Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that control gene expression by recognizing and hybridizing to a specific sequence generally located in the 3΄ untranslated region (UTR) of targeted mRNAs. miRNA-induced inhibition of translation occurs during the initiation step, most probably at the level of ribosome scanning. In this process, the RNA-induced silencing complex interacts both with PABP and the 43S pre-initiation complex to disrupt scanning of the 40S ribosome. However, in some specific cases, miRNAs can stimulate translation. Although the mechanism of miRNA-mediated upregulation is unknown, it appears that the poly(A) tail and the lack of availability of the TNRC6 proteins are amongst major determinants. The genomic RNA of the Hepatitis C Virus is uncapped, non-polyadenylated and harbors a peculiar internal ribosome entry site (IRES) that binds the ribosome directly to the AUG codon. Thus, we have exploited the unique properties of the HCV IRES and other related IRESes (HCV-like) to study how translation initiation can be modulated by miRNAs on these elements. Here, we report that miRNA binding to the 3΄ UTR can stimulate translation of a reporter gene given that its expression is driven by an HCV-like IRES and that it lacks a poly(A) tail at its 3΄ extremity.
INTRODUCTION
Translation initiation in eukaryotes is a highly ordered and regulated process that begins with the binding of eukaryotic initiation factors (eIFs) to the m7GTP cap structure present at the 5΄ end of Pol II transcripts (1,2). The cap moiety is bound by the eIF4F complex composed of eIF4E, eIF4G and eIF4A to promote recruitment of the small 40S ribosomal subunit (3). eIF4G is a large protein that serves as a scaffold for the complex; it contains an eIF4E binding site in its N-terminus whilst its middle domain, called MIF-4G, interacts with eIF4A and eIF3 (4,5). The N-terminus of eIF4G also interacts with the poly(A) binding protein (PABP) which is bound to the poly(A) tail at the 3΄ end of the mRNA to induce pseudo-circularization of the transcript during translation (6). The 5΄ untranslated region (5΄ UTR) of cellular transcripts often exhibits secondary structures that need to be unwound by RNA helicases in order to recruit the 43S pre-initiation complex (PIC) composed of the small 40S ribosomal subunit, eIF3 and eIF2 associated with the tRNA (7). More particularly, the ATP-dependent RNA helicase eIF4A is essential to prepare the mRNA for ribosomal binding and scanning (2,8).
Some RNA viruses and cellular transcripts can initiate protein synthesis through an alternative process that does not require prior binding of eIFs to the cap structure. Instead, these RNAs contain a particular sequence called internal ribosome entry site (IRES) (9,10). By interacting with the 40S ribosomal subunit and a limited set of eIFs, IRES elements can initiate translation independently from the 5΄ end of the mRNA. On the basis of their structure and the pool of initiation factors that they require, IRESes are classified in different types. As such, type I and II need all initiation factors except for eIF4E and the N-terminal part of eIF4G, type III IRESes do not require any members of the eIF4F complex nor PABP while type IV IRESes do not need any of the eIFs (9,10).
In addition to intrinsic features of the transcript and the use of eIFs, translation initiation in eukaryotes can also be regulated by small (18–25 nt) RNAs that are called miRNAs (11,12). Over the last few years, miRNAs have emerged as essential regulators of gene expression in many biological processes such as development, cell metabolism or defence against pathogens (11,13). miRNAs usually bind to the 3΄ UTR of target transcripts through partial base-pairing (14) and serve as guide to scaffold the RNA induced silencing complex (RISC). In mammals, the RISC is principally composed of an Argonaute protein that binds both to the miRNA and TNRC6 proteins (15–17). The association of the RISC to the target mRNA triggers, in most cases, the repression of translation followed by deadenylation and transcript degradation (18–20). This mechanism requires, at least, the 5΄ cap and the 3΄ poly(A) tail and their associated binding proteins, eIF4F and PABP (21–30). In fact, the RISC has been shown to disrupt the physical interaction between eIF4F complex and PABP (25,28–30) or to prevent eIF4F assembly (31–35). Recently, we, and others, have shown that the main mechanism that leads to translation repression targets the 40S ribosomal scanning step (36–38). In this process, the CCR4:NOT complex recruited by TNRC6 (28,39–41) could lock the helicase eIF4A on the 5΄ UTR to inhibit ribosomal scanning (12,37). This could explain why translation repression mediated by miRNAs is less effective on IRES-containing mRNAs that require a minimal length of ribosomal scanning such as the EMCV and HCV-like IRESes that initiate translation directly at the AUG codon (38,42).
It is noteworthy that, under particular conditions, miRNAs have been described to stimulate translation. This was first shown in G0 quiescent cells but also in Xenopus oocytes and in Drosophila embryo extracts (43–48). In the case of Drosophila embryo extracts (43), upregulation of translation is mediated by dAgo2 which, unlike dAgo1, is not able to bind GW182 (TNRC6 in mammals) (49). Even though the composition of drosophila RISC is not exactly similar to the mammalian one, these data are consistent with those obtained by Vasudevan et al. in mammalian cells blocked in G0. Indeed, during this phase of the cell cycle, there is a reduced expression of TNRC6 which is replaced, within the RISC, by FXR1 (Fragile-X mental Retardation 1) (46–48). Moreover, in these quiescent cells, mRNAs were shown to contain a very short poly(A) tail suggesting that upregulation of translation does not require, or requires less, PABP.
The regulation of translation by miRNAs (either repression or stimulation) therefore appears to be determined by specific intrinsic features of the mRNA (presence/absence of a cap/poly(A)), the mechanism used for ribosomal binding and scanning, together with the requirement, or not, for some components of the translational apparatus.
In agreement with such a hypothesis, the mechanism of translation stimulation of the HCV IRES by miR-122 represents an interesting case. Indeed, the genomic RNA (gRNA) of the hepatitis C virus (HCV), a member of the Flaviviridae family, is uncapped and non polyadenylated. It harbors an IRES within its 5΄ UTR which is capable of interacting directly with the small 40S ribosomal subunit and eIF3 independently of cap-recognition, PABP binding and ribosome scanning (50–57). Replication of HCV in hepatocytes is tightly regulated by miR-122 that interacts with two binding sites localized in domain I of the HCV IRES. Binding of miR-122 recruits Ago on the HCV IRES and stabilizes the gRNA by protecting it against Xrn1 5΄-3΄ exonucleases (58–60). In addition to increasing RNA stability, miR-122 enhances IRES activity by an unknown mechanism that leads to more efficient 40S ribosomal recruitment to the entry site (61–64).
These unique structural and translational features of the HCV IRES paved the way for this present study and we have made the assumption that upregulation of HCV IRES expression may also occur under different conditions than those involving miR-122 binding to the 5΄ UTR. Therefore, we have exploited the unique features of the HCV IRES as a paradigm to study the general mechanism of miRNA stimulation. As such, we have conducted our study in a standardized system such as HeLa cells (that are devoid of miR-122) with HCV IRES-driven reporter constructs that bear let-7 target sites in their 3΄ UTR. In this context, let-7 binding leads to a strong stimulation of the expression from the HCV IRES. Such an effect could also be recapitulated with a miRNA from another family (miR-451) suggesting that it is the presence of the RISC at the 3΄ UTR rather than the miRNA sequence which is required for upregulation. These effects are not due to changes in mRNA stability and are exclusively exerted at the level of translation with a magnitude of stimulation comparable to that described for miR-122-binding. These data were extended to other viral families that harbor a HCV-like IRES resulting in a similar response to miRNA binding at the 3΄ UTR. Interestingly, we also show that the presence of the poly(A) tail abolished the stimulation of HCV IRES expression upon miRNA binding, suggesting a cross-talk between the 5΄ and 3΄ extremities. Finally, by using a tethered experimental approach, we show that translation stimulation can be reproduced on an artificial IRES when placed in the context of an uncapped and non-polyadenylated setting.
MATERIALS AND METHODS
DNA plasmid constructs
The 80 nucleotides region containing the T7 promoter of the pGEM-Renilla vector was deleted by PvuII/BamHI digestion and the resulting vector—thereafter named pRenilla—was used to insert the different 5΄ UTRs (65). The human β-globin 5΄-UTR with the authentic initiation codon was obtained by hybridizing two synthetic oligodeoxyribonucleotides and cloned into the double-digested vector (EcoRV and BamHI restriction sites and T7 promoter were added with the oligonucleotides) generating the pGlobin-Renilla vector. The HCV, Avian Encephalomyelitis virus (AEV) or Seneca Valley virus (SVV) IRESes were obtained by polymerase chain reaction (PCR) using respectively the pDC HCV vector (66), the AEV plasmid (67) and SVV+55 construct (68), as templates. PCR products were digested by EcoRV and BamHI and cloned in the double-digested vector (T7 promoter and restriction sites were added by PCR) generating pHCV/AEV/SVV-Renilla luciferase vectors. To generate the pHCV miR-122 mut-Renilla plasmid, the seed region of each of the two miR-122 binding sites in the HCV IRES was mutated using directed mutagenesis. Design of the tethered construct was performed by amplification of the 4-BoxB sequence from a plasmid kindly provided by Dr Niels Gehring (University of Cologne, Germany) with specific oligonucleotides containing PvuII and BamHI restriction sites, and cloned in the 5΄ UTR of pRenilla vector using PvuII/BamHI restrictions sites, respectively. let-7 target sites derived from the lin-41 3΄ UTR of Caenorhabditis elegans (69–72) and miR-451 target sites were generated by hybridizing two synthetic oligodeoxyribonucleotides that contained the target motifs separated by the natural let-7a spacer and cloned into the 3΄UTR of the digested (HindIII) pRenilla vector (25). These target sites were amplified by PCR and sequentially cloned in the EcoRV and XbaI restriction sites to produce a construct containing 1 (for the 2 let-7 site constructs), or 2 (for the 4 let-7 sites constructs) let-7 target sites in tandem that are separated by a 27 nucleotide spacer and cloned in the 3΄UTR of the Renilla luciferase gene.
