Abstract
TIA-1 (T-cell restricted intracellular antigen-1) is an RNA-binding protein involved in splicing and translational repression. It mainly interacts with RNA via its second and third RNA recognition motifs (RRMs), with specificity for U-rich sequences directed by RRM2. It has recently been shown that RRM3 also contributes to binding, with preferential binding for C-rich sequences. Here we designed UC-rich and CU-rich 10-nt sequences for engagement of both RRM2 and RRM3 and demonstrated that the TIA-1 RRM23 construct preferentially binds the UC-rich RNA ligand (5΄-UUUUUACUCC-3΄). Interestingly, this binding depends on the presence of Lys274 that is C-terminal to RRM3 and binding to equivalent DNA sequences occurs with similar affinity. Small-angle X-ray scattering was used to demonstrate that, upon complex formation with target RNA or DNA, TIA-1 RRM23 adopts a compact structure, showing that both RRMs engage with the target 10-nt sequences to form the complex. We also report the crystal structure of TIA-1 RRM2 in complex with DNA to 2.3 Å resolution providing the first atomic resolution structure of any TIA protein RRM in complex with oligonucleotide. Together our data support a specific mode of TIA-1 RRM23 interaction with target oligonucleotides consistent with the role of TIA-1 in binding RNA to regulate gene expression.
INTRODUCTION
TIA-1 (T-cell restricted intracellular antigen-1) and TIAR (TIA-1 related) are ubiquitous RNA-binding proteins that modulate RNA metabolism at several levels effecting diverse functions in cell survival and inflammatory responses (1,2). They are located in both the cytoplasm and the nucleus and continuously shuttle between these two compartments (3). In the nucleus, TIA-1 and TIAR are modulators of alternative splicing by binding to U-rich stretches near 5΄-splice sites of target mRNAs, including those encoding FGFR2, CFTR, FAS, COL2A1, human calcitonin and TIA-1/TIAR themselves (4–8). Best characterized is the nuclear role of TIA-1 in promoting the inclusion of exon 6 in FAS pre-mRNA resulting in an apoptotic form of FAS (9,10). TIA proteins also interact with DNA, potentially providing a link between transcription and splicing (7,11). In the cytoplasm, TIA-1 and TIAR mediate translational repression of target mRNAs via binding to target RNA motifs present in their untranslated regions. These target sequences are generally A/U- or C/U-rich and through them TIA-1/TIAR have been shown to repress the expression of inflammatory mediators COX-2 (12), TNF-α (1) as well as HMPP-13 (13) and mRNA containing 5΄-TOP (5΄-terminal oligopyrimidine) elements (14). TIA-1 and TIAR are also key proteins in the formation of stress granules. Under conditions of stress, including ER stress, heat shock, hypoxia, exposure to arsenite or viral infection, TIA proteins, along with their bound mRNA, are sequestered into stress granules where mRNA translation is temporarily suspended until the stress is resolved and the stress granules are disassembled (15–17). Most recently, TIA proteins have also been identified as playing a role in miRNA regulation (18).
Each of these functions involves the specific recognition of target oligonucleotide sequences by TIA-1 and TIAR. The basis of RNA recognition by TIA proteins has hence been a major focus of their characterization. TIA-1 and TIAR (and their isoforms that all share ∼90% identity in the RNA-binding regions) possess three RNA-recognition motif (RRM) domains through which they interact with single-stranded oligonucleotides, followed by a C-terminal prion-related domain (PRD) that is responsible for their participation in stress granule formation (Figure 1) (19). Early experiments (employing in vitro selection from an RNA library and filter-binding assays) determined that the preferred target of TIA-1 and TIAR proteins are U-rich sequences (20). Subsequently, in vivo studies identifying cellular targets of TIA proteins (using co-immunoprecipitation and iCLIP methods) revealed that, in addition to U-rich sequences, TIA proteins also target mRNAs containing C-rich motifs (14,21–23). These findings were supported by biophysical studies that showed that TIAR RRM domains are capable of binding both U-rich RNA and UC-rich RNA sequences with nanomolar binding affinities (22).
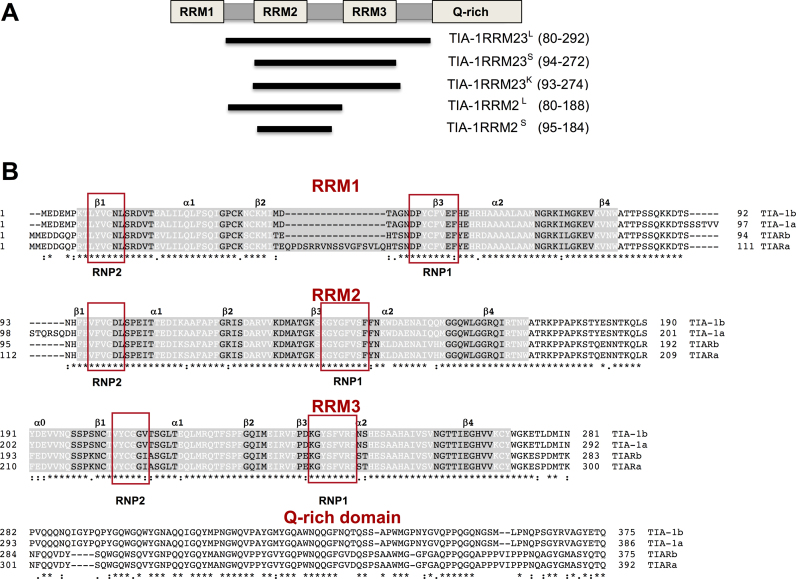
Figure 1.
TIA-1b protein constructs and alignment with other TIA protein isoforms (A) TIA-1 protein domain structure schematic showing the constructs used in this study. (B) Sequence alignment of TIA isoforms indicating the secondary structural elements in white font, and the position of conserved RNP motifs in red boxes.
Further insight into oligonucleotide recognition by TIA-1 and TIAR has been provided by studies of the individual contributions of the three RRM domains to target binding. RRM domains are the most abundant RNA binding motif and are well known for their ability to bind most commonly 3- to 5-nt stretches of single-stranded oligonucleotide (linear stretches or loop structures) in tandem to effect binding (24). The RRMs of a single protein can contribute differentially to the overall oligonucleotide binding, both in terms of affinity (across a nanomolar to micromolar binding range) and specificity (from highly specific to highly promiscuous). Previous work established that TIA protein RRM2 is predominantly responsible for the observed U-rich RNA binding, with negligible contributions from RRM1 or RRM3, although RRM3 was shown to have some RNA-binding capacity (20). It was subsequently found that RRM1 of TIAR preferentially binds to DNA and likely plays a role in the process of transcriptional activation (11). The contribution of RRM3 to RNA binding remained mysterious with studies using isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR) showing only very small contributions to binding to U-rich sequences (22,25). In order to determine whether the contributions to TIA protein binding by RRM3 might depend upon recognition of a specific sequence (other than U-rich RNA) a nuclear magnetic resonance (NMR) approach based on a scaffold-independent analysis was used (26). Interestingly a C-rich RNA motif (5΄-ACUCC-3΄) was identified and confirmed to be bound by TIA-1 RRM3 using SPR. Thus it was recognized that, while RRM3 was able to contribute to binding of U-rich sequences, its physiological role was likely to elicit TIA protein binding specificity toward UC-rich sequences observed to be physiological targets such as FAS pre-mRNA and TOP elements (9,14).
The current study thus investigates the binding of TIA-1 RRM23 (the tandem RRM2 and RRM3 domains of TIA-1) to designed oligonucleotide sequences that combine the U-rich target of RRM2 and the C-rich target of RRM3. We discovered the importance of the RRM23 construct design, with amino acids outside the strictly defined RRM domains making important contributions to binding affinity. We confirmed that binding to target RNA sequences involves both RRM2 and RRM3 and that binding to equivalent DNA sequences occurs with similar affinities. Importantly we demonstrated that binding of TIA-1 RRM23 to a ‘UC-rich’ RNA sequence (in which the U-rich sequence is 5΄ to the C-rich sequence) occurs with significantly higher affinity than to a ‘CU-rich’ RNA sequence (in which the U-rich sequence is 3΄ to the C-rich sequence). This indicates that the TIA-1 RRM23 preferentially binds to target oligonucleotides with tandem RRMs arranged in a 5΄ to 3΄ direction. We also present small-angle X-ray scattering (SAXS) data that provide structural information for the TIA-1 RRM23 construct in complex with target oligonucleotides. Lastly, we present the X-ray crystallography structure of TIA-1 RRM2 in complex with oligonucleotide that provides, for the first time, atomic resolution structural information for the recognition of U-rich sequences by RRM2. Together these findings further our understanding of TIA-1 RNA binding and recognition of target UC-rich sequences.
