Abstract
The zinc finger protein CTCF has been invoked in establishing boundaries between genes, thereby controlling spatial and temporal enhancer activities. However, there is limited genetic evidence to support the concept that these boundaries restrict the search space of enhancers. We have addressed this question in the casein locus containing five mammary and two non-mammary genes under the control of at least seven putative enhancers. We have identified two CTCF binding sites flanking the locus and two associated with a super-enhancer. Individual deletion of these sites from the mouse genome did not alter expression of any of the genes. However, deletion of the border CTCF site separating the Csn1s1 mammary enhancer from neighboring genes resulted in the activation of Sult1d1 at a distance of more than 95 kb but not the more proximal and silent Sult1e1 gene. Loss of this CTCF site led to de novo interactions between the Sult1d1 promoter and several enhancers in the casein locus. Our study demonstrates that only one out of the four CTCF sites in the casein locus had a measurable in vivo activity. Studies on additional loci are needed to determine the biological role of CTCF sites associated with enhancers.
INTRODUCTION
Enhancers activate genes over great distances, likely by serving as hubs for multiple transcription factors (1,2). We recently identified 440 mammary-specific super-enhancers that are bound by the cytokine-sensing signal transducer and activator of transcription (STAT) 5 and other mammary-enriched transcription factors (3). They are also decorated by histones carrying active modifications, such as acetylation of Histone 3 Lysine 27 (H3K27ac) (3,4). These super-enhancers are associated with genes that are expressed preferentially or exclusively in mammary epithelium and are activated up to 1000-fold during pregnancy (3,4). To establish and maintain tightly regulated genetic programs it is essential to ensure that enhancers activate only their respective target genes and to avoid inadvertent spillover to non-target genes. Inappropriate spreading of enhancer activities could result in the deregulation of neighboring non-target genes with potentially adverse effects on cell homeostasis (5–10). At this point, it is not known how the activity of mammary-specific enhancers and super-enhancers is restricted to their respective genes and does not expand to juxtaposed genes that are either expressed specifically in non-mammary cells or across cell types at much lower levels. Understanding the rules defining the search space of enhancers and shedding light on the gene-enhancer-specificity conundrum are at the core of many molecular studies.
It has been suggested that the genome is partitioned into topologically associated domains (TADs) that confine enhancers and their respective target genes and shield them from non-target promoters (11). Such neighborhoods are frequently composed of chromatin loops that are anchored by the zinc finger protein CCCTC-binding factor (CTCF). Insulating genetic units in such neighborhoods would ensure their biological confinement (12,13). Cell-specific super-enhancer domains consist of super-enhancers and their associated genes are located within chromatin loops (14). They are part of topologically associated domains, which are thought to control cell identity and cell-specific functions (11,14).
The insulator protein CTCF, together with the cohesion subunits SMC1 and SMC3, has been invoked in establishing DNA loops and thereby creating functional boundaries separating genetic units (14–18). Moreover, CTCF is thought to control enhancer activities and their functional isolation from genes outside their respective domains (14–18). However, genuine in vivo evidence to support these roles is limited (19–22), as studies have been conducted mainly in cell lines and embryonic stem cells (ESCs) (14,23–25). In ESCs, loss of CTCF binding sites can cause the activation of silent genes situated outside super-enhancer domains (14). However, it should be noted that genes induced in these studies were generally expressed at very low levels, i.e. below 1 Fragments Per Kilobase Of Exon Per Million Fragments Mapped (FPKM) (14,26). In contrast, deletion of CTCF sites associated with higher expressed genes does not result in an additional induction. Thus, it is not clear to what extent CTCF sites shield specific gene classes from enhancers to avoid inadvertent enhancer exposure. Genetic studies in mice have been inconclusive because they are largely based on mice carrying deletions of the Ctcf gene itself rather than on deletions of specific CTCF binding sites (20,21,27,28). Several studies have focused on the contribution of specific CTCF sites in the regulation of immunoglobulin T cell receptor gene rearrangements, but expression analyses had not been conducted (21,22,29). Additional information on the role of CTCF in regulating gene expression comes from studies where the disruption of chromatin loops results in developmental defects and the transition to cancer, possibly as a result of regulatory spillover to neighboring genes (19,30,31).
Here, we have addressed pertinent questions surrounding the biology of CTCF sites in a locus thriving on cytokine-sensing mammary-specific enhancers and super-enhancers. The casein locus contains five genes expressed almost exclusively in mammary epithelium and induced up to 1000-fold during pregnancy (32–34). This locus also harbors at least two genes, Odam and Prr27, which are expressed in non-mammary cells. Specifically, we have asked to what extent CTCF sites shield highly active enhancers from neighboring non-target genes, some of which are widely expressed in non-mammary cells at low levels or are even silent in mammary tissue. We have also asked whether CTCF binding associated with a super-enhancer is required for its activity. Toward this end, we have identified four CTCF binding sites in the mouse casein locus and deleted them individually using CRISPR/Cas9 genome editing.
MATERIALS AND METHODS
Mice
All animals were bred along the guidelines of the NIH and all animal experiments were performed according to the Animal Care and Use Committee (ACUC) of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The CRISPR/Cas9 system was used to generate mutant mice with CTCF binding site deletions from C57BL/6 mice (Charles River) by the transgenic core of the National Heart, Lung, and Blood Institute (NHLBI). Single-guide RNA (sgRNA) constructs were designed to specifically target the individual CTCF binding sites in the casein (Csn) locus (http://crispr.mit.edu/). Target-specific sgRNA and Cas9 mRNA were in vitro transcribed and microinjected into the cytoplasm of fertilized eggs for founder mouse production. To screen for homozygous mice, all mice were genotyped by PCR amplification with genomic DNA from mouse tails and sequencing (Macrogen).
Chromatin immunoprecipitation sequencing (ChIP-seq) and data analysis
Frozen-stored mammary tissues collected at lactation day 1 (L1) were ground into powder. Chromatin was fixed with formaldehyde (1% final concentration) for 15 min at room temperature, and then quenched with glycine (0.125 M final concentration). Samples were processed as previously described (35). The following antibodies were used for ChIP-seq: CTCF (Millipore, 07-729 and Abcam, ab70303), SMC1 (Bethyl, A300-055A), STAT5A (Santa Cruz Biotechnology, sc-1081), GR (Thermo Scientific, PA1-511A), H3K27ac (Abcam, ab4729) and RNA polymerase II (Abcam, ab5408). Libraries for next-generation sequencing were prepared and sequenced with a HiSeq 2500 instrument (Illumina) (35). ChIP-seq data analysis was done as described (36), except for CTCF samples, where the parameters -m 3 and –best were used for Bowtie aligner (37) (version 1.1.2).
Immunoprecipitated DNA was used to ChIP-Real-time quantitative PCR (qRT-PCR) and analyzed by real-time PCR with SYBR green supermix (Biorad). The PCR product was amplified using the following specific primer pairs for the promoter of Sult1d1, forward, 5΄-TTAATGGAAGCCTCGTTCTCTG-3΄ and reverse 5΄-AACATCCACAGCACCTTCTC-3΄.
Total RNA-seq analysis
Total RNA-seq read quality control was done using Trimmomatic (38) (version 0.33). Reads were subsequently aligned with Bowtie aligner (37) (version 1.1.2) using paired end mode.
