Abstract
MicroRNA 26a (miR-26a) reduces cell viability in several cancers, indicating that miR-26a could be used as a therapeutic option in patients. We demonstrate that miR-26a not only inhibits G1-S cell cycle transition and promotes apoptosis, as previously described, but also regulates multiple cell cycle checkpoints. We show that sustained miR-26a over-expression in both breast cancer (BC) cell lines and mouse embryonic fibroblasts (MEFs) induces oversized cells containing either a single-large nucleus or two nuclei, indicating defects in mitosis and cytokinesis. Additionally, we demonstrate that miR-26a induces aneuploidy and centrosome defects and enhances tumorigenesis. Mechanistically, it acts by targeting G1-S transition genes as well as genes involved in mitosis and cytokinesis such as CHFR, LARP1 and YWHAE. Importantly, we show that only the re-expression of CHFR in miR-26a over-expressing cells partially rescues normal mitosis and impairs the tumorigenesis exerted by miR-26a, indicating that CHFR represents an important miR-26a target in the regulation of such phenotypes. We propose that miR-26a delivery might not be a viable therapeutic strategy due to the potential deleterious oncogenic activity of this miRNA.
INTRODUCTION
Errors in the regulation of DNA synthesis, DNA repair, cell-cycle checkpoint progression, chromosome segregation and completion of cytokinesis can lead to genomic instability which promotes cancer development and progression (1). When such instability affects the number or structure of chromosomes it is referred to as chromosomal instability (CIN) (2). CIN is known to significantly contribute to aneuploidy, which is a common driver of many cancers (3). Aneuploidy can be caused by defects in mitotic checkpoints, chromosome cohesion and mitotic spindle as well as merotelic attachment of kinetochores (4). Mitotic checkpoints ensure that once all the chromosomes are aligned on the metaphase plate they are all properly attached to the kinetochores (5). In addition, this checkpoint induces symmetrical tension across the chromosomes, permitting proper formation of a bipolar mitotic spindle, and a correct separation of the sister chromatids. It is extremely important therefore to have a full understanding of the processes and factors that ensure a smooth and error-free progression through all of the stages of the cell cycle.
CHFR has been recently described as a novel mitotic checkpoint protein playing a crucial role during the prophase stage of M-phase (6). It has been shown to delay metaphase entry for cells that experience mitotic stress through preventing chromosome condensation (7) by stopping accumulation of Cyclin B1 in the nucleus (8). CHFR has been shown to be epigenetically inactivated in a number of malignancies including oesophageal, lung and breast cancers (9–11).
MicroRNAs (miRNAs) are small RNA molecules able to post-transcriptionally inhibit gene expression (12). Numerous studies have described a role of miRNAs in cancer and metastatic progression functioning as either tumor suppressors or oncogenes (13).
miR-26a is an abundant ubiquitously expressed miRNA which has an important role in various cancers such as breast (14,15), lung (16) and glioma (17). It acts by inhibiting the G1-S cell cycle transition directly by regulating multiple specific targets such as Chk1, Wee1 (18), EZ2H (19) and RB1 (20). miR-26a transient transfection also inhibits anchorage-independent growth and induces cell-cycle arrest and apoptosis in breast cancer (BC), targeting oncogenes such as MTDH and EZH2 (21). These findings suggest that the over-expression of miR-26a mimic in cancer patients may block cell proliferation and could be considered as a therapeutic option.
Recently, an increasing interest has developed around the therapeutic potential of miRNAs in the cancer clinic (22–24). However, this approach requires particular caution given that a single miRNA can affect multiple transcripts (25), indicating that a comprehensive evaluation of the genes regulated by a specific miRNA in a particular tissue is warranted to enable a better understanding of its therapeutic potential, mechanism of action and potential side effects associated with. As such, we, and others have demonstrated that miRNAs can modulate cellular responses through a complex network of positive and negative feedback loops to confer robustness to regulative processes (26–28). This indicates that either over-expression or down-regulation of single miRNAs could confer deleterious phonotypical aberrations.
Consistent with this hypothesis, we show here that miR-26a over-expression in cells does not only inhibit G1-S transition as previously shown, but also mitosis and cytokinesis. Furthermore, we show that miR-26a expression also mediates comparable phenotypes in embryonic mouse fibroblasts (MEFs), suggesting that these miR-26a-mediated regulative mechanisms have relevance to physiological processes other than tumorigenesis and are conserved across species. In aggregate, we demonstrate that sustained over-expression of miR-26a in BC initially inhibited cell proliferation, but later promoted defects in chromosome segregation and mitosis leading to chromosomal instability (CIN) and increased tumorigenesis. This indicates that the administration of miR-26a or mimetic to cancer patients could have significant detrimental consequences.
MATERIALS AND METHODS
Cell culture
Breast cancer cell lines (MCF-7, MDA-MB-231, SK-BR-3, T-47D, ZR-75-1) and mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium (DMEM). Both were supplemented with 10% fecal calf serum (FCS), 1% penicillin/streptomycin and 2% glutamine. Stable transfected MDA-MB-231 cell clones were expanded in G-418 (Roche Applied Science, Burgess Hill, UK).
Plasmid constructions
miR-26a sponge was constructed by annealing, purifying and cloning oligonucleotides containing six tandem bulged miRNA binding motifs, into the HindIII and BamHI sites of the pEGFP-C1 plasmid (Contech, Saint-Germain-en-Laye, France), as 3΄UTR of the EGFP mRNA. The CHFR, LARP1, YWHAE and MLC1 3΄UTRs cloned into pLightSwitch_3΄UTR GoClone vectors were used (Switch Gear Genomics, Menlo Park, CA, USA). The indicated mutagenized or deleted miR-26a seed-containing luciferase reporter vector was created with a QuickChange II or II XL Site-Directed Mutagenesis Kit (Agilent Technology, Edinburgh, UK) according to the manufacturer's instructions. All plasmid sequences were verified to be free of mutations by direct sequencing.
RNA isolation and real time-qPCR assays
Total RNA from cultured cells was extracted using TRIzol reagent following the manufacturer instructions (Invitrogen). For relative quantification of either mRNA or mature microRNA levels, RT-qPCRs were performed as previously described (29).
Illumina RNA-seq and analysis
Two micrograms of total RNA from each sample was used to produce cDNA libraries from polyA enriched RNA using the True-seq RNA preparation kit (Illumina) according to the manufacturer's instructions. Paired end sequences (reads) 100 nt in length were then generated using a HiSeq 2000 instrument (Illumina). Fastq files containing the sequenced reads, obtained at the end of the sequencing, were mapped to the University of California at Santa Cruz (UCSC) human genome (hg19 assembly) with TopHat version 1.4.1 (http://tophat.cbcb.umd.edu), using default settings. The mapped bam files obtained at the end of the runs were loaded on the Partek Genomic Suite (Partek Incorporated, USA) for quantification of Reads Per Kilobase of transcript per Million mapped reads (RPKM) normalized values and analysis. RPKMs represent the computed values. Significant change in gene expression was selected applying a Chi squared test.
