Abstract
Spt5 is a conserved and essential transcriptional regulator that binds directly to RNA polymerase and is involved in transcription elongation, polymerase pausing and various co-transcriptional processes. To investigate the role of Spt5 in non-coding transcription, we used the unicellular model Paramecium tetraurelia. In this ciliate, development is controlled by epigenetic mechanisms that use different classes of non-coding RNAs to target DNA elimination. We identified two SPT5 genes. One (STP5v) is involved in vegetative growth, while the other (SPT5m) is essential for sexual reproduction. We focused our study on SPT5m, expressed at meiosis and associated with germline nuclei during sexual processes. Upon Spt5m depletion, we observed absence of scnRNAs, piRNA-like 25 nt small RNAs produced at meiosis. The scnRNAs are a temporal copy of the germline genome and play a key role in programming DNA elimination. Moreover, Spt5m depletion abolishes elimination of all germline-limited sequences, including sequences whose excision was previously shown to be scnRNA-independent. This suggests that in addition to scnRNA production, Spt5 is involved in setting some as yet uncharacterized epigenetic information at meiosis. Our study establishes that Spt5m is crucial for developmental genome rearrangements and necessary for scnRNA production.
INTRODUCTION
Analysis of the rapidly growing genomic and transcriptomic data from high-throughput sequencing is increasing awareness of the importance of non-coding RNAs (ncRNAs). Small and long non-coding RNAs bound to effector proteins guide chromatin- and DNA-modifying enzymes to genomic loci and introduce dynamic changes of chromatin state (1,2). The transcriptional silencing pathways that give rise to those ncRNAs are well conserved among eukaryotes, however they differ regarding short non-coding RNA (sRNA) biosynthesis pathways or composition of the effector complexes (3). Regardless of the fact that ncRNAs rarely share evolutionary origins or molecular mechanisms of action, all of them (or their precursors) are produced by RNA polymerase. We are exploring the hypothesis that components of the RNA polymerase complex may contribute directly to the production of ncRNAs as shown recently for some factors (4–8). We noticed that the gene encoding Spt5/NusG transcription elongation factor, which is conserved and essential across Bacteria, Archaea and Eukarya (9), is upregulated at the time when ncRNA are massively produced in Paramecium tetraurelia. Considering its multiple roles in regulation of transcription, we decided that Spt5 would be a good candidate for a protein involved in ncRNA synthesis. In eukaryotes and Archaea, Spt5 is associated with a small zinc finger protein, Spt4 (10,11), and in some higher eukaryotes the Spt4/Spt5 heterodimer forms a complex with negative elongation factor NELF and takes part in a phenomenon of promoter-proximal pausing (12). Spt5 is associated with the body of all actively transcribed genes (13–16), provides comprehensive control over transcription elongation—stimulatory or inhibitory—and co-transcriptional events such as control of chromatin state and RNA processing (17,18). Spt5 interacts with activation-induced cytidine deaminase (AID) and targets it to sites of RNA Polymerase II stalling, where AID can access ssDNA and create U:G mismatches involved in antibody gene diversification (19). In plants, the Spt5-like factor RDM3/KTF1 mediates transcriptional gene silencing, acting as an effector of the RNA-mediated DNA methylation pathway (20,21). Spt5 binds directly to RNA polymerase by interaction of its N-terminal NGN (NusG-N-homology) domain with the RNA polymerase coiled-coil motif, near the active center of the enzyme (22). The other parts of eukaryotic Spt5 - multiple KOWs and the C-terminal domain—are thought to be responsible for interaction with other proteins and newly synthesized RNA molecules (23,24). In this report, we used P. tetraurelia, a model organism in which developmental DNA elimination involves non-coding RNAs and heterochromatin formation, to investigate Spt5 function in these epigenetic processes.
Paramecium tetraurelia harbors two kinds of nuclei in a unique cytoplasm: two diploid germline micronuclei (MIC) and the highly polyploid (800n) somatic macronucleus (MAC). The MAC genome is responsible for gene expression. The MIC's transcriptional activity is manifest exclusively during sexual processes, when the maternal MAC is lost and a new MAC emerges from the MIC as a result of meiosis, karyogamy and mitotic divisions of the zygotic nucleus. During this process, a reproducible program of genome rearrangements is executed (25).The genome is amplified and chromosomes are fragmented (26). At the same time the genome is stripped of germline specific sequences such as minisatellites, transposons and ∼45 000 short, single copy internal eliminated sequences (IESs) distributed throughout the MAC-destined part of the genome (27).
Genome rearrangements are subject to epigenetic regulation mediated by sRNA that are different from 23 nt-long siRNA (28). During meiosis, as a result of genome-wide transcription of the MIC and cleavage of the transcripts by the Dicer-like proteins Dcl2/Dcl3, development-specific 25-nt scnRNAs are produced (29,30). The scnRNAs constitute a temporal copy of the germline genome and are thought to be bound by Piwi proteins (31). In the current model of genome scanning (32), the scnRNAs are transported to the maternal MAC where a fraction of them probably interact with homologous maternal non-coding transcripts (33). This process allows selection of scnRNAs that have no counterparts in the MAC. The selected scnRNAs are transported to the developing new MAC and are proposed to interact with nascent, TFIIS4-dependent transcripts (34) to target elimination of the homologous sequences by PiggyMac (Pgm), a domesticated piggyBac transposase (35). A second class of development specific small RNAs, the iesRNAs (<25–30 nt), is produced in the developing new MACs by another Dicer-like protein (Dcl5). The iesRNAs are thought to help finish IES removal (30). The result of genome scanning is faithful transmission of the rearrangement pattern of the maternal MAC across sexual generations. This genome rearrangement system provides defense against parasitic DNA, which is not only silenced but physically eliminated, and is also exploited for the regulation of cellular genes (36).
Chromatin state is important for the genome rearrangement program. Excision of repeated sequences and of a majority of the unique-copy IESs (∼70%) is guided by H3K9 and H3K27 trimethylation that depends on the histone methyltransferase Ezl1 (37). While scnRNAs are necessary for H3K9me3 and H3K27me3 accumulation, not all Ezl1-dependent IESs require TFIIS4 or Dcl2/Dcl3 proteins and scnRNAs for their excision (37,38). Furthermore, about 1/3 of the IESs require none of these factors for their excision (37,38).
Here, we report the identification and functional characterization of two Spt5-encoding genes in P. tetraurelia. Expression of one of them, SPT5m, is not only indispensable for production of the development-specific scnRNAs in the meiotic germline nucleus, but turns out to be required for excision of all germline-limited sequences, including all IESs, underscoring an essential role for Spt5 in the epigenetic genome rearrangement program.
