Abstract
Germline mutations in SPRTN cause Ruijs–Aalfs syndrome (RJALS), a disorder characterized by genome instability, progeria and early onset hepatocellular carcinoma. Spartan, the protein encoded by SPRTN, is a nuclear metalloprotease that is involved in the repair of DNA–protein crosslinks (DPCs). Although Sprtn hypomorphic mice recapitulate key progeroid phenotypes of RJALS, whether this model expressing low amounts of Spartan is prone to DPC repair defects and spontaneous tumors is unknown. Here, we showed that the livers of Sprtn hypomorphic mice accumulate DPCs containing Topoisomerase 1 covalently linked to DNA. Furthermore, these mice exhibited DNA damage, aneuploidy and spontaneous tumorigenesis in the liver. Collectively, these findings provide evidence that partial loss of Spartan impairs DPC repair and tumor suppression.
INTRODUCTION
Germline mutations in SPRTN cause Ruijs–Aalfs syndrome (RJALS), a human autosomal-recessive disorder characterized by genome instability, progeria and early onset hepatocellular carcinoma (1,2). Our previous work demonstrated that, while complete knockout of Sprtn in mice was embryonically lethal, Sprtn hypomorphic mice are viable despite reduced Spartan expression (3). Sprtn hypomorphic mice, however, developed premature aging phenotypes and chromosomal instability (3), thereby recapitulating some of the findings in RJALS patients. Currently, it is unknown whether Sprtn insufficiency causes tumor susceptibility in Sprtn hypomorphic mice and why RJALS patients are particularly prone to liver tumors.
Spartan was initially identified as a regulator of translesion synthesis (TLS), a specialized replication system that utilizes low fidelity TLS polymerases to bypass certain DNA lesions without their actual repair (4,5). Spartan localizes to DNA lesions through its PCNA-interacting peptide motif and UBZ4, a zinc-finger ubiquitin-binding domain in the C-terminus (6–12). Consistent with the role of Spartan in TLS regulation, bypass of UV-induced DNA lesions was impaired in mouse embryonic fibroblasts (MEFs) after Sprtn knockout (3). However, Sprtn knockout MEFs develop even more severe phenotypes, including incomplete DNA replication, accumulation of abnormal chromosomes and cell death (3), suggesting that Spartan may have TLS-independent functions. Consistent with this possibility, S. cerevisiae Wss1, which has a similar domain structure to Spartan, plays a role in the repair of DNA–protein crosslinks (DPCs) (13,14), which are toxic DNA lesions that block DNA replication and transcription and cause genome instability (15–19). Accordingly, the similarity between Wss1 and Spartan raised the possibility that Spartan might also participate in DPC repair.
Indeed, while this manuscript was in preparation, four studies implicated Spartan in DPC repair (20–23). These studies found that Spartan is a DNA-dependent metalloprotease that prevents accumulation of DPCs (20–23) and a constitutive component of the replication machinery that repairs DPCs in a DNA replication-coupled manner (21). These findings, which are reminiscent of the replication-coupled DPC repair previously reported in Xenopus egg extracts (24), clearly demonstrate the role of proteolytic events in DPC repair. The physiological relevance of this protease-mediated DPC repair process, however, requires further investigation.
DPCs involving Topoisomerase 1 (Top1) have been proposed as one of the substrates of Spartan in human cells (21). Top1 is an essential and abundant enzyme that relieves DNA torsional strain during DNA replication and transcription (25–27). Top1 cuts one strand of DNA, allows rotation around the intact strand and then religates the single-strand break (28). When re-ligation is blocked by adjacent DNA lesions or by chemotherapeutic agents such as camptothecin (CPT), Top1 is trapped as a covalent adduct between the 3΄ end of the nicked DNA and the Top1 active site tyrosine. Commonly referred to as a Top1 cleavage complex (Top1cc), this bulky DPC can block advancing replication forks, ultimately resulting in toxic DNA damage such as DNA double-strand breaks and cell death (16–18,29). Top1ccs can be repaired by a mechanism involving tyrosyl-DNA phosphodiesterase 1 (Tdp1), which hydrolyzes the covalent bond between the Top1 catalytic tyrosine and the DNA (29,30). The fact that a cell expresses specialized enzymes for repairing Top1ccs highlights the importance of their repair. It is, therefore, crucial to understand the role of Spartan in Top1cc repair in tissues as well as isolated cells.
In this study, we investigate whether Sprtn insufficiency predisposes mice to tumor formation, particularly in the liver. We also study the function of Spartan in the repair of DPCs, especially Top1ccs, in mouse cells and tissues. Our findings identify Spartan as a component of Top1cc repair in mice, demonstrate its role as a tumor suppressor and link Spartan deficiency with Top1cc accumulation and tumorigenesis in hepatocytes.
MATERIALS AND METHODS
Sprtn-targeted mice and analyses of tumorigenesis
Sprtn hypomorphic mice (SprtnH/H) have been described previously (3). Mice were euthanized by carbon dioxide inhalation and all major organs were screened for spontaneous overt tumors. For histology analysis of liver tumors, formalin-fixed paraffin-embedded tumor samples were stained with hematoxylin and eosin and examined by a mouse pathologist. All of the animal procedures were approved by Mayo Clinic Institutional Animal Care and Use Committee.
Cell culture
Sprtn F/F; Cre-ERT2 MEFs were described earlier (3). SprtnH/– MEFs were generated as described previously (3) and immortalized by retroviral expression of SV40 T-antigen (31). MEFs were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. SprtnF/F; Cre-ERT2 MEFs were treated for 2 days with 2 μM 4-hydroxytamoxifen (4-OHT) dissolved in methanol (MeOH) to induce Cre-mediated conversion of the floxed allele to the knockout allele (3). In parallel, MEFs were treated with the same amount of MeOH as a vehicle control.
Plasmids, RNA interference and viral infection
Complementary DNA fragments encoding wild-type or the E112A mutant of human Spartan (8) were expressed using the retroviral vector pMSCV-puro. Wild-type human Spartan (full-length) was also expressed with a C-terminal 3xFlag tag using pMSCV-puro. The SprT domain of human Spartan (amino acids 1–219) (8) was expressed with a C-terminal 3xFlag/2xNLS tag using the retroviral vector pBabe-puro. For stable knockdown of Tdp1, a short hairpin RNA (shRNA) against Tdp1 (shTdp1) or a non-targeting shRNA (shControl) was expressed using the lentiviral vector pLKO.1-puro. The shRNA sequences were: shControl, 5΄-CAACAAGAUGAAGAGCACCAA-3΄ and shTdp1, 5΄-UAAAGUCCUGCACCCGUACAG-3΄. Knockdown of Tdp1 was confirmed by quantitative reverse transcription-PCR. Retroviruses and lentiviruses were produced by cotransfection of viral and packaging plasmids in the human embryonic kidney cell line 293T (ATCC, CRL-11268). Cells were infected with virus-containing media in the presence of 2 μg ml−1 polybrene and selected with 3 μg ml−1 puromycin (Sigma).
Quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was purified from cells using an RNeasy Kit (Qiagen) and cDNA was generated using oligo(dT) primers and SuperScript III (Invitrogen) according to the supplier's instructions. qPCR was performed in triplicate for each sample using a CFX96 PCR machine (Bio-Rad) and iTaq Universal SYBR Green Supermix (Bio-Rad). The following primers were used: Tdp1, forward primer (5΄-TGG ACG CTT TCA AGG AAG TC-3΄) and reverse primer (5΄-CCG ATG CTT GAG AAC TGA CC-3΄); Gapdh, forward primer (5΄-AGA ACA TCA TCC CTG CAT CC-3΄) and reverse primer (5΄-CAC ATT GGG GGT AGG AAC AC-3΄); and Sprtn, forward primer (5΄-GGA CCT TGT AGA GAC TCT TTT G-3΄) and reverse primer (5΄-CCT CAT CAT GGA AAG TGT GG-3΄) (3). Sprtn and Tdp1 mRNA levels were normalized to Gapdh.
