhelm D., Browne C., Paavonen K., Karnezis T., Shayan R., Downes M. et al. . Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008; 456:643–647. [DOI] [PubMed] [Google Scholar]
- 3. Liu F., Walmsley M., Rodaway A., Patient R.. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr. Biol. 2008; 18:1234–1240. [DOI] [PubMed] [Google Scholar]
- 4. De Val S., Black B.L.. Transcriptional control of endothelial cell development. Dev. Cell. 2009; 16:180–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meadows S.M., Salanga M.C., Krieg P.A.. Kruppel-like factor 2 cooperates with the ETS family protein ERG to activate Flk1 expression during vascular development. Development. 2009; 136:1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Risau W., Flamme I.. Vasculogenesis. Annu. Rev. Cell Dev. Biol. 1995; 11:73–91. [DOI] [PubMed] [Google Scholar]
- 7. Yamamizu K., Kawasaki K., Katayama S., Watabe T., Yamashita J.K.. Enhancement of vascular progenitor potential by protein kinase A through dual induction of Flk-1 and Neuropilin-1. Blood. 2009; 114:3707–3716. [DOI] [PubMed] [Google Scholar]
- 8. Takeuchi J.K., Bruneau B.G.. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009; 459:708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sekiya S., Suzuki A.. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011; 475:390–393. [DOI] [PubMed] [Google Scholar]
- 10. Takahashi K., Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 11. Ginsberg M., James D., Ding B.S., Nolan D., Geng F., Butler J.M., Schachterle W., Pulijaal V.R., Mathew S., Chasen S.T. et al. . Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFbeta suppression. Cell. 2012; 151:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morita R., Suzuki M., Kasahara H., Shimizu N., Shichita T., Sekiya T., Kimura A., Sasaki K., Yasukawa H., Yoshimura A.. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanki Y., Kohro T., Jiang S., Tsutsumi S., Mimura I., Suehiro J., Wada Y., Ohta Y., Ihara S., Iwanari H. et al. . Epigenetically coordinated GATA2 binding is necessary for endothelium-specific endomucin expression. EMBO J. 2011; 30:2582–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bowles J., Schepers G., Koopman P.. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000; 227:239–255. [DOI] [PubMed] [Google Scholar]
- 16. Niimi T., Hayashi Y., Futaki S., Sekiguchi K.. SOX7 and SOX17 regulate the parietal endoderm-specific enhancer activity of mouse laminin alpha1 gene. J. Biol. Chem. 2004; 279:38055–38061. [DOI] [PubMed] [Google Scholar]
- 17. Hosking B.M., Muscat G.E., Koopman P.A., Dowhan D.H., Dunn T.L.. Trans-activation and DNA-binding properties of the transcription factor, Sox-18. Nucleic Acids Res. 1995; 23:2626–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakamoto Y., Hara K., Kanai-Azuma M., Matsui T., Miura Y., Tsunekawa N., Kurohmaru M., Saijoh Y., Koopman P., Kanai Y.. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem. Biophys. Res. Commun. 2007; 360:539–544. [DOI] [PubMed] [Google Scholar]
- 19. Yamashita J., Itoh H., Hirashima M., Ogawa M., Nishikawa S., Yurugi T., Naito M., Nakao K., Nishikawa S.. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000; 408:92–96. [DOI] [PubMed] [Google Scholar]
- 20. Yamamizu K., Fujihara M., Tachibana M., Katayama S., Takahashi A., Hara E., Imai H., Shinkai Y., Yamashita J.K.. Protein kinase A determines timing of early differentiation through epigenetic regulation with G9a. Cell Stem Cell. 2012; 10:759–770. [DOI] [PubMed] [Google Scholar]
- 21. Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P.. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003; 4:249–264. [DOI] [PubMed] [Google Scholar]
- 22. Johnson W.E., Li C., Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8:118–127. [DOI] [PubMed] [Google Scholar]
- 23. Tozawa H., Kanki Y., Suehiro J., Tsutsumi S., Kohro T., Wada Y., Aburatani H., Aird W.C., Kodama T., Minami T.. Genome-wide approaches reveal functional interleukin-4-inducible STAT6 binding to the vascular cell adhesion molecule 1 promoter. Mol. Cell. Biol. 2011; 31:2196–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N.. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008; 33:61–73. [DOI] [PubMed] [Google Scholar]