RNA isolation and quantitative real-time PCR (qRT–PCR)
Total RNA was extracted from frozen mouse mammary tissue of WT (n = 6) and mutants (A, n = 8; B, n = 7; C and D, n = 6) using homogenizer and the PureLink RNA Mini kit according to the manufacturer's instructions (Invitrogen). Total RNA (2 μg) was reverse transcribed for 1 h at 50°C using 50 mM oligo dT and 1 μl (50 international units) SuperScript II (Invitrogen) in a 20-μl reaction. One microliter of the reverse transcription product was PCR-amplified using the primer pairs. Quantitative real-time PCR (qRT-PCR) was performed using the TaqMan probe (Csn1s1, Mm01160593_m1; Csn2, Mm04207885_m1; Csn1s2a, Mm00839343_m1; Csn1s2b, Mm00839674_m1; Csn3, Mm02581554_m1; Wap, Mm00839913_m1; Sult1d1, Mm00502035_m1; Sult1e1, Mm00499178_m1; Odam, Mm02581573_m1; Cabs1, Mm02344036_m1; Smr3a, Mm01964237_s1; Smr2, Mm00491149_m1; Gapdh, Mm99999915_g1, ThermoFisher) on the CFX384 Real-Time PCR Detection System (Bio-Rad) according to the manufacturer's instructions. PCR conditions were 95°C for 30 s, 94°C for 15 s, and 60°C for 30 s for 40 cycles. All reactions were done in triplicate and normalized to the housekeeping gene Gapdh. Relative differences in PCR results were calculated using the comparative cycle threshold (CT) method.
To detect the mammary-specific Sult1d1 transcript, qRT-PCR was performed using the designed probes (forward, 5΄-TTCGATATTAAAGTGAATCTAAGGAGAAAG-3΄, reverse, 5΄-TGAGTTTCAGGAGATTTCTCATTTTC-3΄; control, forward, 5΄-TCCAGTGGCTTCTACAAATCC-3΄, reverse, 5΄-GTCTGTATCTTCCTGGCGTG-3΄, Integrated DNA technologies).
5΄ rapid amplification of cDNA ends (RACE) assay
RACE assay was done using the FirstChoice RLM-RACE Kit (Ambion) according to the manufacturer's instructions. The following primers were used in the nested PCRs in the assay: outer primer 5΄-ATAAGCTCCATGAATGGTACTCGTTTG-3΄; inner primer 5΄-CAGTATTTCACTGACCCAAGTTGTTC-3΄. The PCR products were then purified and sequenced to identify the 5΄ end of the mammary-specific Sult1d1 transcript.
Chromosome conformation capture (3C)
The 3C protocol was adapted from published methods (39,40). Frozen-stored mammary tissues collected at lactation day 1 (L1) were ground into powder. Chromatin was fixed with formaldehyde (final 1%) for 15 min at room temperature, and then was quenched with glycine (final 0.125 M). Pellets were lysed in lysis buffer (10 mM Tris–HCI pH 8.0, 10 mM NaCl, 0.5% Nonidet P-40) containing Phenylmethylsulfonyl fluoride (PMSF, 100mM) and protease inhibitors, incubated on ice for 30 min, and dounced using pre-chilled glass homogenizer, followed by another 10 min incubation on ice. After removal of supernatant, nuclei pellets were re-suspended in restriction endonuclease buffer 2 (NEB; 50 mM NaCl,10 mM Tris–HCl,10 mM MgCl2,1 mM dithiothreitol; New Englnad Biolabs). SDS was added to a final concentration of 0.2% and samples was incubated for 1 h at 37°C. 2% Triton X-100 was added, followed incubation for 1 h at 37°C. Samples were incubated with 400 units each of restriction enzymes BamHI (New Englnad Biolabs) and were incubated overnight at 37°C. To enzyme inactivation SDS was added (final 1%) and incubated for 30 min at 65°C, followed by SDS-sequestration with 1% Triton X-100 at 37°C for 1 h, T4 ligase (New England Biolabs) was added to each sample and incubated for 1 h at 16°C. Ligated samples were treated overnight with proteinase K (20 mg/ml, Invitrogen) at 65°C and 1 h at 37°C with RNase A (10 mg/ml, Thermo Fisher Scientific), and DNA fragments were purified by phenol–chloroform method. Samples were analyzed by qRT-PCR using SYBR green supermix (Biorad) on the CFX384 Real-Time PCR Detection System (Bio-Rad). The primers used were CTCF site A 5΄-AGCTCAAGGACTTTCTAGGACATTG-3΄ and CTCF site C 5΄-CTCCCAGAGACAAAGCCACC-3΄. Interaction frequencies were normalized to the values of an internal control.
Circular chromosome conformation capture (4C)-seq and data analysis
The 4C protocol was adapted from published methods (41,42). DNA fragments from 3C were digested with NlaIII (New England Biolabs) overnight at 37°C and ligated using T4 ligase overnight at 16°C. DNA was purified by phenol/chloroform method and then amplified with site-specific primers linked to the Illumina DNA adaptors. Libraries for next generation sequencing were prepared and sequenced with HiSeq 2500 (Illumina). The sequencing data were processed using the 4C-ker (43). All 4C-seq images were generated using 4Cker R package with k = 4.
Data
The RNA-seq data for WT controls at p6 and L1 as well as p18 were obtained under GSE37646 (4,34) and GSE92931 in the Gene Expression Omnibus (GEO). ChIP-seq data of wild-type mammary tissue at p13 and L1 were obtained under GSE74826. CTCF ChIP-seq data for ESCs, T cell, kidney, liver and lung were obtained under GSE28247 and GSE40918.
Statistical analyses
Data were presented as standard deviation in each group and were evaluated with a two-tailed unpaired t test using PRISM GraphPad. Statistical significance was obtained by comparing the measures from wild type and each mutant group. A value of P < 0.05 was considered statistically significant.
