Abstract
A major gap in our understanding of ribosome assembly is knowing the precise function of each of the ∼200 assembly factors. The steps in subunit assembly in which these factors participate have been examined for the most part by depleting each protein from cells. Depletion of the assembly factor Erb1 prevents stable assembly of seven other interdependent assembly factors with pre-60S subunits, resulting in turnover of early preribosomes, before the ITS1 spacer can be removed from 27SA3 pre-rRNA. To investigate more specific functions of Erb1, we constructed eight internal deletions of 40–60 amino acid residues each, spanning the amino-terminal half of Erb1. The erb1Δ161–200 and erb1Δ201–245 deletion mutations block a later step than depletion of Erb1, namely cleavage of the C2 site that initiates removal of the ITS2 spacer. Two other remodeling events fail to occur in these erb1 mutants: association of twelve different assembly factors with domain V of 25S rRNA, including the neighborhood surrounding the peptidyl transferase center, and stable association of ribosomal proteins with rRNA surrounding the polypeptide exit tunnel. This suggests that successful initiation of construction of these functional centers is a checkpoint for committing to spacer removal.
INTRODUCTION
Ribosome assembly in eukaryotes involves concerted molecular events of pre-ribosomal RNA (pre-rRNA) processing, modification, and folding, together with ribosomal protein (r-protein) binding, coordinated by the timely entry, action, and exit of ∼200 trans-acting assembly factors (AFs) and >75 small nucleolar RNAs (snoRNAs) (1–3). Most of these AFs are conserved across eukaryotes and essential for cell growth, due to the central role of ribosome function in cellular homeostasis. The AFs include a number of NTPases thought to drive or report ribonucleoprotein (RNP)-remodeling events (4–6). However, the majority of yeast AFs do not contain any predicted enzymatic motifs (1). Thus, they are assumed to participate in structuring rRNA as hubs of protein–protein or protein–RNA interactions, or to act as conduits for communicating structural changes across different RNP neighborhoods within assembling ribosomes. The precise mechanisms by which the non-enzymatic AFs drive assembly forward are only beginning to be understood. This is because their roles in assembly have been characterized primarily by genetically depleting each of them using conditional promoters (e.g. GAL1, TET). While these studies have provided valuable insights into certain aspects of how AFs facilitate ribosome assembly, this approach suffers from a drawback common to studying multicomponent systems using null or conditional mutants. The absence or depletion of a protein abolishes all of its contacts within its multi-molecular complex. This generates a mutant phenotype that is the sum total of all disrupted interactions, often resulting in turnover of preribosomes. A second potential shortcoming of using such conditional null mutants is that they can only reveal the first step in ribosome assembly for which the depleted protein is required.
In the yeast Saccharomyces cerevisiae, ribosome assembly begins in the nucleolus with transcription of pre-rRNA containing sequences for mature 18S, 5.8S and 25S rRNAs, separated and flanked by internal and external transcribed spacers (ITS and ETS), respectively (Supplementary Figure S1). The nascent rRNA is initially packaged together with snoRNPs and a number of r-proteins and AFs to form a 90S preribosome (1,7). Following cleavage at the A2 site, the resulting pre-40S and pre-60S subunits follow separate maturation pathways. Pre-40S subunits (43S preribosomes) are rapidly exported to the cytoplasm. Pre-60S subunits (66S preribosomes) are produced by completion of transcription of 27SA2 pre-rRNA and association of r-proteins and AFs. These 66S preribosomes proceed through a complex maturation pathway as they transit through the nucleolus and nucleoplasm before export into the cytoplasm. Both 43S and 66S preribosomes then undergo final steps of maturation in the cytoplasm, before entering the pool of translationally competent 40S and 60S subunits.
A number of AFs (Erb1, Nop7, Ytm1, Rlp7, Cic1, Nop15, Rrp1, Drs1, Has1, Ebp2, Brx1 and Pwp1) were originally implicated in removal of the ITS1 spacer sequence in 27SA3 pre-rRNA during early stages of 66S pre-rRNP maturation, because their depletion results in accumulation of 27SA3 pre-rRNA and a reduction in its immediate downstream product, 27SB pre-rRNA. Due to this phenotype, these factors have been referred to as ‘A3 factors’ (8–23). However, more recent work indicates that these A3 factors are only indirectly involved in this step in 60S subunit biogenesis, by helping to stabilize preribosomes in which 27SA3 pre-rRNA processing occurs (17). These proteins form a network of direct interactions with each other (Supplementary Figure S2), and consequently exhibit interdependence for association into preribosomes. Depletion of one of the A3 cluster proteins affects stable association of other A3 AFs, leading to turnover of preribosomes before the ITS1 spacer can be removed from 27SA3 pre-rRNA. Thus, any additional downstream assembly defects are masked in these depletion mutants. Therefore, we reasoned that directed mutagenesis of these proteins would allow us to reveal more specific functions, if any, of A3 AFs in 60S ribosomal subunit assembly.
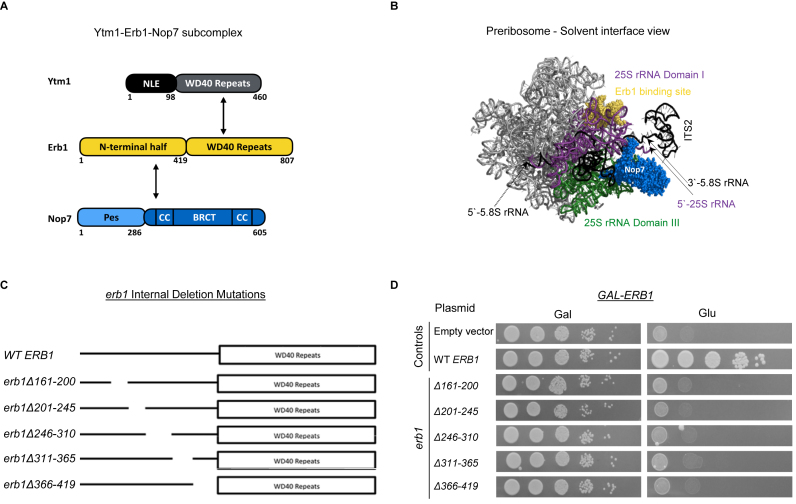
Here, we focused on one of these interdependent AFs, Erb1, to investigate more specific roles that it might play, beyond stabilizing early preribosomes. Erb1 is the central component of a heterotrimeric subcomplex with two other ‘A3’ AFs, Ytm1 and Nop7 (Figure 1A) (22–24). The Ytm1–Erb1–Nop7 subcomplex forms an extensive protein–protein and protein–RNA interaction network within early ribosome assembly intermediates in the nucleolus, thereby potentially serving as a hub for stabilizing or remodeling RNP neighborhoods during 60S subunit biogenesis (Supplementary Figure S2). Erb1 and Ytm1 interact directly with each other through their conserved WD40 motifs (25,26), and Nop7 binds to the amino-terminal half of Erb1 (Figure 1A) (25). Erb1 and Nop7 also contact 25S rRNA in domains I and III, respectively (Figure 1B) (27,28). Thus, this network of protein–protein and protein–RNA interactions might help to establish the tertiary structure of the 25S rRNA domain I/III interface, located adjacent to the ITS2 spacer. Consistent with this idea, mutations in Ytm1 that disrupt Erb1-Ytm1 interactions affect the stable association of Ytm1 and Erb1 with preribosomes, leading to accumulation of 27SA3 pre-rRNA and turnover of preribosomes (22,23,26).
Figure 1.
Distinct portions of the N-terminal half of Erb1 are required for growth. (A) Schematic representation of the heterotrimeric subcomplex formed between Ytm1, Erb1 and Nop7. (B) PyMOL structure depicting the Erb1 binding site (yellow) in 25S rRNA domain I (purple), and the Nop7 protein (blue) bound to 5.8S rRNA/25S rRNA domain III (green) in preribosomes (PDB ID: 3JCT). (C) Schematic representation of internal deletions of highly conserved residues in the N-terminal half of Erb1. (D) Growth of GAL-ERB1 strains containing plasmids encoding wildtype Erb1 or mutant erb1 proteins under the control of the ERB1 promoter, was assayed by spotting ten-fold serial dilutions on solid media containing galactose (C-Trp+Gal) or glucose (C-Trp+Glu). The GAL-ERB1 strain containing the empty vector (pRS314) is included as a negative control.
