Abstract
RNA and protein components of the spliceosome work together to identify the 5΄ splice site, the 3΄ splice site, and the branchsite (BS) of nascent pre-mRNA. SF3b1 plays a key role in recruiting the U2 snRNP to the BS. Mutations in human SF3b1 have been linked to many diseases such as myelodysplasia (MDS) and cancer. We have used SF3b1 mutations associated with MDS to interrogate the role of the yeast ortholog, Hsh155, in BS selection and splicing. These alleles change how the spliceosome recognizes the BS and alter splicing when nonconsensus nucleotides are present at the −2, −1 and +1 positions relative to the branchpoint adenosine. This indicates that changes in BS usage observed in humans with SF3b1 mutations may result from perturbation of a conserved mechanism of BS recognition. Notably, different HSH155 alleles elicit disparate effects on splicing: some increase the fidelity of BS selection while others decrease fidelity. Our data support a model wherein conformational changes in SF3b1 promote U2 association with the BS independently of the action of the DEAD-box ATPase Prp5. We propose that SF3b1 functions to stabilize weak U2/BS duplexes to drive spliceosome assembly and splicing.
INTRODUCTION
The spliceosome is emerging as a potential therapeutic target and a potent driver of human disease (1,2). While defects in the splicing machinery have previously been implicated in spinal muscular atrophies (3) and some forms of retinitis pigmentosa (4–6), recent evidence suggests strong links between the splicing machinery and cancer (7). The spliceosome is an intricate molecular machine composed of 5 U-rich small nuclear ribonucleoproteins (the U1, U2, U4, U5, U6 snRNPs) that function in concert with numerous other splicing factors to excise introns from nascent pre-mRNA (8,9). Mutations in several snRNP proteins are implicated in a variety of cancers, while the splicing machinery in general appears to be critical for proliferation of c-MYC associated cancers (10) as well as DNA repair through the ATM signaling pathway (11).
Among splicing factors implicated in disease, the U2 snRNP protein SF3b1 is of particular interest since SF3b1 mutation is strongly correlated with cancers such as uveal melanoma, chronic lymphocytic leukemia (CLL) and myelodysplastic syndromes (MDS) (12–14). Many of the same mutations are associated with different diseases arising from distinct cell lineages (15). Bioinformatic analysis has shown that SF3b1 mutations are correlated with changes in alternative splicing, often due to the selection of cryptic, upstream 3΄ SS (16). Recent experiments have pointed to alternative BS usage by the spliceosome instigating cryptic 3΄ SS activation (17–19); however, the mechanisms by which SF3b1 mutations can influence usage of one BS or 3΄ SS over another are unclear.
SF3b1 is the largest protein of the SF3 complex, which itself is a component of the U2 snRNP. U2 is recruited to introns early in spliceosome assembly and subsequent ATP-dependent transitions result in basepairing of the U2 snRNA to the branchsite (BS) in the pre-spliceosome or spliceosome A complex (Figure 1A) (20). These transitions require the DEAD-box helicase Prp5/DDX46 (21). U2 then undergoes dramatic conformational changes during splicing resulting in basepairing between the U2 and U6 snRNAs to form the catalytic core of the spliceosome (9).
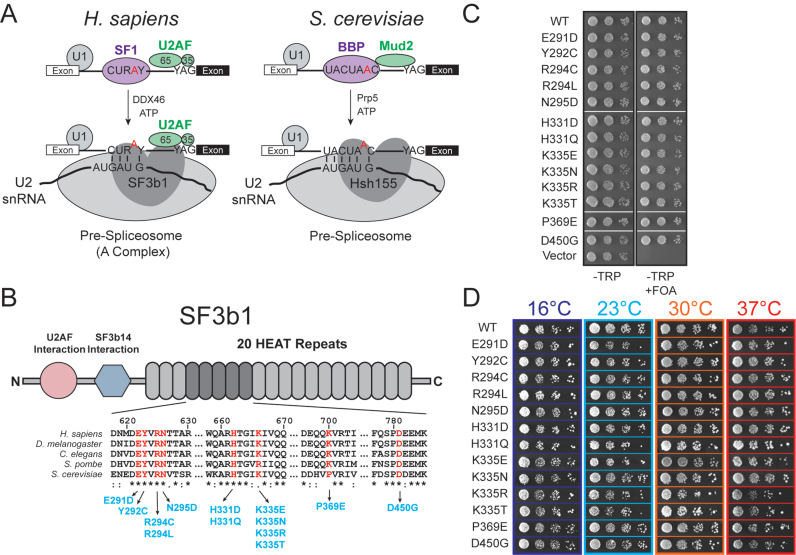
Figure 1.
MDS alleles of Hsh155 do not affect proliferation in yeast. (A) Schematic comparison of pre-spliceosome formation in S. cerevisiae and H. sapiens. Hsh155/SF3b1 function as part of the U2 snRNP, interacting with the BS/U2 snRNA duplex and downstream intronic RNA. (B) (Top) Schematic primary structure of SF3b1, with regions known to interact with other splicing factors indicated. (Bottom) Alignment of sequences from H. sapiens, D. melanogaster, C. elegans, S. pombe and S. cerevisae. Positions found to be frequently mutated in MDS and CLL are shown in red and the amino acid numbering corresponds to H. sapiens SF3b1. The most frequently occurring mutations at those positions are shown in blue with the numbering for S. cerevisiae Hsh155. (C) Haploid yeast expressing only HSH155MDS alleles are viable when plated on FOA. (D) Representative temperature sensitivity growth assays of Hsh155MDS strains plated on YPD. No growth defects are observed in haploid strains expressing only Hsh155MDS plated on YPD at 16, 25, 30 or 37°C. Successive 10-fold dilutions of a OD600 = 0.5 culture are shown.
SF3b1 crosslinks both up- and downstream of the BS in the spliceosome A complex, underlying a role in stabilizing the U2 snRNA/BS duplex and positioning protein factors within the spliceosome that interact with this duplex (22,23). Recent structures of the catalytically activated (Bact) yeast spliceosome (24,25) and the isolated SF3b complex (26) have revealed the molecular architecture of both human and yeast SF3b1/Hsh155 and other components of the SF3b complex. Hsh155 directly contacts the U2 snRNA/BS duplex and may help stabilize the bulged branchpoint adenosine. Missense mutations found in MDS map to the surface of the HEAT-repeat domain of SF3b1 in the region that interacts with the intron between the BS and 3΄ SS and nearby the DEAH-box helicase Prp2. This region of SF3b1 is highly conserved among eukaryotes, suggesting its function within the spliceosome is also conserved (Figure 1B).
SF3b1 is also the target of several antitumor compounds, such as spliceostatin A (27), pladienolide B (28) and herboxidiene (29). The antitumor compound E7107 targets SF3b1 to block ATP-dependent A complex formation as well as a conformational change in U2 that exposes the snRNA region responsible for basepairing to the BS (30). SF3b1 must undergo additional conformational changes during splicing in order to release the U2/BS duplex. Prior to 5΄ splice site (SS) cleavage, Prp2 remodels the spliceosomal active site, resulting in juxtaposition of the 5΄ SS and BS as well as a decrease in affinity between the entire SF3 complex, including SF3b1, and the catalytic spliceosome (31–34). Despite this reduced affinity, SF3b1 still influences splicing chemistry, as pladienolide B binds to SF3b1 to both prevent spliceosome assembly and inhibit exon ligation (35). Together, these data from the E7107 and pladienolide B splicing inhibitors suggest that U2 and SF3b1 may undergo similar conformational changes during assembly of the spliceosome and catalysis.
To investigate the impact of SF3b1 on the molecular mechanisms of splicing, we have incorporated naturally occurring human MDS alleles into the yeast SF3b1 ortholog and studied their impact on the well-characterized yeast spliceosome. In vivo splicing assays in combination with an MDS allele-centered yeast two-hybrid (Y2H) screen have allowed us to define the consequences of mutation of a core U2 snRNP protein on both splicing and the association of essential splicing factors. SF3b1 mutations alter usage of nonconsensus BS containing substitutions at the same positions impacted by mutation of the DEAD-box ATPase Prp5; however, the mechanisms by which mutation of these two splicing factors influence BS usage are distinct. Moreover, the Y2H screen also suggests that SF3b1 is a central hub for recruitment of splicing factors to the spliceosome active site, and we show that MDS mutations can interact genetically with Prp2 mutants. Combined, these results suggest that branchsite selection arises from balancing the opposing activities of SF3b1 and Prp5 during spliceosome assembly.
MATERIALS AND METHODS
Saccharomyces cerevisiae strains used in these studies were derived from 46α (kind gift of David Brow), BJ2168 or ySSC026 (kind gift of Soo-Chen Cheng) (36,37). Supplemental Tables S1 and S2 contain detailed lists of strains and plasmids. Yeast transformation and growth was carried out using standard techniques and media (38).
Site-directed mutagenesis
Point mutants were generated using inverse polymerase chain reaction (PCR) with Phusion DNA polymerase (New England Biolabs; Ipswich, MA, USA). The PCR was performed for 16 cycles using primers with the desired mutations incorporated at the 5΄ ends. PCR products were treated with DpnI (New England Biolabs; Ipswich, MA) to remove template, 5΄ phosphorylated and self-ligated using T4 polynucleotide kinase (New England Biolabs) and T4 DNA ligase (New England Biolabs; Ipswich, MA) and transformed into Top10 competent cells. Individual colonies were screened by sequencing to identify the desired mutation.
Temperature growth assays
Yeast strains expressing WT or mutant HSH155 alleles were grown to mid-log phase in YPD, the OD was adjusted to OD600 = 0.5 and equal volumes were spotted onto YPD plates. Plates were incubated at the indicated temperature and scored after 3 days growth at 23°C, 30°C, 37°C and 10 days at 16°C.
ACT1-CUP1 copper assays
ACT1-CUP1 reporters and growth assays have been described previously (36). Briefly, yeast strains expressing WT or mutant proteins and ACT1-CUP1 reporters were grown to mid-log phase in the appropriate media to maintain selection for the plasmids, adjusted to OD600 = 0.5 and equal volumes were spotted onto plates containing 0, 0.025, 0.05, 0.075, 0.1, 0.15, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0, 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2.0, 2.25 or 2.5 mM CuSO4. Plates were scored after 3 days growth at 30°C.