RESULTS
Identification of enhancers and CTCF sites in the mouse Casein locus
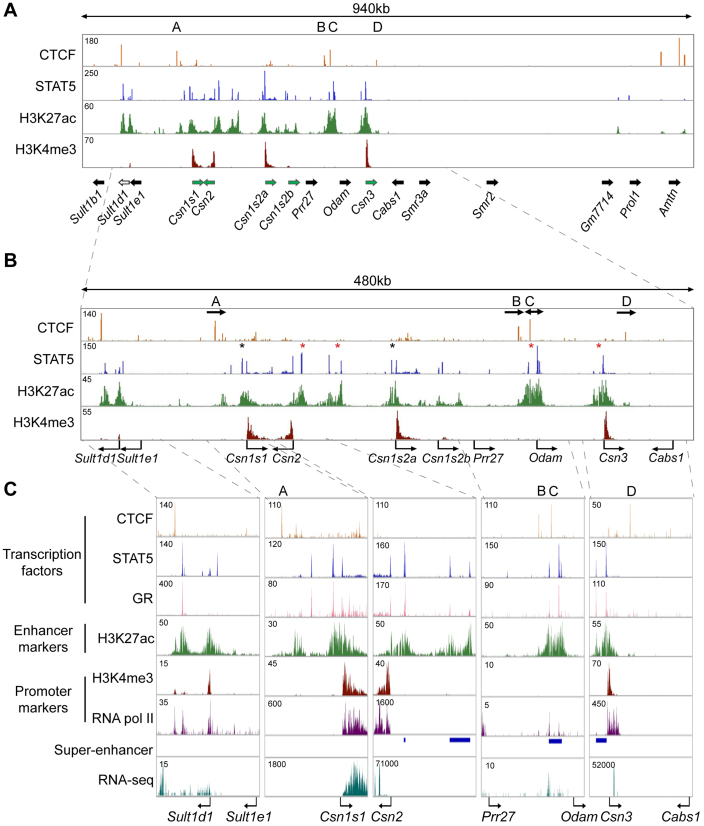
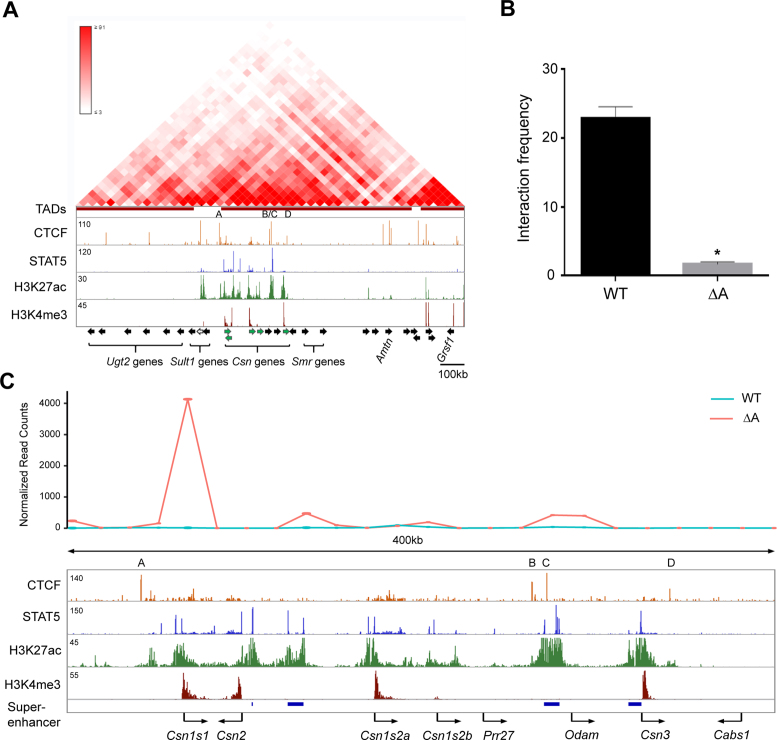
To investigate whether CTCF sites serve as genetic borders, ensuring that enhancers are confined to their respective target genes and shielding juxtaposed non-target genes from undesired transcriptional stimuli, we focused on the mouse casein (Csn) locus (Figure 1A). This locus encodes five casein genes that are expressed almost exclusively in mammary epithelium and are induced up to 1000-fold during pregnancy (4,34,44), likely as the result of enhancers and possibly super-enhancers that are established by the transcription factor STAT5 (3). In contrast, expression of non-mammary genes within the extended Csn locus, including members of the sulfotransferase family (Sult1), the Odontogenic ameloblast-associated protein (Odam) and Cabs1, was very low or not detected (Table 1) (4).
Figure 1.
Genomic features of the Csn locus during lactation. (A–C) ChIP-seq data for CTCF, STAT5, H3K27ac and H3K4me3 provided structural information on the locus (upper panel). The orientation of genes (panel A) is indicated by solid arrows. Green arrows, genes highly expressed in mammary tissue; black arrows, genes not expressed, or at very low levels, in mammary tissue; white arrow, Sult1d1, which is expressed at low levels in mammary tissue and induced upon deletion of CTCF site A. Four STAT5-based super-enhancers (red asterisks in the panel B and blue bars in panel C) and two solitary enhancers (black asterisks in the panel B) were identified. The presence of Pol II and total RNA transcripts was associated with transcriptional units and enhancers (panel C). Arrows in panel B indicate the orientations of CTCF binding sites.
Table 1. mRNA levels of mammary-specific Csn genes and non-mammary genes in the extended Csn locus during pregnancy and lactation.
| Gene | p6 (FPKM) | L1 (FPKM) | Fold change | |
|---|---|---|---|---|
| Mammary-specific genes | Csn1s1 | 1,200 | 87,950 | 74 |
| Csn2 | 3,100 | 193,500 | 63 | |
| Csn1s2a | 1,250 | 164,000 | 132 | |
| Csn1s2b | 1 | 13,600 | 10,490 | |
| Csn3 | 220 | 57,850 | 264 | |
| Non-mammary genes | Sult1b1 | 0 | 0 | n/a |
| Sult1d1 | 0.1 | 6 | 60 | |
| Sult1e1 | 0 | 0 | n/a | |
| Prr27 | 0 | 0 | n/a | |
| Odam | 0.07 | 2 | 28 | |
| Cabs1 | 0 | 0 | n/a | |
| Smr3a | 0 | 0 | n/a | |
| Smr2 | 0 | 0 | n/a |
mRNA levels of mammary-specific genes and non-mammary genes under the control of lactogenic hormones were measured by RNA-seq (4) at day 6 of pregnancy (p6) and day 1 of lactation (L1). Mammary-specific Csn genes were induced up to several thousand-fold during pregnancy, whereas genes not specific to mammary tissue, within or outside the Csn, locus were induced <60-fold. n/a, no activation.
We have identified 440 mammary-specific super-enhancers, which are characterized by the binding of the prolactin-sensing transcription factor STAT5, Mediator complex subunit 1 (MED1) and the Glucocorticoid Receptor (GR) and by the presence of H3K27ac active enhancer marks (3). Four super-enhancers are located within the extended Csn locus, which is bracketed by Csn1s1 and Csn3 (Figure 1A and B). Two super-enhancers are located between the Csn2 and Csn1s2a genes, one upstream of the Csn3 gene and one between the Odam and Prr27 genes (Figure 1B). Solitary enhancers were detected upstream of the Csn1s1 and Csn1s2a genes, and enhancer marks were present within the Csn1s2b gene. There was no direct correlation between the presence of solitary enhancers and super-enhancers and expression levels of the juxtaposed casein genes (Table 1). Importantly, despite the presence of a mammary-specific super-enhancer between the Odam and Prr27 genes neither one was expressed at significant levels in mammary tissue. The Sult1d1 gene, located 97 kb upstream of Csn1s1, contains a mammary enhancer and is expressed in mammary tissue at low levels (Table 1). In contrast, the Sult1e1 gene located more proximal to the Csn1s1 gene is not expressed in mammary tissue. The Cabs1 gene is located 56 kb outside of Csn3 and is not expressed in mammary tissue (Table 1). The combined presence of tri-methylation of H3K4 (H3K4me3) marks and RNA polymerase II (RNA pol II) loading reflected gene activity within the Csn locus (Figure 1). These data also suggest that structurally equivalent solitary enhancers and super-enhancers within the Csn locus likely bestow greatly different activities upon their target genes. Notably, the mammary super-enhancer located between Odam and Prr27, has limited consequence on the activation of neighboring genes. Csn1s2b, which displays enhancer marks within an intron, is expressed at a level of less than 10% of the juxtaposed Csn1s2a gene (Table 1). Moreover, the degree of induction of individual Csn genes during pregnancy did not correlate with the presence of enhancers and super-enhancers (Table 1).
