Abstract
Introns represent almost half of the human genome, although they are eliminated from transcripts through RNA splicing. Yet, different classes of non-canonical miRNAs have been proposed to originate directly from intron splicing. Here, we considered the alternative splicing of introns as an interesting source of miRNAs, compatible with a developmental switch. We report computational prediction of new Short Intron-Derived ncRNAs (SID), defined as precursors of smaller ncRNAs like miRNAs and snoRNAs produced directly by splicing, and tested their dependence on each key factor in canonical or alternative miRNAs biogenesis (Drosha, DGCR8, DBR1, snRNP70, U2AF65, PRP8, Dicer, Ago2). We found that about half of predicted SID rely on debranching of the excised intron-lariat by the enzyme DBR1, as proposed for mirtrons. However, we identified new classes of SID for which miRNAs biogenesis may rely on intermingling between canonical and alternative pathways. We validated selected SID as putative miRNAs precursors and identified new endogenous miRNAs produced by non-canonical pathways, including one hosted in the first intron of SRA (Steroid Receptor RNA activator). Consistent with increased SRA intron retention during myogenic differentiation, release of SRA intron and its associated mature miRNA decreased in cells from healthy subjects but not from myotonic dystrophy patients with splicing defects.
INTRODUCTION
The discovery of non-coding RNAs (ncRNA) and the variety of molecular processes in which they have been implicated suggest that they increase and diversify the amount of regulatory molecules available in the cell. In contrast to housekeeping or infrastructural ncRNAs, which are normally constitutively expressed and required for normal function and viability of the cell, regulatory ncRNAs are expressed in response to external stimuli or at particular stages of development and cell differentiation, and can affect the expression of other genes at the level of transcription or translation (1). Among those, microRNAs (miRNA) are a class of naturally occurring small ncRNAs, about 20–25 nucleotides (nt) in length, which have been identified in almost all eukaryotic cells. MiRNAs are post-transcriptional regulators that bind to complementary sequences on target mRNA, usually resulting in translational repression or target degradation and thus, gene silencing (2). The human genome may encode over 1000 miRNAs, targeting ∼60% of gene products in mammals. Therefore, because they affect gene regulation and are often deregulated in human diseases, their systematic identification has been the focus of many experimental and computational analyses [for reviews see (3,4)].
Canonical miRNAs are generated in a two-step processing pathway, mediated by two major enzymatic complexes containing the RNAse III-family of endonucleases Drosha and Dicer. Drosha, together with DGCR8, is part of the microprocessor multiprotein complex that mediates nuclear processing of the primary miRNA into stem–loop precursors of ∼60–70 nt (pre-miRNA). Exportin-5 (XPO5) mediates the nuclear export of correctly processed miRNA precursors. In the cytoplasm, the pre-miRNA is cleaved by Dicer into the mature 20–25 nt miRNA, which is then incorporated as single-stranded RNA into a ribonucleoprotein complex containing Argonaute 2 (Ago2) protein, known as the RNA-induced silencing complex (RISC). This RISC complex directs the miRNA to its target mRNA leading to its translational repression or its degradation [for a review see (5)].
It has also been postulated that regulatory RNA molecules could originate from the introns of protein-coding genes as functional by-products (6–8). Several recent studies indeed uncovered an atypical pathway to generate miRNA precursors in a way that bypasses the Drosha/DGCR8 complex (9–16). Instead, the pre-miRNA-like hairpins are produced by the action of the splicing machinery followed by lariat-debranching by the enzyme DBR1. The 5΄- and/or the 3΄-tails around the hairpin are then trimmed by the RNA exosome (12). The mirtron pathway merges with the canonical miRNA pathway at hairpin export by XPO5, and subsequent processing of hairpins by Dicer. These unusual miRNA precursors are called mirtrons owing to their embedding into introns of coding and non-protein coding genes (9,14,16). Only a few mirtrons have been described to date although they have been shown to exist from drosophila to humans (9–16). More recently, two other unconventional pathways have been described; the simtron pathway involves a small nuclear ribonucleoprotein (snRNP), which is part of the spliceosome complex snRNP70 that directly recruits Drosha on the stem–loop hairpin formed by the pre-mRNA, independently of DGCR8 (17,18). After cleavage by Drosha, pre-miRNAs are exported to the cytoplasm. The agotron pathway is more intricate given that the spliced introns are processed by Ago2 directly in the nucleus. Hence, miRNAs generated by the agotron pathway are processed independently of Drosha and Dicer (19). Interestingly, several miRNAs have been described to be processed from snoRNAs (20) or tRNAs (21) [for a review see (22)] and the description of non-canonical pathways has accumulated over the years (15,17,19,23–26).
As we just discussed above, many new unconventional miRNAs, and most if not all small nucleolar RNAs (snoRNAs), are produced from introns through splicing mechanisms. Although introns, which represent about half of the genome in mammals, have long been considered as ‘junk’ and ‘dark matter’ since they are quickly degraded within minutes following their excision, an increasing number of studies have shown the importance of introns in producing small ncRNAs. The possibility that there are more to be discovered, expressed at lower levels or in more specialized cellular contexts, calls for the exploitation of genome sequencing information to accelerate their discovery and ease their structural characterization.
We propose herein the bioinformatic identification of human candidates as intron-derived small ncRNAs precursors through data mining of publically available information provided by high-throughput sequencing projects. We used the term SID as an acronym for Short Intron-Derived ncRNAs precursors of smaller ncRNAs like miRNAs and snoRNAs regardless of their biogenesis pathway but produced directly from splicing. Hence, this term may group precursors of snoRNAs that are already known to originate from splicing of introns (27,28), and non-canonical precursors of miRNAs like agotrons and mirtrons (15,19) or other classes of splicing-dependent miRNA precursors yet to be identified.
We focused on small introns (shorter than 200 nt) since they are more prone to alternative splicing (29). We provide a list of 56 new human SID, extracted from databases of alternative splicing events and matching entries in databases of human small RNAs. We further experimentally validated that these SID candidates were indeed detectable, in a panel of six cell lines and primary cells, as discrete and persistent entities. Among the highest detectable candidates in human cells, we validated six of them, which indeed produced a miRNA with variable dependence on known factors of miRNAs pathways. We employed several independent approaches to characterize the miRNA of one of these new SID that is hosted in the first intron of SRA (Steroid Receptor RNA activator). Of particular interest, we already showed that alternative splicing serves to increase the transcriptional output of the SRA1 gene, by producing a protein-coding gene when this intron is excised or a functional non-coding RNA when this intron is retained, disrupting the open reading frame (ORF), concomitant with myogenic differentiation (30,31). We now show that splicing of intron 1 of SRA generates a new miRNA with high levels in undifferentiated myogenic progenitors.
As a whole, this work represents the first experimental validation of human endogenous SID and identified new classes of small ncRNAs of intron origin. It also puts forward that alternative splicing might represent an attractive source of small regulatory RNAs compatible with a tissue-specific or differentiation-stage specific role, which will provide new actors or biomarkers deregulated in disease.
MATERIALS AND METHODS
Data mining and data sets
Datasets of ‘Human Introns’, ‘Alternative Events’, ‘CpG island’, ‘SNP’ and ‘Genes’ were retrieved from the UCSC table browser, using ‘Genes and gene prediction’, ‘Alt events’, ‘Regulation’ and ‘RefSeq Genes’ tracks from the hg19 assembly covering the whole human genome. Using Galaxy and ‘Operate on Genomic Intervals’ tool, we selected introns shorter than 200 nucleotides and 100% covered in ‘Alternative Events’ datasets. Location of intron retention relative to the start and stop codons was obtained by genomic coordinate comparison (from ‘Genes’ dataset). Palindrome formation of selected introns was assessed using ‘palindrome search’ tool (parameters: minimum length of palindrome: 18; maximum length of palindrome: 28; Maximum gap between repeated regions: >100; number of mismatches allowed: <6). Finally, putative SID (Short Intron-Derived ncRNA) and non-SID introns were manually curated to remove potential redundancy. All excluded introns (1713), i.e. which did not present the ability to form a palindrome, were used to construct the NI (normal introns) dataset. The miRNA dataset (1426) was downloaded from miRBase (http://www.mirbase.org/; release 17), the snoRNA dataset (402) was downloaded from snoRNABase (https://www-snorna.biotoul.fr; v3) and a random dataset (10000) was generated using the Random DNA Sequence Generator software. Data for small RNA-seq was obtained from ENCODE/Cold Spring Harbor Lab track from the UCSC table browser (GEO DataSet number GSE24565; part of the BioProject 30709) and consist in NextGen sequencing information for RNAs between 20–200 nt in size, isolated from RNA samples from tissues or sub-cellular compartments of cell lines (K562, GM12878 and prostate cells).
Datasets analysis
All tools were from The European Molecular Biology Open Software Suite EMBOSS. GC content was calculated using ‘infoseq’; CpG islands were mapped using ‘CpG islands’ track from UCSC browser. Single Nucleotide Polymorphisms were searched using the dbSNP132 track from UCSC browser. RNALfold, which calculates local stable secondary structures of RNAs, was used with default parameters. Minimum free energies (MFE, kcal/mol) were corrected relative to the size of the transcripts.
Software and databases
Software and web servers used were as follows: The Mfold web server v2.3, http://mfold.rna.albany.edu; UCSC genome Browser, http://genome.ucsc.edu; Galaxy, http://main.g2.bx.psu.edu; Genomatix Software Suite v2.1, http://www.genomatix.de; Venny, http://bioinfogp.cnb.csic.es; Random DNA sequence generator, http://users-birc.au.dk; miRBase v17; http://www.mirbase.org; snoRNABase v3, https://www-snorna.biotoul.fr; miRNA Primer Design Tool, http://genomics.dote.hu:8080/mirnadesigntool; RNALfold, http://www.tbi.univie.ac.at.
