Abstract
Cockayne syndrome protein B (CSB) belongs to the SWI2/SNF2 ATP-dependent chromatin remodeler family, and CSB is the only ATP-dependent chromatin remodeler essential for transcription-coupled nucleotide excision DNA repair. CSB alone remodels nucleosomes ∼10-fold slower than the ACF remodeling complex. Strikingly, NAP1-like histone chaperones interact with CSB and greatly enhance CSB-mediated chromatin remodeling. While chromatin remodeling by CSB and NAP1-like proteins is crucial for efficient transcription-coupled DNA repair, the mechanism by which NAP1-like proteins enhance chromatin remodeling by CSB remains unknown. Here we studied CSB's DNA-binding and nucleosome-remodeling activities at the single molecule level in real time. We also determined how the NAP1L1 chaperone modulates these activities. We found that CSB interacts with DNA in two principle ways: by simple binding and a more complex association that involves gross DNA distortion. Remarkably, NAP1L1 suppresses both these interactions. Additionally, we demonstrate that nucleosome remodeling by CSB consists of three distinct phases: activation, translocation and pausing, similar to ACF. Importantly, we found that NAP1L1 promotes CSB-mediated remodeling by accelerating both activation and translocation. Additionally, NAP1L1 increases CSB processivity by decreasing the pausing probability during translocation. Our study, therefore, uncovers the different steps of CSB-mediated chromatin remodeling that can be regulated by NAP1L1.
INTRODUCTION
ATP-dependent chromatin remodelers belong to the SWI2/SNF2 ATPase family and are conserved from yeast to humans (1–3). Some of these proteins have demonstrated DNA- and nucleosome-stimulated ATP hydrolysis activities. The energy derived from ATP hydrolysis can be used to alter DNA–histone or other DNA–protein contacts, leading to changes in chromatin structure (3). Therefore, these proteins play critical roles in regulating fundamental nuclear processes, such as transcription and DNA repair (3). Cockayne syndrome complementation group B protein (CSB) is an ATP-dependent chromatin remodeler that is essential for transcription-coupled DNA repair, a process that preferentially removes bulky DNA lesions that stall transcription, such as those created by UV irradiation (4,5). ATP hydrolysis by CSB is essential for its localization to sites of DNA lesion-stalled transcription, the first step of transcription-coupled DNA repair, and chromatin remodeling by CSB is critical for the efficient completion of transcription-coupled DNA repair. CSB also plays critical roles in transcription regulation and base excision DNA repair, as well as other less well-characterized processes (6–9).
In the absence of DNA lesions, CSB interacts dynamically with DNA in cells. Using protein fractionation procedures, it was shown that only ∼10% of CSB remains tightly associated with chromatin, which is in strong contrast to other well studied ATP-dependent chromatin remodelers, such as BRG1 (10). However, upon UV-irradiation, ∼90% of CSB co-fractionates with chromatin, which is consistent with the observation that UV irradiation stabilizes CSB–chromatin interactions at sites of DNA lesion-stalled transcription (10,11). The dynamic CSB–DNA interaction has been proposed to facilitate DNA scanning for DNA lesion-stalled transcription (10,12).
In addition to its DNA and nucleosome stimulated ATP hydrolysis activities, CSB can alter the conformation of the DNA duplex (13,14). In vitro assays have revealed that CSB can actively wrap and unwrap DNA around itself (13). It was suggested that this DNA wrapping and unwrapping activities of CSB may account for CSB's function in altering DNA–protein interactions in cells (12,15,16). Biochemical studies have also reported that CSB can facilitate the annealing and exchange of single-stranded DNA, independent of ATP hydrolysis (17).
CSB has been tested directly for its ability to alter nucleosome structure (14,18). CSB was shown to alter nucleosome structure in an ATP-dependent manner in vitro. However, as revealed by restriction enzyme accessibility assays, CSB remodels nucleosomes with a maximal rate that is 10 times slower than that of the well-characterized human ATP-dependent chromatin remodeling complex ACF (18). Strikingly, in combination with its binding partners, NAP1-like histone chaperones (NAP1L1 or NAP1L4), CSB remodels nucleosomes with a maximal rate similar to that of ACF (18). Most importantly, chromatin remodeling by CSB and NAP1-like histone chaperones is critical for efficient transcription-coupled DNA repair (18). Additionally, the repositioning of nucleosomes in cells by CSB is associated with changes in gene expression. These as well as other observations highlight the significance of ATP-dependent nucleosome remodeling by CSB (8). However, the underlying mechanism by which CSB remodels chromatin and how the NAP1-like histone chaperons enhance the remodeling activity of CSB remain unclear. In this study, we used single-molecule techniques, including Fluorescence Resonance Energy Transfer (FRET) and Protein Induced Fluorescence Enhancement (PIFE) (19) to characterize, in real time, CSB–DNA interactions, chromatin-remodeling by CSB and the mechanisms by which NAP1L1 alters CSB's activities.
MATERIALS AND METHODS
DNA and nucleosome preparation
All DNA fragments used in this study were 228 bp in length and contained the 601 nucleosome positioning sequence (Supplementary Figure S1). DNA fragments were generated by PCR and purified from a 5% polyacrylamide gel (20). Mononucleosomes were designed to have a 78-bp spacer on the entry side and a 3-bp spacer on the exit side. The internal-labeled Cy3 DNA was generated using a primer with a Cy3 label at the 76th base and a biotin at the 5΄ end. The end-labeled Cy3 DNA was generated using a primer with a Cy3 label at the 5΄ end and a second primer containing biotin at the 5΄ end. The DNA fragment used to examine DNA distortion by CSB contained a FRET pair, prepared using one primer labeled with Cy3 at the 5΄ end, and a second primer labeled with Cy5 at the 76th base and a biotin at the 5΄ end. The singly labeled DNA fragment used to examine the nucleosome remodeling activity of CSB was generated using a primer end-labeled with Cy5 at the 5΄ end and a second primer containing biotin at the 5΄ end. Mononucleosomes containing Cy3-labeled H2A were prepared as described (21). Mononucleosomes were reconstituted using salt gradient dialysis (21). DNA sequence is shown in Supplementary Figure S1.