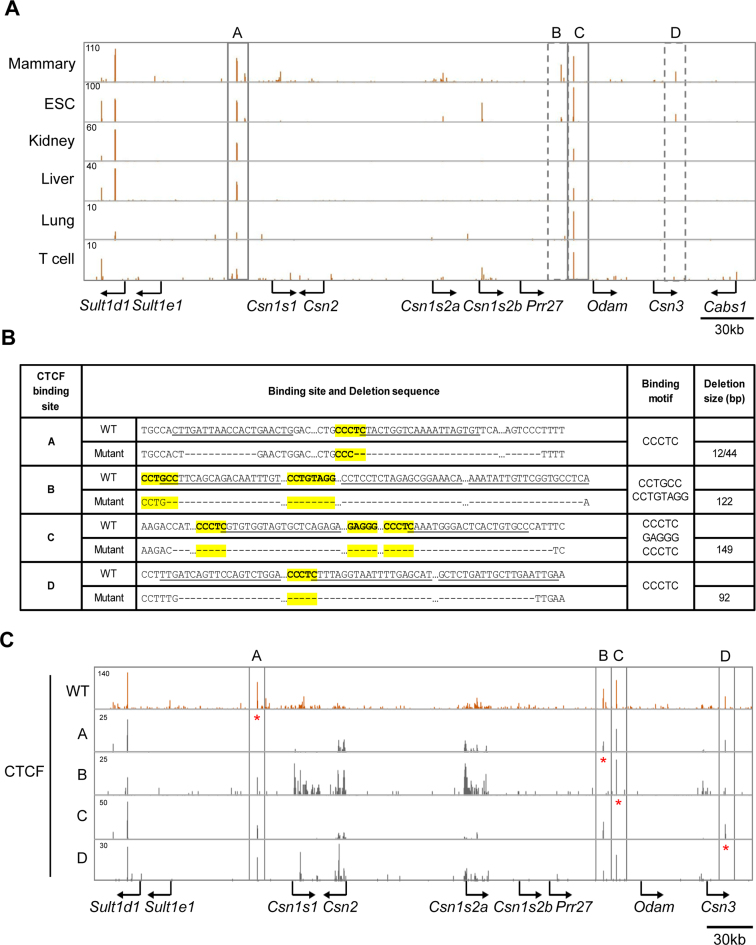
To explore the possibility that the activities of the different enhancers and super-enhancers within the Csn locus are controlled by specific CTCF binding sites, we performed CTCF ChIP-seq analyses on mammary tissue during lactation when expression of Csn genes is highest. We have identified four specific sites bound by CTCF (named A, B, C and D) in the Csn locus (Figure 1). While binding to sites A and C was also observed in most, if not all, cell types, binding to sites B and D was preferentially detected in mammary tissue but also in ES cells (Figure 2A). Sites A and D contain one bona fide CTCF motif (CCCTC) each and site C contains three motifs in both orientations within 40 bp (Figure 2B). In contrast site B does not contain a classical motif. Based on their orientation (Figure 1B) and current knowledge chromatin loops could be formed between sites A and C as well as between sites B and C. While sites A and D flank the Csn locus and are not associated with any additional features reflective of regulatory elements, sites B and C are associated with H3K27ac marks, indicative that they are part of the super-enhancer located 5΄ of Odam (Figure 1B and C).
Figure 2.
Identification of CTCF binding sites in the Csn locus and generation of mice lacking individual sites. (A) Binding of CTCF to sites A and C was detected across non-mammary cells (rectangles with solid lines) while CTCF binding to sites B and D was preferentially observed in mammary tissue (rectangles with dotted lines). (B) Sequences of the four CTCF binding sites in the mouse genome and upon CRISPR/Cas9 gene editing. CTCF binding motifs are depicted in yellow and their sequences are in bold. While sites A, C and D contain classical motifs (CCCTC), site B contains an extended motif (CCN(C/G)N(A/C/T)G(A/G)(T/G)GG(C/A/T)(G/A)(C/G) (69,70). sgRNA sequences are underlined, and deleted nucleotides are indicated as dashes. CTCF binding sequences and deletion sizes are shown. (C) CTCF ChIP-seq data confirmed loss of CTCF binding in mutant mammary tissues.
Impact of CTCF site deletions on the regulation of the Csn locus
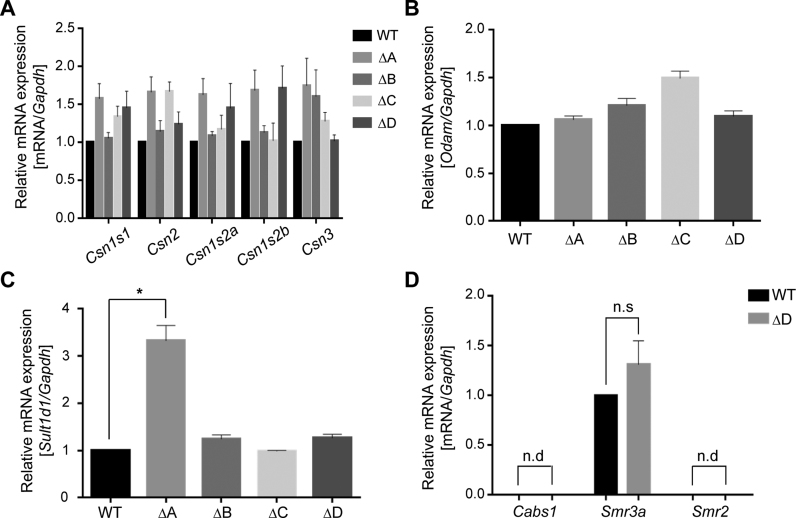
Based on our ChIP-seq data, we hypothesized that CTCF sites A and D might participate in generating genetic boundaries at the Csn locus, encapsulating its highly active enhancers and super-enhancers and shielding neighboring non-mammary genes from their influence. We also hypothesized that sites B and C, which are associated with the most expansive super-enhancer within the Csn locus could contribute to the establishment and function of this regulatory region controlling the activity of the locus. To test these hypotheses, we employed CRISPR/Cas9 gene editing and generated mice lacking individually each of the four CTCF binding sites (Figure 2B and Supplementary Figure S1). The deletion of site C spanned all three CCCTC motifs. Loss of CTCF binding was verified by ChIP-seq (Figure 2C). To determine the biological function of these CTCF binding sites in regulating activity of the Csn locus, we performed real time quantitative reverse transcription PCR (qRT-PCR) for the five Csn genes, the Sult and Cabs1 genes outside the locus and the Odam gene located within the locus (Figure 3). The absence of individual CTCF binding sites did not significantly alter the expression of any of the Csn genes in mammary tissue (Figure 3A), suggesting that their presence is inconsequential for enhancer activities within this locus. The promoter of Odam, a gene preferentially expressed in molar tissue (45) and at low levels in mammary tissue, is located 13 kb from CTCF site C that is part of a putative mammary super-enhancer (Figure 1). This opened the possibility that site C is critical for shielding super-enhancer activity from Odam. However, loss of site C did not enhance or otherwise affect Odam expression (Figure 3B). In contrast, expression of Sult1d1, which is located 73 kb outside CTCF site A, was induced approximately three-fold in mammary tissue lacking site A but not in any of the other mutants (Figure 3C). Sult1e1, which is located proximal to site A, i.e. closer to the nearest Csn enhancer, was not activated in mutant mammary tissue (data not shown). Notably, while Sult1d1 is associated with H3K27ac activating marks and expressed at low levels (6 FPKM) in lactating mammary tissue, the silent Sult1e1 gene does not display any enhancer features (Figure 1A). Deletion of CTCF site D, located at the Csn3 border of the Csn locus did not result in the activation of the outside genes Cabs1, Smr3a or Smr2 in mammary tissue (Figure 3D). In addition, expression of these genes was unaltered in salivary tissue (Supplementary Figure S2). This is evidence that these genes, even in the absence of CTCF sites, are not accessible to the Csn3 super-enhancer. As Sult1e1, Cabs1 and Smr2 are silent in mammary tissue and do not feature any enhancer marks. Sult1d1 encodes a sulfotransferase and its function in the biology of mammary tissue is not known. Lactation performance was not affected in mice overexpressing Sult1d1 (Supplementary Figure S3).