Transfections, reporter assays and cell treatments
For long-term miRNA or siRNA over-expression, cells were plated in 6-well plates at 30% confluence, and transfected with the miRNA mimics (5 nM) using HiPerfect Transfection Reagent (Qiagen, Crawley, UK). After 72 h of transfection, cells were split and re-transfected with additional miRNAs mimics; this protocol was repeated every 3 days for up to 9 days. For transient (48 h or 72 h) cell transfection, cells were plated in 6-well plates at 50% confluence and transfected with miRNA mimics (5 nM) and HiPerfect. For co-expression of miRNA precursor mimics and CHFR-, LARP1- and/or YWHAE-expressing vectors, MCF-7 and MDA-MB-231 cells were transfected as described above with miRNA mimics for 9 days, and co-transfection with gene-expression vectors was performed for the last 3 days (MDA) or 6 days (MCF-7) of the experiment. Transfection was done using Lipofectamine 2000 (Life Technologies Ltd, Paisley, UK).
For 3΄UTR reporter assays, cells were co-transfected with the indicated miRNA precursors and the specific 3΄UTR GoClone reporters (Switchgear Genomics Menlo Park, CA, USA) in 24-well plates and lysed using a passive lysis buffer (Promega, Southampton, UK). Next, the lysates were processed with the LightSwitch Assay System (Switchgear Genomics Menlo Park, CA, USA) according to manufacturer's instructions. Luciferase activity detection was performed using a GLOMAX 96 Microplate luminometer (Promega).
For nocodazole treatment, cells were treated with nocodazole (50 nM) in normal growth medium. After 16 h, cells were allowed to enter mitosis by replacing nocodazole-containing growth medium with normal growth medium for 1 h prior to fixation.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and permealised in 0.2% Triton-X-100 for 30 min. F-Actin was detected with phalloidin-Alexa Fluor® 488 (Invitrogen) and nuclei were visualized with DAPI (Invitrogen). For visualization of mitotic features visualisation, cells were co-stained with γ-tubulin antibody (1:500, ab11316) and α-tubulin antibody (1:1000, 79026, Sigma-Aldrich) and imaged using an EVOS® FL Cell imaging system (Life Technologies).
Metaphase spread preparations and flow cytometry analysis
To prepare metaphase spreads, cell were treated with colcemid (50 ng/ml, GIBCO BRL) for 4 h at 37°C. Cells were then collected, washed with phosphate-buffered saline and incubated with 75 mM KCl at 37°C for 15 min. After fixing in Carnoy's solution (75% methanol, 25% acetic acid), cells were dropped in 20 μl-aliquots onto microscope slides and stained them with 5% Giemsa solution. Metaphase spreads were imaged under light microscopy and chromosome numbers determined. For fluorescence-activated cell-sorting analysis, cells were collected, fixed in ice-cold methanol and incubated with RNase (50 μg/ml) and propidium iodide (PI, 100 μg/ml). DNA contents were determined using a FACSCanto II flow citometer (BD Biosciences).
Soft agar clonogenic assays
After transfection, cells were harvested by trypsinisation, counted with a Scepter™ Handheld automated cell counter (Millipore) and resuspended in a 0.3% agarose/growth medium solution (1 × 105 cells/ml). Two milliliter of cell/agarose mixture was added in each well of a 6-well plate pre-coated with 0.6% agarose in duplicate per condition. Cells were incubated under normal growth conditions for 3–4 weeks to allow anchorage-independent cell growth. Colonies over 75 nm in diameter were imaged by light microscopy and then stained with 0.5 mg/ml of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Invitrogen, Life Technologies Ltd, Paisley, UK) in ddH2O after incubation at normal cell growth conditions overnight. Each well was then photographed to visualize MTT-stained colony formation.
Analysis of miR-26a expression in patient samples
Analysis of miR-26a expression from TCGA and Buffa Camps Breast GSE22216 was performed using SurvMicro (30).
Statistical analysis
Data are presented as mean ± standard error of the mean (S.E.M.) calculated using Graph Prism software. Student's t-test and two-way ANOVA test were used for comparison, with P < 0.05 considered significant.
RESULTS
miR-26a over-expression leads to formation of large, binucleated cells and increased the number of micronuclei
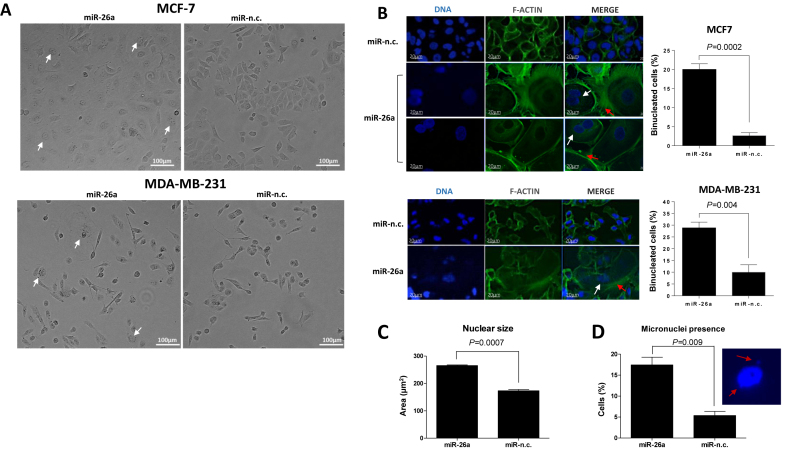
Transient over-expression of miRNA mimics in cancer cell lines for 24–72 h is a commonly used procedure to assess the effect of specific miRNAs on cancer cell phenotypes. MCF-7 and MDA-MB-231 express similar levels of miR-26a (Supplementary Figure S1). Since transient miR-26a over-expression has been shown to reduce MCF-7 and MDA-MB-231 cancer cell properties (14), we questioned whether a sustained miR-26a expression to better mimic cancer treatment would have a similar effect on cancer cells. To this end, we maintained the over-expression of miR-26a for 9 days in both MCF-7 and MDA-MB-231 following evaluation of cellular morphology. Unexpectedly, continuous over-expression of miR-26a induced formation of large cells with aberrant size and/or number of nuclei (Figure 1A–C). MCF-7 and MDA-MB-231 cells derived from the long-term miR-26a over-expression had either a single large nucleus or two nuclei compared to negative control (miR-n.c) expressing cells (Figure 1A–C) indicating that miR-26a promotes both mitosis and/or cytokinesis defects. In addition, such miR-26a over-expression significantly increased the number of micronuclei (Figure 1D) suggesting possible aberrations in spindle dynamics (31,32). Moreover, phalloidin staining demonstrated that miR-26a induced extensive reorganisation of F-actin in cortical microspikes (Figure 1B) that has been linked to impaired cytokinesis (33) and which may explain the increased number of binucleated cells. To further confirm our observations, we extended our investigation to three breast cancer cell lines. Accordingly, over-expression of miR-26a in SK-BR-3, T-47D and ZR-75-1 breast cancer cells yielded increased nuclear size accompanied by an increased number of chromosomes (Supplementary Figure S1B) providing a definitive verification of the observed miR-26a long term expression related phenotype in breast cancer cells.
Figure 1.
