Abstract
The type II arginine methyltransferase PRMT5 is responsible for the symmetric dimethylation of histone to generate the H3R8me2s and H4R3me2s marks, which correlate with the repression of transcription. However, the protein level of a number of genes (MEP50, CCND1, MYC, HIF1a, MTIF and CDKN1B) are reported to be downregulated by the loss of PRMT5, while their mRNA levels remain unchanged, which is counterintuitive for PRMT5's proposed role as a transcription repressor. We noticed that the majority of the genes regulated by PRMT5, at the posttranscriptional level, express mRNA containing an internal ribosome entry site (IRES). Using an IRES-dependent reporter system, we established that PRMT5 facilitates the translation of a subset of IRES-containing genes. The heterogeneous nuclear ribonucleoprotein, hnRNP A1, is an IRES transacting factor (ITAF) that regulates the IRES-dependent translation of Cyclin D1 and c-Myc. We showed that hnRNP A1 is methylated by PRMT5 on two residues, R218 and R225, and that this methylation facilitates the interaction of hnRNP A1 with IRES RNA to promote IRES-dependent translation. This study defines a new role for PRMT5 regulation of cellular protein levels, which goes beyond the known functions of PRMT5 as a transcription and splicing regulator.
INTRODUCTION
Arginine methylation is a prevalent posttranslational modification, and roughly 0.3% of all arginine residues in cellular proteins are methylated (1). Three types of methylarginine species exist: monomethylarginine (MMA), asymmetric dimethylarginine (ADMA) and symmetric dimethylarginine (SDMA). To date, nine protein arginine methyltransferases (PRMTs) have been identified that lay down these three different methyl marks (PRMT1-9) (2). PRMT5 is a type II methyltransferase that is responsible for depositing the majority of the SDMA modification in the cell. It generates the H2AR3me2s, H3R8me2s and H4R3me2s histone marks, which are generally believed to associate with repressed gene expression (3–7). However, in certain contexts PRMT5 also functions as a transcription activator (8–10).
A number of gene products are regulated by PRMT5 at the protein level, while their mRNA levels remain unchanged, suggesting a role for PRMT5 in posttranslational regulation. These include MEP50, CCND1, MYC, HIF1a, MITF and CDKN1B (11–13). The mechanism of the posttranscriptional regulation by PRMT5 is not fully understood, although it has a very important role in splicing (14–16). There are many additional cellular processes, apart from splicing, that regulate the fate of mRNAs and proteins after transcription, including mRNA export, translation, and targeted mRNA and protein degradation. Interestingly, four of the six reported genes regulated by PRMT5 posttranscriptionally (CCND1, MYC, HIF1a, CDKN1B) contain an internal ribosome entry site (IRES) within the 5΄ untranslated region (UTR) of their mRNA (17–20). IRESes are secondary structures usually formed within the 5΄ UTR of mRNAs, which recruit the 40S ribosome to mRNA and initiate translation independent of the m7GpppG cap (21,22). Cap-independent translation requires a reduced subset of initiation factors (as compared to cap-dependent translation) and accounts for ∼5% of initiation events.
It has long been known that the heterogeneous nuclear ribonucleoproteins (hnRNPs) undergo extensive posttranslational modification by PRMTs (23,24). The hnRNPs execute a multitude of functions related to RNA metabolism including splicing, export, localization, stability and translation (25), which could be regulated by these posttranslational modifications. hnRNP A1 is a well-studied member of the heterogeneous nuclear ribonucleoprotein family (hnRNP family) (26), and it has been implicated in aspects of translation control. Indeed, hnRNP A1 is reported to regulate the IRES-dependent translation of CCND1 and MYC genes (27) as well as additional IRES-containing genes (28–30). Post-translational modifications such as phosphorylation and methylation on hnRNP A1 have been studied. While phosphorylation of hnRNP A1 at Ser199 has been demonstrated to inhibit IRES-dependent translation (27), the phosphorylation at Ser4/6 sites is required for the shuttling of hnRNPA1 between nucleus and cytoplasm and thus regulates the downstream IRES-dependent translation (31). In addition, PRMT1 is known to be responsible for ADMA deposition on a number of distinct sites in the Glycine/Arginine-Rich (GAR) motif of hnRNP A1 in both mammals (32) and yeast (33). In yeast, this methylation of hnRNP A1 facilitates its nuclear export (33). However, the role of arginine methylation of hnRNP A1 in mammals is not clear.
In this study, we show that PRMT5 is responsible for regulating the IRES-dependent translation of CCND1, MYC, HIF1a and ESR1 genes. hnRNP A1 is methylated by PRMT5 on two arginine sites in its GAR motif, and this SDMA deposition is required for its ability to promote IRES-dependent translation by regulating RNA binding.
MATERIALS AND METHODS
Cell culture and plasmids
HeLa, 293T and MCF-7 cells were originally bought from ATCC. PRMT5flox/flox MEFs were immortalized with PRMT5flox/flox mouse embryo fibroblasts from Dr Ernesto Guccione (16). hnRNP A1 knockout HeLa cells were a generous gift from Dr Benjamin Chen (34). All cell lines were cultured in high glucose Dulbecco's Modified Eagle Medium, supplemented with 10% FBS, 1% NEAA and 1% penicillin/streptomycin solution, at 37°C with 5% CO2. IRES reporter constructs pRF, pRmycF, pRhifF were a generous gift from Dr Gregory Goodall (19); and pRccnd1F and pResr1F were generated by inserting the cDNAs of 5΄ UTR of human CCND1 or nucleotide 293–814 on ESR1 mRNA (GI number: 182192) into pRF vector. The primers used for the plasmid constructions were:
CCND1-IRES-F, GCTGAATTCCACACGGACTACAGGGGAGTTTT;
CCND1-IRES-R, CGGCCATGGGGCTGGGGCTCTTCCTGGGC;
ESR1-IRES-F, GCTGAATTCACCATGACCCTCCACACCAAAGC;
ESR1-IRES-R, CGGCCATGGTGGTACTGGCCAATCTTTCTCTG.
Myc.PRMT5 plasmid were a generous gift from Dr. Stephane Richard. GFP-tagged hnRNP A1 and GST-tagged hnRNP A1 plasmids were generated by inserting the coding region of human hnRNP A1 into peGFP-C1 or pGEX-6p-1 vectors. Primers used for the constructions were:
GFP-A1-F, CGCAGATCTATGTCTAAGTCAGAGTCTCCTAA;
GFP-A1-R, CGCGAATTCTTAAAATCTTCTGCCACTGCCAT;
GST-A1-F, CGCGAATTCTCTAAGTCAGAGTCTCCTAAAG;
GST-A1-R, CGCCTCGAGTTAAAATCTTCTGCCACTGCCA.
Plasmids for hnRNP A1 mutations were generated with a site-directed mutagenesis kit.