The release of AFs from preribosomes also might play important roles in driving assembly forward, e.g. by triggering structural transitions or acting as timers to signal progress through the pathway. Ytm1 and Erb1 are removed from preribosomes containing 27SB pre-rRNA, prior to exit of 66S particles from the nucleolus, by the AAA-ATPase Rea1 (29). The MIDO domain in the N-terminal portion of Ytm1 is specifically targeted by the MIDAS domain of the Rea1 for stripping Ytm1 and Erb1 from preribosomes (25,26,30). ATP hydrolysis by the hexameric AAA-motor head domain of Rea1 generates mechanochemical forces to drive this release (25). Specific alterations in the network of interactions among Nop7, Erb1 and Ytm1 during this release step may be important to transduce forces to the preribosome to drive downstream steps. Indeed, when release of Ytm1 and Erb1 by Rea1 is blocked, removal of the ITS2 spacer in 27SBS pre-rRNA does not occur (29). It is not known whether or how Erb1 plays a role in facilitating its release together with Ytm1, or in removal of the ITS2 spacer. Because the Nop7-subcomplex proteins show a high degree of evolutionary conservation, and participate in a similar step of ribosome assembly in humans, it is likely that the mechanisms by which these proteins drive assembly are conserved from yeast to humans (10,12,13,22,24,29–34).
In order to investigate the functions of Erb1 in more detail, we constructed a series of internal deletions across its conserved N-terminal half (Figure 1C and Supplementary Figure S3), and assayed the effects of these mutations on cell growth, pre-rRNA processing, preribosome composition and pre-rRNA folding. We identified two erb1 mutants in which Erb1 could stably associate with preribosomes, allowing us to uncover a role for Erb1 in the removal of the ITS2 spacer. Further analysis of these erb1 mutants demonstrated roles of Erb1 in two other remodeling events: organization of domain V of 25S rRNA on the subunit interface, and folding of 5.8S rRNA positioned between domains I and III of 25S rRNA. Evaluation of many other mutants defective in ITS2 spacer removal reveals identical phenotypes (35–39), leading us to conclude that there is a common set of remodeling events required to initiate ITS2 spacer removal.
MATERIALS AND METHODS
Yeast strains and plasmids
Saccharomyces cerevisiae strains used in this study are listed in Supplemental Table S1. Promoter replacements and epitope tagging in the genome were performed by standard procedures (40). Plasmids containing different promoter or epitope-tag fusions of ERB1 were generated by standard techniques or by Gateway cloning (41), and introduced into yeast strains by transformation. Deletions of distinct portions of ERB1 were done using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) and verified by DNA sequencing. For spotting assays, strains expressing wildtype ERB1 or mutant erb1 genes were grown in galactose-containing selective media and diluted to an OD600 of ∼0.8 (4 × 107 cells/ml). Cells were then spotted as ten-fold serial dilutions up to 10−5 on solid selective media containing galactose or glucose, and incubated at 30°C. Although each mutant is presented on individual panels (Figure 1D), all strains were spotted at the same time on identical batches of media on one or two plates, together with positive and negative controls (likewise for Supplementary Figures S4A and C and S5). For pre-rRNA processing assays and purification of preribosomes, yeast strains were grown in selective media containing galactose, shifted to selective media containing glucose for 16 h to deplete endogenous wildtype Erb1, and harvested in mid-log phase of growth, unless specified otherwise.
Yeast two-hybrid analysis
Plasmids expressing YTM1 fused at its N-terminus to GAL4-AD (activation domain), and wildtype or mutant ERB1 fused at its N-terminus to GAL4-DBD (DNA binding domain) were transformed into yeast two-hybrid strains PJ69-4α and PJ69-4A, respectively (42). These yeast strains were mated to test for protein–protein interactions, assayed by activation of GAL-HIS3 or GAL-ADE2 reporter genes, as described previously (22).
Steady-state analysis of pre-rRNA processing
Total RNA was isolated from yeast lysates by acid phenol:chloroform:isoamyl alcohol (Ambion) extraction. Steady-state levels of precursor and mature rRNAs were assayed by primer extension and northern blotting as previously described (16). Oligonucleotide probes used in this study are available upon request.
Affinity purification of preribosomes
Single step affinity purification of pre-60S particles was performed using yeast extracts prepared with TNM150 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 0.1% NP40 and 5 mM ß-mercaptoethanol). Whole-cell lysates were incubated with immunoglobulin G (IgG)-coated Dynabeads (Invitrogen) to purify preribosomes using TAP-tagged AFs as baits (30 μl beads/150 ml of mid-log phase yeast cultures). Preribosomes were eluted by cleavage of the TAP tag with 10 U AcTEV protease (Life Technologies). Proteins were separated on 4–12% NuPAGE or 4–20% Tris–HCl SDS-PAGE gels (Life Technologies). Silver staining and western blotting were performed according to standard procedures. The anti-Nop7 and anti-rpL5 antibodies were generated in the Woolford laboratory, and the anti-DBD antibody was purchased from Santa Cruz Biotech Inc. (sc-577). Sources of other antibodies are listed in the Acknowledgements.
Analysis of preribosomes by iTRAQ mass spectrometry
Preribosomes were purified from yeast cell lysates prepared from 1 l of cell cultures using TAP-tagged AFs Rpf2 or Nsa2 as baits, as described above. IgG-coated beads with bound preribosomes were washed and subjected to AcTEV protease treatment in TNM150 buffer without detergent. Purified preribosomes were TCA precipitated, and dried pellets were sent to Penn State Hershey Core Research Facilities for trypsin digestion and multiplex labeling with iTRAQ reagents (Applied Biosystems). Peptides were separated by 2D liquid chromatography and parent ions were identified on a Sciex/ABI 5800 MALDI-TOF mass spectrometer. Proteins identified with >95% confidence were used for data analysis. For each pair-wise comparison, data were normalized to the change in ratio of the bait protein. For Rpf2-TAP iTRAQ, the average fold change in each protein is reported. Normalized ratios of fold changes were averaged and used to calculate the standard error of the mean. We focused on proteins with a fold change greater than two and refer to these as significant changes. Whenever possible, these results were confirmed by western blotting. Processed iTRAQ data are presented in Supplementary Tables S2 (Supplementary Figure S8), S3 (Supplementary Figure S10), and S4 (Figure 4C and Supplementary Figure S7).
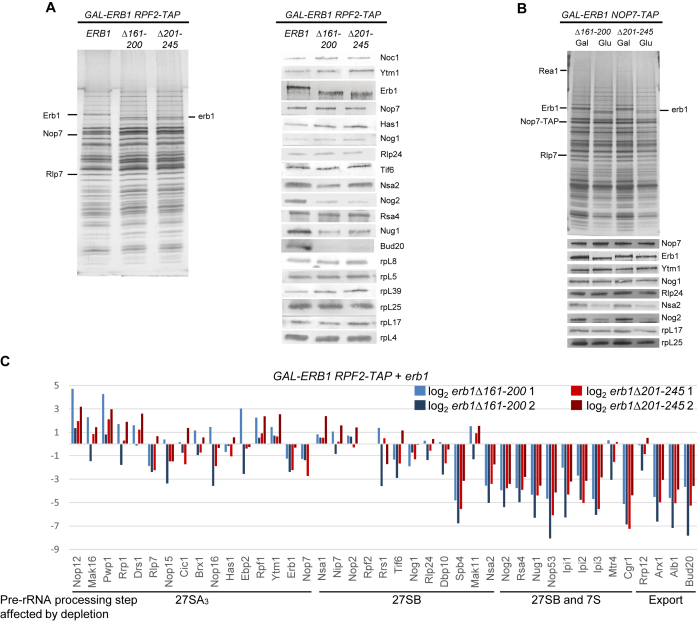
Figure 4.
