Abstract
Trinucleotide repeats are a source of genome instability, causing replication fork stalling, chromosome fragility, and impaired repair. Specialized helicases play an important role in unwinding DNA structures to maintain genome stability. The Srs2 helicase unwinds DNA hairpins, facilitates replication, and prevents repeat instability and fragility. However, since Srs2 is a multifunctional protein with helicase activity and the ability to displace Rad51 recombinase, it was unclear which functions were required for its various protective roles. Here, using SRS2 separation-of-function alleles, we show that in the absence of Srs2 recruitment to PCNA or in helicase-deficient mutants, breakage at a CAG/CTG repeat increases. We conclude that Srs2 interaction with PCNA allows the helicase activity to unwind fork-blocking CAG/CTG hairpin structures to prevent breaks. Independently of PCNA binding, Srs2 also displaces Rad51 from nascent strands to prevent recombination-dependent repeat expansions and contractions. By 2D gel electrophoresis, we detect two different kinds of structured intermediates or joint molecules (JMs). Some JMs are Rad51-independent and exhibit properties of reversed forks, including being processed by the Exo1 nuclease. In addition, in a helicase-deficient mutant, Rad51-dependent JMs are detected, probably corresponding to recombination between sisters. These results clarify the many roles of Srs2 in facilitating replication through fork-blocking hairpin lesions.
INTRODUCTION
Aside from the various known exogenous factors such as ultraviolet light, base modifying chemicals and ionizing radiation that can negatively impact genome stability (1,2), it is recognized that genome instability and disease can often result from endogenous replication-blocking sources such as DNA secondary structures (3–5). Recent evidence supports a role for specialized helicases to faithfully copy through replication-blocking structures and maintain genome stability; known examples include Rrm3 helicase for programmed protein blocks (6), Pif1 for G-quadruplex sequences (7–9) and Srs2 for hairpin sequences (10–14).
Triplet repeat expansions are known replication-blocking lesions and have been implicated in many hereditary neurodegenerative diseases (15–17). A common property shared among disease-causing repeat sequences is their ability to form non-B DNA conformations in single stranded DNA. CAG/CTG repeats, whose expansion causes Huntington's disease, myotonic dystrophy, and multiple spinocerebellar ataxias, are capable of forming hairpin structures (18–20). These structure-forming sequences can interfere with cellular processes such as replication, transcription, recombination and repair, causing stalled forks, nicks, gaps or double-strand breaks in the DNA (3,16,21,22). Paradoxically, the processes of replication fork restart or DNA repair must then occur within the repeat DNA, providing opportunity for expansions or contractions during DNA synthesis or end processing.
In Saccharomyces cerevisiae, it has been shown that the Srs2 helicase is a key protein involved in unwinding CAG/CTG hairpin structures (10,13,14). The Srs2 helicase is a member of superfamily 1 helicases with 3΄ to 5΄ polarity and homology to the helicase domains of Escherichia coli RepA and UvrD (23–25). Unlike RepA and UvrD, Srs2 has an extended C-terminal region that contains domains that facilitate protein-protein interactions, including with the Rad51 recombinase (26,27) and the DNA replication clamp PCNA (28,29). Srs2 is required to prevent expansions of short (CAG/CTG)13–25 repeats (14) and longer (CAG/CTG)55–70 repeats (11). The role of Srs2 in preventing repeat expansions is specific for hairpin-forming triplet repeats, because srs2 mutants did not exhibit increased instability of unstructured repeat DNA (14). Furthermore, Srs2 was able to facilitate replication through a fork stall caused by a (CGG/CCG)40 repeat in vivo, but it did not act on other types of replication barriers such as a G-quadruplex-forming sequence or a protein-mediated stall (10). This ability relies on both its ATP-dependent helicase activity and its interaction with PCNA, but not on ability to interact with and displace Rad51 (10). Thus, Srs2 seems to be uniquely positioned to unwind hairpin structures at an advancing fork and thereby facilitate replication through triplet repeats. Recently, it was shown that the RTEL1 helicase in human cells can perform some of the same functions as Srs2 in both human and yeast cells, including CAG and CTG hairpin unwinding and prevention of CAG expansions and repeat fragility (30).
Another important Srs2 function is the ability to dismantle Rad51-mediated filament formation (26,27,31). Previous work has shown that recombinational repair can be a source of triplet repeat expansions (22,32–34). For longer (CAG/CTG)55-70 repeats, expansions and contractions in cells deleted for SRS2 were suppressed by deletion of RAD51 (11), suggesting that repeat instability in the srs2Δ background was due to excess sister chromatid recombination.
In a previous study, we identified two other roles for Srs2 at expanded CAG/CTG repeat tracts (11). First, we showed that Srs2 prevented fragility of a (CAG)70 or (CTG)70 tract integrated on a yeast chromosome (i.e. in both orientations with respect to replication). Secondly, by analysis of replication intermediates, we identified structured DNA molecules, which we termed joint molecules (JMs), that appeared when a (CTG)55 tract was replicated in wild-type cells. JMs were dependent on the repeat tract and also on Srs2, since they were severely reduced in a srs2Δ background, but were not significantly reduced in the absence of Rad51. We previously proposed that these JMs represented reversed forks (11) based on the appearance of the JMs as a cone structure emanating from the location of the repeat tract on the Y arc of replication intermediates, dependence on replication, independence from Rad51, and known predisposition of CTG repeats to undergo fork reversal in a plasmid assay (35). The formation of such structures in wild-type cells suggested that fork reversal may be a way to replicate through a repeat structure without fork breakage, recombination, or changes in the number of repeat units. JMs could also represent a mixture of reversed forks and other kinds of structured recombination intermediates that would show aberrant migration due to hairpin structures formed by CAG/CTG trinucleotide repeats (11). The reduction of the joint molecules in srs2Δ cells suggested a role for Srs2 in formation or stabilization of reversed forks in vivo.
Because Srs2 is a multi-functional protein, it was not clear which activity was involved in each of its roles. In order to gain a better understanding of the role of Srs2 at CAG repeats and in genome maintenance in general, here we take advantage of multiple well characterized Srs2 domain mutants missing one or more of the protein's functions (26,36–39). We were able to determine that there is a differential requirement for Srs2 helicase activity, PCNA binding, and Rad51 displacement functions in repeat instability, fragility, and formation of JMs. The Srs2–PCNA interaction is required to prevent repeat fragility but dispensable for preventing repeat instability, whereas Rad51 displacement is vital for prevention of instability, but not fragility. JMs appear to be mostly reversed forks that occur independently of Srs2 and are processed by Exo1, but can also include recombination intermediates in the absence of Srs2 helicase activity. We also investigated the role of Mph1 and Rad5 in maintaining CAG repeats and found that they play a modest role in (CAG)70 repeat maintenance and do not appear to play an active role in JM formation in vivo. Our results reveal important roles for the Srs2 protein in preventing fork breakdown and recombination, and in facilitating fork restart at structure-forming barriers in the genome.
MATERIALS AND METHODS
Strains
All strains used for instability and fragility experiments were the isogenic BY4705 or BY4742 backgrounds (Supplementary Table S1) and contained a yeast artificial chromosome containing either no repeat, a (CAG)70 or a (CTG)70 tract as previously described (33,40). Gene knockouts were generated by directed gene replacement and confirmed by PCR for absence of the target gene and presence of the marker gene at the target locus. Key mutants were also checked via Southern using DIG-high prime labeling and detection (Roche) using probes hybridizing to marker genes to confirm proper integration location. Srs2 domain mutants were created by PCR-mediated gene replacement using yeast genomic DNA or plasmids as templates: srs2-(1–860), srs2-(875–902Δ) and srs2-(1–998) were obtained from Patrick Sung (36), and srs2-K41R and srs2-3KR were obtained from Hannah Klein (36,39,41,42). All inserted mutations were confirmed by PCR and sequencing; primer sequences available upon request. The presence of the YAC and the length of the CAG or CTG tract was confirmed by PCR amplification followed by size analysis of the product as described below. CAG or CTG orientations are named after the sequence on the lagging-strand template.