Protein expression, purification and labeling
N-terminally Flag-tagged CSB was expressed in SF9 cells using the baculovirus overexpression system, as described previously (18). N-terminally Flag-tagged NAP1L1 was cloned into pET22b (Novagen) and overexpressed in Rosetta 2(DE3) cells (Novagen). Both proteins were purified using anti-Flag affinity chromatography as previously described (22). To prepare Cy3-labeled CSB, purified CSB and Cy3 maleimide (GE healthcare) were mixed at a 1:3 molar ratio and incubated at 4°C for 4.5 h. Unbound Cy3 was removed using Amicon centrifugal filters with a MWCO of 3kDa (Millipore). After free Cy3 removal, the molar ratio of CSB to Cy3 was determined to be about 1:1, indicating the molar degree of the labeling efficiency was ∼100%.
Single molecule fluorescence experiments
Quartz slides and coverslips were cleaned with piranha solution (mixture of 3:1 concentrated sulfuric acid:30% [v/v] hydrogen peroxide solution), siliconized and coated with PEG (m-PEG-5000, Laysan Bio) and biotin-PEG (biotin-PEG-5000, Laysan Bio). A flow cell was assembled by combining a cover slip and a quartz slide with double-sided adhesive tape (3M). For convenient buffer exchange, polyethylene tubes (PE50; Becton Dickinson) were connected to the flow cell. DNA substrates were immobilized on a PEGylated surface using biotin–streptavidin conjugation. Single-molecule experiments were performed in an imaging buffer (12 mM HEPES pH 7.9, 10 mM Tris–HCl pH 7.5, 60 mM KCl, 4 mM MgCl2, 3% glycerol) containing a gloxy oxygen scavenging system (a mixture of glucose oxidase (1 mg/ml, Sigma), catalase (0.04 mg/ml, Sigma), glucose (0.4% (w/v), Sigma), and Trolox (2 mM, Sigma)). Cy3 and Cy5 were alternatively excited with 532 and 633 nm lasers using the ALEX (Alternative Laser EXcitation) technique (23,24), and single-molecule fluorescence images were synchronously taken with the excitation laser switching at a frame rate of 10 Hz using a home-built prism-type total internal reflection fluorescence microscope equipped with an electron-multiplying charge coupled device camera (Ixon DV897; Andor Technology). Experimental temperature was maintained at 30°C using a temperature control system (Live Cell Instruments).
Analyses of single-molecule FRET and single-molecule PIFE data
For experiments using doubly labeled DNA, only molecules with both Cy3 and Cy5 were selected for data analysis, which was facilitated by the ALEX technique (23,24). The intensity traces of Cy3 and Cy5 were analyzed using a custom made Matlab (MathWorks) script. The FRET efficiencies were calculated from the donor (ID) and acceptor (IA) fluorescence intensities as EFRET = IA/(IA + ID).
To develop single-molecule PIFE histograms, individual traces were normalized to the protein-unbound intensity, which is the averaged intensity during the first 20 s. Changes in PIFE were obtained by collecting data points of the entire trace (∼2 min) after CSB or CSB/NAP1 injection. If photobleaching occurred during observation, data points after photobleaching were removed from the analysis. In general, photobleaching often occurs after ∼300 s, which is much longer than the time interval of our PIFE data collection (start at 20 s – stop at ∼ 120 s). The DNA distortion is defined as FRET efficiency greater than 0.25. DNA-distortion rate constants were calculated as 1/Tnd. Tnd is the time in the non-distorted state, which is determined by fitting histograms of cumulative dwell-time in the non-distorted state to single-exponential decay functions. DNA-distortion reversal rate constants were calculated as 1/Td. Td is the time in the distorted states, which is determined by fitting histograms of cumulative dwell-time in the distorted state to single-exponential decay functions.
RESULTS
CSB interacts with DNA in two principle ways
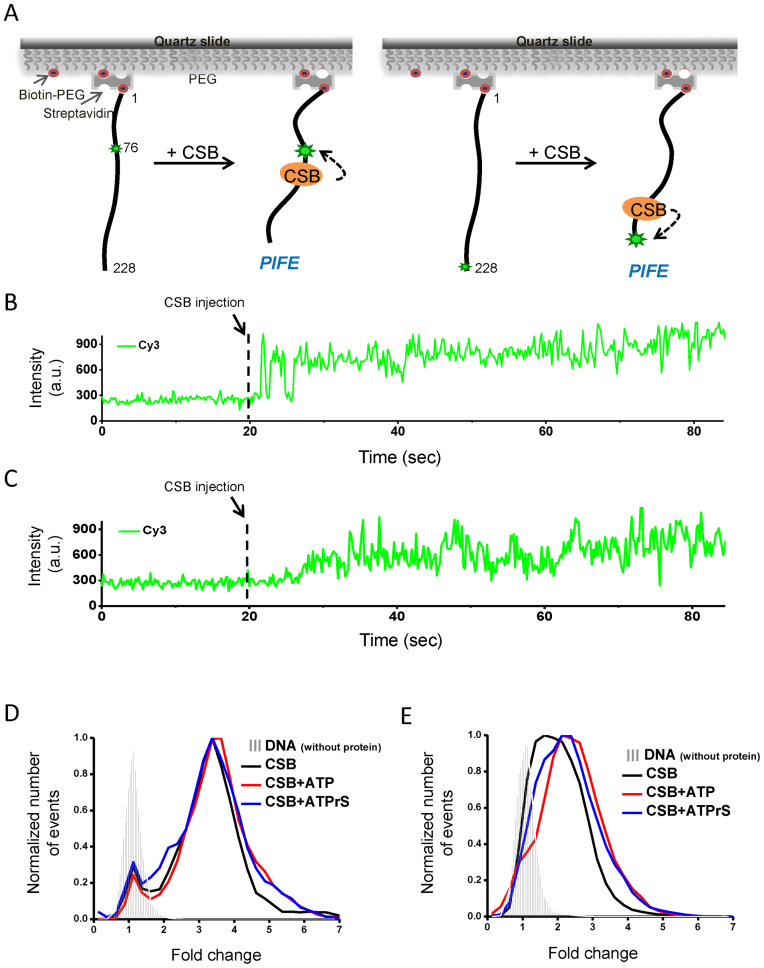
To monitor CSB–DNA interactions at the singe-molecule level, we employed PIFE assays (19). A 228-bp DNA fragment singly labeled with Cy3 at positions 76 or 228 (Supplementary Figure S1) was immobilized on a polyethylene glycol coated (PEGylated) quartz surface (Figure 1A). Purified CSB (Supplementary Figure S2) was injected into the detection chamber, and single-molecule fluorescence imaging was performed in real time. As soon as CSB was injected, a nearly instantaneous increase in the Cy3 signal was detected, a phenomenon called PIFE (Figure 1B and C) (19). PIFE was observed for almost 100% of the traces when CSB was mixed with either internal-labeled or end-labeled DNA substrates, revealing that CSB binds to internal and end positions equally well. Although the amplitude of the PIFE signals generated by internal binding were slightly greater than end binding, this was likely due to the different labeling methods: Cy3 was attached to the 5΄ phosphate at the end position and Cy3 was attached to the base at the internal site.