The 3΄ X region from HCV was amplified from the HCV genotype 1b and was cloned in the EcoRV site of the globin-renilla and HCV-renilla constructs either in the sense or antisense orientation. The Firefly coding plasmid (pFirefly) was constructed by cloning the Firefly luciferase coding region into the pGlobin-Renilla vector digested by BamHI and EcoRV (thus releasing the Renilla luciferase coding region) (25). For transcripts containing a poly(A) tail with different lengths, poly-adenines and poly-thymines oligonucleotides were annealed and cloned at the 3΄ end of the pRenilla vector using XbaI and EcoRI restriction enzymes.
Sequences of MIF-4G domain and Ch2 have been amplified by PCR from a HeLa cDNA library using specific primers containing EcoRI/NotI restriction sites and cloned into the lambda-N-HA and HA pCi-neo vectors (kindly provided by Dr Niels Gehring, University of Cologne) digested by EcoRI and NotI.
In vitro transcription
For in vitro transcriptions, the pRenilla plasmids were either digested by XbaI (non-polyadenylated RNAs) or EcoRI (polyadenylated RNAs) and used as templates in a transcription mixture containing either m7GpppG (to obtain capped transcripts) or ApppG cap analog (to obtain uncapped transcripts). Capped RNAs were obtained by using 2 μg of linear DNA template, 20 U of T7 RNA polymerase (Promega Co., Madison, WI, USA), 40 U of RNAsin (Promega Co, Madison, WI, USA), 10 mM of rCTP, rATP, rUTP, the rGTP concentration was reduced to 0.48 mM, 30 mM DTT in transcription buffer [40 mM Tris–HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine and 10 mM NaCl]. The transcription reaction was carried out at 37°C for 3 h and RNAs were precipitated in cold 4 M NaAc, washed with EtOH 70% and resuspended in water. The integrity of the RNAs was checked by electrophoresis on non-denaturating agarose gels and their concentration was quantified by spectrophotometry at 260 nm using Nanodrop (NanoDrop Technologies, Wilmington, Delaware, USA).
Cell cultures
HeLa and BHK cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% of fetal bovine serum and 1% of Penistrepomycin antibiotics.
DNA transfection
The pCi-neo vectors (500 ng) were transfected into HeLa cells (48 well plates) using JetPEI (Polyplus-transfecion Inc., New York, NY, USA) 24 h prior of the RNA containing 4-BoxB transfection following manufacturer indications.
Pre-miRNA and anti-miRNA transfections
Pre-miRNAs coding for miR-451 or let-7 or a negative control pre-miR (10 nmol final) (Ambion-Thermo Fisher Scientific, Waltham, MA, USA) were transfected 24 h prior to transfection of the RNA reporter gene by using Interferin (Polyplus-transfecion Inc., New York, NY, USA) following manufacturer indications.
Anti-miR against let-7 or negative control (cel-miR-67) anti-miR (miRIDIAN microRNA Hairpin Inhibitors-Dharmacon- GE Healthcare, Little Chalfont, UK) were transfected 9 h prior to transfection of the reporter RNA by using Interferin following manufacturer indications.
RNA transfections and measure of the luciferase activities
Lipofection: Renilla mRNAs (0, 25 pmol) and Firefly mRNAs (0,125 pmol) were transfected into HeLa cells (48 well plates, 80% confluence) using Trans-It transfection kit (Mirus Bio LLC, Madison, WI, USA) following manufacturer indications. One hundred fifty minutes post-transfection, cells were lysed and Renilla and Firefly luciferase activities were measured using Renilla/Firefly Assay System (Promega Co., Madison, WI, USA) on a GloMax luminometer (Promega Co., Madison, WI, USA).
Electroporation: 5 × 105 HeLa cells in suspension were electroporated twice at 1005 V, during 35 ms with the Neon Transfection System (Thermo Fisher Scientific, Waltham, MA, USA), incubated during 1 h at 37°C and split in two pools to measure luciferase activities and to extract cytoplasmic RNAs.
Determination of translational efficiency: for all experiments, translational efficiency was expressed as a percentage of Renilla luciferase activity normalized by Firefly luciferase activity. To determine the miRNA effect, translation efficiency of RNA bearing miRNA binding sites in sense orientation is expressed as the percentage of translation efficiency of control RNA with miRNA binding sites in antisense orientation (set at 100%).
Extraction of cytoplasmic RNAs
A total of 3.105 electroporated HeLa cells were lysed in 100 μl of RLNa buffer (10 mM Tris–HCl (pH 8), 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.5% NP40 and 10 U/ml of RNaseIN (Promega Co., Madison, WI, USA). The lysate was cleared to remove membrane debris. Tris-Reagent (Sigma-Aldrich, St Louis, MO, USA) was added to the lysate and cytoplasmic RNAs were extracted following the protocol provided by the manufacturer. Cytoplasmic RNAs were treated with RQ1 DNase (Promega Co., Madison, WI, USA) to avoid DNA contamination and purified with isopropanol and washed with 70% EtOH.
Quantitative RT-PCR
Reverse transcription of 200 ng of cytoplasmic RNAs was performed using qScript kit (Quanta Biosciences, Gaithersburg, MD, USA). mRNA quantification was performed by quantitative PCR using a 20 μl reaction, which was prepared with 5 μl of template cDNA (1/10 diluted first), 10 μl of Fast Start Universal SYBR Green Premix (Roche, Bâle, Suisse), 0.2 μM of each primer and subjected to amplification using a StepOne fluorescence thermocycler (Applied Biosystems, Foster City, CA, USA) under the following conditions: 10 min at 94°C for initial denaturation, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s and elongation at 72°C for 30 s. This program was followed by a melting curve analysis in order to verify the specificity of the PCR product. Renilla luciferase (Renilla forward AGGTGAAGTTCGTCGTCCAACATTATC/ Renilla reverse GAAACTTCTTGGCACCTTCAACAATAGC) was amplified in parallel with the endogenous housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (GAPDH forward (Human) TCCACCACCCTGTTGCTGTAG/ GAPDH reverse (Human) ACCCACTCCTCCACCTTTGA). The relative copy numbers of Renilla cDNAs were compared to GAPDH using x–ΔCt (where x corresponds to the experimentally calculated amplification efficiency of each primer couple).
RESULTS
HCV IRES translation is stimulated by let-7 in HeLa cells
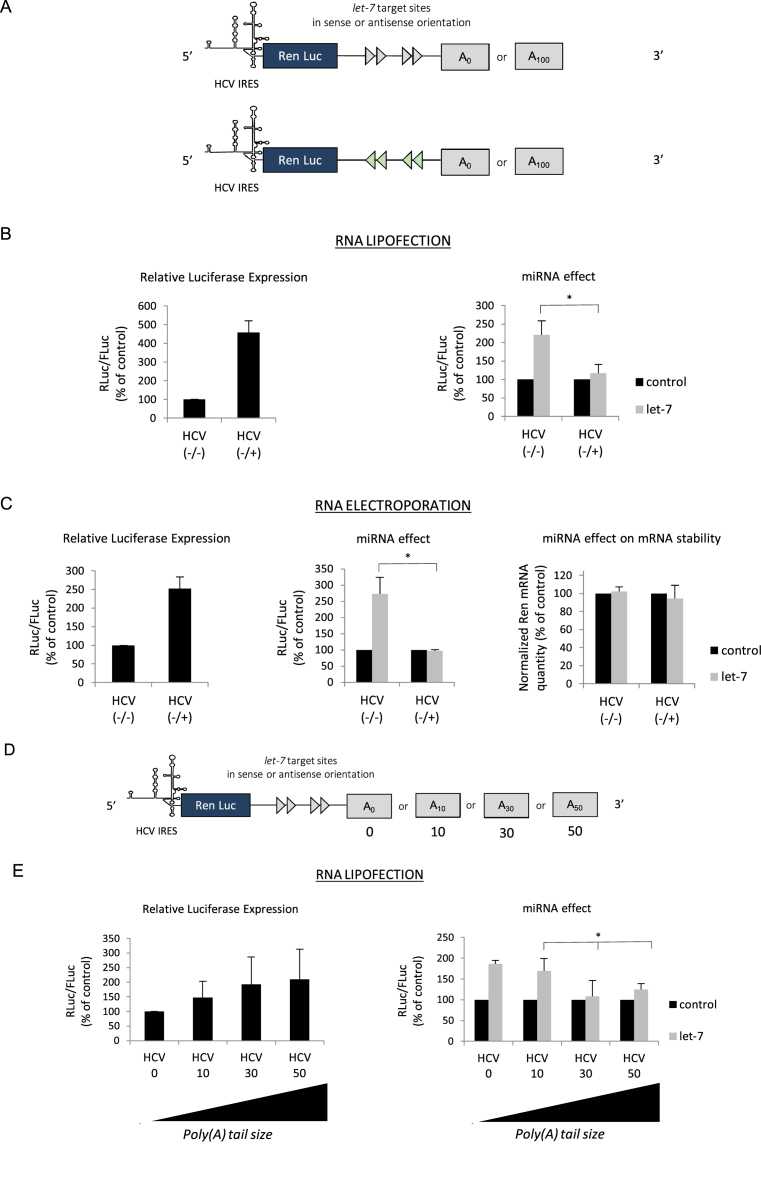
The initial aim of this work was to measure the effect of binding a miRNA on the 3΄ UTR of a reporter construct whose translation is driven by the HCV IRES. For this, we have used an experimental strategy based on the transfection of reporter mRNAs that contain the Renilla luciferase coding region followed by a 3΄ UTR in which 4 let-7 target sites derived from the Caenorhabditis elegans lin-41 gene (69–72) were inserted either in the sense, or antisense orientation (Figure 1A). Expression of luciferase is driven by 5΄ UTRs that were either derived from a cap-dependent cellular transcript (β-globin) or from the HCV IRES. Rationale for inserting let-7 sites in the 3΄ UTR was dictated by the fact that this miRNA is the most commonly studied (69–73) and is expressed in virtually all cell types (74,75). As it has been shown that the effects of miRNAs can greatly vary depending on the promoter and the transfection method used for the reporter gene (70,76), we have deliberately chosen to use mRNA transfection in cultured cells. This presents several advantages: it is promoter independent, it allows expression of capped, uncapped, polyadenylated or non-polyadenylated transcripts in all possible combinations and allow to measure gene expression in a short time frame ( less than 3 h post-transfection). In addition, for most of the experiments, RNAs were transfected both by lipofection and electroporation. As such, it should be noted that, contrary to lipofection which leads to the storage of large amounts of transfected transcripts in intracellular vesicles that are not accessible to the translational machinery, electroporation delivers the RNAs directly to the cytoplasm and thus allows the precise quantification of Renilla reporter RNAs by RT-qPCR. Protein synthesis is then quantified by measuring Renilla and Firefly luciferase activities, and both translation efficiency and miRNA effects are plotted on individual bar graphs (see Materials and Methods).