Analysis of CAG/CTG repeat stability and fragility
Fragility and instability assays were done as previously described (11). Briefly, single colonies were grown on yeast complete (YC) media lacking uracil and leucine to maintain YAC selection. Tract lengths were confirmed by PCR amplification from cells from these starting colonies using primers flanking the repeat. Cells were resuspended in YC-Leu liquid media, grown at 30°C for 6–8 divisions, and plated on 5-FOA selective media. The rate of 5-FOAR was calculated by the method of the maximum likelihood using FALCOR software; a minimum of three replicates were performed. YAC end loss was confirmed in a subset of FOAR colonies to be 94–100% for (CAG/CTG)70 YACs in both wild-type and srs2 strain backgrounds (30–50 independent clones checked for each). The stability of the repeat tract was analyzed via PCR amplification of the repeat using newCAGfor and newCAGrev primers from at least 150 daughter colonies that grew on YC-Leu total cell count plates. Amplicons were visualized using high resolution 2% Metaphor gels (Lonza), which can accurately resolve ±9 bp (three repeats) in the relevant size range; repeat size estimates were made by subtracting the unique sequence length of 159 bp and dividing by three. Sample sizes were chosen to be large enough to obtain statistical significance for a 3-fold difference for instability and a 2-fold difference for fragility. Fragility assay values that were outliers by the Grubbs outlier test were excluded.
Analysis of replication intermediates by 2D gels
(CTG)98 repeats were integrated at the ARG2 locus on chromosome X, as previously described (11). All mutant strains were built in the S288C genetic background. The repeat tract is replicated by ARS1010, an early efficient origin located 7 kb telomere-proximal to the repeat tract. The closest centromere-proximal origin ARS1111 is located 29 kb away from the repeat tract. Therefore, (CTG)98 are replicated such that the CTG sequence is on the lagging-strand template. Cells were grown overnight at 23°C, in 200 ml YPD cultures, centrifuged, washed, resuspended in 800 ml fresh YPD medium at a concentration of 0.8–0.9 × 107 cells per ml, and grown for another hour at 23°C. Subsequently, 10 mg alpha factor were added to the culture for 2.5 h at 23°C. When 90% of the cells were arrested in G1 by microscope observation, the culture was washed and resuspended in 200 ml fresh YPD medium preheated at 23°C. Progression of S phase was followed by microscope observation and confirmed by FACS analysis on a MACSQuant Analyser. Cells were harvested after 30 (wild-type only), 40, 60 and 90 min, and killed by addition of sodium azide (0.1% final concentration). Total genomic DNA was extracted by the CTAB procedure (43) with the following modifications: Cells were frozen overnight at −80°C, thawed on ice, resuspended in 2 ml water, and 2.5 ml solution I (2% CTAB, 1.4 M NaCl, 100 mM Tris–HCl pH 7.6, 25 mM EDTA pH 8.0) was added. Then, 167 μl Zymolyase 100T (Seikagaku, 30 mg/ml) and 5 μl DTT (2M) were added and incubated 1 h at 30°C. DNA extraction and gel set up were performed as described in (43). DNA was transferred overnight in 10× SSC on a charged nylon membrane (Sigma) and UV crosslinked in a Stratagene Stratalinker. Hybridization was performed with a 750 bp randomly primed probe, corresponding to the 5΄ end of the ARG2 gene. Detection of radioactive signals were performed on a Fujifilm FLA-9000, after 4–7 days exposure. Quantifications were made using the ImageQuant software. Individual shapes were drawn around each structure (Y arc, cone, pause, linear DNA). Shapes with identical areas were used in a uniformly unlabeled part of the gel for background calculation. After signal correction by background subtraction, intensity of JMs was determined by: (Cone signal/Y arc signal)/(Cone area/Y arc area). In the wild-type strain, this ratio is 0.5 on average, meaning that the cone contains half as much signal per square millimeter as compared to the Y arc. Only gels with a good Y arc signal to background ratio (>1.2) were used in quantifications. Statistical analysis was performed with the R package (G. Millot, 2011, Comprendre et réaliser les tests statistiques à l’aide de R. Manuel de biostatistique. de boek ed. 2° edition). Statistical tests were performed on pooled data collected at all time points, for each strain.
RESULTS
Srs2 ATPase and Rad51 displacement activities are required to prevent CAG/CTG repeat instability
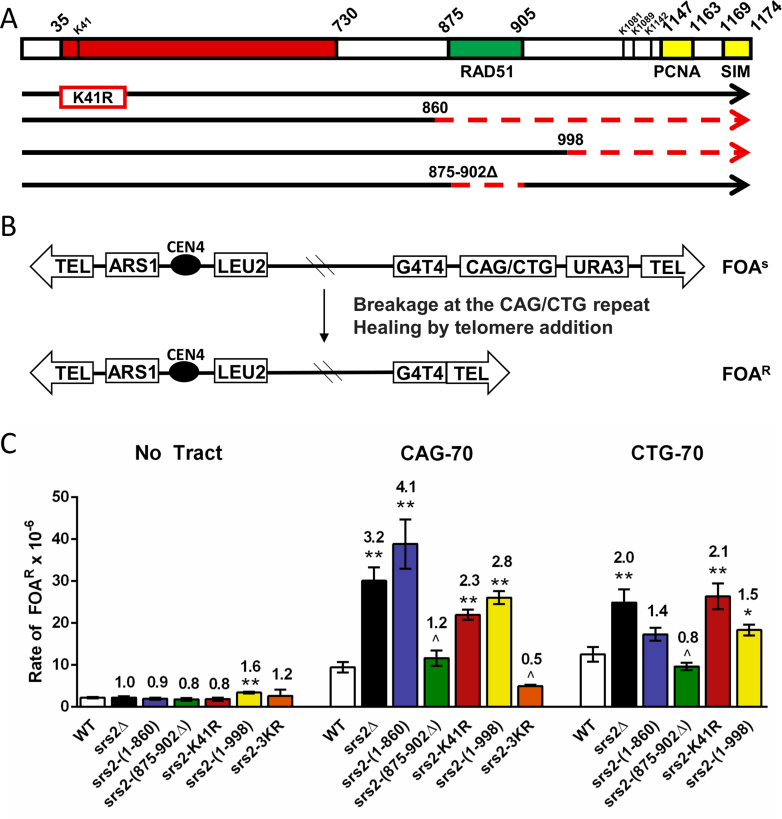
Srs2 is a multifunctional protein with three major functional domains: a helicase domain, a Rad51 interacting domain, and a PCNA interacting domain (Figure 1A). Two motifs that mediate interaction with PCNA have been defined within residues 1147–1163, and C-terminal residues 1169–1174 comprise a SUMO interacting motif (SIM) that cooperates with the adjacent PCNA interacting site in binding to sumoylated PCNA (44,42) (Figure 1). Thus, Srs2 can interact with both SUMOylated and non-SUMOylated PCNA. In addition, Srs2 lysines 1081, 1089 and 1142 have been shown to be SUMOylated which can negatively affect synthesis-dependent strand annealing (SDSA) and protein interactions with the Srs2 SIM domain (41,45). Other proteins that interact with defined Srs2 regions include Mre11 (residues 848–1175) and Mus81 (residues 783–860) (46,47). Previously characterized domain mutants that have at least one of the functions of Srs2 attenuated (26,36–38) (Figure 1A) were used to investigate whether there was a differential requirement for these domains in preventing repeat instability. The endogenous SRS2 gene was replaced with each of these untagged domain mutants at the SRS2 chromosomal locus. Western blot analysis confirmed that all mutants were expressed at levels comparable to the wild-type protein, with the notable exception of the srs2-(1–860) mutant, which showed ∼2-fold overexpression (Supplementary Figure S1A and S1B). Additionally, cellular localization was tested for the srs2-(1–998) and srs2-(1–860) mutants, as a nuclear localization signal (NLS) was previously mapped to the Srs2 C-terminus (24). Results showed that the srs2-(1–998) protein correctly localizes to the nucleus, however the srs2-(1–860) protein is mis-localized, with punctate staining in the cytoplasm (Supplementary Figure S1C). Yeast strains contained a yeast artificial chromsome (YAC) with (CAG)70 repeats. PCR amplification of the repeat tract was used to determine expansion and contraction frequencies (Table 1).
Figure 1.