Prolonged miR-26a over-expression leads to formation of large bi/mononucleated cells with an increased number of micronuclei. (A) Phase-contrast images of MCF-7 (top panel) and MDA-MB-231 (bottom panel) cells upon transfection with miR-26a and miR-n.c. precursors (5 nM) for 9 days. Scale bar: 100 μm. Arrows indicate multinucleated cells. (B) From left to right: DAPI staining of DNA, Alexa Fluor 488 phalloidin (1:500) staining of F-actin and overlay of the two images of MCF-7 (top panel) or MDA-MB-231 cells (bottom panel) transfected with miR-26a and miR-n.c. precursors (5nM) for 9 days. White arrows indicate aberrant nuclei; red arrows indicate cortical microspikes. Scale bar: 20 μm. The graphs show the percentages of multinucleated cells observed in the two BC cell lines after long term-transfection with the indicated miRNA precursors. 50–100 cells were counted per condition in three independent experiments. Data are mean ± S.E.M. (C) The average size of the nucleus in cells transfected with indicated precursors for 9 days was calculated using ImageJ software; 100 cells were calculated per condition. (D) The number of micronuclei in miR-26a and miR-n.c. transfected cells was calculated following DAPI staining and image acquisition using a confocal microscope. >100 cells were analysed per condition. n = 3.
We also performed a timecourse experiment in MCF-7 and MDA-MB-231 cell lines to assess when changes induced by miR-26a can be observed. Looking at the nuclear size as a hallmark of miR-26a-induced aberrations, we saw that the effect can be seen as early as day 3 post-transfection (Supplementary Figure S1C) with a further increase in the average nuclear size over the following days.
miR-26a over-expression causes aneuploidy in human cancer cells as well as in MEFs
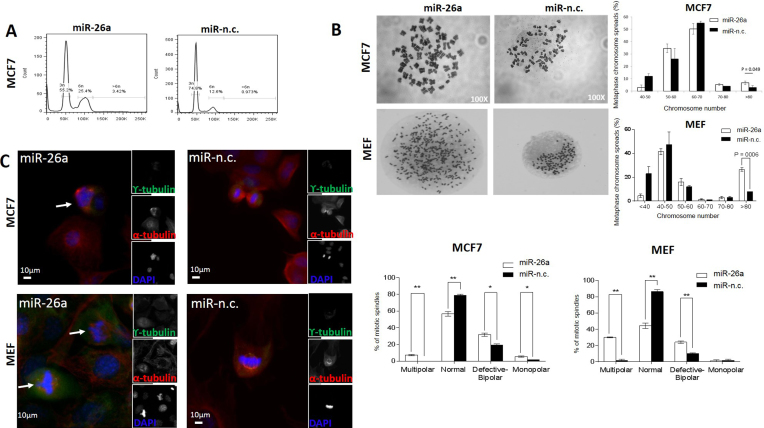
Next, we investigated a possible effect of miR-26a on aneuploidy in BC cell lines (Figure 2 and Supplementary Figure S2). PI staining followed by FACS analysis in MCF-7 cells revealed a threefold increase in aneuploid DNA content in cells transfected with miR-26a precursor for 9 days (>6n; Figure 2A). Similarly, MDA-MB-231 cells expressing miR-26a for 9 days displayed an increased number of cells with abnormally high DNA content (>4n; Supplementary Figure S2A). This observation was further confirmed by data obtained from metaphase chromosome spreads prepared to assess chromosome number more precisely in both cell lines (Figure 2B and Supplementary Figure S2B). Accordingly, both cell lines expressing miR-26a had a significantly higher proportion of cells containing an abnormally high number of chromosomes (>80) compared with control (Figure 2B and Supplementary Figure S2B). In summary, these results showed a significantly higher proportion of cells with an increased chromosome number in miR-26a over-expressing cells, with over 10% of those cells having 80 or more chromosomes in both cell lines. Such state of aneuploidy has been extensively correlated with CIN (2).
Figure 2.
MiR-26a affects cellular DNA content and causes mitotic defects in MCF-7 as well as MEF cells. (A) MCF-7 cells transfected with the indicated miRNA precursors (5 nM) for 9 days were stained with PI for flow cytometry analysis of DNA content. Data are representative of three independent experiments. (B) Representative metaphase chromosome spreads of MCF-7 and MEF cells transfected for 9 days with the indicated miRNA precursors. Magnification: 100×. Percentages of metaphase chromosome spreads containing the indicated ranges of chromosome number. Chromosome number of 100 methaphase spreads was determined per conditions. Data are mean of three independent experiments (P = 0.04, Student's t-test). (C) Cells were transfected with the indicated miRNA precursors (5 nM) for 9 days, synchronized with nocodazole and then released for 1 h to increase mitotic feature observation. Co-immunostaining was performed with γ- and α-tubulin antibodies and chromosomes were stained with DAPI (individual stain is visualized in reduced, gray scale images). Cells were visualized with EVOS® FL Cell imaging system (Life technologies). Scale bar: 10 μm. Arrows show centrosome amplification. The graph on the right shows percentage of mitotic spindle types (multipolar, normal, defective bipolar and monopolar spindles) observed. 80–100 mitotic cells were counted per condition. Data are mean of three independent experiments ± S.E.M. (*P < 0.01, **P ≤ 0.001, ***P ≤ 0.0001, Two-way ANOVA test).
Subsequently, we sought to examine whether the observed phenomena mediated by sustained miR-26a over-expression could also be demonstrated in non-cancerous primary MEFs, which would imply a more general role for miR-26a in regulating other cellular processes. Moreover, we decided to select a murine cell line in order to assess for any cross-species conservation of miR-26a-mediated phenotypic effects. Comparable to human cancer cellular models, sustained miR-26a over-expression caused a significant rise in aneuploid MEFs cells as manifested by an increased proportion of cells with extra chromosomes (>80; Figure 2B).
Over-expression of miR-26a induces centrosome defects
Since chromosome mis-segregation is often linked to centrosome abnormalities, we stained for γ-tubulin centrosome markers to assess mitotic spindle structures and polarity of MCF-7, MDA-MB-231 and MEFs (Figure 2C and Supplementary Figure S2C). Over-expression of miR-26a had a significant impact on centrosome number and polarity status of all the analysed cells which was reflected in an increased number of multipolar, monopolar as well as defective bipolar cells. We observed different polarity configurations in multipolar spindles with three or four distinct poles as well as two, proximally located poles in some cells, plausibly reflecting cellular centrosome clustering activity as an attempt to suppress multipolarity (Figure 2C and Supplementary Figure S2C). In cancer cells, such spindle polarity abnormalities have been described as a major cause of incorrect attachment of kinetochores leading to mis-segregation of chromosomes (34).
miR-26a regulates several genes that modulate different stages of the cell cycle
Our results showed that miR-26a induces formation of oversized cells containing very large, or two nuclei (Figure 1). Moreover, expression of miR-26a mediated aneuploidy and centrosome defects in both human BC cell lines and primary MEFs (Figure 2). In order to identify the genes regulated by miR-26a that might mediate this striking phenotype, we performed RNA-seq in MCF-7 cells following over-expression of miR-26a for 9 days, and in MDA-MB-231 cells in which we had induced stable knock down of miR-26a using a miRNA-sponge strategy (29). We used this approach to identify genes containing miR-26a seed regions that were down-regulated in MCF-7 cells but up-regulated in MDA-MB-231 cells and hence, provide us with a list of likely miR-26a direct targets (Supplementary Table S1).