Immunofluorescence and microscopy
Experiments were performed as described previously (32). Briefly, cells were fixed with 4% paraformaldehyde for 15 min at 4°C, permeabilized for 15 min with 0.25% Triton X-100 at 4°C, and treated with 1% sodium dodecyl sulphate (SDS) for 5 min at room temperature. Cells were then washed five times with wash buffer (0.1% Triton X-100, 0.1% bovine serum albumin (BSA) in phosphate buffered saline (PBS), blocked in 10% milk in 150 mM NaCl and 10 mM Tris-HCl (pH 7.4) and incubated with primary antibodies diluted in 5% normal goat serum overnight at 4°C. Rabbit anti-γH2AX was from Active motif (#39117, 1:750) and mouse anti-Top1cc (1:100) was reported previously (32). After washing five times with wash buffer, cells were stained with Alexa Fluor 488-conjugated (for Top1cc) or Alexa Fluor 568-conjugated (for γH2AX) secondary antibodies (1:1000, Invitrogen) diluted in 5% normal goat serum for 1 h at room temperature. Cells were washed with wash buffer six times, stained with 0.1 μg ml−1 DAPI (for MEFs) or 1 μg ml−1 Hoechst 33258 (for tissue samples) in PBS and mounted with SlowFade Gold (Invitrogen). Images were captured using a LSM 710 scanning confocal microscope with a 100× objective and processed using the ImageJ software and Adobe Photoshop CS5.
For immunostaining of tissues, fresh tissue samples were embedded in optimal cutting temperature compound, frozen in methylbutane at −80°C, sectioned onto slides using a cryostat and immediately fixed with 4% paraformaldehyde for 15 min at 4°C. Using the procedures described above for tissue culture cells, the cells were then permeabilized, treated with SDS and immunostained.
Clonogenic survival assays
For drug sensitivity assays, 500 cells were seeded per well in triplicate into 6-well plates and cultured in the presence of various drugs. Drug-containing media were replenished every other day and cells were cultured for six days. After staining with Coomassie Blue, colonies with more than 50 cells were counted manually (33). Drugs used in clonogenic assays were: camptothecin (Sigma), etoposide (Sigma), PARP inhibitor MK-4827 (ChemieTek) and 5-aza-2΄-deoxycytidine (Sigma).
Flow cytometry
Analyses of DNA content were performed as described previously (3). In brief, cells were harvested and fixed by incubation in 70% ethanol overnight at −20°C. After incubation with 50 μg ml−1 propidium iodide, 10 μg ml−1 RNase A, 0.05% Nonidet P-40 for 30 min, cells were analyzed by flow cytometry using a FACS Canto II (BD Biosciences). Percentages of the cell-cycle phases were calculated using ModFit LT (Verity Software House).
DNA fiber assays
Bypass of UV-induced DNA lesions was examined by DNA fiber assays as described previously (3).
Nuclei isolation from tissues and Western blotting
Isolation of nuclei from mouse tissues was performed using a previously described method (34). Briefly, fresh tissues were rinsed with STM buffer (250 mM sucrose, 50 mM Tris-HCl (pH 7.4), 5 mM MgSO4), minced and homogenized in STM buffer with freshly added 1 mM phenylmethylsulfonyl fluoride (PMSF) (STM/PMSF). After filtering, sedimentation (800 × g for 15 min), and washing with STM/PMSF buffer, the nuclei were resuspended in DSM (2.1 M sucrose, 50 mM Tris-HCl (pH 7.4), 5 mM MgSO4)/PMSF buffer, layered over cushions of the same buffer and centrifuged for 60 min at 20 000 rpm. The nuclei were resuspended in STM/PMSF buffer, layered again over DSM/PMSF buffer and sedimented for another 30 min.
Isolated nuclei were solubilized in alkylation buffer (6 M guanidine hydrochloride, 250 mM Tris-HCl (pH 8.5), 10 mM EDTA) supplemented with 1 mM PMSF and 1% (v/v) 2-mercaptoethanol, sonicated, reacted with iodoacetamide and dialyzed sequentially into 4 M urea and 0.1% SDS as previously described (35). After lyophilization, samples were resuspended in SDS sample buffer (4 M urea, 2% SDS, 62.5 mM Tris-HCl (pH 6.8) and 1 mM EDTA) and heated to 65°C for 20 min. Aliquots containing 20 μg protein were separated by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes and probed with antibodies. Anti-Top1 (C-21) antibody was a gift from Y-C Cheng (Yale University, New Haven, CT, USA). Antibodies against Histone H1 (#61201, 1:5000) were from Active Motif.
To analyze Spartan protein levels in MEFs, cells were lysed in NP-40 lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 10% Glycerol) supplemented with protease inhibitor mix (Sigma). Supernatants containing solubilized proteins were recovered after centrifugation (12 000 rpm, 15 min). Aliquots containing 30 μg of protein were separated by SDS-PAGE, transferred to nitrocellulose membranes and probed with antibodies. Anti-Flag (#F1804, 1:2000) and anti-β-actin (#A5316, 1:5000) antibodies were purchased from Sigma. Mouse anti-human Spartan antibodies were reported previously (9). Antibodies against POLD1 (#sc-17 777, 1:250) and PCNA (#sc-56, 1:2000) were from Santa Cruz Biotechnology. Anti-POLD3 (#A301-244A, 1:1000) and anti-H2A (#07-146, 1:2000) antibodies were from Bethyl Laboratory and EMD Millipore, respectively.
CPT- induced Top1 degradation assay
CPT-induced Top1 degradation was assayed using a previously described method (36,37) with slight modification. Briefly, MEFs grown in 100-mm tissue culture plates were treated with CPT as indicated in the figure legend, incubated for 30 min in drug-free medium, lysed in alkaline lysis buffer (200 mM NaOH, 2 mM EDTA), neutralized with 1.5 M Tris-HCl, pH 8.0 and mixed with 10 × S7 nuclease buffer (50 mM MgCl2, 50 mM CaCl2, 5 mM dithiothreitol, 1 mM EDTA and a protease inhibitor mixture). Cell lysates were incubated with 100 units of staphylococcal S7 nuclease (Sigma) on ice for 20 min, which allowed Top1 release from DNA. Reactions were terminated by the addition of 20 mM EGTA. After a brief sonication, aliquots containing 12 μg of protein were separated by SDS-PAGE, transferred to nitrocellulose membranes and probed with anti-Top1 antibody (C-21, BD Biosciences #556597, 1:2000) and anti-β-actin antibody (#A5316, Sigma, 1:5000).
iPOND (Isolation of proteins on nascent DNA)
iPOND experiments were performed as described previously (38). Briefly, cells were treated with 10 μM EdU (Invitrogen) for 15 or 20 min to label nascent DNA, and chased with 10 μM thymidine for 10 and 30 min. Harvested cells were fixed with 1% formaldehyde for 20 min, permeabilized for 30 min with 0.25% Triton X-100 at room temperature followed by 90 min of click reaction, in which biotin is conjugated to EdU in EdU-labeled DNA. Cells were then lysed in RIPA lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 1 mM EDTA, 0.5% sodium deoxycholate) supplemented with protease inhibitor mix and PMSF followed by sonication (20 s ON, 40 s OFF for 4 cycles). Supernatants containing solubilized DNA-protein complexes were recovered after centrifugation (12 000 rpm, 15 min). To purify the EdU-labeled DNA–protein complexes, recovered samples were incubated with streptavidin–agarose beads overnight at 4°C. After washing the beads once with lysis buffer, once with 1 M NaCl, once with PBS and twice with lysis buffer, the precipitated proteins in DNA–protein complexes were eluted by boiling in 2xLDS (Lithium dodecyl sulfate) sample buffer and analyzed by Western blotting.