Effect of SRS2 domain mutants on CAG/CTG repeat fragility. (A) Schematic of Srs2 domain mutants used in this study. The red box represents the helicase domain, the green box indicates the Rad51 interacting domain, and the yellow boxes indicate PCNA interacting domains (SIM, SUMO Interacting Domain). Dotted red lines represent deletions, numbered according to amino acid position of Srs2. The K41R mutation renders Srs2 ATPase dead, no helicase or translocase activity is possible. Srs2-(1–860) retains helicase function but cannot interact with Rad51 or PCNA. Srs2-(1–998) lacks PCNA interaction but has intact helicase and translocase activities. Srs2-(875–902Δ) retains helicase activity and PCNA interaction but has impaired Rad51 interaction. Srs2-3KR has the three indicated lysine residues mutated to arginine. (B) Schematic of the YAC system. A breakage event occurring within or near the repeat can result in the loss of the URA3 marker gene and healing at the G4T4 telomere seed, leading to strains becoming resistant to 5-fluoroorotic acid (5-FOA). (C) Rate of FOAR x 10−6 is shown. Data represents the average of at least three experiments (numbers in Supplementary Table S3). Error bars represent SEM. No tract: no repeat tract on YAC, CAG-70: (CAG)70 repeats on the lagging strand template, CTG-70: (CTG)70 repeats on the lagging strand template (unpaired Student's t-test comparing averages to wild-type (WT) *P < 0.05; **P < 0.01; srs2-(1–860) (CTG)70P value is 0.06).
Table 1. (CAG)70 Instability on YAC in Srs2 domain mutants.
| Strain name | No. colonies analyzed | % Expansions | Fold over wild-type | % Contractions | Fold over wild-type |
|---|---|---|---|---|---|
| Wild-type | 269 | 1.1 | 1.0 | 2.6 | 1.0 |
| srs2Δ | 231 | 5.6* | 5.1* | 6.5* | 2.5* |
| rad51Δ | 184 | 4.9* | 4.5* | 13.6* | 5.2* |
| srs2-(1-860) | 152 | 5.3* | 4.8* | 12.5* | 4.8* |
| srs2-(875-902Δ) | 162 | 4.3* | 3.6* | 9.9* | 3.8* |
| srs2-K41R | 201 | 5.0* | 4.5* | 9.5* | 3.7* |
| srs2-(1-998) | 122 | 1.6 | 1.5 | 3.3 | 1.3 |
| srs2-3KR | 157 | 1.9 | 1.7 | 5.1 | 2.0 |
| srs2-K41R rad51Δ | 174 | 1.2 | 1.1 | 1.7 | 0.7 |
*P < 0.05 compared to wild-type using Fisher's Exact Test.
First, we tested the srs2-K41R mutant, which is ATPase dead and therefore lacking both helicase and Rad51 displacement activities. The CAG orientation exhibits both elevated expansions and contractions in srs2Δ strains (11). A significant increase in both expansion and contraction frequencies was observed in srs2-K41R strains (4.5- and 3.7-fold over wild-type, respectively), similar to the frequencies in srs2Δ strains (Table 1, Supplementary Table S2). To distinguish between the helicase and Rad51 displacement activities, we tested the srs2-(875–902Δ) mutant, which retains the helicase domain but lacks most of the Rad51 binding domain and is defective for Rad51 displacement in vitro, exhibiting reduced anti-recombinase activity in vivo (36) (Figure 1A). The srs2-(875–902Δ) mutant also had a significant increase in both repeat expansions and contractions, 3.6 and 3.8 over wild-type respectively, similar to srs2-K41R (Table 1). These results suggest that inability to bind and displace Rad51 is the main cause of the increased level of CAG instability in srs2Δ strains. The fact that the srs2-(875–902Δ) expansion phenotype is slightly less than either the srs2Δ or srs2-K41R strains could be because this mutant appears to retain some level of anti-recombinase function in vivo, as it cannot suppress the MMSS of a rad18Δ mutant (36). We previously showed that CAG instability was Rad51 dependent in an srs2 null background (11). To confirm that this suppression also occurred in a background defective in Rad51 displacement, repeat instability was tested in the srs2-K41R rad51Δ double mutant. As predicted, CAG expansions and contractions were both suppressed to wild-type levels (Table 1). Together, these data support the conclusion that (CAG)70 repeat instability arises through homologous recombination (HR) and that Srs2 prevents this instability by antagonizing HR via its Rad51 displacement activity. As described previously (48), deletion of the RAD51 gene causes its own instability phenotype, thus other non-HR pathways can also cause repeat instability. The suppression of instability to wild-type levels in the double mutant suggests that the presence of the ATPase dead Srs2 protein may prevent this alternative pathway, perhaps by preventing access to the replication fork (49).
In contrast to the Rad51 displacement deficient mutants, instability in the srs2-(1–998) mutant, which retains both helicase and Rad51 displacement activity but is missing the PCNA interaction domain, is similar to wild type (Table 1). Therefore PCNA interaction is apparently not required to prevent repeat instability. This result suggests that expansions and contractions occur during a process where Srs2 is acting independently from its interaction with PCNA, for example at a post-replication stage.
The srs2-(1–860) mutant, which retains the helicase domain but lacks the entire C terminus of the protein, including the Rad51, Mre11 and PCNA interaction domains, exhibited a significant increase in both expansions and contractions. In the srs2-(1–860) mutant the contraction frequency was even higher than in the srs2Δ strain (12.5% compared to 6.5%), suggesting this mutant may have a dominant-negative effect. The presence of a wild-type SRS2 gene in a heterozygous diploid suppressed the MMS sensitivity and partially suppressed repeat instability of the srs2-(1–860) mutation, though a 3-fold increase in expansions remained (Supplementary Figure S2). Given the overexpression and mislocalization of the mutant protein (Supplementary Figure S1), and the known toxicity of Srs2 overexpression (26,49), it seems likely that the mutant protein is triggering an abnormal cellular condition that results in repeat instability.
Srs2 helicase activity and PCNA interaction are required to prevent CAG/CTG repeat fragility
To determine the requirements for preventing chromosomal fragility, we tested the rate of 5-FOA-resistance (5-FOAR) in both CAG and CTG orientations utilizing a previously described breakage assay that measures YAC end loss (11) (Figure 1B).
We hypothesized that the srs2-K41R strain would exhibit an increase in repeat fragility because the mutant protein lacks both helicase and Rad51 displacement activity. Indeed, a significant 2.3-fold increase in (CAG)70 and 2.1-fold increase in (CTG)70 fragility compared to wild-type were observed, statistically equivalent to the rates for the srs2Δ strain (Figure 1C, Supplementary Table S3). As we observed previously (11), deleting SRS2 had a greater effect on fragility of tracts in the CAG orientation. To distinguish which activity was important, we tested the srs2-(875–902Δ) mutant lacking Rad51 binding. We saw no increase in fragility in either orientation, indicating that, in contrast to the result observed for instability, the Rad51 displacement activity is dispensable for preventing fragility. Further, this result implicates the helicase activity of Srs2 in preventing both CAG and CTG fragility.
To determine the contribution of PCNA interaction, we tested the srs2-(1–998) mutant. A significant increase in (CAG)70 and (CTG)70 fragility were observed, comparable to a complete deletion of SRS2, especially for the CAG orientation (Figure 1C). This increase is not due to loss of the Srs2 SUMOylation sites in the C-terminus, as mutation of those sites to non-modifiable residues did not have the same effect (Figure 1C, srs2-3KR). Therefore, Srs2 interaction with PCNA is most likely the crucial factor needed to prevent repeat fragility. The srs2-(1–860) mutant also exhibited a significant increase of (CAG)70 fragility. We conclude that Srs2 PCNA interaction and helicase activity are both needed to prevent fragility of expanded CAG/CTG repeats, probably via helicase unwinding of hairpin structures at the replication fork. Thus prevention of repeat fragility and instability are mediated by different functions of the Srs2 protein.