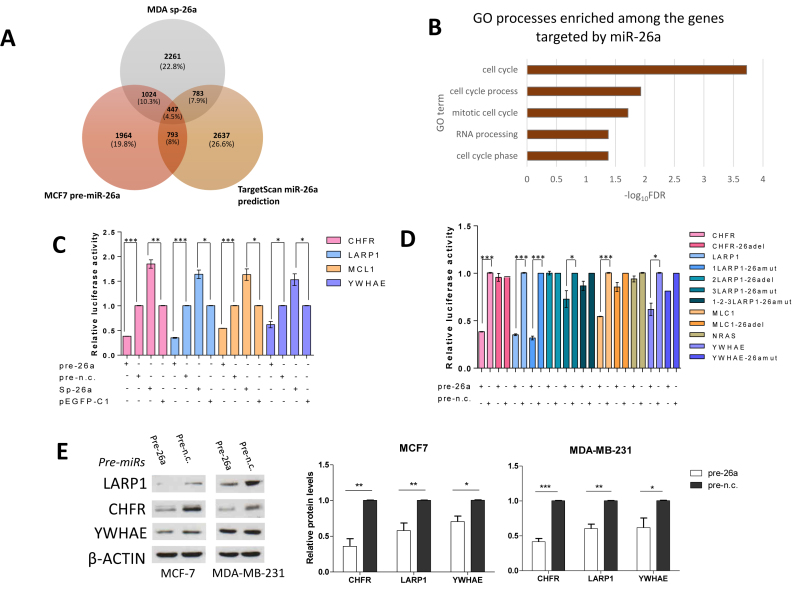
Sequencing our samples (MCF-7 cells transfected with miR-26a, with miR-negative control (miR-n.c.), MDA-MB-231 cells stably expressing p-EGFP-C1 empty vector and expressing miR-26a sponge vector) produced 27, 29, 25 and 33 million reads respectively, which were then mapped to the Hg19 human reference genome. miR-26a expression in MCF-7 cells affected the level of 7.4% of genes, whereas in MDA-MB-231 cells stably transfected with the sponge vector, gene expression changes were observed in 25.6% of genes (Supplementary Table S1). To enrich for genes that were directly regulated by miR-26a, we focused on those transcripts that were down-regulated in MCF-7 cells over-expressing miR-26a that overlapped with those transcripts that were up-regulated in MDA-MB-231 cells stably transfected with a miR-26a sponge construct (sp-26a; Supplementary Figure S3A) and only if they had miR-26a recognition sites predicted by TargetScan analysis (Figure 3A; Supplementary Table S2). We considered a cut-off expression change of 1.2-fold sufficient since miRNAs regulate transcripts by promoting destabilization through deadenylation (35,36) but mediate a stronger effect on protein translation (37), higher fold changes would result in omitting many relevant targets. Furthermore, the impact of miRNAs on gene targets is variable and is usually mild (38,39). To validate our analysis, we performed a thorough literature search and used the miRTarBase database (40) to identify experimentally confirmed targets of miR-26a. Eighty-one percent of previously described miR-26a gene targets were among the intersections representing those genes down-regulated in MCF-7 and/or up-regulated in MDA-MB-231, and those in the TargetScan list (Figure 3A and Supplementary Table S2) indicating the reliability of our methods. In terms of genes involved in cell cycle progression, our overlap list contained previously validated targets of miR-26a, including CCNE2 (41) and RB1 (42). Whilst our analysis identified a large number of miR-26a genes previously demonstrated to be involved in G1-S transition, a surprising finding was that the regulation of cell cycle was not restricted to G1-S transition but also mitotic cell-cycle was amongst the highest scoring GO terms among the regulated genes (Figure 3B) implying a possible undescribed role of this miRNA in the regulation of mitosis separate to its role in G1-S transition.
Figure 3.
Three mitosis-related genes are novel miR-26a targets. (A) Venn diagram representing the number of possible miR-26a targets identified as an area of overlap between transcripts downregulated in MCF cells over-expressing miR-26a, MDA-MB-231 cells with reduced miR-26a level and TargetScan-predicted miR-26a target genes. (B) Chart showing the top five significantly enriched GO terms found with DAVID bioinformatic tool amongst the possible miR-26a targets identified as described in panel A. (C and D) MiR-26a targets CHFR, LARP1, MCL1 and YWHAE by directly interacting with their relative 3΄UTRs. Relative luciferase activity levels were measured after 24 h from co-transfection of MCF-7 cells with the indicated 3΄UTR-luciferase reporter constructs either with miR-26a or miR-n.c. precurors (100 nM) or with mi-26a-sponge construct or pEGFP-C1 parental control (150 ng). Panel D shows luciferase activity levels in cells expressing mutant 3΄UTRs which impair miR-26a binding. Data are mean of three independent experiments (each of them performed in triplicate) ± S.E.M. (*P < 0.05,**P ≤ 0.006,***P ≤ 0.0005). (E) Western blots showing CHFR, LARP1 and YWHAE levels after transfection for 9 days of the two cell lines with the indicated miRNA precursors (5 nM) and relative densitometric quantifications. β-Actin was used as a loading control. Fold changes in protein expression levels normalised for β-actin using ImageJ software are shown underneath each relative protein plot (*P < 0.01, **P ≤ 0.001, ***P ≤ 0.0001).
miR-26a targets CHFR, LARP1 and YWHAE directly interacting with their 3΄ UTRs
Considering the enrichment for miR-26a-regulated cell cycle and mitotic genes among GO terms that we identified (Figure 3B) and the related phenotypic effects derived from miR-26a over-expression in cell lines (Figures 1 and 2), we selected three genes with important functions in mitosis (CHFR, LARP1, YWHAE) (43–45), and cytokinesis (LARP1, YWHAE) (44,46–47) to validate the regulative interaction of miR-26a with the complementary seed region located in the three prime untranslated regions (3΄UTRs) of these transcripts. We firstly used RT-qPCR to confirm that CHFR, LARP1, YWHAE and the positive control MCL1 (14) were regulated by miR-26a (Supplementary Figure S3B). Accordingly, the selected transcripts were down-regulated by miR-26a over-expression in MCF-7 cells and, conversely, up-regulated in MDA-MB-231 cells stably expressing a sponge vector for miR-26a (sp-26a; Supplementary Figure S3B). Next, we confirmed that the chosen targets were important for mitosis-related phenotypes in BC cells, by silencing each of them in MCF-7 and MDA-MB-231 and assessing effects on mitotic apparatus structure (Supplementary Figures S4 and S5, respectively).