MATERIALS AND METHODS
Construction and injections of GFP fusion transgenes
Plasmids pSPT5v-GFP and pSPT5m-GFP encoding C-terminal GFP (Green Fluorescent Protein) fusions to SPT5v and SPT5m, respectively, were obtained by an overlapping polymerase chain reaction (PCR) method (39) in pCRscript vector (Invitrogen). Constructs contain putative promoter regions, open reading frame and putative terminator (genomic coordinates of cloned fragments: SPT5m - 164290..166209 of the acc. no. CAAL01001700; SPT5v - 70136..72841 of the acc. no CAAL01001624). The eGFP coding sequence (40) preceded by a flexible linker (12 aa) was inserted directly before the stop codon of each SPT5 gene. Linearized plasmids carrying GFP fusion transgenes were microinjected into the MAC of vegetative 51 nd7-1 cells, as described previously (40).
Gene silencing
RNAi was performed as described in (34,41). All experiments were carried out with P. tetraurelia strain 51new (42). All RNAi plasmids are derivatives of vector L4440 (43) and carry a fragment of the target gene inserted between two convergent T7 promoters (genomic coordinates of silencing inserts: SPT5m -165576..166166 of the acc. no. CAAL01001700; SPT5v - 72016..72675 of the acc. no CAAL01001624). In principle, cross-silencing between these genes is not possible as they do not share any stretches of 23 identical nucleotides that could give rise to siRNA targeting the other gene. To monitor RNAi phenotypes during vegetative growth, 3–15 cells were placed into 200 μl of freshly induced silencing medium. As a control, the same number of cells was transferred to silencing medium containing induced Escherichia coli harboring ND7- or ICL7a-silencing plasmids (p0ND7c (44) and pICL7a (45), respectively), which target non-essential genes, or standard Klebsiella pneumoniae (Kp) medium. After 24 h (and 48 h), each clone was replicated by transferring a single cell to 200 μl of fresh medium. Each day, the cells were counted in each microculture to evaluate their growth rate. For all replicate experiments, we calculated the average growth rate as well as cell lethality in each silencing medium. We used the data obtained for 60 cell lines on average. In order to check the RNAi phenotype of sexual progeny, autogamy was induced by starvation of the cell cultures silenced either for a non-essential control gene or for SPT5m and the survival was checked following transfer of individual autogamous cells to standard medium. Genomic DNA and total RNA samples were extracted at different time points of the autogamy time-course from ∼400 000 Paramecium cells (35).
sRNA sequencing
Purification, sequencing and analysis of sRNAs from control and Spt5m-depleted cells were carried out as previously described (34). Briefly, the 20–30 nt sRNA reads (accession SRP068457) were filtered for known contaminants (Paramecium rDNA, mitochondrial DNA, feeding bacteria genomes and L4440 feeding vector sequences). In addition, the 23 nt siRNA reads that map to the RNAi targets were removed. The filtered reads were mapped to reference MAC and MAC+IES genomes. Read counts were normalized using the total number of filtered reads. A previously published sRNA dataset for Dcl2/3-depleted cells (30) (accessions SRR907874- SRR907877) was processed in the same way. Accession numbers of all samples are displayed in Supplementary Table S5.
IES retention
For genome-wide evaluation of IES retention in Spt5m-depleted cells, DNA from a cell fraction enriched in late stage developing MACs was subjected to Illumina paired-end sequencing, as previously described (34). This SPT5m dataset (SRP068457) and a contrlol dataset (ERX466735) were then used to measure IES retention using the ParTIES package, using the boundary score method (46). We used the mean of the left and right boundary score for each IES.
The count data for the experimental and control datasets was then used to test statistically if an IES is retained to the same extent in experimental and control samples. An IES is considered as significantly retained if at least one of the two boundaries passed the statistical test with a P-value below 0.05.
Sequence complexity
To estimate the effects of Spt5m depletion on the retention of germline-limited sequences other than IESs, we aligned the reads from the SPT5m (acc.no SAMN04358097), PGM (ERA137444), DCL2/3 (SRR2015146) and Control (wild-type genome; ERA309409, Sample SAMEA2518987) datasets to contigs assembled from the PGM dataset (27), a proxy for the germline genome. The PGM contigs (after removal of contigs smaller than 1 kb) contain 91 Mb. For each sample, the complexity was determined by mapping the reads to the PGM contigs and then calculating the sum of all regions of the PGM contigs covered by a minimum of 2 RPKM (reads per kb per million mapped reads). Regions not covered by the Control are considered to be germline-limited. Alignment was performed by paired-end read mapping with BWA version 0.7.8, using default parameters (47).
Reference genomes
The following reference genomes (27) were used in the different analyses for read mapping and are available from http://paramecium.cgm.cnrs-gif.fr/download/fasta/assemblies: MAC strain 51 reference (ptetraurelia_mac_51.fa); MAC strain 51+IES reference (ptetraurelia_mac_51_with_ies.fa); PGM contigs (ptetraurelia_PGM_k51_ctg.fa), a proxy for the MIC genome.
Identification of Spt5 proteins and tree construction
HMMER v3.1b2 (48) was used (hmmsearch, with default parameters) to search protein databases in fasta format (UniProt or, for Paramecium biaurelia, Paramecium sexaurelia and Paramecium caudatum, species-specific databases (49,50)) with PF03439, the PFAM Spt5-NGN domain (51). The Spt5 amino acid sequences were aligned with MUSCLE v3.8.31 (52) and a tree was constructed using the BioNJ algorithm (53) implemented in SeaView v4.3.3 (54), with the ‘Poisson and Kimura’ protein distance and 1000 bootstrap replicates.