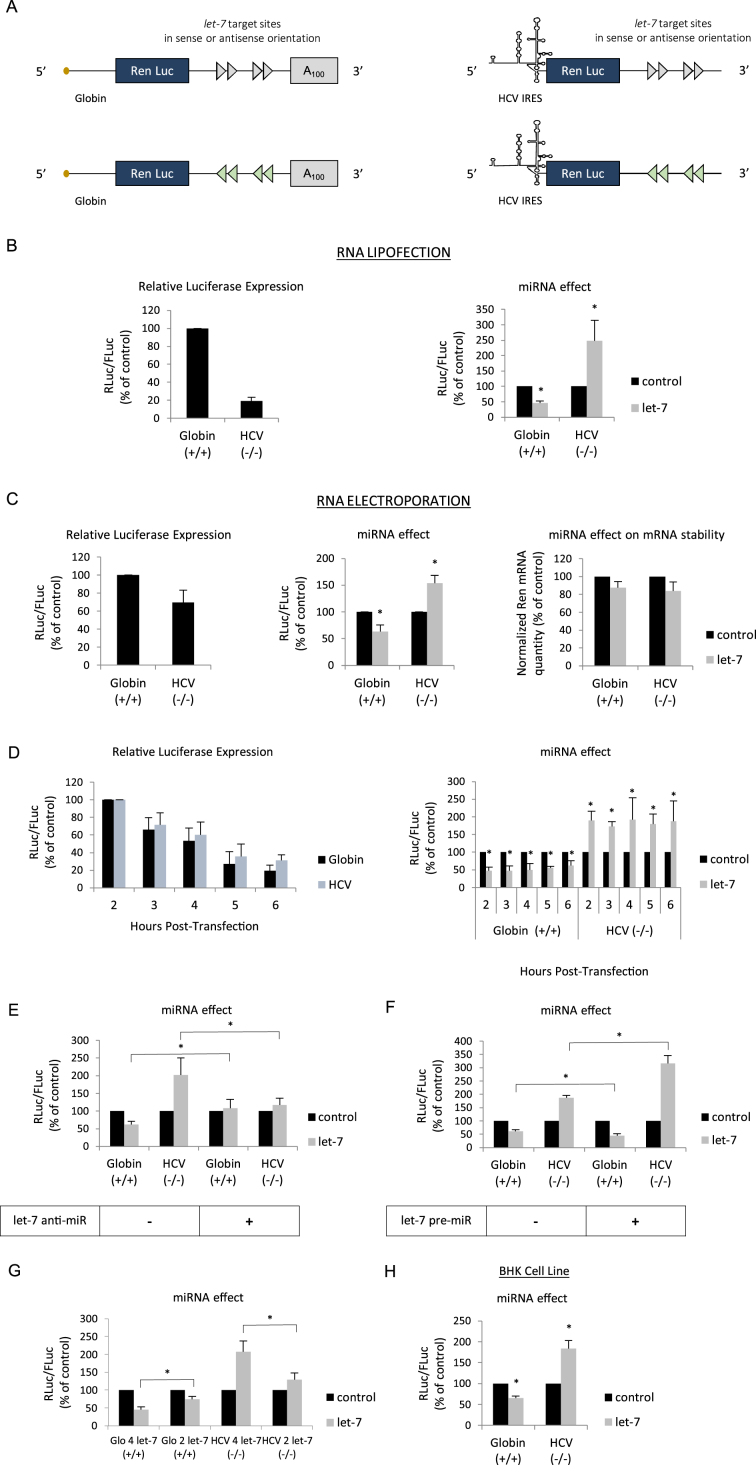
Figure 1.
(A) Schematic representation of the reporter microRNAs (mRNAs) used in this study. The mRNAs were obtained by in vitro transcription and harbor the Renilla luciferase coding region which is driven by either the globin 5΄ UTR or the HCV IRES as indicated. The 3΄ UTR contains 4 let-7 target sites in a sense (blue arrows) or antisense orientation (red arrows) and is terminated, or not, by a stretch of 100 adenine residues (A100). (B) Capped/polyadenylated (+/+) and uncapped/non polyadenylated (-/-) Renilla luciferase mRNAs driven by the β-globin 5΄ UTR and the HCV IRES respectively were transfected together with a control Firefly luciferase mRNA in HeLa cells by lipofection (see Materials and Methods). Luciferase assays were performed 150 min post transfection. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation (no miRNA effect). Right panel shows the miRNA effect which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). (C) The same constructs as above were electroporated together with a control Firefly luciferase mRNA in HeLa cells and luciferase activities were measured after 1 h. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Middle panel shows the miRNA effect which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Right panel shows the quantification of cytoplasmic Renilla luciferase mRNAs by quantitative RT-PCR. Results are normalized to an internal endogenous mRNA (GAPDH) and are expressed as a percentage of the control (construct containing the let-7 sites in the antisense orientation (black bars), set to 100%). (D) Hepatitis C virus (HCV) (black bars) and β-globin 5΄ UTRs (blue bars) driven mRNA constructs were lipofected in HeLa cells and luciferase activities were determined at 2, 3, 4, 5 and 6 h post transfection. Left panel shows the relative luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Right panel shows the miRNA effect that is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). (E) mRNA constructs described above were lipofected with an anti-miR control or an anti-miR specific to let-7 before determination of luciferase activities. The graph shows the miRNA effect that is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). (F) mRNA constructs described above were lipofected with a pre-miR control or a let-7 pre-miR one day before determination of luciferase activities. The graph shows the miRNA effect which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). (G) Capped/polyadenylated (+/+) driven by the β-globin 5΄ UTR and containing either 2 or 4 let-7 sites in their 3΄ UTRs were electroporated with uncapped/non polyadenylated (-/-) Renilla luciferase mRNAs driven by HCV IRES and containing either 2 or 4 let-7 sites in their 3΄ UTRs. A control Firefly luciferase mRNA was also electroporated in HeLa cells and luciferase activities were measured after 1 h. Data were plotted and normalized as a percentage of the control (constructs with the 2 or 4 let-7 sites in the antisense orientation) that was set to 100%. (E) mRNA constructs described above were lipofected in BHK cell line in a similar experimental design than described in (B). The graph shows the miRNA effect which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Error bars correspond to SD obtained from, at least, three different experiments. * corresponds to P-value < 0.05 obtained with a non-parametric Wilcoxon–Mann–Withney test.
HeLa cells were used to transfect the capped and polydenylated globin mRNA together with the HCV IRES containing transcript both in its uncapped and non-polyadenylated form (as depicted in Figure 1A). Luciferase activity was analyzed 150 min post-transfection and expressed as a ratio of Renilla over the Firefly activity of a co-transfected control reporter gene (see Materials and Methods). In the absence of any miRNA binding sites, translation from the globin transcript was more efficient than that from the HCV IRES one (Figure 1B, left panel). However, when both transcripts harbored the let-7 binding sites, expression from the globin 5΄ UTR was markedly decreased whereas HCV IRES-driven initiation was strongly enhanced (Figure 1B, right panel). Essentially similar results were obtained by using electroporation to transfect the RNAs into the cells although the magnitude of repression and stimulation was less pronounced (Figure 1C). Nevertheless, it clearly appears that cap-dependent globin translation was diminished whereas HCV IRES-driven initiation was enhanced by the presence of the let-7 sites (Figure 1C, middle panel). Such an effect could not be attributed to differences in RNA degradation as both transcripts remained stable over the period of incubation as shown by RT-qPCR (Figure 1C, right panel). In order to verify that this effect was specific to the HCV IRES, we have repeated essentially the same experiment using a class II IRES (e.g. EMCV) and we found no evidence of translational stimulation (Supplementary Figure S1A). In addition, changing the time of incubation from 2 to 6 h post-transfection resulted in a sharp decrease of translational efficiency for both mRNAs (Figure 1D, left panel) but did not modify the global effect of let-7 on either the repression of globin translation or stimulation of the HCV IRES (Figure 1D, right panel). Furthermore, the addition of an anti-miR, which specifically blocked let-7, abolished the inhibition of globin translation and dramatically reduced the stimulatory effect on HCV IRES expression (Figure 1E). Conversely, transfection of a pre-miRNA (pre-miR) coding for let-7 resulted in a more pronounced repression of globin translation together with further enhancement of expression driven by the HCV IRES (Figure 1F). We then decided to see whether the number of let-7 sites on the target mRNA can influence the magnitude of translational stimulation. For this, we have transfected reporter constructs containing the globin 5΄ UTR or the HCV IRES and which contain either 2, or 4 let-7 sites in their 3΄ UTRs. The data are presented in Figure 1G and show that both the level of inhibition and stimulation was correlated to the number of let-7 sites present. Finally, the two reporter RNAs were transfected in BHK cells where the effects of let-7 were essentially similar to those observed in HeLa cells indicating that these effects were not restricted to the HeLa cell lineage (Figure 1H). Taken together, these data indicate that binding of let-7 to the 3΄ UTR can stimulate translation driven by the HCV IRES.