ICE assays
In vivo complexing of enzyme (ICE) experiments for detecting Top1cc were performed as described (32). Briefly, MEFs grown in 150-mm tissue culture plates were lysed in lysis buffer (1% (w/v) sarkosyl in 1 M Tris-HCl pH 8.0, 0.1 M EDTA), layered on top of a CsCl gradient and sedimented at 31 000 rpm for 21 h at 22°C to separate DNA–protein complexes from free proteins. Fractions (0.5 ml) were collected from the bottom of the gradients. Using a slot blotting apparatus, 0.05 ml aliquots from each fraction were immobilized onto nitrocellulose membranes and probed with anti-Top1 antibody (C-21, BD Biosciences #556597, 1:2000) and anti-PCNA antibody (#sc-56, Santa Cruz Biotechnology, 1:2000).
Fluorescence in situ hybridization (FISH)
Interphase FISH analysis was performed as previously described (39). Briefly, tissues were cut into small pieces and minced in 1 ml of PBS containing 0.5% BSA and 2 mM EDTA using a gentleMACS Dissociator (Miltenyi Biotec). Twenty microliters of 7 mg ml−1 Liberase TM (Roche, #05401127001) were added, and the cell suspension was incubated at 37°C for 30 min. After subsequent dissociation using gentleMACS Dissociator, cells were collected with 5 ml PBS and separated from undigested tissues using a 70-μm cell strainer. Cells were then collected by centrifugation (1000 rpm, 5 min) and resuspended in 1 ml of PBS containing 0.5% BSA and 2 mM EDTA. Following suspension, 40 μl of methanol:glacial acetic acid (3:1) fixative was added. After two washes with 5 ml of fixative, cells were resuspended in 50–100 μl fixative, dropped onto positively charged microscope slides and labeled using FISH probes for chromosomes 4 and 7. At least 100 cells were analyzed per sample.
Statistics
GraphPad 5 Prism software was used to graph the data. Statistical significance was determined either by a two-tailed unpaired t-test or a Fisher's exact test, where specified. All graphs are indicated with the significance as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
RESULTS
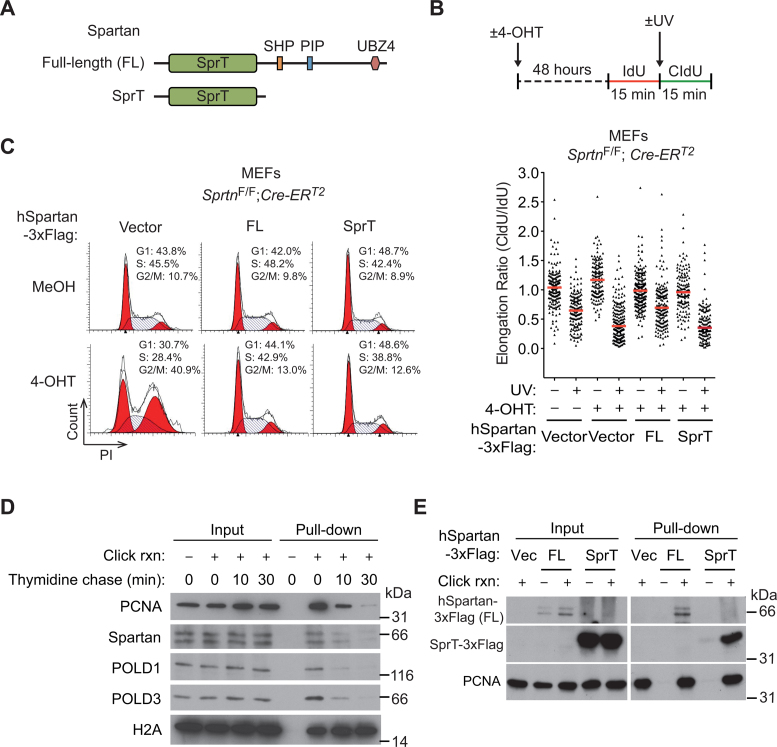
The SprT domain of Spartan is sufficient to support the normal cell cycle, but not lesion bypass.
Conditional Sprtn knockout in MEFs causes impaired TLS of UV-induced DNA lesions and accumulation in late S and G2 phases of the cell cycle (3). Whether the TLS deficiency causes the observed cell-cycle defects in Sprtn knockout cells is not known. To address this question, we compared the ability of full-length Spartan and the N-terminal half of Spartan, which contains the SprT domain, to restore TLS and cell-cycle progression in Sprtn knockout MEFs (Figure 1A). Ectopic expression of full-length human Spartan, but not the isolated SprT domain, restored TLS of UV-induced lesions in Sprtn knockout cells (Figure 1A and B and Supplementary Figure S1), indicating the necessity of the C-terminal half for the TLS function of Spartan (Figure 1A). In contrast, expression of the N-terminal half of Spartan relieved arrest of cells in late S/G2 and restored the normal cell-cycle distribution (Figure 1C). These results indicate that the cell-cycle function of Spartan is separable from the TLS function, and suggest that Spartan has a TLS-independent function that is critical for completing DNA replication in S phase. Consistent with the role of Spartan in DNA replication, iPOND, a method that analyzes proteins isolated with nascent DNA (38), revealed that Spartan localizes at sites of DNA replication and travels with replication machinery components such as POLD1, POLD3 (subunits of DNA polymerase δ) and PCNA (Figure 1D). Given that the SprT domain was sufficient for normal cell-cycle progression, we next asked whether the SprT domain alone can localize to replication forks. Notably, the isolated SprT domain, like full-length Spartan, was recovered with nascent DNA (Figure 1E), indicating the sufficiency of the SprT domain for localization to replication forks.
Figure 1.
The SprT domain is sufficient to suppress the cell-cycle defects, but not the lesion bypass deficiency, in Sprtn knockout cells. (A) Schematic representation of full-length and truncated Spartan proteins used in this study. The truncated Spartan mutant, denoted as SprT, contains the SprT domain but lacks the C-terminal half. SprT, a zinc metalloprotease-like domain; SHP, a p97/VCP-interacting motif; PIP, PCNA-interacting peptide; UBZ4, ubiquitin-binding zinc-finger 4. (B) (Upper panel) Schematic representation of DNA fiber assays. To visualize ongoing replication, mouse embryonic fibroblasts (MEFs) treated with MeOH or 4-OHT for 48 h were sequentially labeled with IdU and CldU with or without UV irradiation (40 J m−2) between the labeling. (Lower panel) DNA fiber assays were performed in SprtnF/F; Cre-ERT2 MEFs expressing full-length human Spartan (FL) or the SprT domain only (amino acids 1–219). Distribution of replication forks at different CldU/IdU ratios is shown. At least 100 fibers were scored for each sample. Horizontal red lines indicate median values. (C) Cell-cycle profiling of SprtnF/F; Cre-ERT2 MEFs expressing full-length human Spartan (FL) or the SprT domain only (amino acids 1–219). Cells were treated with MeOH or 4-OHT for 48 h, stained with PI and analyzed by flow cytometry. (D) Isolation of proteins on nascent DNA (iPOND) assays showing the localization of Spartan at replication forks. 293T cells were pulsed with EdU for 15 min and then chased with thymidine for 10 and 30 min. Eluted proteins were separated by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted for the indicated proteins. PCNA, POLD1 and POLD3 are shown as controls for replisome proteins that are enriched only at replication forks. Histone H2A is shown as a chromatin protein that is not enriched at replication forks. (E) iPOND assays showing that the isolated SprT domain of Spartan can localized at the fork. iPOND was performed in 293T cells expressing full-length human Spartan (FL) or the SprT domain only after pulse labeling with EdU for 20 min. Proteins at EdU-labeled nascent DNA were isolated and assessed using Western blotting. PCNA is shown as a control for replisome protein.