RESULTS
Two Spt5-encoding genes in Paramecium
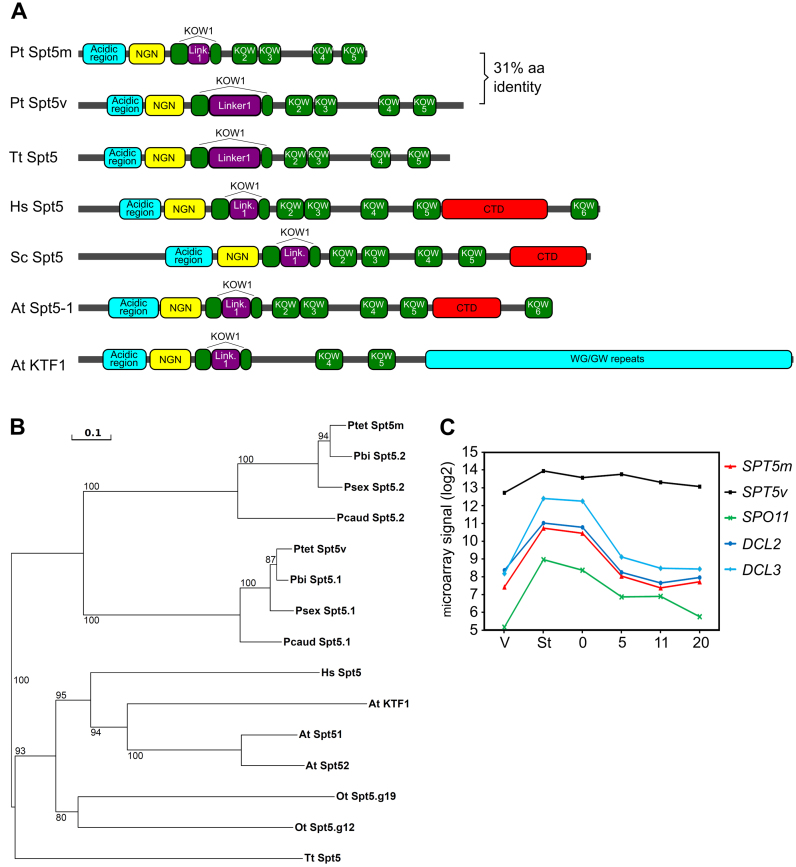
Putative homologs of elongation factor Spt5 were identified by a BLAST search of the P. tetraurelia macronuclear genome (55), using the sequence of the human Spt5 protein as query. A PFAM domain search (51) showed that both of the putative Paramecium homologs contain the conserved Spt5-NGN domain (PF03439). The overall structure of the putative Paramecium Spt5 proteins reveals known characteristics of Spt5 (56): an N-terminal acidic region, a single NGN domain flanked by KOW domains and four additional KOW domains (Figure 1A and Supplementary Figure S1). The predicted secondary structure of the Paramecium Spt5-NGN domain appears to be conserved since we found the same order of β-sheets and α-helices as in Spt5 proteins from plants, animals and archaea (Supplementary Figure S1A). However, these proteins lack a classical CTD domain, that may contribute to Spt5 regulation via phosphorylation (24). Spt5v alone contains tyrosine residues close to the C-terminus that potentially may be phosphorylated (Supplementary Figure S1). Considering the fact that CTD was shown previously not to be essential for cell survival in HeLa cells (57), the importance of CTD in eukaryotic Spt5 proteins and the regulation by phosphorylation of Paramecium Spt5 proteins are open questions.
Figure 1.
Spt5 proteins in Paramecium. (A) Predicted protein domains (colored boxes) for Paramecium tetraurelia, Tetrahymena thermophila, Homo sapiens, Saccharomyces cerevisiae and Arabidopsis thaliana Spt5 proteins. (B) Neighbor-joining tree of Spt5 proteins from ciliates based on the alignment of entire protein sequences (461 positions in the final dataset) and the following parameters: bootstrap 1000 (bootstrap values displayed next to the branches) and Poisson correction. Ptet, P. tetraurelia; Pbi, Paramecium biaurelia; Psex, Paramecium sexaurelia; Pcaud, Paramecium caudatum; Ot, Oxytricha trifallax; Tt, T. thermophila, human (Hs) and arabidopsis (At). (C) Autogamy time-course of gene expression for P. tetraurelia SPT5 (GSPATG00013468001, GSPATG00023145001), DCL2 (GSPATG00008494001), DCL3 (GSPATG00027456001) and SPO11 (GSPATG00009108001). The Y-axis shows the log2 signals from custom microarrays (55,59).
We also found two putative Spt5 proteins in P. caudatum, P. sexaurelia and P. biaurelia (49,50). We used the Spt5 sequences from the four Paramecium species, the ciliates Oxytricha trifallax and Tetrahymena thermophila, human and Arabidopsis to build a neighbor-joining tree (Figure 1B). The tree topology indicates that SPT5 gene duplications in Arabidopsis and in Oxytricha occurred independently of each other and of the Paramecium duplication. Furthermore, Paramecium SPT5 genes appeared before the divergence of P. caudatum and Paramecium aurelia, so the origin of the Paramecium Spt5 proteins, which share only 31% amino acid identity, can be attributed to a gene (or whole genome) duplication that occurred before the two most recent whole genome duplications characterized in this lineage (50,58). Comparison of Spt5v with Spt5m (Supplementary Figure S1B) shows that these proteins share domain composition and secondary structure despite their divergent sequences. Spt5v is longer mainly due to N- and C-terminal unstructured regions that are absent in Spt5m, as well as an unusually long Linker1 within KOW1.
Distinct gene expression and protein localization
The two P. tetraurelia SPT5 genes have very different expression profiles (Figure 1C), according to the transcriptome data available in the P. tetraurelia microarray resource (55,59). Since one of the genes—GSPATG00013468001—is strongly expressed during vegetative growth, we named it SPT5v for ‘vegetative’. The other gene—GSPATG00023145001—is differentially expressed during sexual processes. Since the maximum of its expression is reached at early stages, most likely during meiosis, as confirmed by the profiles of genes known to have meiotic functions in Paramecium, SPO11 (35) and DCL2 (29,30), we named this gene SPT5m for ‘meiosis’.
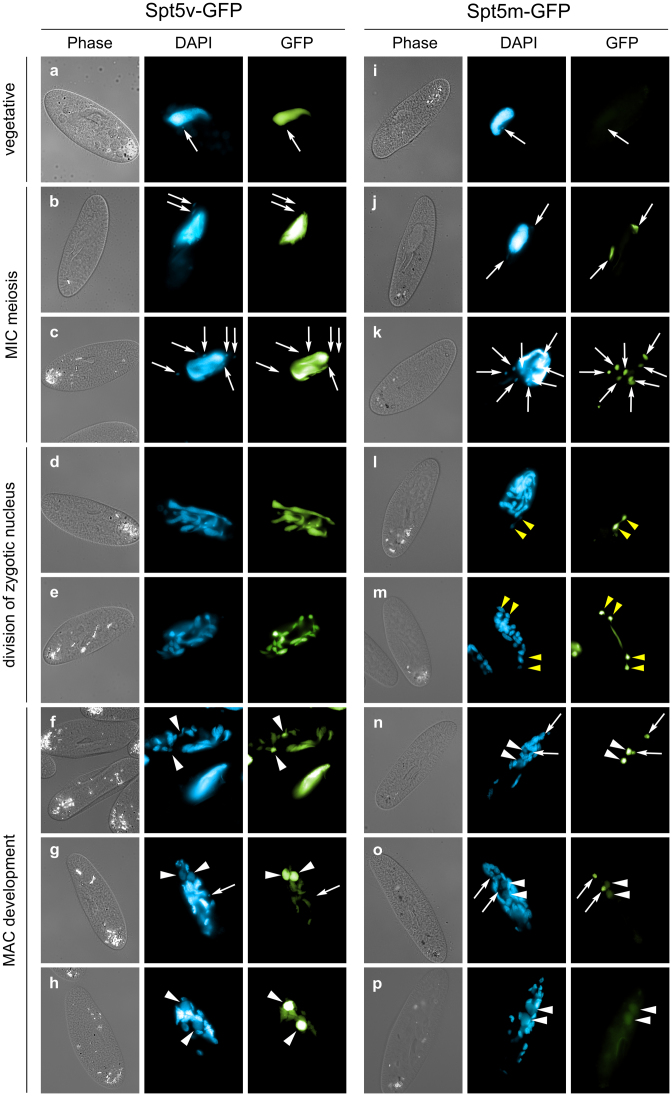
In order to visualize the relationship between Spt5v and Spt5m proteins and nuclear compartments, Paramecium cells were transformed with constructs encoding a C-terminal GFP fusion for each protein. GFP fluorescence was monitored in vegetative cells and during the sexual process of autogamy (self-fertilization). Spt5v-GFP signal was detected in the MACs of vegetative cells (Figure 2, panel a), in MACs undergoing fragmentation and in MAC fragments during autogamy (Figure 2, panels d and e). Spt5v-GFP localizes in the new MACs as soon as they start to differentiate (Figure 2, panel f). As new MACs grow (Figure 2, panel g and h) the GFP signal accumulates in new MACs and almost disappears from fragments of the old MAC. Our observations clearly indicate that Spt5v-GFP is not connected with germline nuclei at any stage and support the hypothesis that Spt5v is important for the expression of the somatic genome.