Figure 3.
In vivo consequences upon the deletion of individual CTCF binding sites. (A) Individual deletion of CTCF binding sites A to D did not significantly affect mRNA levels of the five Csn genes in lactating mammary tissue. qRT-PCR data were normalized to Gapdh levels. Results are shown as the means ± S.E.M. of independent biological replicates (WT, C and D, n = 6; A, n = 8; B, n = 7). A two-tailed unpaired t test was used to evaluate the statistical significance of differences between WT mice and each mutant group. All genes weren't shown the significant difference (at least two-fold) in each mutant group compared to WT. (B) Expression of Odam located within the Csn locus was not affected by any of the four mutants. (C) Sult1d1 expression was induced three-fold in mammary tissue lacking CTCF site A. Sult1d1 expression was not affected by the other mutants. Asterisk indicates P < 0.05 in two-tailed unpaired t test of each mutant group compared to WT. (D) The expression of the Smr3a gene did not change in D mutant using qRT-PCR. The Cabs1 and Smr2 genes were silent. Data were normalized to Gapdh levels. Results are shown as the means ± S.E.M. of independent biological replicates *P < 0.05 (WT and D, n = 6). n.s., not significant; n.d., not detected.
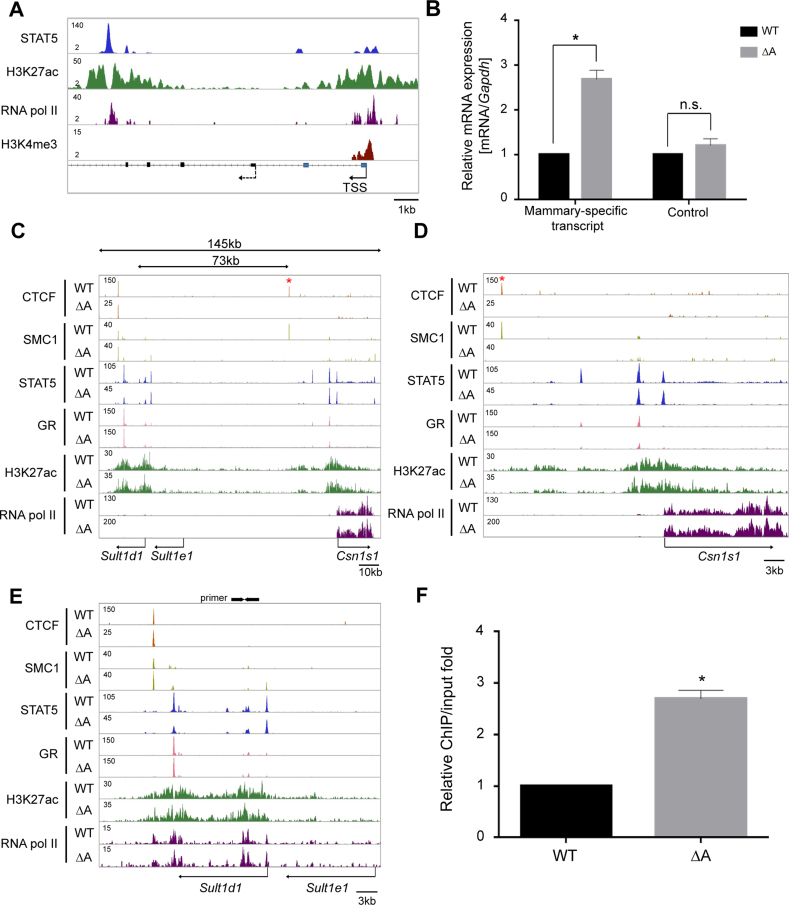
The first exon of the Sult1d1 gene in the reference genome did not coincide with mammary H3K4me3 marks and the presence of RNA pol II (Figure 4A) suggesting the existence of an additional, mammary-specific, promoter. We therefore performed 5΄ RACE experiments on mammary RNA and detected additional Sult1d1 exons (Figure 4A). Based on these findings expression of Sult1d1 is controlled by a mammary-specific promoter, different from the one used in other cell types. qRT-PCR from the 5΄ exon further validated the three-fold increase of Sult1d1 mRNA in mammary tissue lacking CTCF site A (Figure 4B). The putative intronic Sult1d1 mammary-specific enhancer is characterized by STAT5 binding, H3K27ac marks and RNA pol II occupancy (Figure 4A).
Figure 4.
Functional significance of the border CTCF site A on the regulation of outside genes. (A) Identification of the mammary-specific promoter of Sult1d1. The position of H3K4me3 marks and 5΄ RACE from mammary RNA led to the discovery of a mammary-specific promoter. The mammary-specific TSS is marked by a solid arrow and the TSS in other tissues by a dotted arrow. Exons from the reference genome are marked as black rectangles and mammary-specific exons are marked in blue. (B) Mammary-specific transcript levels (qRT-PCR) of Sult1d1 increased significantly in mice lacking CTCF site A. Data were normalized to Gapdh levels. Results are shown as the means ± S.E.M. of independent biological replicates *P < 0.05 (WT and A, n = 4). n.s., not significant. (C–E) ChIP-seq profiles at the Csn1s1 and Sult1d1 locus in lactating mammary tissue from WT and mutant mice. The data for CTCF, STAT5A, and H3K27ac ChIP-seq are representatives of biological replicates. TF binding and histone marks were equivalent in WT and mutant mammary tissue. The red asterisk (panels C and D) indicates CTCF site A and arrows (panel E) indicate the primer set for ChIP-qPCR. (F) RNA pol II binding to chromatin of the mammary-specific Sult1d1 promoter in wild type and mutant mammary tissues. Binding activity of RNA pol II, as measured by ChIP-qPCR, at the Sult1d1 promoter was increased 2-fold in mutant tissue. Results are shown as the means ± S.E.M. of independent biological replicates *P < 0.05 (WT and A, n = 3).
