Abstract
Background:
Viruses and atypical pathogens can cause significant respiratory illness in immunocompromised patients. Multiplex polymerase chain reaction (MPCR) has improved the diagnostic yield of pathogens, and it is easier to identify the co-infections also. The present study was done to evaluate the performance of MPCR on bronchoalveolar lavage (BAL) samples in immunocompromised patients.
Methods:
Atotal of 177 BAL specimens collected over a 19 months period from immunocompromised patients with respiratory illness were analyzed with the MPCR and aerobic culture. Patients were divided into four according to the pathogens. Category V (only viral), Category NV (nonviral, i.e., bacteria and atypical), Category M (mixed, i.e., both viral and nonviral pathogen), and Category UK (unknown etiology).
Results:
MPCR identified the causative pathogen in 59.3% of patients while culture could identify only in 37.8% of patients. Most frequent etiological agent was Klebsiella pneumoniae (32%), followed by cytomegalovirus (21%), and Pneumocystis jirovecii (10%). Numbers of patients in each category were Category V (9.6%), Category NV (43.5%), Category M (19.8%), and Category UK (27.1%). Mortality was significantly higher in patients of Category M having mixed infections.
Conclusion:
MPCR is highly sensitive and rapid tool which can be considered in the routine diagnostic algorithm of respiratory illness in immunocompromised patients.
Keywords: Cytomegalovirus, immunocompromised patients, multiplex polymerase chain reaction, Pneumocystis jirovecii
INTRODUCTION
The number of immunocompromised patients has increased during the past few decades, reflecting increased use of immunosuppressive medications, and improved diagnostic techniques. The important causes of immunosuppression are solid organ transplantation, hematopoietic stem cells transplantation, cancer therapy, HIV infection, autoimmune diseases, primary immunodeficiencies, long-term steroid use, diabetes mellitus, and chronic debilitating illness.[1,2] The immunocompromised patients are more prone to various infections as compared to the immunocompetent ones. The organisms which are of little native virulence in the immunocompetent, can cause life-threatening complications in immunocompromised. Out of all, the infectious complications, respiratory complications are the most common because lungs can be a portal of entry for wide range of microorganisms. Respiratory infections comprise about 75% of pulmonary complications and are a major cause of morbidity and mortality in these patients.[3,4] The number of pathogens capable of causing the pulmonary infections is increasing. Earlier, only the bacterial pathogens were considered significant, but now, the atypical pathogens and viruses are increasingly being documented.[5] Pathogens of greatest concern in immunocompromised are Pneumocystis jirovecii and cytomegalovirus (CMV).[6] P. jirovecii is primarily associated with HIV infection but clusters of cases have also been seen in solid organ transplant recipients and patients with hematological malignancies.[7,8,9] Coinfections are common in the immunocompromised. CMV pneumonitis often develops in association with other causes of pneumonia, especially the P. jirovecii. Subsequent secondary bacterial pneumonia can also develop.[10] Besides these two, other important viral causes of respiratory infections in immunocompromised are respiratory syncytial virus (RSV), influenza virus (Flu), adenovirus (AdV), human metapneumovirus (hMPV), parainfluenza virus, and human rhinovirus.[11,12,13] Prompt accurate diagnosis of pulmonary infections by these pathogens is crucial not only because of the high morbidity and mortality associated with them but also to avoid unnecessary use of antibiotics. Respiratory illness by viruses remains under diagnosed, the main reason being the nonspecific clinical presentation and difficult diagnosis. Now in recent years, with the introduction of molecular-based tests, diagnostic yield of viruses have improved up to 35%.[14] These nucleic acid amplification tests are highly sensitive and rapid. With the multiplex technology, it is easier to identify the coinfections also. Multiplex polymerase chain reaction (MPCR) may not only facilitate the optimal clinical management of immunocompromised patients but also prevent their transmission.[15] The present study was done to evaluate the performance of respiratory MPCR on bronchoalveolar lavage (BAL) samples in immunocompromised patients admitted with respiratory illness.