RNA analysis
Yeast were grown in selective liquid media until OD600 reached 0.5–0.8. Cells (10 OD600 units) were harvested by centrifugation, and total cellular RNA was isolated using a MasterPure Yeast RNA Purification Kit (Epicentre BioTechnologies; Madison, WI) according to the vendor's instructions. Primer extensions reactions were performed using SuperScriptIII reverse transcriptase (ThermoFisher Scientific; Waltham, MA) and the primers YAC6 (5΄-GGCACTCATGACCTTC-3΄) and yU6 (5΄-GAACTGCTGATCATCTCTG-3΄) (39). Primer extension reactions contained 10 μg total cellular RNA, 10 U Superscript III, and 400 nM each [32P]-end-labeled primer. Assembled extension reactions were incubated at 55°C for 1 h and extension products were analyzed using denaturing PAGE (7% acrylamide:bisacrylamide (19:1), 8M urea, 1× TBE). Gels were then transferred to BioRad filter paper, dried, and exposed to a PhosphorImager screen. The PhosphorImager screen was imaged using a Typhoon FLA 9000 biomolecular imager (GE Healthcare Life Sciences; Chicago, IL, USA) and band intensities were quantified using ImageJ software.
Yeast-two hybrid assays
The Hsh155 open reading frame (ORF) was cloned into pGADT7 (Clontech) generating a GAL4-activation domain-Hsh155 fusion. The ORFs of Bud13, Clf1, Cus1, Cus2, Hsh49, Mud2, Prp2, Prp5, Prp11, Prp22, Prp28, Prp43 and Ysf3 were fused to the C-terminus of the Gal4-DNA binding domain in plasmid pGBKT7. Each pair of plasmids was transformed into the S. cerevisiae strain Y2H GOLD, which has the Gal4 UAS upstream of the HIS3 and ADE2 loci. Expression of fusion proteins was confirmed by western blotting and plasmids were assayed for autoactivation in combination with empty vectors (i.e. pGADT7 without anything cloned into the vector). Interactions were examined by growth on media lacking histidine, leucine and tryptophan. Briefly, Y2H expressing both fusions were grown in media lacking leucine and tryptophan to maintain selection for the plasmids. Ten-fold serial dilutions beginning with OD600 = 0.5 was plated onto solid media and plates were scored after 3 days incubation at 30°C.
TCA precipitation and western blotting
Total protein was isolated by tricholoracetic acid (TCA) precipitation (40). Yeast were grown in selective media until reaching OD600 = 0.5–1.0. Ten OD600 units were harvested by centrifugation, and cell pellets were washed with 20% TCA. After washing, pellets were suspended in 20% TCA and subjected to mechanical lysis using glass beads. Glass beads were removed and 5% TCA was added to achieve a final concentration of ∼10% TCA and precipitated proteins were collected by centrifugation. Pellets were washed with 70% ethanol, followed by solublization in 1 M Tris pH 8.0 and subsequent SDS-PAGE.
For western blotting, one volume of 2× Laemmli Buffer was added to TCA-precipitated total protein or soluble yeast whole cell extract and the sample was denatured by incubation at 95°C for 5 min. Centrifugation was used to remove insoluble material and the resulting supernatant was resolved on 4–20% Criterion TGX midi protein gel (200 V for 1 h; Bio-Rad; Hercules, CA, USA). Proteins were subsequently transferred to a nitrocellulose membrane for blotting (30 min, 100 V, 4°C). The membrane was blocked using 5% (w/v) non-fat dry milk dissolved in 1× TBST and probed using the appropriate antibodies. Blots were developed using Clarity Western ECL substrate (Biorad; Hercules, CA) and imaged using an Imagequant LAS 4000 Imager (GE Healthcare Life Sciences; Chicago, IL, USA). The Prp8 antibody was a kind gift of Soo-Chen Cheng. Antibodies against the HA and c-myc tags were conjugated to horseradish peroxidase (HRP) and obtained from Sigma Aldrich and ThermoFisher Scienitific, respectively. V5 antibody was purchased from Bio-Rad AbD Serotech (Hercules, CA). Goat α-rabbit-HRP and goat α-mouse-HRP secondary antibodies Bio-rad AbD Serotech (Hercules, CA).
RESULTS
Given that cancer-causing mutations in human SF3b1 have been implicated in altering BS selection by the spliceosome (18), we reasoned that a library of SF3b1 mutations could be used to produce a set of alleles in yeast that would allow us to dissect the role of the protein. The majority of SF3b1 mutations associated with MDS and other diseases cluster within a region corresponding to the C-terminal HEAT repeats of the protein, specifically repeats four through nine (Figure 1B). This region is highly conserved (>50% identical) between humans and the yeast SF3b1 ortholog, Hsh155. We deleted the chromosomal HSH155 gene and maintained yeast viability by expression of wild type (WT) Hsh155 from a low-copy URA3/CEN6-containing plasmid. We then generated yeast strains expressing only the MDS alleles by transformation of the WT/URA3 yeast with TRP1/CEN6-containing plasmids with MDS mutant alleles and subsequent 5-FOA selection of the resulting transformants. Because the most frequently mutated position in human disease, K700, corresponds to P369 in yeast, we generated both P369K and P369E alleles. Additionally, we also incorporated two disease alleles (corresponding to G409E and K410N in Hsh155) that have so far only been observed in patients diagnosed with CLL but not MDS (41). All transformants were viable when grown on 5-FOA-containing media and the genotypes were confirmed by plasmid rescue and DNA sequencing (Figure 1C and Supplemental Figure S1B). In total, we generated a library of 17 isogenic strains containing either WT or one of 16 different missense mutations corresponding to MDS and CLL disease alleles (collectively labeled Hsh155MDS; Figure 1B, Supplemental Figure S1A, and Supplemental Table S1).
Disease alleles do not affect cellular proliferation in yeast
We initially screened the mutant yeast strains for defects in proliferation or temperature sensitivity, which has often been observed upon mutation of the splicing machinery. All of the mutant yeast strains were viable when expressing only mutant Hsh155. Measurement of doubling times in liquid culture at 30°C also showed no significant differences between the mutant and WT strains (Supplemental Figure S1C). When the growth of each strain was assayed at different temperatures ranging from 16 to 37°C, we detected no discernable difference between any of the mutants and the WT control (Figure 1D and Supplemental Figure S1D). These data suggest that HSH155MDS alleles do not result in general defects in proliferation. As a consequence, MDS mutant Hsh155 proteins are functional and mutations likely do not cause general disruption of pre-mRNA splicing in yeast.
MDS mutations alter the splicing of pre-mRNAs with nonconsensus branchsites
We next assayed our Hsh155MDS mutant library using the ACT1-CUP1 splicing reporter to evaluate the capacity of each mutant to splice pre-mRNA. This assay utilizes a reporter plasmid expressing the CUP1 copper resistance gene fused to an intron-containing portion of the actin (ACT1) pre-mRNA (Figure 2A) (36). Expression and proper splicing of this reporter gene confers growth in the presence of Cu2+, with the maximum concentration of Cu2+ upon which the yeast grow proportional to the extent of ACT1-CUP1 pre-mRNA splicing.
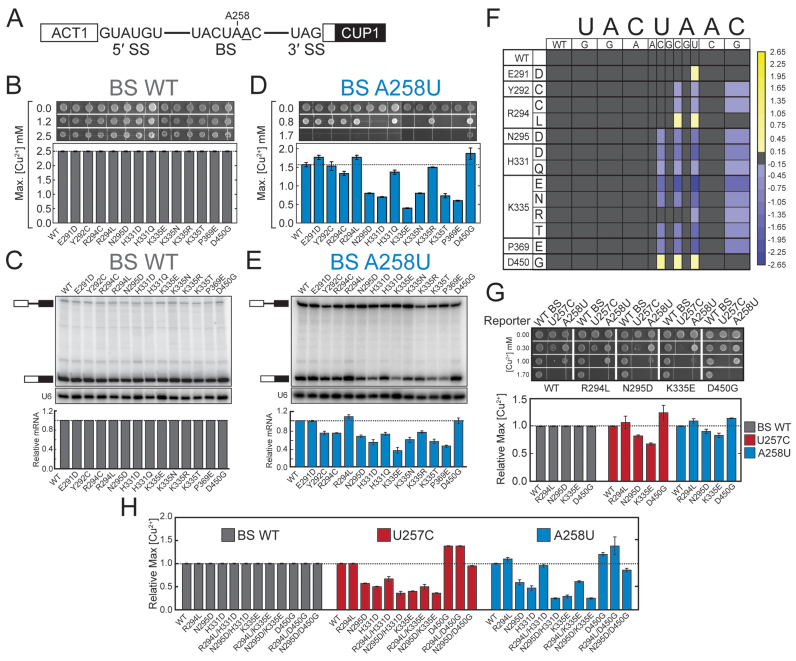
Figure 2.
MDS mutations alter the splicing of introns with nonconsensus BS sequences. (A) Schematic representation of the ACT1-CUP1 reporter pre-mRNA. The consensus sequences of the yeast 5΄ SS, BS, and 3΄ SS are shown. The position of A258 is noted and the branchpoint adenosine is underlined. (B) Cu2+ growth assay of strains carrying an ACT1-CUP1 reporter plasmid with a consensus intron. Representative images are shown at the top and the maximum [Cu2+] at which growth was observed is plotted below. (C) Determination of ACT1-CUP1 reporter RNA levels by primer extension from isolated total yeast RNA. (Top) Positions of the pre-mRNA and mRNA are noted in the primer extension polyacrylamide gel. (Middle) Primer extension analysis of the U6 snRNA was used as an internal control and analyzed on the same gel as shown in the top panel. (Bottom) Quantification of the amount of ACT1-CUP1 mRNA after normalization to U6 for each strain. U6 bands are taken from the same gel and contrast has been adjusted. (D) Cu2+ growth assay of strains carrying an ACT1-CUP1 reporter plasmid with a A258U nonconsensus BS. (E) Determination of A258U ACT1-CUP1 reporter RNA levels by primer extension from isolated total yeast RNA. (F) Heatmap summarizing mutant ACT1-CUP1 reporter data for all BS reporters tested. Plotted data represent the log2 transform of the ratio of the maximum [Cu2+] at which growth was observed for the indicated Hsh155MDS mutant to the maximum [Cu2+] at which growth was observed for Hsh155WT. Purple colors indicate decreased growth relative to Hsh155WT, and yellow colors indicate improved growth. (G) Cu2+ growth assay of merodiploid strains expressing the indicated HSH155MDS allele from a plasmid in addition to the chromosomal copy of Hsh155WT for the WT, U257C and A258U ACT1-CUP1 splicing reporters. (H) Cu2+ growth assay of strains expressing Hsh155 proteins harboring multiple MDS mutations for the WT, U257C, and A258U ACT1-CUP1 splicing reporters. In panels B, D-E, and G-H, each bar represents the average of three independent experiments, and error bars represent the standard deviation.