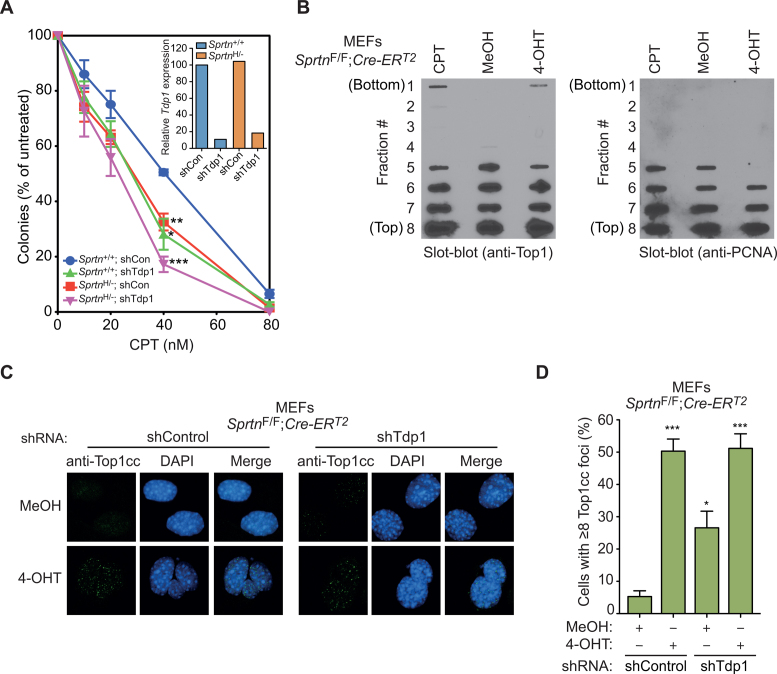
Reduced Sprtn expression sensitizes cells to camptothecin
In exploring the TLS-independent function of Spartan, we turned our attention to the recent report that Wss1, a Spartan ortholog in S. cerevisiae, plays a role in the repair of DPCs, including Top1ccs (13,40). To assess whether Spartan plays a similar role in Top1cc repair in vertebrates, we examined the possibility that Spartan counteracts the toxic effect of Top1ccs induced by CPT in mammalian cells and compared the effect to knockdown of Tdp1, a phosphodiesterase implicated in enzymatic removal of Top1ccs. Because Sprtn knockout is lethal in MEFs (3), we used Sprtn hypomorphic MEFs (SprtnH/–), which express reduced levels of Spartan but exhibit a normal cell-cycle distribution (3) (Supplementary Figure S2A and S2B). SprtnH/– MEFs displayed increased CPT sensitivity compared to Sprtn+/+ MEFs (Figure 2A), suggesting a possible role for Spartan in the repair of Top1ccs. Moreover, the degree of sensitization to CPT in SprtnH/– MEFs was similar to that of Tdp1 knockdown in Sprtn+/+ MEFs, indicating that Spartan is as important as Tdp1 in counteracting Top1cc toxicity.
Figure 2.
Sprtn H/− MEFs exhibit increased sensitivity to camptothecin and Sprtn knockout causes accumulation of Top1ccs in MEFs. (A) Clonogenic survival assays. Sprtn+/+ and SprtnH/− MEFs, with or without Tdp1 knockdown, were cultured in the presence of the indicated camptothecin (CPT) concentration for 6 days while drug-containing media were replenished every other day. Cells were stained with Coomassie Blue and colonies with more than 50 cells were counted. Values are mean ± s.e.m. of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed unpaired t-test) relative to Sprtn+/+ shControl. The inset shows qPCR analyses of Tdp1 mRNA levels in the indicated MEFs. Values were normalized to Gapdh and presented relative to Sprtn+/+ shControl. (B) Detection of Top1ccs by in vivo complexing of enzyme (ICE) assays. SprtnF/F; Cre-ERT2 MEFs were treated with MeOH or 4-OHT for 48 h and Top1ccs were separated from free Top1 by cesium chloride gradients. Fractions (#1-8) were deposited onto nitrocellulose membranes and probed with antibody against Top1 (left) or PCNA (right). Cells treated with 5 μM CPT for 1 h are shown as a positive control for Top1ccs. (C) Top1cc focus formation. The indicated MEFs treated with MeOH or 4-OHT for 48 h were stained with anti-Top1cc to detect Top1cc accumulation. DNA was co-stained with DAPI to visualize nuclei. (D) Quantitation of Top1cc foci. Experiments were performed as in C. At least 100 cells were scored for Top1cc foci and percentages of cells with 8 or more foci are shown. Values are mean ± s.e.m. of three independent experiments. *P < 0.05; ***P < 0.001 (two-tailed unpaired t-test) relative to the MeOH-treated shControl samples.
In addition to Top1cc processing by Tdp1, proteasome-dependent Top1 degradation has been reported (36,37,41,42). To assess whether Spartan participates in this proteasomal pathway, Spartan-proficient and conditional Sprtn-knockout MEFs were treated for up to 4 h with 25 μM CPT (36,37,41,42) and subjected to immunoblotting for Top1. Top1 degradation was observed only at high CPT concentrations and was not altered in Sprtn–/– MEFs (Supplementary Figure S2C), arguing against a role for Spartan in CPT-induced proteasomal degradation of Top1.
In contrast to CPT, no increases in sensitivity were observed in SprtnH/– MEFs treated with the following DPC-forming drugs: etoposide (which traps Topoisomerase 2-DNA covalent complexes), MK-4827 (which enhances poly(ADP-ribose) polymerase 1-DNA interactions, including covalent adducts) and 5-aza-2΄-deoxycytidine (which causes DNA methyltransferase-DNA covalent adducts) (43,44) (Supplementary Figure S2D–F). These results indicate that Spartan is particularly important in combating toxic lesions generated by Top1 poisons.
Spartan loss causes accumulation of Top1ccs
To test whether Spartan participates in the repair of Top1ccs, we examined the accumulation of Top1ccs in MEFs after Sprtn deletion. In initial experiments, we used ICE assays, which separate covalent protein–DNA complexes from free proteins on CsCl gradients. These assays detected increased Top1, but not PCNA, in DNA-containing gradient fractions after the Sprtn gene was conditionally deleted in MEFs (Figure 2B). These results indicate that Spartan loss increases the covalent binding of Top1 to DNA.
To verify this finding using another experimental approach, we stained MEFs with an antibody that specifically detects Top1ccs (32) (Supplementary Figure S3A). Sprtn knockout resulted in punctate staining of nuclei with this antibody (Figure 2C and D), a pattern similar to that observed with CPT (32). In agreement with the role of Tdp1 in Top1cc repair (29), Tdp1 knockdown also caused increased accumulation of Top1ccs in Spartan-proficient MEFs (Figure 2C and D and Supplementary Figure S3B). Interestingly, depletion of Tdp1 in Sprtn knockout cells did not cause further Top1cc accumulation compared to Sprtn knockout alone (Figure 2C and D). Collectively, these results suggest that Spartan plays an important role in the repair of Top1ccs in a pathway that may also involve Tdp1.