MATERIALS AND METHODS
Expression and purification of the TIA-1 RRM23 domains
DNA segments containing the TIA-1 central domains, TIA-1 RRM23L (residues 80–292) and TIA-1 RRM23S (residues 94–272), were cloned into pETM-11 vector; TIA-1 RRM23K (residues 93–274) was cloned into pETtrx 1a vector (European Molecular Biology Laboratory, Heidelberg, Germany). TIA-1 RRM2L (residues 80–188) was prepared as previously reported (27). The proteins were expressed in the Escherichia coli strain Rosetta 2 (DE3). The culture was grown until an OD600 of 0.6–0.8 was reached, and induced with 0.5 mM IPTG and grown at 18°C overnight. The cells were harvested by centrifugation and resuspended in ice-cold lysis buffer (50 mM HEPES pH 7.4, 300 mM NaCl, 5% glycerol). The cells were disrupted using an Avestin cell homogenizer. The N-terminal poly-histidine (His-tag) protein constructs were purified using Ni-NTA agarose (QIAGEN). The His tag was removed by incubating protein with TEV protease (Promega) overnight at 4°C followed by a second round of Ni-NTA affinity chromatography to separate the solubility tags from the cleaved protein. The final purification step was achieved by size-exclusion chromatography using a Superdex 75 16/60 column (GE healthcare) in storage buffer (20 mM HEPES pH 7.0, 100 mM NaCl, 3% glycerol). All purified constructs were concentrated and kept at −80°C. The purity of protein obtained was >95% as determined from Coomassie Blue staining following sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
Preparation of 15N-labeled TIA-1 RRM2
15N labeled TIA-1 RRM2S (residues 95–184) was prepared for NMR studies as previously described (28). 15N labeled RRM2S was expressed in E. coli BL21(DE3) cells as a His-tagged protein and was purified by nickel affinity chromatography (Ni Sepharose 6 Fast Flow, GE Healthcare) according to earlier protocols (26,29). The His-tag was cleaved using TEV protease (AcTEV Protease, Life Technologies) and removed by a second nickel affinity chromatography purification. The protein was dialyzed into 20 mM potassium phosphate buffer containing 50 mM KCl (pH 5) and concentrated to 70 μM. 5΄TTTTT3΄ DNA oligonucleotide was chemically synthesized (IDT, Integrated DNA Technologies; 1 mmol synthesis) and solubilized in 20 mM potassium phosphate buffer containing 50 mM KCl (pH 5.0) to a final concentration of 2 mM.
Fluorescence anisotropy experiments
The oligonucleotides used for fluorescence anisotropy experiments were 5΄-ACUCCUUUUU-3΄ (CU-rich RNA); 5΄-ACTCCTTTTT-3΄ (CT-rich DNA); 5΄-AUUCUCCUUU-3΄ (FAS RNA sequence); 5΄-ATTCTCCTTT-3΄ (FAS DNA sequence); 5΄-UUUUUACUCC-3΄ (UC-rich RNA); 5΄-TTTTTACTCC-3΄ (TC-rich DNA). The oligonucleotides were chemically synthesized carrying a 5΄ fluorescein label and supplied as HPLC purified oligonucleotides (RNA from Dharmacon research and DNA from Invitrogen). The RNA was deprotected according to the manufacturer's instructions. Both RNA and DNA were diluted to a final concentration of 1 μM in 10 mM HEPES pH 7, 100 mM NaCl. Purified TIA-1 RRM23 protein (10 mM HEPES pH 7, 100 mM NaCl, 3% glycerol) was added at increasing concentrations to a constant concentration of fluorescein-labeled RNA/DNA (1 nM). Experiments were performed in triplicate using a PHERAstar plate reader (BMG LabTech). Data were analyzed using 1:1 Langmuir interaction model to determine the affinities (KD) of protein–RNA/DNA interactions.
SAXS analysis
TIA-1 RRM23K protein both in apo-form and in a 1:1.2 molar ratio with 10-mer 5΄-UUUUUACUCC-3΄ (UC-rich RNA), 5΄-TTTTTACTCC-3΄ (TC-rich DNA), 5΄-ACUCCUUUUU-3΄ (CU-rich RNA) and 5΄-ACTCCTTTTT-3΄ (CT-rich DNA) were centrifuged at 13 200 × g for 15 min before data collection. SAXS data were collected at the SAXS-WAXS beamline at the Australian Synchrotron. Samples at various concentrations (0.625, 1.25, 2.5, 5.0 mg/ml protein) were loaded as volumes of 60 μl into a 96-well plate (Axygen) and sealed with silicone sealing mat. The samples were run as sequentially using an autoloader system with each sample taken up into the quartz capillary using an autoloader. After analysis the sample was returned back into the 96-well plate. The SAXS data reduction was performed using software ‘scatterBrain’ developed at the Australian Synchrotron. The SAXS data averaging, buffer blank subtraction, and further processing were performed using the program PRIMUS and ATSAS software (30). DAMMIF (31) was used to calculate 15 ab intio dummy atom models from scattering curve. The models were superposed, merged and filtered using the program DAMAVER (32).
Crystallization and data collection
The purified proteins (TIA-1 RRM23L and TIA-1 RRM23K) were concentrated to 630 μM using an Eppendorf concentrator with a molecular weight cutoff limit of 10 K at 4°C. TIA-1 RRM23 complexes were formed with 5΄-ACTCCTTTTT-3΄ and 5΄-ATTCTCCTTT-3΄ oligonucleotides (Dharmacon research) to a final protein:RNA ratio of 1:1.2. The sitting drop method was employed for crystallization trials by mixing a 1:1 ratio of sample and reservoir solution using screens including MIDAS, PGA (Molecular Dimension), PACT (QIAGEN), Natrix (Sigma) and PEG/Ion (Hampton Research). Crystallization experiments were set up at the Monash Macromolecular Crystallization Facility (Monash University). TIA-1 RRM23/DNA complex crystals were obtained for TIA-1 RRM23L combined with 5΄-ACTCCTTTTT-3΄ in 25% (W/V) polyethylene glycol 1500, 10% (V/V) malonate-imidazole-borate, pH 8 (from the PACT screen). TIA-1 RRM23K complexes were formed with 5΄-ACTCCTTTTT-3΄ and 5΄-ATTCTCCTTT-3΄ oligonucleotides (Dharmacon Research) at the final protein:oligonucleotide ratio of 1:1.2. Both crystallized in 0.1 M Potassium Chloride, 25 w/v Sokalan CP-7, 0.1 M HEPES pH 7. The crystals were flash-cooled in liquid nitrogen prior to data collection.
X-ray data were collected at the high-throughput micro-focus MX2 beamline at the Australian Synchrotron at a wavelength of 0.954 Å using an ADSC quantum 315r detector and BLU-ICE software for data acquisition (33). The diffraction data were integrated and scaled using XDSME (34). The reported crystal structure of the apo-TIA-1 RRM2 (PDB ID: 3BS9) was used as a search model for molecular replacement using the program PHASER (35). Several rounds of structure refinement were completed using PHENIX (36) and COOT (37). The data collection and refinement statistics are summarized in Table 1. Figures were produced using PYMOL (38).
Table 1. Data collection and refinement statistics.
| TIA-1 RRM2_TTT | |
|---|---|
| Data collection | |
| Space group | P 65 |
| Cell dimensions | |
| a, b, c (Å) | 45.43 45.43 84.90 |
| α, β, γ (°) | 90 90 120 |
| Resolution (Å) | 22.97-2.31 (2.44-2.31) |
| R sym | 4.5 (62.1) |
| I/σI | 35.07 (5.17) |
| Completeness (%) | 99.9% |
| Mutiplicity | 12.6 |
| CC1/2 | 100 (97.1) |
| Refinement | |
| Resolution (Å) | 2.31 |
| No. reflections | 4381 (441) |
| R work | 0.2277 (0.3179) |
| R free | 0.2605 (0.3509) |
| No. atoms | 681 |
| Protein residues | 80 |
| DNA residues | 3 |
| Water | 5 |
| Ramachandran favored (%) | 96 |
| Ramachandran outliers (%) | 0 |
| RMS bond lengths (Å) | 0.005 |
| RMS bond angles (°) | 0.91 |
NMR spectroscopy
Two-dimensional (2D) 1H-15N HSQC experiments were acquired for the free and DNA-bound TIA-1 RRM2S domain (1:1.2 ratio) at 283 K on a 500 MHz Bruker AVANCE III spectrometer provided with a cryoprobe (CITIUS NMR services, University of Sevilla). A 30 μM sample of 15N-labeled RRM2 was prepared in 20 mM potassium phosphate buffer and 50 mM KCl containing 90% H2O and 10% D2O at pH 5. For the DNA-bound complex, also performed at pH 5.0, a 5΄-TTTTT-3΄ oligonucleotide was added to the previous sample at a final concentration of 36 μM. The spectral windows of 2D 1H-15N HSQC were fitted to 15 ppm (1H) and 145 ppm (15N), centered at 4.7 and 152.5 ppm, respectively. An additional NMR spectrum using a 15N spectral window with the same width (145 ppm) but centered at 162.5 ppm was recorded in order to demonstrate that the 1J (N-H) couplings of His96 imidazole group were not aliased.
RESULTS
Binding of TIA-1 RRM23 to RNA and DNA
Although early work suggested that TIA-1 binding to target RNA occurs primarily through RRM2 interactions with U-rich sequences, more recent work has established a role for RRM3. In particular, a pentameric C-rich consensus motif was established for TIA-1 RRM3 binding (26). We therefore set out to characterize the way in which these two adjacent RRMs interact with target RNAs comprised of both U-rich and C-rich regions. The ‘CU-rich’ sequence 5΄-ACUCCUUUUU-3΄ consists of a pentamer of the RRM3 consensus sequence (ACUCC) followed by a poly-U pentamer consistent with established RRM2 RNA-binding data. These are arranged such that RRM23 binding can occur in the 3΄ to 5΄ direction from RRM2 to RRM3 (i.e. 3΄-5΄ / N-C) as seen for many tandem RRM interactions with RNA that have been structurally characterized (e.g. PDB IDs: 1CVJ, 4ED5, 1FJE). The ‘UC-rich’ sequence 5΄-UUUUUACUCC-3΄, in contrast, positions the pentamer consensus sequences in the reverse order.