HCV IRES translation is also stimulated by miR-451
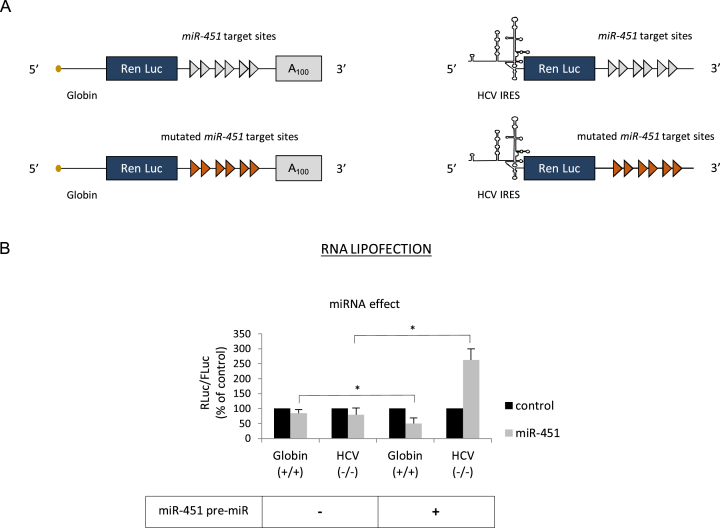
The next step was to investigate whether HCV IRES stimulation can be reproduced with a miRNA that belongs to a different family than let-7. For this, we have chosen to test the effect of miR-451 that was first identified as a major actor in erythropoiesis differentiation (77,78). Due to its specificity in the erythroid lineage, miR-451 is virtually absent from HeLa cells as previously described and verified by RT-qPCR (data not shown) (75). Furthermore, previous work from our laboratory has shown that miR-451 recapitulates all aspects of the miRNA response in vitro (38,65). Thus, we have replaced the let-7 sites on the 3΄ UTR of the luciferase reporter constructs with six target sites for miR-451, or the mutated sites that were not able to bind miR-451 anymore (Figure 2A); these sites were validated to be functional in our previous studies (25,38). A pre-miRNA encoding miR-451 was co-transfected into HeLa cells together with two reporter genes containing the globin 5΄ UTR or the HCV IRES in a similar experimental setting than described in Figure 1. Processing and expression of this pre-miRNA was checked in HeLa cells (data not shown). This resulted in the inhibition of translation from the globin 5΄ UTR in the presence of miR-451 whereas, at the same time, activity of the HCV IRES was enhanced (Figure 2B). Interestingly, no effect could be observed on any of the RNAs that were co-expressed with an unspecific pre-miR control (not specific of the miR-451 sites) indicating that variation in gene expression was the consequence of the binding of miR-451 on its targets. Above all, this result indicates that HCV IRES stimulation is not particular to let-7 and can be reproduced by other miRNAs, even if they are expressed exogenously in a form of a pre-miRNA.
Figure 2.
(A) Schematic cartoon of the transcripts used. The capped and polyadenylated mRNA is driven by globin 5΄ UTR and the uncapped and non-polyadenylated mRNA is driven by the HCV IRES. For both constructs, the 3΄ UTR contains 6 miR-451 target sites (blue arrows) or six miR-451 sites that were mutated in a way that prevents the binding of miR-451 (red arrows). (B) Capped/polyadenylated and uncapped/non polyadenylated Renilla luciferase mRNAs driven by the β-globin and the HCV IRES, respectively, were lipofected together with a control Firefly luciferase mRNA in HeLa cells that were previously transfected with a pre-miR control or a pre-miR expressing miR-451 (see Materials and Methods). Luciferase assays were performed after 150 min. The graph shows the miRNA effect that is given by the difference of expression from constructs that bear the mutated miR-451 target sites in the antisense orientation (black bars, control) and in the sense orientation (grey bars, miR-451). Results are expressed as a percentage of the control (construct bearing the mutated miR-451 sites (black bars), set to 100%). Error bars correspond to SD obtained from, at least, three different experiments. * corresponds to P-value < 0.05 obtained with a non-parametric Wilcoxon–Mann–Withney test.
Stimulation of the HCV IRES is independent from miR-122
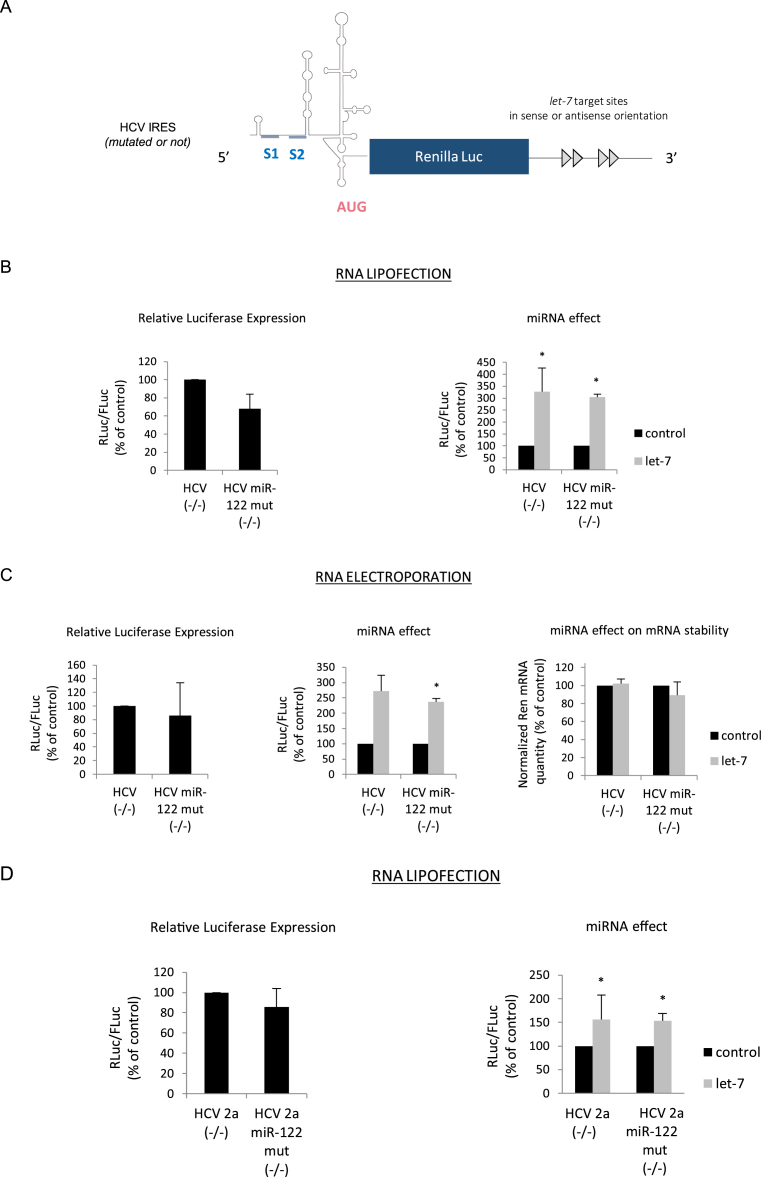
It is well established that HCV replication is stimulated by the binding of miRNA-122 which is specifically expressed in the liver (74,79,80). This miRNA principally binds two sites (S1 and S2) that are located within domain I of the HCV IRES (Figure 3A) (80,81). Although the effects of miR-122 are pleiotropic on viral replication, its binding to S1 and S2 was shown to stimulate HCV IRES activity (61,62,64). Given these facts, it cannot be formerly ruled out that the presence of these miR-122 binding sites on the HCV IRES could play a role in the stimulation of translation of the HCV IRES observed in the context of our study. Although it has been previously reported that miR-122 is virtually absent from HeLa cells (75), it still may be possible that the presence of the miR-122 target sites could play a role in translation stimulation. Thus, to directly address this issue, the two seeds S1 and S2 were mutated (Figure 3A) and the resulting transcripts were translated together with a control RNA harboring the wild-type HCV IRES. These RNAs were introduced into HeLa cells by lipofection (Figure 3B) or electroporation (Figure 3C). Mutations of the S1 and S2 sites did not radically impact translational efficiency in the absence of any let-7 functional sites (Figure 3B and C, left panel), which is consistent with the fact that domain I is not critical for IRES activity (55). However, the presence of the let-7 target sites within the 3΄ UTR of the reporter construct enhanced HCV translation whether the S1 and S2 seeds were mutated or not; this was observed when the RNAs were lipofected and electroporated (Figure 3B and C, right and middle panel, respectively). Interestingly, quantification of the transcripts by RT-qPCR revealed that RNA stability was not affected by the binding of the let-7 miRNAs neither in the context of the wild type (WT) nor within the miR-122 mutated transcripts (Figure 3C, right panel). In order to further confirm these results, we have also used an HCV IRES derived from the HCV 2a genotype that was cloned within the same luciferase reporter backbone. Once again, mutations of the S1 and S2 sites did not affect translational efficiency (Figure 3D, left panel) and binding of let-7 to its target sites resulted in a similar stimulation of expression of both the WT and the S1/S2 mutated IRESes (Figure 3D, right panel). Taken together, these data indicate that let-7 activates the HCV IRES in HeLa cells by a mechanism that is independent from miR-122.
Figure 3.