Assembly factors that bind to domain V fail to associate with preribosomes in the erb1Δ161–200 and erb1Δ201–245 mutants. The protein composition of preribosomes purified from erb1 mutants using TAP-tagged AF Rpf2 (A) or Nop7 (B) as bait was assayed by SDS-PAGE, followed by silver staining. Western blotting was used to assay levels of select AFs and r-proteins in the preribosomes. (C) Relative changes in levels of large subunit AFs (mutant:WT) found in Rpf2-TAP particles were determined by semi-quantitative iTRAQ mass spectrometry. The ratios were normalized to levels of the bait protein, and the fold change of two biological replicates per mutant is present in log2 scale.
Chemical probing of RNA structure in vivo
GAL1-3HA-ERB1 cells containing plasmids expressing wild-type or mutant erb1 protein from the pOBD2 vector were grown to an OD600 of 0.6, washed with PBS and treated with 100 mM 2-methyl nicotinic acid imidazolide (NAI) for 20 min at 30°C. Total RNA was extracted with acid phenol:chloroform:isoamyl alcohol (Ambion). Primer extensions of 5 μg of total RNA extracted from NAI-treated cells were performed using Transcriptor Reverse Transcriptase (Roche Diagnostics) with oligonucleotides designed to base-pair with the ITS2 sequence in 27S pre-rRNA (20).
Molecular visualizations
The structures of pre-60S (3JCT) and 60S (4V88) ribosomal subunits were obtained from Protein Data Base (PDB), and visualized using PyMOL molecular visualization software (28,43).
RESULTS
Conserved regions in the N-terminal half of Erb1 are essential for growth
The domain architecture of Erb1 contains two prominent features: (i) an N-terminal half (residues 1–419), and (ii) a C-terminal half containing WD40 repeats that form a donut–shaped structure (residues 420–807) (Figure 1A). Both of these domains are essential for cell growth and ribosome assembly (Supplementary Figure S4C) (25). We decided not to focus on the C-terminal half of Erb1, because mutations in this region affect removal of the ITS1 spacer from 27SA3 pre-rRNA by perturbing Erb1-Ytm1 interaction and their stable association with preribosomes (22,23,26). To search for additional functions of Erb1 in 60S ribosomal subunit assembly, we targeted its relatively unexplored N-terminal half for mutagenesis. We constructed eight consecutive internal deletions of 40–60 amino acid residues across the N-terminal portion of Erb1 (Figure 1C), and introduced plasmids expressing them into a yeast strain in which genomic ERB1 was placed under the control of a glucose-repressible GAL1 promoter (erb1: GAL-3HA-ERB1). Thus, when shifted from galactose-containing medium to glucose-containing medium, GAL promoter-driven wild-type ERB1 expression was shut off, allowing us to assay effects associated with expression of only the plasmid-borne erb1 mutant alleles.
Since Erb1 is a nuclear protein, we predicted that mutations affecting its nuclear localization would resemble depletion of Erb1 (10,34). Therefore, we expressed the mutant erb1 proteins as fusions containing a nuclear localization sequence (NLS) (DBD-ERB1), or without an NLS (GFP-ERB1 or untagged ERB1), to distinguish mutant phenotypes due to the inability of mutant erb1 protein to enter the nucleus. These tags did not affect the function of the wild-type ERB1 gene; growth and pre-rRNA processing resembled that of a strain expressing untagged Erb1 from its endogenous promoter (Supplementary Figure S4).
Deletions within the relatively less conserved N-terminal 150 amino acids of Erb1 (erb1Δ3-50, erb1Δ51-100 and erb1Δ101-160) did not affect growth or pre-rRNA processing when NLS-containing erb1 fusion constructs were used (Supplementary Figure S5 and data not shown). However, deletion mutant erb1Δ51–100 lacking a fused NLS (GFP-erb1Δ51–100) failed to grow in glucose, suggesting that this region might be required for nuclear localization of Erb1 (Supplementary Figure S5). In contrast, the erb1Δ161–200, erb1Δ201–245, erb1Δ246–310, erb1Δ311–365 and erb1Δ366–419 mutants failed to grow either in the presence or the absence of an NLS (Figure 1D and Supplementary Figure S5). Thus, we focused on these NLS-independent inviable mutants. Most of the remaining experiments were done using both endogenous Erb1 and DBD fusions to Erb1, and identical results were obtained. Unless indicated otherwise, results are shown for endogenous constructs.
27SB pre-rRNA processing is affected in the erb1Δ161–200 and erb1Δ201–245 mutants
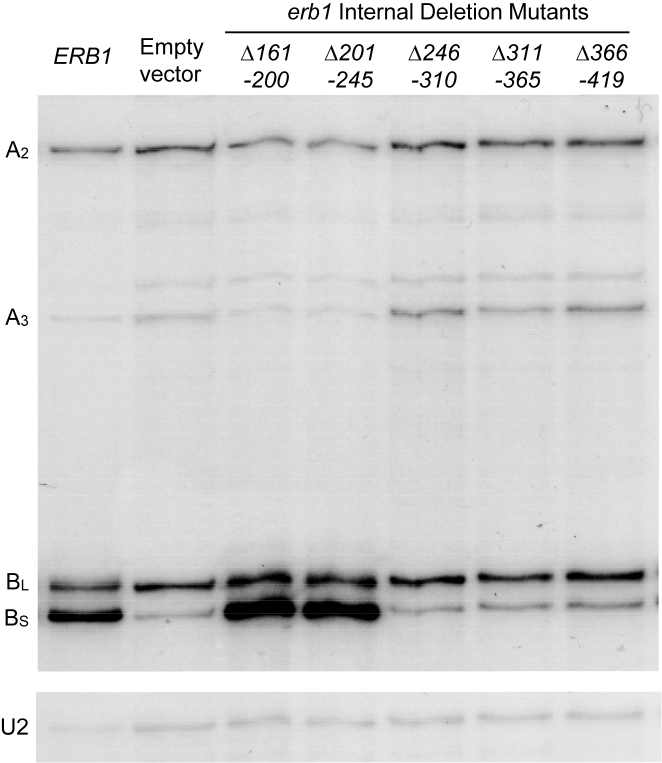
To examine effects of the lethal erb1 mutations on pre-rRNA processing, we assayed levels of pre-rRNA intermediates. Primer extension indicated that the erb1Δ246–310, erb1Δ311–365, and erb1Δ366–419 mutants accumulated 27SA3 pre-rRNA, similar to the conditional null GAL-ERB1 mutant (Figure 2). In contrast, the erb1Δ161–200 and erb1Δ201–245 mutants accumulated 27SB pre-rRNA containing the uncleaved ITS2 spacer (Figure 2). The block in cleavage at the C2 site in ITS2 in these mutants was confirmed by northern blotting, which demonstrated an accumulation of 27SB pre-rRNA and decreased levels of 7S pre-rRNA, relative to wild-type amounts (data not shown). This is the first evidence for a role of Erb1 in ITS2 removal in S. cerevisiae. Bop1, the mammalian homolog of Erb1, was previously implicated in ITS2 processing, but it was not known whether this was due to species-specific functional specialization of Bop1 (31,32). Not surprisingly, the Bop1Δ mutant that is blocked in ITS2 cleavage lacks N-terminal residues 1–231, portions of which are absent in the yeast erb1Δ161–200 and erb1Δ201–245 mutants. Together, these results support evolutionarily conserved functions for different domains of Erb1/Bop1 in the removal of the ITS1 and ITS2 spacers from pre-rRNA during ribosome assembly in eukaryotes.
Figure 2.
The erb1Δ161–200 and erb1Δ201–245 mutants are defective in processing of 27SB pre-rRNA. Total RNA was extracted from GAL-ERB1 strains carrying plasmids containing no insert (pRS314), wildtype ERB1 or erb1 mutant alleles grown in C-Trp+Gal media and shifted to C-Trp+Glu, and assayed by primer extension. Amounts of U2 snRNA assayed in the same primer extension reaction were used as the loading control.