Consistent with the proliferation data in Figure 1, all of the Hsh155MDS strains grew equally well in the presence of Cu2+ while expressing an ACT1-CUP1 reporter with consensus splice sites (Figure 2B and Supplemental Figure S1E). To probe ACT1-CUP1 pre-mRNA and mRNA levels directly, total cellular RNA was isolated from each strain and primer extension reactions were performed. In all cases we observed the spliced ACT1-CUP1 mRNA as the predominant species and only small amounts of unspliced pre-mRNA (Figure 2C). Taken together these data indicate that the splicing of introns containing consensus splice sites is not affected by these mutations of Hsh155.
To investigate if MDS alleles would alter the splicing of nonconsensus introns, we combined our mutant library with an ACT1-CUP1 reporter incorporating a single substitution in the BS sequence (i.e. A258U: UACUuAC, substitution in lowercase; Figure 2A). In contrast to our results with the consensus ACT1-CUP1 reporter, yeast strains transformed with the A258U reporter no longer grew equally well in the presence of Cu2+ (Figure 2D). Most strains (e.g. Hsh155K335E) could only support growth at lower levels of Cu2+ than Hsh155WT. However, some mutants grew more robustly than Hsh155WT and supported growth at high Cu2+ levels (the E291D, R294L and D450G mutants). To validate that the changes in growth are correlated with changes in pre-mRNA splicing, we isolated total RNA from each strain and characterized the relative amounts of spliced and unspliced reporter by primer extension. The general trends observed in the Cu2+ growth assay with the A258U reporter are recapitulated with the primer extension assay with the strains showing the greatest growth inhibition also showing the smallest accumulation of spliced mRNA (Figure 2E). Thus, MDS variants of Hsh155 alter splicing of introns containing the nonconsensus BS substitution A258U but not the consensus BS.
To assess whether or not the splicing of introns with BS substitutions other than A258U is impacted by MDS mutations, we singly transformed each member of our missense library with ten additional ACT1-CUP1 reporters encoding at least one substitution at each position within the BS. We then tested each strain to determine the extent of growth on Cu2+-containing media. Given the size of the resultant data set, we created a heatmap showing the growth of each strain with each reporter as the log2 transform of the ratio of the maximum [Cu2+] tolerated by the Hsh155 mutant strain relative to the maximum [Cu2+] tolerated by the WT strain (Figure 2F).
The data show a striking and highly specific impact of MDS alleles on the splicing of introns containing substitutions at positions −2, −1 and +1 relative to the branchpoint adenosine (i.e. substitutions at U257, A258 and C260). Every MDS allele tested in our library altered the splicing of at least one of the ACT1-CUP1 reporters with substitutions at these positions. As with the A258U reporter, most of the MDS alleles tested showed impaired growth on Cu2+ relative to WT for other BS reporters and a corresponding decrease in mRNA by primer extension (purple boxes, Figure 2F and Supplemental Figure S3A-C). Splicing of reporters with substitutions immediately 5΄ of the branchpoint (−1, A258) was strongly affected by MDS alleles, with A258U showing effects with every missense Hsh155 mutant tested. However, not all substitutions at A258 impacted splicing equally: the A258G substitution showed no change between the WT and MDS alleles while the A258C mutation was nearly as impactful as A258U. Many but not all MDS alleles that showed decreased growth relative to WT with the A258U reporter also showed decreased growth with substitutions at the −2 and +1 positions (U257C and C260G, respectively). The Hsh155P369E mutation corresponding to the frequently observed K700E MDS allele was more disruptive than incorporation of the lysine found in SF3b1 at that position (Hsh155P369K) (Figure 2F and Supplemental Figure S1D). However, less frequently observed MDS alleles affect yeast growth more significantly than either P369 mutation (cf. Hsh155P369E vs. Hsh155K335E). Together these results show that Hsh155MDS alleles impact the splicing of introns containing nonconsensus nucleotides at the −2, −1 and +1 BS positions, these alleles are most sensitive to transversions at the −1 position, and the most common result is a decrease in splicing of introns with these nonconsensus BS.
The majority of the SF3b1 mutations tested in our ACT1-CUP1 assay have been implicated in both CLL and MDS. Despite the fact that many of the same mutations are found in both diseases, the prognostic outcome for an MDS patient differs greatly from a CLL patient, with SF3b1 mutation being favorable in MDS and unfavorable in CLL (15,42). We sought to further understand this disparity by investigating the mutations G409E and K410N, which have thus far only been linked to CLL. Like mutations associated with both diseases, combination of the CLL-specific mutations with ACT1-CUP1 reporters bearing nonconsensus BS revealed that Hsh155G409E and Hsh155K410N only affect substitutions at the −2, −1 and +1 position of the BS (Supplemental Figure S1E). These results suggest that while different mutations in SF3b1 in humans are correlated with distinct cancers, the mechanism of action in yeast for the mutations in the HEAT repeat is likely the same.
While most of the MDS mutants grew less well than Hsh155WT with BS-substituted ACT1-CUP1 reporters, a few alleles exhibited the opposite effect and showed increased growth on Cu2+ relative to Hsh155WT. The strains Hsh155E291D, Hsh155R294L, and Hsh155D450G all showed increased growth in the presence of Cu2+ compared to Hsh155WT (Figure 2F; yellow boxes). While Hsh155E291D only displayed this phenotype with the A258U reporter, both Hsh155R294L and Hsh155D450G showed increased growth with multiple ACT1-CUP1 reporters and were sensitive to both the A258U and A258C substitutions. Hsh155D450G displayed the broadest impact on splicing, affecting growth in yeast with reporters containing substitutions at U257 and A258 (Figure 2A and F). Strikingly, a single position mutated to different amino acids yielded opposite phenotypes. While Hsh155R294L showed increased growth with the A258U and A258C reporters, Hsh155R294C showed a decrease in growth using these same reporters. Combined with the results described above, these experiments demonstrate that MDS alleles can increase or decrease splicing of an intron containing BS substitutions at the −2, −1 or +1 positions and that different missense mutations of the same amino acid can have opposite effects.
It is possible that mutations in HSH155 are destabilizing and lead to changes in nonconsensus intron splicing by reducing the concentration of the protein in cells. To test this, we generated strains with three copies of the HA epitope at the C-terminus of Hsh155WT as well as two of the Hsh155MDS mutants showing the strongest phenotypes in our Cu2+ growth assay (Hsh155K335E and Hsh155D450G) and assayed protein levels by western blot (Supplemental Figure S2). All mutants showed similar levels of Hsh155 relative to both Prp8 and Prp5, suggesting that the mutations do not affect Hsh155 expression. Additionally, we generated merodiploid strains expressing both mutated and wild-type Hsh155 to determine whether the effect of MDS mutants on splicing the U257C and A258U reporters is dominant or recessive. In all cases tested, the effect of expressing Hsh155 with MDS mutations alone is recapitulated in the merodiploid strains, including the small effect of the R294L mutation on splicing the U257C reporter (Figure 2G). However, the magnitudes of the changes in splicing are less than what is observed when expressing only the mutant copy, which is consistent with the incorporation of both isoforms into functional spliceosomes and indicates that Hsh155MDS mutations are semi-dominant. These data show that Hsh155 plays an active role in BS selection and mutations associated with MDS compromise the ability of Hsh155 to act during splicing.
The effects of Hsh155MDS mutations are additive
We further explored the effect of these mutations by generating additional strains bearing multiple Hsh155MDS mutations and assaying them for altered reporter splicing. For this, we chose the mutations R294L and N295D (which decrease and increase growth in ACT1-CUP1 reporter assays, respectively) and individually combined them with the mutations H331D, K335E, and D450G to generate six additional strains. When tested using ACT1-CUP1 reporters with substitutions in the BS at the −2 or −1 position, Hsh155 double mutants displayed additive effects (Figure 2H). For example, the Hsh155R294L/H331D double mutant strain was less tolerant of Cu2+ than Hsh155R294L alone and the Hsh155R294L/D450G double mutant was more tolerant than Hsh155R294L alone. The same additive trend was also observed for the N295D mutation when combined with H331D or D450G. The Hsh155N295D/K335E double mutant strain was the only variant to deviate from this trend, but this may be the result of being unable to further reduce splicing and Cu2+ tolerance in a strain already severely impaired by the K335E mutation. Interestingly, these double mutant strains still showed no changes in splicing consensus intron reporters, further supporting the notion that MDS mutations give rise to change by altering the splicing of specific nonconsensus introns rather than by causing a general pre-mRNA splicing defect.
Hsh155MDS mutations do not alter splicing of nonconsensus 5΄ or 3΄ splice sites and do not affect cryptic 3΄ SS discrimination
To investigate if the impact of MDS alleles is limited to BS substitutions, we tested eight additional ACT1-CUP1 reporters with single nucleotide substitutions in the consensus 5΄ splice site (5΄ SS) or 3΄ splice site (3΄ SS). In all cases, yeast strains with MDS alleles grew to levels equivalent to Hsh155WT in the presence of Cu2+ (Figure 3A and B), supporting the notion that splicing of reporters with mutations at these sites are not affected by MDS alleles. This is consistent with SF3b1/Hsh155 primarily functioning near the BS and at nearby, downstream sequences.
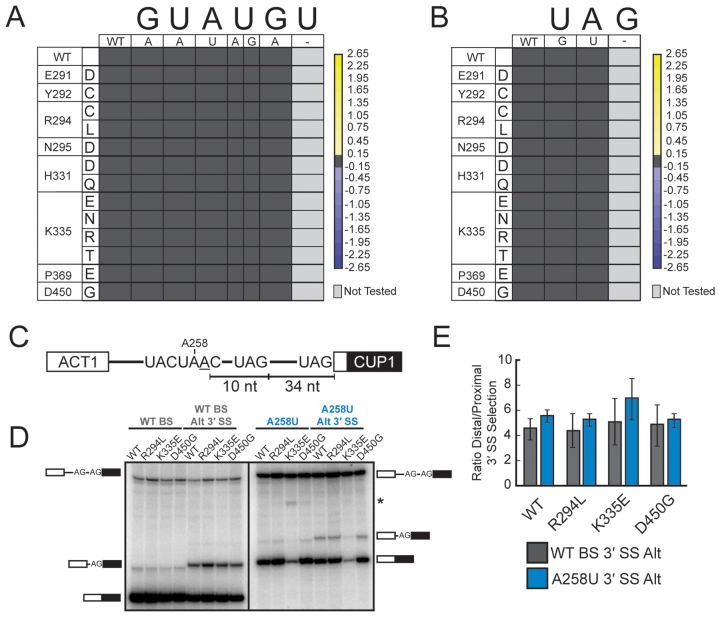
Figure 3.