(A) Schematic cartoon of the uncapped and non-polyadenylated mRNA reporter genes whose translation is driven by the HCV IRES. The positions of the 2 miR-122 binding sites are indicated on the figure and denoted S1 and S2. All constructs contain either 4 let-7 binding sites in the sense or antisense orientation within the 3΄ UTR. (B) Uncapped and non polyadenylated luciferase mRNAs driven by the HCV IRES or the HCV IRES in which S1 and S2 were mutated (miR-122 mut) were transfected together with a control Firefly luciferase mRNA in HeLa cells by lipofection. Luciferase assays were performed after 150 min. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Right panel shows the miRNA effect that is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). (C) The same constructs as above were electroporated in HeLa cells for 1 h before reading of the luciferase activities. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation, set to 100%). Middle panel shows the miRNA effect which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Right panel shows the quantification of cytoplasmic Renilla luciferase transcripts by quantitative RT-PCR. Results are normalized to an internal mRNA (GAPDH) and mRNA concentration is expressed as a percentage of the control (construct containing the let-7 sites in the antisense orientation (black bars), set to 100%). (D) Similar experimental procedure than in (B) except that the HCV IRES was derived from HCV genotype 2a. Error bars correspond to SD obtained from, at least, three different experiments. * corresponds to P-value < 0.05 obtained with a non-parametric Wilcoxon–Mann–Withney test.
let-7 stimulates HCV-like IRESes translation initiation
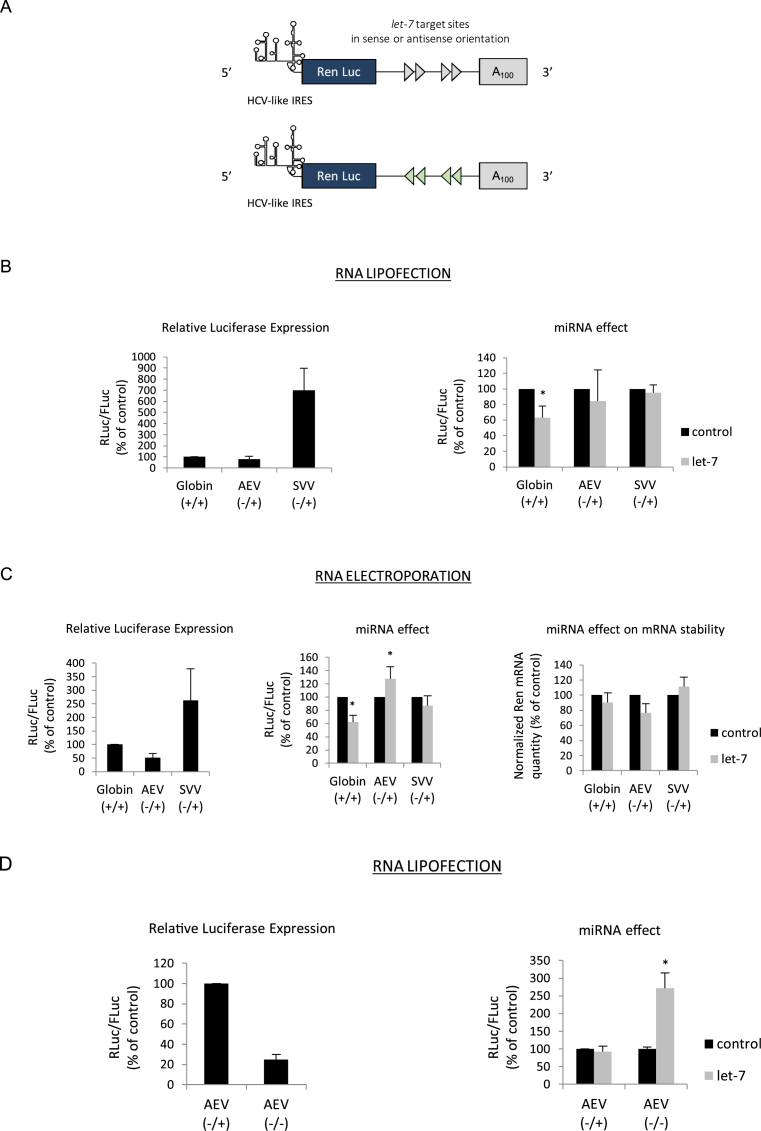
The HCV IRES is the prototype member of a family of ‘HCV-like IRESes’ which are defined by the use of a minimal set of initiation factors and the presence of a pseudoknot at the initiation site that allows ribosome recruitment with no need for ribosomal scanning (50–57). Members of the HCV-like IRESes are not only found within Flaviviridae but also in other unrelated viral families such as Picornaviruses. As such, the SVV (68) and AEV (67), both members of the picornaviral family contain a HCV-like type of IRES within their 5΄ UTRs. However, the genomic RNAs of these viruses are uncapped but harbor a poly(A) tail at their 3΄ end (67,68). Both of theses IRESes, derived from AEV and SVV, were cloned in the luciferase reporter backbone with, or without, the let-7 target sites at their 3΄ UTRs (Figure 4A). The resulting in vitro transcribed mRNAs were introduced in HeLa cells by lipofection (Figure 4B) or electroporation (Figure 4C) together with RNAs containing the globin 5΄ UTR as control. It is noteworthy that each construct was expressed as it is found in its natural context regarding the presence of a cap and/or a poly (A) tail: e.g. capped and polyadenylated for globin (+/+) and uncapped/polyadenylated (-/+) for both AEV and SVV. Translational efficiency driven by the SVV IRES was very high as it had been previously described (Figure 4B and C, left panel) (38). However, and unlike the situation with HCV (see data obtained previously), expression from both picornaviral IRESes was not stimulated in the context of the RNA harboring the let-7 sites (Figure 4B and C, right and middle panel, respectively). Quantitative RT-PCR analysis after electroporation revealed no significant difference in the RNA stability of the SVV and AEV containing constructs (Figure 4C, right panel). This lack of stimulation of the SVV and AEV IRESes was rather puzzling as we would have expected to observe a similar effect of the let-7 miRNA due to functional homologies with the HCV IRES. However, it is noteworthy that the AEV/SVV IRES containing constructs harbored a poly(A) tail in the context of our experiment whereas constructs containing the HCV IRES did not. Thus, we wondered whether the presence of this poly(A) tail at the 3΄ end could have any effect on translational stimulation. To address this issue, translation from both the uncapped/polyadenylated (-/+) and uncapped/non polyadenylated (-/-) forms of the AEV-containing RNAs was assessed (Figure 4D). It is interesting to note that the removal of the poly(A) tail resulted in a strong decrease of translational efficiency driven by the AEV IRES (Figure 4D, left panel). More importantly, we also observed a switch from non- to highly-stimulated translation of transcripts harboring the let-7 target sites (Figure 4D, right panel). It should be noted that we have also looked at the effect of removing the poly(A) tail on a capped globin RNA and we found that it did not result in translation stimulation (Supplementary Figure S1B). Therefore, these results indicate that HCV-like IRESes can also be stimulated by the binding of miRNAs but only in the absence of a 3΄ poly(A) tail.
Figure 4.
(A) Schematic cartoon of the mRNA reporters whose translation is driven by the AEV or SVV IRESes and which contain four let-7 sites in either sense (blue arrows) or antisense (red arrows) orientation. Please note that the constructs harbor a 100 nucleotides poly(A) tail after in vitro transcription. (B) Capped/polyadenylated (+/+) and uncapped/polyadenylated (-/+) luciferase mRNAs driven by the β-globin (Globin +/+), the AEV (AEV -/+) or the SVV (SVV -/+) IRESes respectively were transfected together with a control Firefly luciferase mRNA in HeLa cells by lipofection (see Materials and Methods). Luciferase assays were performed after 150 min. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Right panel shows the miRNA effect that is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). (C) The same constructs as above were electroporated in HeLa cells for 1 h before reading of the luciferase activities. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Middle panel shows the miRNA effect given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). Right panel shows the quantification of cytoplasmic Renilla luciferase transcripts by quantitative RT-PCR. Results are normalized to an internal mRNA (GAPDH) and are expressed as a percentage of the control (construct containing the let-7 sites in the antisense orientation, (black bars)). (D) Uncapped/polyadenylated (AEV -/+) and uncapped/non-polyadenylated (AEV-/-) constructs driven by the AEV IRES were lipofected together with a Firefly Luciferase mRNA in HeLa cells (see Materials and Methods). Luciferase assays were performed after 150 min. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Right panel shows the miRNA effect which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). Error bars correspond to SD obtained from, at least, three different experiments. * corresponds to P-value < 0.05 obtained with a non-parametric Wilcoxon–Mann–Withney test.
Stimulation of HCV IRES translation by let-7 is abolished by the presence of a poly(A) tail
Data obtained above pointed out to a specific role of the poly(A) tail in IRES-driven translational stimulation by miRNAs. Although the genomic RNA of HCV does not naturally harbor a poly(A) tail at its 3΄ end, it has been previously shown that the artificial addition of a poly(A) tail stimulates translation driven by the HCV IRES suggesting that it can exploit this feature to promote translation by a, yet, unknown mechanism (55,82). This prompted us to investigate further the effects of poly(A) tail addition on HCV IRES translation (Figure 5A). The polyadenylated (-/+) and non-polyadenylated (-/-) constructs were introduced in HeLa cells by lipofection (Figure 5B) or electroporation (Figure 5C). This first confirmed that addition of the poly(A) tail enhanced significantly translation driven by the HCV IRES in the context of our experimental setting (Figure 5B and C, left panel, compare HCV -/+ with HCV -/-). However, polyadenylated RNAs were not stimulated by the binding of let-7 (Figure 5B, left panel and Figure 5C, middle panel), a situation reminiscent to that observed for the AEV IRES (Figure 4). Once again, differences between non-polyadenylated and polyadenylated mRNAs translation cannot be attributed to variations in RNA stability (Figure 5C, right panel). This confirms that the poly(A) tail interferes with the stimulation of the HCV IRES by miRNAs. To investigate this further, we have added different lengths (0, 10, 30 or 50) of poly(A) residues to the 3΄ end of the HCV IRES-driven luciferase reporter gene (with or without the let-7 sites) (Figure 5D). This resulted in a positive correlation between the length of adenine residues added and translation efficiency (Figure 5E, left panel). However, when we looked at the consequences on miRNA stimulation, there was an opposite trend with a negative correlation between the addition of adenine residues and the potential of stimulation by let-7; this stimulatory effect was virtually abolished by appending 30, or more, adenosines (Figure 5E, right panel).