Figure 2.
Localization of Spt5-GFP fusion proteins. For both SPT5v-GFP and SPT5m-GFP transgenes, representative images illustrate different developmental stages. Panels a and i show vegetative cells. Successive stages of autogamy are shown in the following panels: panels b and j—meiotic crescent stage; panels c and k—cells with eight haploid nuclei resulting from meiosis II (only six nuclei are visible in panel C); panels d, e, l and m—fragmentation of old MAC; panel L—first division of the zygotic nucleus; panel M—second division of the zygotic nucleus; panels f and n—early MAC development; panels g, h, o and p—late MAC development. In all panels, white arrows point at MICs (some were omitted when MICs were not clearly distinguishable by DAPI staining), white arrowheads indicate new MACs. Yellow arrowheads in panels L and M point to division products of the zygotic nucleus. The Spt5v-GFP fusion (left side) localizes to old, then new MACs. The Spt5m-GFP (right side) localizes to meiotic MICs, the zygotic nucleus as well as products of division of the zygotic nucleus.
In accordance with the microarray data, Spt5m-GFP is not detected during vegetative growth (Figure 2, panel i). At the beginning of autogamy, GFP signal was detected in meiotic MICs (Figure 2, panel j) and then in the eight haploid products of the meiotic divisions (Figure 2, panel k). Spt5m-GFP is present in the zygotic nucleus arising from self-fertilization and in the products of its division by mitosis (Figure 2, panel l and m). The protein stays in the new MICs and the new MACs during their early development (Figure 2, panel n and o) and finally GFP fluorescence disappears from both compartments (Figure 2, panel p). Thus Spt5m is clearly associated with germline nuclei during sexual processes.
Spt5v is important during vegetative growth while Spt5m is essential for development
To investigate Spt5 function, we knocked-down the expression of SPT5v or SPT5m using RNA interference (60). As the two genes are completely different at the nucleotide level, cross-RNAi is not possible. First, we silenced both genes during vegetative growth and observed cell survival, division rate and cell morphology over a 3-day period, as previously described (41). Cells subjected to SPT5m RNAi grew normally (similar to control cells) and were able to make approximately four divisions per day. On the contrary, Spt5v-depleted cells divided only twice after 24 h of silencing and gradually decreased their division rate day after day and eventually practically stopped cell division (Supplementary Figure S2). At the same time, lethality was quite low—only 13% of cells on average died every day. RT-PCR analysis confirmed that exposure to RNAi leads to significant reduction of SPT5v mRNA level after 32–48 h of silencing (Supplementary Figure S4A).
Secondly, we studied the influence of SPT5m expression on the progression of the sexual cycle by letting cells starve and enter autogamy in silencing medium. We were unable to perform this analysis for SPT5v as—given the slow-growth phenotype—it was not possible to control the cell cycle. Northern blot and RT-PCR analysis showed that when SPT5m was silenced, its mRNA level decreased while SPT5v was constitutively expressed (Supplementary Figure S4C and D). RNAi against SPT5m led to a severe lethality phenotype in post-autogamous progeny, with only ∼1% of survival (Supplementary Table S1). The cells were unable to proliferate after the sexual process—they died before or just after the first (karyonidal) division. Cytological observation of DAPI-stained cells confirmed that Spt5m-depleted cells are able to undergo meiosis and that new MACs are formed and amplify DNA normally (Supplementary Figure S3) and exhibit transcriptional activity (Supplementary Figure S5). Similar results were obtained during conjugation—SPT5m silencing led to 89% lethality in post-conjugation progeny (Supplementary Table S2), even though conjugation proceeded normally without delay in couple formation, couple separation or karyonidal division. We conclude that Spt5m is essential for sexual reproduction and development, as reported in metazoans (15,61,62).
Spt5m is necessary for scnRNA accumulation
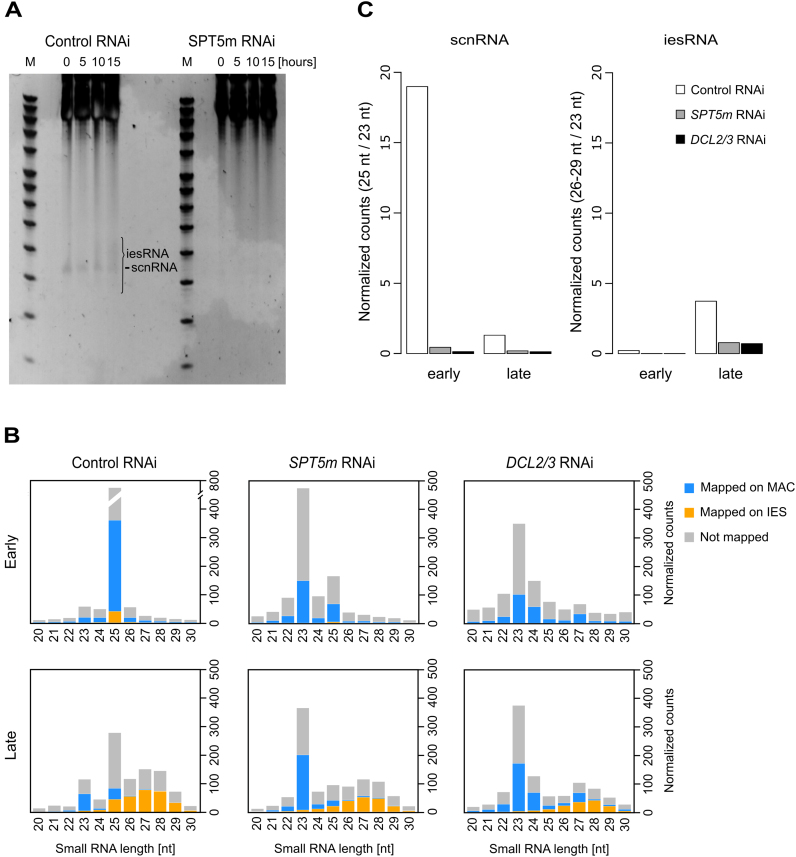
In order to see whether Spt5m depletion affects sRNA levels, we first used PAGE coupled to SYBR gold staining. Both scnRNA (25 nt) and iesRNA (26–29 nt) were visible in the control autogamy time series, however no sRNA could be detected at any time after Spt5m depletion (Figure 3A). In order to confirm this result, we used high-throughput sRNA sequencing. We compared sRNA populations present in control and SPT5m-silenced cells at two time-points: early autogamy (T0, when old MAC fragments can be seen in ∼50% of cells which are thus post-meiotic; T0 is operationally defined as the starting point of development; for details see Supplementary Figure S4B and review (63)) and a later time-point (T15, 15 h after T0, when new developing MACs can be seen in ∼100% of cells) corresponding to development of the new MACs (Figure 3B and Supplementary Table S3). We normalized the read counts (cf. ‘Materials and Methods’ section), and compared the relative amounts of scnRNAs and iesRNAs compared to the 23 nt siRNAs assumed to remain constant (Figure 3C).