MDS mutations do not affect the splicing of introns containing nonconsensus 5΄ SS and 3΄ SS or 3΄ SS selection. (A) Heatmap summarizing mutant ACT1-CUP1 reporter data for all 5΄ SS substitution reporters tested. Data were normalized and the heatmap generated as in Figure 2F. No changes in 5΄ SS usage were observed. (B) Heatmap summarizing mutant ACT1-CUP1 reporter data for all 3΄ SS substitution reporters tested. Data were normalized and the heatmap generated as in Figure 2F. No changes in 3΄ SS usage were observed. (C) Schematic representation of the ACT1-CUP1 reporters used to evaluate cryptic 3΄ SS selection. The cryptic 3΄ SS is located 10 nt downstream of the branchpoint adenosine and 34 nt upstream of the canonical 3΄ SS. Reporters containing both a consensus BS and an A258U substitution were used. (D) Primer extension and PAGE analysis of spliced products of the ACT1-CUP1 reporters shown in (C) from total RNA isolated from the given yeast strains. Positions of the pre-mRNA and mRNA products are noted. The reporter containing the A258U nonconsensus BS also contains a larger 3΄ exon leading to shift in electrophoretic mobility between the consensus and nonconsensus reporter RNAs. The asterisk (*) indicates an unknown band that was not reproducible. (E) Quantification of the data shown in (D) for 3΄ SS usage by the Hsh155WT and given Hsh155MDS strains. Bars represent the average of three independent experiments, and error bars represent the standard deviation.
To evaluate directly whether Hsh155MDS mutants are intrinsically impaired at discriminating against cryptic 3΄ SS, we employed an ACT1-CUP1 reporter mutated to include a second consensus 3΄ SS 10 nucleotides (nt) downstream of the branchpoint adenosine and 34 nt upstream of the canonical 3΄ SS (Figure 3C) (43). We tested for use of the proximal and distal 3΄ SS in Hsh155WT, Hsh155R294L, Hsh155K335E and Hsh155D450G mutant strains by primer extension (Figure 3D, left panel). We observed very little change in 3΄ SS discrimination. Further testing of these strains with a reporter bearing both the A258U BS substitution and a cryptic 3΄ SS also showed similar ratios of 3΄ SS usage between HSH155 alleles (Figure 3D, right panel). Together, our ACT1-CUP1 reporter data support the idea that MDS alleles likely do not affect 5΄ or 3΄ SS usage or discrimination between cryptic and bona fide 3΄ SS. Changes in cryptic 3΄ SS usage observed in humans with MDS may instead arise from a defect in the ability of the spliceosome to utilize weak BS, leading to alternative positioning of U2 on the intron and selection of a different BS.
Hsh155 interacts with multiple components of the splicing machinery
Our experiments using the ACT1-CUP1 reporter reveal that SF3b1 mutations alter usage of nonconsensus BS. Recent structures have implicated the mutated HEAT repeats in direct binding of RNA downstream of the U2/BS duplex (24,25). It is possible that mutation of these HEAT repeats either directly or indirectly distort the conformation of Hsh155/SF3b1 thereby altering contacts with other components of the spliceosome and leading to the observed pre-mRNA splicing changes. To test this idea, we used a yeast two-hybrid assay to screen for altered interactions upon mutation of Hsh155. A number of proteins that interact with Hsh155 have previously been identified by Y2H (44), and we assayed these identified interactions in combination with MDS mutations (Figure 4A; representative images in Supplemental Figure S4). Since SF3b1 has recently been implicated in influencing steps after pre-spliceosome formation (35), we also included a number of other factors that interact with the spliceosome during splicing.
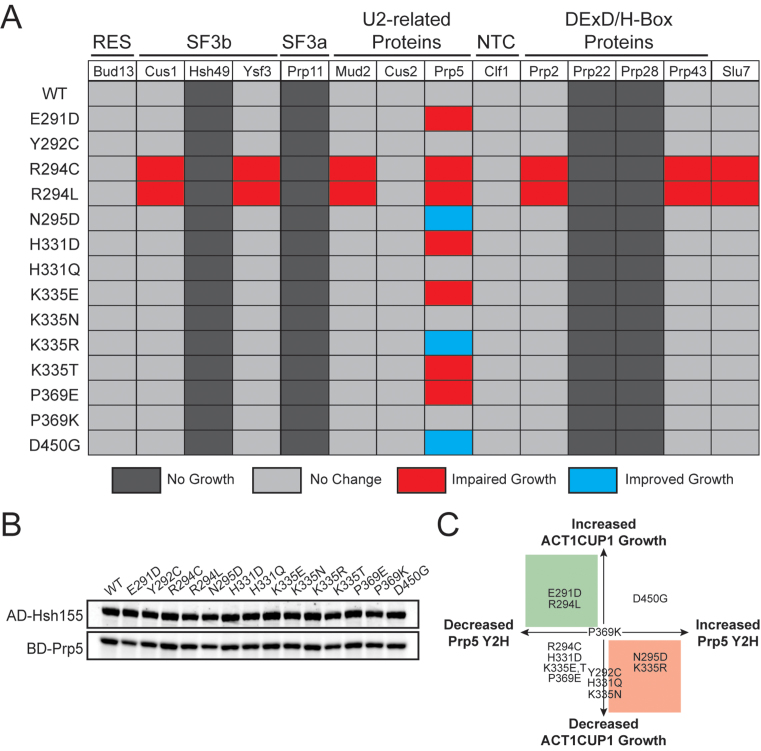
Figure 4.
MDS mutations perturb interactions between Hsh155 and Prp5 but leave most other interactions intact. (A) Pseudo-heatmap showing the observed Y2H interactions of Hsh155 upon incorporation of MDS mutations with the given splicing factors. Red indicates an impaired growth relative to Hsh155WT when plated on media that is selective for the Y2H interaction (-His dropout media), blue indicates improved growth, and light grey indicates no change. Dark grey indicates no observable Y2H interaction. (B) Representative western blot confirming expression of the fusion proteins to HSH155MDS used in the Y2H assay. Expression of each potential interacting partner was also confirmed by western blotting and expression of Prp5 is also shown as a representative example. (C) Graphical representation of the relationship between changes in yeast growth observed with the BS A258U ACT1-CUP1 splicing reporter (Figure 2D) and altered interactions observed by Y2H (Figure 4A). Shaded areas represent predictions made from a previously described model for Prp5-based BS fidelity in which retention of Prp5 leads to increased fidelity (red) and weakening of the Prp5 interaction leads to relaxed fidelity (green) (37).
Hsh155 was fused to the GAL4 activation domain (AD) while each potential interacting protein was fused to the GAL4 DNA binding domain (BD). We confirmed expression of each AD-Hsh155 mutant by western blotting, and all mutants expressed equally well in the Y2H strain (Figure 4B). Similarly, we confirmed expression of potential interaction partners and only the fusions that were shown to express by western blotting were included in the assay. We screened 15 alleles of Hsh155 against 15 components of the splicing machinery, for a total of 225 potential interactions.
The Y2H screen using an AD-Hsh155WT fusion confirmed previously known interactions with Bud13, Clf1, Cus2, Mud2, and Prp5 as well as identified new potential binding partners. Novel Y2H interactions were detected between Hsh155 and the SF3b components Cus1 and Ysf3. We did not observe any Y2H interaction between AD-Hsh155WT and either the SF3a protein Prp11 or SF3b protein Hsh49. These results suggest that the AD-Hsh155 Y2H assay is reporting on a subset of protein-protein interactions occurring within U2 or the spliceosome.
The Y2H screen also identified previously unknown interactions between Hsh155 and Prp2, Prp43, and Slu7. Prp2 and Prp43 are both spliceosomal DEAH-box ATPases (32), while Slu7 is a second step factor important for selection of 3΄ SS (43). Our observed interaction between Hsh155 and Prp2 agrees with the role of Prp2 in activation and remodeling of the U2/U6 active site (which involves destabilization of SF3) as well as recent cryo-electron microscopy (cryo-EM) structures of spliceosomes (24,25,32–34,45). To our knowledge, a Y2H interaction between Hsh155 and Prp43 has not previously been reported. Prp43 has multiple roles in the splicing cycle and is responsible for disassembly of lariat–intron product complexes as well as spliceosomes rejected by proofreading mechanisms (46,47). Prp43 may interact with Hsh155 to gain access to the U2/U6 active site during disassembly (48). We observed no interaction between AD-Hsh155WT and the DEAD-box ATPase Prp28 or DEAH-box ATPase Prp22. This is consistent with Prp28 and Prp22 acting on the spliceosome at regions other than the BS: Prp28 isomerizes interactions between the 5΄ SS and U1 and U6, while Prp22 promotes mRNA release and crosslinks to the 3΄ exon (32). Together these results suggest that SF3b1 may interact with a subset of spliceosomal ATPases that need to function at or near the U2/BS pairing region.
Interactions with Hsh155 remain intact upon inclusion of SF3b1 disease alleles with the exception of Prp5
HSH155MDS alleles altered a small subset of the Y2H interactions while leaving most others unaffected (Figure 4A). None of the MDS mutations changed interactions between Hsh155 and Bud13, Cus2, or Clf1. The R294C and R294L mutations disrupted interactions among the greatest number of splicing factors, including components of U2 snRNP (Cus1, Ysf3), factors involved in early spliceosome assembly (Mud2 and Prp5) and factors involved in spliceosome activation, catalysis, or disassembly (Prp2, Slu7 and Prp43, respectively) (Figure 4A). The disruptions caused by missense mutations of R294 could be due to changes in Hsh155 structure that influence several binding sites or interactions, a result possibly amplified in the context of the Y2H assay. In support of this idea, transformation and subsequent 5-FOA selection of the HSH155 shuffle strain with the AD-Hsh155R294L plasmid resulted in viable yeast, showing that AD-Hsh155R294L is active for splicing notwithstanding these altered Y2H interactions (Supplementary Figure S5). Surprisingly, both R294L and R294C disrupted identical sets of interactions despite these alleles showing opposite phenotypes in our ACT1-CUP1 reporter assay (Figure 2F). This suggests that while R294L and R294C disturb binding of many of the same splicing factors, the mutations likely alter Hsh155 structure in unique ways.
Aside from the R294C and R294L mutations, interactions between the other HSH155MDS alleles and the 3΄ SS selection factor Slu7 remained intact (Figure 4A). This indicates that while a molecular signature of MDS in humans is selection of cryptic 3΄ SS, disruption of the interaction between Hsh155 and Slu7 is not likely to be a major driver of the process in yeast. Supporting this conclusion is our observation that 3΄ SS choice in the ACT1-CUP1 assay is unaffected even by the Hsh155R294L mutation (Figure 3C–E).