Figure 5.
(A) Schematic cartoon of the uncapped mRNA Renilla reporter genes whose translation is driven by the HCV IRES. Please note that the construct can be transcribed with, or without a poly(A) tail of 100 adenosines. (B) Uncapped/non polyadenylated (HCV -/-) and uncapped/polyadenylated (HCV -/+) Renilla luciferase mRNAs driven by the HCV IRES were transfected together with a Firefly Luciferase mRNA in HeLa cells by lipofection. Luciferase assays were performed after 150 min. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Right panel shows the miRNA effect which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). (C) The same constructs as above were electroporated in HeLa cells for 1 h before reading of the luciferase activities. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Left panel shows the translational activity for the constructs that bear the let-7 target sites in the antisense orientation. Middle panel shows the miRNA effect that is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation, set to 100%). Right panel shows the quantification of cytoplasmic Renilla luciferase transcripts by quantitative RT-PCR. Results are normalized to an internal mRNA (GAPDH) and are expressed as a percentage of the control (construct containing the let-7 sites in the antisense orientation (black bars), set to 100%). (D) Schematic cartoon of the uncapped mRNA reporter gene whose translation is driven by the HCV IRES. Please note that the construct can be transcribed without (HCV 0) or with a stretch of 10 (HCV 10), 30 (HCV 30) or 50 (HCV 50) adenosine residues as indicated. (E) The luciferase constructs driven by the HCV IRES depicted in (A) were lipofected together with a Firefly Luciferase mRNA in HeLa cells. Luciferase assays were performed after 150 min. Left panel shows the relative Luciferase expression level (ratio RLuc/FLuc) for the constructs that bear the let-7 target sites in the antisense orientation. Right panel shows the miRNA effect, which is given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). Error bars correspond to SD obtained from, at least, three different experiments. * corresponds to P-value < 0.05 obtained with a non-parametric Wilcoxon–Mann–Withney test.
Translation driven from a minimal artificial IRES is stimulated by let-7 miRNA
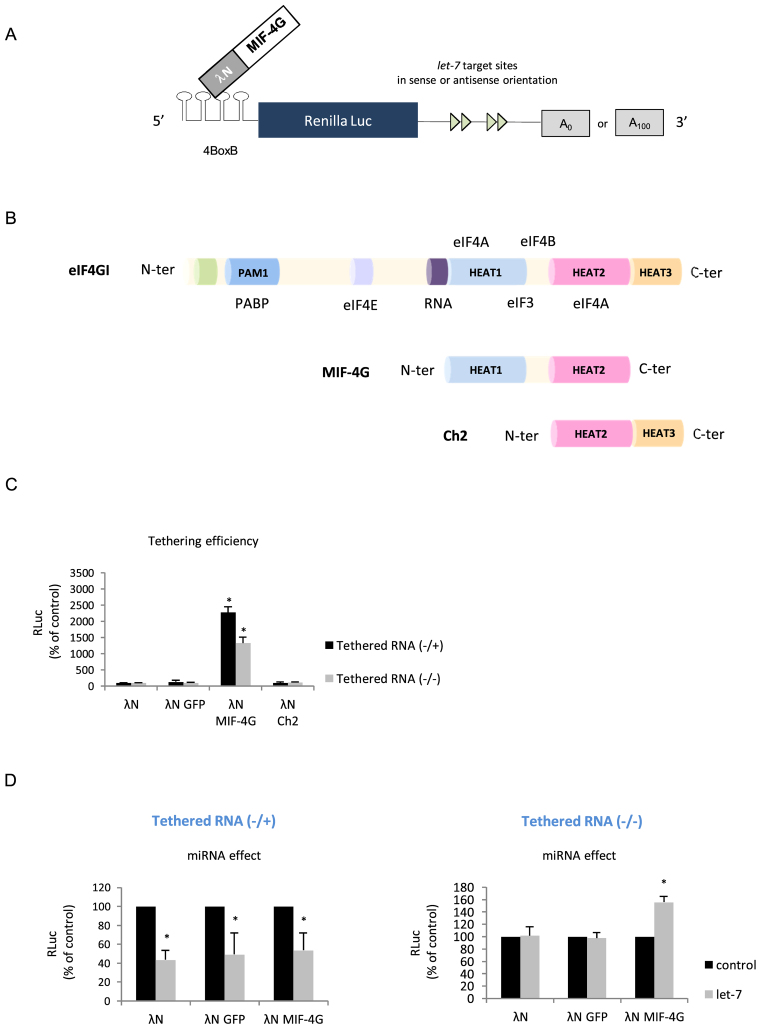
Data obtained so far suggest that translational stimulation by miRNA requires, at least, two essential features that are: (i) the presence of a HCV-like IRES in the 5΄ UTR and (ii) the absence of a poly(A) tail at the 3΄ end of the reporter mRNA. HCV-like IRESes exhibit peculiar features that provide them with a unique mechanism of ribosomal recruitment (9,67,68). As such, they only need a limited set of initiation factors to bind the ribosome to the AUG with no requirement for ribosomal scanning. Interestingly, recent data have shown that the mechanism of miRNA translation repression takes place during the ribosomal scanning step through the targeting of some initiation factors such as eIF4E, eIF4A and PABP (33–38,83). Thus, it suggests that the composition of the pre-initiation complex and the interplay between initiation factors, the cap and the poly(A) tail appear to be essentials for the miRNA response. In the light of these data, it can be postulated that translation stimulation of HCV-like IRESes by miRNAs may reflect the fact that these IRESes are able to recruit a minimal pre-initiation complex and to bind it directly to the AUG initiation site with no involvement of ribosomal scanning. Thus, our next goal was to investigate whether translation stimulation could also occur on an artificial system devoid of any of the viral sequences but which recapitulates features of the HCV-like IRESes. In order to address this question, we have decided to undertake a tethering approach (84) in which the middle domain of eIF4G (MIF-4G) is used to recruit a minimal pre-initiation complex at the 5΄ end of a reporter gene (Figure 6A). The MIF-4G domain contains the binding site for eIF3 and was previously shown to be the minimal fragment sufficient to promote binding of the ribosome onto the mRNA (3,5,4,85) (Figure 6B). As such, it mimics the role of an artificial IRES; this approach has been successfully used previously to address the role of initiation factors in ribosome binding (5). By using the lambdaN protein, we have tethered the MIF domain of eIF4G to the 5΄ UTR of a construct containing the 4-BoxB recognized by the lambdaN protein (84). This construct is uncapped at its 5΄ extremity, contains the Renilla luciferase as reporter gene followed by four let-7 sites in a sense, or an antisense, orientation and can be polyadenylated or not (Figure 6A). We first checked that tethering of the MIF domain of eIF4G could efficiently drive translation of this reporter mRNA. For this, we have also tethered lambda N alone, lambda N fused to the GFP protein (lambda N-GFP), lambda N fused to the Ch2 fragment of eIF4G or lambda N fused to MIF-4G (lambda N-MIF-4G) (Figure 6B). Data are presented in Figure 6C and show that no Renilla luciferase activity was detected unless the MIF domain of eIF4G was co-expressed. Polyadenylation of the transcript enhanced translation but was not absolutely required for activity (compare 4-BoxB -/+ with 4-BoxB -/-) (Figure 6C). Having shown that translation can be driven by the MIF-4G domain alone, we have then looked at translation of the reporter mRNAs that contain, or not, the let-7 binding sites in their 3΄ UTR (Figure 6D). Interestingly, this revealed that binding of let-7 to its target sites can stimulate translation driven by lambda N-MIF-4G only if the transcript lacked the poly(A) tail (Figure 6D, right panel, polyA-). In the presence of the poly(A) tail, we observed a rather strong repression of translation (Figure 6D, left panel, polyA+). It is noteworthy that transfection of MIF-4G alone (not appended to lambda N) could not stimulate translation of the reporter RNA (data not shown). Taken together, these data show that miRNA can stimulate translation of an unpolyadenylated reporter gene whose translation is driven by an artifical IRES element.
Figure 6.
(A) Schematic diagram of the experimental method for the tethering approach. An RNA segment composed of 4-BoxB was fused at the uncapped 5΄ end of the Renilla luciferase reporter gene containing 4 let-7 sites in the sense or antisense orientation in its 3΄ UTR. The mRNA can be produced with or without a poly(A) tail. A fusion protein made with the lambda-N fragment fused with the MIF or the Ch2 domains of eIF4G is co-expressed with the Renilla luciferase mRNA. (B) Schematic cartoon of the initiation factor eIF4G and its domains of interaction with PABP, eIF4E, ARN, eIF4A, eIF3 as indicated. The MIF domain of eIF4G and the Ch-2 fragment is also represented below. (C) HeLa cells were transfected with cDNA plasmids coding for lambda N, lambda N-GFP, lambda N-Ch2 or lambda N-MIF-4G proteins by lipofection 24 h prior to transfection of the uncapped 4-BoxB Renilla luciferase mRNA either in its polyadenylated (4-BoxB-Renilla -/+) or non-polyadenylated (4-BoxB-Renilla -/-) form. Luciferase activities were measured 3 h post transfection to determine translational activity for the polyadenylated and non-polyadenylated constructs that bear the let-7 target sites in the antisense orientation as indicated on the figure. (D) Panels summarize the miRNA effects for both tethered RNA constructs (left panel: 4-BoxB -/+ and right panel: 4-BoxB -/-) which are given by the difference of expression from constructs containing the let-7 sites in the antisense orientation (black bars) with those in the sense orientation (grey bars). Results are expressed as a percentage of the control (construct bearing the let-7 sites in the antisense orientation (black bars), set to 100%). Error bars correspond to SD obtained from, at least, three different experiments. * corresponds to P-value < 0.05 obtained with a non-parametric Wilcoxon–Mann–Withney test.