Western blotting and Antibodies
Western blotting was performed with standard protocol. Briefly, cells were washed with ice-cold PBS buffer and lysed with RIPA buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, 1% sodium deoxycholate, 5 mM EDTA), supplemented with protease inhibitor cocktail (Roche). After loading to PAGE-gel for electrophoresis, proteins were transferred to PVDF membrane and sequentially incubated in 5% milk in PBST (PBS + 0.05% Tween 20), primary antibody diluted in PBST, and secondary antibody diluted in PBST. Then the membrane was incubated with ECL reagents and developed with X-ray films. The antibodies used in this study included anti-PRMT5 (active Motif), anti-MEP50 (Active Motif), anti-Cyclin D1 (Abcam), anti-c-Myc (Abcam), anti-HIF-1α (Santa Cruz), anti-ERα (Santa Cruz), anti-HSP70 (Santa Cruz), anti-pan-SDMA (collaboration with Cell Signaling Technologies), anti-β-actin (Sigma), anti-hnRNP A1 (Santa Cruz), anti-pan-ADMA (collaboration with Cell Signaling Technologies) and anti-GFP (Santa Cruz for WB, and Life Technologies for IP).
RT-qPCR
Total RNA was extracted from MEFs or MCF-7 cells using the TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Total mRNAs were reverse transcribed with a SuperScript III Reverse Transcriptase kit (Invitrogen). cDNAs were amplified with the primers listed in supplementary materials.
Luciferase assay
Luciferase reporter transfected cells were collected and the luciferase activities (firefly luciferase and renilla luciferase) were measured using the Dual Luciferease kit (Promega) according to the manufacturer's instructions.
In vitro Methylation by PRMT5
293T cells were transfected with Myc.PRMT5 plasmids for 24 h. Cells were then washed with ice-cold PBS and lysed with mild buffer (50 mM Tris–HCl pH7.5, 150 mM NaCl, 0.5% NP-40, 1% sodium deoxycholate, 5 mM EDTA) supplemented with protease inhibitor cocktail. Two microgram of anti-Myc tag antibodies (Sigma) were added to 500 μl cell lysate and rotated at 4°C overnight. Twenty five microliters protein A/G beads were added and incubated for 1 h. Then the beads were pelleted and washed three times with mild buffer. Myc.PRMT5 bound beads were split into 6–10 vials. Three microgram of recombinant proteins, as well as 1 μl 3H-AdoMet, were added to each vial and incubated at 37°C for 1.5 h. The mix of reaction was then boiled at 95°C with SDS loading buffer. Supernatant was subjected to PAGE electrophoresis and the proteins were transferred to PVDF membrane. Then the PVDF membrane was sprayed with autoradiography enhancer and the radioactivity was detected with X-ray films.
RNA pull-down assay
PRMT5flox/flox MEFs were treated with or without 2 μM 4-hydroxytamoxifen for 8 days. Cell lysates were prepared in hypotonic lysis buffer containing 10 mM HEPES pH7.5, 10 mM potassium acetate, 1.5 mM magnesium acetate, 2.5 mM DTT, 0.05% NP-40, 10 mM NaF and 1 mM sodium orthovanadate, supplemented with protease inhibitor cocktail, and homogenized by sonication. Lysates were pre-cleared by adding 25 μl Dynabeads MyOne Streptavidin T1 (Life Technologies) and 15 μg/ml yeast tRNA. Biotinylated RNAs were prepared by in vitro transcription with T7 RNA polymerase, and followed by biotin labeling using RNA 3΄-end Biotinylation kit (Thermo Scientific). 2–3 μg of biotinylated RNAs as well as 2 mM ribonucleoside vanadyl complex (VRC) were added to the cleared cell lysates and incubated at 4°C for 2 h. Twenty five microliters of Dynabeads MyOne Streptavidin T1 were added to the cell lysates and incubated at 4°C for 1 h. The beads were pelleted and washed 3 times in mild buffer, supplemented with protease inhibitor cocktail and 2 mM VRC. The protein and RNA complex was then boiled with SDS loading buffer and subjected to Western blotting.
RNA immunoprecipitation
PRMT5flox/flox MEFs were treated with or without 2 μM 4-hydroxytamoxifen for 8 days. The cells were then lysed with RIP buffer (50 mM Tris–HCl pH7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate) supplemented with protease inhibitor cocktail and 2 mM VRC, and homogenized by sonication. Cell lysates were cleared by centrifugation and the supernatant was pre-cleared by adding 25 μl Dynabeads protein A (Life Technologies), as well as 20 μg/ml yeast tRNA for 1 h. Cleared supernatant was incubated with 2 μg normal mouse IgG or anti-hnRNP A1 antibodies at 4°C overnight. Twenty five microliters of Dynabeads protein A was added to immunoprecipitate the RNA/protein complex. The beads were washed three times with washing buffer I (50 mM Tris–HCl pH7.5, 1 M NaCl, 1% NP-40, 1% sodium deoxycholate, 2 mM VRC), followed by three more washes in washing buffer II (50 mM Tris–HCl pH7.5, 1 M NaCl, 1% NP-40, 1% sodium deoxycholate, 2 mM VRC, 1 M Urea). The immunoprecipitated complex was eluted from beads by adding 100 μl of elution buffer (100 mM Tris–HCl pH 8.0, 10 mM EDTA, 1% SDS). Protease K was added into the RNA sample and incubated at 42°C for 1 h followed by 65°C for 1 h. RNA was then extracted with RNeasy mini kit (Qiagen) and applied to RT-qPCR analysis.
Statistics analyses
GraphPad Prism6 software was used for analysis of P-values based on at least two independent experiments in three independent PCR reactions or luciferase activity assays. The two-tailed unpaired t test was used to compare the differences between two groups. P values <0.05 were considered statistically significant, *P < 0.05, **P < 0.01 and ***P < 0.001.