The majority of HSH155 mutant alleles (10 of 14) altered Y2H interactions to Prp5, implying that many MDS mutations either directly or indirectly influence interactions between these two proteins during spliceosome assembly. Interestingly, previous work has shown that Prp5 mutations also alter BS fidelity at the same positions flanking the branchpoint adenosine that we observe to be altered by the MDS alleles (Figure 2F) (49). Taken together, the Y2H data support the notion that most protein-protein interactions between Hsh155 and other splicing factors are unaffected by Hsh155MDS. The major exception is the interaction between Hsh155 and Prp5.
Hsh155MDS mutations affect splicing in a manner distinct from Prp5 proofreading
Since Prp5 plays essential roles during pre-spliceosome assembly and U2/BS pairing, we tested whether the altered interactions between Hsh155 and Prp5 were the cause of the observed changes in BS usage. We noticed that results from the Y2H screen with Prp5 do not directly correlate with those from the ACT1-CUP1 assay (Figure 4C). For example, HSH155MDS alleles that decrease growth in the ACT1-CUP1 assay with the nonconsensus A258U BS reporter show a variety of effects in Hsh155/Prp5 in the Y2H assay. This suggests that changes in BS usage arise from more complicated mechanisms than simply disrupting or strengthening the interactions between Hsh155 and Prp5. To resolve this, we investigated the known roles of Prp5 to look for impacts on these functions by MDS mutations.
In an ATP-dependent role, Prp5 has been proposed to displace Cus2 from the U2 snRNP to permit U2 association with the pre-mRNA (Figure 5A) (50,51). We hypothesized that mutation of Hsh155 could be impacting Cus2 displacement by Prp5, and that this could lead to defects in spliceosome assembly and the observed changes in ACT1-CUP1 splicing. To investigate this, we generated MDS strains with CUS2 deleted and assayed them in an ACT1-CUP1 reporter assay using the A258U BS mutant. All CUS2Δ strains grew equally well as strains with intact CUS2 (Figure 5B). This shows that the observed changes in pre-mRNA splicing do not result from changes in Cus2 displacement by Prp5 during pre-spliceosome formation.
Figure 5.
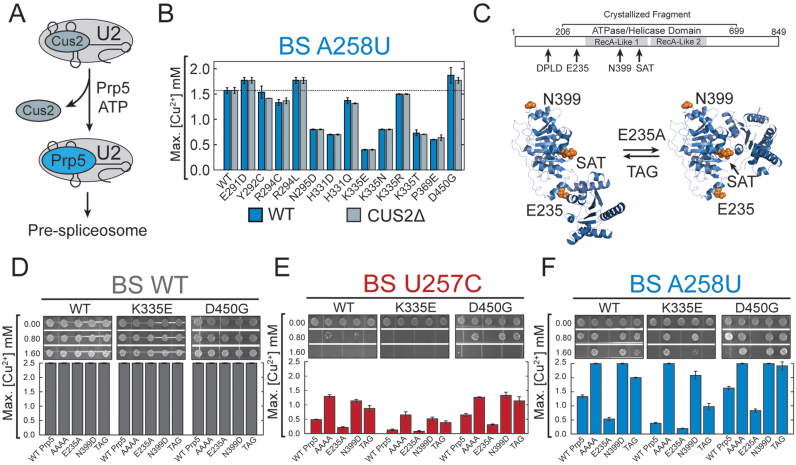
Hsh155 MDS mutants affect BS fidelity at a different step than Prp5 proofreading. (A) Cartoon depicting the proposed ATP-dependent function of Prp5 in displacement of Cus2 from the U2 snRNP during spliceosome assembly. (B) Comparison of Cu2+ growth assays using the nonconsensus A258U BS substitution ACT1-CUP1 reporter pre-mRNA between strains containing and lacking the Cus2 protein. No significant differences were observed between the two strains. (C) (Top) Schematic of Prp5 structure indicating the positions of Prp5 mutations used in this assay relative to the two DEAD-box RecA-like domains and the fragment of Prp5 whose structure has been determined by X-ray crystallography. (Bottom) Prp5 has been proposed to undergo a conformational change to promote splicing. The open structure (left) represents the structure determined by X-ray crystallography (pdb 4LJY) while the closed structure (right) is believed to be necessary for ATP hydrolysis and was modeled based on structures of other DEAD-box proteins (coordinates for the closed structure were obtained from Yong-Zhen Xu and Charles Query) (53). Positions of the Prp5 mutations used in this study are noted. The E235A mutation is believed to favor the closed conformation while the TAG mutation of the SAT-motif is believed to favor the open conformation. It is unclear if the N399D mutation used here would impact conformational switching. (D) Cu2+ growth assay for strains containing the Hsh155 WT, K335E, or D450G alleles in combination with the given Prp5 mutations (see text for additional explanation of each Prp5 mutation) with the ACT1-CUP1 reporter containing a consensus BS. (E) Cu2+ growth assay for combinations of Hsh155 and Prp5 as in part (D) except the U257C nonconsensus BS ACT1-CUP1 reporter was used. (F) Cu2+ growth assay for combinations of Hsh155 and Prp5 as in part (D) except the A258U nonconsensus BS ACT1-CUP1 reporter was used. In panels B and D–F, bars represent the average of three independent experiments, and error bars represent the standard deviation.
In addition to Cus2 displacement, Prp5 has also been implicated in proofreading at the BS during spliceosome assembly although the mechanism remains unclear (49,52). A number of mutations in Prp5 change the splicing of introns with BS substitutions and previous work has suggested that these may function in part by altering interactions between Prp5 and other splicing factors or by modulating Prp5 transitions between open and closed conformations (Figure 5C) (37,53). For example, alanine mutation of the N-terminal DPLD motif of Prp5 (AAAA) disrupts the interaction with U2/SF3b and causes greatly improved splicing of nonconsensus reporter substrates in vivo (54). The Prp5 mutation E235A disrupts the open conformation of the protein and diminishes splicing of nonconsensus reporters, while mutation of the Prp5 DEAD-box SAT motif to TAG may disrupt the closed conformation and improve splicing of nonconsensus reporters (Figure 5C) (53). The Prp5 mutation N399D also increases splicing of nonconsensus reporters; however, its mechanism is unclear (49). It has recently been proposed that all of these Prp5 mutations ultimately impact splicing by influencing how Prp5 is retained on the pre-spliceosome (37). In this model, Prp5 ensures BS fidelity by recognizing mispairing between the U2 snRNA and the intron BS and preventing tri-snRNP recruitment in the presence of a mismatch. Prp5 mutants with higher affinity for the pre-spliceosome (e.g. Prp5E235A) impair nonconsensus BS usage by retention of Prp5 in the pre-spliceosome and preventing tri-snRNP addition. Opposing mutants (e.g. Prp5AAAA, Prp5TAG and Prp5N399D) promote Prp5 release and progression of spliceosome assembly.
We next investigated the outcome of combining Prp5 mutations with MDS alleles during splicing. For this, we employed the MDS alleles HSH155K335E and HSH155D450G because these alleles show opposing effects in BS usage and interaction with Prp5 (Figures 2C and F; 4A). We generated strains expressing each combination of Prp5 and Hsh155 mutations and tested them in ACT1-CUP1 reporter assays using a consensus intron (Figure 5D). No differences were observed for any combination of Hsh155/Prp5 mutations for the WT ACT1-CUP1 reporter. When Hsh155/Prp5 mutant strains were tested in combination with the U257C and A258U BS substitution reporters, we observed that the Prp5 mutations AAAA, N399D, and TAG improved growth on Cu2+ while E235A diminished growth regardless of the Hsh155 background (Figure 5E and F). However, the MDS alleles of HSH155 still affected growth, as strains with Hsh155K335E showed generally diminished growth relative to Hsh155WT and Hsh155D450G strains showed improved growth irrespective of the PRP5 allele. This suggests that the mechanism of action of the Hsh155 mutations is independent from the mechanism of Prp5 mutation: in our assays, Hsh155 establishes a baseline level of BS usage that Prp5 mutations either can raise or lower.
To further evaluate the effects of Prp5 mutations on interactions between Prp5 and Hsh155, we expanded our Y2H assay to include the Prp5AAAA, Prp5E235A, Prp5N399D, and Prp5TAG mutants. We confirmed expression of all BD-Prp5 variants by western blot (Supplemental Figure S6A). BD-Prp5AAAA shows a complete loss of interaction with all AD-Hsh155 variants by Y2H (Supplemental Figure S6B). This result is consistent with earlier reports that showed that this region of Prp5 is important for the interaction of Prp5 with the SF3b complex (54). The BD-Prp5TAG mutant also decreased the interaction with Hsh155. These data support the model that the Prp5AAAA and Prp5TAG mutations improve nonconsensus BS usage by weakening the interaction between Prp5 and other splicing factors. Interestingly, BD-Prp5E235A and BD-Prp5N399D mutants showed only minor changes in growth relative to BD-Prp5WT in Y2H assays despite the strong influence these mutations have on BS usage in ACT1-CUP1 reporter assays. The Prp5E235A mutant modestly improved growth relative to Prp5WT for a number of Hsh155 mutations (e.g. WT, H331D, K335E, etc.) while Prp5N399D showed slightly impaired growth (e.g. WT, H331D, K335N, etc.). The directions of these changes are consistent with Prp5E235A and Prp5N399D interacting with the pre-spliceosome with different affinities to impact BS usage (37), and our data support Prp5E235A having higher affinity than Prp5N399D.
Importantly, the growth pattern of the HSH155MDS alleles relative to one another was maintained independent of the Prp5 mutation. For example, AD-Hsh155N295D grew better than AD-Hsh155WT and AD-Hsh155H331D grew worse than AD-Hsh155WT in all instances. While mutation of BD-Prp5 changed the Y2H interaction with AD-Hsh155WT and all Hsh155 alleles equivalently, the AD-Hsh155MDS variants showed distinct changes in Y2H interactions with BD-Prp5. Our results from the ACT1-CUP1 splicing reporter and Y2H assays argue that MDS alleles influence BS usage at a step distinct from that influenced by Prp5 mutations.