Figure 3.
Analysis of sRNA populations in SPT5m-RNAi cells. (A) Gel electrophoresis of sRNA from SPT5m-silenced cells and control silencing. Total RNA samples corresponding to the T0, T5, T10 and T15 time-points were run on a denaturing 15% polyacrylamide-urea gel. After electrophoresis the gel was stained with SYBR Gold (Invitrogen). M: DNA Low Molecular Weight Marker (USB). The ∼25 nt signal that corresponds to the fraction of scnRNAs (29) is labeled. In the control, at the T15 time-point, additional bands corresponding to 26–30 nt iesRNAs are present (indicated by a bracket). In SPT5m-silenced samples neither scnRNAs nor iesRNAs can be seen. (B) Small RNA libraries corresponding to early and late development time-points were sequenced and mapped to the reference genomes (Paramecium tetraurelia MAC reference genome and MAC+IES reference genome). Results obtained for a control culture (cells silenced for ND7 gene expression) are shown in the left hand panel, SPT5m-silencing in the middle panel and DCL2/3-silencing (from (30)) in the right hand panel. Stacked bar plots show the normalized number of sRNA reads that match the MAC genome (blue), annotated IESs (yellow) or were not mapped (gray). (C) Read counts obtained during sequencing of small RNA libraries corresponding to early and late development time-points were normalized. Bar plots show the relative amounts of sRNA reads for control (white), SPT5-RNAi (gray) and DCL2/3-RNAi (black, for details of see (30)).The left hand panel corresponds to scnRNA and shows the number of 25 nt reads divided by the number of 23 nt siRNA reads for each sample. Right hand panel shows iesRNA reads (26–29 nt) in relation to 23 nt reads. Since iesRNAs are supposed to be produced by Dcl5 from excised IESs (30), their appearance in the late sample could be reduced owing to reduced IES excision in SPT5 and DCL2/3-RNAi.
At early autogamy, 25-nt scnRNAs constitute a majority of the sRNAs in the control sample, as reported previously (29,30), (Figure 3B). Indeed, in our control the scnRNAs are much more abundant than 23 nt sRNAs (ratio of 19). However, the situation is strikingly different after Spt5m depletion as the ratio is only 0.22, a 40-fold decrease (Figure 3C). This disappearance of scnRNAs is comparable to that found after depletion of Dcl2/Dcl3 (Figure 3C), proteins known to be required for scnRNA biogenesis (29,30) (Figure 3B). Spt5m may thus be involved in an early phase of scnRNA production, as a putative component of the transcriptional machinery.
The level of iesRNAs is also reduced under SPT5m-RNAi, although the effect is less striking than for the scnRNAs. In the control, the iesRNAs at the late time point are nearly four times more abundant than the 23 nt sRNAs. Upon Spt5m depletion, the ratio is reduced to 0.78, a more than 4-fold reduction (Figure 3C): we observe a similar reduction after Dcl2/Dcl3 depletion. Thus, Spt5m depletion shows very similar effects on developmental sRNA levels as Dcl2/Dcl3 depletion.
Spt5m is required for Dcl2/3-independent IES excision
The fact that sRNA profiles obtained after Spt5m depletion are very similar to those obtained after Dcl2/3 depletion (29,30) suggests that Spt5m and Dcl2/3 function in the same pathway. To see whether Spt5m depletion has the same effect as Dcl2/3 depletion on developmental DNA elimination, we performed high-throughput sequencing of DNA enriched in new MACs that developed under SPT5m-RNAi. We first estimated the effects of Spt5m depletion on the retention of germline-limited sequences other than IESs, by comparing sequence complexity obtained upon SPT5m-RNAi with sequence complexity upon PGM-RNAi. The latter provides a proxy for the MIC genome, since the Pgm domesticated transposase is required for essentially all DNA elimination (35). As expected, 99.6% of germline-specific sequences are retained after SPT5m silencing (Table 1). This value is similar to that found after Dcl2/3 depletion (Table 1), in agreement with previous work showing that scnRNAs are required for all non-IES developmental DNA elimination (37).
Table 1. Sequence complexity in control (wild-type developing genome), PGM KD, SPT5m KD and DCL2/3 KD.
| Dataset | PGM | SPT5m | DCL2/3 | Control |
|---|---|---|---|---|
| PGM | 89.00 Mb | 88.64 Mb | 88.74 Mb | 76.09 Mb |
| 100.0% | 99.6% | 99.71% | 85.5% | |
| PGM not Control | 12.91 Mb | 12.66 Mb | 12.66 Mb | 0.00 Mb |
| 100.0% | 98.05% | 98.04% | 0.0% |
The 91 Mb PGM assembly was used as a proxy for the germline genome, as explained in the text. Samples of Illumina paired-end reads were mapped to the assembly and regions covered by at least 2 RPKM (reads per kilobase per million mapped reads) were scored as explained in ‘Materials and Methods’ section. The stringency of this cutoff explains the value of 89 Mb found for the PGM sample itself. The ‘PGM’ reference contains contigs covered by the PGM dataset. The ‘PGM not Control’ contains contigs covered by the PGM dataset but not by the control (wild-type) dataset, representing the MIC restricted regions. Each column indicates the sum of the lengths of contigs covered by the given dataset.