Cell culture
Primary human satellite cells (LHCN-M2, myoblast, MB) and their in vitro differentiated myotubes (MT) counterpart, HEK-293, K562, MCF-7 and MDA-MB-231 cells were grown as previously described (30,31). RNA from primary human satellite cells isolated from muscle biopsies of three foetuses showing clinical symptoms of the congenital Myotonic Dystrophy type 1 (DM1), and from one immortalized cell line (DM11), all carrying more than 2000 CTG repeats, were a gift from Denis Furling (Myology Institute, Paris, France) and were used previously (31,32).
Antibodies
Primary antibodies used were directed against Drosha (ab12286, Abcam), DGCR8 (ab90579, Abcam), DBR1 (sc-99369, Santa Cruz), snRNP70 (sc-9571 c-18, Santa Cruz), PRP8 (sc-55534 F-6, Santa Cruz), U2AF65 (sc-48804 H-300, Santa Cruz), Dicer (sc-56651), Ago2 (sc-32877 H-300, Santa Cruz) and γ-Tubulin (T6557, Sigma).
Plasmids and constructs
pSicoR human Dicer1 (ID 14763), pSicoR human Drosha1 (ID 14766) and pSicoR human DGCR8-1 (ID 14769) were requested from Addgene (www.addgene.org). Short hairpin RNAs (shRNA) directed against human DBR1 (DBR1-1, DBR1-2 and DBR1-3), snRNP70+PRP8, U2AF65+PRP8, Ago2 (Ago2-1 and Ago2-2) and Luciferase were produced using MessageMuter™ shRNA production kit (Epicentre Biotechnologies) and in vitro transcribed using T7 RiboMax large-scale production system (Promega) following manufacturer's instructions. Sequences are depicted in Supplemental Table S2.
Knockdown of snRNP70 or U2AF65 was combined to that of the major splicing factor PRP8, since interference of snRNP70 or U2AF65 alone was insufficient to reduce splicing significantly in another study (18).
RNA preparation
Total RNA was isolated using Trizol reagent (Life Technologies) according to the manufacturer's instructions and as previously described (31,33). Short and long RNAs were purified using mirVana™ miRNA Isolation Kit (Life Technologies) according to instructions.
Oligonucleotide array
Oligonucleotides, 45–55 nt in length and complementary to candidate SID, at a final concentration of 10 μM, were spotted on GeneScreen Plus (PerkinElmer) membranes using a 96 well manifold system (BioRad). To confirm the specificity of hybridization, we included a series of control oligonucleotides on the array. Briefly, RNU6-1 (U6) oligonucleotide served as loading control, two randomly-generated oligonucleotides were used to set the baseline signal and two oligonucleotides matching constitutive exon 4 of GAPDH and exon 4 of SRA mRNA were used as size fractionation controls. All oligonucleotides were first phosphorylated using 1U Polynucleotide kinase (New England Biolabs) per microgram of oligonucleotide for 2h at 37°C followed by enzyme inactivation by addition of 1 vol. of 0.5 M EDTA.
Membranes were briefly washed once with 1× MOPS–NaOH pH 7 and once with 0.1× SSC, 0.125 N NaOH. Oligonucleotides were denaturated in 500 mM NaOH before being diluted in 0.1× SSC, 0.125 N NaOH and spotted onto membranes. Membranes were further washed again twice in 1× MOPS–NaOH pH 7. Spotted oligonucleotides were cross-linked to the membrane using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) method described in (34). Arrays were not stored but used directly after spotting.
Five micrograms of small RNAs freshly isolated from cell lines, transfected or not with shRNA, were preheated at 80°C for 3 min, cooled on ice, and labelled overnight at 37°C using 1 U poly (A) polymerase and 50 μCi 32P-α-ATP in the presence of 40 U RNAse Out (Life Technologies).
Membranes were pre-hybridized in Ultrahyb®-oligo (Life Technologies) at 42°C for at least 30 min followed by an overnight hybridization in the same solution containing the RNA probe. Following hybridization, membranes were washed twice with 2× SSC/0.5% SDS at 42°C and once at 1× SSC/0.5% SDS. Membranes were exposed to a phosphor storage screen, scanned using Phosphor Imager (Fujifilm), and hybridization signals were quantified using Multi Gauge v3 (Fujifilm). Membranes were never stripped and used only once.
Hybridization signals for each spot of the array and background values at eight empty spots were measured. Averaged hybridization signal at empty spots was subtracted from each other spot signal and normalized using the U6 signal. Signals from spots with random oligonucleotides were averaged and served as baseline value. Signals from candidate SID from among duplicate spots and triplicate arrays were averaged and considered positive when being over the baseline value. Hybridization with RNA from the different cell types was performed three times (n = 3), or four times (n = 4) following RNA interference in K562 cells.
Northern blot
RNA northern blot using short RNA fractions were performed as described by Pall and Hamilton (34) using 5 μg of small RNAs purified as described above and probed with the 32P-γ-ATP radio-labelled oligonucleotides used in the macro-array.
Pocket-sized RNA-seq
The method was fully described in reference (35). Sequences used to produce human SRA intron 1 or luciferase baits are provided in Supplemental Table S2.
RT-PCR, RT-qPCR and stem–loop RT-qPCR
RNA was isolated as described above and reverse transcribed as described previously (30,31,33,36,37). Primers used for amplification are described in Supplemental Table S2. Radio-labeled PCR products were separated on denaturating poly-acrylamide/urea gels as previously described (38). Following electrophoresis, the gels were dried and exposed to a phosphor storage screen, and scanned as above (n = 2). Quantitative PCR was carried out using the LightCycler® 480 SYBR Green I master on the Light cycler 480 real-time PCR system (Roche Diagnostics). For stem–loop RT-qPCR, reverse transcriptase reactions were performed replacing random hexamer primers by stem–loop primers. All reactions, including no-template controls and RT-minus controls, were run in duplicate (n = 3). Relative expression of genes was calculated by using the comparative CT method (30,31), i.e. 2−((shMock sample – shMock control) – (shTarget sample – shTarget control)). Expression of immunoprecipitated RNAs was obtained by calculating the ‘relative to the irrelevant antibody’ method, i.e. 2−(Ct specific Ab – Ct irrelevant Ab) and was expressed as fold enrichment (n = 2). Mature miRNA-specific primers were designed using miRNA Primer Design Tool Website and normalization was performed relative to the signal of RNU6-1 (U6) amplification. All primers used in qPCR experiments had efficiencies >98%, much more above the MIQE requirement.
RNA and protein immunoprecipitation
Cells were lysed, RNA and proteins were extracted and quantified as described previously (31) and used for native RNA immunoprecipitation (RIP) or protein immunoprecipitation experiments, respectively. Protein extracts (1 mg) were incubated with the appropriate antibody (1 μg/mg of proteins) for 2 h at 4°C as described earlier (33,36). Total proteins were analyzed by SDS-PAGE and immunoblotting as previously described (36) or when appropriate, co-precipitated RNA was extracted using Trizol method, reverse transcribed and PCR amplified as described above (n = 2).
Transfection experiments
Plasmid constructs and shRNA were transiently transfected (n = 3) using jetPEI™ and interferin reagents (PolyPlus transfection), respectively, following the manufacturer's instructions and as previously described (31).
RESULTS
Bioinformatic prediction of candidate Short Intron-Derived ncRNA (SID)
In order to systematically identify SID in the human genome database we used the workflow outlined in the Material and Methods section and in Supplementary Figure S1, starting from 326, 537 introns extracted from UCSC table browser. Since 100% of pre-miRNA hairpins (calculated from miRBase) and 95% of snoRNA (calculated from snoRNABase) are not >200 nt in length, our computational strategy retained only introns <200 nt but >50 nt so they can form a hairpin compatible with the size of a pre-miRNA or a snoRNA. This restriction has also been chosen for further experimental validation after RNA size fractionation that imposes a cut-off around 150–200 nt. Although splicing of introns is a prerequisite to produce mirtrons and snoRNAs, we reasoned that alternative splicing events would be of prime interest to account for their regulatory functions during cellular differentiation or to integrate external or environmental signals. Therefore, the 28 118 extracted introns of 50–200 nt in size were further confronted with 92 222 alternative splicing events from UCSC (39), of which 6450 represent retained introns. This first step led to the identification of 2147 short introns subjected to alternative splicing. In addition to effective splicing and intron length, structural characteristics are also important to define a SID. We thus selected 218 unique introns shorter than 200 nucleotides, which contained inverted repeats necessary for the formation of a stem–loop structure. These short hairpin introns, unlike typical introns that are rapidly degraded following splicing (40), are much more stable in the cell. Thus, out of these 218 introns, we retained introns that matched collections of small RNAs of 20–200 nt in length isolated from tissues or sub-cellular compartments from ENCODE cell lines (http://genome.ucsc.edu/ENCODE/) and deep sequencing of small RNA-seq samples from ENCODE project (GEO Series accession number GSE24565), potentially containing both discrete and stable introns or mature miRNAs.