TIA-1 RRM23 constructs were initially cloned, including a ‘long’ sequence (TIA-1 RRM23L; residues 80–292), and a ‘short’ sequence (TIA-1 RRM23S; residues 94–272) (Figure 1A). Note that we selected TIA-1 isoform b, which lacks an insertion between RRM 1 and 2 compared to isoform a; (Figure 1B). The latter ‘short’ sequence was designed to span only the well-defined secondary structural elements of RRM2 and RRM3, as well as the linker between them, according to their determined three-dimensional apo-structures (PDB IDs: 2MJN, 2DGO, 2RNE, 3BS9). It was anticipated that this construct would be more amenable to crystallization. These constructs were readily overexpressed and purified for biophysical study.
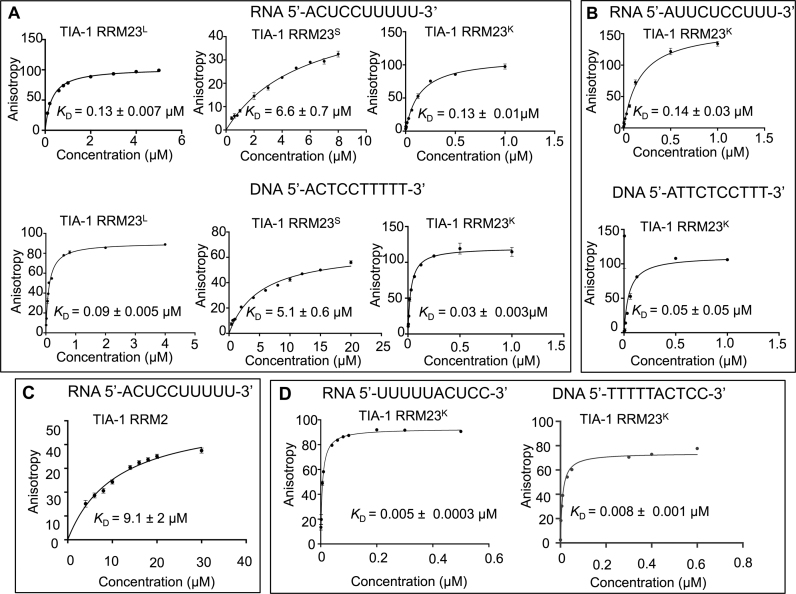
Fluorescence anisotropy was used to determine the binding affinity of TIA-1 RRM23L and RRM23S to the 5΄-FITC-labeled CU-rich RNA sequence: 5΄-ACUCCUUUUU-3΄ as well as the equivalent CT-rich DNA sequence: 5΄-ACTCCTTTTT-3΄ for comparison. Figure 2A shows the binding curves obtained for the interactions as well as the determined binding affinities. Interestingly, while the TIA-1 RRM23L construct bound to the RNA with an equilibrium dissociation constant of KD = 0.13 ± 0.007 μM, the TIA-1 RRM23S construct bound with a lower affinity KD = 6.6 ± 0.7 μM. Binding to DNA was of slightly higher affinity, but also showed this trend of loss of affinity by the shorter construct. Together this demonstrated that TIA-1 RRM23 binds to both RNA and DNA target sequences with micromolar affinity and that residues N- or C-terminal to the well-defined RRM regions are important in contributing to oligonucleotide binding.
Figure 2.
Fluorescence anisotropy binding curves for TIA-1 RRM23 interactions with RNA and DNA sequences. (A) The binding of TIA-1 RRM23L, TIA-1 RRM23S and TIA-1 RRM23K to CU-rich RNA (top) and CT-rich DNA (bottom), (B) TIA-1 RRM23K binding to a FAS-derived RNA sequence (top) and the corresponding DNA sequence (bottom), (C) TIA-1 RRM2 L binding to UC-rich RNA and (D) TIA-1 RRM23K binding to UC-rich RNA and TC-rich DNA. Increasing concentrations of protein were added to a constant amount of the fluorescent RNA and DNA. Data points represent triplicate samples and the errors shown are standard errors of the mean (SEM). The binding data were fit by the 1:1 Langmuir binding model to derive the KD.
Previous studies of TIAR RRM2 demonstrated the importance of a lysine residue immediately C-terminal to the well-defined secondary structure of RRM2 (39) leading us to consider whether Lys274, that is located 2 amino acid residues C-terminal to the TIA-1 RRM3 domain might similarly be important to binding by RRM3. We therefore prepared a third construct, TIA-1 RRM23K (residues 93–274), that includes Lys274 and repeated the binding experiments as shown in Figure 2A. The inclusion of the extra residues completely restored the RNA and DNA binding of the TIA-1 RRM23K construct to KD = 0.13 μM ± 0.01 and KD = 0.03 μM ± 0.003 respectively, demonstrating that the higher binding affinity observed for TIA-1 RRM23L was due to these extra N- and C-terminal residues—most likely due to the contribution of Lys274 to oligonucleotide interactions.
It was also of interest to measure the binding affinity of the TIA-1 RRM23K construct for a physiological sequence for comparison with the consensus motif derived sequence. We therefore measured the binding of TIA-1 RRM23K construct to a sequence (5΄-AUUCUCUUU-3΄) derived from a region of FAS pre-mRNA that has previously been reported to be bound by TIA-1 (40) as well as its DNA equivalent. Figure 2B shows the fluorescence anisotropy-derived binding curves to 5΄-FITC labeled RNA and DNA sequences. The binding affinities to these sequences were almost identical to those determined for the consensus motif-derived sequences, with KD = 0.14 μM ± 0.03 for RNA and a slightly higher affinity of KD = 0.05 μM ± 0.05 for DNA.
In order to determine the contribution of TIA-1 RRM3 toward binding to the CU-rich RNA sequence, the binding of TIA-1 RRM2 (residues 80–188) was tested with 5΄-ACUCCUUUUU-3΄ RNA. The KD calculated for the isolated RRM2 with CU-rich RNA was 9.1 ± 2 μM, as compared to TIA-1 RRM23L with a KD = 0.13 μM (Figure 2C), confirming that RRM3 makes a significant contribution in increasing the affinity toward binding to RNA.
Oligonucleotide binding by TIA-1 RRM23 occurs in a 5΄ to 3΄ orientation
Finally we examined whether oligonucleotide sequences in which the two pentamer target sequences were reversed in order showed any difference in their affinity of binding by TIA-1 RRM23K. Thus we performed fluorescence anisotropy experiments with the 5΄-FITC-labeled UC-rich RNA sequence: 5΄-UUUUUACUCC-3΄ as well as the equivalent DNA TC-rich sequence: 5΄-TTTTTACTCC-3΄ for comparison (Figure 2D). To our surprise, binding by TIA-1 RRM23K was enhanced to KD = 0.005 ± 0.0003 μM and KD = 0.008 ± 0.001 μM respectively—both with higher affinity compared to the RNA and DNA sequences in which the RRM3 target motif was placed 5΄ to the RRM2 target motif. In order to confirm this unexpected result, we also used ITC to measure the affinity of TIA-1 RRM23K for non-FITC labeled UC-rich RNA sequence (UUUUUACUCC) compared to the CU-rich sequence (ACUCCUUUUU). Consistent with the fluorescence anisotropy binding experiments, we found that TIA-1 RRM23K binding to the UC-rich RNA was significantly higher (KD = 0.012 ± 0.002 μM) than binding to the CU-rich sequence (KD = 0.07 ± 0.003 μM) (Supplementary Figure S1 and Table S1). This finding suggests that TIA-1 RRM23 adopts a more favorable interaction with oligonucleotides when tandem binding is in the parallel 5΄ to 3΄ orientation (i.e. 5΄-3΄/ N-C).
To further verify the 5΄-3΄ orientation of the binding of TIA-1 RRM23 to DNA, paramagnetic NMR spectroscopy was performed by labeling the thiol group at 5΄ of a TC-rich DNA (5΄thiolTC-rich DNA) with the paramagnetic tag 3-(2-Iodoacetamido)-proxyl (PROXYL) (Supplementary Figure S2A). If TIA-1 RRM23 binds to the TC-rich DNA in a parallel orientation (5΄-3΄ / N-C), the distance of the paramagnetic spin label to RRM2 should be shorter than that to RRM3. This fact would lead to a higher paramagnetic effect in resonances from RRM2 than those from RRM3. In order to test this, 15N HSQC spectra were recorded upon binding TIA-1 RRM23 to the diamagnetic (5΄thiolTC-rich DNA) and paramagnetic (PROXYL-modified) DNA. Although a general paramagnetic effect was observed for all TIA-1 RRM23 resonances involved in DNA binding due to the short length of the DNA molecule, the effect was substantially more pronounced in TIA-1 RRM2 (Supplementary Figure S2B). We therefore quantified intensity changes of the NMR signals in the DNA binding regions (RNP1 and RNP2 in the RRMs (Supplementary Figure S2C)) upon spin labeling. Consistent with a 5΄-3΄/N-C orientation, the resonances with a higher drop in their intensity under paramagnetic conditions were included in the RNP2 and RNP1 of RRM2 (stretches Val97-Leu102 and Lys136-Ser142) and the surroundings RNP2 residues Ser103 and Glu105 (Supplementary Figure S2D).