Mph1 and Rad5 play a lesser role in preventing (CAG)70 instability and fragility
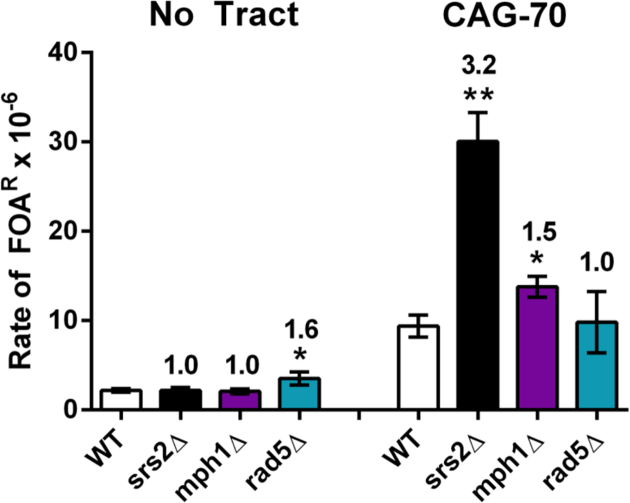
In addition to Srs2, other helicases could be involved in regulation of recombination or fork restart including Rad5, Mph1 and Sgs1. The Sgs1 helicase was shown to have a role in preventing (CAG/CTG)70 repeat instability and fragility (11), however the effect of Mph1 and Rad5 on (CAG)70 repeats had not been examined.
Mph1 is a 3΄ to 5΄ helicase homologous to FANCM that regulates recombination by unwinding Rad51 D-loops (50) and processes Holliday junction intermediates through branch migration (51). Cells deleted for Mph1 exhibited an increase in both repeat contractions and expansions, although not statistically different from wild-type (Table 2). Deletion of Mph1 resulted in a rate of (CAG)70 breakage that is also slightly elevated compared to wild-type (1.5 fold, P = 0.024, Figure 2). Altogether, Mph1 appears to have a role, albeit minor, in preventing chromosomal instability and fragility at CAG repeats.
Table 2. (CAG)70 Instability on YAC in srs2Δ, mph1Δ, rad5Δ mutants.
| Strain name | No. colonies analyzed | % Expansions | Fold over wild-type | % Contractions | Fold over wild-type |
|---|---|---|---|---|---|
| wild-type | 269 | 1.1 | 1.0 | 2.6 | 1.0 |
| srs2Δ | 231 | 5.6* | 5.1* | 6.5* | 2.5* |
| mph1Δ | 290 | 2.8 | 2.5 | 5.2 | 2.0 |
| rad5Δ | 190 | 2.1 | 1.9 | 4.7 | 1.8 |
*P < 0.05 compared to wild-type using Fisher's Exact Test.
Figure 2.
Impact of MPH1 or RAD5 deletion on (CAG)70 repeat fragility. Rate of FOAR × 10−6 is shown. Data represents the average of at least three experiments (Supplementary Table S3). Error bars represent SEM. No tract: no repeat tract, CAG-70: (CAG)70 repeats on YAC. (Unpaired student t-test comparing averages to wild-type (WT) *P < 0.05; **P < 0.01).
Rad5 is a DNA-dependent ATPase and E3 ubiquitin ligase involved in the error-free branch of DNA damage bypass. The Rad5/Mms2/Ubc13 complex polyubiquitylates PCNA to facilitate template switching (52). Previous studies have shown that in the absence of Rad5, there is a reduction in instability of several types of non-hairpin forming repeats including poly(GT)29 (53), (ATTCT)60 (54), and (GAA)100 (55). Deletion of RAD5 led to a 4–6-fold increase in expansion rate in short (CAG/CTG)13-25 repeat tracts (12) and a 3-fold increase in longer (CAG)85 tracts (56). Deletion of RAD5 in our experiments led to a slight increase in repeat expansions and contractions, but not statistically different from wild type (Table 2). It did not lead to significant changes in CAG repeat fragility (Figure 2). We conclude that Rad5 does not play a major role in maintenance of (CAG)70 repeats, at least in a wild-type background.
Analysis of replication and recombination intermediates by 2D gel electrophoresis in Srs2 domain mutants
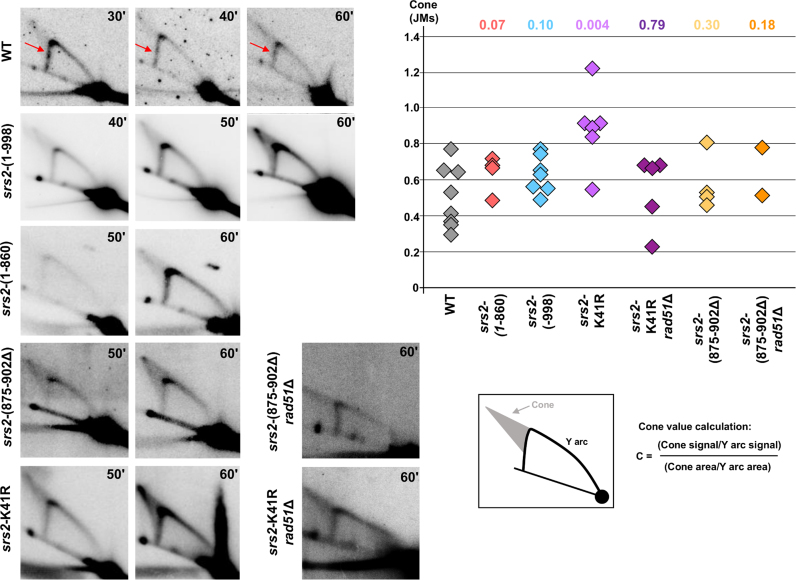
In our previous study, we identified SRS2-dependent joint molecules (JMs) whose formation occurred during replication of a (CTG)55 trinucleotide repeat tract (11). Here, replication of a (CTG)98 repeat tract was analyzed by 2D gel electrophoresis in the srs2 domain mutants.
In wild-type cells, a signal corresponding to replication fork stalling is detected on the descending Y arc, at the place where the triplet repeats are integrated (Figure 3). Such a pausing signal was not detected as clearly with shorter repeats (11). We conclude that (CTG)98 repeats stall forks more efficiently than (CTG)55 repeats, likely because this longer repeat tract has a higher probability of hairpin structure formation. Interestingly, the JMs are somewhat less prominent for the CTG98 tract compared to (CTG)55, suggesting there could be an inverse relationship between a stable fork stall and JMs, a conclusion also reached in a recent study (57).
Figure 3.
Representative 2D gels for wild-type and srs2 mutant strains. Times at which cells were collected after alpha-factor release are indicated in the top right corner of each gel. Quantifications of the ratio of cone (JMs) over Y arc signals obtained for each individual gel in a particular background are shown on the accompanying graph. P values compared to wild-type (Mann–Whitney Wilcoxon rank test) are listed above the quantifications. Arrows point to the site where replication pauses are clearly visible. Quantified pause signals and cone values for individual gels are given in Supplementary Table S4.
In order to identify which part of the Srs2 protein was responsible for JM formation, the JM signal intensity was quantified in comparison to the Y arc in the wild-type strain and the different srs2 mutants (Supplementary Table S4). In the srs2-(1–998) mutant, the amount of JMs is not different from wild-type (Figure 3). We conclude that the C-terminal part of the protein, interacting with PCNA, is not required for JM formation or maintenance.
If the decrease in JMs in the srs2Δ strain was due to the helicase activity of Srs2 actively reversing hairpin-stalled forks, they should also decrease in a srs2 helicase deficient strain. Surprisingly, JMs were significantly increased in the srs2-K41R mutant (Figure 3). We note that the signal is especially high at the tip of the cone in this background, where X-shaped recombination intermediates are known to migrate. Since the ATPase is needed for both the helicase and the Rad51 displacement activity, the increase in JMs in the srs2-K41R mutant could be due to increased Rad51 binding and a resultant increase in recombination structures. To test this idea, replication intermediates were visualized in a srs2-K41R rad51Δ double mutant. Notably, the JMs decreased to wild-type level in the double mutant (Figure 3). Thus, in the srs2-K41R mutant, JMs include recombination intermediates that run in the same area as structured DNA that is not Rad51-dependent. This data is consistent with the result that repeat instability in the srs2-K41R background is recombination dependent (Table 1). Since the JMs that remain in the srs2-K41R rad51Δ double mutant are not Rad51-dependent recombination intermediates, they likely represent reversed forks. If so, since the K41R mutation is helicase dead, reversed forks are not created by active Srs2 helicase activity.