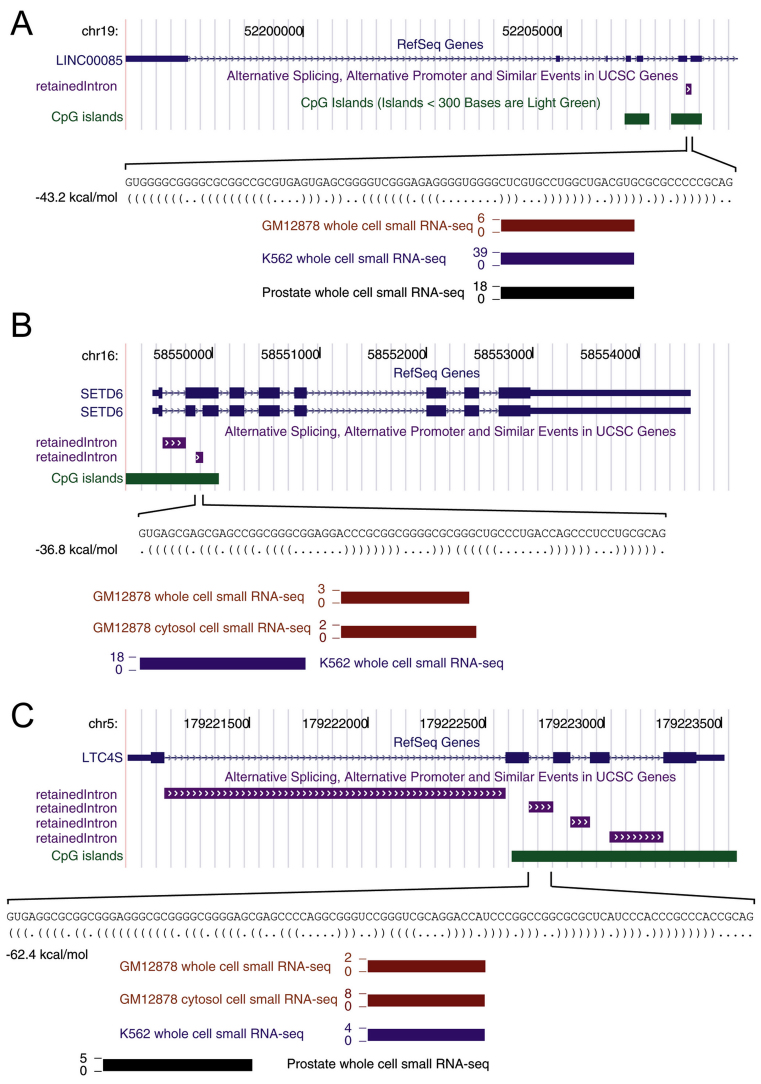
Overall, we identified 56 introns that are compatible in length, structure and sequence, with the formation of SID hairpins and for which small RNAs exist in small RNAs databases (Figure 1 and Supplementary Table S1).
Figure 1.
typical example of SID candidates. (A) Schematic representation of the human LINC00085 gene located on chromosome 19 (chr19:52, 196, 593-52, 208, 443 described as SID #39 in Supplementary Table S1). The sixth intron was identified as a retained intron in database. In this particular example, the gene considered was described as a non-coding RNA (NR_024330). (B) Schematic representation of the human SETD6 gene located on chromosome 16 (chr16:58, 549, 383-58, 554, 456 described as SID #8 in Supplementary Table S1). The second intron was identified as a retained intron in database. In this particular example, the two transcript variants 1 and 2 have been already described as NM_001160305 (containing the intron in frame with the ORF) and NM_024860, respectively. (C) Schematic representation of the human LTC4S gene located on chromosome 5 (chr5:179, 220, 986-179, 223, 512 described as SID #47 in Supplementary Table S1). Although its four introns were described as retained introns in database, only the second intron was compatible in size. (A–C) A zoom capture is presented below each gene map including small RNA-seq data obtained from ENCODE in three cell lines (GM12878, K562 and prostate cells and fractionated data when available). Sequence and structure determined using RNALfold are also indicated.
Structural features of candidate Short Intron-Derived ncRNA (SID)
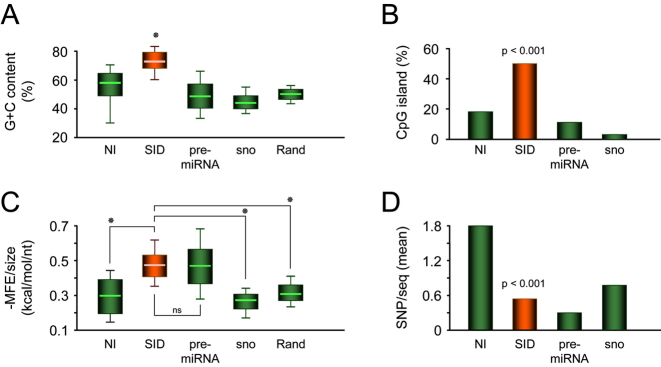
We then analyzed structural features of the putative 56 new human SID compared to the excluded normal introns (NI) and to annotated pre-miRNAs and snoRNAs extracted from miRBase v17 and snoRNABase v3, respectively. We showed that human SID exhibit a higher G+C content (Figure 2A) and preferentially reside within CpG island regions (Figure 2B). Furthermore, the stability of their secondary structure is higher than that of NI, snoRNAs (41) or random sequences as determined using RNALfold, and is comparable to that of annotated pre-miRNAs (Figure 2C). Altogether, these findings suggest that formation of a pre-miRNA-like structure is not a common feature of introns but is a structural characteristic of both canonical pre-miRNAs and SID. Interestingly, searching for genomic common single-nucleotide polymorphism (SNP), we found that the mean number of SNP per intron is almost three-times higher in NI than in SID (Figure 2D). These results are consistent with a higher selective pressure on SID than on NI, equivalent to that on pre-miRNA (42) and snoRNAs (43), and suggestive of the functional importance of these sequences.
Figure 2.
Structural and sequence features of newly identified SID. (A) G+C content of NI (introns; n = 1713), SID (n = 56), pre-miRNAs (from miRBase v17; n = 1426), snoRNAs (sno, from snoRNABase v3; n = 402), and random sequences (generated as described in Materials and Methods; n = 10 000), all with similar nucleotide length, were analyzed and plotted as box-and-whisker diagrams. (B) Percentage of sequences that mapped in an annotated CpG island region. (C) Corrected MFE (minimum free energy) was calculated for each sequence by dividing the absolute value obtained from RNALfold program (see Materials and Methods) by the corresponding sequence length. Data were plotted as box-and-whisker diagrams. (D) Total number of SNP was calculated for each dataset using Galaxy tools and the dbSNP132 track and a mean of SNP per sequence was then plotted. (A and B) * mean P < 0.001 (Student's t-test). (C and D) Statistical analyses were performed using Chi-squared test.
Our approach was further validated with the finding that previously identified human mirtrons (44) like pre-miR-1234 (SID #44) and pre-miR-6743 (SID #26) were present in our data set (Supplementary Table S1). Moreover, two of the identified host genes, TCIRG1 (T-Cell, Immune Regulator 1, ATPase, H+ Transporting, Lysosomal V0 Subunit A3) hosting SID #12 and CPSF1 (Cleavage and polyadenylation specificity factor 1) hosting SID #44 and #46, were also host genes for agotrons (19).
Experimental validation of SID candidates
To test whether the 56 potentially new human SID could be detected as discrete entities, we developed low-density oligonucleotide arrays hybridized with RNA extracted from cultured cells and fractionated in size (Supplementary Figure S2). Oligonucleotides matching the 56 SID were spotted as duplicates. Positive controls for hybridization were designed to detect highly expressed RNU6-1 snRNA (U6) and the hsa-miR-21 expressed in most tumour cell lines. Negative controls consisted of randomly chosen oligonucleotides. Oligonucleotides matching exons of housekeeping mRNAs served as controls for size fractionation since exon-containing mRNAs should not be found in the small RNA fraction. Arrays were hybridized using radio-labelled RNA probes extracted from six different human cell types and fractionated to retain RNAs shorter than 200 nt in length. Hybridization signals were quantified and normalized as described in the Material and Methods section. Out of the 56 candidates, 36 showed signals above baseline levels in at least one cell type, representing our best candidates as new human SID (Supplementary Table S1). Although SID candidates were selected using a specific workflow that integrates alternative splicing events, we first validated expression of their host gene as well as alternative splicing of their RNA precursors as measured by detection of intron-retaining or -spliced isoforms, using radioactive RT-PCR (Supplementary Figures S3 and S4). All but 3 (#1, #27, #47) of the 36 SID candidates showed retention and excision events, even weak, in at least one of the six chosen cell lines. Candidates #6 and #8 showed intermediate amplification products in all cell lines studied and #36 showed intermediate amplification products in MDA-MB-231 cells (Supplementary Figures S3), consistent with alternative acceptor or donor splicing sites already reported for candidates #6 and #8, respectively (http://fasterdb.ens-lyon.fr). Cloning and sequencing of candidate #36 intermediate product also identified an alternative non-canonical (GT→GA) donor splicing site (Supplementary Figures S5).
As a whole, 33 SID (Supplementary Table S1) were detectable in at least one of the six cell lines, and showed variable levels consistent with introns originating from alternative splicing events being cell-type- or differentiation-stage- specific. For example, SID #10, #24 and #39 showed higher levels in ERalpha-negative, highly invasive, fibroblast-like MDA-MB-231 than in ERalpha-positive, weakly invasive, luminal epithelial-like MCF-7. In contrast, SID #42 showed higher levels in MCF-7 cells than in MDA-MB-231. Similarly, SID #39 and #43 showed higher levels in undifferentiated human myoblasts than in their differentiated counterpart, whereas SID #8 and #53 increased during myoblast differentiation. Some SID, such as #10, #16, #24 and #39, were detected in all six cell types at high levels. In contrast, high levels of #42 and #46 were a hallmark of cancer cells whatever the origin of the tumour, whereas levels of #21 were low in cancer cell lines compared to normal myogenic cells.