METHODS
Patients
We retrospectively studied immunocompromised patients with respiratory illness admitted to our hospital from January 2014 to July 2015. Immunocompromised status was defined by the presence of malignancy, transplantation (solid organ and bone marrow transplantation), autoimmune disease, long-term steroid use, chronic debilitating illness, primary immunodeficiency, and immunosuppressive therapy. For the purpose of the present study, only patients in whom a BAL sample was available for respiratory MPCR and culture were included. A total of 177 patients were considered for evaluation.
Demographic and clinical data
In all cases, the following variables were recorded: age, gender, clinical area, any underlying lung disease, clinical symptoms, days of symptoms, predisposing immunocompromised condition, complete blood count, C-reactive protein, and length of hospital stay and final outcome.
Microbiological methods
Respiratory MPCR and aerobic culture were done on all the BAL samples. PCR was done with the commercial multiplex FTD Respiratory pathogens 33 Kit. Kit was used according to manufacturer's instructions (Fast Track Diagnostics, Junglinster, Luxembourg) following total nucleic acid extraction performed by QIAsymphony (Qiagen, Hilden, Germany). Following pathogens were detected; Flu A, B, and C; parainfluenza virus 1, 2, 3, and 4; corona virus NL63, 229E, OC43, and HKU1; hMPV A and B rhinovirus, RSVs A and B; AdV, enterovirus, parechovirus, bocavirus; CMV, P. jirovecii, Mycoplasma pneumoniae, Chlamydia pneumoniae, Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Moraxella catarrhalis, Bordetella spp., Klebsiella pneumoniae, Legionella species, and Salmonella species.
Category definition according to the pathogen
Four categories were defined according to the pathogens detected by MPCR and culture. Category V (Only viral), Category NV (nonviral i.e., bacteria and atypical), Category M (mixedi.e., both viral and nonviral pathogen), and Category UK (unknown etiology).
Statistical analysis
Statistical analysis was performed using the Statistical Package for the Social Sciences system (IBM SPSS Statistics, Version 24.0. Armonk, NY: IBM Corp.). Quantitative characteristics for each of the four groups were analyzed by mean and standard deviation. Comparison among the four categories was made by one-way ANOVA and Chi-square tests. P ≤0.05 was considered statistically significant.
RESULTS
Patient characteristics
Of the 177 immunocompromised patients admitted during the study, 61 (34.5%) were suffering from end-stage renal disease, 42 (23.7%) had undergone transplantation (23 kidneys, 16 livers and 3 hematopoietic stem cell transplantation), 39 (22%) had neoplastic disease, 15 (8.5%) had autoimmune disease, and 20 (11%) were receiving immunosuppressive therapy for some other reasons. Of the 177, 126 (71%) were male and 51 (29%) were female. A total of 59 (80.3%) patients were admitted to the Intensive Care Units (ICUs).
Pathogen identification by polymerase chain reaction and culture
Out of 177 patients, pathogens were identified by MPCR in 105 (59.3%) patients while culture could identify the causative pathogen only in 67 (37.8%) patients. Hence, as compared to aerobic culture, pathogen identification rate of MPCR was significantly higher (P = 0.0001).
Category wise distribution
According to the pathogen identified by MPCR and culture, 177 patients were divided into four categories as shown in Table 1. Pathogens identified in various categories are summarized in Table 2 and Figure 1. Bacteria and atypical agents were detected in 111 (62.7%) while viruses were detected in 51 (28.8%) of cases. Most frequent etiological agents was K. pneumoniae in 32% (n = 56, monomicrobial 29 and polymicrobial 27), followed by CMV in 21% (n = 37, mono microbial 11 and poly microbial 26), and P. jirovecii in 10% (n = 18, monomicrobial 5 and polymicrobial 13) of patients. Most frequent mixed infection seen was CMV + P. jirovecii + K. pneumoniae.
Table 1.
Category-wise distribution of patients

Table 2.
Spectrum of pathogens in various categories

Figure 1.