MDS mutations show genetic interactions with a Prp2 ATPase mutant
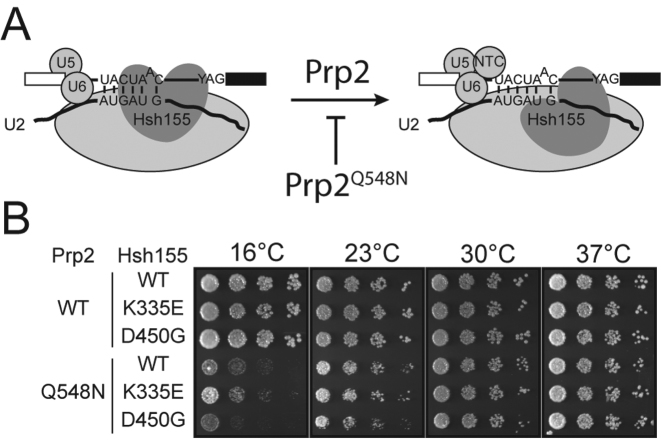
To investigate whether MDS mutations can impact splicing at steps subsequent to assembly, we looked for genetic interactions with Prp2. Prp2 is responsible for destabilizing the SF3b complex from the U2/BS duplex to allow further steps in splicing to occur (Figure 6A), likely resulting in release of the U2 snRNA/BS duplex so that it may enter the spliceosome active site (45). We generated strains with the Prp2 ATPase mutation Q548N in backgrounds with two different Hsh155MDS mutations (K335E and D450G). Prp2Q548N confers cold sensitivity (cs) to yeast, likely due to poor ATPase and/or helicase activity (34). Like Prp5, Prp2 is a fidelity factor and its function is correlated with proper basepairing between U2 and U6 snRNAs. Prp2Q548N suppresses lethal mutations in U2 that perturb U2/U6 helix Ia basepairing, suggesting that efficient ATPase activity of Prp2 prevents catalytic activation of spliceosomes with improperly formed active sites (34).
Figure 6.
MDS mutations interact genetically with a Prp2 mutation. (A) Cartoon schematic of Prp2-dependent activation of the spliceosome. Prp2 is believed to destabilize Hsh155 as well as the rest of the SF3b complex from interacting with the BS. The PRP2Q548N allele likely stalls this process at low temperatures (34). (B) Representative temperature sensitivity growth assays of the given Hsh155 variants in combination with Prp2WT or Prp2Q548N when plated on YPD at the given temperatures. Hsh155K335E partially suppresses Prp2Q548N and Hsh155D450G enhances cold sensitivity.
Strains expressing Prp2Q548N grew better at 16°C in the presence of Hsh155K335E than with Hsh155WT, while Prp2Q548N in the presence of Hsh155D450G grew slightly worse. (Figure 6B). These data indicate that MDS mutations such as Hsh155K335E that impair the splicing of reporter pre-mRNAs containing BS substitutions also partially suppress Prp2Q548N cold sensitivity. This suggests that Hsh155K335E-containing SF3 complexes may be partially destabilized and thus aid Prp2Q548N activity, possibly by altering the structure of the SF3 complex during a Prp2-dependent step in spliceosome activation. Mutations that improve splicing (Hsh155D450G) may do the opposite. Significantly, since BS sequences sensitive to Hsh155K335E and Hsh155D450G mutations (Figure 2) are extremely rare in yeast introns (if they are present at all) (55–57), it is likely that the difference in cold sensitivity for Prp2Q548N strains is due to changes in the splicing of introns containing consensus BS (or nonconcensus BS other than those affected by Hsh155MDS in our ACT1-CUP1 assay). MDS mutations may alter the stability of yeast U2 proteins at the BS in general and this altered stability of the Bact spliceosome can in turn modulate the requirement for Prp2. HSH155MDS alleles might function at multiple steps in splicing and in response to different sequence elements within the BS during each step.
DISCUSSION
Mounting evidence has implicated mutations in the splicing machinery as potent drivers of human disease (7). Among splicing factors that have been linked to disease, the essential and conserved U2 component SF3b1 has been found to be frequently mutated (12–14,41). We sought to understand the role of SF3b1 during splicing and the impact that MDS mutations can have on the function of the protein. We find that mutations associated with MDS and CLL alter the usage of certain nonconsensus BS without affecting the splicing of introns with consensus BS. Different mutations result in disparate changes in BS usage, and these mutations by themselves do not change 3΄ SS selection. Furthermore, these mutations appear to modify BS usage by a novel mechanism, potentially by disrupting Hsh155/SF3b1 conformations that stabilize weak U2:BS RNA duplexes formed on nonconsensus introns (Figure 7A, B). We show that the interaction network of Hsh155 is largely unperturbed by MDS mutations with the exception of Prp5; however, Prp5-dependent proofreading is not driving changes in BS usage. Finally, we show that mutations in Hsh155 have a genetic interaction with cs Prp2, suggesting a role for the protein in stabilizing the SF3b complex at the U2/BS duplex until activation. Together, these data suggest that SF3b1 mutations may cause disease through disruption of a BS recognition step conserved between yeast and humans that impacts how nonconsensus splice sites are utilized by the spliceosome.
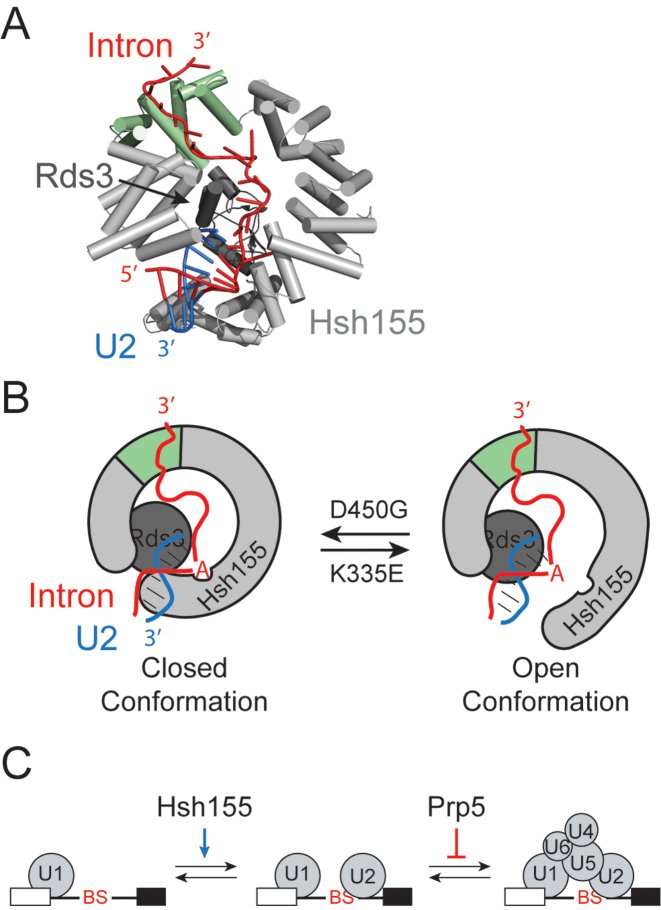
Figure 7.
Models for SF3b1/Hsh155 function in BS duplex stabilization during splicing. (A) Cartoon representation of Hsh155 (light grey and green) and Rds3 (dark gray) bound to the U2 snRNA/BS duplex from the cryo-EM structure of the yeast Bact spliceosome (25). The region of Hsh155 containing the MDS mutations studied here is shown in green. (B) Model for SF3b1/Hsh155 action at the BS. In addition to the structure shown in (A), Hsh155 must also exist in a conformation that permits release of the U2 snRNA/BS RNA duplex and splicing. MDS mutations impact Hsh155 conformation and lead to changes that affect recognition and stabilization of the BS duplex. Mutations that increase splicing of nonconsensus BS (e.g. Hsh155D450G) could stabilize the ‘closed’ or BS duplex bound form whereas mutations that inhibit splicing (e.g. Hsh155K335E) could favor an open form that is necessary for splicing catalysis but does not help stabilize a mismatched duplex during spliceosome assembly. (C) Model for opposing activities of SF3b1/Hsh155 and Prp5 during splicing. SF3b1 functions to stabilize U2 snRNA/BS duplex formation, particularly at nonconsensus or weak BS. Prp5 proofreading opposes this function to enforce BS fidelity by blocking tri-snRNP association. The relative activities of SF3b1/Hsh155 and Prp5 at particular BS may be used to promote or inhibit spliceosome assembly.
SF3b1/Hsh155 interacts with numerous splicing factors during splicing
In both yeast and humans, Hsh155/SF3b1 directly contacts the pre-mRNA substrate in the region near the BS and is probably present throughout splicing (8,23). This places Hsh155/SF3b1 in a position to influence how BS are selected during assembly as well as later steps in catalysis. One mechanism by which Hsh155/SF3b1 could influence splicing is by regulating the recruitment and retention of spliceosome proteins. Consistent with this role is prior Y2H data showing interactions between Hsh155 and numerous splicing factors (44), and our data identifying novel Y2H interactions between Hsh155 and Prp2, Prp43 and Slu7 (Figure 4A). Our Y2H results confirm recently observed crosslinks between Hsh155 and a Prp43 variant in BactΔprp2 spliceosomes (48). Our Y2H data additionally suggest that destabilization of the SF3 complex by Prp2 may occur in part through direct contact between Prp2 and Hsh155, in agreement with recent cryo-EM structures (24,25). We speculate that changes in Slu7 function due to altered interaction with Hsh155/SF3b1 may in turn explain how small molecules that bind SF3b1 also impact exon ligation in human spliceosomes, since Slu7 has previously been implicated in 3΄ SS selection in both yeast and humans (43,58,59). Thus, by modulating interactions between the spliceosome and transiently associated splicing factors, Hsh155/SF3b1 could potentially regulate spliceosome assembly via Prp5, spliceosome activation via Prp2, 3΄ SS selection via Slu7, and spliceosome disassembly or discard via Prp43. Hsh155/SF3b1 may act as a general hub on the spliceosome for proteins needing BS access throughout splicing. This hypothesis is intriguing because the N-terminus of SF3b1 in humans contains numerous ULM regions that interact with additional partners not found in yeast (60). These additional factors could modulate constitutive or alternative splicing by binding to and acting through SF3b1.
Hsh155/SF3b1 functions to stabilize the U2/BS duplex
Accurate recognition of splice sites is essential for maintaining the integrity of a spliced mRNA, and the spliceosome has evolved numerous mechanisms to ensure high fidelity at nearly every stage of splicing. Many spliceosome proofreading mechanisms rely on coupling the activity of DExH ATPases with the stalling or discard of spliceosomes (32). For example, one mechanism proposed for proofreading BS selection involves recognition of mispairing between the BS and U2 snRNA by Prp5 (49). Mispairing triggers Prp5 retention on the spliceosome, thereby blocking subsequent assembly steps (37). Our data suggest the functions of Prp5 are unaffected by MDS mutations. First, deletion of Cus2 (which is believed to be removed from U2 by ATP-dependent Prp5 activity) showed no changes in reporter RNA splicing, suggesting that MDS mutations do not act through retention of Cus2. Additionally, Prp5 mutations and MDS alleles of Hsh155 are not epistatic. Prp5 mutations known to affect fidelity still impact splicing when used in combination with MDS alleles, suggesting these mutants act at different times during splicing. Based on these data and our Y2H results, we propose that Hsh155/SF3b1 modulates BS usage in a manner distinct from Prp5. We believe that a function of Hsh155 is to confer stability to weak duplexes, thereby improving spliceosome assembly and splicing on introns containing weak BS.