RESULTS
PRMT5 regulates IRES-containing genes at the posttranscriptional level
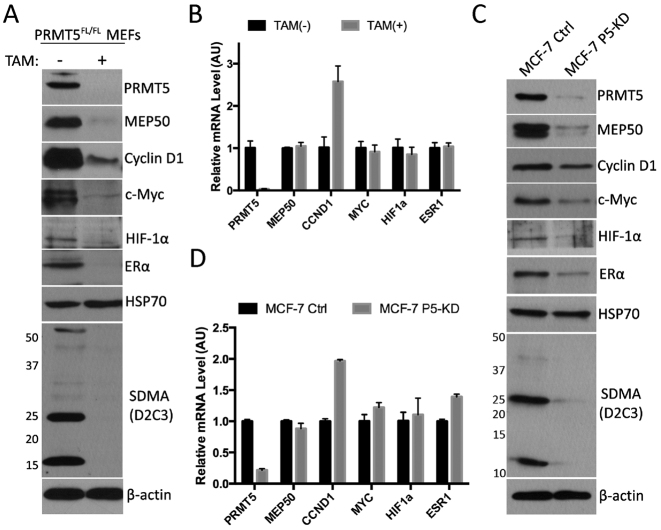
It has been reported that the protein levels, but not the mRNA level, of Mep50, Ccnd1, Myc, Hif1a, Mitf and Cdkn1B genes are regulated by PRMT5 (11–13), suggesting that PRMT5 has a general role on posttranscriptional regulation. To confirm these reported findings, we took advantage of an inducible PRMT5 knockout mouse fibroblast cell line, which was generated from the mouse embryos harboring PRMT5flox/flox allele and ER-Cre (16). Upon treatment with 4-hydroxytamoxifen (TAM), the Cre recombinase translocates into the nucleus, and abolishes the expression of PRMT5 gene by excising an exon. To avoid potential effect of TAM on gene expression, PRMT5flox/flox MEFs were treated for 10 days to disrupt the PRMT5 locus, and then cultured in regular DMEM for another 3 days. The efficiency of PRMT5 knockout and the subsequent loss of substrate methylation (αSDMA) were confirmed by Western blot analysis (Figure 1A). The protein levels and mRNA levels of Mep50 (Wdr77), Cyclin D1 (Ccnd1), c-Myc (Myc), Hif-1α (Hif1a), as well as Erα (Esr1) were detected by western blot analysis and RT-qPCR, respectively (Figure 1A and B). As expected, the protein levels of Mep50, Cyclin D1, c-Myc and Hif-1α are downregulated by loss of PRMT5, whereas the mRNA levels remain unchanged or even upregulated (Ccnd1). Interestingly, Erα, which contains an IRES in the 5΄ UTR of its mRNA (35), is also downregulated by loss of PRMT5 at the protein level but not at the mRNA level. To determine whether the regulation of the above genes is prevalent and not an idiosyncrasy of MEFs, we performed similar experiments in human MCF-7 cells. MCF-7 cell lines stably expressing control shRNA or PRMT5 shRNA are generated by lentivirus transduction, and the protein levels or mRNA levels of the above genes were detected (Figure 1C and D). Consistently, with the loss of PRMT5, the protein levels but not the mRNA levels of MEP50, CYCLIN D1, c-MYC, HIF-1α and ERα were downregulated. Noticeably, HSP70, which is another IRES-containing gene (36), is not regulated by PRMT5. Together, these results confirmed that PRMT5 could regulate Mep50, Cyclin D1, c-Myc, Hif-1α and Erα at the posttranscriptional level, and that this regulation is observed in both mouse and human cell lines.
Figure 1. The protein and mRNA levels of IRES-containing genes in PRMT5 deficient MEFs and MCF-7 cells. (A and B) PRMT5flox/flox MEFs were treated with or without 2 μM tamoxifen for 10 days for PRMT5 depletion, and cultured in regular DMEM for another 3 days to minimize the effect of tamoxifen (TAM) on transcription. Cells were then harvested and lysed for western blotting for the indicated proteins (A) or subjected to RNA purification and RT-qPCR for the relative levels of indicated genes (B). (C and D) MCF-7 cells were transducted with lentiviruses encoding control shRNA or PRMT5 shRNA, and selected with puromycin to generate stable cell lines (Ctrl and P5-KD lines). MCF-7 cells stably expressing control shRNA or PRMT5 shRNA were then harvested and lysed for Western blotting for the indicated proteins (C) or subjected to RNA purification and RT-qPCR for the relative levels ofindicated genes (D).
Methyltransferase activity is required for PRMT5 to regulate the IRES-containing genes
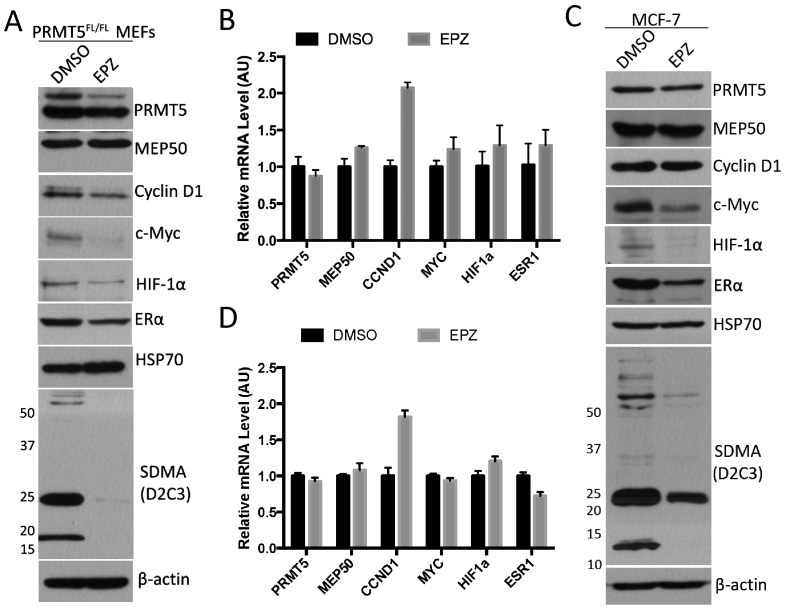
Wild type PRMT5 and its enzyme-dead mutant have been reported to rescue MEP50 protein levels to similar degrees in PRMT5 knockdown cells, suggesting that methyltransferase activity is not required for the posttranslational regulation of MEP50 (37). To determine whether the methyltransferase activity is required for the observed regulation, we utilized a recently developed PRMT5-specific small compound inhibitor, EPZ015666 (38), and assayed the protein level as well as the mRNA level of the same set of genes tested above. The PRMT5 inhibitor did not significantly affect the expression of PRMT5, but dramatically inhibited the methylation of its substrates in both MEFs and MCF-7 cells (Figure 2A–D). The inhibition of PRMT5 activity, similarly regulated Cyclin D1, c-Myc, Hif-1α and Erα at the protein level, but not at the mRNA level. Importantly, the level of Mep50 is not altered by PRMT5 inhibitor-treatment, suggesting that the mechanism that PRMT5 regulates the protein level of Mep50 is different from that of the other genes. It should be noted that in MCF-7 cells (Figure 2C and D), while the mRNA level of CCND1 was consistently upregulated (almost doubled) by PRMT5 inhibition, its protein level remained equal, suggesting that there is some degree of PRMT5 enzyme-dependent posttranscriptional regulation of CCND1. Thus, the methyltransferase activity of PRMT5 is required for its posttranscriptional regulation of Cyclin D1, c-Myc, Hif-1α and Erα, but not Mep50.
Figure 2.
Effect of PRMT5 inhibitor on the protein and mRNA levels of IRES-containing genes in MEFs and MCF-7 cells. (A and B) PRMT5flox/flox MEFs were treated with DMSO or 5 μM PRMT5 inhibitor (EPZ015666, EPZ for short) for 6 days, and harvested for Western blotting for the indicated proteins (C) or RT-qPCR for the mRNA levels of the indicated genes (D). (C and D) MCF-7 cells were treated with DMSO or PRMT5 inhibitor for 6 days, and harvested for Western blotting (C) or RT-qPCR for the indicated genes (D).