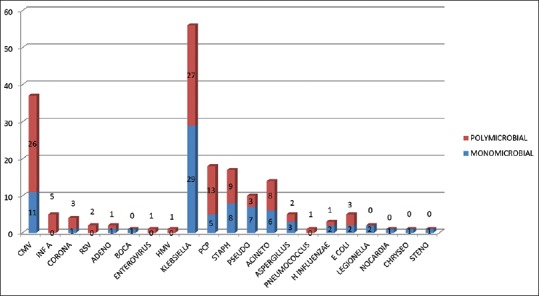
Spectrum of pathogens detected
Correlation between cytomegalovirus deoxyribonucleic acid in bronchoalveolar lavage and blood
Out of 37 patients, positive for CMV deoxyribonucleic acid (DNA) in BAL fluid, we were able to check CMV DNA in blood samples of 22 patients (Qiagen). Of the 22 patients, 19 (86.3%) were positive for CMV DNA in blood sample with a mean of 575650 DNA copies/ml (95% confidence interval 72%–100%).
Patient characteristics comparison according to category
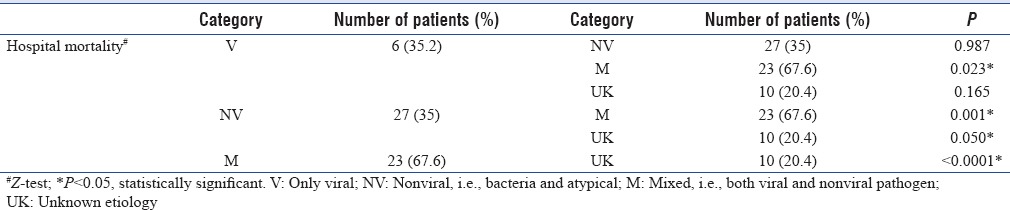
Categories were compared for various clinical characteristics [Table 3]. No significant difference were found in age, gender, ICU admission, preexisting lung disease, days of symptoms, predisposing immunocompromised condition, clinical symptoms, and laboratory investigations. There were significant difference among groups for length of stay in hospital and the final outcome. Total hospital stay was significant higher in Category NV, and mortality was significantly higher in patients of Category M with mixed infections. Further, comparison was done among the categories for the significant parameters [Tables 4 and 5]. Total hospital stay was significantly high in patients of Category NV as compared to Category UK only. It was not significant among patients with viral, nonviral, and mixed infections. However, mortality was significantly high in Category M as compared to all other categories. It was also significantly high in Category NV when compared to UK.
Table 3.
Patients characteristics according to the pathogen isolated
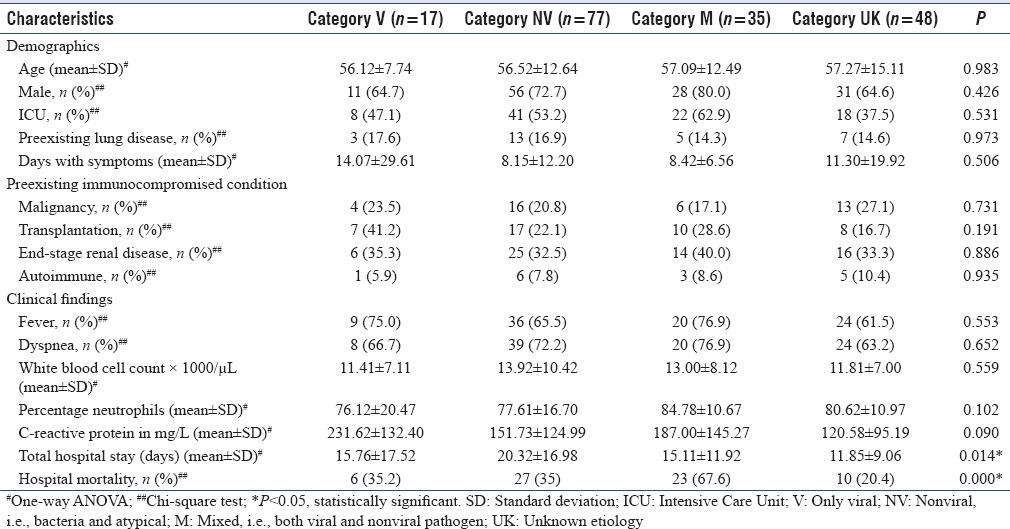
Table 4.