Next, we examined the possibility that ablation of CTCF sites associated with enhancers leads to the expansion of active histone marks and altered binding of STAT5 and GR, key regulators of the Csn locus. The focus was on CTCF site A as its loss resulted in increased expression of the neighboring Sult1d1. We performed ChIP-seq for H3K27ac, RNA pol II, STAT5 and GR using mammary tissue from wild type and mutants. First, ChIP-seq validated the absence of CTCF and SMC1 binding at the mutant site (Figure 4C). Binding of STAT5 and GR at the Csn1s1 and Sult1d1 enhancers was not affected (Figure 4C), suggesting that the structural establishment of enhancers is independent of CTCF and its potential role of establishing insulated neighborhoods. Notably, loss of site A did not result in the expansion of H3K27ac marks associated with the Csn1s1 enhancer (Figure 4D). Although expression of Sult1d1 increased three-fold in the absence of CTCF site A, the H3K27ac landscape as well as STAT5 binding at this locus were not altered (Figure 4E). However, ChIP-qPCR experiments demonstrated increased binding of RNA pol II at the Sult1d1 promoter in mammary tissue lacking CTCF site A (Figure 4F) but not to the Sult1e1 promoter. Transcription factor binding and epigenomic features at the super-enhancer surrounding CTCF site C were unaffected in the absence of site C (Supplementary Figure S4). These results suggest that only genes already expressed, even at low levels, can be further induced upon removal of CTCF sites separating them from enhancers.
CTCF site restricts promoter–enhancer interactions
Deletion of CTCF site A, which separates the highly active Csn1s1 enhancer from Sult1d1 at a distance of ∼95 kb, results in a three-fold induction of Sult1d1. To determine whether this was the result of a de novo interaction between the Sult1d1 promoter and Csn1s1 enhancer, or possibly other enhancers within the Csn locus, we conducted 3C and 4C experiments (Figure 5). Hi-C and 4C data (46,47) suggest an interaction between CTCF sites A and C (Figure 5A). 3C experiments demonstrated that interactions between the CTCF sites A and C decreased approximately 10-fold in mammary tissue upon loss of CTCF site A (Figure 5B).
Figure 5.
Interactions between individual CTCF sites within the casein locus (A, B) and between the Sult1d1 promoter and casein enhancers. (A) Hi-C data from mESC (46,47) define TADs and ChIP-seq data from mammary gland in the casein locus. (B) Interactions between CTCF sites A and site C were detected by 3C in WT mammary tissue and greatly reduced upon deletion of CTCF site A. Results are shown as means ± S.E.M. of independent biological replicates (WT and A, n = 3). A two-tailed unpaired t test was used to evaluate the statistical significance of differences between WT and A mutant: *P < 0.05. (C) The Sult1d1 promoter, as anchor, interacted with several Csn enhancers as determined by 4C in mammary tissue lacking CTCF site A.
It can be hypothesized that CTCF sites limit the search space of enhancers for putative target promoters. We therefore investigated whether loss of CTCF site A enabled additional enhancers within the casein locus to interact with the Sult1d1 gene. 4C experiments were performed with an anchor at the Sul1d1 promoter. Indeed, upon deletion of site A additional de novo interactions between the Sult1d1 gene and the casein locus were detected (Figure 5C). In addition to the one with the Csn1s1 gene, interactions with putative super-enhancer associated with the Csn2 gene and the Odam region were detected. Interactions were also detected with H3K27ac active marks at the Csn1s2b gene.
DISCUSSION
CTCF and its role in organizing chromatin to compartmentalize regulatory units have been extensively studied in cell culture (14,26,48–50), ESCs (14,23–25) and to a lesser extent in mice (19–22,28). However, there is a dearth of knowledge on its genuine role in controlling cell-specific and hormone-sensing enhancers and shielding them from non-target genes. Here we demonstrate that individually the four CTCF binding sites associated with mammary-specific enhancers and super-enhancers (3) within the extended casein locus do not play any measurable role in the regulation and expression of the five Csn genes and the embedded non-mammary Odam gene. However, one CTCF site bordering the Csn locus has the capacity to shield an outside gene from Csn enhancers. While loss of this site results in the induction of an already active gene at a distance of more than 95 kb, a silent gene located more proximal to the enhancer fails to be activated. These findings suggest that mammary-specific enhancers have the capacity to stimulate only already activate promoters but not silent ones. In support of this, silent genes located at the other end of the Csn locus are also not activated after deletion of the CTCF site separating them from mammary enhancers. This mechanism is distinct from that observed in the HoxA locus in ESCs where CTCF sites insulate heterochromatin and their loss results in the displacement of repressive chromatin and the activation or repression of selected outside genes (25). It is also different from the concept that loss of CTCF sites flanking super-enhancers can activate silent or very low expressed genes (14,26). It is well possible that cell-restricted enhancers employ a different strategy to search for target genes than more commonly used enhancers. Our study also support findings from tissue culture cells and ESCs that only a minority of CTCF sites play a role as insulators (51) and ablation of several CTCF sites linked to insulated neighborhoods might be required to obtain changes in gene expression (14).
Although sequences encompassing CTCF sites have been deleted from the mouse genome (19–22,28) the consequences on the expression of specific genes has not been investigated in depth. Ablation of two CTCF sites in the mouse Ig heavy chain (IgH) locus resulted in aberrant V(D)J recombination (22,52) but the consequences on the expression of specific genes was not reported. Ablation of sequences including CTCF sites within the TCR locus also resulted in aberrant V(D)J recombination (53,54). Evidence that loss of CTCF binding can influence the expression of genes outside specific chromatin loops comes from a study in which de novo methylation of a CTCF site in the mouse genome results in loss of CTCF binding and altered expression of an outside gene (55). Based on our study we suggest that CTCF sites can insulate already active genes from mammary-specific enhancers. However, silent genes are not awakened upon the deletion of CTCF sites separating them from active mammary enhancers. Notably, loss of the border CTCF site not only resulted in de novo interactions between the Sult1d1 and the nearest Csn enhancer but also with enhancers at a distance of more than 200 kb. It is not clear which of these interacting Csn enhancers is responsible for the activation of Sult1d1. It is also not understood whether searches for regulatory partners are initiated by promoters, enhancers or both.
The function of CTCF sites has been studied in embryonic stem cells, human embryonal carcinoma cell lines and the Drosophila Bithorax complex within the Hox locus (25,56–58). Deletion of CTCF sites in the HoxC cluster resulted in an aberrant activation of some Hoxc genes and the establishment of H3K4me3 marks on these genes (25,58). However, the absolute degree of activation and the association with H3K27ac marks is not clear. Hoxd genes share their enhancers during embryonic development (59). Similar to the Hoxd gene cluster, Csn genes are closely spaced and probably share the four cytokine-sensing super-enhancers. CTCF sites within enhancers have been proposed to control their activity (14,23), albeit genetic evidence is sparse. Deletion of the CTCF site central to a core casein super-enhancer, possibly the one required for the activation of this locus (60) did not result in a measurable expression change of any of the five casein genes. Based on our study in mice and others in tissue culture cells (14,26,48–50), ESCs (14,23–25) and primary fibroblasts (61), including the deletion of the Sox2 enhancer (14,23), CTCF sites do not play a prominent role in the regulation of enhancer activity.