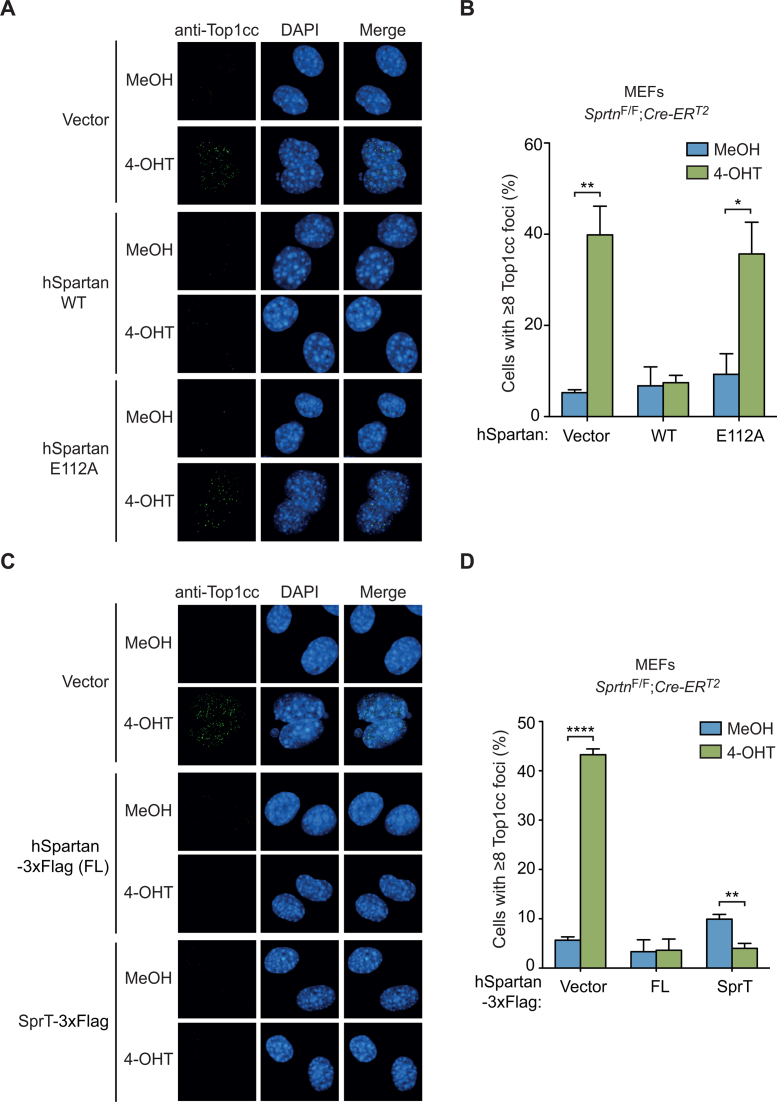
The SprT domain is necessary and sufficient for preventing Top1cc accumulation
Spartan contains a metalloprotease domain SprT (Figure 1A). To test whether the SprT domain plays a role in the repair of Top1ccs, we expressed either wild-type human Spartan or SpartanE112A, which harbors a point mutation in the SprT metalloprotease active site (8), in conditional Sprtn knockout MEFs (Supplementary Figure S4). Ectopic expression of wild-type human Spartan, but not SpartanE112A, suppressed the accumulation of Top1ccs in Sprtn–/– cells (Figure 3A and B), suggesting a role for catalytically active metalloprotease in Top1cc repair. To further evaluate the role of the SprT domain, we next asked whether the SprT domain itself is sufficient to suppress Top1cc accumulation. Notably, introduction of just the SprT domain suppressed Top1cc accumulation in Sprtn–/– cells as effectively as wild-type Spartan (Figure 3C and D and Supplementary Figure S1). These results demonstrate that the metalloprotease SprT domain alone is sufficient for Top1cc repair, and suggest that a catalytically active SprT domain is necessary for Top1cc removal.
Figure 3.
The SprT domain of Spartan is necessary and sufficient to suppress Top1cc accumulation in Sprtn knockout MEFs. (A) Top1cc focus formation in SprtnF/F; Cre-ERT2 MEFs expressing wild-type human Spartan or the E112A mutant. Cells were treated with MeOH or 4-OHT for 48 h, and stained with anti-Top1cc antibodies and DAPI. (B) Quantitation of cells containing Top1cc foci. At least 100 cells were scored for Top1cc foci and percentages of cells with 8 or more foci are shown. Values are mean ± s.e.m. of four independent experiments. (C) Top1cc focus formation in SprtnF/F; Cre-ERT2 MEFs expressing full-length (FL) human Spartan or the SprT domain (amino acids 1–219). Cells were treated as in A. (D) Quantitation of cells containing Top1cc foci. Cells were scored as in B. Values are mean ± s.e.m. of three independent experiments. *P < 0.05; **P < 0.01; ****P < 0.0001 (two-tailed unpaired t-test).
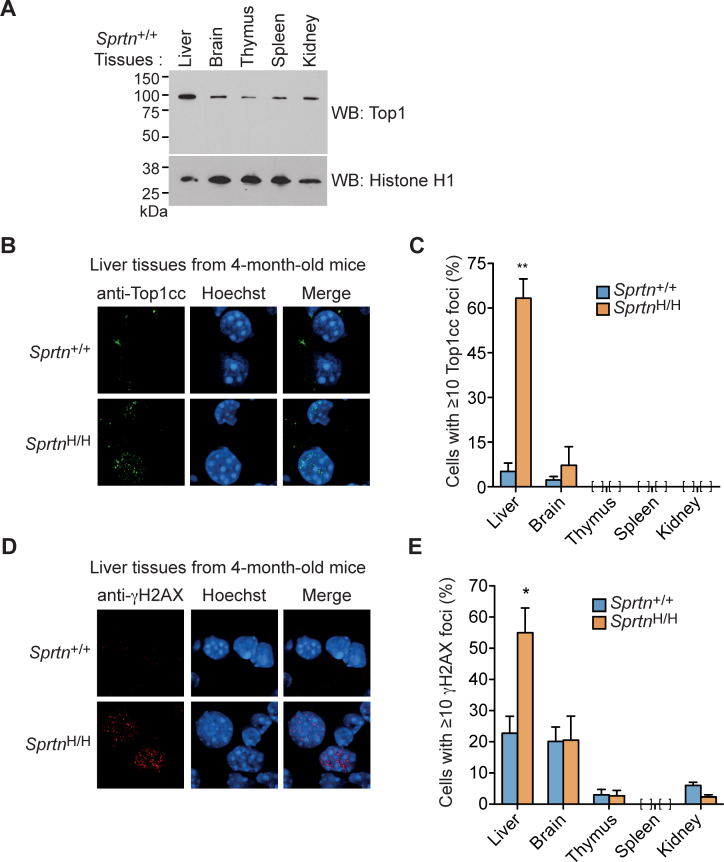
Accumulation of Top1ccs in the liver of SprtnH/H mice
We next asked whether Top1cc lesions accumulate in Sprtn hypomorphic mouse tissues. For these studies, we focused on the liver because (i) humans with germline Sprtn mutations develop hepatocellular carcinoma (1,2), implying that the liver is specifically affected by Spartan deficiency; and (ii) Top1 (∼100 kDa) is highly expressed in nuclei from the mouse liver compared to other tissues (Figure 4A). As shown in Figure 4B, Top1ccs were readily detected in the livers of 4-month-old SprtnH/H mice, indicating that SprtnH/H livers exhibit impaired Top1cc repair at an early age. In contrast, few if any Top1cc foci were observed in other tissues including brain, thymus, spleen and kidney from Sprtn+/+ and SprtnH/H mice (Figure 4C), consistent with lower Top1 expression in these tissues compared to the liver (Figure 4A). In addition, 4-month-old SprtnH/H mice displayed increased levels of γH2AX foci, a DNA damage marker, in the liver but not in other tissues (Figure 4D and E). These findings suggest that Sprtn insufficiency causes Top1cc accumulation in the liver at an early age, accompanied by DNA damage. Collectively, these results in mouse liver (Figure 4) identify a physiological context in which Spartan plays a rate-limiting role in Top1cc repair.
Figure 4.