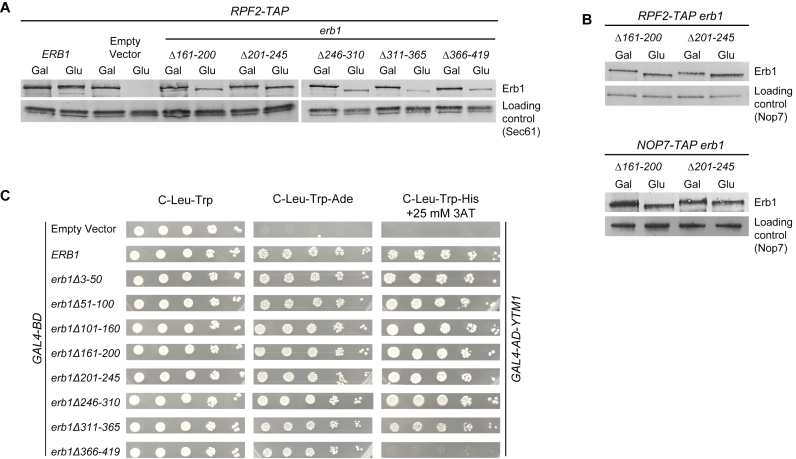
The erb1 mutant proteins erb1Δ161–200 and erb1Δ201–245 assemble stably into preribosomes
To investigate the ability of erb1 mutant proteins to enter preribosomes, we used TAP-tagged AFs Nop7 and Rpf2 to affinity purify assembly intermediates from erb1 mutants, and assayed for the presence of mutant erb1 proteins by western blotting. Nop7 and Rpf2 were used as baits because they assemble into early 66S preribosomes and dissociate during late nuclear steps of 60S subunit assembly, and thus can be used to broadly survey nuclear pre-60S subunits (9,27,44). The erb1Δ246–310, erb1Δ311–365 and erb1Δ366–419 mutant proteins associated poorly or not at all with preribosomes (Supplementary Figure S6 and data not shown), although these mutant proteins were stably expressed (Figure 3A). Consistent with lack of efficient assembly of these erb1 mutant proteins into preribosomes, the SDS-PAGE profile of proteins in preribosomes purified from cells expressing these alleles resembled that from Erb1 depleted strains (data not shown). In contrast, the erb1Δ161–200 and erb1Δ201–245 mutant proteins were both stably expressed (Figure 3A) and efficiently assembled into preribosomes (Figure 3B). Thus, in the remaining work we focused on the erb1Δ161–200 and erb1Δ201–245 mutants, which seemed likely to reveal new functions for Erb1.
Figure 3.
The erb1Δ161–200 and erb1Δ201–245 mutant proteins assemble into preribosomes and interact with Ytm1 protein. (A) Stable expression of each of the mutant erb1 proteins was assayed by western blotting of whole cell extracts using antibodies against Erb1. Sec 61 was used as a loading control. (B) The ability of mutant erb1 proteins to assemble into pre-60S ribosomes was assayed using TAP-tagged AFs Rpf2 or Nop7 as baits to purify preribosomes, followed by western blotting. (C) The yeast two-hybrid assay for interaction of Erb1 with Ytm1. Ten-fold serial dilutions of yeast were spotted on C-Leu-Trp, C-Leu-Trp-Ade or C-Leu-Trp-His + 25 mM 3AT media. Wildtype Erb1 and Ytm1 proteins exhibit a positive interaction, allowing yeast cells to grow in the absence of adenine or histidine. The strength of interaction was assayed by the ability of strains to grow in C-Leu-Trp-His medium containing 25 mM 3AT.
Interactions between Erb1 and Ytm1 or Nop7 may play important roles in 60S subunit assembly, e.g. to propagate forces to the preribosome when Ytm1 and Erb1 are removed (25,29). To test whether the interaction of erb1Δ161–200 or erb1Δ201–245 with Ytm1 was perturbed, we used the yeast two-hybrid system (Figure 3C). All of the erb1 internal deletion mutant proteins, except for erb1Δ366–419, exhibited wildtype interactions with Ytm1, as indicated by a stringent criterion; the growth of yeast two-hybrid reporter strains on C-His media containing 25 mM 3AT.
The erb1Δ161–200 and erb1Δ201–245 mutations affect stable association with preribosomes of AFs required for ITS2 spacer removal
To investigate assembly defects in the erbΔ161–200 and erb1Δ201–245 mutants blocked in removal of the ITS2 spacer, we examined the composition of preribosomes purified using TAP-tagged AF Rpf2 or Nop7 as baits. In addition to SDS-PAGE and western blotting (Figure 4A and B), we used semi-quantitative mass spectrometry (iTRAQ) to pinpoint changes in protein composition of mutant preribosomes (Figure 4C and Supplementary Figures S7 and S8). Compared to wildtype preribosomes, the mutant pre-rRNPs contained increased amounts or showed no significant changes in amounts of early-acting AFs (Supplementary Figures S7 and S8B). The A3 AFs (Pwp1, Ebp2, Brx1, Cic1, Rlp7, Nop15, Nop12, Drs1, Has1, Erb1, Ytm1 and Nop7) also were marginally increased or showed no changes (Figure 4B and C) (8–24). Likewise, AFs that are required for cleavage at the C2 site in ITS2 but enter preribosomes early in the pathway, into pre-rRNPs containing 27SA2 pre-rRNA (Nop2, Rrs1, Tif6, Nog1 and Rlp24), did not change significantly (Figure 4B and C) (35,45–50). In contrast, there were decreased amounts of other AFs that enter preribosomes immediately prior to C2 cleavage (Nsa2, Nug1, Rsa4, Nop53, Nog2, Ipi1, Ipi2/Rix1, Ipi3), one AF that assembles into early preribosomes containing 27SA2 pre-rRNA (Spb4), and two AFs whose entry times are not yet known (Cgr1 and Spb1) (20,51–58). Interestingly, all of these factors that were present in decreased amounts in preribosomes, with the exception of Spb4, have been shown to act on or bind (directly or in multi-protein complexes) to domain V of 25S rRNA (28,59–62). In addition, the silver-stained SDS-PAGE gel of pre-ribosomal proteins co-purifying with Nop7-TAP showed a decrease in intensity of the band corresponding to the AAA-ATPase Rea1 (Figure 4B).
Erb1 is required for proper folding of 5.8S rRNA at the interface of domains I and III of 25S rRNA, and for binding of r-proteins to 5.8S rRNA
Since the erb1Δ161–200 and erb1Δ201–245 mutations block cleavage of the C2 site in ITS2, we hypothesized that Erb1 could play a role in structuring the ITS2 spacer RNA. Therefore, we examined effects of these mutations on pre-rRNA folding in vivo, using chemical modification with NAI followed by primer extension (SHAPE) (63). However, nucleotides in ITS2 showed relatively few modifications in the erb1Δ161–200 and erb1Δ201–245 mutants, indicating that ITS2 RNA structure is largely intact (Supplementary Figure S9). Consistent with this observation, the levels of AFs Nop15, Rlp7 and Cic1 that specifically bind to ITS2 (27,28,64,65) showed no significant changes in preribosomes purified from these mutants (Figure 4C and Supplementary Figure S8A).