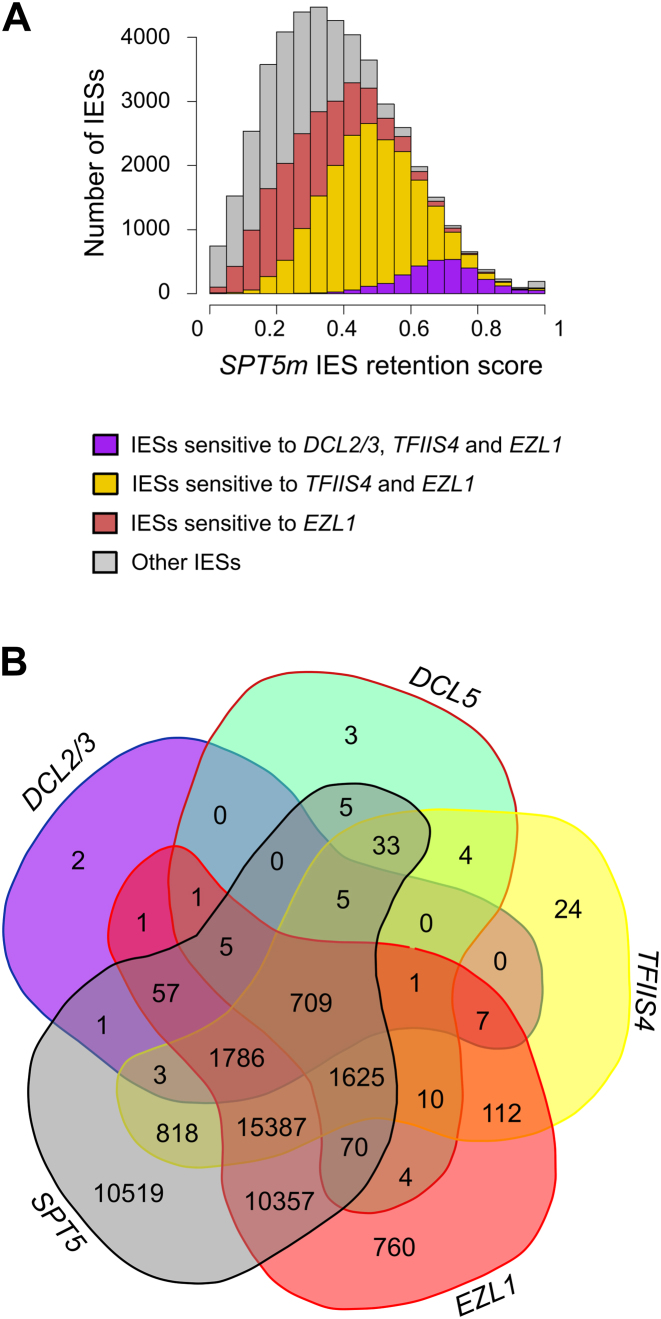
We obtained an unexpected result when we analyzed IES retention: excision of all IESs seems to be altered and 92% (∼41 000) of the IESs are significantly retained in the developing MAC after SPT5m-RNAi, compared to only 7% after DCL2/3-RNAi (37,38) (Supplementary Table S4 and Figure S6) or ∼45% after silencing of TFIIS4 (34). We observed a broad and non-normal (Shapiro test, P < 10−16) distribution of IES retention scores (mean 0.38, median 0.36) (Figure 4). We further found that the IES retention scores measured after the depletion of other factors are correlated with the Spt5m retention scores: Dcl2/3-sensitive IESs have the highest Spt5m retention scores (spearman correlation coefficient, scc = 0.58), followed by TFIIS4-sensitive IESs (scc = 0.76) and then by EZL1-sensitive IESs (scc = 0.64) (Figure 4 and Supplementary Figure S7). There is however no correlation between Spt5m retention scores and genomic location (intragenic, intergenic, position on scaffold) of the IESs and there is only a weak correlation with IES size (Supplementary Figure S8). Thus, Spt5 depletion affects excision of nearly all IESs. Moreover, the effects are graded and well-correlated with the effects of epigenetic factors known to program IES excision.
Figure 4.
Analysis of IES excision in SPT5m-silenced cells. (A) Retention score distribution determined by re-sequencing DNA extracted from a cell fraction enriched in new MACs, after SPT5m silencing. IESs with a given SPT5m retention score that are sensitive to DCL2/3, TFIIS4 or EZL1 as determined using published data (34,37,38) are indicated. Bar coloring: purple, DCL2/3 + TFIIS + EZL1; yellow, TFIIS4 + EZL1; red, EZL1. (B) Venn diagram showing significantly retained IESs after SPT5m, EZL1, TFIIS4, DCL2/3 or DCL5 silencing. Details can be found in Supplementary Table S4.
DISCUSSION
Throughout eukaryotes, transcription allows production of different classes of coding and non-coding RNAs in coordination with other biological processes (splicing, RNA nuclear export, chromatin modification, etc.). Among the numerous proteins involved, we have studied the transcription elongation factor Spt5. We have shown here that Paramecium species have two SPT5 genes specialized with regard to their expression and localization patterns: Spt5v expressed during vegetative growth and Spt5m dedicated to meiosis and development. The presence of more than one SPT5 gene is rare in eukaryotes and has been reported so far only for plants and ciliates (5,13).
Our results suggest that the localization in the somatic nucleus and the function of Spt5v, whose depletion leads to a slow growth phenotype, is similar to that of Spt5 during vegetative growth in other eukaryotes: yeast cold-sensitive SPT5 mutant exhibited slow-growth (10), silencing of SPT5 in mammalian HeLa cells caused reduced division rate and apoptosis (57) and in Arabidopsis thaliana reduction of fresh weight was observed (13). We conclude that Spt5v is an important transcription factor in Paramecium that may play a key role in expression of the somatic genome.
In contrast, Spt5m is expressed starting at meiosis and is localized in the germline MICs, their haploid meiotic products and the zygotic nucleus after karyogamy. It is known that in Paramecium, meiosis initiates the program of defense against transposable elements (TEs), known as genome scanning (63). Genome scanning implicates generalized transcription of the germline MIC genome that results in 25 nt scnRNAs. scnRNA maturation from putative dsRNA precursors requires the Dicer-like proteins Dcl2 and Dcl3. The working model for genome scanning predicts that sequences absent from the maternal MAC will be targeted for deletion by the scnRNAs during development (63). The absence of scnRNA owing to Dcl2/Dcl3 or Spt5m depletion impairs the deletion of the bulk of germline-limited sequences, including known TEs (27), in accordance with the model. This leads us to hypothesize that Spt5m is involved in the generalized transcription of the germline genome at meiosis. We cannot exclude the possibility that the effect on scnRNA production we observe upon SPT5m-RNAi is indirect. At the time of meiosis, some Spt5m-dependent mRNA might be produced from the MIC genome and a protein product from those transcripts could be responsible for regulation of non-coding RNA transcription.