Inspection of the 3΄UTRs of CHFR, MCL1 and YWHAE indicated a single miR-26a binding site for these transcripts with three binding sites for LARP1 (Supplementary Figure S3C). Subsequently, we confirmed the direct regulation of these targets by miR-26a, using 3΄UTR luciferase assays following miR-26a over-expression or down-regulation (Figure 3C). Furthermore, site-directed mutagenesis for the miR-26a binding sites located along the 3΄UTRs of these transcripts indicated that miR-26a regulated these transcripts directly, via interaction with the identified miR-26a binding sites, except for one of the three LARP1 sites (Figure 3D). Moreover, LARP1, CHFR and YWHAE protein levels were significantly reduced upon miR-26a over-expression in both MCF-7 and MDA-MB-231 (Figure 3E), and levels of the potential targets were increased in both cell lines expressing the sponge constructs (Supplementary Figure S3D).
Restoration of CHFR expression in cells over-expressing miR-26a partially rescues miR-26a-induced mitotic phenotypes
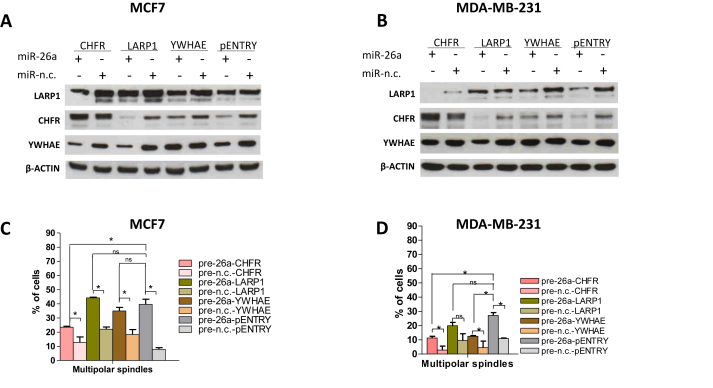
To validate the impact of the identified mitosis-related miR-26a targets on mitotic spindle formation and polarity mediated by miR-26a, we over-expressed each corresponding coding region for 72 h (MDA-MB-231) or 144 h (MCF-7) in cells transfected with either miR-26a or miR-n.c. precursor for 9 days (Figure 4). Next, we performed Western Blotting experiments to verify the expression levels of CHFR, LARP1 and YWHAE after transfection of the two cell lines with the indicated miRNA precursors and the indicated plasmid cDNA vectors (Figure 4A and B). miR-26a reduced endogenous protein levels because of the presence of intact 3΄UTRs in their corresponding mRNA transcripts, whereas the reintroduction of selected, exogenous targets rescued the expression of their corresponding proteins as they bypass the miRNA-regulatory stage, in both MCF-7 and MDA-MB-231 (Figure 4A and B).
Figure 4.
Restoration of specific miR-26a targets expression in miR-26a-transfected cells partially rescues miR-26a-induced mitotic phenotypes. (A and B) Representative Western blots showing CHFR, LARP1 and YWHAE levels after transfection of the two cell lines with the indicated miRNA precursors (5 nM) for 9 days and with the indicated plasmid vectors for 72 h. β-Actin was used as a loading control; pENTRY was used as an empty plasmid control. Graphs showing percentage of cells with multipolar mitotic spindles observed in MCF-7 (C) or MDA-MB-231 (D) cells transfected for 9 days with the indicated precursors (5 nM) and for 72 h (MDA) or 144 h (MCF-7) with the indicated vectors (2 μg). 80–100 mitotic cells were counted per condition. Data are mean of three independent experiments ± S.E.M. (*P < 0.05,**P ≤ 0.01,***P ≤ 0.001, ns: non-significant, Student's t-test.
Since we had demonstrated that knockdown of each one of the targets affected chromosome separation during mitosis (Supplementary Figures S4A and S5A) and produced an increased proportion of cells with an aberrantly high number of chromosomes (Supplementary Figures S4B and S5B), we subsequently looked at the effect that re-introduction of the targets had on the mitotic spindle in cells over-expressing miR-26a. The overall trend for MCF-7 (Figure 4C) and MDA-MB-231 (Figure 4D) was a decreased number of cells with multipolar spindles. Interestingly, only CHFR re-expression partially rescued the induction of multipolarity exerted by miR-26a in both MCF-7 and MDA-MB-231 cells (Figure 4C and D). Surprisingly, YWHAE re-expression in miR-26a over-expressing MDA-MB-231, but not in MCF-7 cells, partially rescued the cellular multipolarity induced by miR-26a indicating that this mechanism may be tissue or cell-type specific.
Long-term miR-26a over-expression enhances the tumorigenic potential of MCF-7 cells
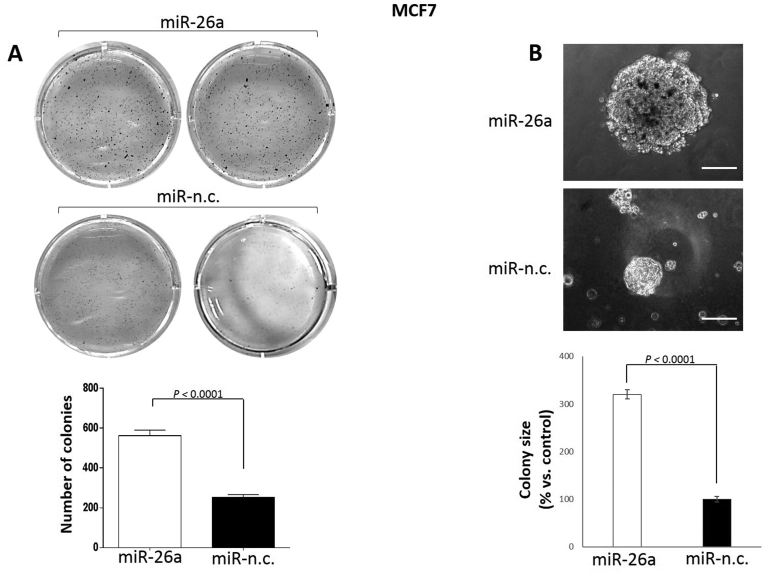
Having established that miR-26a targets mitotic related genes leading to disruption of both the mitotic spindle and chromosome segregation, we decided to investigate the tumorigenic potential of MCF-7 cells transfected with this miRNA. To this end, we performed soft agar clonogenic assays to examine the ability of these cells to spontaneously form colonies (Figure 5A) and provide an index of increased tumorigenesis. We observed a significant increase in the number of colonies formed by the miR-26a over-expressing cells (Figure 5B). Furthermore, the average size of the colonies formed by miR-26a-transfected cells were over three times greater compared to control samples (Figure 5B). Based on these observations we concluded that the continuous over-expression of miR-26a increases tumorigenicity of MCF-7 cells.
Figure 5.
Long-term miR-26a over-expression enhances tumorigenic potential of MCF-7 cells. (A) Spontaneous colony formation by MCF-7 cells transfected for 9 days with the indicated miRNA precursors and plated on soft agar substrates in 6-well plates. After 3–4 weeks cells were fixed and stained with MTT. Number of colonies were quantified by using ImageJ software (bottom graph). (B) Single colonies formed as described in (A) were visualized by light microscopy before colony staining with MTT and the size was quantified by using ImageJ tool (bottom graph). Magnification: 20×. Triplicates per conditions were assessed in three independent experiments. Data are mean ± S.E.M. (P < 0.0001, Student's t-test).