Figure 1.
CSB–DNA interactions revealed by single-molecule PIFE. (A) Experimental scheme. A 228-bp, biotinylated DNA fragment labeled with Cy3 either at the internal nucleotide position 76 or at the end position 228 was immobilized to biotinylated PEG using streptavidin (small green stars). The binding of CSB in the vicinity of the Cy3 fluorophore can lead to PIFE (large green stars). (B and C) Binding reactions were initiated by injecting 33 nM CSB into the detection chamber at 20 s (dashed line). (B) Shown is a representative time trace using internal-labeled Cy3. (C) Same as in (B) except the DNA is end-labeled with Cy3. (D and E) PIFE distributions in the presence of ATP and ATPγS. CSB was used at 33 nM. ATP and ATPγS were used at 1 mM. Data for the histograms were generated from the cumulative Cy3 signals over the entire observation time normalized to the average Cy3 intensity of the first 20 s, before protein injection. Small changes in PIFE observed in the absence of protein (gray strips) were due to buffer exchange. (D) Internal-Cy3 DNA substrate and (E) end-Cy3 DNA substrate.
We next determined if ATP binding or hydrolysis altered CSB-induced PIFE. Changes in PIFE were determined by first normalizing fluorescence intensity after protein injection to the average intensity of the first 20 s before injection. We then generated PIFE histograms using all data points of the entire trace after protein injection (Figure 1D and E). At a saturating concentration (1 mM) of ATP or ATPγS (a non-hydrolyzable ATP analog), the PIFE distributions appeared similar to that of CSB alone (Figure 1D and E), indicating that the interaction between CSB and DNA revealed by PIFE is largely ATP-independent.
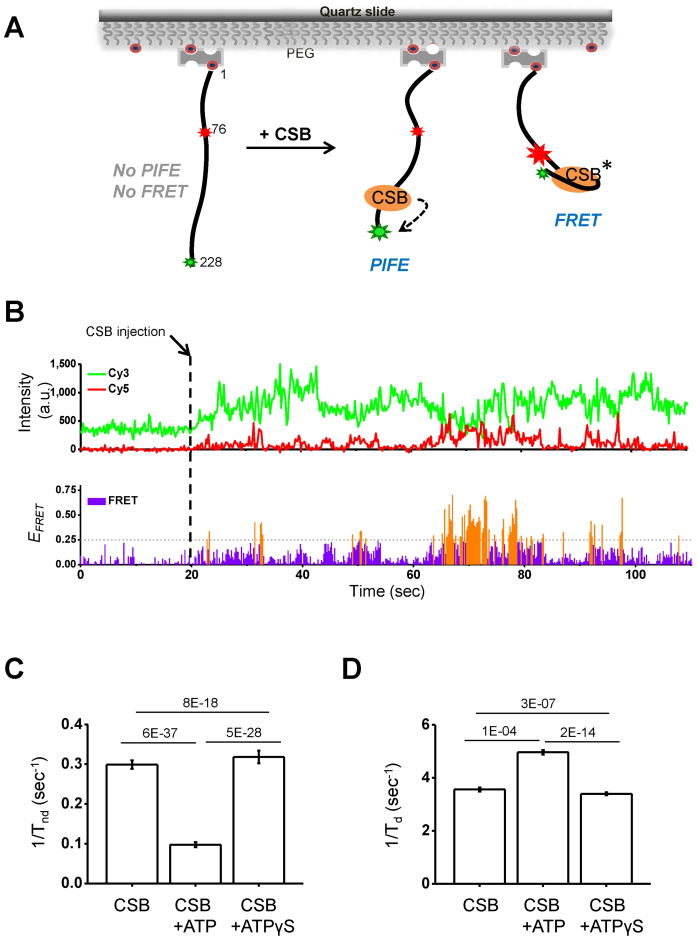
CSB has been suggested to actively wrap and unwrap DNA, and this activity was suggested to alter DNA–protein interactions (13). To test if CSB could alter DNA conformation, we employed single molecule FRET assays (25), using a 228-bp DNA fragment that contained a Cy3–Cy5 FRET pair located at positions 228 and 76, respectively (Figure 2A and Supplementary Figure S1). Shown in Figure 2B are examples of the fluorescence traces. Individual Cy3 and Cy5 signal intensities are shown in the top trace, and FRET signals are shown in the lower trace. We detected frequent FRET signals after adding CSB, and Cy3-PIFE was again seen for almost 100% of the traces. This increase in FRET after CSB addition indicates a shortening of the distance between the Cy3 and Cy5 fluorophores resulting from CSB–DNA interactions. We refer to this increase in FRET as CSB-induced gross DNA distortion. Together, our results indicate at least two types of CSB–DNA interactions occur in the absence of ATP: a rapid binding of CSB to DNA, as revealed by PIFE, and occasional gross DNA distortion, as revealed by FRET.
Figure 2.
CSB–DNA interactions revealed by single molecule PIFE and FRET. (A) Experimental scheme. A 228-bp, biotinylated DNA fragment labeled with a FRET donor (Cy3, small green star) and acceptor (Cy5, small red star) was immobilized to biotinylated PEG using streptavidin. The binding of CSB in the vicinity of the Cy3 fluorophore can lead to PIFE (large green star). Additionally, if the interaction of CSB with DNA brings the Cy3 and Cy5 fluorophores close together, FRET will be detected (large red star). The asterisk indicates that the number of CSB molecules needed to induce FRET is unknown. (B) Representative time traces. Binding reactions were initiated by adding 33 nM CSB into the detection chamber at 20 s (dashed line). PIFE and FRET signals were observed simultaneously. (Top) Fluorescence intensities of Cy3 (green, ICy3) and Cy5 (red, ICy5). An increase in ICy3 after protein injection is the result of PIFE. An increase in ICy5 is the result of FRET. (Bottom) The sum of ICy3 and ICy5 was normalized to the average of the sum before CSB injection. FRET efficiencies (EFRET) = ICy5/(ICy3 + ICy5,). The threshold for a positive FRET event was set at 0.25 (highlighted in orange). (C and D) Effects of ATP on CSB–DNA interactions. CSB was used at 33 nM. ATP and ATPγS were used at 1 mM. DNA distortion and distortion reversal rate constants were derived from data shown in Supplementary Figure S3. (C) DNA-distortion rate constants were calculated as 1/Tnd, where Tnd = duration in the non-distorted state. (D) Distortion reversal rate constants were calculated as 1/Td, where Td = duration in the distorted state. Error bars represent curve fitting errors. One-sample t-tests with equal variance were used to calculate P-values, as shown above the plots.