Biogenesis of miRNAs from SID candidates
In order to decipher the pathways involved in the biogenesis of miRNAs from SID, we used the 19 candidates that showed signals above baseline levels in the ENCODE common cell line K562, in which we knocked down all major players involved in miRNAs, mirtrons, simtrons and snoRNAs biogenesis, using RNA interference (see Material and Methods section and Supplementary Table S2). Size fractionated RNAs extracted from K562 were radio-labelled and then used as probes on low-density oligonucleotide arrays to assess the impact of knock down assays on individual SID levels. RT-PCR and western blots controlling efficiency of RNA interference are shown in Supplementary Figure S6A and B, respectively. We used shRNA directed against luciferase gene as a mock control (Supplementary Table S2).
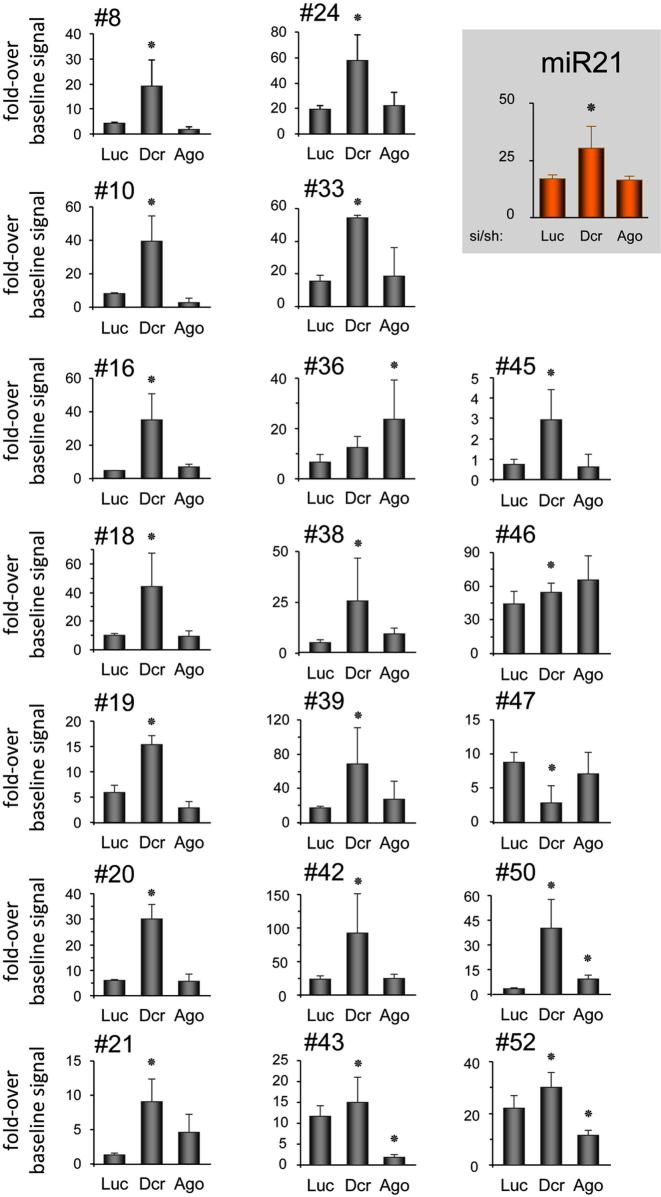
We first performed experiments to knock down Dicer and Ago2 (Figure 3). In addition to controls for size fractionation already mentioned earlier for the experimental validation of SID candidates, the increased signal following Dicer knockdown suggests that only SID, i.e. precursors of smaller ncRNAs that accumulate as the result of their impaired processing into smaller ncRNAs, are detectable by the method. For all SID candidates but 2 (#36 and #47), we observed an accumulation of SID in conditions of reduced levels of Dicer protein (Figure 3), suggesting that Dicer is indeed involved in SID maturation into smaller RNAs as shown for canonical miRNAs. As expected, decreased expression of Ago2 did not affect levels of most SID, although increased levels of SID #36 might be explained by the previously described dicing activity of Ago2 that can replace Dicer in pre-miRNA processing (24,25).
Figure 3.
Role of Dicer and Ago2 in biogenesis of SID. Oligonucleotide arrays developed in Supplementary Figure S2, and spotted with the SID that shows a signal above baseline in K562 cells (Supplementary Table S1) were hybridized with radio-labeled small RNA isolated from K562 cells in which Dicer, Ago2 or Luc were knocked down (n = 4). Hybridization signals were plotted as a fold-over baseline signal as described in Material and Methods section. Luc, control luciferase knockdown; Dcr, Dicer knockdown; Ago, Ago2 knockdown. The hsa-miR-21 (miR21) was used as positive control. Significant differences were assessed using Student's t-test (*P < 0.05). Error bars represent standard error at the mean (SEM).
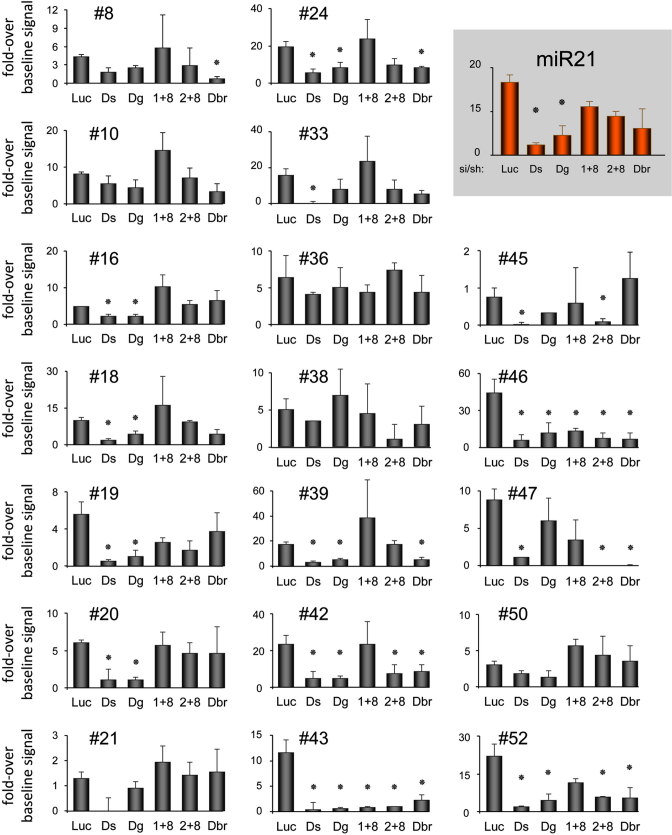
We then used shRNAs directed against Drosha and DGCR8 that compose the microprocessor (45), snRNA70+PRP8 and U2AF65+PRP8 that are components of the splicing machinery (18), and DBR1 that debranches intron lariats (46) (Figure 4). The 19 SID showed distinct dependency for one to several of these proteins, allowing to classify them in five different categories: (i) the members of the first category correspond to classical pre-miRNAs and showed dependency on the microprocessor only. It included pre-miR-21, used here as a control, and SID #16, #18, #19 and #20; (ii) the second category was composed of one mirtron candidate (#8), as it showed dependency on the debranching enzyme DBR1 but not on the microprocessor components or snRNP70+PRP8 and U2AF65+PRP8; (iii) SID #45 was dependent on Drosha and snRNP proteins, as shown previously for simtrons (17,18); (iv) a new category of candidates, including #24, #39, #42, #43, #46, #47 and #52, showed dependency on both the microprocessor (Drosha and DGCR8) and DBR1; (v) unclassified SID for which no dependency could clearly and/or significantly be determined, corresponding to SID #10, #21, #33, #36, #38 and #50.
Figure 4.
Role of Drosha, DGCR8, snRNP70+PRP8, U2AF65+PRP8 and DBR1 in biogenesis of SID. Oligonucleotide arrays developed in Supplementary Figure S2, and spotted with the SID that shows a signal above baseline in K562 cells (Supplementary Table S1) were hybridized with radio-labelled small RNA isolated from K562 cells in which Drosha, DGCR8, snRNP70+PRP8, U2AF65+PRP8, DBR1 or Luc were knocked down (n = 4). Hybridization signals were plotted as a fold-over baseline signal as described in Material and Methods section. Luc, control luciferase knockdown; Ds, Drosha knockdown; Dg, DGCR8 knockdown; 1+8, snRNP70+PRP8 knockdown; 2+8, U2AF65+PRP8 knockdown; Db, DBR1 knockdown. The hsa-miR-21 (miR21) was used as positive control. Significant differences were assessed using Student's t-test (*P < 0.05). Error bars represent standard error at the mean (SEM).
Physical interaction of SID candidates with key factors involved in biogenesis of miRNAs
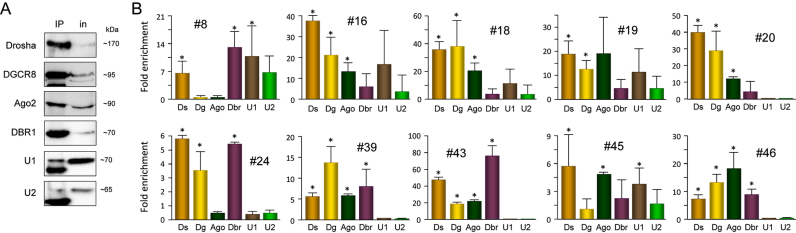
Because knock down experiments could lead to indirect effects, and to test whether proteins involved in these pathways act directly on the processing of SID into miRNAs (Figure 5), we performed native RIP to detect interactions of a given SID with proteins implicated in the canonical miRNA pathway (microprocessor proteins Drosha and DGCR8), in intron-lariat debranching (DBR1) or in splicing (snRNP70, component of the U1 complex; U2AF65, component of the U2 complex). We also assessed the interaction of SID with Ago2, which is a prerequisite to suggest loading into the RISC complex and hence, to suggest a regulatory function for the resulting miRNA. Western blots controlling efficiency of immunoprecipitation assays are shown in Figure 5A. Results shown in Figure 5B are expressed as a fold enrichment relative to immunoprecipitation using an irrelevant antibody.