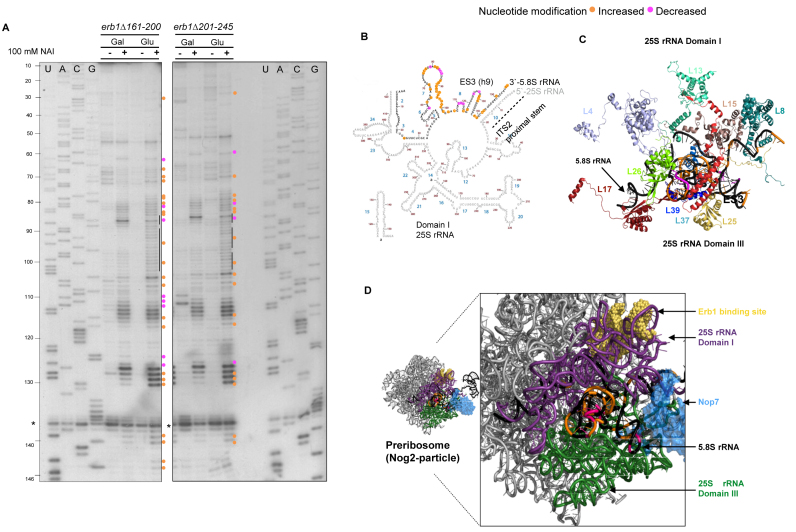
Erb1 and Nop7 bind to domains I and III of 25S rRNA, respectively, at positions where these domains exhibit significant contact with each other (Figure 5D) (27,28). Thus, it is possible that the heterotrimeric Ytm1–Erb1–Nop7 subcomplex could bridge these domains together to establish rRNA tertiary structure, or serve as a hub for conveying RNP remodeling across the domain I/III interface of 25S rRNA during ribosome assembly. Since 5.8S rRNA is sandwiched between domains I and III of 25S rRNA, we reasoned that its folding can be used as an indicator of rRNA folding in the domain I/III interface. Folding of 5.8S rRNA was assayed by chemical probing with NAI and primer extension (64) using an oligonucleotide probe that hybridizes to the 5΄-end of ITS2 (20), allowing us to identify changes in 5.8S rRNA structure specifically in preribosomes. In contrast to ITS2 RNA, 5.8S rRNA showed extensive modifications in the two erb1 mutants compared to wildtype cells, specifically at the binding sites of r-proteins L17, L35, L37, L39, L25 and L26 (Figure 5). The presence of these r-proteins is required for C2 cleavage, and their binding is tightened during or after removal of ITS2 (17,38,39,66–68). Consistent with this observation, the amounts of these r-proteins copurifying with Nsa2-TAP-purified preribosomes were reduced in erb1Δ161–200 and erb1Δ201–245, indicating that these r-proteins are not stably bound, perhaps because they fail to transition from weak, initial contacts to form stable, final interactions with rRNA (Supplementary Figures S10B and D).
Figure 5.
5.8S rRNA is not folded properly in the erb1Δ161–200 and erb1Δ201–245 mutants. (A) In vivo SHAPE assay of 5.8S rRNA structure in GAL-ERB1 + erb1 mutants grown in galactose, or grown in galactose and shifted to glucose. Nucleotides with increased or decreased modification are indicated with orange and pink circles, respectively. RNA isolated from cells treated with DMSO alone were used as controls (indicated by –). The sequencing ladders are shown and nucleotide positions are indicated (U, A, C, G) for 5.8S rRNA. The common stop (*) in pre-rRNA was used to assess loading. (B) Locations of nucleotides modified in the erb1 mutants are displayed on the 5.8S rRNA/domain I of 25S rRNA structure (www.ribovision.org). Nucleotide numbers (brown) and helices (blue) in 5.8S rRNA are marked. (C) PyMOL representation of modified residues in 5.8S rRNA with the r-proteins that bind it (PDB ID: 4V88). (D) The 5.8S rRNA is sandwiched between 25S rRNA domains I (purple) and III (green) (PDB ID: 3JCT) (28). The structure of Nop7 protein (blue) and the rRNA binding site of Erb1 (yellow) are also shown.
Do the erb1Δ161-200 and erb1Δ201-245 mutations prevent release of Ytm1 and Erb1 from preribosomes?
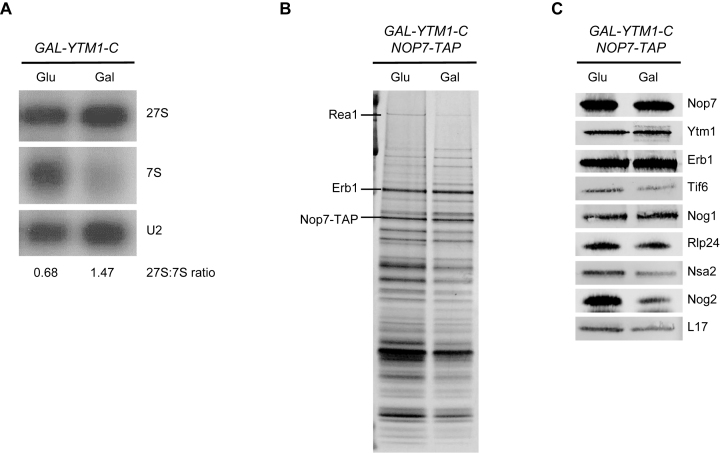
The YTM1E80A mutation prevents interaction of the AAA-ATPase Rea1 with Ytm1 and consequently blocks release of Ytm1 and Erb1 from preribosomes (29). This mutation also prevents removal of the ITS2 spacer, causes an accumulation of early AFs in preribosomes, and blocks efficient association of AFs with domain V of 25S rRNA (29). We observed the same three phenotypes upon expression of the Ytm1-C mutant protein lacking the entire N-terminal domain of Ytm1 required for interaction with Rea1 (Figure 6). Because the erb1Δ161–200 and erb1Δ201–245 mutations cause these same phenotypes, including decreased assembly of Rea1, we considered the possibility that the primary defect of these two erb1 mutations might be preventing the function and/or release of Ytm1 and Erb1 from preribosomes. However, we did not observe significant changes in the amounts of Ytm1 or Erb1 in preribosomes isolated from either erb1 mutant, using Rpf2-TAP or Nop7-TAP as baits (Figure 4 and Supplementary Figures S8 and S10), which suggests that release of Erb1 and Ytm1 is not blocked in these mutants.
Figure 6.
Removal of the ITS2 spacer is blocked in the YTM1-C mutant. (A) Pre-rRNA processing in the dominant negative mutant YTM1-C was assayed by northern blotting. U2 snRNA was used as the loading control. The protein composition of preribosomes purified from the YTM1-C mutant using TAP-tagged AF Nop7 as bait was assayed by SDS-PAGE (B), and western blotting (C).
DISCUSSION
In this study, we uncovered new functions for the Nop7-subcomplex protein Erb1 in 60S ribosomal subunit assembly. We assayed internal deletions of ∼50 residue-long intervals in the N-terminal domain of Erb1 (1–419). The first 160 amino acid residues that are predicted to be disordered (Supplementary Figure S3), are not essential for ribosome assembly, except for a stretch potentially involved in nuclear localization of Erb1 (amino acid residues 51–100). Erb1 proteins lacking residues 246–310, 311–365 or 366–419 are expressed but do not efficiently associate with preribosomes, which prevents the stable association of the other interdependent A3 AFs with preribosomes. This results in the same phenotype as Erb1 depletion: destabilization of early assembly intermediates before ITS1 can be removed from 27SA3 pre-rRNA (17). In contrast, the erb1Δ161–200 and erb1Δ201–245 mutant proteins can assemble into preribosomes, but cleavage at the C2 site in ITS2 is blocked. We observed two other defects in these two erb1 mutants: (1) AFs that bind 25S rRNA domain V and are themselves required for cleavage and processing of 27SB pre-rRNA fail to stably associate with preribosomes. (2) The ITS2 proximal stem and 5.8S rRNA fail to fold properly. Consistent with this, we also observed that stable assembly of r-proteins that bind this neighborhood is affected in preribosomes purified from these two erb1 mutants.
Erb1 plays a role in structuring 25S rRNA domain V, including the peptidyl transferase center
Analysis of preribosomes from the erb1Δ161–200 and erb1Δ201–245 mutants revealed sixteen AFs whose levels decreased (Figure 4 and Supplementary Figure S8). Twelve of these proteins are required for either C2 cleavage (Nsa2, Spb1, Spb4, Cgr1) or both C2 cleavage and subsequent exonucleolytic processing of ITS2 (Nog2, Nug1, Rsa4, Nop53, Ipi1, Ipi2/Rix1, Ipi3, Rea1) (53,54,56,69–73). Sda1 is necessary only for the downstream step, processing of 7S pre-rRNA (74). The roles in pre-rRNA processing for Arx1, Alb1 and Bud20, remain to be determined.