The livers of SprtnH/H mice exhibit increased accumulation of Top1ccs. (A) Expression levels of nuclear Top1 in the indicated tissues of 10-month-old female Sprtn+/+ mice as assessed by anti-Top1 Western blotting. Histone H1 is shown as a loading control. (B) Immunohistochemistry of Top1ccs in the liver of 4-month-old mice. Cryosections of the liver from Sprtn+/+ and SprtnH/H mice were stained with anti-Top1cc. DNA was stained with Hoechst to visualize nuclei. (C) Quantitation of cells containing Top1cc foci in the indicated tissues of 4-month-old mice. At least 100 cells were scored for Top1cc foci in Sprtn+/+ and SprtnH/H mice. Percentages of cells with 10 or more foci are shown. Values are mean ± s.e.m. (n = 3). No cells were scored as positive for Top1cc foci in the thymus, spleen and kidney. (D) Immunohistochemistry of γH2AX in the liver of 4-month-old mice. Cryosections of the liver from Sprtn+/+ and SprtnH/H mice were stained with anti-γH2AX. DNA was stained with Hoechst to visualize nuclei. (E) Quantitation of cells containing γH2AX foci in the indicated tissues of 4-month-old mice. At least 100 cells were scored for γH2AX foci and percentages of cells with 10 or more foci are shown. Values are mean ± s.e.m. (n = 3). No cells were scored as positive for γH2AX foci in the spleen. *P < 0.05; **P < 0.01 (two-tailed unpaired t-test).
Sprtn insufficiency drives tumorigenesis in mice, particularly in the liver
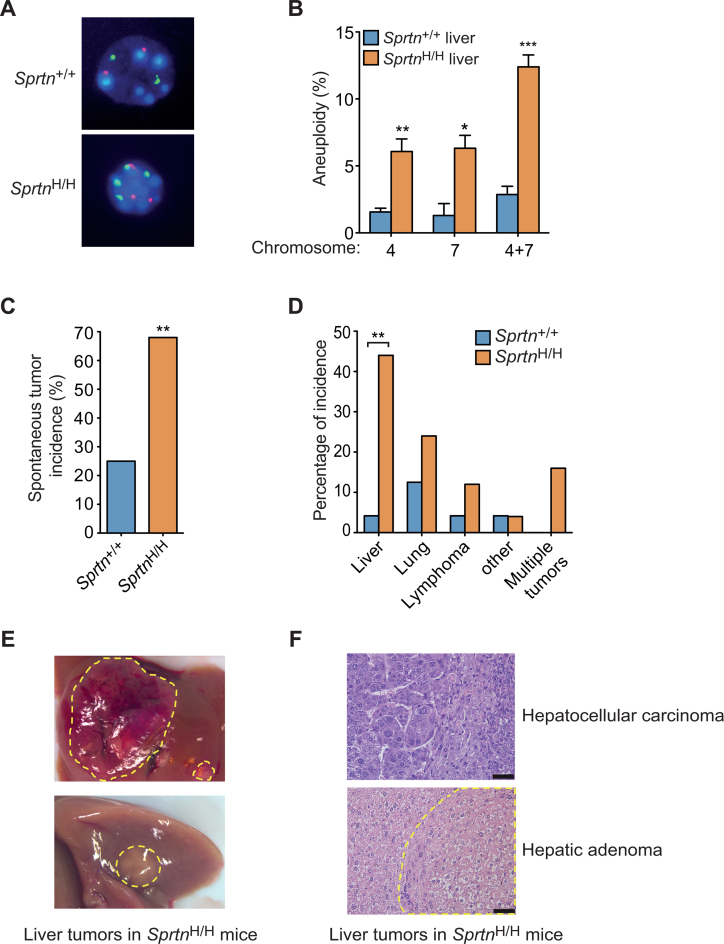
Because cells isolated from Sprtn hypomorphic mice exhibited chromosome instability (3), we next examined whether the livers of these mice have abnormal numbers of chromosomes. FISH analyses revealed that livers in SprtnH/H mice are more aneuploid than livers from Sprtn+/+ mice (Figure 5A and B). Given that increased aneuploidy and chromosome instability have been commonly linked with cancer predisposition (45–48), we also asked whether Sprtn insufficiency renders mice more prone to tumorigenesis. To address this question, we established cohorts of Sprtn+/+ and SprtnH/H mice and screened them for overt tumor formation at 22–25 months of age. Nearly 70% of SprtnH/H mice displayed spontaneous tumors, a significant increase in the tumor incidence from 25% in Sprtn+/+ mice (Figure 5C). This result indicates that reduced Spartan expression promotes spontaneous tumor formation in mice. Notably, the types of tumors were different in Sprtn+/+ and SprtnH/H mice (Figure 5D). The livers of SprtnH/H mice, which showed persistent accumulation of Top1ccs (Supplementary Figure S5A and S5B), were the primary site of tumor formation, although the size and number of tumors per liver varied among the mice (Figure 5D and E). Histology of the tumors indicated that most of the liver tumors were hepatic adenomas, characterized by nodular collections of hepatocytes with bland cytology and normal trabecular architecture. In one of eight nodules examined, liver plates were widened (3–4 cells thick) and hepatocytes were pleomorphic, supporting a diagnosis of hepatocellular carcinoma (Figure 5F). Collectively, these results indicate that Sprtn insufficiency predisposes mice to spontaneous tumorigenesis, particularly in the liver.
Figure 5.
Sprtn insufficiency promotes spontaneous tumorigenesis. (A) Images of interphase fluorescence in situ hybridization (FISH) for chromosomes 4 (red) and 7 (green). Liver cells from 22-month-old male Sprtn+/+ and SprtnH/H mice are shown. Because normal liver cells can be polyploid, only cells that contain odd numbers of chromosomes 4 or 7 were scored as aneuploid. Note that liver cells from Sprtn+/+ mice have normal ploidy, whereas liver cells from SprtnH/H mice exhibit aneuploidy with three copies of chromosomes 4. (B) Quantitation of aneuploidy in liver cells from Sprtn+/+ and SprtnH/H mice. Experiments were performed as in A. At least 100 cells were scored for aneuploidy and percentages of aneuploid cells are shown. Values are mean ± s.e.m. (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed unpaired t-test). (C) Spontaneous tumor incidence of 22- to 25-month old Sprtn+/+ and SprtnH/H mice. (Sprtn+/+, n = 24; SprtnH/H, n = 25). **P < 0.01 (Fisher's exact test). (D) Spectrum of spontaneous tumor types in 22- to 25-month old Sprtn+/+ and SprtnH/H mice. **P < 0.01 (Fisher's exact test). (E) Images of spontaneous liver tumors in 22-month-old SprtnH/H mice. Tumors are indicated by dotted yellow lines. (F) Hematoxylin and eosin-stained histological images of spontaneous liver tumors (hepatocellular carcinoma and hepatic adenoma) from SprtnH/H mice. Dotted yellow lines indicate hepatic adenoma. Scale bars, 50 μm.
DISCUSSION
Our previous study demonstrated that reduced Spartan levels in mice caused premature aging phenotypes, including lordokyphosis, cataracts and senescence (3). Here, we demonstrate that Sprtn hypomorphic mice are also prone to spontaneous tumorigenesis as they age further (Figure 5C). Both premature aging and the cancer susceptibility seen in the Sprtn hypomorphic mice recapitulate phenotypes observed in patients with RJALS. Consistent with the notion that RJALS is a genome instability syndrome, DNA damage and abnormal numbers of chromosomes are observed in Sprtn hypomorphic mice (3). To our knowledge these results provide the first evidence that Sprtn insufficiency in mice recapitulates all of the major phenotypes of RJALS in humans.