Figure 5.
SID interactions with miRNA pathway key factors. (A) Native RIP was performed using the indicated antibodies and the effective protein precipitation was confirmed by western blotting. IP, immunoprecipitation; in, 5% input. (B) Co-precipitated RNAs were extracted as described in Material and Methods section (n = 2). Quantitative PCR were performed using specific primer pairs (Supplementary Table S2) to amplify SID #8, #16, #18, #19, #20, #24, #39, #43, #45 and #46. Amount of immunoprecipitated material is expressed as fold enrichment compared to irrelevant antibody. Ds, Drosha; Dg, DGCR8; Ago, Ago2; Dbr, DBR1; U1, snRNP70; U2, U2AF65. Significant differences with irrelevant antibody were assessed using Student's t-test (*P < 0.05). Error bars represent standard error at the mean (SEM).
As anticipated from data described above, SID #16, #18, #19 and #20 were significantly associated with Drosha and DGCR8, but not with DBR1 nor snRNP70 or U2AF65. Conversely, SID #45, which we classified as a simtron-like, showed a significant association with Drosha and snRNP70 as expected (17,18). SID #43, which we have demonstrated the dependence on Drosha and DBR1 by RNA interference (Figure 4), was indeed associated with these two proteins. However, SID #8 that we classified as a mirtron according to its dependency on DBR1 but not on the microprocessor (14), unexpectedly co-precipitated with Drosha and snRNP70 as well as with DBR1.
Because of the direct interactions described above, SID #8, that we defined as a mirtron, should be referred to as a mirtron-like RNA since it also interacts with snRNP70, which has never been described in the case of mirtrons. Similarly, SID #45, which we classified as a simtron, should be referred to as a simtron-like because of its interaction with Ago2 that is dispensable in the simtron pathway (17). Of note, it is not so surprising to find the full SID associated with the Ago2 complex since Dicer, Ago2 and TAR RNA-binding protein 2 (TRBP2) that compose the RISC-loading complex (RLC) are first loaded together on the stem–loop before cleavage of the loop by Dicer and incorporation of the guide-strand into the RISC complex and repression of the target RNAs (47–50).
Data from Figures 3, 4 and 5 relative to the biogenesis of SID candidates (interference experiments and native RIP assays) are summarized in Supplementary Table S3 in which we propose a categorization of SID candidates.
Mature miRNAs originating from SID candidates
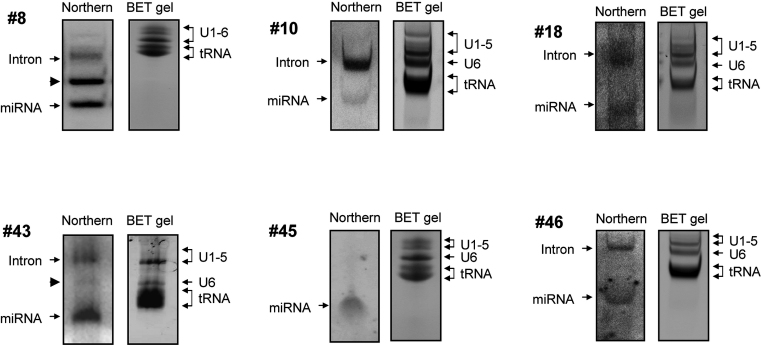
In order to confirm that SID candidates are genuine precursors of smaller ncRNAs, in particular mature miRNAs, we developed northern blot dedicated to the detection of endogenous small RNAs (see Material and Methods section). We have arbitrarily selected SID candidates from each category described above, i.e. #8, #10, #18, #43, #45 and #46. As shown in Figure 6, northern blot analyses further revealed that endogenous miRNAs originating from these six SID were indeed detectable in the small fraction of RNAs. These results strongly suggest that these introns meet the requirements to be classified as new human SID. For reasons that are not clear yet, the intermediate product corresponding to the hairpin (pre-miRNA) has never been detected in this study, except for SID #8 and SID #43. Likewise, many pre-miRNAs have not been detected in other studies either, like for hsa-miR-148a (51), hsa-miR-17 (52) or mmu-miR-103 (53). It must also be emphasized that only the mature small RNA with the expected size for miRNAs was detected using a probe against SID #45 that we classified as a simtron-like. Indeed, in the case of simtrons, the primary sequence (pri-miRNA) does not correspond to the intron alone but to the pre-messenger RNA (which is larger than 200 nt and therefore eliminated from the size fractionation).
Figure 6.
Small RNA isolated from HEK-293 cells were analysed by northern blot using specific radio-labelled probes complementary to SID #8, #10, #18, #43, #45 and #46. The corresponding acrylamide gels stained with ethidium bromide are shown. The upper and lower bands corresponding to the SID and its miRNA product, respectively, are indicated by small arrows, and the intermediate product for #8 and #43 is indicated by an arrow-head.
Characterization of a new mature miRNA processed from SID #43
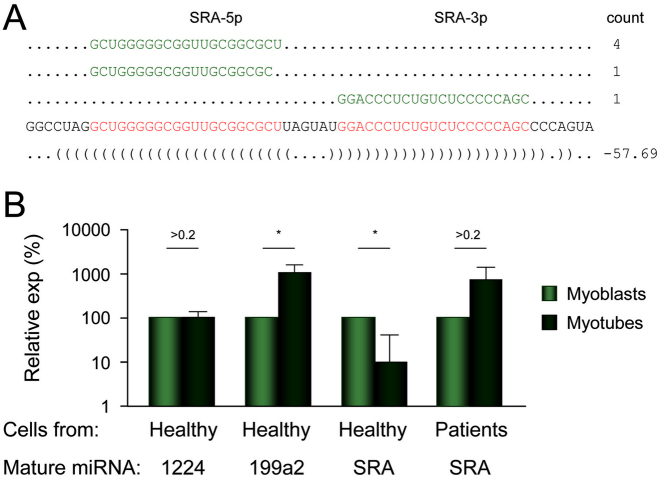
SID #43 corresponds to intron 1 of SRA1 (Steroid Receptor RNA Activator 1) gene for which we already reported the importance of alternative splicing in other contexts (30,31,38,54). In order to clone and sequence the exact mature sequence of the miRNA processed from intron 1 of SRA1, we used the method that we recently developed (35) based on in vitro transcription, RNA pull down and adapted RACE-PCR techniques. As shown in Figure 7A, we were able to characterize the mature SRA miRNA, allowing to asses its expression profile in myoblasts and myotubes using stem–loop RT-qPCR. We used hsa-miR-1224 and hsa-miR-199a2 as positive controls. During myogenic differentiation of human healthy myoblasts to myotubes, we observed the previously reported increase in miR-199a2 during myogenic differentiation (55). Consistent with the previously described decrease in SID #43 levels during the course of myoblast differentiation (Supplementary Table S1), levels of the mature miRNA derived from SID #43 also decreased as measured by stem–loop RT-qPCR (Figure 7B). It is worth noting that the decrease in both SID #43 and mature miRNA levels was consistent with the increased retention of intron 1 in long SRA isoforms that we recently reported during this process (31). We also detected the mature miRNA that derive from SID #43 in cells isolated from DM1 patients (see Material and Methods) by stem–loop RT-qPCR (Figure 7B). However, and in contrast to what was observed in muscle satellite cells from healthy donors, we did not detect changes in the levels of mature SRA miRNA, consistent with the previously identified defect in alternative splicing of SRA intron 1 in DM1 cells (31).
Figure 7.
A novel miRNA is produced from SID #43 (SRA intron 1). (A) Out of the 16 clones that were sequenced, the six corresponding to SRA intron 1 are positioned. (B) Relative expression of indicated mature miRNA amplified by stem loop RT-qPCR in undifferentiated or differentiated human myogenic cells (n = 3). 1224, mature miR-1224; 199a2, mature miR-199a2; SRA, mature SRA-miRNA; exp, expression; Healthy, cells from healthy control; Patients, cells from DM1 patients (n = 3). Significant differences were assessed using Student's t-test (*P < 0.05). Error bars represent standard error at the mean (SEM).
Prediction of target genes of mature miRNAs produced from SID #8 and SID #43
Because the prediction of a stem–loop secondary structure is not sufficient to predict a miRNA seed sequence without cloning and sequencing of the mature miRNA as performed above for SRA miRNA (from SID #43), we mined the large amount of publically available small RNA-seq data to identify the most likely sequence for a mature miRNA produced from candidate SID. It was only in the case of SID #8 that we found enough sequencing reads to delineate a miRNA sequence (Supplementary Figure S7). Using sequences of miRNAs from SID #43 characterized above and #8 inferred from RNA-seq data, we predicted target genes as described in the legend of Supplementary Figure S8 and assessed their expression levels using RNA-seq data from human cell lines. Interestingly, in cell lines expressing high levels of the candidate mature miRNAs originating from SID #8 (MCF7) or SID #43 (MB), expression levels of the predicted target genes were lower than in cell lines in which the miRNA was not expressed or at low levels (HUVEC for SID #8 and MT for SID #43). Such anti-correlated expression profiles between miRNAs and their predicted target genes suggest that miRNAs produced from candidate SID identified in this study are genuine regulatory small ncRNAs.