DISCUSSION
Many reports have shed light on the importance of the intrinsic features of the mRNA such as the 5΄ cap and the 3΄ poly(A) tail and their associated binding proteins (eIF4F complex and PABP) as being critical for miRNA induced translational repression to take place (21–30). Recently, using an in vitro system, we have demonstrated that miRNAs target the 43S ribosomal scanning along the 5΄ UTR (38). In addition to their primary role in repressing gene expression, miRNAs have also been involved in posttranscriptional upregulation under certain physiological conditions such as G0 cell quiescence, developing germ cells, cell cycle arrest upon serum deprivation and contact inhibition (44–48). Several lines of evidence suggest that translation stimulation may rely on the composition of the RISC complex together with some intrinsic features of the mRNA such as the presence/absence of the poly(A) tail. In their works, Vasudevan et al. pointed out that the absence of a poly(A) tail on the targeted transcript is one of the distinctive features that leads to RISC induced translation upregulation (44–48). Indeed, the lack of poly(A) tail results in the absence of incorporation of the TNRC6 protein into the RISC and, consequently, the stimulation of translation.
Interestingly, HCV IRES translation initiation does not require cap-recognition and ribosome scanning which are features targeted by miRNA to repress translation initiation and the HCV gRNA does not harbor a poly(A) tail at its 3΄ end (55). Therefore, HCV translation initiation exhibits unique characteristics that are compatible with a possible stimulation by miRNA binding on the 3΄ UTR of mRNA. This set the rationale of this study that consisted of using the HCV IRES as a paradigm to investigate the molecular mechanism of translation upregulation by miRNAs. As such, we have deliberately chosen to conduct this work in non-permissive cell lines and with a spectrum of artificial constructs that only possess the IRES element from HCV and HCV-like viral genomes.
We have first investigated variations in gene expression of Renilla-based reporter constructs driven by the HCV IRES (genotype 1b) and harboring 4 let-7 target sites in their 3΄ UTR and we were able to show that this resulted in an important stimulation of gene expression, due to an upregulation of HCV IRES translation without any significant variations in mRNA stability (Figure 1). Similar results were obtained in the BHK cell line (Figure 1). Moreover, upregulation of HCV IRES translation was also evidenced by replacing let-7 with miR-451 target sites followed by exogenous addition of the latter in HeLa cells (Figure 2). This shows that HCV IRES translation stimulation could be triggered by the binding of any miRNAs and further suggests an effect mediated by the RISC complex rather than by the binding of the miRNA per se.
Upregulation of HCV IRES dependent expression has been previously described to occur following the binding of miR-122 on two sites (S1 and S2) localized within domain I of the HCV IRES (61–64). Upon binding to its target sites, miR-122 protects the gRNA from XrnI 5΄-3΄ degradation and promotes IRES-dependent translation (58–60). However, data reported herein describe a distinct mechanism of translation stimulation as it occurs with different miRNAs that target the 3΄ UTR of a reporter gene whose translation is driven by the HCV IRES (Figures 1 and 2). In addition, the mechanism occurs in HeLa cells that are devoid of miR-122 (data not shown) and with an HCV IRES mutated in the two binding sites (Figure 3). It is noteworthy that such an effect is not dependent on the nature of the 3΄ UTR as its replacement by the authentic 3΄ X region of the HCV genome did not change the stimulatory effect of adding let-7 sites given that translation was indeed driven by the HCV IRES (see Supplementary Figure S2).
By using the AEV IRES that belongs to the picornaviral family but harbors a ‘HCV-like IRES’ (67), we could show that upregulation of translation was dependent on the absence of a poly(A) tail (Figure 4) suggesting that the presence of a poly(A) tail interferes with stimulation. Such an effect could also be evidenced with the HCV IRES by adding a stretch of 10 to 100 adenosines to the 3΄ end of the reporter RNA (Figure 5). In this particular case, stimulation by let-7 binding was lost by the addition of 30 adenosine residues onwards (Figure 5). Interestingly, PABP needs a minimum of 27 adenosines to bind to the poly(A) tail (86), which suggests that the protein may play a role in this mechanism although it is certainly not the only factor needed. Interestingly, the use of a type II IRES (e.g. EMCV), together with a capped but non-polyadenylated globin construct (Supplementary Figure S1A and B, respectively) showed that neither of these two constructs were stimulated by the binding of the let-7 miRNAs on their 3΄ UTRs. Taken together, our data show that upregulation of gene expression by miRNAs relies on two essential parameters that are the presence of an HCV-like IRES and the absence of a poly(A) tail at the 3΄ end.
Finally, we have modulated the composition of the entering pre-initiation complex at the 5΄ end to reproduce an internal entry of ribosome without requiring ribosome scanning, cap recognition and poly(A) tail as it occurs naturally with the HCV IRES. As such, tethering of the MIF domain of eIF4G, which binds to eIF4A, eIF4B and eIF3 (4,5), is sufficient to deliver the ribosomal subunit to the mRNA (Figure 6). Interestingly, we found that further addition of let-7 sites to this MIF-4G-driven construct resulted in the upregulation of translation only if the reporter RNA lacked a poly(A) tail (Figure 6). Therefore, the use of this artificial construct indicates that translation stimulation is not unique to viral IRESes but could be extended to a general mechanism under conditions where the ribosome is recruited to the 5΄ end of the mRNA with a limited set of initiation factors. This suggests a cross-talk between the RISC complex at the 3΄ end of the reporter gene and the RNA structures at the 5΄ end. To be competent for stimulation, these structures at the 5΄ end must be originated from HCV-like IRESes (e.g. HCV, AEV or SVV) or able to recruit a 40S ribosomal subunit independently from the cap (Figure 6) and with a very limited set of initiation factors, the composition of which should be determined in a near future.
Additionally, our experimental data clearly point out to the importance of the absence of a poly(A) tail in this process. It is tempting to speculate that the lack of poly(A) tail would consequently prevent the incorporation of the TNRC6 protein into the RISC and its replacement by other(s) protein(s) such as FXR1 as previously described in G0 cells (44–48). Thus, the presence of FXR1 could be a way to divert the RISC from its repressive role to become a translational enhancer, probably by increasing the binding of the 40S ribosomal subunit to the IRES. Such a role for RISC in promoting ribosomal binding is not that provocative considering that Ago2 was originally described as an initiation factor (eIF2C) able to stimulate ternary complex formation onto the 40S subunit in the presence of mRNA (87). Finally, it is clear that this mechanism of HCV translation activation by miRNAs bound at the 3΄ end should not be opposed to the process of miR-122 stimulation already described. Rather, it should be considered as novel mechanism that occurs, but is not restricted to HCV IRES translation initiation. In a broader prospect, diversion of the repressive function of the miRNAs could represent a novel viral strategy to hijack antiviral cellular defence to promote viral protein synthesis for better replication.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Niels Gehring for plasmid gifts and Dr Marlène Dreux, Dr Patrice André and Dr François-Loïc Cosset for donating cell lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche sur le SIDA et les Hépatites virales [ANRS]; Ministère de la Recherche et de l’Enseignement Supérieur Français [to C.M.]; Cluster Infectiologie Région Rhône-Alpes and Fondation pour la Recherche Médicale (FRM) [to T.L.]; Conicyt-Chile and Agence Nationale pour la Recherche contre le SIDA et les Hépatites Virales [to R.S.R.]. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1. Aitken C.E., Lorsch J.R.. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012; 19:568–576. [DOI] [PubMed] [Google Scholar]
- 2. Lomakin I.B., Steitz T.A.. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013; 500:307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Merrick W.C. eIF4F: a retrospective. J. Biol. Chem. 2015; 290:24091–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohlmann T., Rau M., Pain V.M., Morley S.J.. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996; 15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 5. De Gregorio E., Preiss T., Hentze M.W.. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5΄ end-dependent. RNA. 1998; 4:828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imataka H., Gradi A., Sonenberg N.. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998; 17:7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graifer D., Karpova G.. Interaction of tRNA with eukaryotic ribosome. Int. J. Mol. Sci. 2015; 16:7173–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 9. Balvay L., Rifo R.S., Ricci E.P., Decimo D., Ohlmann T.. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta. 2009; 1789:542–557. [DOI] [PubMed] [Google Scholar]
- 10. Lozano G., Martínez-Salas E.. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015; 12:113–120. [DOI] [PubMed] [Google Scholar]
- 11. Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015; 87:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jonas S., Izaurralde E.. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015; 16:421–433. [DOI] [PubMed] [Google Scholar]
- 13. Hicks J., Liu H.-C.. Involvement of eukaryotic small RNA pathways in host defense and viral pathogenesis. Viruses. 2013; 5:2659–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartel D.P. MicroRNA target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lian S.L., Li S., Abadal G.X., Pauley B.A., Fritzler M.J., Chan E.K.L.. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009; 15:804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfaff J., Hennig J., Herzog F., Aebersold R., Sattler M., Niessing D., Meister G.. Structural features of Argonaute–GW182 protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3770–E3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takimoto K., Wakiyama M., Yokoyama S.. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009; 15:1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bazzini A.A., Lee M.T., Giraldez A.J.. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012; 336:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Béthune J., Artus-Revel C.G., Filipowicz W.. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012; 13:716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Djuranovic S., Nahvi A., Green R.. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012; 336:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Humphreys D.T., Westman B.J., Martin D.I.K., Preiss T.. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16961–16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mathonnet G., Fabian M.R., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S. et al. . MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007; 317:1764–1767. [DOI] [PubMed] [Google Scholar]
- 23. Moretti F., Kaiser C., Zdanowicz-Specht A., Hentze M.W.. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 2012; 19:603–608. [DOI] [PubMed] [Google Scholar]
- 24. Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W.. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005; 309:1573–1576. [DOI] [PubMed] [Google Scholar]