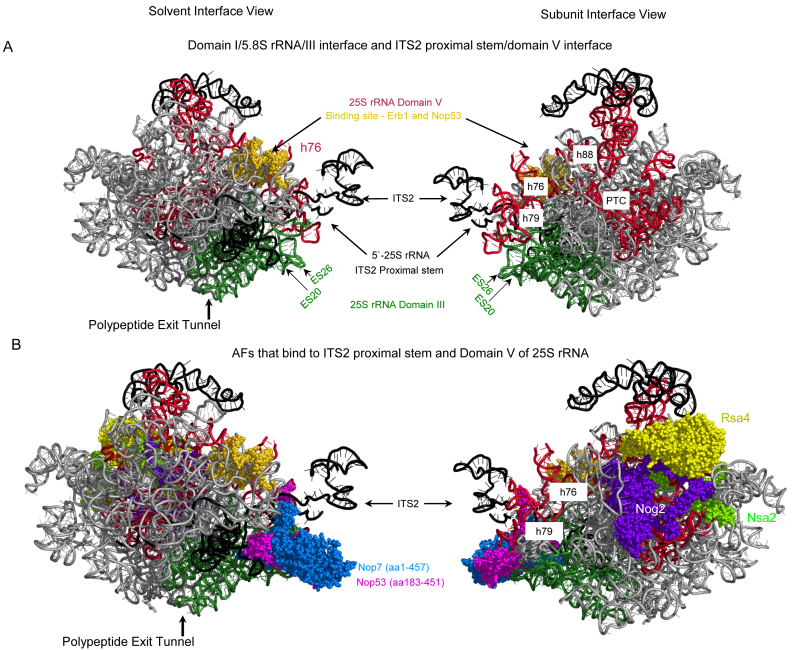
Recent high-resolution cryo-EM structures and protein–RNA crosslinking studies revealed that Nsa2, Nog2, Nug1 and Rsa4 bind to domain V of 25S rRNA in or immediately surrounding what will become the peptidyl transferase center of the 60S subunit (28,59–62) (Figure 7B). The AF Spb1 is a methyltransferase responsible for modification of G2922 in 27S pre-rRNA (75) and thus must contact the nascent peptidyl transferase center. Nop53 and Sda1 also have binding sites in domain V of 25S rRNA, and Ipi1, Ip12/Rix1, Ip13 and Rea1 bind to each other and Sda1 (28,61). We conclude that in the erb1 mutants, the structure of 25S rRNA domain V is not configured properly to enable optimal binding and/or recruitment of these AFs required for C2 cleavage. Alternatively, domain V of 25S rRNA might not fold properly as a result of the failure of these proteins to stably assemble into preribosomes. Thus, in wildtype cells, the stable association of these AFs with domain V in 27S pre-rRNA requires the function of Erb1, and might be necessary for reconfiguring this domain of rRNA to facilitate C2 cleavage in the ITS2 spacer (Figure 7).
Figure 7.
rRNA and AF neighborhoods affected in the erb1Δ161–200 and erb1Δ201–245 mutants. (A) Images of preribosomes showing the rRNA structural elements contacted by Erb1. Helices h76 and h88 in domain V (red) of 25S rRNA contact the binding site of Erb1 and Nop53 (yellow spheres) in domain I of 25S rRNA (27,28,81). The 3΄-end of 5.8S rRNA base-pairs with the 5΄-end of 25S rRNA in domain I to form the ITS2 proximal stem, which is also in close proximity with helix h79 (ES31). The 5΄-portion of 5.8S rRNA lines the polypeptide exit tunnel. (B) Nop7 binds to domain III of 25S rRNA (ES26), the ITS2 proximal stem, and h71 in domain V of 25S rRNA (27,28). The AFs Nsa2, Nog2, and Rsa4, whose association with preribosomes is perturbed in erb1Δ161–200 and erb1Δ201–245 mutants, bind next to the peptidyl transferase center (PTC) in domain V. Nop53 binds domain V of 25S rRNA and ITS2 proximal stem, adjacent to Nop7. Only partial structures of Nop7 and Nop53 are resolved in cryo-EM structures of Nog2-purified preribosomes (PDB ID: 3JCT) (28).
Erb1 is required for structuring 5.8S rRNA, including the polypeptide exit tunnel
Proper folding of ITS2, and formation of the ITS2 proximal stem via base-pairing between the 5΄-end of 25S rRNA and the 3΄-end of 5.8S rRNA, are required for cleavage at the C2 site (76–78). The structure of ITS2 is largely intact in the erb1Δ161–200 and erb1Δ201–245 mutants (Supplementary Figure S9). However, changes in the pattern of chemical modifications of 5.8S rRNA and decreased levels of r-proteins that bind to it suggest an inability to correctly structure the ITS2 proximal stem and 5.8S rRNA in these erb1 mutants (Figure 5, and Supplementary Figure S10). Depletion of r-proteins that bind to 5.8S rRNA or mutations affecting their rRNA contacts also impair ITS2 cleavage (17,38,39,66–68). Most of these r-proteins are located around the exit site of the polypeptide exit tunnel in mature 60S subunits and, thus might play important roles in construction of the tunnel and its exit site. While these r-proteins first associate with early ribosome assembly intermediates, they are thought to transition from encounter complexes to form more stable contacts with preribosomes just before or during cleavage at the C2 site in ITS2 (17,38,39,68). Thus, the erb1 mutations could be preventing this transition to stabilize the binding of r-proteins to the polypeptide exit tunnel neighborhood in the domain III/5.8S rRNA interface.
In preribosomes purified using Rpf2-TAP, we did not observe many significant changes in amounts of r-proteins associated with pre-ribosomes, indicating their entry and association is not affected during earlier stages of 60S subunit assembly, i.e. prior to ITS2 cleavage (Figure 4 and Supplementary Figure S8C). Conversely, in particles purified using the bait Nsa2-TAP, we observed a striking, selective decrease in amounts of r-proteins that bind to the interface formed between 25S rRNA domains I, III and 5.8S rRNA (Supplementary Figures S10B and D). In the erb1 mutants, Nsa2, which enters 27SB pre-rRNA containing particles (69), is weakly bound to preribosomes. By using Nsa2 as bait, we captured pre-ribosomes at the stage when their assembly is blocked, revealing the effects of the erb1Δ161–200 and erb1Δ201–245 mutations on the r-proteins that bind 5.8S rRNA. Since the erb1Δ161–200 and erb1Δ201–245 mutations affect stable binding of 5.8S rRNA binding r-proteins to Nsa2-TAP purified pre-ribosomes, and not to Rpf2-TAP purified particles, we conclude that these r-proteins bind preribosomes during early steps of 60S ribosomal subunit assembly, but fall off from preribosomes in the erb1 mutants failing to undergo ITS2 cleavage, due to their inability to form final contacts with the rRNA.
Do the erb1Δ161–200 and erb1Δ201–245 mutations affect other remodeling events?
Erb1 has an extensive molecular interaction network including a number of proteins involved in C2 cleavage (Supplementary Figure S2). Thus, the erb1Δ161–200 and erb1Δ201–245 mutations could be perturbing one or more of these networks in a way that prevents remodeling events required for C2 cleavage. Several observations suggest that these erb1 mutations could block removal of Erb1 and Ytm1 from preribosomes: (i) Ytm1 and most of Erb1 seem likely to be released from preribosomes before Nog2 assembles and before C2 cleavage occurs. In support of this idea, in wild type cells, Ytm1 is undetectable and amounts of Erb1 are diminished significantly in preribosomes purified using Nog2-TAP compared to those purified using earlier assembly intermediates, such as Nsa1-TAP (Hailey Brown and Joe Park, Personal Communication). In addition, Nog2 enters pre-60S subunits before cleavage at the C2 site. (ii) Mutations in Ytm1 that prevent its release from preribosomes block C2 cleavage (Figure 6) (29). (iii) Rea1, which is necessary for the release of Ytm1 from preribosomes, does not copurify with preribosomes from the erb1Δ161–200 and erb1Δ201–245 mutants. Furthermore, both the YTM1 mutants and the erb1Δ161–200 and erb1Δ201–245 mutants affect assembly of domain V containing the PTC and the exit of the PET.