DISCUSSION
Alternative splicing is the first phenomenon that made scientists realise that genomic complexity is not proportional to the number of protein-coding genes. More recently, advances in high throughput transcriptome analysis highlighted the pervasive nature of transcription of mammalian genomes and revealed an ever-growing number of non protein-coding yet functional transcripts, which amount parallels that of genomes complexity (6,56). Furthermore, the recent characterization of RNAs for which both coding capacity and activity as functional RNAs have been reported, adds an additional degree of complexity (30,31,57,58). We recently proposed that intron retention contributes to the diversification of the information carried by genes by producing functional RNA instead of a protein product (58). This concept was exemplified by SRA1, for which transcripts exist as coding and non-coding isoforms, through alternative splicing of the first intron (30,31). Adding to the complexity, a number of functional ncRNAs are known to be hosted within introns (59). Although many of them can be produced independently of the transcription of their host gene, others are produced in a post-splicing manner, suggesting that alternative splicing might also serve to produce regulatory RNAs (60). For example, snoRNAs and miRNAs can originate from the processing of debranched introns after their excision from the pre-mRNA (16,59,61–63). Recent work reported that a subset of these spliced and debranched introns indeed has the ability to form hairpin structures compatible with their export to the cytoplasm and further processing into miRNAs by Dicer in the case of mirtrons (9,15), or bypassing Dicer in the case of agotrons (19). Thus, a single transcription unit could generate multiple molecules including proteins, long or smaller regulatory ncRNAs such as miRNAs, depending upon the need of the cell to respond to particular environmental conditions.
We report here the bioinformatic prediction of SID RNAs, for which short introns produced by alternative splicing and their complementary miRNAs could be extracted from public databases. Owing to their splicing origin and key structural attributes, SID are indeed well suited for a relatively efficient bioinformatic prediction and distinction from the bulk of introns. Based on comparative genomics and computational search for conserved intronic hairpins, earlier studies identified a few human mirtron candidates (9,16). Here, we proposed a bioinformatic approach coupled with experimental validation to identify potential new human SID and characterize their biogenesis pathway. Although miRNAs could originate from constitutive or alternative splicing events, we decided to focus on the latter case. Even though less abundant and hence enabling easier experimental validation, this was intended to consider their potential contribution to cell fate and identity.
Previous predictions of human mirtrons and intronic miRNAs were based on conservation and hairpin formation (9,11,13,14,16,44,64–70) but never on splicing events. Despite these approaches being different from the one proposed herein, we also computationally identified SID producing hsa-miR-6743 (#26) and hsa-miR-1234 (#44), previously described as mirtrons (66). Although levels of these SID were too weak for further experimental validation, we now show that they originate from an alternative splicing event of the transcripts RIC8A (Resistance to Inhibitors of Cholinesterase 8 homolog A) and CPSF1 (Cleavage and Polyadenylation Specificity Factor), respectively. CPSF1 gene encodes the largest subunit of the CPSF complex, a multi-subunit complex that plays a central role in 3΄ processing of pre-mRNA (71). The gene locus appears to be very complex in that it contains around 50 introns, of which, interestingly, 11 are alternatively spliced (72). We identified an additional SID produced by alternative splicing of CPSF1 (#46), which is distinct from the previously reported SID producing hsa-miR-1234 (9), hsa-miR-939 (16), and an agotron (19) also hosted in this gene. It is not surprising to find multiple SID in genes containing high numbers of introns, which in addition are subject to alternative splicing. Along the same lines, it is not surprising to find a higher density of SID on chromosome 19 (12/56) that contains the highest density of genes (8,73), but more importantly, the highest number of genes with short introns (http://www.ncbi.nlm.nih.gov/genome) compatible with their processing and formation of stable hairpin structures (29). Like CPSF1, chromosome 19 appears to host both SID and canonical miRNAs precursors as it contains the largest cluster of pre-miRNAs identified in mammals, where at least 46 different pre-miRNAs were found within a 100-kb region (74).
We did not restrict our search to introns of protein-coding genes since the possibility that non-coding RNAs might also produce shorter RNAs has already been proposed (59,75,76) as exemplified by SNHG (Small Nucleolar RNA Host Gene) family of genes and GAS5 (Growth Arrest Specific 5) that are non-coding multi-snoRNAs hosting genes (77), or by MIR17HG also known as the miR-17/92 cluster (78). Consistent with this, we identified SID candidates #39, #16 and #43 respectively hosted in LINC00085 (long intergenic non-protein coding RNA 85, NR_024330), RHBG (Rh family, B glycoprotein, which variant 2 is a non-coding isoform, NR_026549) and SRA1 [which we previously described as both non-coding RNAs and mRNA isoforms, see ref. (30,31,38,54)]. Interestingly, RHBG, similarly to what was shown for SRA, could represent a new bifunctional RNA since retention of its intron disrupts the ORF whereas, as for classical alternatively spliced mRNA, splicing preserves the ORF (30,31,58). In both cases, we have now shown that alternative splicing of introns not only favours the production of a protein product, but also promotes the formation of a miRNA. Adding a degree of complexity, alternative donor and acceptor splicing sites might also contribute to the diversification of the RNA forms produced, i.e. mRNA, long ncRNA and/or short ncRNA, as it might be the case for SID #6, #8 and #36. Whether the shorter SID generated by alternative donor or acceptor sites can also produces a miRNA in certain cellular contexts remains to be tested.
Although bioinformatic prediction of human SID eased their identification based on conservation, splicing origin or structural features [see refs. (9,11,13,14,16,44,64,65,67) and the present study], computational analysis is likely to generate large amounts of false positive predictions. Indeed, out of the first 19 mirtrons previously identified (9), 3 were experimentally tested and one of them (hsa-miR-1233 precursor) was suggested not to be a mirtron or a simtron (68). Thus, it stresses the need to experimentally test these SID candidates and characterize their biogenesis pathways.
Our primary goal was to identify SID originating from alternative splicing events being cell-type- or differentiation-stage- specific. However, we identified intronic miRNAs, which biogenesis should not rely in theory on splicing mechanisms. Indeed, intronic miRNAs are either transcribed independently of, or share promoter with, their host gene (79–82), but in both cases transcripts that correspond to pri-miRNAs should be much longer than the intron alone. However, SID #18, which we defined as a miRNA, had the exact size of the spliced intron, suggesting that, at least for this candidate, splicing mechanism was a prerequisite to produce the corresponding intronic miRNA. Apparent discrepancy also exists if we consider the levels of SID-hosting transcripts isoforms (Supplementary Figures S3 and S4) and that of the SID candidates (summarized in Supplementary Table S1). However, the fact that only SID and not mature miRNAs are detected on the oligonucleotide arrays could explain this discrepancy for SID that are rapidly processed in a given cellular context. In addition, the stability and fate, and hence the detectable levels, of the spliced intron are independent of that of the longer transcripts from which they derived. Such example already exists with EGFL7 (epidermal growth factor domain 7) that hosts the hsa-miR-126 in intron 6. EGFL7 was shown to be highly expressed in lung and to a lesser extent in heart whereas the opposite was observed for hsa-miR-126 [(83) and http://www.microrna.org].
In agreement with splicing proposed to be required for the production of non-canonical precursors of miRNAs, we report the experimentally-tested dependence of several SID on components of the splicing machinery (snRNP70+PRP8, U2AF65+PRP8) and of the microprocessor (Drosha, DGCR8), as exemplified by SID #8, #43, #45 and #46. Several other studies have reported a crosstalk between spliceosome and microprocessor. One striking example comes from plants, along which production of ath-miR-163 in Arabidopsis thaliana is favoured by components of the snRNP70 that potentiate miRNA biogenesis by hiding the proximal polyA signal and thus prevent polyadenylation and premature cleavage/degradation of the pri-miRNA (84). Other examples of such interplay have been reported in mammals, but almost invariably represent a competition between components of the spliceosome and those of the microprocessor (61,84–86). Interestingly, Drosha and DGCR8 were found in association with the spliceosome machinery in a supraspliceosome context (87), where knockdown of Drosha resulted in increased splicing whereas inhibition of the spliceosome enhanced miRNA production (87).
To date, the canonical pathway (Drosha/DGCR8–XPO5–Dicer–Ago2) is considered as the main biogenesis pathway for the majority of miRNAs (88). In the last few years, alternative pathways for miRNA biogenesis have also been reported. The first identified, the mirtron pathway, bypasses the cleavage by the microprocessor (9,15) since spliced-introns, first debranched by DBR1, are directly used as pre-miRNAs and exported in the cytoplasm by XPO5. The simtron pathway (17,18) uses the snRNP70 small nuclear ribonucleoprotein to recruit Drosha that will directly process the stem–loop structure from the pre-mRNA to produce the pre-miRNA. This mechanism is independent of DGCR8 that usually recognizes pri-miRNAs and directs Drosha RNAse III activity to release the pre-miRNA hairpin (89). We now report that these pathways are likely to be more complicated and inter-connected than previously expected, at least for miRNAs produced from short introns (SID). We described four intronic pre-miRNAs, 1 simtron-like and 1 mirtron-like, but also several other SID which biogenesis relies on both microprocessor (Drosha and DGCR8) and debranching enzyme (DBR1). Nonetheless, we did not exclude the possibility that miRNA originating from SID could be produced through multiple pathways, either depending of the host-gene expression/regulation, or relying on the intron excision/retention balance.