Comparison of mean value of total hospital stay among various categories
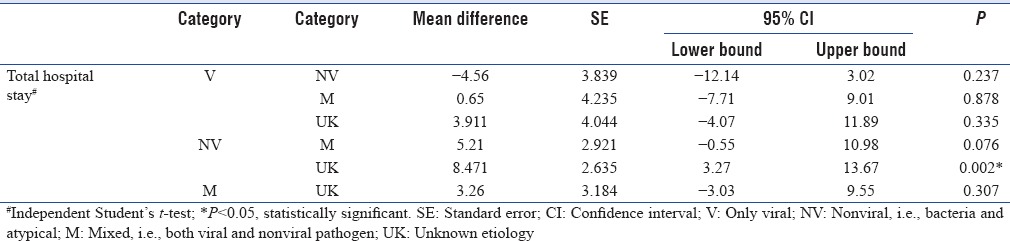
Table 5.
Comparison of final outcome among various categories
DISCUSSION
The present study was done on 177 immunocompromised patients to evaluate the role of MPCR in diagnosis of respiratory illness. Following observations were made:
Multiplex polymerase chain reaction has higher pathogen identification rate as compared to culture alone
In our study, causative pathogen was identified by MPCR in 59.3% of patients while culture could identify only in 37.8% of cases. MPCR is targeting the 33 pathogens simultaneously, but aerobic culture done in routine can only identify the easily cultivable bacteria. Hence, if MPCR is routinely implemented in the diagnostic protocol of respiratory illness in immunocompromised patients, causative pathogen will be identified in significantly higher number of patients. Baudel et al. showed similar type of results where MPCR was positive in 66%, and culture was positive in only 40% of patients.[16] Nevertheless, it has some limitations. First, it cannot quantify the pathogen load so it is difficult to differentiate colonization from infection. Second, it cannot give the antibiotic sensitivity for which we have to rely on culture only. But still, it is a highly sensitive tool in immunocompromised patients where traditional methods for diagnosis of virus and atypical organisms are very cumbersome, and bacteria are also difficult to recover even in lung biopsy.[17]
Viral and atypical organisms are important cause of respiratory illness in immunocompromised patients
In our study, viruses were detected in 28.8% of cases. It is in line with the previous studies where the prevalence of viruses in BAL has been reported from 3.6% to 68.2%.[13,18,19,20,21] K. pneumoniae was the most frequent pathogen obtained from 32% of cases, followed by CMV (21%), and P. jirovecii (10%). In agreement with our results, there are studies where CMV has been reported as the most common viral cause of respiratory illness in immunocompromised patients.[22,23] For P. jirovecii, high prevalence has been reported in HIV-negative immunocompromised patients. It can be highly fulminant leading to mortality as high as 50%. Although PCR is not definitive diagnosis, it cannot differentiate colonization from infection, but still, this is the most useful tool in non-HIV immunocompromised patients where organism load is too low to be detected by staining methods. Its negative predictive value is as high as 100%.[24,25,26] Other viruses detected in our study were Flu A, corona virus, RSV, AdV, bocavirus, enterovirus, and hMPV. These viruses have been reported as an important cause of respiratory illness in immunocompromised patients.[27,28]
Cytomegalovirus deoxyribonucleic acid in bronchoalveolar lavage correlates well with the cytomegalovirus deoxyribonucleic acid in the blood in immunocompromised patients
CMV is a significant pathogen in immunocompromised patients associated with high morbidity and mortality. The definitive diagnosis is difficult as it requires transbronchial or open lung biopsy specimen. It is too risky to be performed routinely in all clinically suspected patients. Culture is also difficult to interpret and time-consuming. Now, in recent years, interest is focused on rapid molecular techniques. PCR is also not confirmatory as it can represent either viral shedding or pulmonary disease. However, if done in clinically suspected immunocompromised patients, it is most sensitive and rapid method. In our study, 21% of patients were positive for CMV DNA in BAL. Chemaly et al. have found that high CMV viral load in BAL in lung transplant recipients is strongly associated with CMV pneumonitis.[29] Our test has limitations as it was not quantitative. To obviate this, we have considered only those BAL positive for CMV DNA which were having cycling threshold value below 30. We correlated it with CMV viral load in the blood samples, and 86.3% of patients were also positive for CMV DNA in blood sample with a mean of 575,650 DNA copies/ml. According to various studies, CMV DNA in blood correlates well with CMV pneumonitis.[29,30,31] CMV DNA detection in BAL can give useful information if done in clinically suspected immunocompromised patients.