TIA-1 RRM23 engages with target RNA and DNA to form a compact structure
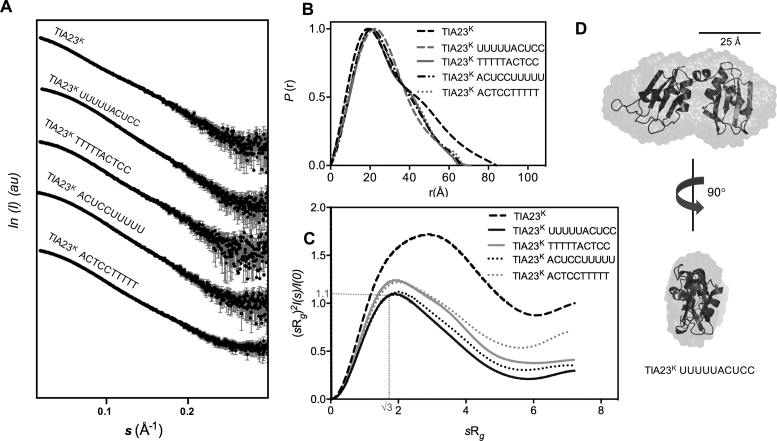
In order to obtain structural information, SAXS was used to characterize the solution structure of TIA-1 RRM23K, in apo-form and in complex with each of the consensus sequence derived oligonucleotides: 5΄-ACUCCUUUUU-3΄ (CU-rich RNA), 5΄-ACTCCTTTTT-3΄ (CT-rich DNA), 5΄-UUUUUACUCC-3΄ (UC-rich RNA) and 5΄-TTTTTACTCC-3΄ (TC-rich DNA). SAXS data were collected at the SAXS/WAXS beamline of the Australian Synchrotron for both the apo-TIA-1 RRM23K and the protein in complex with oligonucleotide over a range of concentrations (0.6, 1.25, 2.5 and 5.0 mg/ml). The radius of gyration (Rg) estimated from the Guinier region of the scattering data showed fairly consistent values at the three lower concentrations, but increased above 2.5 mg/ml suggesting that aggregation was taking place above this concentration (Supplementary Figure S3). The scattering curves obtained at 2.5 mg/ml protein are shown in Figure 3A and were used for further analysis.
Figure 3.
SAXS analysis of apo-TIA-1 RRM23K, TIA-1 RRM23K/RNA and TIA-1 RRM23K/DNA complexes. (A) The scattering intensity (I) is plotted in arbitrary units (au) versus the forward scattering vector (s) in the units of inverse Å for TIA-1 RRM23K (abbreviated TIA23K) before and after being combined with RNA and DNA sequences (sequences as indicated). The error bars in gray indicate the mean ± SD. (B) P(r) profiles calculated from the scattering data are plotted against r in the units of Å. (C) Normalized Kratky Plots for TIA-1 RRM23K calculated from the scattering data are shown across units of sRg. (D) Ab initio shape reconstruction determined for the TIA-1RRM23K /UUUUUACUCC complex using DAMMIF (NSD = 0.66 ± 0.04) (31). Superposed are ribbon representations of TIA-1 RRM2 and RRM3 derived from the NMR structure PDB ID: 2MJN to show the relative scale of the ab initio shape.
In the case of the apo-TIA-1 RRM23K protein, the pairwise density distribution function (P(r)) plot showed multi-modal features tailing toward a maximum dimension (Rmax) of 84 Å (Figure 3B and Table 2). The normalized Kratky plot showed a non-parabolic curve that did not completely return to zero and had a maxima not placed at 1.1 for a value of sRg = √3 (Figure 3C). This pattern is indicative of the presence of both globular and partially disordered regions in the protein (41). These data are thus consistent with apo-TIA-1 RRM23K behaving in solution as a pair of RRM domains connected by a flexible linker, as would be expected.
Table 2. SAXS data collection and scattering parameters.
| TIA-1 RRM23K | TIA-1 RRM23K +UC-rich RNA | TIA-1RRM23K +TC-rich DNA | TIA-1 RRM23K +CU-rich RNA | TIA-1RRM23K +CT-rich DNA | |
|---|---|---|---|---|---|
| Data collection | scatterBrain | scatterBrain | scatterBrain | scatterBrain | scatterBrain |
| Wavelength (Å) | 1.03320 | 1.03320 | 1.03320 | 1.03320 | 1.03320 |
| q range (Å−1) | 0.006–0.35 | 0.006–0.35 | 0.006–0.35 | 0.006–0.35 | 0.006–0.35 |
| Exposure time (s)a | 2 | 2 | 2 | 2 | 2 |
| Concentration (mg ml−1) | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| Temperature (K) | 288 | 288 | 288 | 288 | 288 |
| Structural parameters b | |||||
| I(0) (cm−1) [from P(r)] | 0.04 | 0.06 | 0.04 | 0.06 | 0.06 |
| Rg (Å) [from P(r)] | 24.6 | 20.9 | 21.4 | 21.5 | 21.8 |
| Rg (Å) [from Guinier] | 23.1 | 20.8 | 21.1 | 21.7 | 21.8 |
| I(0) (cm−1) [from Guinier] | 0.04 | 0.06 | 0.04 | 0.06 | 0.06 |
| Rmax (Å) | 84 | 65 | 71 | 66 | 67 |
| Porod volume estimate (Å3) | 25 332 | 26 481 | 32 006 | 29 869 | 30 591 |
| Molecular mass determination b | |||||
| Mr [Porod volume] | 18.8 kDa | 17.6 kDa | 21.3 kDa | 18.5 kDa | 21.8 kDa |
| Calculated Mr from sequence | 20.2 kDa | 23.7 kDa | 23.1 kDa | 23.7 kDa | 23.1 kDa |
| Data processing | PRIMUS | PRIMUS | PRIMUS | PRIMUS | PRIMUS |
aFifteen frames of 2 s exposure were recorded for each sample, and averaged.
bReported for a 2.5 mg/ml measurement.
Scattering curves obtained for TIA-1 RRM23K in complex with the UC-rich RNA showed a distinct change of shape indicative of the formation of a more compact and less flexible structure upon binding to the oligonucleotide. The P(r) plot is unimodal with some tailing toward an Rmax value of 65 Å (Figure 3B and Table 2). The shape of the normalized Kratky plot is parabolic with maxima close to 1.1 for a value of sRg = √3 and returns to low values, consistent with the formation of a well-defined complex (Figure 3C). In order to visualize the shape of TIA-1 RRM23K in complex with oligonucleotide, DAMMIF (31) was used to calculate the three-dimensional, ab initio shape reconstruction of TIA-1 RRM23K in complex with the UC-rich RNA (Figure 3D). It reveals a slightly elongated oblate shape consistent with two RRMs positioned adjacent to one another. In the case of TIA-1 RRM23K in complex with TC-rich DNA, the scattering and P(r) plot look very similar, indicative of a compact complex also formed with the DNA (Figure 3A and B). The normalized Kratky plot, however, shows higher values across sRg, indicative of a slightly increased flexibility of this complex with DNA (Figure 3C).
Scattering curves and P(r) plots obtained for TIA-1 RRM23K in complex with the CU-rich RNA and CT-rich DNA also showed similar profiles (Figure 3A and B), indicating that when TIA-1 RRM23K binds to these lower affinity oligonucleotides, it still adopts a more compact and less flexible structure in solution. Interestingly, the normalized Kratky plots reveal subtle differences for these complexes compared to the complexes formed with the UC-rich RNA and TC-rich DNA counterparts. In both cases the curves are slightly higher beyond sRg = √3 values, indicative of a subtle increase in flexibility or difference in shape of these complexes (Figure 3C). Together this indicates that binding by both RRMs still takes place to these oligonucleotides in which the target consensus sequences for RRM2 and RRM3 are arranged in a 3΄ to 5΄ direction; however the complexes formed do have a slight difference in their shape or flexibility.
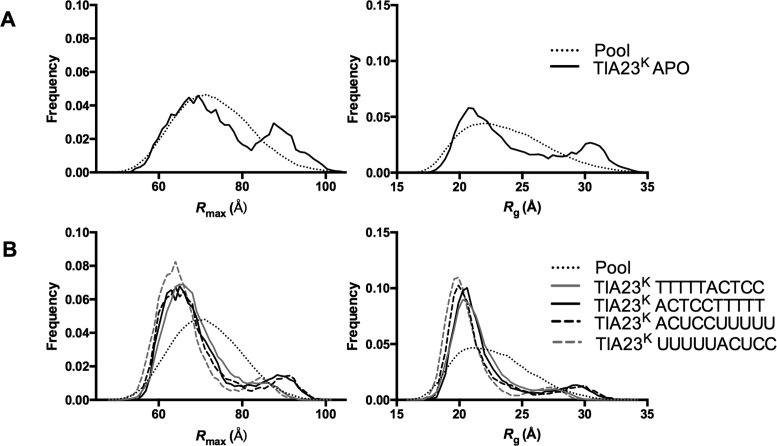
The differences between the scattering data can be further evaluated using the ensemble optimization method (42). This method involves the in silico generation of a large pool of molecular models of the system from which a minimal subset are selected that together predict the scattering curve. For this the NMR model of TIA-1 RRM23 (PDB ID: 2MJN) was used as a starting point with or without RNA modeled across the face of RRM2. A total of 10 000 models were generated in which each of the RRMs was held as a rigid body and the linker allowed to randomly explore conformational space using a Monte Carlo algorithm. Figure 4A shows the frequency distribution of molecular models of apo-TIA-1 RRM23K in the pool plotted against Rmax and Rg. Superposed is a frequency distribution indicating the distribution of selected models that together best predicted the apo-TIA-1 RRM23K scattering data (χ2 = 0.34). In this case, two populations of models are apparent—one with a small average Rmax and Rg, and the other with an average Rmax and Rg at the longest and largest end of the pool. This is a reflection of the dynamics of the system with TIA-1 RRM23K existing in equilibrium between more compact and more extended forms.