PRMT5 regulates the IRES-dependent translation
After a specific mRNA is transcribed, many cellular processes determine its fate and the level of its corresponding protein. Among the five genes identified as posttranscriptionally regulated by PRMT5 (Figure 1), four of them (the exception is Mep50) have been reported to harbor an IRES structure in their mRNA (17–19,35), which is likely not a coincidence. Moreover, the methyltransferase activity of PRMT5 is required for its regulation of all the four IRES-containing genes, but not for MEP50. We thus hypothesized that CYCLIN D1, c-MYC, HIF-1α and ERα are regulated by PRMT5 at the level of cap-independent IRES-dependent translation, and MEP50 protein levels are regulated by a distinct, and as yet unknown, mechanism. To test this hypothesis, we took advantage of a series of IRES-dependent bicistronic reporter constructs (Figure 3A) and the ratio of firefly over renilla luciferase activity represents the level of IRES-dependent translation. Control or PRMT5 KD MCF-7 cells were transfected with the constructs harboring CCND1, MYC, HIF1a and ESR1 IRESes, and the IRES-dependent translation level of each gene was tested (Figure 3B). Loss of PRMT5 repressed the activity of all four IRESes. To further determine whether this regulation requires PRMT5 methyltransferase activity, MCF-7 cells were first treated with EPZ015666 and then transfected with the IRES-dependent reporters (Figure 3C). Inhibition of PRMT5 activity significantly reduced the translation level of all the tested IRESes. With these results, we conclude that PRMT5 can regulate the IRES-dependent translation of CCND1, MYC, HIF1a and ESR1.
Figure 3.
Effect of PRMT5 on IRES-dependent translation. (A) Schematic representation of bicistronic reporter constructs with different IRESes. (B) MCF-7 Ctrl and MCF-7 P5-KD cells were transfected with indicated IRES-dependent reporters, respectively. The firefly and renilla luciferase activities were measured and the ratios of firefly luciferase activity over renilla luciferase activity were calculated. (C) MCF-7 cells were treated with DMSO or 5 μM EPZ015666 for 5 days and then transfected with indicated IRES-dependent reporters for 24 hours. Cells were then lysed to determine the firefly and renilla luciferase activities. The ratios of firefly luciferase activity over renilla luciferase activity were calculated.
hnRNP A1 is a substrate of PRMT5
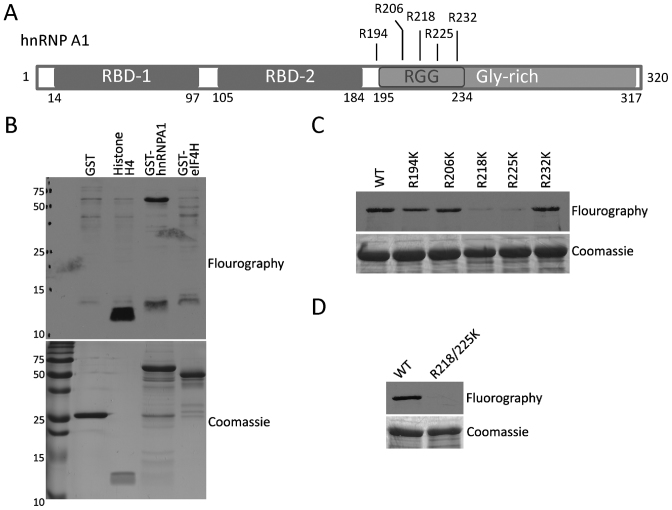
The hnRNP A1 protein is known to regulate the IRES-dependent translation of a number of genes, including CCND1 and MYC (27–30). Moreover, publicly available curated mass spectrometry data (phosphosite.org) reveals a hotspot for arginine methylation within a GAR motif found roughly in the middle of the protein (Figure 4A), which is further supported by a focused study on the hnRNP A/B paralog family (39). Five arginine residues are heavily dimethylated in this patch, and PRMT1 is known to methylate full-length recombinant hnRNP A1 (32). PRMT1 and PRMT5 share the ability to methylate GAR motifs (40,41), making hnRNP A1 a potential substrate for PRMT5 as well. eIF4H is another protein reported to regulate CCND1 and MYC IRESes (42). Using an in vitro methylation approach, we found that PRMT5 could methylate hnRNP A1 but not eIF4H (Figure 4B). To identify the site(s) of hnRNP A1 methylation by PRMT5, we mutated the five mass spec-identified arginine sites to lysine (Figure 4A), and again performed an in vitro methylation assay. This experiment identified R218 and R225 as significant sites of methylation (Figure 4C), and the double-sites mutation totally abolished the methylation by PRMT5 (Figure 4D).
Figure 4.
PRMT5 methylates hnRNP A1 in vitro. (A) Schematic representation of hnRNP A1 domains as well as the reported di-methylated sites. (B) Myc.PRMT5 complex were immunoprecipitated from 293T cells overexpressing Myc-tagged PRMT5, then mixed with 3H-AdoMet and GST, recombinant Histone H4 or indicated GST-fusion proteins, for the in vitro methylation assay by PRMT5. (C) GST-hnRNP A1 wild type or single site mutated proteins were applied to in vitro methylation by PRMT5. (D) GST-hnRNP A1 wild type protein or the double mutant R218/225K was subjected to in vitro methylation by PRMT5. In all cases, the recombinant proteins were subjected to PAGE electrophoresis and stained with Coomassie blue as a loading control.
The regulation of IRES-dependent translation by PRMT5 is dependent on hnRNP A1
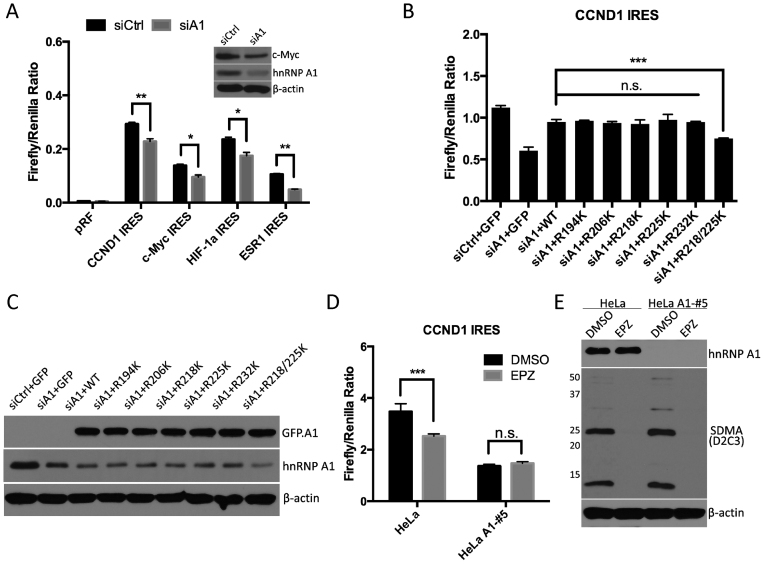
To study the role of SDMA modified hnRNP A1 in the IRES-dependent translation control, we first confirmed the role of hnRNP A1 in the IRES-dependent translation, by knocking-down hnRNP A1 and transfecting the four different IRES-dependent reporters into 293T cells (Figure 5A). Reduced levels of hnRNP A1 significantly suppressed the activities of CCND1 and MYC IRESes, as previously reported (27–30). Moreover, loss of hnRNP A1 also significantly suppressed the activities of HIF1a and ESR1 IRESes. To determine whether SDMA modification of hnRNP A1 is required for its regulatory function, GFP-tagged hnRNP A1 wild type or single-site mutants, as well as the double-site mutant (R218/225K) were transfected into 293T cells, 24 h post hnRNP A1 siRNA transfection. CCND1 IRES reporter was also transfected to monitor the IRES-dependent translation (Figure 5B and C). While the rescue with hnRNP A1 mutations at R218 or R225 displaced no defect in IRES activity, as compared with wild type hnRNP A1, the mutant of both R218/R225 failed to rescue the IRES-dependent translation, suggesting that at least one methylation arginine site (either R218 or R225) is required to support IRES-dependent translation.