Mixed infection leads to more severe disease as compared to bacteria or virus alone
In our study, 34 (19.8%) patients were having mixed infections. A variable range from 14% to 38% has been reported in the previous studies.[32,33] Most frequent pathogens associated with mixed infections were CMV and P. jirovecii. This is in concordance with several published studies.[27,34,35] Mortality was significantly higher in patients with mixed infections as compared to other categories. Previous studies have shown the variable outcome among patients with mixed infections. Some showing more severe clinical disease while other showing no association. Mortalities were not compared significantly in these studies as total number of deaths was less.[32,33,36,37,38] Our study differs in that we only studied immunocompromised patients so combinations of pathogens observed in mixed infections are different. To the best of our knowledge, this is the first study to evaluate the outcome of mixed infection (categorized by MPCR and culture) in immunocompromised patients. To confirm our observation, further more studies are needed.
CONCLUSION
Results of the present study demonstrate that viruses and atypical pathogens can be significant respiratory pathogens in immunocompromised patients leading to coinfections and high mortality. MPCR is highly sensitive and rapid tool which should be routinely implemented for the diagnosis of respiratory illness in immunocompromised patients.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Franquet T. High-resolution computed tomography (HRCT) of lung infections in non-AIDS immunocompromised patients. Eur Radiol. 2006;16:707–18. doi: 10.1007/s00330-005-0008-1. [DOI] [PubMed] [Google Scholar]
- 2.Jepson SL, Pakkal M, Bajaj A, Raj V. Pulmonary complications in the non-HIV immunocompromised patient. Clin Radiol. 2012;67:1001–10. doi: 10.1016/j.crad.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Cereser L, Zuiani C, Graziani G, Girometti R, Como G, Zaja F, et al. Impact of clinical data on chest radiography sensitivity in detecting pulmonary abnormalities in immunocompromised patients with suspected pneumonia. Radiol Med. 2010;115:205–14. doi: 10.1007/s11547-009-0433-3. [DOI] [PubMed] [Google Scholar]
- 4.Safadi AR, Soubani AO. Diagnostic approach of pulmonary disease in the HIV negative immunocompromised host. Eur J Intern Med. 2009;20:268–79. doi: 10.1016/j.ejim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Corti M, Palmero D, Eiguchi K. Respiratory infections in immunocompromised patients. Curr Opin Pulm Med. 2009;15:209–17. doi: 10.1097/MCP.0b013e328329bd2c. [DOI] [PubMed] [Google Scholar]
- 6.Baughman RP. The lung in the immunocompromised patient. Infectious complications Part 1. Respiration. 1999;66:95–109. doi: 10.1159/000029349. [DOI] [PubMed] [Google Scholar]
- 7.Bollée G, Sarfati C, Thiéry G, Bergeron A, de Miranda S, Menotti J, et al. Clinical picture of Pneumocystis jiroveci pneumonia in cancer patients. Chest. 2007;132:1305–10. doi: 10.1378/chest.07-0223. [DOI] [PubMed] [Google Scholar]
- 8.Monnet X, Vidal-Petiot E, Osman D, Hamzaoui O, Durrbach A, Goujard C, et al. Critical care management and outcome of severe Pneumocystis pneumonia in patients with and without HIV infection. Crit Care. 2008;12:R28. doi: 10.1186/cc6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansharamani NG, Garland R, Delaney D, Koziel H. Management and outcome patterns for adult Pneumocystis carinii pneumonia, 1985 to 1995: Comparison of HIV-associated cases to other immunocompromised states. Chest. 2000;118:704–11. doi: 10.1378/chest.118.3.704. [DOI] [PubMed] [Google Scholar]