Figure 4.
Ensemble optimization analysis of apo-TIA-1 RRM23K, TIA-1 RRM23K/RNA and TIA-1 RRM23K/DNA complexes. Rmax and Rg distributions from the best fitting ensembles were calculated using ensemble optimization method (42). (A) The distribution of the pool of 10 000 models (dotted line) and the selected best fitting ensemble (solid line) are shown for apo-TIA-1 RRM23K (abbreviated TIA23K APO). (B) The distribution of the pool of 10 000 models (dotted line) and the selected best fitting ensemble are shown for TIA-1 RRM23K complexes with RNA and DNA sequences (as indicated in the key).
Figure 4B shows the frequency distributions of selected molecular models of TIA-1 RRM23K bound to oligonucleotide that predict the scattering curves derived for TIA-1 RRM23K bound to the four different oligonucleotides (χ2 ≤ 0.44 in all cases; fits shown in Supplementary Figure S4). In all cases there were two populations selected—a main population at the low end of Rmax and Rg indicative of a compact structure, and a much smaller population at the longer and larger end of these parameters indicative of the presence of some elongated molecules. This again likely reflects an equilibrium between compact TIA-1 RRM23K bound to oligonucleotide and partially unbound species but, in contrast to the apo-TIA-1 RRM23K structure, with the equilibrium lying well toward the compact, fully bound structure. Interestingly, upon close examination of the subset of elongated structures it can be seen that, in the case of TIA-1 RRM23K in complex with the CU-rich RNA and CT-rich DNA, the selected ensemble is shifted further to the right than in the cases of TIA-1 RRM23K in complex with UC-rich RNA and TC-rich DNA. In other words, the data suggest that the complexes formed with the higher affinity UC-rich RNA and TC-rich DNA have slightly different structures compared to the complexes formed with the lower affinity CU-rich RNA and CT-rich DNA.
SAXS experiments were also conducted with TIA-1 RRM23L and the CU-rich and CT-rich oligonucleotides in order to see whether TIA-1 RRM23L construct by itself and in complex with oligonucleotide behaves similarly to TIA-1 RRM23K. SAXS analysis showed that apo-TIA-1 RRM23L behaves as an extended molecule in solution. Upon complex formation with CU-rich oligonucleotide a more compact arrangement of two RRM domains was observed (Supplementary Figure S5 and Table S2). These data are consistent with the TIA-1 RRM23L construct also binding the target oligonucleotides with both RRMs engaged to form a stable complex.
Crystal structure of TIA-1 RRM2 in complex with T-rich DNA
Having established that TIA-1 RRM23 forms a complex with the 10-mer RNA and DNA, with both RRMs engaged, we were interested to examine the structural basis for TIA-1 recognition of the target nucleotide sequence. TIA-1 RRM23L and TIA-1 RRM23K were combined with CU-rich and UC-rich RNA and equivalent DNA sequences and subjected to crystallization trials. TIA-1 RRM23L co-crystallized with 5΄-ACTCCTTTTT-3΄ DNA, forming crystals that diffracted to 2.3 Å resolution. These crystals were determined to belong to space group P 65, with unit cell dimensions a = b = 45.43 Å and c = 84.9 Å. The structure was solved by molecular replacement, using the structure of the RRM2 domain of TIA-1 (PDB ID: 3BS9) as a search model.
Unexpectedly, only a single RRM domain with bound nucleotide was visible in the electron density. The density was consistent with the presence of the RRM2 sequence and not RRM3 suggesting that protein cleavage had occurred resulting in an RRM2 product that had crystallized with the bound oligonucleotide. To investigate this possibility, a crystal was picked from the well, washed in fresh buffer and examined using SDS-PAGE. Indeed, a smaller band corresponding to the molecular weight of a single RRM predominated, suggesting that proteolytic cleavage of RRM23 had occurred, and that the single RRM2, in complex with the oligonucleotide, had preferentially crystallized (Supplementary Figure S6). Repeated efforts to form crystals using the TIA-1 RRM23K construct in the presence of protease inhibitors resulted in two further crystals being formed with 5΄-ACTCCTTTTT-3΄ and 5΄-ATTCTCCTTT-3΄ DNA (that diffracted to 2.9 and 2.8 Å resolution respectively and the same space group and unit cell dimensions as above). Upon solving their structures, the electron density was found, once more, to contain only RRM2 bound to oligonucleotide, with no sign of RRM3. The structures obtained, nevertheless, represent the first example of a TIA protein RRM structure solved in the presence of oligonucleotide. They therefore present a unique opportunity to observe the detailed interactions via which TIA-1 RRM2 recognizes target oligonucleotide.
The refinement of the 2.3 Å resolution TIA-1 RRM2/DNA structure was completed using PHENIX (36). The refined model consists of 80 amino acids (Residues 92–171 could be seen in the electron density), three nucleotides and four ordered water molecules. The nucleotide density in the model was consistent with three thymine residues out of ten DNA nucleotides. No density was observed on either side, likely due to flexibility of the DNA outside the bound region. The refined model has good statistics with an R-factor of 0.2277 (free R-factor 0.2605). The Ramachandran plot showed all backbone torsion angles ϕ-ψ within the allowed regions and >98% of the residues were in the energetically most favored regions (Table 1).
The structural basis of nucleotide binding by TIA-1 RRM2
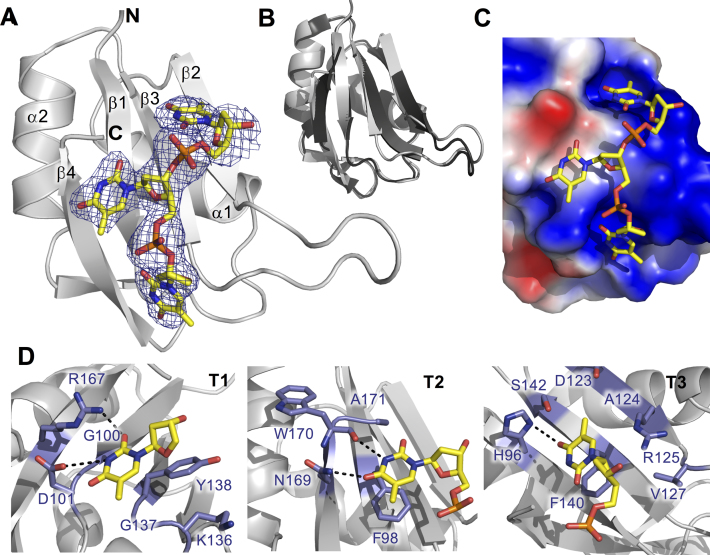
The structure of TIA-1 RRM2/TTT complex is shown in Figure 5A. RRM2 adopts the classical βαββαβ topology with four β-sheets packed against two α-helices. The RMSD of Cα atoms compared with the structure of apo-RRM2 (PDB ID: 3BS9) is 0.441 (calculated for 71 Cα out of 78), with the main difference between the molecules occurring in the loop region between β-strands 2 and 3 (Figure 5B). The nucleotides are positioned against the β-sheet face of the TIA-1 RRM2 as is the classical nucleotide binding face for RRMs (43). The surface is contoured, providing shallow binding pockets for each nucleotide and a predominantly positively charged surface accommodating the oligonucleotide backbone (Figure 5C and Supplementary Figure S7). The base interactions are mainly van der Waals or hydrophobic, but aromatic stacking and several specific hydrogen bonds are also observed. The two conserved phenylalanine residues (Phe98 and Phe140) located in the RNP-2 and RNP-1 sub motifs of RRM2 interact directly with Thy2 and Thy3 through aromatic base stacking. Another RNP-2 residue, Asp101, forms a hydrogen bond with the Thy1 N3 atom and Arg167 interacts with the Thy1 O4 atom (Figure 5D, left). In the case of Thy2, the Asn169 Nδ2 and Trp170 backbone CO, located in β4 of RRM2, form hydrogen bonds with Thy2 O4 and N3 atoms respectively (Figure 5D, middle). Thirdly, in the case of Thy3, the His96 Nδ1 hydrogen bonds with the Thy3 O4 and the Arg125 guanidinium group is positioned to form electrostatic or water mediated interactions with the oligonucleotide backbone (Figure 5D, right). It should be noted that the hydrogen bond and electrostatic interactions listed above could not take place with other bases, with the exception of cytosine in position 1, where it could potentially make similar interactions with Arg167 via its O4 atom and engage Asp101 via its NH2 group. Altogether, therefore, the structure reveals the interactions that contribute to the affinity of nucleotide binding to TIA-1 RRM2 and also helps to explain the preferential binding of TIA-1 RRM2 for uracil and thymine nucleotides.