Figure 5.
hnRNP A1 methylation is required for the translational regulation of IRES-dependent genes by PRMT5. (A) 293T cells were transfected with control siRNA (siCtrl) or siRNA to hnRNP A1 (siA1). After 24 h, indicated IRES-dependent reporters were transfected into each group of cells. The luciferase activities were measured after 24 h and the ratios of firefly luciferase activity over renilla luciferase activity were calculated. A representative western blotting was shown to confirm the knockdown of hnRNP A1. (B and C) 293T cells were transfected with control siRNA or siRNA to hnRNP A1. 24 h later, CCND1 IRES reporter construct was co-transfected with GFP-tagged hnRNP A1 wild type or indicated mutants into siA1-transfected cells. GFP plasmid was used to normalize the total transfected DNA. The luciferase activities were measured the following day, and the ratios of firefly over renilla luciferase activity were calculated (B). The knockdown of hnRNP A1, as well as rescued expression of GFP-tagged hnRNP A1, was shown by Western blots (C). (D and E) Wild type HeLa cells or hnRNP A1 knockout HeLa cells were treated with DMSO or 5 μM EPZ015666 for 4 days. On day 3, cells were transfected with CCND1 IRES reporter construct. At the end of day 4, half of the cells were harvested for luciferase activity assay. The ratios of firefly over renilla luciferase activity were calculated (D). The other half was lysed with RIPA buffer and applied to Western blotting against hnRNP A1 and pan-SDMA substrates (E).
PRMT5 methylates a large number of substrates, and many of them possess the capacity for RNA binding (43–45). It is thus likely that loss of PRMT5 activity will have pleiotropic effects on RNA metabolism. To attempt to understand the significance of hnRNP A1, versus other IRES transacting factor (ITAF) in PRMT5 regulated IRES-dependent translation, we investigated the effects of the PRMT5 inhibitor on IRES-dependent translation in the absence of hnRNP A1. hnRNP A1 wild type and knockout HeLa cells (generated by CRISPR-mediated deletion (34)), were treated with DMSO or EPZ015666, and the CCND1 IRES reporter was used to monitor the IRES-dependent translation (Figure 5D and E). While in wild type HeLa cells, inhibition of PRMT5 activity resulted in downregulated IRES-dependent translation; in the hnRNP A1 knockout cells, inhibition of PRMT5 activity has no effect on IRES-dependent translation. These findings suggest that hnRNP A1 is the single most important ITAF substrate for PRMT5, and its methylation is a critical regulatory node for IRES-dependent translation.
Symmetric dimethylation of hnRNP A1 enhances its binding to target IRESes
The subcellular localization of ITAFs is believed to play a critical role in their regulation of IRES-dependent translation (46). Hyperphosphorylation induced by osmotic stress leads to the cytoplasmic accumulation of hnRNP A1, through the p38 stress-signaling pathway (47,48). hnRNP A1 phosphorylation, induced by IL-6 through the same pathway, altered its localization and enhanced the IRES-dependent translation of c-Myc (49). Also, the nucleocytoplasmic shuttling of RNA binding proteins is known to be regulated by arginine methylation (33,50). To investigate the possibility that PRMT5 loss results in aberrant shuttling of hnRNP A1, we performed immunofluorescent staining and cell fractionation studies for hnRNP A1 in the presence and absence of PRMT5. Clearly, hnRNP A1 subcellular localization is not affected by the loss of PRMT5 (Supplementary Figure S1). We thus investigated other mechanisms by which PRMT5 methylation of hnRNP A1 might impact IRES-dependent translation.
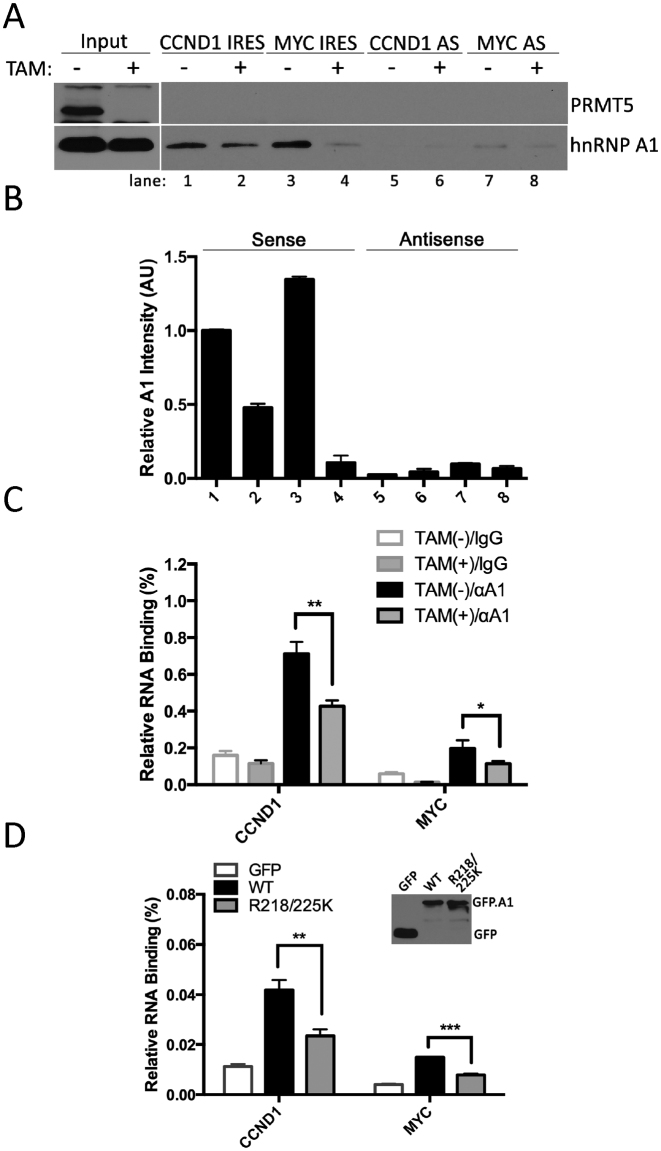
hnRNP A1 has two RNA recognition motifs (RRM), which are also referred to as RNA binding domains (RBD-1 and RBD-2, Figure 4A). In addition, hnRNP A1 harbors a GAR motif, which is heavily arginine methylated, adjacent to RBD-2. GAR motifs also possess the capacity to interact with RNA (41). We thus asked whether the arginine sites methylated by PRMT5, i.e. Arg218 and Arg225, had any effect on IRES binding. To this end, biotinylation RNA representing the IRESes of CCND1 and MYC were synthesized and used to pull-down hnRNP A1 from PRMT5 wild type or knockout MEFs. The loss of PRMT5 resulted in a significant reduction of hnRNP A1 binding to both the CCND1 and MYC IRES, indicating that symmetric dimethylation on hnRNP A1 enhances its ability to target IRES RNA structures (Figure 6A and B). As a control, hnRNP A1 bound to antisense IRESes weakly, and the methylation on hnRNP A1 did not affect the binding of CCND1 and MYC antisense IRESes (Figure 6A and B). To further confirm the effect of methylation on RNA binding, an RNA immunoprecipitation (RIP) experiment was performed; hnRNP A1/RNAs complex was immunoprecipitated from PRMT5 wild type or knockout MEFs. The levels of CCND1 and MYC mRNAs are detected by RT-qPCR (Figure 6C). Independently, this confirms that with the loss of symmetric dimethylation, the ability of hnRNP A1 to engage its target mRNAs is much reduced. Moreover, we transfected GFP-tagged wild type hnRNP A1 and its double-site mutant into hnRNP A1 knockout HeLa cells, and again using a RIP approach detected the levels of CCND1 and MYC mRNAs bound to wild type and double-site mutated hnRNP A1 (Figure 6D). Mutated hnRNP A1 showed much less affinity to both CCND1 and MYC mRNAs, comparing with the wild type.