- 10.Japanese Respiratory Society. Pneumonia in immunocompromised patients. Respirology. 2009;14(Suppl 2):S44–50. doi: 10.1111/j.1440-1843.2009.01576.x. [DOI] [PubMed] [Google Scholar]
- 11.Tregoning JS, Schwarze J. Respiratory viral infections in infants: Causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brittain-Long R, Westin J, Olofsson S, Lindh M, Andersson LM. Prospective evaluation of a novel multiplex real-time PCR assay for detection of fifteen respiratory pathogens-duration of symptoms significantly affects detection rate. J Clin Virol. 2010;47:263–7. doi: 10.1016/j.jcv.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren L, Gonzalez R, Wang Z, Xiang Z, Wang Y, Zhou H, et al. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005-2007. Clin Microbiol Infect. 2009;15:1146–53. doi: 10.1111/j.1469-0691.2009.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcos MA, Esperatti M, Torres A. Viral pneumonia. Curr Opin Infect Dis. 2009;22:143–7. doi: 10.1097/QCO.0b013e328328cf65. [DOI] [PubMed] [Google Scholar]
- 15.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baudel JL, Tankovic J, Dahoumane R, Carrat F, Galbois A, Ait-Oufella H, et al. Multiplex PCR performed of bronchoalveolar lavage fluid increases pathogen identification rate in critically ill patients with pneumonia: A pilot study. Ann Intensive Care. 2014;4:35. doi: 10.1186/s13613-014-0035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiatt JR, Gong H, Mulder DG, Ramming KP. The value of open lung biopsy in the immunosuppressed patient. Surgery. 1982;92:285–91. [PubMed] [Google Scholar]
- 18.Vu DL, Bridevaux PO, Aubert JD, Soccal PM, Kaiser L. Respiratory viruses in lung transplant recipients: A critical review and pooled analysis of clinical studies. Am J Transplant. 2011;11:1071–8. doi: 10.1111/j.1600-6143.2011.03490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soccal PM, Aubert JD, Bridevaux PO, Garbino J, Thomas Y, Rochat T, et al. Upper and lower respiratory tract viral infections and acute graft rejection in lung transplant recipients. Clin Infect Dis. 2010;51:163–70. doi: 10.1086/653529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar D, Husain S, Chen MH, Moussa G, Himsworth D, Manuel O, et al. A prospective molecular surveillance study evaluating the clinical impact of community-acquired respiratory viruses in lung transplant recipients. Transplantation. 2010;89:1028–33. doi: 10.1097/TP.0b013e3181d05a71. [DOI] [PubMed] [Google Scholar]
- 21.Gambarino S, Mantovani S, Astegiano S, Libertucci D, Solidoro P, Baldi S, et al. Lower respiratory tract viral infections in hospitalized adult patients. Minerva Med. 2009;100:349–55. [PubMed] [Google Scholar]
- 22.Barton TD, Blumberg EA. Viral pneumonias other than cytomegalovirus in transplant recipients. Clin Chest Med. 2005;26:707–20, viii. doi: 10.1016/j.ccm.2005.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Giraldo A, Sialer S, Esperatti M, Torres A. Viral-reactivated pneumonia during mechanical ventilation: Is there need for antiviral treatment? Front Pharmacol. 2011;2:66. doi: 10.3389/fphar.2011.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azoulay E, Bergeron A, Chevret S, Bele N, Schlemmer B, Menotti J. Polymerase chain reaction for diagnosing pneumocystis pneumonia in non-HIV immunocompromised patients with pulmonary infiltrates. Chest. 2009;135:655–61. doi: 10.1378/chest.08-1309. [DOI] [PubMed] [Google Scholar]