Among the new SID that we experimentally validated, we focused on the interesting case of the miRNA contained in the first intron of SRA RNA. SRA RNA was first described as a non-coding RNA (90) that acts as a transcriptional co-activator of nuclear receptors [For recent reviews see refs. (91,92)]. Since then, we described new isoforms produced by alternative splicing (30,31,54). SRA RNA was then proposed as the first member of a new class of bifunctional RNAs since both a protein and a non-coding RNA can be produced from the same genetic entity (30,31,54). We now report that intron 1 of SRA is detectable as a discrete entity in all 6 cell types that we have tested, although less abundantly in differentiated primary myotubes. Besides, we have recently found that coding (spliced intron 1) SRA isoforms were more abundant in differentiated human myogenic cells whereas non-coding (intron 1 retaining) SRA isoforms increased during myogenic differentiation and enhanced myogenic differentiation and myogenic conversion of non-muscle cells through the co-activation of MyoD activity (31). Interestingly, the amount of intron 1 and derived-mature miRNA were also higher in undifferentiated myoblasts suggesting that during myogenic differentiation, splicing of SRA intron 1 parallels the production of a SID. We previously showed that induction of muscle differentiation was not accompanied by a change in the ratio between non-coding and coding SRA isoforms in DM1 cells compared to non-affected muscle cells (31). We showed here that the levels of intron 1 of SRA were not affected during the course of differentiation of DM1 cells. While we highlighted a role of non-coding and coding SRA isoforms in the differentiation of normal cells (31), a role of the SID (intron 1) and more likely of the mature miRNA derived from it, cannot be excluded. Indeed, the impact of the SRA-mature miRNA could be direct (by inhibiting the non coding SRA isoforms retaining intron 1) or indirect (through other targets remaining to be identified). Interestingly, among the top five predicted target genes, and although not much data is available on FBXO41 (F box protein 41) or PRX (Periaxin) in the literature, the three other candidates have a potential link with myoblast differentiation. DAGLA (Diacylglycerol Lipase Alpha) expression is significantly increased upon myogenic differentiation in vitro (93). NECTIN1 (Nectin Cell Adhesion Molecule 1) belongs to the family of cellular adhesion molecules that play central roles in cell adhesion, cell motility, proliferation and survival, and contributes to the morphogenesis and differentiation of many cell types and tissues (94,95). Increased levels of MECP2 (MEthyl CpG binding Protein 2) during myogenic differentiation have been linked to global heterochromatin rearrangements that occur during and are a prerequisite for myogenic terminal differentiation (96). Of course, the whole range of miRNA targets originating from the SID identified in this study still remains to be identified in a given cellular context.
As a whole, we present evidence for the existence of new classes of Short Intron-Derived ncRNAs that we termed SID, that regroup agotron, mirtron and simtron RNAs. In addition, we uncovered new types of miRNAs which biogenesis appears to be splicing-dependant but nevertheless closely connected to the classical miRNA pathway.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jean-François Ouimette and Hélène Quach for valuable comments on the manuscript, members of the team for critical reading and Drs Vincent Mouly and Denis Furling (Myology Institute, Paris, France) for providing primary human myogenic cells and RNAs.
Footnotes
Present address: Sabrina Forveille, Metabolomics and Cell Biology Platforms, Gustave Roussy Cancer Campus, Villejuif, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
AFM (Association Française contre les Myopathies) [16055]; ANR (Agence Nationale pour la Recherche), LNCC (Ligue Nationale Contre le Cancer) and Gefluc (to C.F. team). Funding for open access charge: AFM (Association Française contre les Myopathies).
Conflict of interest statement. None declared.
REFERENCES
- 1. Clark M.B., Mattick J.S.. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011; 22:366–376. [DOI] [PubMed] [Google Scholar]
- 2. He L., Hannon G.J.. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004; 5:522–531. [DOI] [PubMed] [Google Scholar]
- 3. Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011; 12:846–860. [DOI] [PubMed] [Google Scholar]
- 4. Pais H., Moxon S., Dalmay T., Moulton V.. Small RNA discovery and characterisation in eukaryotes using high-throughput approaches. Adv. Exp. Med. Biol. 2011; 722:239–254. [DOI] [PubMed] [Google Scholar]
- 5. Kawamata T., Tomari Y.. Making RISC. Trends Biochem. Sci. 2010; 35:368–376. [DOI] [PubMed] [Google Scholar]
- 6. Mattick J.S., Gagen M.J.. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol. 2001; 18:1611–1630. [DOI] [PubMed] [Google Scholar]
- 7. Mattick J.S. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001; 2:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hube F., Francastel C.. Mammalian introns: when the junk generates molecular diversity. Int. J. Mol Sci. 2015; 16:4429–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berezikov E., Chung W.J., Willis J., Cuppen E., Lai E.C.. Mammalian mirtron genes. Mol. Cell. 2007; 28:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berezikov E., Liu N., Flynt A.S., Hodges E., Rooks M., Hannon G.J., Lai E.C.. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat. Genet. 2010; 42:6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung W.J., Agius P., Westholm J.O., Chen M., Okamura K., Robine N., Leslie C.S., Lai E.C.. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 2011; 21:286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flynt A.S., Greimann J.C., Chung W.J., Lima C.D., Lai E.C.. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell. 2010; 38:900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glazov E.A., Kongsuwan K., Assavalapsakul W., Horwood P.F., Mitter N., Mahony T.J.. Repertoire of bovine miRNA and miRNA-like small regulatory RNAs expressed upon viral infection. PLoS One. 2009; 4:e6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C.. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007; 130:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruby J.G., Jan C.H., Bartel D.P.. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007; 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valen E., Preker P., Andersen P.R., Zhao X., Chen Y., Ender C., Dueck A., Meister G., Sandelin A., Jensen T.H.. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat. Struct. Mol. Biol. 2011; 18:1075–1082. [DOI] [PubMed] [Google Scholar]
- 17. Havens M.A., Reich A.A., Duelli D.M., Hastings M.L.. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012; 40:4626–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Janas M.M., Khaled M., Schubert S., Bernstein J.G., Golan D., Veguilla R.A., Fisher D.E., Shomron N., Levy C., Novina C.D.. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011; 7:e1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansen T.B., Veno M.T., Jensen T.I., Schaefer A., Damgaard C.K., Kjems J.. Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 2016; 7:11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G.. A human snoRNA with microRNA-like functions. Mol. Cell. 2008; 32:519–528. [DOI] [PubMed] [Google Scholar]
- 21. Lee Y.S., Shibata Y., Malhotra A., Dutta A.. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009; 23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdelfattah A.M., Park C., Choi M.Y.. Update on non-canonical microRNAs. Biomol. Concepts. 2014; 5:275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jha A., Panzade G., Pandey R., Shankar R.. A legion of potential regulatory sRNAs exists beyond the typical microRNAs microcosm. Nucleic Acids Res. 2015; 43:8713–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010; 328:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J.. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010; 465:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Havens M.A., Reich A.A., Hastings M.L.. Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet. 2014; 10:e1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J., Maxwell E.S.. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res. 1990; 18:6565–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mattick J.S., Makunin I.V.. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005; 14:R121–R132.Spec No 1. [DOI] [PubMed] [Google Scholar]
- 29. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 30. Hube F., Guo J., Chooniedass-Kothari S., Cooper C., Hamedani M.K., Dibrov A.A., Blanchard A.A., Wang X., Deng G., Myal Y. et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006; 25:418–428. [DOI] [PubMed] [Google Scholar]
- 31. Hube F., Velasco G., Rollin J., Furling D., Francastel C.. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011; 39:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Furling D., Coiffier L., Mouly V., Barbet J.P., St Guily J.L., Taneja K., Gourdon G., Junien C., Butler-Browne G.S.. Defective satellite cells in congenital myotonic dystrophy. Hum. Mol. Genet. 2001; 10:2079–2087. [DOI] [PubMed] [Google Scholar]
- 33. Velasco G., Hube F., Rollin J., Neuillet D., Philippe C., Bouzinba-Segard H., Galvani A., Viegas-Pequignot E., Francastel C.. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc. Natl. Acad. Sci. U.S.A. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pall G.S., Hamilton A.J.. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008; 3:1077–1084. [DOI] [PubMed] [Google Scholar]
- 35. Hube F., Francastel C.. “Pocket-sized RNA-Seq": a method to capture new mature microRNA produced from a genomic region of interest. Non-Coding RNA. 2015; 1:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferri F., Bouzinba-Segard H., Velasco G., Hube F., Francastel C.. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009; 37:5071–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Varkonyi-Gasic E., Hellens R.P.. Quantitative stem–loop RT-PCR for detection of microRNAs. Methods Mol. Biol. 2011; 744:145–157. [DOI] [PubMed] [Google Scholar]
- 38. Cooper C., Guo J., Yan Y., Chooniedass-Kothari S., Hube F., Hamedani M.K., Murphy L.C., Myal Y., Leygue E.. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009; 37:4518–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugnet C.W., Kent W.J., Ares M. Jr., Haussler D.. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004; 66–77. [DOI] [PubMed] [Google Scholar]