Replication intermediates were also visualized in the srs2-(875–902Δ) mutant, lacking efficient Rad51 displacement activity but retaining helicase activity. The srs2-(875–902Δ) mutant showed no difference in the level of JM structures compared to wild-type, despite the expectation that recombination intermediates might increase in this background (Figure 3). In the double mutant srs2-(875–902Δ) rad51Δ, the JM amount is indistinguishable from both the wild type and the srs2-(875–902Δ) single mutant, showing that they are generally not Rad51-dependent recombination intermediates. We conclude that the presence of the helicase activity combined with PCNA binding is sufficient to unwind hairpin structures encountered at the fork to prevent undue accumulation of recombination intermediates, though apparently not to prevent all excess recombination, as repeat instability was increased in this background.
Because none of the Srs2 domain mutants showed a reduction in JMs as was observed at a much shorter (CTG)55 tract (11), we created a srs2Δ in the (CTG)98 strain used for the current experiments. Surprisingly, with this longer repeat, JM structures were still present in the srs2Δ background at a level similar to the wild-type strain (Figure 4). These results confirm that the Srs2 protein is not required for JM formation when forks stall at a (CTG)98 repeat tract, and shed light on why none of the domain mutants eliminated JM formation. A pause was also visible at the 90’ time point in the srs2Δ mutant, indicating that the presence of the Srs2 protein is not essential to transiently stall forks at long chromosome-borne CTG repeats (Wilcoxon test P-value of 0.14 compared to wild-type).
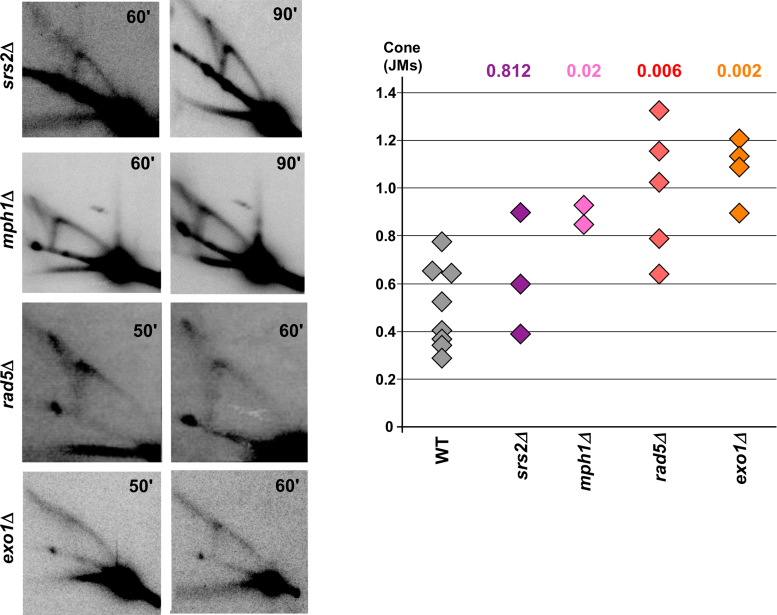
Figure 4.
Representative 2D gels for srs2Δ, mph1Δ, rad5Δ and exo1Δ strains. Times at which cells were collected after alpha-factor release are indicated in the top right corner of each gel. Quantifications of the ratio of cone (JMs) over Y arc signals are shown on the accompanying graph; P values compared to wild-type (Mann–Whitney Wilcoxon rank test) are listed above the respective mutant and in Supplementary Table S4.
Altogether, our results suggest that structured intermediates (JMs) formed during replication of a long CAG/CTG repeat in a wild-type strain do not require Srs2 for their formation, but are influenced by Srs2 activity. In particular, loss of the anti-recombinase activity does not modify the overall amount of JMs, suggesting that Rad51 is not directly involved in JM formation at a CAG tract, consistent with the finding that JMs were not measurably reduced in the rad51Δ background (11). Fork-coupled helicase activity appears to be important both for preventing fork breakdown and for preventing excess recombination intermediates, but not for causing fork reversal.
Effect of Rad5, Mph1 and Exo1 on CAG repeat replication
In vitro studies have shown that both Mph1 and Rad5 can convert model fork substrates to a four stranded reversed fork-like structure (51,58–61). Thus, it has been speculated that these proteins may catalyze fork reversal in vivo. To test the effect of these proteins at the (CTG)98 tract, replication intermediates were analyzed in mph1Δ and rad5Δ strains. Interestingly, JMs are significantly increased in both mph1Δ and rad5Δ strains (Figure 4), showing that neither protein is required to generate JMs. Since Mph1 is expected to reduce recombination intermediates by unwinding Rad51-formed D-loops, the observed increase in JMs in mph1Δ may correspond to recombination intermediates that are running in the area of the cone, similarly to what was observed in the srs2-K41R mutant. However the increase of JMs in the rad5Δ strain, which should decrease template switching, was unexpected. The increase is mostly due to the spot at the tip of the spike, where Holliday junctions are expected to run, suggesting a shift to recombination structures could be occurring in this mutant.
To further investigate the nature of the JMs, replication intermediates were visualized in an exo1Δ mutant. In an in vivo fork reversal model in bacteriophage T4, it was shown that unprocessed reversed forks run as a spike off the Y arc in exonuclease deficient strains, which is converted to a cone structure in wild-type strains (62). Exo1 has also been shown to process forks stalled by nucleotide depletion or a protein barrier (63,64). At the (CTG)98 repeat, deletion of EXO1 resulted in a reduction of the cone signal and an increase in structured molecules migrating as a spike eliminating from the top of the Y arc (Figure 4). This data supports the idea that in wild-type cells, when recombination is controlled, the cone mostly represents resected reversed forks.
DISCUSSION
Srs2 prevents (CAG/CTG)70 instability and fragility using different activities
By using Srs2 domain mutants, we investigated how the different Srs2 functions contribute to the maintenance of triplet repeats. We show that the (CAG/CTG)70 expansion and contraction phenotypes can be genetically uncoupled from the fragility phenotype. From our experiments, Rad51 displacement activity appears to be critical and PCNA interaction dispensable for preventing repeat expansions and contractions, whereas the C-terminal PCNA interaction domain and helicase activity are critical for preventing (CAG/CTG)70 breakage.
Since no significant increase in expansions or contractions were observed in the srs2-(1–998) mutant, it is likely that repeat instability is primarily arising from aberrant recombination. This conclusion is reinforced by the observation of increased instability in all the mutants with defective Rad51 displacement, as well as a reduction of instability levels in the srs2-K41R rad51Δ double mutant. These data strengthen our previous conclusion (11) that recombination is a major source of triplet repeat instability if not properly controlled. We note that Srs2 unwinding of hairpins on the nascent lagging strand has also been proposed as a pathway to prevent repeat expansions (3,65), and our data do not rule out this mechanism. Indeed, CAG expansions are slightly higher in srs2Δ strains compared to srs2 (875–902Δ), which could be accounted for by this second pathway.
Intriguingly, the srs2-(1–998) allele had no effect on instability, suggesting that Srs2 prevents instability independently from replication fork progression. Evidence supports that recruitment of Srs2 to repair centers occurs independently from replication, as Srs2 can localize to Rad54 foci in the absence of PCNA interaction (66). Perhaps the interaction between Srs2 and Rad51 or other repair proteins can recruit Srs2 to presynaptic filaments (31), as other work has also shown Srs2 can prevent recombination independently from its PCNA interaction (37). Our data suggest that Srs2 antirecombinase function may not operate directly at a stalled fork.
Roles of Srs2 in the formation of reversed forks and other joint molecules
Our previous study using a shorter (CTG)55 repeat showed the appearance of a cone structure emanating from the location of the repeat tract on the Y arc of replication intermediates that was dependent on both the repeat tract and replication, but independent of Rad51 (11). In the current study using a longer (CTG)98 repeat, we were able to see a more distinct pause signal in wild-type cells, showing that expanded CAG/CTG repeats on a eukaryotic chromosome pause replication, consistent with other recent results (57). Even though the pausing signal was reduced overall in the srs2 mutants compared to wild-type (Supplementary Table S4) it is still visible in some cases, indicating that the pause is not dependent on the presence of the full-length Srs2 protein. This specific question could not be addressed previously, since no clear pausing signal could be detected with shorter repeats.