We then sought to determine the rate at which the gross DNA distortion and DNA distortion reversal occurred, and whether or not these processes were ATP dependent. We first calculated the cumulative time each DNA molecule remained in the non-distorted state during the entire observation time (Tnd). DNA-distortion rate constants were then calculated as 1/Tnd. As shown in Figure 2C and Supplementary Figure S3, the inclusion of ATP in the reaction decreased the rate of DNA distortion by CSB, whereas ATPγS had little or no effect. We also determined the rate constant for gross DNA distortion reversal, which was calculated as 1/Td, where Td represents the duration in the distorted state. As shown in Figure 2D and Supplementary Figure S3, ATP and ATPγS had little or no effect on the rate of DNA distortion reversal. Together, these results indicate that ATP hydrolysis by CSB reduces the propensity of CSB to distort DNA.
NAP1L1 reduces CSB–DNA interactions and leads to more ordered CSB–DNA binding and release events
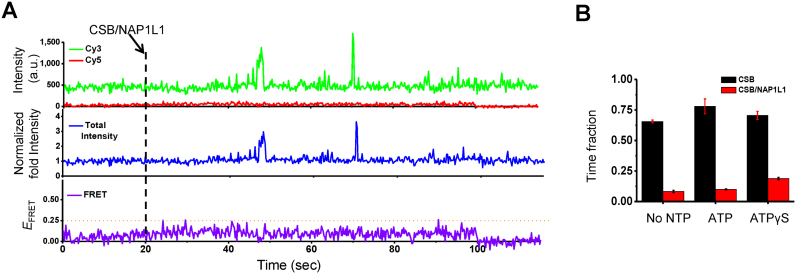
We next studied the effects of NAP1L1 on CSB–DNA interactions. For these experiments, CSB and NAP1L1 were mixed at a molar ratio of 1 to 2, respectively, for 2 min before being injected into the detection chamber, which already contained DNA dual-labeled with Cy3 and Cy5. In strong contrast to CSB alone (Figure 2B), CSB and NAP1L1 together induced rare and brief PIFE and no FRET during the first 100 seconds after protein injection (Figure 3A). Notably, PIFE induced by CSB/NAP1L1 displayed well-defined borders, likely indicating better-defined DNA-binding and release events, in strong contrast to CSB alone. In addition, NAP1L1 alone did not generate PIFE signals (data not shown). Moreover, in contrast to CSB alone, which showed PIFE for ∼100% of the traces, in the presence of NAP1L1, the number of traces that showed any PIFE within a 2-min observation window decreased to 50–60%, indicating that NAP1L1 decreased the interaction of CSB with naked DNA. To determine the fraction of time that CSB or CSB/NAP1L1 remained near the fluorophores, we calculated the cumulative time for signals showing PIFE >2.0 of normalized intensity divided by the total observation time (∼2 min). As shown in Figure 3B, the presence of NAP1L1 shortened the time fraction CSB stayed near the fluorophore by ∼80%, suggesting that NAP1L1 also promotes the dissociation of CSB from DNA. Neither ATP nor ATPγS affected the activity of NAP1L1 on the CSB–DNA interaction (Figure 3B). Similar effects were observed when singly labeled DNA substrates were used (Supplementary Figure S4).
Figure 3.
Effect of NAP1L1 on CSB–DNA interactions. (A) Representative time trace showing the interaction of CSB with DNA in the presence of NAP1L1. 33 nM of CSB was incubated with 66 nM of NAP1L1 in imaging buffer for 2 minute and then injected into the detection chamber at 20 s, as indicated by the dashed line. Only rare and transient increases in PIFE and no changes in FRET values were observed for the first 100 seconds after CSB and NAP1L1 addition. (B) The fraction of time that CSB or CSB/NAP1L1 was in the DNA-bound state (defined as >2-fold normalized PIFE intensity, which is the sum of Cy3 and Cy5 intensities for the first 100 s after protein injection divided by the average intensity for the first 20 s before protein injection). ATP and ATPγS were used at 1 mM. Error bars represent standard deviation from two experiments and each with 60 molecules.
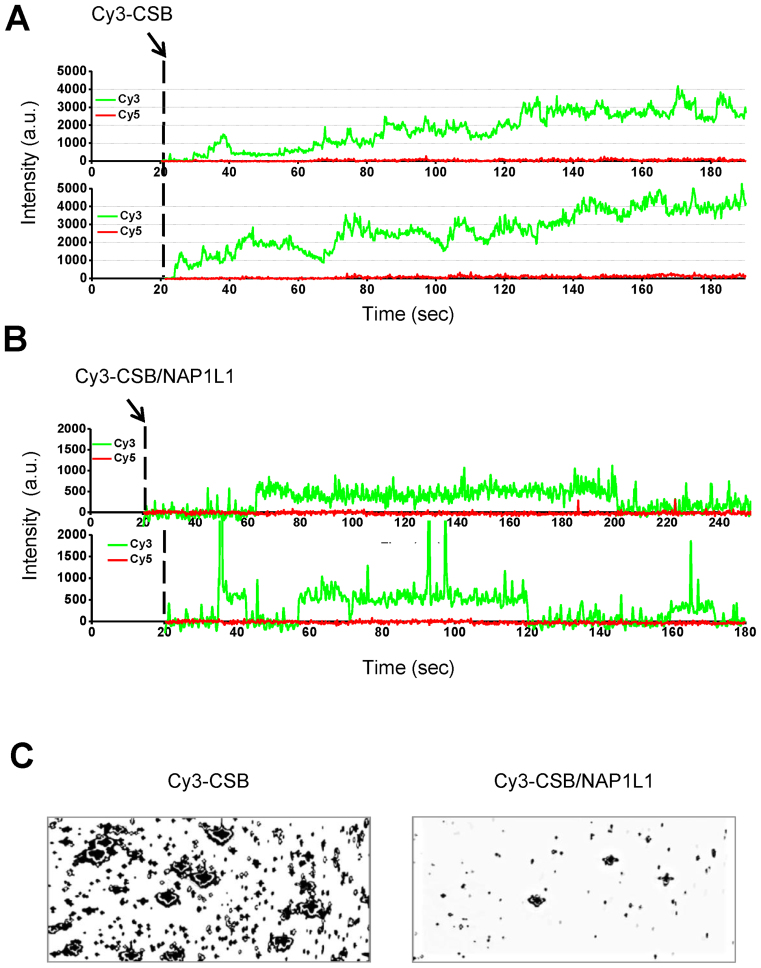
To gain more insight into the nature of the CSB–DNA interaction and how NAP1L1 alters that interaction, we directly monitored CSB–DNA interactions in real time, by imaging fluorescence signals after injecting Cy3-labeled CSB into the detection chamber, which already contained Cy5 end-labeled DNA (Figure 4). Due to high background, the experiments were performed at a relatively low CSB concentration (<10 nM). After CSB injection, we observed a gradual increase in the Cy3 signal, indicating that multiple CSB molecules associated with the DNA fragment over time (Figure 4A). To understand if NAP1L1 alters this behavior, we monitored DNA binding of Cy3-labeled CSB in the presence of NAP1L1. As shown in Figure 4B, the Cy3 signal in the presence of NAP1L1 was, in general, lower and more uniform in intensity than in the absence of NAP1L1, and a gradual increase in the Cy3 signal was not observed. Furthermore, in the presence of NAP1L1, the Cy3 signals again displayed better-defined borders (bound versus non-bound states).