- 40. Moore M.J. Nuclear RNA turnover. Cell. 2002; 108:431–434. [DOI] [PubMed] [Google Scholar]
- 41. Zhang B., Han D., Korostelev Y., Yan Z., Shao N., Khrameeva E., Velichkovsky B.M., Chen Y.P., Gelfand M.S., Khaitovich P.. Changes in snoRNA and snRNA abundance in the human, chimpanzee, macaque, and mouse brain. Genome Biol. Evol. 2016; 8:840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quach H., Barreiro L.B., Laval G., Zidane N., Patin E., Kidd K.K., Kidd J.R., Bouchier C., Veuille M., Antoniewski C. et al. Signatures of purifying and local positive selection in human miRNAs. Am. J. Hum. Genet. 2009; 84:316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhartiya D., Talwar J., Hasija Y., Scaria V.. Systematic curation and analysis of genomic variations and their potential functional consequences in snoRNA loci. Hum. Mutat. 2012; 33:E2367–E2374. [DOI] [PubMed] [Google Scholar]
- 44. Wen J., Ladewig E., Shenker S., Mohammed J., Lai E.C.. Analysis of nearly one thousand mammalian mirtrons reveals novel features of dicer substrates. PLoS Comput. Biol. 2015; 11:e1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R.. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004; 432:235–240. [DOI] [PubMed] [Google Scholar]
- 46. Kim H.C., Kim G.M., Yang J.M., Ki J.W.. Cloning, expression, and complementation test of the RNA lariat debranching enzyme cDNA from mouse. Mol. Cell. 2001; 11:198–203. [PubMed] [Google Scholar]
- 47. MacRae I.J., Ma E., Zhou M., Robinson C.V., Doudna J.A.. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R.. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005; 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005; 30:106–114. [DOI] [PubMed] [Google Scholar]
- 50. Maniataki E., Mourelatos Z.. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005; 19:2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kozlowska E., Krzyzosiak W.J., Koscianska E.. Regulation of huntingtin gene expression by miRNA-137, -214, -148a, and their respective isomiRs. Int. J. Mol. Sci. 2013; 14:16999–17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jin H.Y., Gonzalez-Martin A., Miletic A.V., Lai M., Knight S., Sabouri-Ghomi M., Head S.R., Macauley M.S., Rickert R.C., Xiao C.. Transfection of microRNA mimics should be used with caution. Front. Genet. 2015; 6:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ohno M., Shibata C., Kishikawa T., Yoshikawa T., Takata A., Kojima K., Akanuma M., Kang Y.J., Yoshida H., Otsuka M. et al. The flavonoid apigenin improves glucose tolerance through inhibition of microRNA maturation in miRNA103 transgenic mice. Sci. Rep. 2013; 3:2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chooniedass-Kothari S., Emberley E., Hamedani M.K., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E.. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004; 566:43–47. [DOI] [PubMed] [Google Scholar]
- 55. Jung S.Y., Malovannaya A., Wei J., O’Malley B.W., Qin J.. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol. Endocrinol. 2005; 19:2451–2465. [DOI] [PubMed] [Google Scholar]
- 56. Kapusta A., Feschotte C.. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014; 30:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ulveling D., Francastel C., Hube F.. When one is better than two: RNA with dual functions. Biochimie. 2011; 93:633–644. [DOI] [PubMed] [Google Scholar]
- 58. Ulveling D., Francastel C., Hube F.. Identification of potentially new bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie. 2011; 93:2024–2027. [DOI] [PubMed] [Google Scholar]
- 59. Mattick J.S., Makunin I.V.. Non-coding RNA. Hum. Mol. Genet. 2006; 15:R17–R29.Spec No 1. [DOI] [PubMed] [Google Scholar]
- 60. Rearick D., Prakash A., McSweeny A., Shepard S.S., Fedorova L., Fedorov A.. Critical association of ncRNA with introns. Nucleic Acids Res. 2011; 39:2357–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brown J.W., Marshall D.F., Echeverria M.. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008; 13:335–342. [DOI] [PubMed] [Google Scholar]
- 62. Jung C.H., Hansen M.A., Makunin I.V., Korbie D.J., Mattick J.S.. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC. Genomics. 2010; 11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou H., Lin K.. Excess of microRNAs in large and very 5΄ biased introns. Biochem. Biophys. Res. Commun. 2008; 368:709–715. [DOI] [PubMed] [Google Scholar]
- 64. Chen D., Meng Y., Yuan C., Bai L., Huang D., Lv S., Wu P., Chen L.L., Chen M.. Plant siRNAs from introns mediate DNA methylation of host genes. RNA. 2011; 17:1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Joshi P.K., Gupta D., Nandal U.K., Khan Y., Mukherjee S.K., Sanan-Mishra N.. Identification of mirtrons in rice using MirtronPred: a tool for predicting plant mirtrons. Genomics. 2012; 99:370–375. [DOI] [PubMed] [Google Scholar]
- 66. Ladewig E., Okamura K., Flynt A.S., Westholm J.O., Lai E.C.. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 2012; 22:1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Meng Y., Shao C.. Large-scale identification of mirtrons in Arabidopsis and rice. PLoS One. 2012; 7:e31163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schamberger A., Sarkadi B., Orban T.I.. Human mirtrons can express functional microRNAs simultaneously from both arms in a flanking exon-independent manner. RNA. Biol. 2012; 9:1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. St L.G., Shtokalo D., Tackett M.R., Yang Z., Eremina T., Wahlestedt C., Urcuqui-Inc Seilheimer B., McCaffrey T.A., Kapranov P.. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC. Genomics. 2012; 13:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhu Q.H., Spriggs A., Matthew L., Fan L., Kennedy G., Gubler F., Helliwell C.. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008; 18:1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mandel C.R., Bai Y., Tong L.. Protein factors in pre-mRNA 3΄-end processing. Cell Mol. Life Sci. 2008; 65:1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thierry-Mieg D., Thierry-Mieg J.. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006; 7(Suppl 1):S12–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grimwood J., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M. et al. The DNA sequence and biology of human chromosome 19. Nature. 2004; 428:529–535. [DOI] [PubMed] [Google Scholar]
- 74. Bortolin-Cavaille M.L., Dance M., Weber M., Cavaille J.. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009; 37:3464–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mercer T.R., Dinger M.E., Bracken C.P., Kolle G., Szubert J.M., Korbie D.J., skarian-Amiri M.E., Gardiner B.B., Goodall G.J., Grimmond S.M. et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010; 20:1639–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mercer T.R., Gerhardt D.J., Dinger M.E., Crawford J., Trapnell C., Jeddeloh J.A., Mattick J.S., Rinn J.L.. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2011; 30:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Williams G.T., Mourtada-Maarabouni M., Farzaneh F.. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans. 2011; 39:482–486. [DOI] [PubMed] [Google Scholar]
- 78. de P.L., Yao E., Callier P., Faivre L., Drouin V., Cariou S., Van H.A., Genevieve D., Goldenberg A., Oufadem M. et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat. Genet. 2011; 43:1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 80. Kim Y.K., Kim V.N.. Processing of intronic microRNAs. EMBO J. 2007; 26:775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Monteys A.M., Spengler R.M., Wan J., Tecedor L., Lennox K.A., Xing Y., Davidson B.L.. Structure and activity of putative intronic miRNA promoters. RNA. 2010; 16:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ramalingam P., Palanichamy J.K., Singh A., Das P., Bhagat M., Kassab M.A., Sinha S., Chattopadhyay P.. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA. 2014; 20:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fitch M.J., Campagnolo L., Kuhnert F., Stuhlmann H.. Egfl7, a novel epidermal growth factor-domain gene expressed in endothelial cells. Dev. Dyn. 2004; 230:316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Szweykowska-Kulinska Z., Jarmolowski A., Vazquez F.. The crosstalk between plant microRNA biogenesis factors and the spliceosome. Plant Signal. Behav. 2013; 8:e26955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mattioli C., Pianigiani G., Pagani F.. Cross talk between spliceosome and microprocessor defines the fate of pre-mRNA. Wiley. Interdiscip. Rev. RNA. 2014; 5:647–658. [DOI] [PubMed] [Google Scholar]
- 86. Melamed Z., Levy A., shwal-Fluss R., Lev-Maor G., Mekahel K., Atias N., Gilad S., Sharan R., Levy C., Kadener S. et al. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol. Cell. 2013; 50:869–881. [DOI] [PubMed] [Google Scholar]
- 87. Agranat-Tamir L., Shomron N., Sperling J., Sperling R.. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 2014; 42:4640–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 89. Seitz H., Zamore P.D.. Rethinking the microprocessor. Cell. 2006; 125:827–829. [DOI] [PubMed] [Google Scholar]
- 90. Lanz R.B., McKenna N.J., Onate S.A., Albrecht U., Wong J., Tsai S.Y., Tsai M.J., O’Malley B.W.. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999; 97:17–27. [DOI] [PubMed] [Google Scholar]
- 91. Colley S.M., Leedman P.J.. Steroid receptor RNA activator - a nuclear receptor coregulator with multiple partners: Insights and challenges. Biochimie. 2011; 93:1966–1972. [DOI] [PubMed] [Google Scholar]
- 92. Cooper C., Vincett D., Yan Y., Hamedani M.K., Myal Y., Leygue E.. Steroid receptor RNA activator bi-faceted genetic system: heads or tails. Biochimie. 2011; 93:1973–1980. [DOI] [PubMed] [Google Scholar]
- 93. Zhao D., Pond A., Watkins B., Gerrard D., Wen Y., Kuang S., Hannon K.. Peripheral endocannabinoids regulate skeletal muscle development and maintenance. Eur. J. Transl. Myol. 2010; 1:167–179. [Google Scholar]
- 94. Takai Y., Ikeda W., Ogita H., Rikitake Y.. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu. Rev. Cell Dev. Biol. 2008; 24:309–342. [DOI] [PubMed] [Google Scholar]
- 95. Takai Y., Miyoshi J., Ikeda W., Ogita H.. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol Cell Biol. 2008; 9:603–615. [DOI] [PubMed] [Google Scholar]
- 96. Brero A., Easwaran H.P., Nowak D., Grunewald I., Cremer T., Leonhardt H., Cardoso M.C.. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005; 169:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.