Figure 6.
Methylation of hnRNP A1 by PRMT5 facilitates recognition of IRES-containing RNAs. PRMT5flox/flox MEFs were treated with 2 μM tamoxifen (TAM) for 8 days. (A) The cell lysates from the treated and untreated cells were incubated overnight with biotin-labeled CCND1 or MYC IRES RNA or their control antisense RNAs. RNA complexes were pulled down by streptavidin-conjugated beads and subjected to Western blotting with anti-hnRNP A1 or anti-PRMT5 antibodies. (B) The relative IRES-bound hnRNP A1 signal intensity from Figure 6A was calculated by densitometry. Signal intensity of hnRNP A1 from lane 1 was set as 1. (C) Cell lysates from tamoxifen treated or untreated cells were incubated with normal mouse IgG or anti-hnRNP A1 antibody in the presence of RNase inhibitors overnight, and immunoprecipitated with protein A/G conjugated beads. Bound RNAs were then eluted, purified and subjected to RT-qPCR for CCND1 and MYC mRNAs. (D) Wild type or double-site mutant of GFP-hnRNP A1 constructs were transfected into hnRNP A1 knockout HeLa cells. The levels of CCND1 and MYC mRNAs bound to wild type or mutated hnRNP A1 were detected by RT-qPCR, following RNA immunoprecipitation with anti-GFP antibody.
DISCUSSION
How does PRMT5 regulate MEP50 protein level?
MEP50 is a critical co-factor in the PRMT5 methylosome (51–53). A number of studies in frogs, mice and man have identified a requirement of PRMT5 for MEP50 protein expression (9,11,37), and visa versa (53,54). We initiated this study to try and determine the mechanism of this reciprocal regulation between MEP50 and PRMT5. We confirmed that PRMT5 could regulate the protein level, but not mRNA level of its co-factor (Figure 1). However, surprisingly, the methyltransferase activity of PRMT5 is not required for this regulation (Figure 2). PRMT5 is well studied as a transcriptional co-regulator, both negatively (3,5–7) and positively (9,10,55), and as a regulator of alternative splicing (14–16,56,57). However, the methyltransferase activity of PRMT5 is required for all these types of regulation. It is possible that complex formation between PRMT5 and MEP50 is required for the stability of both proteins. However, inhibition of the proteasomal degradation pathway with MG132 or the lysosomal degradation pathway (Supplementary Figure S2A and B) had no effect on stabilizing MEP50 protein levels. Moreover, loss of PRMT5 does not cause a general stress response, which could lead to partial translational shutdown through the phosphorylation of eIF2α on Ser51 (58) (Supplementary Figure S2C). The mechanism by which PRMT5 regulates MEP50 protein level remains unresolved.
PRMT5 regulates IRES-dependent translation
Besides MEP50, we have identified/validated a number of other proteins that are reduced upon PRMT5 loss or small molecule inactivation (Figures 1 and 2), and the mRNA for all four of these proteins encode for the presence of IRES structures. Further analysis revealed that PRMT5 regulated IRES-mediated translation of these four genes (Figure 3). This regulation is mediated in large part, through the methylation of hnRNP A1 (Figures 4 and 5). Different IRES structures recruit different cohorts of IRES transacting factors (ITAF) (59,60). hnRNP A1 is a ITAF, although it is unclear if it regulates translation from all IRESes, or just a subset. Indeed, it is predicted that over one hundred cellular genes contain IRES elements (21). It is thus possible that PRMT5 could regulate the IRES-dependent translation of a large portion of them, through the methylation of hnRNP A1.
ADMA versus SDMA on hnRNP A1
Arginine methylation of hnRNP A1 was first identified more than two decades ago (24). Recombinant hnRNP A1 was then reported to be a good methyl-acceptor for PRMT1, in an in vitro methylation reaction (32). More recently, a focused mass spectrometric study on the hnRNP A/B paralog family identified specific sites of arginine methylation, and it found that hnRNP A1 was primarily asymmetric dimethylation. However, peptides that harbored the residues that we identified as methylated by PRMT5 (R218 & R225) were dimethylated, but did not carry ADMA diagnostic ions, indirectly suggesting that these are sites of SDMA (39). Clearly, the GAR motif of hnRNP A1 is modified with both ADMA and SDMA marks. To investigate a potential role for ADMA in IRES-dependent translation, we treated MCF-7 cells with a recently described type I PRMT inhibitor, MS023, which blocks ADMA methylation but not SDMA methylation (61). Importantly, neither the protein levels of IRES-regulated transcripts nor the IRES-dependent reporters are affected by this small molecule inhibitor (Supplementary Figure S3), suggesting that the asymmetric dimethylation of hnRNP A1 is not required for IRES-dependent translation. Thus, the two different types of methylated hnRNP A1 likely execute distinct functions in RNA metabolism.