Hsh155 could help to stabilize structures within the spliceosome that are ultimately necessary for catalysis. Specifically, SF3b1/Hsh155 may help bolster U2/BS duplexes with mismatches near the branchpoint adenosine at the −2, −1 and +1 positions early during spliceosome assembly and this stabilization may allow for progression to subsequent steps in splicing (Figure 7C). In this model, mutations that impact splicing, such as those found in MDS, are those that affect the ability of SF3b1/Hsh155 to stabilize the U2/BS duplex, with some mutations conferring greater stability (e.g. D450G) than WT and others conferring less (e.g. K335E). Consistent with this hypothesis are our observations that transversions occurring at A258, which immediately flanks the branchpoint at the −1 position, impact splicing in these mutants. These transversions introduce C/U and U/U mismatches within the snRNA/BS duplex. The A258G transition, which can likely form a stable G/U wobble pair with the snRNA, shows no splicing defects. Also consistent with this hypothesis is that SF3b1/Hsh155 mutations do not change the splicing of an intron containing a consensus BS sequence or a sequence with substitution of the branchpoint adenosine with cytidine. This position is not paired with the snRNA and therefore may contribute less to the overall stability of the helix (61). Recognition and proofreading of the branchpoint nucleotide is performed by other splicing factors (branchpoint bridging protein/SF1 during assembly and Prp16 prior to 5΄ SS cleavage) (62–64) while Hsh155 is critical for formation the U2 snRNA/BS duplex. Finally, our results agree with recent structures of the yeast Bact spliceosome. In those structures, the nucleotides of the U2/BS duplex immediately flanking the branchpoint adenosine also make extensive contacts with Hsh155 (24,25); these are the same nucleotide positions shown by our ACT1-CUP1 to be impacted by Hsh155MDS. Thus, these MDS alleles of Hsh155 may change how Hsh155 interacts with the U2/BS duplex in the branchpoint region and ultimately lead to stabilization or destabilization of duplexes containing nearby mismatches.
Our data also show that MDS mutations that impair BS usage can affect mRNA levels to a greater extent than those that improve usage (Figure 2E). The biological function of Hsh155 may be to relax the specificity of the spliceosome and allow it to splice introns with BS that deviate from the consensus sequence and form metastable U2 snRNA/BS duplexes. We propose that this role is of greater necessity in organisms like humans that have introns with poorly conserved splice sites. Indeed, the lack of observable growth phenotypes in yeast expressing MDS alleles is likely the result of the scarcity of nonconsensus BS sequences (Figure 2F) in native yeast introns (55–57). Intriguingly, this mechanism also suggests a finely tuned balance between Hsh155 and Prp5 function to modulate splicing. In this model, Prp5 would counteract Hsh155 activity and reduce nonconsensus BS usage at a step subsequent to Hsh155 binding of the U2 snRNA/BS duplex (Figure 7C). This is consistent with our data that Prp5 and Hsh155 mutations both impact splicing of reporter RNAs when combined (Figure 5D–F). Balancing the competing activities of Hsh155 and Prp5 could be used to regulate splicing of transcripts containing particular BS sequences.
Conformational flexibility in SF3b1 leads to changes in BS usage
Proposed structural changes in SF3b1 could play a role in coupling snRNA/BS duplex recognition and stabilization with conformations that either promote or impede splicing. Indeed, conformational changes in SF3b1 have been previously proposed based on low resolution EM reconstructions and the crystal structure of the human SF3b complex (26,65,66). A number of positions mutated in MDS are involved in intramolecular interactions between helices in the HEAT repeats (26). Directly affecting the conformational equilibrium between multiple states of Hsh155 is one way to achieve disparate effects on splicing. In our model, binding of intronic RNA downstream of the BS to HEAT repeats mutated in MDS helps to select a closed conformation of Hsh155 that will bind and stabilize the U2/BS duplex (Figure 7B). The K335E mutation and others like it impede these processes and favor an open conformation, potentially by interfering with RNA binding or an allosteric switch. Conversely, mutations that improve nonconsensus intron splicing (e.g. D450G) shift the equilibrium towards the closed, duplex-bound state. This model is supported by our observation that MDS mutations have additive effects on splicing, as the shift in equilibrium can be restored by including a second antagonist allele (Figure 2H). We cannot exclude an opposing model in which MDS alleles primarily function by disrupting intronic RNA binding near the site of the mutations. In this case, a stable RNA duplex formed between the U2 snRNA and a consensus BS would overcome the effect of mutating this RNA binding site on Hsh155. However, we do not favor this model, as a functional purpose of RNA binding to this region of Hsh155 is unclear. Moreover, a recent mutational analysis of human SF3b1 revealed no detectable changes in RNA binding by SF3b1 after introduction of a MDS mutation (26). Finally, we note that Hsh155 structural equilibrium may also impact the competition between U2 branchsite stem loop formation and snRNA/BS basepairing to facilitate coupled snRNA and protein conformational changes during splicing (67). Structures of Bact spliceosomes and U2 snRNPs containing MDS mutations would provide interesting insight to the effects these mutations have on the overall structure.
Further evidence supporting the model that conformational change in Hsh155 is an important driver of BS usage comes from our experiments in which MDS alleles were combined with a mutant of Prp2 (Figure 6B). These results are consistent with our model that the K335E mutation favors the open, unbound conformation of Hsh155 and D450G favors the closed, bound conformation Hsh155. The former conformation facilitates Prp2-dependent spliceosome activation, while the latter impedes this step. It is likely that spliceosome activation involves release of contacts between Hsh155 and the U2/BS duplex to allow this RNA to enter the active site and splicing to proceed. As suggested by our data, RNA release and structural transitions in Hsh155 are likely coupled to one another as well as to Prp2 activity. ATP hydrolysis by Prp2 may help to trigger Hsh155 conformational change during activation of the spliceosome.
How usage of a different intronic BS leads to alternative 3΄ SS selection in MDS is not immediately obvious based on sequence predictions or structural models. Consistent with our observation that MDS mutants do not have defects in cryptic 3΄ SS discrimination (Figure 3D, E), recent work has identified that most splice site changes arise from switching of the BS from a ‘weak’ BS to ‘strong’ BS located nearby and upstream of the canonical BS (17,18). Changes in how MDS mutant SF3b1 stabilizes weak U2/BS duplexes could lead to repositioning of the spliceosome to regions of the intron with differing complementarity to the U2 snRNA. Whether BS repositioning occurs during assembly or in spliceosomes through the action of a DEAH-box helicase (e.g. Prp2 or Prp16) is not known. These helicases facilitate sampling of multiple potential BS by the spliceosome (68), and how these BS are sampled and their competitiveness with one another may be influenced by MDS mutations in SF3b1. Altered BS sampling in MDS potentially rationalizes the observation that a weak polypyrmidine (Py) tract is necessary for BS switching in humans. The binding of the splicing factors U2AF65/35 to strong Py tracts could help limit BS sampling of neighboring sequences by the spliceosome during assembly.
The work presented here supports a novel mechanism wherein SF3b1 helps to define the BS during pre-mRNA splicing. Furthermore, we have provided insight into how mutations in a splicing factor can change fundamental functions of the spliceosome. The specific changes in alternative splicing that predispose individuals to MDS is currently unclear. Recent work has shown that the MDS-linked U2AF35 S34F mutation predisposes the cell to transformation through aberrant processing of the ATG7 transcript (69). A similar mechanism may be occurring in MDS patients with mutant SF3b1, wherein only a fraction of the misprocessed transcripts lead to disease. These misprocessed transcripts may be produced by subtle alteration of how BS compete with one another during splicing and/or by how human-specific splicing regulatory proteins interact with SF3b1 to stabilize BS duplexes containing mismatches. It has been speculated that BS switching due to MDS alleles arises from selection of sequences with increased pairing potential to the U2 snRNA (17,18), consistent with our results showing that some of the homologous Hsh155 mutations impair splicing when mismatches between the BS and snRNA are present. This suggests that principles that emerge from understanding how these disease alleles alter splicing in yeast will be informative for studies of human splicing in cancer. Understanding how SF3b1 functions in molecular detail is crucial to remedying defects associated with these processes and for designing novel SF3b1-targeted therapeutics for patients suffering from these malignancies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Charles Query, Soo-Chen Cheng, Jill Wildonger, and Dave Brow for strains, plasmids, and antibodies and Sandy Tretbar and George Luo for technical assistance. We also thank Sam Butcher, Dave Brow, Allison Didychuk, Jon Staley and Betty Craig for careful reading of the manuscript, comments and advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R00 GM086471, R01 GM112735 to A.A.H., T32-GM08349 to T.J.C.]; Shaw Scientist Award [to A.A.H.]; Beckman Young Investigator Awards [to A.A.H.]; startup funding from the University of Wisconsin-Madison, Wisconsin Alumni Research Foundation (WARF) and the Department of Biochemistry [to A.A.H.]; William H. Peterson Fellowship [to T.J.C.]. Funding for open access charge: NIH R01 grant award as well as my start-up funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Anczuków O., Krainer A.R.. Splicing-factor alterations in cancers. RNA. 2016; 22:1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dvinge H., Kim E., Abdel-Wahab O., Bradley R.K.. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer. 2016; 16:413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997; 6:1205–1214. [DOI] [PubMed] [Google Scholar]
- 4. McKie A.B., McHale J.C., Keen T.J., Tarttelin E.E., Goliath R., van Lith-Verhoeven J.J., Greenberg J., Ramesar R.S., Hoyng C.B., Cremers F.P. et al. . Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum. Mol. Genet. 2001; 10:1555–1562. [DOI] [PubMed] [Google Scholar]
- 5. Vithana E.N., Abu-Safieh L., Allen M.J., Carey A., Papaioannou M., Chakarova C., Al-Maghtheh M., Ebenezer N.D., Willis C., Moore A.T. et al. . A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol. Cell. 2001; 8:375–381. [DOI] [PubMed] [Google Scholar]
- 6. Chakarova C.F., Hims M.M., Bolz H., Abu-Safieh L., Patel R.J., Papaioannou M.G., Inglehearn C.F., Keen T.J., Willis C., Moore A.T. et al. . Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2002; 11:87–92. [DOI] [PubMed] [Google Scholar]