- 25. Hayashi-Takanaka Y., Yamagata K., Wakayama T., Stasevich T.J., Kainuma T., Tsurimoto T., Tachibana M., Shinkai Y., Kurumizaka H., Nozaki N. et al. . Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011; 39:6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lattin J.E., Schroder K., Su A.I., Walker J.R., Zhang J., Wiltshire T., Saijo K., Glass C.K., Hume D.A., Kellie S. et al. . Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008; 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voigt P., Tee W.W., Reinberg D.. A double take on bivalent promoters. Genes Dev. 2013; 27:1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adam R.C., Yang H., Rockowitz S., Larsen S.B., Nikolova M., Oristian D.S., Polak L., Kadaja M., Asare A., Zheng D. et al. . Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015; 521:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saint-Andre V., Federation A.J., Lin C.Y., Abraham B.J., Reddy J., Lee T.I., Bradner J.E., Young R.A.. Models of human core transcriptional regulatory circuitries. Genome Res. 2016; 26:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D.. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007; 17:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakaki R., Kang J., Tateno M.. A novel ab initio identification system of transcriptional regulation motifs in genome DNA sequences based on direct comparison scheme of signal/noise distributions. Nucleic Acids Res. 2012; 40:8835–8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al. . Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. . A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 35. Yamamizu K., Yamashita J.K.. Roles of cyclic adenosine monophosphate signaling in endothelial cell differentiation and arterial-venous specification during vascular development. Circ. J. 2011; 75:253–260. [DOI] [PubMed] [Google Scholar]
- 36. Zhang B., Day D.S., Ho J.W., Song L., Cao J., Christodoulou D., Seidman J.G., Crawford G.E., Park P.J., Pu W.T.. A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Res. 2013; 23:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park C., Kim T.M., Malik A.B.. Transcriptional regulation of endothelial cell and vascular development. Circ. Res. 2013; 112:1380–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okita K., Ichisaka T., Yamanaka S.. Generation of germline-competent induced pluripotent stem cells. Nature. 2007; 448:313–317. [DOI] [PubMed] [Google Scholar]
- 39. Kataoka H., Hayashi M., Nakagawa R., Tanaka Y., Izumi N., Nishikawa S., Jakt M.L., Tarui H., Nishikawa S.. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. 2011; 118:6975–6986. [DOI] [PubMed] [Google Scholar]
- 40. Rasmussen T.L., Kweon J., Diekmann M.A., Belema-Bedada F., Song Q., Bowlin K., Shi X., Ferdous A., Li T., Kyba M. et al. . ER71 directs mesodermal fate decisions during embryogenesis. Development. 2011; 138:4801–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shi X., Richard J., Zirbes K.M., Gong W., Lin G., Kyba M., Thomson J.A., Koyano-Nakagawa N., Garry D.J.. Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev Biol. 2014; 389:208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamamizu K., Matsunaga T., Katayama S., Kataoka H., Takayama N., Eto K., Nishikawa S., Yamashita J.K.. PKA/CREB signaling triggers initiation of endothelial and hematopoietic cell differentiation via Etv2 induction. Stem Cells. 2012; 30:687–696. [DOI] [PubMed] [Google Scholar]
- 43. Wei G., Srinivasan R., Cantemir-Stone C.Z., Sharma S.M., Santhanam R., Weinstein M., Muthusamy N., Man A.K., Oshima R.G., Leone G. et al. . Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009; 114:1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Birdsey G.M., Shah A.V., Dufton N., Reynolds L.E., Osuna Almagro L., Yang Y., Aspalter I.M., Khan S.T., Mason J.C., Dejana E. et al. . The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/beta-catenin signaling. Dev. Cell. 2015; 32:82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hsu T., Trojanowska M., Watson D.K.. Ets proteins in biological control and cancer. J. Cell Biochem. 2004; 91:896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsui T., Kanai-Azuma M., Hara K., Matoba S., Hiramatsu R., Kawakami H., Kurohmaru M., Koopman P., Kanai Y.. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J. Cell Sci. 2006; 119:3513–3526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.