Figure 4.
Direct visualization of CSB–DNA complexes. A 228-bp DNA fragment was end-labeled with Cy5 and immobilized to biotinylated PEG using streptavidin. Cy3-labeled CSB was then injected into the detection chamber and binding events were imaged. (A) Representative time traces of Cy3 fluorescence intensity after injection of Cy3-labeled CSB at 20 s (dashed line). In the experimental condition, the fluorescence intensity of a single Cy3 dye was about 250–300 a.u. (B) Representative time traces of Cy3 fluorescence intensity after injection of Cy3-labeled CSB and unlabeled NAP1L1 at 20 s (dashed line). (C) Fluorescence images of Cy3–CSB bound to DNA in the absence (left) or presence (right) of NAP1L1. Unbound proteins were washed out of the detection chamber before imaging.
To capture fluorescence images of Cy3–CSB bound to DNA in the presence or absence of NAP1L1, we incubated Cy3–CSB or Cy3-CSB and NAP1L1 with DNA for 2 min and then removed unbound proteins from the detection chamber. When DNA was incubated with Cy3–CSB alone, spots of high Cy3 intensity were observed, likely representing multiple CSB molecules binding to DNA (Figure 4C, left). However, when DNA was incubated with both Cy3–CSB and NAP1L1, the number of spots with high Cy3-intensity was substantially reduced (Figure 4C, right). These results suggest that CSB may have a tendency to multimerize on a DNA fragment, and that NAP1L1 decreases the tendency for this to happen.
Nucleosome remodeling by CSB contains an activation phase
We next used FRET to study the nucleosome remodeling activity of CSB at single-molecule resolution. To accomplish this, we applied the experimental design developed for studying the human remodeling complex ACF (26). For these assays, the nucleosomal core DNA contained the 601 positioning sequence, and this was flanked by a 78-bp spacer on the entry side and a 3-bp spacer on the exit side. For FRET measurements, histone H2A was labeled with Cy3 and the DNA was labeled with Cy5 on the exit side (Figure 5A). Because there are two copies of histone H2A in a nucleosome, heterogeneous populations of Cy3 labeling are expected. For data analysis, we only used nucleosomes that contained a single Cy3 on the H2A proximal to the Cy5 on the exit side of the DNA, which exhibited a FRET efficiency of ∼0.73 before remodeling (Supplementary Figure S5). The labeled nucleosome and DNA did not compromise the remodeling activity of CSB, as reveled by native polyacrylamide gel electrophoresis (Supplementary Figure S6).
Figure 5.
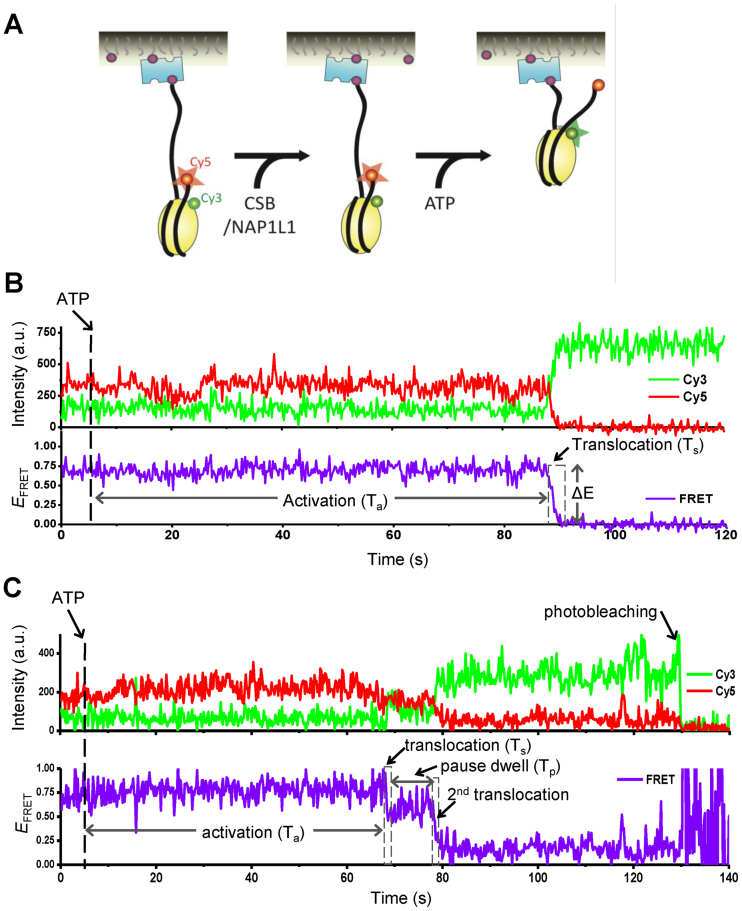
Nucleosome remodeling by CSB and NAP1L1 revealed by single-molecule FRET. (A) Single molecule measurement scheme for nucleosome remodeling by CSB and NAP1L1. The nucleosome had a 3 bp linker on the exit side and a 78 bp linker on the entry side. The nucleosome was labeled with a FRET donor (Cy3, green) on histone H2A (residue 120) and a FRET acceptor (Cy5, red) on the exit side of the DNA. FRET is observed when the donor and acceptor are close together (large red star). An increase in the distance between the FRET probes will result in a decrease in FRET intensity (small red circle). (B and C) Representative fluorescence intensity and corresponding FRET traces displaying nucleosome remodeling by 75 nM CSB and 300 nM NAP1L1. ATP (1 mM) was added into the detection chamber at 5 seconds (dashed line). (B) Remodeling without a pause. Activation time (Ta) indicates the time lapse between ATP injection and the first decrease in the FRET efficiency. The translocation time (Ts) indicates the duration of the first decrease in FRET efficiency (dashed box). (C) Remodeling with a pause. The pause dwell time (Tp) indicates the lifetime of the lower FRET state after the first decrease in FRET efficiency. Ta and Ts are defined as in (B).