Figure 5.
Structure of TIA-1 RRM2/TTT complex (PDB ID: 5ITH) (A) Cartoon representation of the solved TIA-1 RRM2 structure with electron density map indicating the position of the bound oligonucleotide. DNA is shown as sticks, colored by atom type. N-and C-termini as well as secondary structural elements of the protein are labeled. (B) Superposition of the oligonucleotide-bound form of TIA-1 RRM2 (gray) with the apo-structure previously reported (PDB ID: 3BS9). (C) Electrostatic surface contour representation, showing thymine binding pockets across the surface of TIA-1 RRM2 and the oligonucleotide in stick representation. The surface was generated using the Adaptive Poisson-Boltzmann Solver software package (http://www.poissonboltzmann.org). Positive and negative potentials are colored blue and red, respectively. (D) Detail of TIA-1 RRM2 sidechain interactions with thymine in the three oligonucleotide binding sites. Hydrogen bond interactions are highlighted with hashed lines.
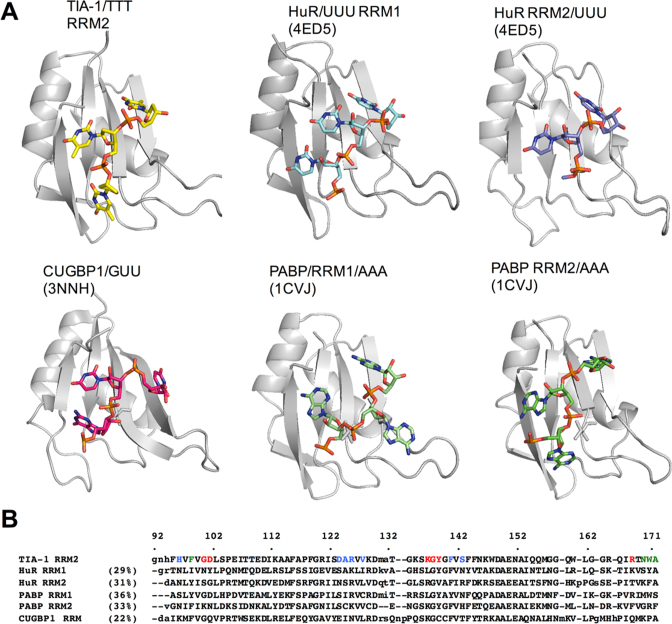
Nucleotide binding by TIA-1 RRM2 bears some resemblance to other characterized RRM/RNA interactions. Figure 6 shows cartoon representations of closely related RRM/RNA structures alongside TIA-1 RRM2/TTT as well as a structure based sequence alignment. The oligonucleotide orientation is the same in each case, with the 5΄-end closest to the C-terminal β-strand, but the precise nucleotide binding positions vary. TIA-1 RRM2 nucleotide binding positions are most similar to those adopted by HuR RRM2/UUU and PABP RRM2/AAA (though there are also similarities between the binding position of T3 in HuR RRM1/UUU and CUGBP1/AUU). Interestingly, however, there is no sequence similarity in the type of amino acid interactions with targeted nucleotide bases, even in the case of HuR RRM2 that also preferentially binds to U-rich sequences. The only exception is the presence of aromatic residues in RNP1 and RNP2 motifs that facilitate nucleotide base stacking interactions.
Figure 6.
Comparison of the TIA-1 RRM2 structure with other RRM/RNA interactions. (A) Structural representations of RRM/oligonucleotide interactions determined for HuR (PDB ID: 4ED5), CUGBP1 (PDB ID: 3NNH) and PCBP (PDB ID: 1CVJ) shown in the same orientation as that for TIA-1 RRM2/TTT. (B) Structure-based sequence alignment of the depicted RRMs. The TIA-1 RRM2 amino acids engaged in interactions with thymine are colored in red (T1), green (T2) and blue (T3).
The interaction of TIA-1 RRM2 His96 with thymine is in agreement with NMR experiments
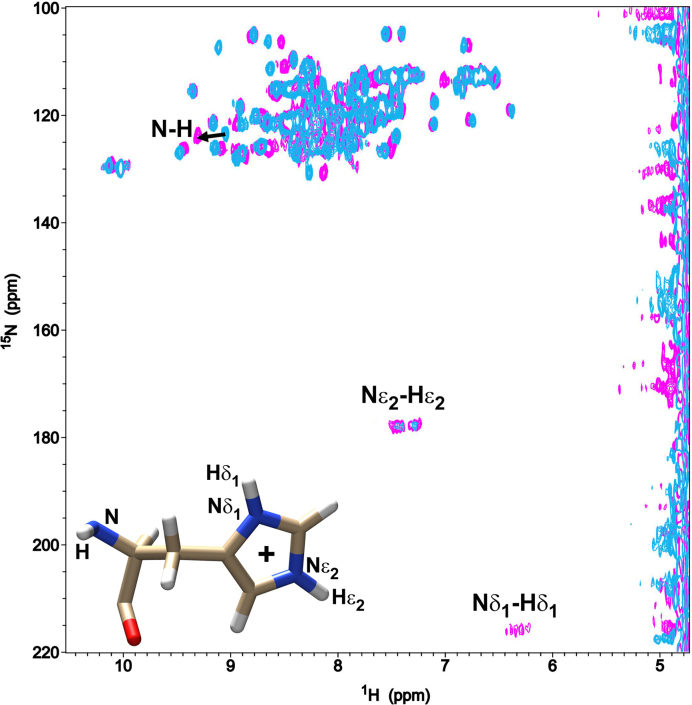
A specific feature revealed by the TIA-1 RRM2 structure bound to oligonucleotide was the hydrogen bond between His96 Nδ1 hydrogen and the Thy2 O4 (Figure 5D, right). This interaction likely explains the previously reported rise in pKa value of His96 that occurs upon oligonucleotide binding by TIA-1 RRM2 (28). The direct hydrogen bonding of His96 Nδ1 would be expected to increase the nucleophilicity of the Nε2 atom of the imidazole and thus raise the pKa. In order to further investigate this interaction we performed 2D 1H-15N HSQC experiments that could provide a direct readout of the protonation state of the TIA-1 RRM2 His96 sidechain and backbone nitrogens before and after forming a complex with a 5΄-TTTTT-3΄ oligonucleotide. At pH 5 (which is well below the His96 pKa value of 5.3 for apo-TIA-1 RRM2 and His96 pKa of 5.6 for TIA-1 RRM2 bound to DNA; (28,29) the 1H-15N HSQC spectra show a 2-fold increase in intensity for the Nε2–Hε2 cross-peaks when the complex with DNA is formed (integrated value of 1.075 for free peaks versus 2.167 for DNA-bound signals in arbitrary units) (Figure 7). This is consistent with its increased nucleophilicity. In addition, The Nδ1–Hδ1 coupling of His96 of TIA-1 RRM2 could not be seen for apo-TIA-1 RRM2, but became detectable upon DNA complex formation, in agreement with the formation of a hydrogen-bond between His96 Nδ1 and Thy3 O4. It was also noted that the His96 backbone NH coupling was shifted upon complex formation with DNA, which is also in agreement with involvement of His96 in the interaction. Together the NMR data corroborate the observed interaction in the crystal structure.
Figure 7.
1 J (N-H) couplings of TIA-1 RRM2 domain. Overlapped 2D 15N-1H HSQC spectra of free TIA-1 RRM2 (cyan) and TIA-1 RRM2 in complex with 5΄-TTTTT-3΄ DNA (magenta). The 1J (N-H) couplings of the single histidine (His96) from TIA-1 RRM2 construct are labeled. The black arrow points out the chemical-shift perturbation of the backbone amide (N-H) resonance of His96 observed upon DNA addition. A schematic structure of a histidine residue identifying different imidazole groups involved in 1J couplings is also shown. H atoms are depicted in white, N atoms in blue, O atoms in red and C atoms in brown.
DISCUSSION
TIA-1 and TIAR are critical proteins for alternative splicing, translation repression and sequestration into stress granules of mRNA targets. The current study has investigated the biophysical basis for oligonucleotide recognition and binding by TIA-1 that underlies these functions. It has been previously established that RRM1 does not contribute significantly to RNA binding, that RRM2 preferentially binds to U-rich oligonucleotides and that RRM3 contributes to binding through its ability to bind to C-rich sequences (20,26). The focus has thus been on the structural basis of binding and recognition by the TIA-1 RRM23 construct to UC-rich and CU-rich RNA and equivalent DNA sequences. For this study 10-nt ssRNA and ssDNA were utilized for all biophysical and structural studies. This would allow for recognition of up to 5-nt by each RRM. A footprint of 10 or 9-nt is in keeping with binding studies by Wang et al., in which 1:1 stoichiometry was observed for TIA-1 RRM23 binding to a 10-nt RNA, but 2:1 stoichiometry was observed for a 16-nt oligonucleotide (40). It is possible, however, that shorter oligonucleotides would be sufficient for recognition and binding by TIA proteins.