DCL2/DCL3 KD impairs scnRNA production and elimination of repeated sequences but does not affect excision of most IESs (30,37,38). How is Paramecium able to target the excision of thousands of unique sequences without sRNAs? Epigenetic factors have been shown to be necessary for Dcl2/Dcl3-dependent and -independent IES excision. These include the transcription elongation factor TFIIS4 (34) and the histone methyltransferase Ezl1 necessary for the acquisition of H3K9me3 and H3K27me3 epigenetic marks (37), both of which localize in the developing MAC. Previous publications showed that all IESs that require Dcl2/Dcl3 also require TFIIS4, and all TFIIS4 IESs also require EZL1. Finally, about a third of known IESs are efficiently excised in the absence of any of these epigenetic factors. It is striking that the IESs with the highest Spt5 retention score are those which require all of these factors. We can picture this situation (Figure 4) as a set of Russian dolls, with Dcl2/Dcl3 the smallest one, inside TFIIS4, inside Ezl1 and with Spt5 being the largest one. This illustration is consistent with the model of evolution of the IES recognition system proposed recently (34,37) and points to a role of Spt5m as the most general factor known to be involved in this process.
Our localization experiments showed that Spt5m appears at meiosis in MICs where and when scnRNAs are produced, well before the appearance of developing MACs. Later in development, an effect of Spt5m depletion is the retention of nearly all IESs. If Spt5m were necessary only for scnRNA production, we would have observed only the retention of the IESs that require Dcl proteins for their complete excision. It is therefore tempting to speculate that Spt5m is not only involved in meiotic germline transcription to provide scnRNA precursors, but also allows modifications, introduced most probably co-transcriptionally, that encode information used later for targeting IES excision. Although we do not yet know how this epigenetic information is encoded—known or unknown histone modifications, DNA base modifications or histone exchange—it clearly must persist through meiosis and fertilization in order to influence genome rearrangements. It is important to note that this potential function of Spt5m may be expressed as well in the post-meiotic MIC and/or in the new MAC as Spt5m-GFP appears in these compartments.
In conclusion, we have shown for the first time in any eukaryote, that an Spt5 homolog is an essential factor for the production of development-specific non-coding RNAs. As Spt5/NusG proteins have been conserved across evolution, it is possible that a similar function exists in other organisms. Our results provide a new perspective on the diverse transcription-dependent and independent roles of Spt5. Furthermore, these results can be considered as a new example of the specialization of transcriptional complexes toward non-coding RNA synthesis, similarly to the action of Mediator complex in Schizosaccharomyces pombe and A. thaliana (6,8), Rpb2 in S. pombe (4) or polymerases dedicated to non-coding RNA production—Pol IV and Pol V in plants (7). Paramecium, as a complex unicellular organism in which different transcriptional programs can be studied independently, thanks to nuclear dimorphism, provides an excellent model for exploration of non-coding transcription. Future work will be required to characterize the transcripts produced, identify the components of the transcriptional machinery and determine their biochemical functions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dorota Adamska from the Andrzej Dziembowski lab for help in Illumina sequencing. We are grateful to Mireille Bétermier for encouragement throughout this study and for useful comments on the manuscript.
Author contributions: J.K.N. conceived the study; J.G. and J.K.N. performed the experiments; O.A., C.D.W. and L.S. performed data analysis; J.G., C.D.W., L.S. and J.K.N. prepared the manuscript.
Footnotes
Present address: Cyril Denby Wilkes, Institut de Biologie et deTechnologies de Saclay (IBITECS), CEA, F-91191 Gif-sur-Yvette Cedex, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Polish National Science Centre [N303 808840, 2015/17/B/NZ2/01173 to J.K.N.]; French National Research Agency [ANR-12-BSV6-0017 “INFERNO" to L.S.]. Funding for open access charge: Centre National de la Recherche Scientifique [European Research Group “Paramecium Genome Dynamics and Evolution"].
Conflict of interest statement. None declared.
REFERENCES
- 1. Holoch D., Moazed D.. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015; 16:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Castel S.E., Martienssen R.A.. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013; 14:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castel S.E., Martienssen R.A.. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013; 14:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kato H., Goto D.B., Martienssen R.A., Urano T., Furukawa K., Murakami Y.. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005; 309:467–469. [DOI] [PubMed] [Google Scholar]
- 5. Khurana J.S., Wang X., Chen X., Perlman D.H., Landweber L.F.. Transcription-independent functions of an RNA polymerase II subunit, Rpb2, during genome rearrangement in the ciliate, oxytricha trifallax. Genetics. 2014; 197:839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oya E., Kato H., Chikashige Y., Tsutsumi C., Hiraoka Y., Murakami Y.. Mediator directs co-transcriptional heterochromatin assembly by RNA interference-dependent and -independent pathways. PLoS Genet. 2013; 9:e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haag J.R., Pikaard C.S.. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011; 12:483–492. [DOI] [PubMed] [Google Scholar]
- 8. Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X.. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011; 30:814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 2012; 417:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartzog G.A., Wada T., Handa H., Winston F.. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998; 12:357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F. et al. . DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998; 12:343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwak H., Lis J.T.. Control of transcriptional elongation. Annu. Rev. Genet. 2013; 47:483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Durr J., Lolas I.B., Sørensen B.B., Schubert V., Houben A., Melzer M., Deutzmann R., Grasser M., Grasser K.D.. The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 2014; 42:4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaplan C.D., Morris J.R., Wu C., Winston F.. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000; 14:2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keegan B.R., Feldman J.L., Lee D.H., Koos D.S., Ho R.K., Stainier D.Y., Yelon D.. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development. 2002; 129:1623–1632. [DOI] [PubMed] [Google Scholar]
- 16. Mayer A., Lidschreiber M., Siebert M., Leike K., Soding J., Cramer P.. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010; 17:1272–1278. [DOI] [PubMed] [Google Scholar]