- 7. Visconte V., Makishima H., Maciejewski J.P., Tiu R.V.. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia. 2012; 26:2447–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Will C.L., Lührmann R.. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wahl M.C., Will C.L., Lührmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 10. Hsu T.Y.T., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S., Echeverria G.V., Sun T., Kurley S.J., Tyagi S. et al. . The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015; 525:384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tresini M., Warmerdam D.O., Kolovos P., Snijder L., Vrouwe M.G., Demmers J.A.A., van IJcken W.F.J., Grosveld F.G., Medema R.H., Hoeijmakers J.H.J. et al. . The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015; 523:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin M., Maßhöfer L., Temming P., Rahmann S., Metz C., Bornfeld N., van de Nes J., Klein-Hitpass L., Hinnebusch A.G., Horsthemke B. et al. . Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013; 45:933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D., Pellagatti A., Wainscoat J.S., Hellstrom-Lindberg E., Gambacorti-Passerini C. et al. . Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011; 365:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quesada V., Conde L., Villamor N., Ordóñez G.R., Jares P., Bassaganyas L., Ramsay A.J., Beà S., Pinyol M., Martínez-Trillos A. et al. . Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011; 44:47–52. [DOI] [PubMed] [Google Scholar]
- 15. Gentien D., Kosmider O., Nguyen-Khac F., Albaud B., Rapinat A., Dumont A.G., Damm F., Popova T., Marais R., Fontenay M. et al. . A common alternative splicing signature is associated with SF3B1 mutations in malignancies from different cell lineages. Leukemia. 2014; 28:1355–1357. [DOI] [PubMed] [Google Scholar]
- 16. DeBoever C., Ghia E.M., Shepard P.J., Rassenti L., Barrett C.L., Jepsen K., Jamieson C.H.M., Carson D., Kipps T.J., Frazer K.A.. Transcriptome sequencing reveals potential mechanism of cryptic 3΄ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 2015; 11:e1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alsafadi S., Houy A., Battistella A., Popova T., Wassef M., Henry E., Tirode F., Constantinou A., Piperno-Neumann S., Roman-Roman S. et al. . Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016; 7:10615–10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D., Bailey S.L., Bhavsar E.B., Chan B., Colla S. et al. . Cancer-associated SF3B1 hotspot mutations induce cryptic 3΄ splice site selection through use of a different branch point. Cell Rep. 2015; 13:1033–1045. [DOI] [PubMed] [Google Scholar]
- 19. Mupo A., Seiler M., Sathiaseelan V., Pance A., Yang Y., Agrawal A.A., Ioria F., Bautista R., Pacharne S., Tzelepis K. et al. . Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia. 2016; 10.1038/leu.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Behrens S.E., Tyc K., Kastner B., Reichelt J., Lührmann R.. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell Biol. 1993; 13:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruby S.W., Chang T.H., Abelson J.. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993; 7:1909–1925. [DOI] [PubMed] [Google Scholar]
- 22. McPheeters D.S., Muhlenkamp P.. Spatial organization of protein-RNA interactions in the branch site-3΄ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 2003; 23:4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider C., Agafonov D.E., Schmitzová J., Hartmuth K., Fabrizio P., Lührmann R.. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3΄ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet. 2015; 11:e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rauhut R., Fabrizio P., Dybkov O., Hartmuth K., Pena V., Chari A., Kumar V., Lee C.-T., Urlaub H., Kastner B. et al. . Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016; 353:1399–1405. [DOI] [PubMed] [Google Scholar]
- 25. Yan C., Wan R., Bai R., Huang G., Shi Y.. Structure of a yeast activated spliceosome at 3.5 A resolution. Science. 2016; 353:904–911. [DOI] [PubMed] [Google Scholar]
- 26. Cretu C., Schmitzová J., Ponce-Salvatierra A., Dybkov O., DeLaurentiis E.I., Sharma K., Will C.L., Urlaub H., Lührmann R., Pena V.. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell. 2016; 64:307–319. [DOI] [PubMed] [Google Scholar]
- 27. Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K., Watanabe H., Kitahara T., Yoshida T., Nakajima H. et al. . Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007; 3:576–583. [DOI] [PubMed] [Google Scholar]
- 28. Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y.. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007; 3:570–575. [DOI] [PubMed] [Google Scholar]
- 29. Hasegawa M., Miura T., Kuzuya K., Inoue A., Won Ki S., Horinouchi S., Yoshida T., Kunoh T., Koseki K., Mino K. et al. . Identification of SAP155 as the Target of GEX1A (Herboxidiene), an antitumor natural product. ACS Chem. Biol. 2011; 6:229–233. [DOI] [PubMed] [Google Scholar]
- 30. Folco E.G., Coil K.E., Reed R.. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011; 25:440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim S.-H., Lin R.-J.. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:888–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y.-C., Cheng S.-C.. Functional roles of DExD/H-box RNA helicases in Pre-mRNA splicing. J. Biomed. Sci. 2015; 22:54–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohrt T., Prior M., Dannenberg J., Odenwalder P., Dybkov O., Rasche N., Schmitzova J., Gregor I., Fabrizio P., Enderlein J. et al. . Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012; 18:1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wlodaver A.M., Staley J.P.. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA. 2014; 20:282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Effenberger K.A., Urabe V.K., Prichard B.E., Ghosh A.K., Jurica M.S.. Interchangeable SF3B1 inhibitors interfere with pre-mRNA splicing at multiple stages. RNA. 2016; 22:350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lesser C.F., Guthrie C.. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993; 133:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang W.-W., Cheng S.-C.. A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence. Genes Dev. 2015; 29:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Amberg D.C., Burke D., Strathern J.N.. Methods in Yeast Genetics. 2005; Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 39. Siatecka M., Reyes J.L., Konarska M.M.. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999; 13:1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keogh M.-C., Kim J.-A., Downey M., Fillingham J., Chowdhury D., Harrison J.C., Onishi M., Datta N., Galicia S., Emili A. et al. . A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2005; 439:497–501. [DOI] [PubMed] [Google Scholar]
- 41. Wan Y., Wu C.J.. SF3B1 mutations in chronic lymphocytic leukemia. Blood. 2013; 121:4627–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rossi D., Spina V., Bomben R., Rasi S., Dal-Bo M., Bruscaggin A., Rossi F.M., Monti S., Degan M., Ciardullo C. et al. . Association between molecular lesions and specific B-cell receptor subsets in chronic lymphocytic leukemia. Blood. 2013; 121:4902–4905. [DOI] [PubMed] [Google Scholar]
- 43. Frank D., Guthrie C.. An essential splicing factor, SLU7, mediates 3΄ splice site choice in yeast. Genes Dev. 1992; 6:2112–2124. [DOI] [PubMed] [Google Scholar]
- 44. Wang Q., He J., Lynn B., Rymond B.C.. Interactions of the yeast SF3b splicing factor. Mol. Cell. Biol. 2005; 25:10745–10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lardelli R.M., Thompson J.X., Yates J.R., Stevens S.W.. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010; 16:516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin A., Schneider S., Schwer B.. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002; 277:17743–17750. [DOI] [PubMed] [Google Scholar]
- 47. Mayas R.M., Maita H., Staley J.P.. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006; 13:482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fourmann J.B., Dybkov O., Agafonov D.E., Tauchert M.J., Urlaub H., Ficner R., Fabrizio P., Lührmann R.. The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. eLife. 2016; 5:e15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu Y.-Z., Query C.C.. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell. 2007; 28:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perriman R., Ares M.. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000; 14:97–107. [PMC free article] [PubMed] [Google Scholar]
- 51. Perriman R., Barta I., Voeltz G.K., Abelson J., Ares M.. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 24:13857–13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dalbadie-McFarland G., Abelson J.. PRP5: a helicase-like protein required for mRNA splicing in yeast. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:4236–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Z.-M., Yang F., Zhang J., Tang Q., Li J., Gu J., Zhou J., Xu Y.-Z.. Crystal Structure of Prp5p Reveals Interdomain Interactions that Impact Spliceosome Assembly. Cell Rep. 2013; 5:1269–1278. [DOI] [PubMed] [Google Scholar]
- 54. Shao W., Kim H.S., Cao Y., Xu Y.Z., Query C.C.. A U1-U2 snRNP Interaction Network during Intron Definition. Mol. Cell. Biol. 2011; 32:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gould G.M., Paggi J.M., Guo Y., Phizicky D.V., Zinshteyn B., Wang E.T., Gilbert W.V., Gifford D.K., Burge C.B.. Identification of new branch points and unconventional introns in Saccharomyces cerevisiae. RNA. 2016; 20:1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qin D., Huang L., Wlodaver A., Andrade J., Staley J.P.. Sequencing of lariat termini in S. cerevisiae reveals 5΄ splice sites, branch points, and novel splicing events. RNA. 2016; 22:237–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grate L., Ares M.. Searching yeast intron data at Ares lab web site. Methods Enzymol. 2002; 350:380–392. [DOI] [PubMed] [Google Scholar]
- 58. Chua K., Reed R.. The RNA splicing factor hSlu7 is required for correct 3΄ splice-site choice. Nature. 1999; 402:207–210. [DOI] [PubMed] [Google Scholar]
- 59. Zhang X., Schwer B.. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 1997; 25:2146–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thickman K.R., Swenson M.C., Kabogo J.M., Gryczynski Z., Kielkopf C.L.. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein–protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 2006; 356:664–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Query C.C., Moore M.J., Sharp P.A.. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994; 8:587–597. [DOI] [PubMed] [Google Scholar]
- 62. Berglund J.A., Chua K., Abovich N., Reed R., Rosbash M.. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997; 89:781–787. [DOI] [PubMed] [Google Scholar]
- 63. Koodathingal P., Novak T., Piccirilli J.A., Staley J.P.. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5΄ splice site cleavage during pre-mRNA splicing. Mol. Cell. 2010; 39:385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Burgess S.M., Guthrie C.. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993; 73:1377–1391. [DOI] [PubMed] [Google Scholar]
- 65. Golas M.M., Sander B., Will C.L., Lührmann R., Stark H.. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol. Cell. 2005; 17:869–883. [DOI] [PubMed] [Google Scholar]
- 66. Rakesh R., Joseph A.P., Bhaskara R.M., Srinivasan N.. Structural and mechanistic insights into human splicing factor SF3b complex derived using an integrated approach guided by the cryo-EM density maps. RNA Biol. 2016; 13:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Perriman R., Ares M.. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol. Cell. 2010; 38:416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Semlow D.R., Blanco M.R., Walter N.G., Staley J.P.. Spliceosomal DEAH-Box ATPases remodel Pre- mRNA to activate alternative splice sites. Cell. 2016; 164:985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Park S.M., Ou J., Chamberlain L., Simone T.M., Yang H., Virbasius C-M., Ali A.M., Zhu L.J., Mukherjee S., Raza A. et al. . U2AF35 (S34F) promotes transformation by directing aberrant ATG7 Pre-mRNA 3΄ end formation. Mol. Cell. 2016; 62:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.