However, several other results are inconsistent with the hypothesis that the erb1Δ161–200 and erb1Δ201–245 mutations affect release of Ytm1 and Erb1: (i) We did not observe increased amounts of Ytm1 or Erb1 in preribosomes isolated from these erb1 mutants. (ii) The exceptionally strong yeast two-hybrid interactions between wild type Erb1 and Ytm1 were not affected by the erb1Δ161–200 and erb1Δ201–245 mutations in the N-terminal portion of Erb1. This is not surprising, as it has been shown that Erb1 and Ytm1 interact through their C-terminal WD40 repeat domains (25,26). Thus, it seems likely that these deletions of the N-terminal portion of Erb1 might trigger effects on the PTC and PET by other means, perhaps through disrupted interactions with Nop7 or its other interaction partners (Supplementary Figure S2). Consistent with this idea, Nop7 is thought to interact with the N-terminal portion of Erb1 (25,26).
Because Erb1 binds domain I and Nop7 binds domain III, the Erb1–Nop7 interaction can provide a structural bridge between these domains in 25S rRNA (Figure 7B) (27,28). The 5.8S rRNA and the ITS2 proximal stem are positioned in between these domains. Therefore, rearrangements of Nop7–pre-rRNA interactions caused by the erb1Δ161–200 and erb1Δ201–245 mutations might prevent restructuring of the ITS2 proximal stem neighborhood necessary for ITS2 processing. In support of this idea, deletion of helices ES20 and ES26 contacted by Nop7 in domain III of 25S rRNA blocks ITS2 cleavage (Figure 7A) (79).
Preribosomes purified from erb1 mutants contain lower amounts of the AF Nop53 (Figure 4C), which binds to the domain I/III interface and functions as an adaptor to recruit Mtr4, a component of the exosome that processes ITS2 (28,80,81). Nop53 cross-links to the same helices in domain I of 25S rRNA as Erb1 (helix 51), and binds the 5.8S rRNA/domain III interface next to Nop7 (ES26) (Figure 7B). Because the binding sites on rRNA of the later-entering AF Nop53 overlap with those of Erb1 (h51) and Nop7 (ES26) that assemble early in the pathway, it is likely that distortions of the neighborhood containing Erb1 and Nop7 might impact proper binding of Nop53 with domains I and III of 25S rRNA before C2 cleavage.
A consistent pattern of remodeling defects emerges from analysis of multiple mutants blocked in cleavage of the C2 site
In addition to Erb1, at least 24 other AFs or r-proteins are necessary or important for cleavage of the C2 site in ITS2 (reviewed in (1)). The effects of depleting these proteins on preribosome composition have been examined by western blotting and, in some cases, quantified in greater detail by iTRAQ or SILAC mass spectrometry (35–39). In each case, we observe phenotypes quite similar to those of the erb1 mutants: (i) AFs that bind domain V of 25S rRNA are diminished, and (ii) 5.8S rRNA structure is perturbed, as r-proteins that bind to the domain I/5.8SrRNA/III interface fail to stably associate with preribosomes. These common effects of mutations that block C2 cleavage indicate that in wildtype cells several remodeling events are necessary to create a proper substrate in preribosomes for cleavage at the C2 site: binding of multiple AFs to domain V of 25S rRNA, and stabilization of the domain I/ III interface including 5.8S rRNA.
The coordination between these remodeling events involving construction of the peptidyl transferase center and the polypeptide exit tunnel suggests that there is a critical checkpoint for successful initiation of construction of functional centers before committing to the irreversible step of spacer removal during biogenesis of the large 60S ribosomal subunit.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gordon Rule, C. Joel McManus, Jeffrey Brodsky, Jelena Jakovljevic, Madhumitha Ramesh, Karen Kormuth, Beril Tutuncuoglu, Amber LaPeruta and Dan Wilson for critical discussions and reading of the manuscript. We also thank Jill Dembowski for help with iTRAQ data analysis. We are grateful to the following people for their generous gifts of antibodies: Jesus de la Cruz and Patrick Linder (Erb1, Ytm1, Has1 and Rsa4), Janine Maddock (Nog1), Cosmin Saveanu and Michelline Fromont-Racine (Rlp24, Nsa2, Tif6 and Nog2), Arlen Johnson (rpL8), Sabine Rospert (rpL17), K. Siegers (rpL25), Vikram Panse (Nug1 and Bud20), Maurice Swanson (rpL39), Herbert Tschochner (Noc1), Lasse Lindahl (rpL4) and Jeffrey Brodsky (Sec 61).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 GM028301 to J.L.W.]. Funding for open access charge: NIH [R01 GM028301 to J.L.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Woolford J.L., Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nerurkar P., Altvater M., Gerhardy S., Schütz S., Fischer U., Weirich C., Panse V.G.. Eukaryotic ribosome assembly and nuclear export. Int. Rev. Cell. Mol. Biol. 2015; 319:107–140. [DOI] [PubMed] [Google Scholar]
- 3. Sharma S., Lafontaine D.L.J.. ‘View from a bridge’: A new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015; 40:560–575. [DOI] [PubMed] [Google Scholar]
- 4. Strunk B.S., Karbstein K.. Powering through ribosome assembly. RNA. 2009; 15:2083–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kressler D., Hurt E., Baßler J.. Driving ribosome assembly. Biochim. Biophys. Acta. 2010; 1803:673–683. [DOI] [PubMed] [Google Scholar]
- 6. Kressler D., Hurt E., Bergler H., Baßler J.. The power of AAA-ATPases on the road of pre-60S ribosome maturation – molecular machines that strip pre-ribosomal particles. Biochim. Biophys. Acta. 2012; 1823:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernández-Pevida A., Kressler D., de la Cruz J.. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscipl. Rev.: RNA. 2015; 6:191–209. [DOI] [PubMed] [Google Scholar]
- 8. Ripmaster T.L., Vaughn G.P., Woolford J.L.. A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. PNAS. 1992; 89:11131–11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C.J. et al. . Composition and functional characterization of yeast 66S ribosome assembly intermediates. 2001; 8:505–515. [DOI] [PubMed] [Google Scholar]
- 10. Pestov D.G., Stockelman M.G., Strezoska Ž, Lau L.F.. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 2001; 29:3621–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu K., Wu P., Aris J.P.. Nucleolar protein Nop12p participates in synthesis of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2001; 29:2938–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oeffinger M., Leung A., Lamond A., Tollervey D., Lueng A.. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA. 2002; 8:626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adams C.C., Jakovljevic J., Roman J., Harnpicharnchai P., Woolford J.L.. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002; 8:150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oeffinger M., Tollervey D.. Yeast Nop15p is an RNA‐binding protein required for pre‐rRNA processing and cytokinesis. EMBO J. 2003; 22:6573–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fatica A., Oeffinger M., Tollervey D., Bozzoni I.. Cic1p/Nsa3p is required for synthesis and nuclear export of 60S ribosomal subunits. RNA. 2003; 9:1431–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horsey E.W., Jakovljevic J., Miles T.D., Harnpicharnchai P., Woolford J.L.. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004; 10:813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sahasranaman A., Dembowski J., Strahler J., Andrews P., Maddock J., Woolford J.L.. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011; 30:4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimoji K., Jakovljevic J., Tsuchihashi K., Umeki Y., Wan K., Kawasaki S., Talkish J., Woolford J.L., Mizuta K.. Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:4574–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Talkish J., Campbell I.W., Sahasranaman A., Jakovljevic J., Woolford J.L.. Ribosome assembly factors Pwp1 and Nop12 are important for folding of 5.8S rRNA during ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2014; 34:1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dembowski J.A., Kuo B., Woolford J.L.. Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res. 2013; 41:7889–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talkish J., Biedka S., Jakovljevic J., Zhang J., Tang L., Strahler J.R., Andrews P.C., Maddock J.R., Woolford J.L.. Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis. RNA. 2016; 22:852–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miles T.D., Jakovljevic J., Horsey E.W., Harnpicharnchai P., Tang L., Woolford J.L.. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005; 25:10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang L., Sahasranaman A., Jakovljevic J., Schleifman E., Woolford J.L.. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol. Biol. Cell. 2008; 19:2844–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hölzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Hiddemann W., Bornkamm G.W. et al. . Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005; 170:367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thoms M., Ahmed Y.L., Maddi K., Hurt E., Sinning I.. Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res. 2016; 44:926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wegrecki M., Rodríguez-Galán O., de la Cruz J., Bravo J.. The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two β-propellers during the assembly of large ribosomal subunits in eukaryotes. Nucleic Acids Res. 2015; 43:11017–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Granneman S., Petfalski E., Tollervey D.. A cluster of ribosome synthesis factors regulate pre‐rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011; 30:4006–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu S., Tutuncuoglu B., Yan K., Brown H., Zhang Y., Tan D., Gamalinda M., Yuan Y., Li Z., Jakovljevic J. et al. . Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016; 534:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baßler J., Kallas M., Pertschy B., Ulbrich C., Thoms M., Hurt E.. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell. 2010; 38:712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romes E.M., Sobhany M., Stanley R.E.. The crystal structure of the Ubiquitin-like domain of ribosome assembly factor Ytm1 and characterization of its interaction with the AAA-ATPase midasin. J. Biol. Chem. 2016; 291:882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strezoska Ž., Pestov D.G., Lau L.F.. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 2000; 20:5516–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strezoska Ž., Pestov D.G., Lau L.F.. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 2002; 277:29617–29625. [DOI] [PubMed] [Google Scholar]