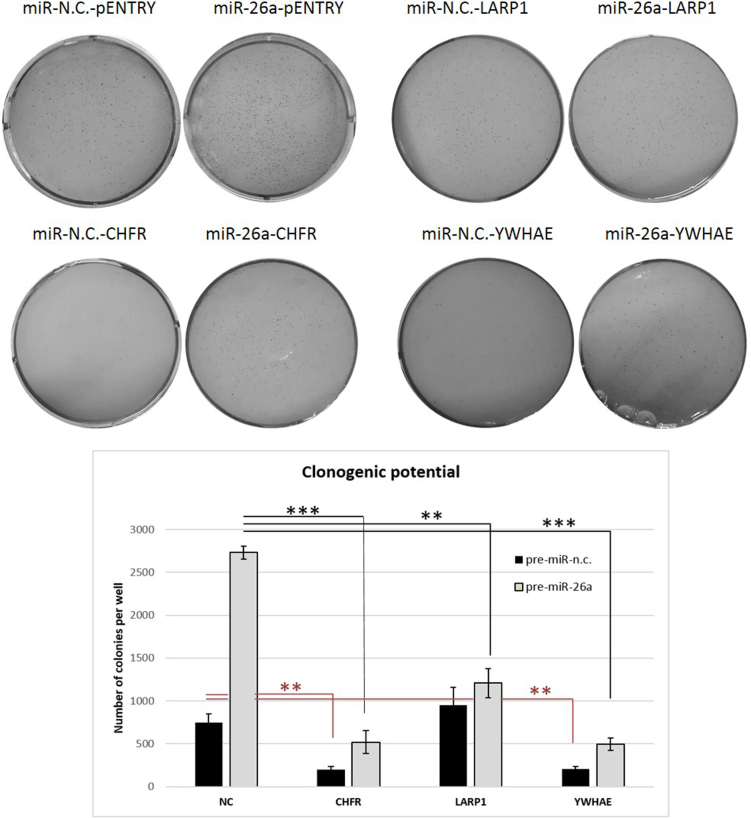
Restoration of CHFR expression in miR-26a-transfected MCF-7 cells partially rescues clonogenic potential of MCF-7 cells
Next, we evaluated whether re-establishment of normal levels of CHFR, LARP1 and YWHAE proteins in cells transfected with miR-26a would reduce the clonogenic properties of MCF-7 cells transfected with miR-26a (Figure 6). Only CHFR re-expression in miR-26a over-expressing cells partly rescued tumorigenesis induced by miR-26a indicating that CHFR represents the crucial miR-26a target for the modulation of tumorigenic potential. We also observed that CHFR was the only target whose expression significantly reduced the miR-26a-induced enlargement of cellular nuclei (Supplementary Figure S6), implying its important role in downstream mediation of miR-26a actions.
Figure 6.
Restoration of CHFR expression in miR-26a-transfected cells partially rescues clonogenic abilities of MCF-7 cells. MCF-7 cells were transfected with the indicated miRNA precursors (5 nM) for 9 days and with the indicated plasmid vectors for 72 h, and subjected to soft-agar clonogenic assays. After 3–4 weeks, colonies were imaged by light microscopy and counted by using ImageJ software. Data are mean of two independent experiments performed in duplicate ± S.E.M. (*P < 0.05,**P ≤ 0.01,***P ≤ 0.001, Student's t-test).
miR-26a could be highly expressed in either high or low risk BC patients
Since we demonstrated that the over-expression of miR-26a had both oncogenic and tumor suppressor effects such as inhibition of cell proliferation, activation of mitotic defects and increased tumorigenesis we investigated the levels of miR-26a in high- and low-risk BC patients. To this end we examined miR-26a expression in selected BC datasets analysing a total of 1660 BC samples [Buffa Camps datasets (GSE22216) 210 samples, TCGA Breast Invasive Carcinoma (Illumina GA) 322 samples and TCGA Breast Invasive Carcinoma (Illumina HiSeq) 528 samples]. We found that miR-26a expression was significantly higher in low risk BC patients compared with high risk BC patients in the 850 samples obtained from both TCGA datasets (Supplementary Figures S7A and S7B), but was significantly higher in high risk BC patients compared to low risk BC patients in the Buffa Camps (GSE22216) BC datasets (Supplementary Figure S7C) confirming that high or low levels could influence cancer formation.
DISCUSSION
The activity of miRNAs can be efficiently and specifically inhibited in vivo using chemically modified, stable complementary oligonucleotides. Conversely, synthetic molecules that mimic tumor-suppressor miRNAs could be delivered to patients in order to restore the expression of a lost onco-suppressor miRNA. Due to these characteristics it has been widely proposed that anti-miRNAs or miRNA mimics could be used to therapeutically modulate the activity of ‘onco’-miRNAs or tumor-suppressor miRNAs, respectively. Nevertheless, because miRNAs act by targeting multiple transcripts through negative and/or positive feedback loops to confer robustness of regulative networks (26–28), we hypothesize that long term over-expression of candidate tumor suppressor miRNAs could have deleterious effects.
miR-26a is an ubiquitously expressed, abundant miRNA that has been proposed to have tumor suppressing roles in various tumors, including breast and liver cancers (15,21,48). Mechanistically, it induces cell cycle arrest and apoptosis by modulating both cell cycle-related genes such as CCND2, CCNE2 (48) and anti-apoptotic genes such as MCL1 (14). Consistent with this hypothesis, the in vivo delivery of miR-26a using vectors based on an adeno-associated virus, repressed liver tumor growth in mouse models (48). Notably, both miR-26a genes are located in fragile genomic sites that can be deleted (49) or amplified (17) in cancer indicating that it could also act as an oncogenic miRNA.
Based on these observations, we aimed to investigate the phenotypic effects of the sustained over-expression of miR-26a in MCF-7 and MDA-MB-231 BC cell lines for which its transient over-expression has been demonstrated to induce cell cycle arrest and apoptosis (14). Surprisingly, long-term over-expression of miR-26a in these cells induced abnormally large nuclei and/or binucleated cells as well as increased the presence of micronuclei (Figure 1). In addition, the cells displayed reorganization of actin filaments in cortical microspikes which indicate cytokinesis defects (33). Based on the observed phenotypes we propose that miR-26a modulates mitosis and cytokinesis in addition to modulating G1-S cell cycle progression (15,21,48). This indicates that, therapeutically, although miR-26a could halt the proliferation of a large number of cells, a considerable portion could develop mitotic defects, aneuploidy and chromosomal instability, ultimately leading to tumorigenesis. Importantly, we also demonstrated miR-26a-induced mitotic defects in MEFs, indicating that miR-26a regulated mitotic phenotypes are not restricted to tumor cells and, in addition, are conserved across species. Moreover, elevated miR-26a levels increased DNA content of both BC cells and MEFs, which is reflected in our findings of a significantly higher percentage of aneuploid cells compared with a control population (Figure 2A, B and Supplementary Figure S1A, B). We also demonstrated that over-expression on miR-26a in MCF-7 cells, MDA-MD-231 cells and MEFs, led to a significantly higher proportion of abnormal mitotic spindles (Figure 2C and Supplementary Figure S2C), a feature which has been shown to induce a variety of cancers (50–52).