In addition to paused replication, we observed a cone structure that appeared in replication intermediates from both wild-type and mutant strains, confirming that it is an inherent characteristic of CAG repeat replication. The precise molecular nature of these JMs is unclear, but their migration pattern is strongly reminiscent of structures detected in other systems that were concluded to be reversed replication forks (62,67–69). Other authors have suggested that the cone-signal could be a mixture of replication molecules in which initiation occurs randomly outside the locus being analyzed, leading to double Y structures with various points of fork convergence (70). In the present case, we find this hypothesis unlikely, since the replication origin normally used to replicate this locus is clearly identified (71,72). In addition, to create the double Ys in this scenario, the JMs should increase with more stalling, which is not the case; if anything there may be an inverse correlation. That the cone shifts to a spike in exo1Δ cells further supports the interpretation of reversed forks, as stalled and reversed forks are known to be processed by exonucleases in multiple systems, resulting in a reduced level of diversity and a tighter migration pattern of structured molecules (62–64).
Our former study showed a clear reduction in JMs in the srs2Δ strain (11). Hence, we hypothesized that JMs were formed by Srs2 helicase activity. Here, we found that additional, not fewer JMs were formed in the srs2-K41R ATPase mutant that is lacking both helicase and Rad51 displacement activity. Further analysis indicated that the additional JMs that occur in this mutant are Rad51-dependent, distinctly from those formed in wild-type cells. Therefore the cone likely contains a mixture of structures, which are mostly reversed forks in wild-type cells but can also include recombination intermediates in mutants with disregulated recombination. Consistent with the persistence of JMs in all the srs2 domain mutants tested, the srs2Δ strain also exhibited JMs that formed at the long (CTG)98 tract. The finding of a differential role of the Srs2 protein on short and long repeat tracts was unexpected. We previously found that expanded CTG tracts are mildly toxic to cells lacking Srs2 (11) and in the present work that the cell cycle is delayed in srs2Δ cells with longer repeats. It is known that longer CAG/CTG repeats lead to more cell cycle arrests and relocation to the nuclear pore in yeast (33,73). Differently from the shorter tract, JMs in the (CTG)98 containing srs2Δ cells usually persisted to the 90 min time point. Thus, they may be accumulating in arrested cells that are unable to recover (74), whereas the intermediates at a shorter repeat are usually able to be resolved. The diverse nature of the JM structures indicates that these intermediates may be at various stages of D-loop extension and could also include hairpin structures that affect mobility in the second dimension of gel electrophoresis. The increase in JM formation between the srs2-K41R mutant and the complete deletion of SRS2 points to a dominant negative effect of a catalytically inactive Srs2 protein that can nonetheless bind to PCNA and DNA.
Interestingly, the complete deletion of SRS2 also led to a reduction in structured molecules formed by template switch at a protein-mediated stall (75). However at a CGG repeat that causes a stable fork stall, the srs2-K41R mutation caused a significant increase in fork stalling (10), supporting the idea that fork restart is inhibited in this background. In addition, srs2-(1-998), srs2-(875–902Δ) and srs2-(1–860) mutants, that retain helicase activity but are defective in either PCNA binding or Rad51p-interaction or both, did not exhibit a decrease or increase in JMs compared to wild-type, even though each of these mutants variously affected repeat fragility and instability. These results further suggest that DNA binding in the absence of catalytic activity blocks fork restart and traps the DNA in a reversed fork or recombination structure. This interpretation fits with a recent role identified for Srs2 in inhibiting polymerase δ access to PCNA (76). The Srs2-K41R protein that can still bind PCNA could block Polδ access but also be unable to unwind hairpins, resulting in a persistant reversed fork, increase in recombination intermediates, and the continued presence of the cone on 2D gels.
Altogether, our data support the model that wild-type Srs2 facilitates fork restart by hairpin unwinding to minimize formation of joint molecules, rather than causing fork reversal or stabilizing reversed forks.
Role of Mph1 and Rad5 at long CAG/CTG repeats
Several helicases have been shown to reverse model branched substrates in vitro (see (77) for review). From analysis of replication intermediates at the (CTG)98 repeat in mph1Δ and rad5Δ cells, it appears that neither Mph1 nor Rad5 are needed to form JMs, suggesting that either fork reversal in vivo happens spontaneously when the fork encounters a hairpin barrier, or that we have not yet identified the helicase that is involved in fork reversal in vivo. CAG/CTG and telomere repeats both undergo extensive reversal in a plasmid assay system (35), and may be inherently ‘slippery’ because of the extensive region of homology available in multiple possible alignments. Non-helicase mediated fork reversal can also occur, as RecA can promote fork reversal on branched DNA molecules in the absence of the replisome (78,79), and Rad51 is required for fork reversal at forks stalled by low doses of replication inhibitors in eukaryotic cells (80). We note, however, that at the CAG repeat, neither Rad51 (11) nor Rad52 (data not shown) were required for generation of the observed JMs in otherwise wild-type cells.
In contrast, deletion of either Mph1 or Rad5 increased the level of JMs, suggesting that reversed forks were stabilized and/or converted to recombination intermediates more often than in wild-type cells. Mph1 could prevent instability from occurring at a stalled fork by dismantling D-loops and preventing formation of recombination intermediates, explaining the increase in JMs in its absence. This is consistent with the mild increase in CAG instability in mph1Δ cells (Table 2). Rad5-dependent post-replication repair has been shown to prevent expansion of short CAG repeats in a manner epistatic to Srs2 (12), though unregulated Rad5-dependent template switching can also cause repeat expansions (56). In our case, the absence of Rad5 led to an apparent increase in recombination intermediates and a corresponding mild increase in (CAG)70 instability.
A model for replication through structure-forming CAG/CTG repeats
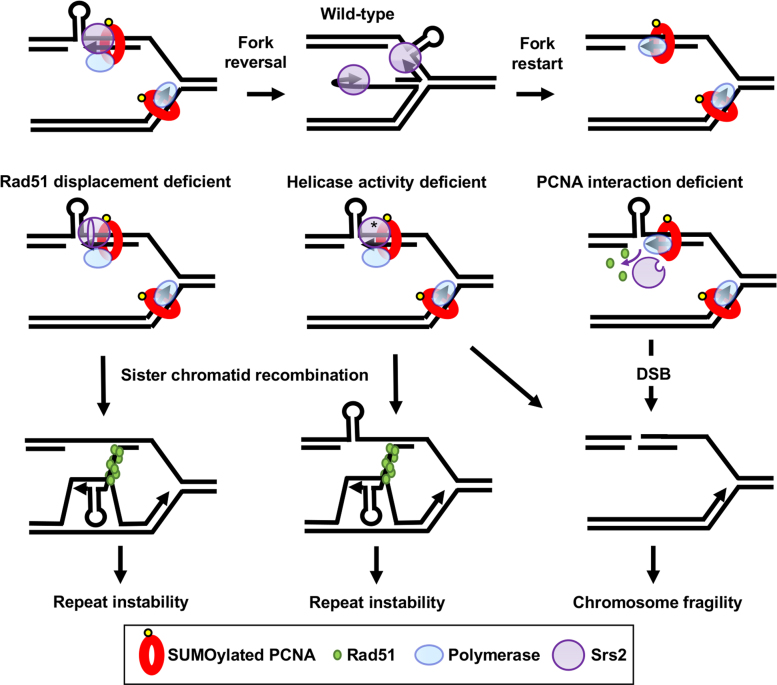
Our current results clarify the dynamic nature of events that can occur in response to replication fork stalling at a structure-forming DNA sequence. In wild-type cells, an expanded CTG repeat on the template strand causes transient fork stalling, which was evident as an accumulation of Y-shaped structures. In addition, Rad51-independent JMs are detected and shift to a spike of X-shaped molecules in an exo1Δ mutant, supporting the conclusion that they are mostly reversed forks (Figure 5, top). Our data suggest that the Srs2 protein locates to the stalled fork via its interaction with PCNA and utilizes its helicase activity to unwind the fork-blocking hairpin structure and facilitate fork restart (Figure 5, top row).
Figure 5.