CTCF sites are part of the 3D architecture of chromatin (18,23,62,63) and the interaction of adjacent CTCF binding sites with underlying convergent oriented motifs results in the establishment of chromatin loops (50,64–66). Of the four CTCF sites in the Csn locus, three contain one or more genuine CCCTC-motifs. Site A and D contain one forward oriented motif and site C contains two forward and one reverse oriented motif. Therefore, it is likely that sites A and C form a loop to restrict enhancer-promoter interactions for four casein genes and avoid spillover of enhancer activity into flanking genes, including Sult1d1. Our data agree with the concept that ablation of a CTCF site can lead to the activation of a gene by altering the chromatin structure and supporting novel promoter–enhancer interactions (21,26,48,50,67). Notably, site A is conserved between mouse and human and mutations have been found in several cancers, suggesting biological relevance (68). Consistent with previous reports on limb development (19), our data provide genetic evidence that CTCF plays a role in restricting inappropriate enhancer-promoter communication. Although the presence of the loop between sites A and C can explain the confinement of several Csn genes and their mammary-specific enhancers, it does not explain why the most prominent putative mammary enhancer located outside this loop does not activate the juxtaposed non-mammary Odam gene. This expansive enhancer is recognized by mammary-enriched transcription factors and extensive H3K27ac marks but ablation of the underlying CTCF site is inconsequential for its structure.
Our data demonstrate de novo interactions between Csn enhancers and the active Sult1d1 gene but not with the silent Sult1e1. Likewise, Capture-C data in embryonic limb development demonstrate that active regions, including enhancers and promoters preferentially interact with each other, while repressive ones are spatially clustered together (59). Taken together, we propose that the establishment of cell-specific enhancer-promoter loops requires the presence of active machineries at both ends.
Taken together our study, based on four CTCF sites within a complex hormone-sensing locus, suggests that only a fraction of these sites has a measurable biological activity. Further studies on cell-specific loci are needed to obtain a more comprehensive picture on the biological significance of CTCF in the in vivo regulation of genes.
ACCESSION NUMBERS
ChIP-seq and 4C-seq data have been deposited under GSE92591 and GSE95757, respectively. All ChIP-seq and 4C-seq data sets have been deposited under GSE92591 in GEO .
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Sumin Oh for initial discussion, the NHLBI transgenic core for generating the CRISPR/Cas9-based mouse mutants and the NIDDK genomics core for next-generation sequencing.
Author contributions: H.K.L. established mutant mouse lines, performed gene expression analyses, ChIP-seq, 3C and 4C experiments and analyzed data. MW processed and analyzed ChIP-seq and RNA-seq data sets. C.W. performed ChIP-seq and 5΄RACE and analyzed 4C data. C.M.Y. identified founder mice and established lines. HES provided advice on 4C and NGS experiments. C.L. generated mutant mice. L.H. conceived and supervised the study and analyzed data. H.K.L., M.W., C.W. and L.H. provided continued key intellectual input throughout the project as it progressed and as new questions developed. L.H. and H.K.L. wrote the manuscript and all authors approved it.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IPR of the NIDDK/NIH; Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea [HI15C1184 to H.K.L.]. Funding for open access charge: IPR of the NIDDK/NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Banerji J., Rusconi S., Schaffner W.. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981; 27:299–308. [DOI] [PubMed] [Google Scholar]
- 2. Ong C.T., Corces V.G.. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011; 12:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shin H.Y., Willi M., Yoo K.H., Zeng X., Wang C., Metser G., Hennighausen L.. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016; 48:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamaji D., Kang K., Robinson G.W., Hennighausen L.. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res. 2013; 41:1622–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shore A.N., Rosen J.M.. Regulation of mammary epithelial cell homeostasis by lncRNAs. Int. J. Biochem. Cell Biol. 2014; 54:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oki M., Kamakaka R.T.. Blockers and barriers to transcription: competing activities. Curr. Opin. Cell Biol. 2002; 14:299–304. [DOI] [PubMed] [Google Scholar]
- 7. Fredriksson N.J., Ny L., Nilsson J.A., Larsson E.. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 2014; 46:1258–1263. [DOI] [PubMed] [Google Scholar]
- 8. Khurana E., Fu Y., Chakravarty D., Demichelis F., Rubin M.A., Gerstein M.. Role of non-coding sequence variants in cancer. Nat. Rev. Genet. 2016; 17:93–108. [DOI] [PubMed] [Google Scholar]
- 9. Stergachis A.B., Neph S., Reynolds A., Humbert R., Miller B., Paige S.L., Vernot B., Cheng J.B., Thurman R.E., Sandstrom R. et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013; 154:888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herz H.M. Enhancer deregulation in cancer and other diseases. BioEssays. 2016; 38:1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hnisz D., Day D.S., Young R.A.. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016; 167:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doyle B., Fudenberg G., Imakaev M., Mirny L.A.. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput. Biol. 2014; 10:e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shlyueva D., Stampfel G., Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 14. Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schuijers J., Lee T.I., Zhao K. et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014; 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holwerda S.J., de Laat W.. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013; 368:20120369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S., Yu N.K., Kaang B.K.. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015; 47:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F.. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004; 430:573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ong C.T., Corces V.G.. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014; 15:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soshnikova N., Montavon T., Leleu M., Galjart N., Duboule D.. Functional analysis of CTCF during mammalian limb development. Dev. Cell. 2010; 19:819–830. [DOI] [PubMed] [Google Scholar]
- 21. Chen L., Carico Z., Shih H.Y., Krangel M.S.. A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRdelta and TCRalpha repertoires. Nat. Immunol. 2015; 16:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin S.G., Guo C., Su A., Zhang Y., Alt F.W.. CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Wit E., Vos E.S., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H., de Laat W.. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015; 60:676–684. [DOI] [PubMed] [Google Scholar]
- 24. Blinka S., Reimer M.H. Jr, Pulakanti K., Rao S.. Super-enhancers at the Nanog locus differentially regulate neighboring pluripotency-associated genes. Cell Rep. 2016; 17:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y. et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015; 162:900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nikolic T., Movita D., Lambers M.E., Ribeiro de Almeida C., Biesta P., Kreefft K., de Bruijn M.J., Bergen I., Galjart N., Boonstra A. et al. The DNA-binding factor Ctcf critically controls gene expression in macrophages. Cell. Mol. Immunol. 2014; 11:58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W.. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006; 20:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu J., Zhang Y., Zhao L., Frock R.L., Du Z., Meyers R.M., Meng F.L., Schatz D.G., Alt F.W.. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell. 2015; 163:947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hnisz D., Weintraub A.S., Day D.S., Valton A.L., Bak R.O., Li C.H., Goldmann J., Lajoie B.R., Fan Z.P., Sigova A.A. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016; 351:1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franke M., Ibrahim D.M., Andrey G., Schwarzer W., Heinrich V., Schopflin R., Kraft K., Kempfer R., Jerkovic I., Chan W.L. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016; 538:265–269. [DOI] [PubMed] [Google Scholar]