For the single-molecule FRET experiments, Cy3/Cy5-labeled mononucleosomes were immobilized on a PEGylated quartz surface and then incubated with CSB or CSB plus NAP1L1 for 2 minutes. The remodeling reaction was initiated by injecting imaging buffer containing ATP into the detection chamber (Figure 5A). Representative FRET traces displaying nucleosome remodeling are shown in Figure 5B and C. As shown in Figure 5B, decreased FRET efficiency was observed after the addition of ATP, indicating an increase in distance between the Cy3 and Cy5 fluorophores. Interestingly, the decrease in FRET efficiency did not occur immediately but with a time delay (Ta) after ATP injection, suggesting that nucleosome remodeling by CSB and CSB/NAP1L1 requires an activation process. After the initiation of nucleosome remodeling, the FRET signal decreased to background levels in seconds (Ts). Occasionally, the decrease occurred in two or more phases that were separated by a long pause (Tp), as observed by an intermediate FRET signal (Figure 5C). Changes in FRET were not observed using the non-hydrolyzable ATP analog ATPγS, indicating that the events were dependent upon ATP-hydrolysis by CSB (data no shown). The decrease in scored FRET events were always accompanied by increased Cy3 signals, arguing against the possibility that the FRET decrease was due to loss of histone H2A–H2B or photobleaching. The decrease in FRET, as shown in Figure 5, could be due to DNA ‘peeling’ off the exit site of the nucleosome. To demonstrate further that CSB can catalyze the translocation of DNA around the histone octamer, we moved the Cy5 probe from the exit site of the DNA to a position near the entry site of the nucleosome (position 68) (Supplementary Figure S7A), as described previously (26,27). If DNA were simply peeled away from the nucleosome at the entry site, then we expect to detect a decrease in FRET signals after adding CSB and ATP. However, when mixing CSB with this nucleosomal substrate, we observed increased FRET signals (Supplementary Figure S7B, arrows), supporting the notion that CSB can change the translational positions of a nucleosome. Furthermore, we observed oscillation of the FRET signal (e.g. Supplementary Figure S7B, lower panel, between 70 and 80 s), likely representing CSB-induced bi-directional movement of the histone octamer on the DNA.
NAP1L1 alters multiple steps of CSB-mediated nucleosome remodeling
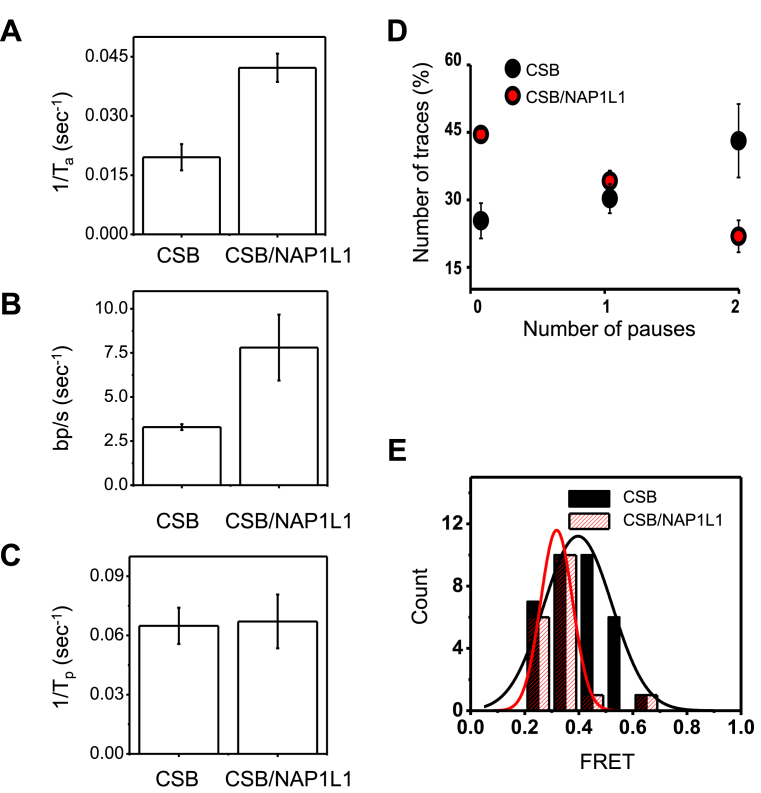
To determine if NAP1L1 alters the nucleosome remodeling activity of CSB, we quantified the activation rate constant, the translocation rate constant, the pause time, the pause frequency and the distribution of FRET values during the first nucleosome translocation event in the presence of a saturating amount of ATP (1 mM) (Figure 6 and Supplementary Figure S8). Of note, no change in FRET was observed when NAP1L1 was incubated with nucleosomes in the presence of ATP (data not shown). Determination of the translocation rate constant was based on the calibration data shown in Supplementary Figure S9 (26). It has been shown that FRET is sensitive enough to detect movement as small as 1 bp (27). Excitingly, we found that NAP1L1 altered multiple steps of CSB-mediated nucleosome remodeling. First, NAP1L1 significantly increased the activation rate as well as the translocation rate of CSB (Figure 6A and B). Second, NAP1L1 did not change the duration of the translocation pauses (Figure 6C) but decreased the frequency of pauses during CSB remodeling (Figure 6D). We also examined the FRET distributions at the end of the first translocation event before pausing. After plotting the data as a histogram and fitting the data to a Gaussian distribution, we found that the peak FRET value, which was 0.73 before remodeling, dropped to ∼0.4 for CSB and ∼0.3 for CSB/NAP1L1 after remodeling (Figure 6E). These FRET values correspond to 4.7-bp and 5.9-bp translocation steps, respectively, indicating that, in the presence of NAP1L1, CSB can move the histone octamer along DNA further, before reaching the first paused state. This observation supports the notion that NAP1L1 decreases pausing during nucleosome remodeling by CSB. Furthermore, in the presence of NAP1L1, CSB displayed a narrower distribution of FRET values with a width of 2-bp, as compared to 4.5-bp for CSB alone, indicating that CSB generates more homogenous remodeled products in the presence of NAP1L1, consistent with results obtained from previous population-based remodeling assays (18).