RNA-seq followed by experimental validations indicated that miR-26a directly regulates the mitosis related genes CHFR, LARP1 and YWHAE, in addition to the previously identified G1-S cell-cycle transition and apoptosis related genes (Supplementary Table S2 and Figure 3) suggesting the mechanism through which miR-26a regulates mitosis. CHFR functions as a mitotic checkpoint protein which responds to mitotic stress and has a role in promoting chromosomal stability (43). Moreover, low levels of its expression have been linked to aneuploidy, increased proliferation rates and more a malignant phenotype in BC (53). On the other hand knockdown of LARP1 has been shown to interfere with chromosome segregation and cause defects in cytokinesis (44). Furthermore, YWHEA, is a member of the 14-3-3 family of phosphor-serine/phospho-threonine-binding proteins, which has not been well-characterised to date. However, members of this protein family have been implicated in the regulation of anchorage-independent growth as well as cytokinesis, with their down-regulation linked to cytokinetic defects (46,47) and the generation of multinucleated cells (54).
We confirmed that the down-regulation of these novel miR-26a targets by siRNAs promoted mitotic defects, indicating that their inhibition by miR-26a over-expression could be implicated in the observed phenotypes. In order to prove this hypothesis, we over-expressed each of the selected miR-26a targets in cells transfected with this miRNA and noted that CHFR demonstrated a great role in rescuing the cells from miR-26a-induced spindle dysfunction in both MCF-7 and MDA-MB-231 cellular models (Figure 4) as well as prevention of nuclear enlargement (Supplementary Figure S6). Reintroduction of LARP1 and YWHAE did not seem to rescue the cells from the miR-26a insult. Conversely, over-expression of LAPR1 and YWHAE for such a prolonged period of time in MCF-7 cells, seemed to have a negative effect on spindle morphology (increased number of aberrant spindles in the absence of pre-miR-26a). In case of LARP1, its over-expression has previously been shown to be correlated with poor prognosis in colorectal cancer (55) whilst over-expression of YWHAE in the context of chromosomal stability and cancer has not been described yet. Based on our results, we believe that a balanced expression of both of these proteins is required for their correct function.
All of the identified miR-26a-induced phenotypic changes are linked to chromosomal instability, which in turn is known to increase cellular tumorigenic potential (56). Accordingly, we demonstrated that long-term over-expression of miR-26a increased the number and size of colonies formed by MCF-7 cells (Figure 5), which contrasts with the finding that transient over-expression of this miRNA reduces breast tumor formation (14). Moreover, having performed the rescue experiment (Figure 6), we observed that CHFR as well as LAPR1 and YWHAE seem to have a negative effect on miR-26a-induced clonogenic potential boost. We believe that the global effect of CHFR over-expression seen across all our rescue experiments, including prevention of spindle damage as well as nuclear size reduction, might be due to its specific role as a mitotic checkpoint protein. Whereas LAPR1 and YWHAE act via a different route to reduce clonogenic potential without preventing errors in mitotic processes investigated by us.
All of the obtained data suggests that CHFR is an important miR-26a target for the mitotic defect and tumorigeneis effect exerted by the miRNA.
Finally, analysis of miR-26a levels in BC clinical samples indicated that both low and high expression can be significantly linked with high risk cancer traits (Supplementary Figure S7).
We propose that miR-26a, through its regulation of CHFR, has a role in modulating mitosis and cytokinesis in breast cancer, in addition to its previously identified role as a modulator of cell cycle arrest. This indicates that the therapeutic use of miR-26a might not be viable because its expression in patients could induce aneuploidy and chromosomal instability that are hallmarks of cancer development and progression.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Luis Aragón for critical advice. We also thank Action Against Cancer (AAC), Breast Cancer Now and Worldwide Cancer Research for funding this study.
RNA-seq data associated with this study has been deposited to Gene Expression Omnibus (GEO) with the id: GSE43726.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Action Against Cancer (AAC) [P45714]. Funding for open access charge: Imperial Open Access Fund (Imperial College London).
Conflict of interest statement. None declared.
REFERENCES
- 1. Negrini S., Gorgoulis V.G., Halazonetis T.D.. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010; 11:220–228. [DOI] [PubMed] [Google Scholar]
- 2. Ganem N.J., Godinho S.A., Pellman D.. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009; 460:278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albertson D.G., Collins C., McCormick F., Gray J.W.. Chromosome aberrations in solid tumors. Nat. Genet. 2003; 34:369–376. [DOI] [PubMed] [Google Scholar]
- 4. Orr B., Godek K.M., Compton D.. Aneuploidy. Curr. Biol. 2015; 25:R538–R542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amon A. The spindle checkpoint. Curr. Opin. Genet. Dev. 1999; 9:69–75. [DOI] [PubMed] [Google Scholar]
- 6. Kim J.S., Park Y.Y., Park S.Y., Cho H., Kang D., Cho H.. The auto-ubiquitylation of E3 ubiquitin-protein ligase Chfr at G2 phase is required for accumulation of polo-like kinase 1 and mitotic entry in mammalian cells. J. Biol. Chem. 2011; 286:30615–30623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scolnick D.M., Halazonetis T.D.. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000; 406:430–435. [DOI] [PubMed] [Google Scholar]
- 8. Summers M.K., Bothos J., Halazonetis T.D.. The CHFR mitotic checkpoint protein delays cell cycle progression by excluding Cyclin B1 from the nucleus. Oncogene. 2005; 24:2589–2598. [DOI] [PubMed] [Google Scholar]
- 9. Shibata Y., Haruki N., Kuwabara Y., Ishiguro H., Shinoda N., Sato A., Kimura M., Koyama H., Toyama T., Nishiwaki T. et al. . Chfr expression is downregulated by CpG island hypermethylation in esophageal cancer. Carcinogenesis. 2002; 23:1695–1699. [DOI] [PubMed] [Google Scholar]
- 10. Murria R., Palanca S., de Juan I., Egoavil C., Alenda C., García-Casado Z., Juan M.J., Sánchez A.B., Santaballa A., Chirivella I.. Methylation of tumor suppressor genes is related with copy number aberrations in breast cancer. Am. J. Cancer Res. 2015; 5:375. [PMC free article] [PubMed] [Google Scholar]
- 11. Mizuno K., Osada H., Konishi H., Tatematsu Y., Yatabe Y., Mitsudomi T., Fujii Y., Takahashi T.. Aberrant hypermethylation of the CHFR prophase checkpoint gene in human lung cancers. Oncogene. 2002; 21:2328–2333. [DOI] [PubMed] [Google Scholar]
- 12. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 13. Calin G., Croce C.. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006; 25:6202–6210. [DOI] [PubMed] [Google Scholar]
- 14. Gao J., Li L., Wu M., Liu M., Xie X., Guo J., Tang H., Xie X.. MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS One. 2013; 8:e65138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ichikawa T., Sato F., Terasawa K., Tsuchiya S., Toi M., Tsujimoto G., Shimizu K.. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS One. 2012; 7:e31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu B., Wu X., Liu B., Wang C., Liu Y., Zhou Q., Xu K.. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2012; 1822:1692–1704. [DOI] [PubMed] [Google Scholar]