Arginine methylation and RNA binding
There is no general rule for how arginine methylation will impact the way a protein interacts with RNA; there are examples of arginine methylation promoting, preventing and having no effect on RNA binding. GAR motifs are enriched in RNA-binding proteins and these motifs possess inherent RNA-binding activity (41). GAR motifs have a particularly high-affinity for RNA sequences that form G-quadruplexes (G4) (62,63). The GAR motif of Aven strongly interacts with G4 RNA structures, and its GAR motif is also a target for methylation by PRMT1 (64). These observations make Aven an obvious candidate for RNA-protein interaction regulation by arginine methylation. This hypothesis was tested by the Richard group, and unexpectedly, arginine methylation had no impact on the ability of the Aven GAR motif to interact with G4 RNA (64). The splicing factor, SFPQ, is modified by PRMT1 at a number of sites. UV crosslinking studies, in the presence of global PRMT inhibitors (AdOx and AMI-1), reveal reduced SFPQ binding to mRNA when arginine methylation is blocked (65). In this case, arginine methylation is required for optimal RNA binding. A subset of mRNAs harbor inverted repeated Alu elements (IRAlus) in their 3΄ untranslated regions. p54nrb interacts with these IRAlus and sequesters these mature mRNA in the nucleus, thereby preventing translation. The methylation of p54nrb by CARM1 reduces its ability to bind these RNA elements and promotes translation (66). Here we see a case in which arginine methylation blocks RNA binding. In line with these results, it was found that knockdown of PRMT1 or CARM1 increased the RNA-binding properties of hnRNP UL1 (67), although knockdown of PRMT5 and loss of SDMA impaired the RNA binding function of the splicing factor, SRSF2 (67). In this study, we showed that SDMA on hnRNP A1 promotes its binding to the target IRESes (Figure 6 and Supplementary Figure S4). Although the unmodified recombinant hnRNP A1 is able to bind target RNA in vitro, modifications such as SDMA could provide a regulatory mechanism to fine tune its target preference.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Donghang Cheng for his effort in making the immortalized PRMT5flox/flox MEF line. We also thank Dr Benjamin Chen for providing the hnRNP A1 knockout HeLa cells; Dr Gregory Goodall for the generous gift of the IRES reporter constructs pRF, pRmycF, pRhif and Dr Stephane Richard for the Myc-PRMT5 plasmid.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [DK062248 to M.T.B]. Funding for open access charge: National Institutes of Health [DK062248].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dhar S., Vemulapalli V., Patananan A.N., Huang G.L., Di Lorenzo A., Richard S., Comb M.J., Guo A., Clarke S.G., Bedford M.T.. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 2013; 3:1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang Y., Bedford M.T.. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013; 13:37–50. [DOI] [PubMed] [Google Scholar]
- 3. Pal S., Vishwanath S.N., Erdjument-Bromage H., Tempst P., Sif S.. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004; 24:9630–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pollack B.P., Kotenko S.V., He W., Izotova L.S., Barnoski B.L., Pestka S.. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 1999; 274:31531–31542. [DOI] [PubMed] [Google Scholar]
- 5. Xu X., Hoang S., Mayo M.W., Bekiranov S.. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics. 2010; 11:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hou Z., Peng H., Ayyanathan K., Yan K.P., Langer E.M., Longmore G.D., Rauscher F.J. 3rd. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol. Cell. Biol. 2008; 28:3198–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tae S., Karkhanis V., Velasco K., Yaneva M., Erdjument-Bromage H., Tempst P., Sif S.. Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res. 2011; 39:5424–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Migliori V., Muller J., Phalke S., Low D., Bezzi M., Mok W.C., Sahu S.K., Gunaratne J., Capasso P., Bassi C. et al. . Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 2012; 19:136–144. [DOI] [PubMed] [Google Scholar]
- 9. Chen H., Lorton B., Gupta V., Shechter D.. A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene. 2016; 36:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tarighat S.S., Santhanam R., Frankhouser D., Radomska H.S., Lai H., Anghelina M., Wang H., Huang X., Alinari L., Walker A. et al. . The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2016; 30:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu F., Cheng G., Hamard P.J., Greenblatt S., Wang L., Man N., Perna F., Xu H., Tadi M., Luciani L. et al. . Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J. Clin. Invest. 2015; 125:3532–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lim J.H., Lee Y.M., Lee G., Choi Y.J., Lim B.O., Kim Y.J., Choi D.K., Park J.W.. PRMT5 is essential for the eIF4E-mediated 5΄-cap dependent translation. Biochem. Biophys. Res. Commun. 2014; 452:1016–1021. [DOI] [PubMed] [Google Scholar]
- 13. Nicholas C., Yang J., Peters S.B., Bill M.A., Baiocchi R.A., Yan F., Sif S., Tae S., Gaudio E., Wu X. et al. . PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1.). PLoS One. 2013; 8:e74710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friesen W.J., Paushkin S., Wyce A., Massenet S., Pesiridis G.S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G.. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 2001; 21:8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koh C.M., Bezzi M., Low D.H., Ang W.X., Teo S.X., Gay F.P., Al-Haddawi M., Tan S.Y., Osato M., Sabo A. et al. . MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015; 523:96–100. [DOI] [PubMed] [Google Scholar]
- 16. Bezzi M., Teo S.X., Muller J., Mok W.C., Sahu S.K., Vardy L.A., Bonday Z.Q., Guccione E.. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013; 27:1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi Y., Sharma A., Wu H., Lichtenstein A., Gera J.. Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J. Biol. Chem. 2005; 280:10964–10973. [DOI] [PubMed] [Google Scholar]
- 18. Stoneley M., Paulin F.E., Le Quesne J.P., Chappell S.A., Willis A.E.. C-Myc 5΄ untranslated region contains an internal ribosome entry segment. Oncogene. 1998; 16:423–428. [DOI] [PubMed] [Google Scholar]
- 19. Lang K.J., Kappel A., Goodall G.J.. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002; 13:1792–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miskimins W.K., Wang G., Hawkinson M., Miskimins R.. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 2001; 21:4960–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jackson R.J. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 2013; 5:a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mokrejs M., Masek T., Vopalensky V., Hlubucek P., Delbos P., Pospisek M.. IRESite–a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 2010; 38:D131–D136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Q., Dreyfuss G.. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995; 15:2800–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajpurohit R., Paik W.K., Kim S.. Enzymatic methylation of heterogeneous nuclear ribonucleoprotein in isolated liver nuclei. Biochim. Biophys. Acta. 1992; 1122:183–188. [DOI] [PubMed] [Google Scholar]
- 25. Dreyfuss G., Kim V.N., Kataoka N.. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002; 3:195–205. [DOI] [PubMed] [Google Scholar]
- 26. Jean-Philippe J., Paz S., Caputi M.. hnRNP A1: the Swiss army knife of gene expression. Int. J. Mol. Sci. 2013; 14:18999–19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jo O.D., Martin J., Bernath A., Masri J., Lichtenstein A., Gera J.. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J. Biol. Chem. 2008; 283:23274–23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A.C., Vagner S.. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 2005; 280:4144–4153. [DOI] [PubMed] [Google Scholar]
- 29. Damiano F., Rochira A., Tocci R., Alemanno S., Gnoni A., Siculella L.. hnRNP A1 mediates the activation of the IRES-dependent SREBP-1a mRNA translation in response to endoplasmic reticulum stress. Biochem. J. 2013; 449:543–553. [DOI] [PubMed] [Google Scholar]