Our current study also identifies Top1cc as a DPC lesion that accumulates in the liver when Spartan levels are reduced (Figure 4B and C and Supplementary Figure S5A and S5B). Coupled with the observation that conditional Sprtn knockout in MEFs also causes Top1cc accumulation (Figure 2B–D), which is consistent with a role for Spartan in Top1cc repair, our results in mice support and extend the recent report that Spartan repairs DPCs, including Top1cc, in cultured cell lines (21). Importantly, we demonstrate that Sprtn insufficiency in mice causes Top1cc accumulation in the absence of Top1 poisons (Figure 4B and C and Supplementary Figure S5A and S5B), suggesting that Spartan plays a critical role in keeping the Top1cc levels low even in mice that have not been treated with Top1 poisons. Our findings are consistent with the possibility that DPC repair defects contribute to the phenotypes observed in Sprtn hypomorphic mice and RJALS patients.
The liver is the predominant site of tumor formation in RJALS patients and Sprtn hypomorphic mice (Figure 5D) (1,2). While the mechanism underlying the tissue specificity remains unknown, it is noteworthy that total Top1 protein levels are particularly high in liver nuclei (Figure 4A), raising the possibility that more Top1cc will form in this tissue during normal Top1-mediated catalysis. Moreover, we find that Top1ccs accumulate in the livers of Sprtn hypomorphic mice from an early age (Figure 4B and C). Given that Top1ccs are mutagenic and genome destabilizing in yeast as well as in mammalian cells (49–51), it is tempting to speculate that Top1cc-associated genomic instability might contribute to tumor formation in the livers of Sprtn hypomorphic mice. In addition, given the recent finding that Spartan promotes repair of formaldehyde-induced DPCs (20–23), it is possible that formaldehyde-induced DPCs might contribute to liver tumorigenesis as well. Because the liver is the main organ that processes formaldehyde, Spartan might play a particularly important role in the liver to combat DPCs. Altogether, the liver appears to be more prone to DPC accumulation and, therefore, specifically affected by Spartan inactivation.
An undefined replication-coupled proteolytic event has been described for DPC repair in Xenopus egg extracts, although the protease remains unidentified (24). In this system, replication fork collision with a DPC leads to DPC proteolysis followed by TLS past the remaining peptide–DNA lesion. Consistent with this model, a recent report suggested that Spartan is a constitutive part of the replication machinery (Figure 1D) (21). Our experiments also support this conclusion and extend it further by showing that the isolated SprT domain localizes to replication sites (Figure 1E). These data suggest that the SprT domain associates with the replication machinery independently of the C-terminal PCNA-interacting peptide motif and UBZ4 domain, and the recently reported DNA-binding sites (20,21,52). Consistent with this interpretation, our previous study identified interactions between the Spartan SprT domain and DNA polymerase δ (8). On the other hand, a DNA replication-independent function of Spartan has also been suggested in C. elegans and D. melanogaster (20,53). Thus, it remains unclear whether the Spartan-dependent DPC repair pathway is always coupled with DNA replication. Nevertheless, the present findings further support an emerging role for Spartan in the repair of DPCs.
We also obtained additional insight into the mechanism of Top1cc repair by Spartan using our conditional Sprtn-knockout and Sprtn hypomorphic MEFs. First, we show that the active site in the SprT protease domain is necessary for repair (Figure 3A and B), and that the SprT domain is sufficient for the Top1cc repair in vivo if nuclear localization signals are attached (Figure 3C and D). Thus, although the C-terminus of Spartan contributes to optimal protease activity in vitro (20,21), that portion of the protein is dispensable for the cell cycle and Top1cc repair in intact cells. Residual protease activity of the SprT domain and its intrinsic affinity for the replication machinery might be sufficient to allow DPC repair in vivo. Second, our data show that Top1ccs do not accumulate further when Tdp1 was depleted in Sprtn knockout cells (Figure 2C and D), supporting the recent notion that Spartan and Tdp1 may work in the same Top1cc repair pathway (21). Third, we show that cells with reduced Spartan expression are sensitive to CPT, but not to other DPC-inducing drugs (Figure 2A and Supplementary Figure S2D–F). This suggests that Spartan may not participate in the repair of all DPCs. However, recent studies reported hypersensitivity of Spartan-depleted cells and RJALS patient cells to etoposide (21,22). This discrepancy might be explained if the Sprtn hypomorphic cells used here expressed enough Spartan to repair etoposide-induced DPCs but not Top1ccs. Nonetheless, our data suggest that Top1ccs might be the predominant DPCs that depend on Spartan for clearance. Finally, our data show that Spartan does not participate in CPT-induced proteasomal degradation of Top1 (Supplementary Figure S2C). This suggests that Spartan might be important for Top1cc repair when Top1cc accumulation is under a certain threshold, whereas the proteasome pathway becomes predominant when cells accumulate extremely high levels of Top1cc after treatment with CPT concentrations that are unachievable with clinical drug exposures.
In conclusion, this study identifies Spartan as an important factor in Top1cc repair, particularly in the liver and shows that Spartan is a tumor suppressor. Future work will determine the role of DPCs in premature aging and tumorigenesis in Sprtn hypomorphic mice. Understanding the different functions of Spartan at the molecular level will also help determine how failure to respond to various DNA lesions affects cell viability, progeria and cancer.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Cytogenetics Core Shared Resource for FISH analysis, and the Pathology Research Core for cryosectioning of frozen tissues. The authors thank Larry M. Karnitz for advice and critical reading of the manuscript.
Footnotes
Present addresses:
Hyun-Ja Nam, MOGAM Institute for Biomedical Research, Yongin, Republic of Korea.
Myoung Shin Kim, Bio Institute, Cytogen, Inc., Seoul, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 CA181178 to Y.J.M., R01 CA190423 to S.H.K.]; American Cancer Society Research Scholar Grant [RSG-11-147-01-DMC to Y.J.M.]. Funding for open access charge: National Institutes of Health [R01 CA181178 to Y.J.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lessel D., Vaz B., Halder S., Lockhart P.J., Marinovic-Terzic I., Lopez-Mosqueda J., Philipp M., Sim J.C., Smith K.R., Oehler J. et al. . Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat. Genet. 2014; 46:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ruijs M.W., van Andel R.N., Oshima J., Madan K., Nieuwint A.W., Aalfs C.M.. Atypical progeroid syndrome: an unknown helicase gene defect. Am. J. Med. Genet. A. 2003; 116:295–299. [DOI] [PubMed] [Google Scholar]
- 3. Maskey R.S., Kim M.S., Baker D.J., Childs B., Malureanu L.A., Jeganathan K.B., Machida Y., van Deursen J.M., Machida Y.J.. Spartan deficiency causes genomic instability and progeroid phenotypes. Nat. Commun. 2014; 5:5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang D.J., Cimprich K.A.. DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 2009; 5:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sale J.E., Lehmann A.R., Woodgate R.. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012; 13:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Centore R.C., Yazinski S.A., Tse A., Zou L.. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol. Cell. 2012; 46:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Juhasz S., Balogh D., Hajdu I., Burkovics P., Villamil M.A., Zhuang Z., Haracska L.. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012; 40:10795–10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim M.S., Machida Y., Vashisht A.A., Wohlschlegel J.A., Pang Y.P., Machida Y.J.. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic Acids Res. 2013; 41:1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Machida Y., Kim M.S., Machida Y.J.. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle. 2012; 11:3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis E.J., Lachaud C., Appleton P., Macartney T.J., Nathke I., Rouse J.. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 2012; 19:1093–1100. [DOI] [PubMed] [Google Scholar]