Oligonucleotide binding affinity was found to be dramatically affected when amino acid residues outside of the strictly defined RRM3 were included. In particular, the presence of three extra N- and C-terminal residues enhanced the binding of TIA-RRM23 to RNA and DNA targets by 50- and 100-fold respectively. This enhancement likely reflects the contribution of the Lys274 positive charge to the oligonucleotide binding. Lys274 is positioned at the edge of the oligonucleotide binding face of RRM3 and thus contributes to the electrostatic charge at this site. The interaction is entirely analogous to what was discovered by our group in a separate study of the related TIAR RRM2 domain (39). The TIAR RRM2 domain affinity for oligonucleotide was greatly enhanced by the amino acid residues immediately C-terminal to the strictly defined RRM domain. In particular, Lys176 contributed to a 25- to 100-fold enhancement of binding to RNA and DNA respectively. This lysine is also conserved in TIA-1 (Lys174) and likely also plays the same role in stabilizing oligonucleotide interactions by TIA-1.
Interestingly, TIA-1 RRM23 binding to DNA sequences was found to occur with similar or slightly enhanced binding affinity compared with that to equivalent RNA sequences. We have previously reported that, in the case of TIAR, binding to T-rich DNA occurs with slightly reduced affinity compared with U-rich RNA. This was shown to be due to a combination of affinity loss owing to the absence of the 2΄-hydroxyl groups and an affinity enhancement imparted by the presence of the methyl groups of thymine (44). TIAR and TIA-1 RRMs share high sequence identity of ∼90% for RRMs of TIA-1b and TIARb and it is thus likely that these factors are also at play in TIA-1 oligonucleotide binding. Other RNA-binding proteins are far less promiscuous when it comes to RNA versus DNA binding. In the case of HuR, which is also an AU-rich RNA binding protein with three RRMs, binding to equivalent DNA sequences resulted in a dramatic loss in binding affinity (44). The ability of TIA-1 to bind to DNA may reflect its role in alternative splicing. It has been proposed that TIA proteins interact with single-stranded regions of DNA in chromatin undergoing active transcription facilitating their shuttling to pre-mRNA in preparation for subsequent splicing activity (11).
Having established that the TIA-1 RRM23K construct forms stable interactions with target oligonucleotides, we compared the binding affinity with other similarly composed sequences. The initial CU-rich sequence (5΄-ACUCCUUUUU-3΄) was designed based on established RNA target sequences derived for RRM3 and RRM2 (26). The binding motifs were arranged such that RNA binding could occur in the 3΄ to 5΄ direction from RRM2 to RRM3 as seen for most other tandem RRM constructs that have been structurally investigated in complex with RNA (43). Binding affinity was in the low micromolar range for this oligonucleotide sequence, as it was for a similar sequence derived from the FAS mRNA TIA-1 target motif. Furthermore, the binding affinity of TIA-1 RRM23K was greater than that measured for TIA-1 RRM2 alone, demonstrating that RRM3 contributes to binding. However, we then measured the binding affinity of TIA-1 RRM23K to a sequence in which the RRM target sequences were placed in the reverse order, i.e. a UC-rich RNA sequence (5΄-UUUUUACUCC-‘3) and determined binding in the nanomolar range. This finding suggested that a much more favorable interaction of TIA-1 RRM23K takes place when RRM2 and RRM3 target sequences are positioned in the 5΄ to 3΄ direction, which we subsequently verified using an NMR spectroscopic approach. This arrangement is less usual, but not unprecedented. Tandem RRMs of splicing factor TDP-43 were also found to be arranged in a 5΄ to 3΄ direction upon their solution structure determination by NMR spectroscopy (45).
Small angle X-ray scattering experiments confirmed that the TIA-1 RRM23K construct forms a compact structure in complex with the UC-rich sequence as well as the equivalent TC-rich DNA sequence, again consistent with the engagement of both RRM2 and RRM3 in the oligonucleotide binding interaction. Interestingly, similar scattering data were also observed for TIA-1 RRM23K in complex with the CU-rich RNA and CT-rich DNA sequences—also consistent with a compact structure forming but with a subtle difference in structure or flexibility apparent. The scattering data are similar to that reported for TIA-1 RRM23 binding to a FAS pre-mRNA derived 16mer RNA (40) and TIA-1 RRM123 binding to a longer FAS mRNA sequence (25) that both show that a more compact structure is formed when TIA-1 RRMs engage with RNA.
With this information we trialled crystallization conditions for complexes formed between TIA-1 RRM23K (and TIA-1 RRM23L) both UC-rich and CU-rich RNA sequences and their equivalent DNA sequences. In three cases diffracting crystals were achieved (with DNA rather than RNA) and the structures solved, but in all cases only RRM2 bound to oligonucleotide was present in the crystal. It was rationalized that limited proteolysis that occurred in the crystal trays were selectively crystallized. This occurred despite the presence of complete protease inhibitor tablet in the protein buffer. All three structures were solved, and the one solved to the highest resolution is reported herein. The structure revealed, for the first time, the TIA-1 RRM2 structure engaged with T-rich DNA. The oligonucleotide sequence is positioned across the face of the TIA-1 RRM2 similarly to the interaction between PABP RRM2 and poly-A in terms of the nucleotide binding arrangement. Hydrogen bond interactions were observed that help to explain the preferential binding of U-rich or T-rich oligonucleotides. In particular, an interaction between His96 and the third thymine base provided structural verification of a previous observation that the pKa of this histidine sidechain is impacted by RNA binding (28,29). We further probed this using NMR and observed increased signal intensities of the sidechain N-H signals upon DNA binding, again consistent with the formation of an oligonucleotide interaction with His96.
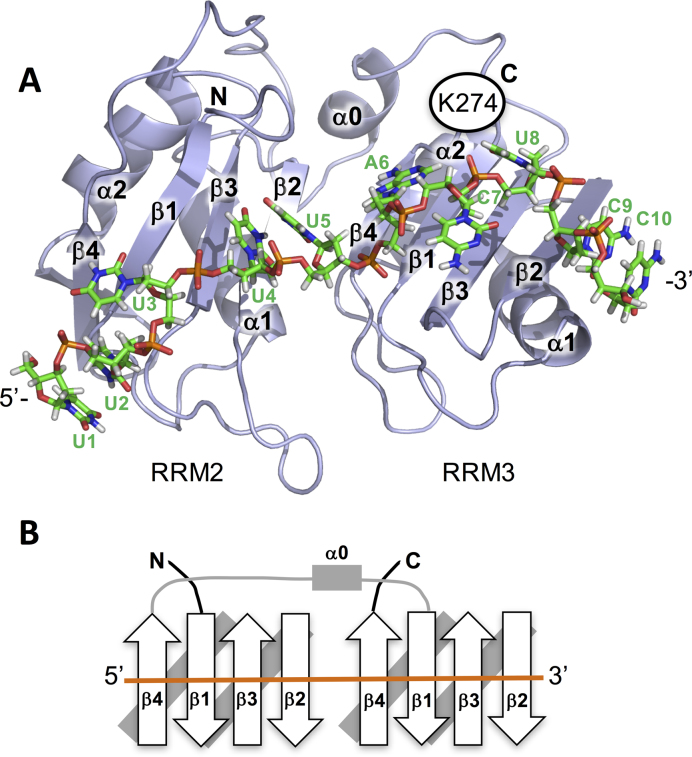
Together the biophysical data permit the construction of a model of the interaction between TIA-1 RRM23 and the UC-rich RNA target (Figure 8A). In this model, RRM2 is positioned at the 5΄-end of the oligonucleotide, engaging with a U-rich pentamer by analogy to the TIA-1 RRM2/TTT crystal structure. RRM3 is positioned at the 3΄-end of the DNA, engaged with the ACTCC sequence. Here the RRM oligonucleotide interaction has been constructed based on the interaction observed between PABP RRM2 and poly-A, as this RRM has the highest similarity to TIA-1 RRM3 of any known RRM that has been structurally characterized with oligonucleotide bound. Positioned in this way, Lys274 that occurs immediately C-terminal to RRM3 is positioned close to the oligonucleotide and could form electrostatic interactions with the phosphate backbone, explaining the importance of its contribution to the binding affinity. The juxtaposition of RRM2 5΄ to RRM3 requires the positioning of the linker region across the RRM2 domain to reach the N-terminal α0-helix of RRM3 (Figure 8B). This α-helix, which occurs in addition to the conventional βαββαβ topology of an RRM (27), is positioned away from the oligonucleotide binding face of RRM3. The linker between RRM2 and RRM3 is long enough to extend between RRM2 and RRM3 without a structural distortion—but it would not be unexpected for a small rearrangement to take place that would reduce any strain imposed of this linker region. Indeed, HSQC titration mapping of the amino acid residues of TIA-1 RRM23 that are perturbed upon oligonucleotide binding show that, as well as perturbations to the oligonucleotide binding faces of RRM2 and RRM3, the α0-helix on RRM3 is also perturbed (40).
Figure 8.
Model of TIA-1 RRM23 bound to a UC-rich RNA sequence. (A) Model based on the TIA-1 RRM2/TTT structure reported herein (PDB ID: 5ITH) positioned next to the TIA-1 RRM3 structure (PDB ID: 2MJN) bound to the oligonucleotide 5΄-UUUUUACUCC-3΄ in a 5΄ to 3΄ direction. The juxtaposition of the two RRMs and the interaction between RRM3 and oligonucleotide is by structural homology to the PABP/poly-A structure (PDB ID:1CVJ). The model demonstrates that the linker region is sufficiently long to reach from the RRM2 C-terminus to the RRM3 C-terminus. (B) Topology diagram of the model highlighting the 5΄ to 3΄ directionality of the TIA-1 RRM23 interaction with target oligonucleotide.