- 33. Grimm T., Hölzel M., Rohrmoser M., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Eick D.. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006; 34:3030–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., Eick D.. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 2007; 27:3682–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L.. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:8646–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lebreton A., Rousselle J.-C., Lenormand P., Namane A., Jacquier A., Fromont-Racine M., Saveanu C.. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008; 36:4988–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohmayer U., Gamalinda M., Sauert M., Ossowski J., Pöll G., Linnemann J., Hierlmeier T., Perez-Fernandez J., Kumcuoglu B., Leger-Silvestre I. et al. . Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One. 2013; 8:e68412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gamalinda M., Jakovljevic J., Babiano R., Talkish J., de la Cruz J., Woolford J.L.. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res. 2013; 41:1965–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gamalinda M., Ohmayer U., Jakovljevic J., Kumcuoglu B., Woolford J., Mbom B., Lin L., Woolford J.L.. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014; 28:198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 41. Alberti S., Gitler A.D., Lindquist S.. A suite of Gateway® cloning vectors for high‐throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007; 24:913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. James P., Halladay J., Craig E.A.. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996; 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M.. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011; 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 44. Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M.P., Hiley S.L., Hughes T., Woolford J.L.. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007; 21:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hong B., Brockenbrough J.S., Wu P., Aris J.P.. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 1997; 17:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Basu U., Si K., Warner J.R., Maitra U.. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 2001; 21:1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hong B., Wu K., Brockenbrough J.S., Wu P., Aris J.P.. Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res. 2001; 29:2927–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saveanu C., Namane A., Gleizes P.-E., Lebreton A., Rousselle J.-C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M.. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003; 23:4449–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jensen B.C., Wang Q., Kifer C.T., Parsons M.. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J. Biol. Chem. 2003; 278:32204–32211. [DOI] [PubMed] [Google Scholar]
- 50. Kallstrom G., Hedges J., Johnson A.. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 2003; 23:4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lebreton A., Rousselle J.-C., Lenormand P., Namane A., Jacquier A., Fromont-Racine M., Saveanu C.. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008; 36:4988–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. García-Gómez J.J., Lebaron S., Froment C., Monsarrat B., Henry Y., de la Cruz J.. Dynamics of the putative RNA helicase Spb4 during ribosome assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 2011; 31:4156–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saveanu C., Bienvenu D., Namane A., Gleizes P.E., Gas N., Jacquier A., Fromont-Racine M.. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001; 20:6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E.. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001; 8:517–529. [DOI] [PubMed] [Google Scholar]
- 55. de la Cruz J, Sanz-Martínez E., Remacha M.. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005; 33:5728–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sydorskyy Y., Dilworth D.J., Halloran B., Yi E.C., Makhnevych T., Wozniak R.W., Aitchison J.D.. Nop53p is a novel nucleolar 60S ribosomal subunit biogenesis protein. Biochem. J. 2005; 388:819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Galani K., Nissan T.A., Petfalski E., Tollervey D., Hurt E.. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 2004; 279:55411–55418. [DOI] [PubMed] [Google Scholar]
- 58. de la Cruz J., Kressler D., Rojo M., Tollervey D., Linder P.. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998; 4:1268–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baßler J., Paternoga H., Holdermann I., Thoms M., Granneman S., Barrio-Garcia C., Nyarko A., Lee W., Stier G., Clark S.A. et al. . A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J. Cell Biol. 2014; 207:481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Matsuo Y., Granneman S., Thoms M., Manikas R.-G., Tollervey D., Hurt E.. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2014; 505:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barrio-Garcia C., Thoms M., Flemming D., Kater L., Berninghausen O., Baßler J., Beckmann R., Hurt E.. Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2016; 23:37–44. [DOI] [PubMed] [Google Scholar]
- 62. Manikas R.-G., Thomson E., Thoms M., Hurt E.. The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 2016; 44:1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spitale R.C., Crisalli P., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y.. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013; 9:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dembowski J.A., Ramesh M., McManus C.J., Woolford J.L.. Identification of the binding site of Rlp7 on assembling 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 2013; 19:1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Babiano R., Badis G., Saveanu C., Namane A., Doyen A., Díaz-Quintana A., Jacquier A., Fromont-Racine M., de la Cruz J.. Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre-60S ribosomal particles. Nucleic Acids Res. 2013; 41:9461–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Beekvelt C.A., de Graaff-Vincent M., Faber A.W., van’t Riet J., Venema J., Raué H.A.. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 2001; 29:5001–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Babiano R., Gamalinda M., Woolford J.L., de la Cruz J.. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol. Cell. Biol. 2012; 32:3228–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Babiano R., de la Cruz J.. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2010; 38:5177–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lebreton A., Saveanu C., Decourty L., Jacquier A., Fromont-Racine M.. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 2006; 281:27099–27108. [DOI] [PubMed] [Google Scholar]
- 70. Kressler D., Rojo M., Linder P., de la Cruz J.. Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 1999; 27:4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moy T.I., Boettner D., Rhodes J.C., Silver P.A., Askew D.S.. Identification of a role for Saccharomyces cerevisiae Cgr1p in pre-rRNA processing and 60S ribosome subunit synthesis. Microbiology. 2002; 148:1081–1090. [DOI] [PubMed] [Google Scholar]
- 72. de la Cruz J, Sanz-Martínez E., Remacha M.. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005; 33:5728–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Galani K., Nissan T.A., Petfalski E., Tollervey D., Hurt E.. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 2004; 279:55411–55418. [DOI] [PubMed] [Google Scholar]
- 74. Dez C., Housley J., Tollervey D.. Surveillance of nuclear-restricted preribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006; 25:1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lapeyre B., Purushothaman S.K.. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004; 16:663–669. [DOI] [PubMed] [Google Scholar]
- 76. Côté C.A., Peculis B.A.. Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res. 2001; 29:2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Peculis B.A., Greer C.L.. The structure of the ITS2-proximal stem is required for pre-rRNA processing in yeast. RNA. 1998; 4:1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Côté C.A., Greer C.L., Peculis B.A.. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA. 2002; 8:786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ramesh M., Woolford J.L.. Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA. 2016; 22:1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Granato D.C., Machado Santelli G.M., Oliveira C.C.. Nop53p interacts with 5.8S rRNA co-transcriptionally, and regulates processing of pre-rRNA by the exosome. FEBS J. 2008; 275:4164–4178. [DOI] [PubMed] [Google Scholar]
- 81. Thoms M., Thomson E., Baßler J., Gnädig M., Griesel S., Hurt E.. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell. 2015; 162:1029–1038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.