- 25. Ricci E.P., Limousin T., Soto-Rifo R., Allison R., Pöyry T., Decimo D., Jackson R.J., Ohlmann T.. Activation of a microRNA response in trans reveals a new role for poly(A) in translational repression. Nucleic Acids Res. 2011; 39:5215–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walters R.W., Bradrick S.S., Gromeier M.. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010; 16:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zdanowicz A., Thermann R., Kowalska J., Jemielity J., Duncan K., Preiss T., Darzynkiewicz E., Hentze M.W.. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol. Cell. 2009; 35:881–888. [DOI] [PubMed] [Google Scholar]
- 28. Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A. et al. . Mammalian miRNA RISC Recruits CAF1 and PABP to Affect PABP-Dependent Deadenylation. Mol. Cell. 2009; 35:868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zekri L., Huntzinger E., Heimstädt S., Izaurralde E.. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of MicroRNA targets and is required for target release. Mol. Cell. Biol. 2009; 29:6220–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zekri L., Kuzuoğlu-Öztürk D., Izaurralde E.. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 2013; 32:1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fukaya T., Iwakawa H.-O., Tomari Y.. MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Mol. Cell. 2014; 56:67–78. [DOI] [PubMed] [Google Scholar]
- 32. Kamenska A., Lu W.-T., Kubacka D., Broomhead H., Minshall N., Bushell M., Standart N.. Human 4E-T represses translation of bound mRNAs and enhances microRNA-mediated silencing. Nucleic Acids Res. 2014; 42:3298–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathys H., Basquin J., Ozgur S., Czarnocki-Cieciura M., Bonneau F., Aartse A., Dziembowski A., Nowotny M., Conti E., Filipowicz W.. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell. 2014; 54:751–765. [DOI] [PubMed] [Google Scholar]
- 34. Rouya C., Siddiqui N., Morita M., Duchaine T.F., Fabian M.R., Sonenberg N.. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA. 2014; 20:1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y., Boland A., Kuzuoğlu-Öztürk D., Bawankar P., Loh B., Chang C.-T., Weichenrieder O., Izaurralde E.. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell. 2014; 54:737–750. [DOI] [PubMed] [Google Scholar]
- 36. Fukao A., Mishima Y., Takizawa N., Oka S., Imataka H., Pelletier J., Sonenberg N., Thoma C., Fujiwara T.. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol. Cell. 2014; 56:79–89. [DOI] [PubMed] [Google Scholar]
- 37. Meijer H.A., Kong Y.W., Lu W.T., Wilczynska A., Spriggs R.V., Robinson S.W., Godfrey J.D., Willis A.E., Bushell M.. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013; 340:82–85. [DOI] [PubMed] [Google Scholar]
- 38. Ricci E.P., Limousin T., Soto-Rifo R., Rubilar P.S., Decimo D., Ohlmann T.. miRNA repression of translation in vitro takes place during 43S ribosomal scanning. Nucleic Acids Res. 2013; 41:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Braun J.E., Huntzinger E., Fauser M., Izaurralde E.. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011; 44:120–133. [DOI] [PubMed] [Google Scholar]
- 40. Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W.. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011; 18:1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huntzinger E., Kuzuoglu-Öztürk D., Braun J.E., Eulalio A., Wohlbold L., Izaurralde E.. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013; 41:978–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bushell M., Wood W., Carpenter G., Pain V.M., Morley S.J., Clemens M.J.. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J. Biol. Chem. 2001; 276:23922–23928. [DOI] [PubMed] [Google Scholar]
- 43. Iwasaki S., Tomari Y.. Argonaute-mediated translational repression (and activation). Fly (Austin). 2009; 3:204–206. [PubMed] [Google Scholar]
- 44. Mortensen R.D., Serra M., Steitz J.A., Vasudevan S.. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA–protein complexes (microRNPs). Proc. Natl. Acad. Sci. U.S.A. 2011; 108:8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Truesdell S.S., Mortensen R.D., Seo M., Schroeder J.C., Lee J.H., LeTonqueze O., Vasudevan S.. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep. 2012; 2:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vasudevan S., Tong Y., Steitz J.A.. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007; 318:1931–1934. [DOI] [PubMed] [Google Scholar]
- 47. Vasudevan S., Tong Y., Steitz J.A.. Cell cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008; 7:1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vasudevan S. Posttranscriptional upregulation by MicroRNAs. Wiley Interdiscip. Rev. RNA. 2012; 3:311–330. [DOI] [PubMed] [Google Scholar]
- 49. Iwasaki S., Kawamata T., Tomari Y.. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell. 2009; 34:58–67. [DOI] [PubMed] [Google Scholar]
- 50. Berry K.E., Waghray S., Doudna J.A.. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010; 16:1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cai Q., Todorovic A., Andaya A., Gao J., Leary J.A., Cate J.H.D.. Distinct regions of human eIF3 are sufficient for binding to the HCV IRES and the 40S ribosomal subunit. J. Mol. Biol. 2010; 403:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Pestova T.V., Hellen C.U.T., Frank J.. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013; 503:539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Locker N., Easton L.E., Lukavsky P.J.. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007; 26:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lukavsky P.J. Structure and function of HCV IRES domains. Virus Res. 2009; 139:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Niepmann M. Hepatitis C virus RNA translation. Curr. Top. Microbiol. Immunol. 2013; 369:143–166. [DOI] [PubMed] [Google Scholar]
- 56. Pérard J., Rasia R., Medenbach J., Ayala I., Boisbouvier J., Drouet E., Baudin F.. Human initiation factor eIF3 subunit b interacts with HCV IRES RNA through its N-terminal RNA recognition motif. FEBS Lett. 2009; 583:70–74. [DOI] [PubMed] [Google Scholar]
- 57. Sun C., Querol-Audí J., Mortimer S.A., Arias-Palomo E., Doudna J.A., Nogales E., Cate J.H.D.. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 2013; 41:7512–7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M.. Competing and noncompeting activities of miR-122 and the 5΄ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li Y., Yamane D., Lemon S.M.. Dissecting the roles of the 5΄ exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J. Virol. 2015; 89:4857–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sedano C.D., Sarnow P.. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014; 16:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Conrad K.D., Giering F., Erfurth C., Neumann A., Fehr C., Meister G., Niepmann M.. microRNA-122 dependent binding of Ago2 protein to hepatitis C virus RNA is associated with enhanced RNA stability and translation stimulation. PLoS One. 2013; 8:e56272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fehr C., Conrad K.D., Niepmann M.. Differential stimulation of hepatitis C virus RNA translation by microRNA-122 in different cell cycle phases. Cell Cycle. 2012; 11:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Henke J.I., Goergen D., Zheng J., Song Y., Schüttler C.G., Fehr C., Jünemann C., Niepmann M.. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008; 27:3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roberts A.P.E., Lewis A.P., Jopling C.L.. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011; 39:7716–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Soto Rifo R., Ricci E.P., Décimo D., Moncorgé O., Ohlmann T.. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007; 35:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Prévôt D., Décimo D., Herbreteau C.H., Roux F., Garin J., Darlix J.-L., Ohlmann T.. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003; 22:1909–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bakhshesh M., Groppelli E., Willcocks M.M., Royall E., Belsham G.J., Roberts L.O.. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 2008; 82:1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Willcocks M.M., Locker N., Gomwalk Z., Royall E., Bakhshesh M., Belsham G.J., Idamakanti N., Burroughs K.D., Reddy P.S., Hallenbeck P.L. et al. . Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES. J. Virol. 2011; 85:4452–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E.. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005; 122:553–563. [DOI] [PubMed] [Google Scholar]
- 70. Lytle J.R., Yario T.A., Steitz J.A.. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5΄ UTR as in the 3΄ UTR. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:9667–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pasquinelli A.E., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Müller P. et al. . Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000; 408:86–89. [DOI] [PubMed] [Google Scholar]
- 72. Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G.. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000; 403:901–906. [DOI] [PubMed] [Google Scholar]
- 73. Roush S., Slack F.J.. The let-7 family of microRNAs. Trends Cell Biol. 2008; 18:505–516. [DOI] [PubMed] [Google Scholar]
- 74. Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T.. Identification of Novel Genes Coding for Small Expressed RNAs. Science. 2001; 294:853–858. [DOI] [PubMed] [Google Scholar]
- 75. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M. et al. . A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell. 2007; 129:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kong Y.W., Cannell I.G., de Moor C.H., Hill K., Garside P.G., Hamilton T.L., Meijer H.A., Dobbyn H.C., Stoneley M., Spriggs K.A. et al. . The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8866–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Masaki S., Ohtsuka R., Abe Y., Muta K., Umemura T.. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem. Biophys. Res. Commun. 2007; 364:509–514. [DOI] [PubMed] [Google Scholar]
- 78. Merkerova M., Belickova M., Bruchova H.. Differential expression of microRNAs in hematopoietic cell lineages. Eur. J. Haematol. 2008; 81:304–310. [DOI] [PubMed] [Google Scholar]
- 79. Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R. et al. . miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004; 1:106–113. [DOI] [PubMed] [Google Scholar]
- 80. Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P.. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005; 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 81. Jopling C.L., Schütz S., Sarnow P.. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008; 4:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bradrick S.S., Walters R.W., Gromeier M.. The hepatitis C virus 3΄-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucl. Acids Res. 2006; 34:1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Andrei M.A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Lührmann R.. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005; 11:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Baron-Benhamou J., Gehring N.H., Kulozik A.E., Hentze M.W.. Using the lambdaN peptide to tether proteins to RNAs. Methods Mol. Biol. 2004; 257:135–154. [DOI] [PubMed] [Google Scholar]
- 85. Villa N., Do A., Hershey J.W.B., Fraser C.S.. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 2013; 288:32932–32940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Baer B.W., Kornberg R.D.. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell Biol. 1983; 96:717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Roy A.L., Chakrabarti D., Datta B., Hileman R.E., Gupta N.K.. Natural mRNA is required for directing Met-tRNA(f) binding to 40S ribosomal subunits in animal cells: involvement of Co-eIF-2A in natural mRNA-directed initiation complex formation. Biochemistry. 1988; 27:8203–8209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.