- 11. Mosbech A., Gibbs-Seymour I., Kagias K., Thorslund T., Beli P., Povlsen L., Nielsen S.V., Smedegaard S., Sedgwick G., Lukas C. et al. . DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 2012; 19:1084–1092. [DOI] [PubMed] [Google Scholar]
- 12. Ghosal G., Leung J.W., Nair B.C., Fong K.W., Chen J.. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J. Biol. Chem. 2012; 287:34225–34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balakirev M.Y., Mullally J.E., Favier A., Assard N., Sulpice E., Lindsey D.F., Rulina A.V., Gidrol X., Wilkinson K.D.. Wss1 metalloprotease partners with Cdc48/Doa1 in processing genotoxic SUMO conjugates. Elife. 2015; 4:e06763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stingele J., Jentsch S.. DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol. 2015; 16:455–460. [DOI] [PubMed] [Google Scholar]
- 15. Barker S., Weinfeld M., Murray D.. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005; 589:111–135. [DOI] [PubMed] [Google Scholar]
- 16. Liu L.F., Desai S.D., Li T.K., Mao Y., Sun M., Sim S.P.. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 2000; 922:1–10. [DOI] [PubMed] [Google Scholar]
- 17. Furuta T., Takemura H., Liao Z.Y., Aune G.J., Redon C., Sedelnikova O.A., Pilch D.R., Rogakou E.P., Celeste A., Chen H.T. et al. . Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 2003; 278:20303–20312. [DOI] [PubMed] [Google Scholar]
- 18. Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006; 6:789–802. [DOI] [PubMed] [Google Scholar]
- 19. Nakano T., Miyamoto-Matsubara M., Shoulkamy M.I., Salem A.M., Pack S.P., Ishimi Y., Ide H.. Translocation and stability of replicative DNA helicases upon encountering DNA-protein cross-links. J. Biol. Chem. 2013; 288:4649–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stingele J., Bellelli R., Alte F., Hewitt G., Sarek G., Maslen S.L., Tsutakawa S.E., Borg A., Kjaer S., Tainer J.A. et al. . Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN. Mol. Cell. 2016; 64:688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaz B., Popovic M., Newman J.A., Fielden J., Aitkenhead H., Halder S., Singh A.N., Vendrell I., Fischer R., Torrecilla I. et al. . Metalloprotease SPRTN/DVC1 orchestrates replication-coupled DNA-Protein crosslink repair. Mol Cell. 2016; 64:704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez-Mosqueda J., Maddi K., Prgomet S., Kalayill S., Marinovic-Terzic I., Terzic J., Dikic I.. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. eLife. 2016; 5:e21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morocz M., Zsigmond E., Toth R., Enyedi M.Z., Pinter L., Haracska L.. DNA-dependent protease activity of human Spartan facilitates replication of DNA-protein crosslink-containing DNA. Nucleic Acids Res. 2017; doi:10.1093/nar/gkw1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duxin J.P., Dewar J.M., Yardimci H., Walter J.C.. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014; 159:346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morham S.G., Kluckman K.D., Voulomanos N., Smithies O.. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell. Biol. 1996; 16:6804–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang C.X., Chen A.D., Gettel N.J., Hsieh T.S.. Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev. Biol. 2000; 222:27–40. [DOI] [PubMed] [Google Scholar]
- 27. Pommier Y., Barcelo J.M., Rao V.A., Sordet O., Jobson A.G., Thibaut L., Miao Z.H., Seiler J.A., Zhang H., Marchand C. et al. . Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2006; 81:179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001; 70:369–413. [DOI] [PubMed] [Google Scholar]
- 29. Pommier Y., Huang S.Y., Gao R., Das B.B., Murai J., Marchand C.. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst). 2014; 19:114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Interthal H., Pouliot J.J., Champoux J.J.. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hahn W.C., Dessain S.K., Brooks M.W., King J.E., Elenbaas B., Sabatini D.M., DeCaprio J.A., Weinberg R.A.. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 2002; 22:2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patel A.G., Flatten K.S., Peterson K.L., Beito T.G., Schneider P.A., Perkins A.L., Harki D.A., Kaufmann S.H.. Immunodetection of human topoisomerase I-DNA covalent complexes. Nucleic Acids Res. 2016; 44:2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joshi P.M., Sutor S.L., Huntoon C.J., Karnitz L.M.. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J. Biol. Chem. 2014; 289:9247–9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaufmann S.H., Gibson W., Shaper J.H.. Characterization of the major polypeptides of the rat liver nuclear envelope. J. Biol. Chem. 1983; 258:2710–2719. [PubMed] [Google Scholar]
- 35. Kaufmann S.H., Svingen P.A., Gore S.D., Armstrong D.K., Cheng Y.C., Rowinsky E.K.. Altered formation of topotecan-stabilized topoisomerase I-DNA adducts in human leukemia cells. Blood. 1997; 89:2098–2104. [PubMed] [Google Scholar]
- 36. Desai S.D., Zhang H., Rodriguez-Bauman A., Yang J.M., Wu X., Gounder M.K., Rubin E.H., Liu L.F.. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol. Cell. Biol. 2003; 23:2341–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin C.P., Ban Y., Lyu Y.L., Liu L.F.. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double strand breaks at arrested replication forks. J. Biol. Chem. 2009; 284:28084–28092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sirbu B.M., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., Cortez D.. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Gene Dev. 2011; 25:1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker D.J., Jin F., Jeganathan K.B., van Deursen J.M.. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009; 16:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stingele J., Schwarz M.S., Bloemeke N., Wolf P.G., Jentsch S.. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014; 158:327–338. [DOI] [PubMed] [Google Scholar]
- 41. Desai S.D., Li T.K., Rodriguez-Bauman A., Rubin E.H., Liu L.F.. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001; 61:5926–5932. [PubMed] [Google Scholar]
- 42. Desai S.D., Liu L.F., Vazquez-Abad D., D’Arpa P.. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J. Biol. Chem. 1997; 272:24159–24164. [DOI] [PubMed] [Google Scholar]
- 43. Ide H., Shoulkamy M.I., Nakano T., Miyamoto-Matsubara M., Salem A.M.. Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res. 2011; 711:113–122. [DOI] [PubMed] [Google Scholar]
- 44. Jones P., Altamura S., Boueres J., Ferrigno F., Fonsi M., Giomini C., Lamartina S., Monteagudo E., Ontoria J.M., Orsale M.V. et al. . Discovery of 2-{4-[(3S)-Piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and-2 mutant tumors. J. Med. Chem. 2009; 52:7170–7185. [DOI] [PubMed] [Google Scholar]
- 45. Holland A.J., Cleveland D.W.. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009; 10:478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ricke R.M., van Deursen J.M.. Aneuploidy in health, disease, and aging. J. Cell Biol. 2013; 201:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ricke R.M., van Ree J.H., van Deursen J.M.. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008; 24:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Venkatesan S., Natarajan A.T., Hande M.P.. Chromosomal instability–mechanisms and consequences. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015; 793:176–184. [DOI] [PubMed] [Google Scholar]
- 49. Hashimoto H., Chatterjee S., Berger N.A.. Mutagenic activity of topoisomerase I inhibitors. Clin. Cancer Res. 1995; 1:369–376. [PubMed] [Google Scholar]
- 50. Andersen S.L., Sloan R.S., Petes T.D., Jinks-Robertson S.. Genome-destabilizing effects associated with top1 loss or accumulation of top1 cleavage complexes in yeast. PLoS Genet. 2015; 11:e1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Balestrieri E., Zanier R., Degrassi F.. Molecular characterisation of camptothecin-induced mutations at the hprt locus in Chinese hamster cells. Mutat. Res. 2001; 476:63–69. [DOI] [PubMed] [Google Scholar]
- 52. Toth A., Hegedus L., Juhasz S., Haracska L., Burkovics P.. The DNA-binding box of human SPARTAN contributes to the targeting of Poleta to DNA damage sites. DNA Repair (Amst). 2017; 49:33–42. [DOI] [PubMed] [Google Scholar]
- 53. Delabaere L., Orsi G.A., Sapey-Triomphe L., Horard B., Couble P., Loppin B.. The Spartan ortholog maternal haploid is required for paternal chromosome integrity in the Drosophila zygote. Curr. Biol. 2014; 24:2281–2287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.