A model to explain how the different domains of the Srs2 protein are involved in repeat instability, chromosomal fragility and JM formation. Top row: in the wild-type situation Srs2 is brought to the fork by PCNA interaction and unwinds hairpins formed by triplet repeats. For simplicity, only a lagging strand template hairpin is shown, though we note that leading strand template or nascent strand hairpins are also possible. Transient fork reversal may facilitate unwinding, or reversal may occur when unwinding fails, forming some JMs in wild-type cells (reversed fork JMs, Rad51 independent). Homologous recombination is discouraged by Srs2-dependent Rad51 displacement, but may occasionally happen. Right: in the absence of interaction with PCNA, Srs2 is not recruited to the fork to unwind template hairpin structures, leading to fork breakage. Srs2 may still displace Rad51, preventing recombination-dependent instability. Center: when Srs2 ATPase activity is absent (*) hairpins will not be unwound, leading to more fork breakdown, and Rad51 is not displaced, leading to increased recombination intermediates. In this case, JMs are increased, consisting of both reversed forks and recombination intermediates. Left: Srs2 missing the Rad51 interaction domain is brought to the fork by PCNA interaction and can unwind secondary structures to prevent fork breakdown, but cannot displace Rad51p. Increased homologous recombination occurs, leading to repeat instability.
In the absence of Srs2–PCNA association or in an ATPase-deficient mutant, fork breakage results. Both helicase and Rad51 displacement are ATPase dependent, but comparison of srs2-(875–902Δ) and srs2-K41R mutants suggests that it is the helicase activity that is required to prevent chromosomal breakage, probably by unwinding hairpin structures (Figure 5, middle replication fork and right pathway). In agreement with this conclusion, both the Srs2 helicase activity and PCNA interaction are required for replication through (CGG/CCG)40 repeats, which form strong replication blocking hairpins (10). We note that in the CAG orientation, CTG template hairpins would be more likely to form on the leading strand template, which appears to be particularly deleterious in the absence of Srs2 unwinding, leading to higher levels of fragility. The human RTEL1 helicase, which can unwind CAG and CTG hairpins, can complement Srs2 in yeast to prevent CAG repeat fragility (30). Since it also associates with SUMOylated PCNA at the replisome to prevent replication fork stalling and telomere fragility (81), RTEL1 may be acting equivalently in human cells.
Independently of PCNA binding, Srs2 also displaces Rad51 from nascent strands to prevent recombination. In the absence of Rad51 displacement, such as in the srs2 mutants lacking ATPase activity or the Rad51 interaction domain, increased recombination occurs, providing an increased probability of repeat expansions and contractions. Instability can result when the nascent strand invades or anneals out of register, or due to hairpin-facilitated slippage during D-loop extension (Figure 5, left pathway). The srs2-K41R mutant led to a visible increase in recombination intermediates by 2D gels. This was not observed in the srs2-(875-902Δ) mutant, most likely because the presence of helicase activity is still allowing hairpin unwinding to limit the amount of recombination intermediates, which is supported by the lack of repeat-induced fragility observed in this mutant.
In summary, our results show that the Srs2 protein plays an important role in facilitating replication at fork-blocking hairpin lesions, both by unwinding structures to prevent fork stalling and breakage, and by blocking sister-chromatid recombination to prevent undue microsatellite instability.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Patrick Sung and Hannah Klein for providing Srs2 domain mutant genes, which were key reagents without which this project would not have been possible, and Xiaofeng Allen Su for contributing the srs2-3KR experiments.
Author contributions: J.N. and R.A. performed the stability and fragility experiments with the help of L.V. and L.S., analyzed the data, and helped write the manuscript. D.V. performed the 2D gel experiments and analysis under the supervision of G-F.R. C.H.F. supervised the whole project, analyzed data, and wrote the manuscript with the help of G.-F.R.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Tufts University, National Institutes of Health [R01GM063066, P01GM105473 to C.H.F]; Tufts Graduate Student Research Award (to J.N.); Association pour la Recherche contre le Cancer [ARC SFI20111203641 to G.-F.R]. Funding for open access charge: National Institutes of Health [GM105473 to C.H.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res. - Rev. Mutat. Res. 2000; 462:129–135. [DOI] [PubMed] [Google Scholar]
- 3. Usdin K., House N.C.M., Freudenreich C.H.. Repeat instability during DNA repair: insights from model systems. Crit. Rev. Biochem. Mol. Biol. 2015; 9238:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang G., Vasquez K.M.. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst.). 2014; 19:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aguilera A., García-Muse T.. Causes of genome instability. Annu. Rev. Genet. 2013; 47:1–32. [DOI] [PubMed] [Google Scholar]
- 6. Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A.. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003; 12:1525–1536. [DOI] [PubMed] [Google Scholar]
- 7. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paeschke K., Capra J.a., Zakian V.A.. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011; 145:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ribeyre C., Lopes J., Boulé J.B., Piazza A., Guédin A., Zakian V.A., Mergny J.L., Nicolas A.. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009; 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anand R.P., Shah K.A., Niu H., Sung P., Mirkin S.M., Freudenreich C.H.. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012; 40:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kerrest A., Anand R.P., Sundararajan R., Bermejo R., Liberi G., Dujon B., Freudenreich C.H., Richard G.-F.. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat. Struct. Mol. Biol. 2009; 16:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daee D.L., Mertz T., Lahue R.S.. Postreplication repair inhibits CAG.CTG repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007; 27:102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhattacharyya S., Lahue R.S.. Srs2 helicase of Saccharomyces cerevisiae selectively unwinds triplet repeat DNA. J. Biol. Chem. 2005; 280:33311–33317. [DOI] [PubMed] [Google Scholar]
- 14. Bhattacharyya S., Lahue R.S.. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 2004; 24:7324–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McMurray C.T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010; 11:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007; 447:932–940. [DOI] [PubMed] [Google Scholar]
- 17. Pearson C.E., Nichol Edamura K., Cleary J.D.. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005; 6:729–742. [DOI] [PubMed] [Google Scholar]
- 18. Liu G., Chen X., Bissler J.J., Sinden R.R., Leffak M.. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010; 6:652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miret J.J., Pessoa-Brandão L., Lahue R.S.. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pearson C.E., Sinden R.R.. Alternative DNA structures within the trinucleotide repeats of the myotonic dystrophy and fragile X locus. Biochemistry. 1996; 35:5041–5053. [DOI] [PubMed] [Google Scholar]
- 21. Richard G.-F., Kerrest A., Dujon B.. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 2008; 72:686–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richard G.F., Pâques F.. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 2000; 1:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hall M.C., Matson S.W.. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 1999; 34:867–877. [DOI] [PubMed] [Google Scholar]
- 24. Rong L., Klein H.L.. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1993; 268:1252–1259. [PubMed] [Google Scholar]
- 25. Aboussekhra A., Chanet R., Zgaga Z., Cassier-Chauvat C., Heude M., Fabre F.. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989; 17:7211–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P.. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003; 423:305–309. [DOI] [PubMed] [Google Scholar]
- 27. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F.. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003; 423:309–312. [DOI] [PubMed] [Google Scholar]
- 28. Watts F.Z. Sumoylation of PCNA: wrestling with recombination at stalled replication forks. DNA Repair (Amst.). 2006; 5:399–403. [DOI] [PubMed] [Google Scholar]
- 29. Papouli E., Chen S., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D.. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005; 19:123–133. [DOI] [PubMed] [Google Scholar]
- 30. Frizzell A., Nguyen J.H.G., Petalcorin M.I.R., Turner K.D., Boulton S.J., Freudenreich C.H., Lahue R.S.. RTEL1 inhibits trinucleotide repeat expansions and fragility. Cell Rep. 2014; 6:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antony E., Tomko E.J., Xiao Q., Krejci L., Lohman T.M., Ellenberger T.. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009; 35:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gellon L., Razidlo D.F., Gleeson O., Verra L., Schulz D., Lahue R.S., Freudenreich C.H.. New functions of Ctf18-RFC in preserving genome stability outside its role in sister chromatid cohesion. PLoS Genet. 2011; 7:e1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sundararajan R., Freudenreich C.H.. Expanded CAG/CTG repeat DNA induces a checkpoint response that impacts cell proliferation in Saccharomyces cerevisiae. PLoS Genet. 2011; 7:e1001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richard G.F., Cyncynatus C., Dujon B.. Contractions and expansions of CAG/CTG trinucleotide repeats occur during ectopic gene conversion in yeast, by a MUS81-independent mechanism. J. Mol. Biol. 2003; 326:769–782. [DOI] [PubMed] [Google Scholar]