- 17. Hartzog G.A., Fu J.. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim. Biophys. Acta. 2012; 1829:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. NandyMazumdar M., Artsimovitch I.. Ubiquitous transcription factors display structural plasticity and diverse functions: NusG proteins—shifting shapes and paradigms. Bioessays. 2015; 37:324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V.. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010; 143:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He X.-J., Hsu Y.-F., Zhu S., Wierzbicki A.T., Pontes O., Pikaard C.S., Liu H.-L., Wang C.-S., Jin H., Zhu J.-K.. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009; 137:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rowley M.J., Avrutsky M.I., Sifuentes C.J., Pereira L., Wierzbicki A.T.. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 2011; 7:e1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirtreiter A., Damsma G.E., Cheung A.C., Klose D., Grohmann D., Vojnic E., Martin A.C., Cramer P., Werner F.. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010; 38:4040–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meyer P.A., Li S., Zhang M., Yamada K., Takagi Y., Hartzog G.A., Fu J.. Structures and functions of the multiple KOW domains of transcription elongation factor Spt5. Mol. Cell. Biol. 2015; 35:3354–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mbogning J., Pagé V., Burston J., Schwenger E., Fisher R.P., Schwer B., Shuman S., Tanny J.C.. Functional interaction of Rpb1 and Spt5 C-terminal domains in co-transcriptional histone modification. Nucleic Acids Res. 2015; 43:9766–9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duharcourt S., Lepère G., Meyer E.. Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 2009; 25:344–350. [DOI] [PubMed] [Google Scholar]
- 26. Le Mouël A., Butler A., Caron F., Meyer E.. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot. Cell. 2003; 2:1076–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arnaiz O., Mathy N., Baudry C., Malinsky S., Aury J.M., Wilkes C.D., Garnier O., Labadie K., Lauderdale B.E., Le Mouël A. et al. . The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012; 8:e1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carradec Q., Gotz U., Arnaiz O., Pouch J., Simon M., Meyer E., Marker S.. Primary and secondary siRNA synthesis triggered by RNAs from food bacteria in the ciliate Paramecium tetraurelia. Nucleic Acids Res. 2015; 43:1818–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lepère G., Nowacki M., Serrano V., Gout J.-F., Guglielmi G., Duharcourt S., Meyer E.. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2009; 37:903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sandoval P.Y., Swart E.C., Arambasic M., Nowacki M.. Functional diversification of dicer-like proteins and small RNAs required for genome sculpting. Dev. Cell. 2014; 28:174–188. [DOI] [PubMed] [Google Scholar]
- 31. Bouhouche K., Gout J.F., Kapusta A., Bétermier M., Meyer E.. Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res. 2011; 39:4249–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coyne R.S., Lhuillier-Akakpo M., Duharcourt S.. RNA-guided DNA rearrangements in ciliates: is the best genome defence a good offence. Biol. Cell Auspices Eur. Cell Biol. Organ. 2012; 104:309–325. [DOI] [PubMed] [Google Scholar]
- 33. Lepère G., Bétermier M., Meyer E., Duharcourt S.. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008; 22:1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maliszewska-Olejniczak K., Gruchota J., Gromadka R., Denby Wilkes C., Arnaiz O., Mathy N., Duharcourt S., Bétermier M., Nowak J.K.. TFIIS-dependent noncoding transcription regulates developmental genome rearrangements. PLoS Genet. 2015; 11:e1005383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baudry C., Malinsky S., Restituito M., Kapusta A., Rosa S., Meyer E., Bétermier M.. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009; 23:2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh D.P., Saudemont B., Guglielmi G., Arnaiz O., Goût J.-F., Prajer M., Potekhin A., Przybòs E., Aubusson-Fleury A., Bhullar S. et al. . Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature. 2014; 509:447–452. [DOI] [PubMed] [Google Scholar]
- 37. Lhuillier-Akakpo M., Frapporti A., Denby Wilkes C., Matelot M., Vervoort M., Sperling L., Duharcourt S.. Local effect of enhancer of zeste-like reveals cooperation of epigenetic and cis-acting determinants for zygotic genome rearrangements. PLoS Genet. 2014; 10:e1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swart E.C., Wilkes C.D., Sandoval P.Y., Arambasic M., Sperling L., Nowacki M.. Genome-wide analysis of genetic and epigenetic control of programmed DNA deletion. Nucleic Acids Res. 2014; 42:8970–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nelson M.D., Fitch D.H.. Overlap extension PCR: an efficient method for transgene construction. Methods Mol. Biol. 2011; 772:459–470. [DOI] [PubMed] [Google Scholar]
- 40. Nowacki M., Zagorski-Ostoja W., Meyer E.. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 2005; 15:1616–1628. [DOI] [PubMed] [Google Scholar]
- 41. Nowak J.K., Gromadka R., Juszczuk M., Jerka-Dziadosz M., Maliszewska K., Mucchielli M.H., Gout J.F., Arnaiz O., Agier N., Tang T. et al. . Functional study of genes essential for autogamy and nuclear reorganization in Paramecium. Eukaryot. Cell. 2011; 10:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gratias A., Bétermier M.. Processing of double-strand breaks is involved in the precise excision of paramecium internal eliminated sequences. Mol. Cell. Biol. 2003; 23:7152–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Timmons L., Fire A.. Specific interference by ingested dsRNA. Nature. 1998; 395:854–854. [DOI] [PubMed] [Google Scholar]
- 44. Garnier O., Serrano V., Duharcourt S., Meyer E.. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 2004; 24:7370–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gogendeau D., Klotz C., Arnaiz O., Malinowska A., Dadlez M., de Loubresse N.G., Ruiz F., Koll F., Beisson J.. Functional diversification of centrins and cell morphological complexity. J. Cell Sci. 2008; 121:65–74. [DOI] [PubMed] [Google Scholar]
- 46. Denby Wilkes C., Arnaiz O., Sperling L.. ParTIES: a toolbox for Paramecium interspersed DNA elimination studies. Bioinformatics. 2016; 32:599–601. [DOI] [PubMed] [Google Scholar]
- 47. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eddy S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGrath C.L., Gout J.-F., Johri P., Doak T.G., Lynch M.. Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 2014; 24:1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McGrath C.L., Gout J.-F., Doak T.G., Yanagi A., Lynch M.. Insights into three whole-genome duplications gleaned from the Paramecium caudatum genome sequence. Genetics. 2014; 197:1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J. et al. . Pfam: the protein families database. Nucleic Acids Res. 2014; 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997; 14:685–695. [DOI] [PubMed] [Google Scholar]
- 54. Gouy M., Guindon S., Gascuel O.. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010; 27:221–224. [DOI] [PubMed] [Google Scholar]
- 55. Arnaiz O., Sperling L.. ParameciumDB in 2011: new tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 2011; 39:D632–D636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ponting C.P. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002; 30:3643–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Komori T., Inukai N., Yamada T., Yamaguchi Y., Handa H.. Role of human transcription elongation factor DSIF in the suppression of senescence and apoptosis. Genes Cells. 2009; 14:343–354. [DOI] [PubMed] [Google Scholar]
- 58. Aury J.-M., Jaillon O., Duret L., Noel B., Jubin C., Porcel B.M., Ségurens B., Daubin V., Anthouard V., Aiach N. et al. . Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006; 444:171–178. [DOI] [PubMed] [Google Scholar]
- 59. Arnaiz O., Goût J.-F., Bétermier M., Bouhouche K., Cohen J., Duret L., Kapusta A., Meyer E., Sperling L.. Gene expression in a paleopolyploid: a transcriptome resource for the ciliate Paramecium tetraurelia. BMC Genomics. 2010; 11:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Galvani A., Sperling L.. RNA interference by feeding in Paramecium. Trends Genet. TIG. 2002; 18:11–12. [DOI] [PubMed] [Google Scholar]
- 61. Shim E.Y., Walker A.K., Shi Y., Blackwell T.K.. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002; 16:2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jennings B.H., Shah S., Yamaguchi Y., Seki M., Phillips R.G., Handa H., Ish-Horowicz D.. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr. Biol. 2004; 14:1680–1684. [DOI] [PubMed] [Google Scholar]
- 63. Betermier M., Duharcourt S.. Programmed rearrangement in ciliates: paramecium. Microbiol. Spectr. 2014; 2, MDNA3-0035-2014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.