- 17. Huse J.T., Brennan C., Hambardzumyan D., Wee B., Pena J., Rouhanifard S.H., Sohn-Lee C., le Sage C., Agami R., Tuschl T., Holland E.C.. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009; 23:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lezina L., Purmessur N., Antonov A., Ivanova T., Karpova E., Krishan K., Ivan M., Aksenova V., Tentler D., Garabadgiu A.. miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. Cell Death Dis. 2013; 4:e953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sander S., Bullinger L., Klapproth K., Fiedler K., Kestler H.A., Barth T.F., Moller P., Stilgenbauer S., Pollack J.R., Wirth T.. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008; 112:4202–4212. [DOI] [PubMed] [Google Scholar]
- 20. Zhu Y., Lu Y., Zhang Q., Liu J.J., Li T.J., Yang J.R., Zeng C., Zhuang S.M.. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012; 40:4615–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang B., Liu X.X., He J.R., Zhou C.X., Guo M., He M., Li M.F., Chen G.Q., Zhao Q.. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis. 2011; 32:2–9. [DOI] [PubMed] [Google Scholar]
- 22. Izquierdo M. Short interfering RNAs as a tool for cancer gene therapy. Cancer Gene Ther. 2005; 12:217–227. [DOI] [PubMed] [Google Scholar]
- 23. Gleave M.E., Monia B.P.. Antisense therapy for cancer. Nat. Rev. Cancer. 2005; 5:468–479. [DOI] [PubMed] [Google Scholar]
- 24. Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M.. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005; 438:685–689. [DOI] [PubMed] [Google Scholar]
- 25. Lewis B.P., Burge C.B., Bartel D.P.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120:15–20. [DOI] [PubMed] [Google Scholar]
- 26. Castellano L., Giamas G., Jacob J., Coombes R.C., Lucchesi W., Thiruchelvam P., Barton G., Jiao L.R., Wait R., Waxman J. et al. . The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15732–15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krell J., Stebbing J., Carissimi C., Dabrowska A.F., de Giorgio A., Frampton A.E., Harding V., Fulci V., Macino G., Colombo T., Castellano L.. TP53 regulates miRNA association with AGO2 to remodel the miRNA-mRNA interaction network. Genome Res. 2016; 26:331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsang J., Zhu J., van Oudenaarden A.. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007; 26:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pellegrino L., Stebbing J., Braga V.M., Frampton A.E., Jacob J., Buluwela L., Jiao L.R., Periyasamy M., Madsen C.D., Caley M.P. et al. . miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 2013; 41:5400–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aguirre-Gamboa R., Trevino V.. SurvMicro: assessment of miRNA-based prognostic signatures for cancer clinical outcomes by multivariate survival analysis. Bioinformatics. 2014; 30:1630–1632. [DOI] [PubMed] [Google Scholar]
- 31. Fenech M., Kirsch-Volders M., Natarajan A.T., Surralles J., Crott J.W., Parry J., Norppa H., Eastmond D.A., Tucker J.D., Thomas P.. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011; 26:125–132. [DOI] [PubMed] [Google Scholar]
- 32. Holy J.M. Curcumin disrupts mitotic spindle structure and induces micronucleation in MCF-7 breast cancer cells. Mut. Res./Genet. Toxicol. Environ. Mutagen. 2002; 518:71–84. [DOI] [PubMed] [Google Scholar]
- 33. Dutartre H., Davoust J., Gorvel J.P., Chavrier P.. Cytokinesis arrest and redistribution of actin-cytoskeleton regulatory components in cells expressing the Rho GTPase CDC42Hs. J. Cell. Sci. 1996; 109:367–377. [DOI] [PubMed] [Google Scholar]
- 34. Silkworth W.T., Nardi I.K., Scholl L.M., Cimini D.. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009; 4:e6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W.. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011; 18:1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F. et al. . miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011; 18:1211–1217. [DOI] [PubMed] [Google Scholar]
- 37. Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W.. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005; 309:1573–1576. [DOI] [PubMed] [Google Scholar]
- 38. Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P.. The impact of microRNAs on protein output. Nature. 2008; 455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N.. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008; 455:58–63. [DOI] [PubMed] [Google Scholar]
- 40. Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M. et al. . miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011; 39:D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kota J., Chivukula R.R., O'Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., Chang T.C., Vivekanandan P., Torbenson M., Clark K.R. et al. . Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009; 137:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim H., Huang W., Jiang X., Pennicooke B., Park P.J., Johnson M.D.. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2183–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu X., Minter-Dykhouse K., Malureanu L., Zhao W., Zhang D., Merkle C. J., Ward I. M., Saya H., Fang G., van Deursen J.. Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 2005; 37:401–406. [DOI] [PubMed] [Google Scholar]
- 44. Burrows C., Abd Latip N., Lam S.J., Carpenter L., Sawicka K., Tzolovsky G., Gabra H., Bushell M., Glover D.M., Willis A.E., Blagden S.P.. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. 2010; 38:5542–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dalal S.N., Yaffe M.B., DeCaprio J.A.. 14-3-3 family members act coordinately to regulate mitotic progression. Cell Cycle. 2004; 3:670–675. [PubMed] [Google Scholar]
- 46. Douglas M.E., Davies T., Joseph N., Mishima M.. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr. Biol. 2010; 20:927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fu H., Subramanian R.R., Masters S.C.. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000; 40:617–647. [DOI] [PubMed] [Google Scholar]
- 48. Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H., Chang T., Vivekanandan P., Torbenson M., Clark K.R.. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009; 137:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M. et al. . Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saunders W. Centrosomal amplification and spindle multipolarity in cancer cells. 2005; 15:25–32. [DOI] [PubMed] [Google Scholar]
- 51. Hayward D.G., Clarke R.B., Faragher A.J., Pillai M.R., Hagan I.M., Fry A.M.. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004; 64:7370–7376. [DOI] [PubMed] [Google Scholar]
- 52. Pihan G.A., Wallace J., Zhou Y., Doxsey S.J.. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003; 63:1398–1404. [PubMed] [Google Scholar]
- 53. Privette L.M., Gonzalez M.E., Ding L., Kleer C.G., Petty E.M.. Altered expression of the early mitotic checkpoint protein, CHFR, in breast cancers: implications for tumor suppression. Cancer Res. 2007; 67:6064–6074. [DOI] [PubMed] [Google Scholar]
- 54. Zhou Q., Kee Y., Poirier C.C., Jelinek C., Osborne J., Divi S., Surcel A., Will M.E., Eggert U.S., Müller-Taubenberger A.. 14-3-3 coordinates microtubules, Rac, and myosin II to control cell mechanics and cytokinesis. Curr. Biol. 2010; 20:1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ye L., Lin S.T., Mi Y.S., Liu Y., Ma Y., Sun H.M., Peng Z.H., Fan J.W.. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumour Biol. 2016; 37:14585–14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou H., Kuang J., Zhong L., Kuo W., Gray J., Sahin A., Brinkley B., Sen S.. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 1998; 20:189–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.