- 30. Kunze M.M., Benz F., Brauss T.F., Lampe S., Weigand J.E., Braun J., Richter F.M., Wittig I., Brune B., Schmid T.. sST2 translation is regulated by FGF2 via an hnRNP A1-mediated IRES-dependent mechanism. Biochim. Biophys. Acta. 2016; 1859:848–859. [DOI] [PubMed] [Google Scholar]
- 31. Roy R., Durie D., Li H., Liu B.Q., Skehel J.M., Mauri F., Cuorvo L.V., Barbareschi M., Guo L., Holcik M. et al. . hnRNPA1 couples nuclear export and translation of specific mRNAs downstream of FGF-2/S6K2 signalling. Nucleic Acids Res. 2014; 42:12483–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nichols R.C., Wang X.W., Tang J., Hamilton B.J., High F.A., Herschman H.R., Rigby W.F.. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 2000; 256:522–532. [DOI] [PubMed] [Google Scholar]
- 33. Shen E.C., Henry M.F., Weiss V.H., Valentini S.R., Silver P.A., Lee M.S.. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998; 12:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sui J., Lin Y.F., Xu K., Lee K.J., Wang D., Chen B.P.. DNA-PKcs phosphorylates hnRNP-A1 to facilitate the RPA-to-POT1 switch and telomere capping after replication. Nucleic Acids Res. 2015; 43:5971–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barraille P., Chinestra P., Bayard F., Faye J.C.. Alternative initiation of translation accounts for a 67/45 kDa dimorphism of the human estrogen receptor ERalpha. Biochem. Biophys. Res. Commun. 1999; 257:84–88. [DOI] [PubMed] [Google Scholar]
- 36. Rubtsova M.P., Sizova D.V., Dmitriev S.E., Ivanov D.S., Prassolov V.S., Shatsky I.N.. Distinctive properties of the 5΄-untranslated region of human hsp70 mRNA. J. Biol. Chem. 2003; 278:22350–22356. [DOI] [PubMed] [Google Scholar]
- 37. Gu Z., Li Y., Lee P., Liu T., Wan C., Wang Z.. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS One. 2012; 7:e44033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan-Penebre E., Kuplast K.G., Majer C.R., Boriack-Sjodin P.A., Wigle T.J., Johnston L.D., Rioux N., Munchhof M.J., Jin L., Jacques S.L. et al. . A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015; 11:432–437. [DOI] [PubMed] [Google Scholar]
- 39. Friend L.R., Landsberg M.J., Nouwens A.S., Wei Y., Rothnagel J.A., Smith R.. Arginine methylation of hnRNP A2 does not directly govern its subcellular localization. PLoS One. 2013; 8:e75669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Branscombe T.L., Frankel A., Lee J.H., Cook J.R., Yang Z., Pestka S., Clarke S.. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 2001; 276:32971–32976. [DOI] [PubMed] [Google Scholar]
- 41. Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 42. Vaysse C., Philippe C., Martineau Y., Quelen C., Hieblot C., Renaud C., Nicaise Y., Desquesnes A., Pannese M., Filleron T. et al. . Key contribution of eIF4H-mediated translational control in tumor promotion. Oncotarget. 2015; 6:39924–39940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bedford M.T., Richard S.. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005; 18:263–272. [DOI] [PubMed] [Google Scholar]
- 44. Bedford M.T., Clarke S.G.. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009; 33:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stopa N., Krebs J.E., Shechter D.. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell. Mol. Life Sci. 2015; 72:2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lewis S.M., Holcik M.. For IRES trans-acting factors, it is all about location. Oncogene. 2008; 27:1033–1035. [DOI] [PubMed] [Google Scholar]
- 47. van der Houven van Oordt, W. Diaz-Meco M.T., Lozano J., Krainer A.R., Moscat J., Caceres J.F.. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 2000; 149:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guil S., Long J.C., Caceres J.F.. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell. Biol. 2006; 26:5744–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi Y., Frost P., Hoang B., Benavides A., Gera J., Lichtenstein A.. IL-6-induced enhancement of c-Myc translation in multiple myeloma cells: critical role of cytoplasmic localization of the rna-binding protein hnRNP A1. J. Biol. Chem. 2011; 286:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tradewell M.L., Yu Z., Tibshirani M., Boulanger M.C., Durham H.D., Richard S.. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum. Mol. Genet. 2012; 21:136–149. [DOI] [PubMed] [Google Scholar]
- 51. Friesen W.J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., Dreyfuss G.. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 2002; 277:8243–8247. [DOI] [PubMed] [Google Scholar]
- 52. Antonysamy S., Bonday Z., Campbell R.M., Doyle B., Druzina Z., Gheyi T., Han B., Jungheim L.N., Qian Y., Rauch C. et al. . Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:17960–17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ho M.C., Wilczek C., Bonanno J.B., Xing L., Seznec J., Matsui T., Carter L.G., Onikubo T., Kumar P.R., Chan M.K. et al. . Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One. 2013; 8:e57008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gonsalvez G.B., Rajendra T.K., Tian L., Matera A.G.. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 2006; 16:1077–1089. [DOI] [PubMed] [Google Scholar]
- 55. Deng X., Shao G., Zhang H.T., Li C., Zhang D., Cheng L., Elzey B.D., Pili R., Ratliff T.L., Huang J. et al. . Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S. et al. . Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:19114–19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sanchez S.E., Petrillo E., Beckwith E.J., Zhang X., Rugnone M.L., Hernando C.E., Cuevas J.C., Godoy Herz M.A., Depetris-Chauvin A., Simpson C.G. et al. . A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010; 468:112–116. [DOI] [PubMed] [Google Scholar]
- 58. Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M.. The integrated stress response. EMBO Rep. 2016; 17:1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Souii A., Ben M’hadheb-Gharbi M., Gharbi J.. Role of RNA structure motifs in IRES-dependent translation initiation of the coxsackievirus B3: new insights for developing live-attenuated strains for vaccines and gene therapy. Mol. Biotechnol. 2013; 55:179–202. [DOI] [PubMed] [Google Scholar]
- 60. Fitzgerald K.D., Semler B.L.. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta. 2009; 1789:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eram M.S., Shen Y., Szewczyk M.M., Wu H., Senisterra G., Li F., Butler K.V., Kaniskan H.U., Speed B.A., dela Sena C. et al. . A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem. Biol. 2016; 11:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Phan A.T., Kuryavyi V., Darnell J.C., Serganov A., Majumdar A., Ilin S., Raslin T., Polonskaia A., Chen C., Clain D. et al. . Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011; 18:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hanakahi L.A., Sun H., Maizels N.. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999; 274:15908–15912. [DOI] [PubMed] [Google Scholar]
- 64. Thandapani P., Song J., Gandin V., Cai Y., Rouleau S.G., Garant J.M., Boisvert F.M., Yu Z., Perreault J.P., Topisirovic I. et al. . Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. Elife. 2015; 4, doi:10.7554/eLife.06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Snijders A.P., Hautbergue G.M., Bloom A., Williamson J.C., Minshull T.C., Phillips H.L., Mihaylov S.R., Gjerde D.T., Hornby D.P., Wilson S.A. et al. . Arginine methylation and citrullination of splicing factor proline- and glutamine-rich (SFPQ/PSF) regulates its association with mRNA. RNA. 2015; 21:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hu S.B., Xiang J.F., Li X., Xu Y., Xue W., Huang M., Wong C.C., Sagum C.A., Bedford M.T., Yang L. et al. . Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev. 2015; 29:630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Larsen S.C., Sylvestersen K.B., Mund A., Lyon D., Mullari M., Madsen M.V., Daniel J.A., Jensen L.J., Nielsen M.L.. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016; 9:rs9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.