Figure 6.
Comparison of chromatin remodeling by CSB in the presence and absence of NAP1L1. (A and B) Quantification of nucleosome remodeling parameters using 75 nM CSB alone or in the presence of 300 nM NAP1L1 and 1 mM ATP. The rates were obtained from the data shown in Supplementary Figures S8. Error bars indicate curve-fitting errors. (A) Rate constants of activation (1/Ta, where Ta is the time required for the first FRET change). One-sample t-tests with equal variance yielded a P-value of 0.004. (B) Translocation rate. To obtain the translocation rate, calibration data in Supplementary Figure S9 were used to convert the FRET change (ΔE) for the first remodeling event to the translocation length, which was then divided by the time duration of the FRET change (Ts). One-sample t-tests with equal variance yielded a P-value of 0.003. (C) The pause dwell time (Tp), which indicates the lifetime of the lower FRET state after the first translocation. One-sample t-tests with equal variance yielded a P-value of 0.6. (D) Pause frequency. Shown are means ± SEM (n = 58 for CSB and n = 61 for CSB/NAP1L1). (E) The distribution of FRET values during the first nucleosome translocation. The data were fit to a single Gaussian function. The resulting center FRET values for CSB and CSB/NAP1 were determined to be 0.4 and 0.3 with standard deviations of 0.32 and 0.14, respectively.
DISCUSSION
The results of this study provide new insights into how CSB interacts with DNA and remodels nucleosomes, and how NAP1L1 alters these processes. Using single-molecule fluorescence approaches, we found that CSB readily binds DNA independently of ATP and without preference for DNA ends or internal sites (Figures 1, 2 and 4). Strikingly, the CSB-interacting histone chaperone, NAP1L1, decreases the binding of CSB to DNA and also promotes the dissociation of DNA-bound CSB (Figures 3 and 4, Supplementary Figure S4). Previously, we demonstrated that the association of CSB with chromatin is dynamic during replicative cell growth and proposed that this dynamic association allows CSB to efficiently scan chromatin for DNA lesion-stalled transcription (10). The results presented here suggest that NAP1L1 may function, in part, to help maintain dynamic CSB–DNA interactions to promote chromatin scanning for its preferred substrates, such as DNA-lesion stalled transcription.
Occasionally, the binding of CSB to DNA leads to a gross distortion of the DNA (Figure 2). Interestingly, unlike the simple DNA interaction described above, this CSB-induced DNA distortion was decreased through ATP hydrolysis by CSB (Figure 2B), which was similar to results obtained from studying CSB–DNA association using scanning force microscopy. In that study, Beerens et al. (13) observed a shortening of contour length of circular DNA upon CSB binding, presumably due to DNA wrapping, and ATP hydrolysis by CSB promoted DNA unwrapping (13). It was suggested that this DNA wrapping and unwrapping activities of CSB may account for CSB's function in altering DNA–protein interaction in cells (13). By directly visualizing the interaction of Cy3-labeled CSB with DNA, we found that multiple CSB molecules can occupy a single DNA fragment and that NAP1L1 decreases such occupancy (Figure 4). We do not yet know if this multimerization is promoted by direct CSB-CSB interactions. Whether or not the DNA distortion that we observed is dependent upon the binding of more than one CSB molecule remains to be determined.
Our single-molecule assays revealed that nucleosome remodeling by CSB, like the human ACF remodeling complex, contains three distinct steps: activation, translocation and translocation pausing (26). Furthermore, preliminary observations indicate that CSB can diffuse along DNA, and that ATP may enhance this activity two-fold (Jong-Bong Lee, personal communication). This ATP-dependent DNA translocase activity of CSB may underlie its nucleosome remodeling activity, as has been proposed for the ISWI remodeler (28), or may be critical to other CSB functions that await to be determined. The activation step and translocation pause are two critical rate-determining steps for nucleosome remodeling by CSB, as translocation without pausing occurs rapidly. Our data indicate that CSB appears to have a translocation rate of ∼2 bp/s, similar to that of ACF (Figure 6B, D and E) (26). However, one major difference is that ACF is much more processive than CSB, as the translocation pauses by ACF were only observed at sub-saturating ATP concentrations, while the translocation pauses that we observed for CSB occurred in the presence of a saturating ATP concentration (1 mM). This difference may reflect fundamental differences in biological functions of CSB and ACF. For example, CSB may only need to move nucleosomes over short distances for efficient DNA repair and transcription regulation, while ACF may need to move nucleosomes over a greater distance for efficient nucleosome spacing. Indeed, we have demonstrated that CSB can reposition nucleosomes locally, near its binding site in cells, as revealed by ChIP-qPCR and MNase-qPCR. Furthermore, these changes in nucleosome position correlate well with changes in gene expression (8). Moreover, chromatin remodeling by CSB is required for step(s) subsequent to DNA lesion recognition (18). This activity may be important for creating an epigenetic landscape for more efficient DNA repair and/or transcription resumption after repair; such alterations could involve DNA interactions with both histone and non-histone protein, such as RNA polymerase II.
Previously, we showed that NAP1L1 interacts with CSB and enhances the rate at which CSB exposes a nucleosomal site by 10-fold, using restriction enzyme accessibility assays (18). Our single molecule experiments revealed that NAP1L1 impacts nucleosome remodeling by CSB at multiple steps of the remodeling process, increasing the rate of activation and translocation by about 2-fold (Figure 6A and B). Significantly, NAP1L1 also makes CSB more processive by decreasing the frequency of translocation pauses (Figure 6D and E). In an earlier study, using bulk nucleosome remodeling assays, we found that CSB generated multiple remodeled products containing histone octamers at different translational positions and that NAP1L1 facilitated CSB to generate more homogenous remodeled products (18), a phenomenon we also observed here using single molecule approaches (Figure 6E).