- 32. Rijnkels M., Freeman-Zadrowski C., Hernandez J., Potluri V., Wang L., Li W., Lemay D.G.. Epigenetic modifications unlock the milk protein gene loci during mouse mammary gland development and differentiation. PLoS One. 2013; 8:e53270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Faucon F., Rebours E., Bevilacqua C., Helbling J.C., Aubert J., Makhzami S., Dhorne-Pollet S., Robin S., Martin P.. Terminal differentiation of goat mammary tissue during pregnancy requires the expression of genes involved in immune functions. Physiol. Genomics. 2009; 40:61–82. [DOI] [PubMed] [Google Scholar]
- 34. Kang K., Yamaji D., Yoo K.H., Robinson G.W., Hennighausen L.. Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Mol. Cell. Biol. 2014; 34:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Metser G., Shin H.Y., Wang C., Yoo K.H., Oh S., Villarino A.V., O'Shea J.J., Kang K., Hennighausen L.. An autoregulatory enhancer controls mammary-specific STAT5 functions. Nucleic Acids Res. 2016; 44:1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Willi M., Yoo K.H., Wang C., Trajanoski Z., Hennighausen L.. Differential cytokine sensitivities of STAT5-dependent enhancers rely on Stat5 autoregulation. Nucleic Acids Res. 2016; 44:10277–10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naumova N., Smith E.M., Zhan Y., Dekker J.. Analysis of long-range chromatin interactions using chromosome conformation capture. Methods. 2012; 58:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hagege H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forne T.. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2007; 2:1722–1733. [DOI] [PubMed] [Google Scholar]
- 41. Stadhouders R., Kolovos P., Brouwer R., Zuin J., van den Heuvel A., Kockx C., Palstra R.J., Wendt K.S., Grosveld F., van Ijcken W. et al. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat. Protoc. 2013; 8:509–524. [DOI] [PubMed] [Google Scholar]
- 42. Gheldof N., Leleu M., Noordermeer D., Rougemont J., Reymond A.. Detecting long-range chromatin interactions using the chromosome conformation capture sequencing (4C-seq) method. Methods Mol. Biol. 2012; 786:211–225. [DOI] [PubMed] [Google Scholar]
- 43. Raviram R., Rocha P.P., Muller C.L., Miraldi E.R., Badri S., Fu Y., Swanzey E., Proudhon C., Snetkova V., Bonneau R. et al. 4C-ker: a method to reproducibly identify genome-wide interactions captured by 4C-Seq experiments. PLoS Computat. Biol. 2016; 12:e1004780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guyette W.A., Matusik R.J., Rosen J.M.. Prolactin-mediated transcriptional and post-transcriptional control of casein gene expression. Cell. 1979; 17:1013–1023. [DOI] [PubMed] [Google Scholar]
- 45. Lee H.K., Park S.J., Oh H.J., Kim J.W., Bae H.S., Park J.C.. Expression pattern, subcellular localization, and functional implications of ODAM in ameloblasts, odontoblasts, osteoblasts, and various cancer cells. Gene Expression Patterns: GEP. 2012; 12:102–108. [DOI] [PubMed] [Google Scholar]
- 46. Cairns J., Freire-Pritchett P., Wingett S.W., Varnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.M., Osborne C. et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016; 17:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang R., Kerschner J.L., Gosalia N., Neems D., Gorsic L.K., Safi A., Crawford G.E., Kosak S.T., Leir S.H., Harris A.. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016; 44:3082–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang J., Liu X., Li D., Shao Z., Cao H., Zhang Y., Trompouki E., Bowman T.V., Zon L.I., Yuan G.C. et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell. 2016; 36:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B. et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015; 163:1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu M., Maurano M.T., Wang H., Qi H., Song C.Z., Navas P.A., Emery D.W., Stamatoyannopoulos J.A., Stamatoyannopoulos G.. Genomic discovery of potent chromatin insulators for human gene therapy. Nat. Biotechnol. 2015; 33:198–203. [DOI] [PubMed] [Google Scholar]
- 52. Guo C., Yoon H.S., Franklin A., Jain S., Ebert A., Cheng H.L., Hansen E., Despo O., Bossen C., Vettermann C. et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011; 477:424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen L., Zhao L., Alt F.W., Krangel M.S.. An ectopic CTCF binding element inhibits Tcrd rearrangement by limiting contact between Vdelta and Ddelta gene segments. J. Immunol. 2016; 197:3188–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao L., Frock R.L., Du Z., Hu J., Chen L., Krangel M.S., Alt F.W.. Orientation-specific RAG activity in chromosomal loop domains contributes to Tcrd V(D)J recombination during T cell development. J. Exp. Med. 2016; 213:1921–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu X.S., Wu H., Ji X., Stelzer Y., Wu X., Czauderna S., Shu J., Dadon D., Young R.A., Jaenisch R.. Editing DNA methylation in the mammalian genome. Cell. 2016; 167:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu M., Zhao G.N., Lv X., Liu G., Wang L.Y., Hao D.L., Wang J., Liu D.P., Liang C.C.. CTCF controls HOXA cluster silencing and mediates PRC2-repressive higher-order chromatin structure in NT2/D1 cells. Mol. Cell. Biol. 2014; 34:3867–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Magbanua J.P., Runneburger E., Russell S., White R.. A variably occupied CTCF binding site in the ultrabithorax gene in the Drosophila bithorax complex. Mol. Cell. Biol. 2015; 35:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Narendra V., Bulajic M., Dekker J., Mazzoni E.O., Reinberg D.. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016; 30:2657–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Andrey G., Schopflin R., Jerkovic I., Heinrich V., Ibrahim D.M., Paliou C., Hochradel M., Timmermann B., Haas S., Vingron M. et al. Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res. 2016; 27:223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rijnkels M., Kabotyanski E., Montazer-Torbati M.B., Hue Beauvais C., Vassetzky Y., Rosen J.M., Devinoy E.. The epigenetic landscape of mammary gland development and functional differentiation. J. Mamm. Gland Biol. Neoplasia. 2010; 15:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fitzpatrick G.V., Pugacheva E.M., Shin J.Y., Abdullaev Z., Yang Y., Khatod K., Lobanenkov V.V., Higgins M.J.. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell. Biol. 2007; 27:2636–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lu Y., Shan G., Xue J., Chen C., Zhang C.. Defining the multivalent functions of CTCF from chromatin state and three-dimensional chromatin interactions. Nucleic Acids Res. 2016; 44:6200–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ferraiuolo M.A., Rousseau M., Miyamoto C., Shenker S., Wang X.Q., Nadler M., Blanchette M., Dostie J.. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010; 38:7472–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gomez-Marin C., Tena J.J., Acemel R.D., Lopez-Mayorga M., Naranjo S., de la Calle-Mustienes E., Maeso I., Beccari L., Aneas I., Vielmas E. et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., Hadjur S.. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015; 10:1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gosalia N., Neems D., Kerschner J.L., Kosak S.T., Harris A.. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 2014; 42:9612–9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ji X., Dadon D.B., Powell B.E., Fan Z.P., Borges-Rivera D., Shachar S., Weintraub A.S., Hnisz D., Pegoraro G., Lee T.I. et al. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell. 2016; 18:262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Essien K., Vigneau S., Apreleva S., Singh L.N., Bartolomei M.S., Hannenhalli S.. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009; 10:R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Plasschaert R.N., Vigneau S., Tempera I., Gupta R., Maksimoska J., Everett L., Davuluri R., Mamorstein R., Lieberman P.M., Schultz D. et al. CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation. Nucleic Acids Res. 2014; 42:774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.