TIA-1 proteins possess varied functions in post-transcriptional gene regulation, including interactions with splicing factors, mRNA translational control and mRNA sequestration into stress granules—all of which involve oligonucleotide binding. The current biophysical and structural studies provide a better understanding of the structural arrangement and requirements for binding to linear RNA and DNA sequences. This lays the foundation for future studies to discover the precise arrangement of the tandem RRMs, the structural basis for RRM3 recognition of C-rich targets and whether there are other elements of oligonucleotide secondary structure that may also be important for binding. Such studies will be important for ultimately understanding the mechanistic role of TIA proteins in their various cellular functions.
ACCESSION NUMBERS
Atomic coordinates and structure factors for TIA-1 RRM23/TTT crystal structure have been deposited in the RCSB PDB under the accession number PDB ID: 5ITH. SAXS data have been deposited in the SASDB under the accession number: SASDBM6 (apo-TIA-1 RRM23); SASDBN6 (TIA-1 RRM23/CU-rich RNA); SASDBP6 (TIA-1 RRM23/CT-rich DNA); SASDBR6 (TIA-1 RRM23/UC-rich RNA); SASDBQ6 (TIA-1 RRM23/TC-rich DNA).
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank all the beamline staff at the SAX/WAX and MX2 beamlines at the Australian Synchrotron, Victoria, Australia. In particular, Dr Tom Caradoc-Davies provided exceptional assistance with data collection. The authors also thank Dr Manuel Angulo and Dr Encarnación Zafra for assistance in the NMR data collection at CITIUS NMR Facility of the University of Seville.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council [APP1105801 to J.A.W., M.C.W., M.G.]; National Health and Medical Research Senior Research Fellowship [FP1079611 to M.C.J.W.]; NIA-IRP, NIH (to M.G.); Andalusian Government [P11-CVI-7216, BIO198]; Spanish Ministry of Science and Innovation [BFU2015-71017-P]. Funding for open access charge: Monash University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Piecyk M., Wax S., Beck A.R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M. et al. . TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000; 19:4154–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V.. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 1999; 274:2322–2326. [DOI] [PubMed] [Google Scholar]
- 3. Zhang T., Delestienne N., Huez G., Kruys V., Gueydan C.. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J. Cell Sci. 2005; 118:5453–5463. [DOI] [PubMed] [Google Scholar]
- 4. Del Gatto-Konczak F., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R.. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5΄ splice site. Mol. Cell. Biol. 2000; 20:6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zuccato E., Buratti E., Stuani C., Baralle F.E., Pagani F.. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 2004; 279:16980–16988. [DOI] [PubMed] [Google Scholar]
- 6. Forch P., Puig O., Kedersha N., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J.. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000; 6:1089–1098. [DOI] [PubMed] [Google Scholar]
- 7. McAlinden A., Liang L., Mukudai Y., Imamura T., Sandell L.J.. Nuclear protein TIA-1 regulates COL2A1 alternative splicing and interacts with precursor mRNA and genomic DNA. J. Biol. Chem. 2007; 282:24444–24454. [DOI] [PubMed] [Google Scholar]
- 8. Zhu H., Hasman R.A., Young K.M., Kedersha N.L., Lou H.. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 2003; 23:5959–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Izquierdo J.M., Majos N., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J.. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005; 19:475–484. [DOI] [PubMed] [Google Scholar]
- 10. Cheng J., Zhou T., Liu C., Shapiro J.P., Brauer M.J., Kiefer M.C., Barr P.J., Mountz J.D.. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994; 263:1759–1762. [DOI] [PubMed] [Google Scholar]
- 11. Suswam E.A., Li Y.Y., Mahtani H., King P.H.. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005; 33:4507–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dixon D.A., Balch G.C., Kedersha N., Anderson P., Zimmerman G.A., Beauchamp R.D., Prescott S.M.. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 2003; 198:475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu Q., Cok S.J., Zeng C., Morrison A.R.. Translational repression of human matrix metalloproteinases-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR). J. Biol. Chem. 2003; 278:1579–1584. [DOI] [PubMed] [Google Scholar]
- 14. Damgaard C.K., Lykke-Andersen J.. Translational coregulation of 5΄TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011; 25:2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waris S., Wilce M.C., Wilce J.A.. RNA recognition and stress granule formation by TIA proteins. Int. J. Mol. Sci. 2014; 15:23377–23388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kedersha N.L., Gupta M., Li W., Miller I., Anderson P.. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999; 147:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anderson P., Kedersha N.. Stressful initiations. J. Cell Sci. 2002; 115:3227–3234. [DOI] [PubMed] [Google Scholar]
- 18. Sanchez-Jimenez C., Carrascoso I., Barrero J., Izquierdo J.M.. Identification of a set of miRNAs differentially expressed in transiently TIA-depleted HeLa cells by genome-wide profiling. BMC Mol. Biol. 2013; 14:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P.. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004; 15:5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dember L.M., Kim N.D., Liu K.Q., Anderson P.. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996; 271:2783–2788. [DOI] [PubMed] [Google Scholar]
- 21. Lopez de Silanes I., Galban S., Martindale J.L., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M.. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005; 25:9520–9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim H.S., Kuwano Y., Zhan M., Pullmann R. Jr, Mazan-Mamczarz K., Li H., Kedersha N., Anderson P., Wilce M.C., Gorospe M. et al. . Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell. Biol. 2007; 27:6806–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Z., Kayikci M., Briese M., Zarnack K., Luscombe N.M., Rot G., Zupan B., Curk T., Ule J.. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010; 8:e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Afroz T., Cienikova Z., Cléry A., Allain F.H.. One, Two, Three, Four! How multiple rrms read the genome sequence. Methods Enzymol. 2015; 558:235–278. [DOI] [PubMed] [Google Scholar]
- 25. Bauer W.J., Heath J., Jenkins J.L., Kielkopf C.L.. Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. J. Mol. Biol. 2012; 415:727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cruz-Gallardo I., Aroca A., Gunzburg M.J., Sivakumaran A., Yoon J.H., Angulo J., Persson C., Gorospe M., Karlsson B.G., Wilce J.A. et al. . The binding of TIA-1 to RNA C-rich sequences is driven by its C-terminal RRM domain. RNA Biol. 2014; 11:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aroca A., Diaz-Quintana A., Diaz-Moreno I.. A structural insight into the C-terminal RNA recognition motifs of T-cell intracellular antigen-1 protein. FEBS Lett. 2011; 585:2958–2964. [DOI] [PubMed] [Google Scholar]
- 28. Cruz-Gallardo I., Del Conte R., Velazquez-Campoy A., Garcia-Maurino S.M., Diaz-Moreno I.. A non-invasive NMR method based on histidine imidazoles to analyze the pH-modulation of protein-nucleic acid interfaces. Chemistry. 2015; 21:7588–7595. [DOI] [PubMed] [Google Scholar]
- 29. Cruz-Gallardo I., Aroca A., Persson C., Karlsson B.G., Diaz-Moreno I.. RNA binding of T-cell intracellular antigen-1 (TIA-1) C-terminal RNA recognition motif is modified by pH conditions. J. Biol. Chem. 2013; 288:25986–25994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I.. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012; 45:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franke D., Svergun D.I.. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009; 42:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Volkov V.V., Svergun D.I.. Uniqueness of ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2003; 36:860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McPhillips T.M., McPhillips S.E., Chiu H.J., Cohen A.E., Deacon A.M., Ellis P.J., Garman E., Gonzalez A., Sauter N.K., Phizackerley R.P. et al. . Blu-ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron. Radiat. 2002; 9:401–406. [DOI] [PubMed] [Google Scholar]
- 34. Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Grell L., Parkin C., Slatest L., Craig P.A.. EZ-Viz, a tool for simplifying molecular viewing in PyMOL. Biochem. Mol. Biol. Educ. 2006; 34:402–407. [DOI] [PubMed] [Google Scholar]
- 39. Kim H.S., Headey S.J., Yoga Y.M., Scanlon M.J., Gorospe M., Wilce M.C., Wilce J.A.. Distinct binding properties of TIAR RRMs and linker region. RNA Biol. 2013; 10:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang I., Hennig J., Jagtap P.K., Sonntag M., Valcarcel J., Sattler M.. Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1. Nucleic Acids Res. 2014; 42:5949–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Receveur-Brechot V., Durand D.. How Random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 2012; 13:55–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bernado P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I.. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007; 129:5656–5664. [DOI] [PubMed] [Google Scholar]
- 43. Clery A., Blatter M., Allain F.H.. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008; 18:290–298. [DOI] [PubMed] [Google Scholar]
- 44. Kim H.S., Wilce M.C., Yoga Y.M., Pendini N.R., Gunzburg M.J., Cowieson N.P., Wilson G.M., Williams B.R., Gorospe M., Wilce J.A.. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011; 39:1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lukavsky P.J., Daujotyte D., Tollervey J.R., Ule J., Stuani C., Buratti E., Baralle F.E., Damberger F.F., Allain F.H.. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol. 2013; 20:1443–1449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.