We would like to propose that NAP1L1 enhances chromatin remodeling by CSB, in part, by decreasing non-productive chromatin associations. This hypothesis is born out of our observation (Figures 3 and 4) that the association of CSB with DNA, as revealed by PIFE and FRET, is largely relieved by NAP1L1 (Figures 3 and 4). By decreasing non-productive chromatin associations, the NAP1 chaperones would allow CSB to sample chromatin more efficiently to find its preferred substrates, such as DNA-lesion stalled transcription. The non-productive association of CSB with DNA might also create a barrier that could impede nucleosome remodeling. The yeast Nap1 protein has been shown to play a similar role with histone proteins, countering non-productive histone–DNA interactions during nucleosome assembly (29). This is accomplished by reversing interactions between H2A–H2B dimers and DNA (29). In addition to the role of human NAP1-like proteins in regulating histone dynamics, human NAP1 also regulate the binding of the CENP-B protein to its cognate CENP-B box in DNA by preventing the nonspecific binding of CENP-B to nucleosomes (30). In this context, our results indicate that NAP1-like proteins can interact with a chromatin remodeler to decrease non-productive DNA and nucleosome associations as well.
Together, our study provides novel insights into how CSB interacts with DNA and remodels nucleosomes, and how NAP1L1 alters these processes to facilitate nucleosome remodeling. It remains to be determined at mechanistic levels how NAP1L1 shortens the activation step, enhances the translocation step and decreases pausing, and how CSB uses its specific nucleosome remodeling strategy to efficiently carry out its biological functions in DNA repair and transcription, as well as other processes in which it has been implicated to function.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Prof. Ji-Joon Song at KAIST for providing histones.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Creative Research Initiative program [2009-0081562 to S.H.]; National Institutes of Health [GM115888 to H.Y.F.]. Funding for open access charge: National Research Foundation of Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1. Flaus A., Martin D.M., Barton G.J., Owen-Hughes T.. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006; 34:2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hopfner K.P., Gerhold C.B., Lakomek K., Wollmann P.. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr. Opin. Struct. Biol. 2012; 22:225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 492:273–304. [DOI] [PubMed] [Google Scholar]
- 4. Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J.H.. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992; 71:939–953. [DOI] [PubMed] [Google Scholar]
- 5. Lake R.J., Fan H.Y.. Structure, function and regulation of CSB: a multi-talented gymnast. Mech. Ageing Dev. 2013; 78:202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balajee A.S., May A., Dianov G.L., Friedberg E.C., Bohr V.A.. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:4306–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Newman J.C., Bailey A.D., Weiner A.M.. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lake R.J., Boetefuer E.L., Tsai P.F., Jeong J., Choi I., Won K.J., Fan H.Y.. The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure. PLoS Genet. 2014; 10:e1004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stevnsner T., Muftuoglu M., Aamann M.D., Bohr V.A.. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 2008; 129:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lake R.J., Geyko A., Hemashettar G., Zhao Y., Fan H.Y.. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell. 2010; 37:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van den Boom V., Citterio E., Hoogstraten D., Zotter A., Egly J.M., van Cappellen W.A., Hoeijmakers J.H., Houtsmuller A.B., Vermeulen W.. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J. Cell Biol. 2004; 166:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lake R.J., Basheer A., Fan H.Y.. Reciprocally regulated chromatin association of Cockayne syndrome protein B and p53 protein. J. Biol. Chem. 2011; 286:34951–34958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beerens N., Hoeijmakers J.H., Kanaar R., Vermeulen W., Wyman C.. The CSB protein actively wraps DNA. J. Biol. Chem. 2005; 280:4722–4729. [DOI] [PubMed] [Google Scholar]
- 14. Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R.E., Hoeijmakers J.H., Vermeulen W.. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000; 20:7643–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berquist B.R., Canugovi C., Sykora P., Wilson D.M. 3rd, Bohr V.A.. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 2012; 40:8392–8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lake R.J., Boetefuer E.L., Won K.-J., Fan H.-Y.. The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res. 2016; 44:2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muftuoglu M., Sharma S., Thorslund T., Stevnsner T., Soerensen M.M., Brosh R.M. Jr, Bohr V.A.. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 2006; 34:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho I., Tsai P.F., Lake R.J., Basheer A., Fan H.Y.. ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 2013; 9:e1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hwang H., Myong S.. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem. Soc. Rev. 2014; 43:1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jung J., Han K.Y., Koh H.R., Lee J., Choi Y.M., Kim C., Kim S.K.. Effect of single-base mutation on activity and folding of 10-23 deoxyribozyme studied by three-color single-molecule ALEX FRET. J. Phys. Chem. B. 2012; 116:3007–3012. [DOI] [PubMed] [Google Scholar]
- 21. Lee S., Hohng S.. An optical trap combined with three-color FRET. J. Am. Chem. Soc. 2013; 135:18260–18263. [DOI] [PubMed] [Google Scholar]
- 22. Fan H.Y., He X., Kingston R.E., Narlikar G.J.. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell. 2003; 11:1311–1322. [DOI] [PubMed] [Google Scholar]
- 23. Kapanidis A.N., Laurence T.A., Lee N.K., Margeat E., Kong X., Weiss S.. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005; 38:523–533. [DOI] [PubMed] [Google Scholar]
- 24. Lee J., Lee S., Ragunathan K., Joo C., Ha T., Hohng S.. Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl. 2010; 49:9922–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy R., Hohng S., Ha T.. A practical guide to single-molecule FRET. Nat. Methods. 2008; 5:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blosser T.R., Yang J.G., Stone M.D., Narlikar G.J., Zhuang X.. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009; 462:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deindl S., Hwang W.L., Hota S.K., Blosser T.R., Prasad P., Bartholomew B., Zhuang X.. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell. 2013; 152:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Whitehouse I., Stockdale C., Flaus A., Szczelkun M.D., Owen-Hughes T.. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 2003; 23:1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andrews A.J., Chen X., Zevin A., Stargell L.A., Luger K.. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell. 2010; 37:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tachiwana H., Miya Y., Shono N., Ohzeki J., Osakabe A., Otake K., Larionov V., Earnshaw W.C., Kimura H., Masumoto H. et al. Nap1 regulates proper CENP-B binding to nucleosomes. Nucleic Acids Res. 2013; 41:2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.