- 35. Fouché N., Özgür S., Roy D., Griffith J.D.. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006; 34:6044–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colavito S., Macris-Kiss M., Seong C., Gleeson O., Greene E.C., Klein H.L., Krejci L., Sung P.. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 2009; 37:6754–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le Breton C., Dupaigne P., Robert T., Le Cam E., Gangloff S., Fabre F., Veaute X.. Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res. 2008; 36:4964–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pfander B., Moldovan G.-L., Sacher M., Hoege C., Jentsch S.. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005; 436:428–433. [DOI] [PubMed] [Google Scholar]
- 39. Krejci L., Macris M., Li Y., Van Komen S., Villemain J., Ellenberger T., Klein H., Sung P.. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 2004; 279:23193–23199. [DOI] [PubMed] [Google Scholar]
- 40. Callahan J.L., Andrews K.J., Zakian V.A., Freudenreich C.H.. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol. 2003; 23:7849–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saponaro M., Callahan D., Zheng X., Krejci L., Haber J.E., Klein H.L., Liberi G.. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 2010; 6:e1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kolesar P., Sarangi P., Altmannova V., Zhao X., Krejci L.. Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 2012; 40:7831–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liberi G., Cotta-Ramusino C., Lopes M., Sogo J., Conti C., Bensimon A., Foiani M.. Methods to study replication fork collapse in budding yeast. Methods Enzymol. 2006; 409:442–462. [DOI] [PubMed] [Google Scholar]
- 44. Armstrong A.A., Mohideen F., Lima C.D.. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012; 483:U59–U99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kolesar P., Altmannova V., Silva S., Lisby M., Krejci L.. Pro-recombination role of Srs2 requires SUMO but is independent of PCNA interaction Peter. J. Biol. Chem. 2016; 291:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiolo I., Carotenuto W., Maffioletti G., Petrini J.H.J., Foiani M., Liberi G.. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 2005; 25:5738–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chavdarova M., Marini V., Sisakova A., Sedlackova H., Vigasova D., Brill S.J., Lisby M., Krejci L.. Srs2 promotes Mus81-Mms4-mediated resolution of recombination intermediates. Nucleic Acids Res. 2015; 43:3626–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sundararajan R., Gellon L., Zunder R.M., Freudenreich C.H.. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010; 184:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. León Ortiz A.M., Reid R.J.D., Dittmar J.C., Rothstein R., Nicolas A.. Srs2 overexpression reveals a helicase-independent role at replication forks that requires diverse cell functions. DNA Repair (Amst.). 2011; 10:506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J.E., Sung P. et al. . Yeast Mphl helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev. 2009; 23:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng X.F., Prakash R., Saro D., Longerich S., Niu H., Sung P.. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair (Amst.). 2011; 10:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Branzei D. Ubiquitin family modifications and template switching. FEBS Lett. 2011; 585:2810–2817. [DOI] [PubMed] [Google Scholar]
- 53. Johnson R.E., Henderson S.T., Petes T.D., Prakash S., Bankmann M., Prakash L.. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 1992; 12:3807–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cherng N., Shishkin A.A., Schlager L.I., Tuck R.H., Sloan L., Matera R., Sarkar P.S., Ashizawa T., Freudenreich C.H., Mirkin S.M.. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shishkin A.A., Voineagu I., Matera R., Cherng N., Chernet B.T., Krasilnikova M.M., Narayanan V., Lobachev K.S., Mirkin S.M.. Large-Scale Expansions of Friedreich's Ataxia GAA Repeats in Yeast. Mol. Cell. 2009; 35:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. House N.C.M., Yang J.H., Walsh S.C., Moy J.M., Freudenreich C.H.. NuA4 initiates dynamic histone H4 acetylation to promote high-fidelity sister chromatid recombination at postreplication gaps. Mol. Cell. 2014; 55:818–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Viterbo D., Michoud G., Mosbach V., Dujon B., Richard G.-F.. Replication stalling and heteroduplex formation within CAG/CTG trinucleotide repeats by mismatch repair. DNA Repair (Amst.). 2016; 42:94–106. [DOI] [PubMed] [Google Scholar]
- 58. Kang Y.H., Munashingha P.R., Lee C.H., Nguyen T.A., Seo Y.S.. Biochemical studies of the Saccharomyces cerevisiae Mph1 helicase on junction-containing DNA structures. Nucleic Acids Res. 2012; 40:2089–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gari K., Décaillet C., Stasiak A.Z., Stasiak A., Constantinou A.. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008; 29:141–148. [DOI] [PubMed] [Google Scholar]
- 60. Sun W., Nandi S., Osman F., Ahn J.S., Jakovleska J., Lorenz A., Whitby M.C.. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008; 32:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blastyák A., Pintér L., Unk I., Prakash L., Prakash S., Haracska L.. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007; 28:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Long D.T., Kreuzer K.N.. Regression supports two mechanisms of fork processing in phage T4. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:6852–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsang E., Miyabe I., Iraqui I., Zheng J., Lambert S.A.E., Carr A.M.. The extent of error-prone replication restart by homologous recombination is controlled by Exo1 and checkpoint proteins. J. Cell Sci. 2014; 127:2983–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopes M., Sogo J., Foiani M.. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005; 17:153–159. [DOI] [PubMed] [Google Scholar]
- 65. Lahue R.S., Slater D.L.. DNA repair and trinucleotide repeat instability. Front Biosci. 2003; 8:s653–s665. [DOI] [PubMed] [Google Scholar]
- 66. Burgess R.C., Lisby M., Altmannova V., Krejci L., Sung P., Rothstein R.. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell Biol. 2009; 185:969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Long D.T., Kreuzer K.N.. Fork regression is an active helicase-driven pathway in bacteriophage T4. EMBO Rep. 2009; 10:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Courcelle J., Donaldson J.R., Chow K.-H., Courcelle C.T.. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003; 299:1064–1067. [DOI] [PubMed] [Google Scholar]
- 69. Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M.. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001; 412:557–561. [DOI] [PubMed] [Google Scholar]
- 70. Schvartzman J.B., Martínez-Robles M.L., López V., Hernández P., Krimer D.B.. 2D gels and their third-dimension potential. Methods. 2012; 57:170–178. [DOI] [PubMed] [Google Scholar]
- 71. Yabuki N., Terashima H., Kitada K.. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002; 7:781–789. [DOI] [PubMed] [Google Scholar]
- 72. Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L.. Replication dynamics of the yeast genome. Science. 2001; 294:115–121. [DOI] [PubMed] [Google Scholar]
- 73. Su X.A., Dion V., Gasser S.M., Freudenreich C.H.. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 2015; 29:1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vaze M.B., Pellicioli A., Lee S.E., Ira G., Liberi G., Arbel-eden A., Foiani M., Haber J.E., Firc I.. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. 2002; 10:373–385. [DOI] [PubMed] [Google Scholar]
- 75. Lambert S., Mizuno K., Blaisonneau J., Martineau S., Chanet R., Fréon K., Murray J.M., Carr A.M., Baldacci G.. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell. 2010; 39:346–359. [DOI] [PubMed] [Google Scholar]
- 76. Burkovics P., Sebesta M., Sisakova A., Plault N., Szukacsov V., Robert T., Pinter L., Marini V., Kolesar P., Haracska L. et al. . Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 2013; 32:742–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Atkinson J., McGlynn P.. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009; 37:3475–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gupta S., Yeeles J.T.P., Marians K.J.. Regression of replication forks stalled by leading-strand template damage: I. Both RecG and RuvAB catalyze regression, but RuvC cleaves the holliday junctions formed by RecG preferentially. J. Biol. Chem. 2014; 289:28376–28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Robu M.E., Inman R.B., Cox M.M.. RecA protein promotes the regression of stalled replication forks in vitro. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M.. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015; 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Vannier J.-B., Sandhu S., Petalcorin M.I.R., Wu X., Nabi Z., Ding H., Boulton S.J.. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013; 342:239–242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.