- 25.Mühlethaler K, Bögli-Stuber K, Wasmer S, von Garnier C, Dumont P, Rauch A, et al. Quantitative PCR to diagnose Pneumocystis pneumonia in immunocompromised non-HIV patients. Eur Respir J. 2012;39:971–8. doi: 10.1183/09031936.00095811. [DOI] [PubMed] [Google Scholar]
- 26.Li MC, Lee NY, Lee CC, Lee HC, Chang CM, Ko WC. Pneumocystis jiroveci pneumonia in immunocompromised patients: Delayed diagnosis and poor outcomes in non-HIV-infected individuals. J Microbiol Immunol Infect. 2014;47:42–7. doi: 10.1016/j.jmii.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 27.Danés C, González-Martín J, Pumarola T, Rañó A, Benito N, Torres A, et al. Pulmonary infiltrates in immunosuppressed patients: Analysis of a diagnostic protocol. J Clin Microbiol. 2002;40:2134–40. doi: 10.1128/JCM.40.6.2134-2140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vigil KJ, Adachi JA, Chemaly RF. Viral pneumonias in immunocompromised adult hosts. Intensive Care Med. 2010;25:307–26. doi: 10.1177/0885066610377969. [DOI] [PubMed] [Google Scholar]
- 29.Chemaly RF, Yen-Lieberman B, Chapman J, Reilly A, Bekele BN, Gordon SM, et al. Clinical utility of cytomegalovirus viral load in bronchoalveolar lavage in lung transplant recipients. Am J Transplant. 2005;5:544–8. doi: 10.1111/j.1600-6143.2005.00747.x. [DOI] [PubMed] [Google Scholar]
- 30.Michaelides A, Liolios L, Glare EM, Spelman DW, Bailey MJ, Walters EH, et al. Increased human cytomegalovirus (HCMV) DNA load in peripheral blood leukocytes after lung transplantation correlates with HCMV pneumonitis. Transplantation. 2001;72:141–7. doi: 10.1097/00007890-200107150-00027. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez JL, Kruger RM, Paranjothi S, Trulock EP, Lynch JP, Hicks C, et al. Relationship of cytomegalovirus viral load in blood to pneumonitis in lung transplant recipients. Transplantation. 2001;72:733–5. doi: 10.1097/00007890-200108270-00030. [DOI] [PubMed] [Google Scholar]
- 32.Jennings LC, Anderson TP, Beynon KA, Chua A, Laing RT, Werno AM, et al. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63:42–8. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 33.Templeton KE, Scheltinga SA, van den Eeden WC, Graffelman AW, van den Broek PJ, Claas EC. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41:345–51. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polaczek MM, Zych J, Oniszh K, Szopinski J, Grudny J, Roszkowski-Sliz K. Pneumocystis pneumonia in HIV-infected patients with cytomegalovirus co-infection. Two case reports and a literature review. Pneumonol Alergol Pol. 2014;82:458–66. doi: 10.5603/PiAP.2014.0060. [DOI] [PubMed] [Google Scholar]
- 35.Vetter M, Battegay M, Trendelenburg M. Primary cytomegalovirus infection with accompanying Pneumocystis jiroveci pneumonia in a patient with large-vessel vasculitis. Infection. 2010;38:331–4. doi: 10.1007/s15010-010-0024-1. [DOI] [PubMed] [Google Scholar]
- 36.Johnstone J, Majumdar SR, Fox JD, Marrie TJ. Viral infection in adults hospitalized with community-acquired pneumonia: Prevalence, pathogens, and presentation. Chest. 2008;134:1141–8. doi: 10.1378/chest.08-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson N, Kalin M, Hedlund J. Clinical impact of combined viral and bacterial infection in patients with community-acquired pneumonia. Scand J Infect Dis. 2011;43:609–15. doi: 10.3109/00365548.2011.570785. [DOI] [PubMed] [Google Scholar]
- 38.Choi SH, Hong SB, Ko GB, Lee Y, Park HJ, Park SY, et al. Viral infection in patients with severe pneumonia requiring intensive care unit admission. Am J Respir Crit Care Med. 2012;186:325–32. doi: 10.1164/rccm.201112-2240OC. [DOI] [PubMed] [Google Scholar]


