Abstract
Polydactyl zinc finger (ZF) proteins have prominent roles in gene regulation and often execute multiple regulatory functions. To understand how these proteins perform varied regulation, we studiedDrosophila Suppressor of Hairy-wing [Su(Hw)], an exemplar multifunctional polydactyl ZF protein. We identified separation-of-function (SOF) alleles that encode proteins disrupted in a single ZF that retain one of the Su(Hw) regulatory activities. Through extended in vitro analyses of the Su(Hw) ZF domain, we show that clusters of ZFs bind individual modules within a compound DNA consensus sequence. Through in vivo analysis of SOF mutants, we find that Su(Hw) genomic sites separate into sequence subclasses comprised of combinations of modules, with subclasses enriched for different chromatin features. These data suggest a Su(Hw) code, wherein DNA binding dictates its cofactor recruitment and regulatory output. We propose that similar DNA codes might be used to confer multiple regulatory functions of other polydactyl ZF proteins.
INTRODUCTION
Cell fate depends upon differential gene expression controlled largely at the transcriptional level. These processes require spatial and temporal coordination of transcription factors that recruit cofactors to regulate RNA polymerase activity (1). Among transcription factors, DNA binding proteins are critical. Several types of metazoan DNA binding proteins exist, with the largest family corresponding to Cys2-His2 ZF proteins (2–4). The hallmark of these proteins is a self-folding ββα domain formed through chelation of a zinc ion (5). Each ZF typically recognizes three nucleotides within a longer DNA binding motif (6). The large size of the Cys2-His2 ZF protein family underscores the importance of this class of DNA binding protein in transcriptional regulation.
Common among the Cys2-His2 ZF protein family are polydactyl proteins with five or more ZFs (2,3). Nearly 40% of the ∼375Drosophila ZF proteins have more than four ZFs (3). Further, nearly half of all human transcription factors are C2H2 ZF proteins (7) that carry an average of 10 ZFs per protein (4). A growing number of polydactyl ZF proteins have been found to confer multiple transcriptional functions (8–11). These observations suggest that regulatory versatility might result from functional plasticity imparted by the presence of many ZFs. Even though Cys2-His2 ZFs are typically used for DNA binding, these domains also support protein-protein or protein–RNA interactions (12,13). In fact, some ZFs simultaneously interact with DNA and another cofactor (12). Defining how individual ZFs work within multi-ZF domains will improve our understanding of the regulatory output of this class of metazoan transcription factors.
Drosophila Suppressor of Hairy-wing [Su(Hw)] is an exemplar multifunctional polydactyl transcription factor. This DNA binding protein contains a 12 ZF domain comprised of 2 C2HC and 10 C2H2 ZFs (Supplementary Table S1). Su(Hw) was first identified for its insulator function, as it is responsible for enhancer blocking of the insulator within the gypsy retrotransposon (14–16). More recent studies revealed that Su(Hw) has non-insulator transcriptional roles (17,18). An activator function was discovered in studies of the endogenous Su(Hw) binding site (SBS) 1A-2. Although 1A-2 demonstrated enhancer blocking activity in transgene assays (19,20), within its natural location, 1A-2 is required for transcriptional activation of the nearby non-coding RNA gene yar (18). Subsequently, a repressor function was discovered in studies of the Su(Hw) requirement in oogenesis (17). Indeed, female sterility of su(Hw) mutants was linked to derepression of neuronal genes in the ovary, particularly the RNA binding protein 9 (Rbp9) gene. A more extensive repressor function of Su(Hw) was suggested by findings that SBSs are primarily located within repressive ‘black’ chromatin (21) and that loss of Su(Hw) is globally associated with derepression of nearby genes (22). Taken together, these data indicate that Su(Hw) is a context-specific transcriptional regulator, with insulator, activator and repressor functions.
Here, we conducted a genetic screen for new su(Hw) alleles to advance our understanding of mechanisms responsible for the multivalency of Su(Hw) transcriptional regulation. This screen identified multiple su(Hw) alleles, including new separation-of-function (SOF) alleles. Molecular characterization of the SOF mutants revealed that these alleles encode full-length Su(Hw) proteins disrupted in a single ZF. Motivated by this discovery, we defined the in vitro and in vivo requirements for each of the twelve ZFs in the Su(Hw) DNA binding domain. These analyses revealed that Su(Hw) uses clusters of ZFs to bind a compound consensus comprised of three sequence modules. Using genome-wide occupancy data, we show that the SOF Su(Hw) mutants bind distinct sequence subclasses of genomic SBSs that are enriched for different chromatin features and cofactors. These data suggest that theDrosophila genome carries a ‘Su(Hw) code’ and predict that how Su(Hw) binds to DNA influences its cofactor recruitment and regulatory output. Our findings add to growing evidence that the regulation of multifunctional polydactyl ZF proteins depends upon a DNA code (11,23,24).
MATERIALS AND METHODS
Drosophila stocks and culture conditions
Flies were raised at 25°C, 70% humidity on standard corn meal/agar medium. Extant su(Hw) alleles were used including four su(Hw) null alleles [su(Hw)2 caused by insertion of a jockey element within the first intron (25,26), su(Hw)Pb [su(Hw)e04061 in Flybase] caused by an insertion of a white marked piggy-bac transposon at the 5΄ end of the second exon, su(Hw)v caused by a deletion encompassing the promoters of su(Hw) and the neighboring essential RpII15 gene (27) and su(Hw)E8 caused by mutation of the codon for a zinc-chelating amino acid in ZF7 (25)] and one hypomorphic allele [su(Hw)f] caused by mutation of the codon for a zinc-chelating amino acid in ZF10 (25).
Mutagenic screen and identification of su(Hw) mutant alleles
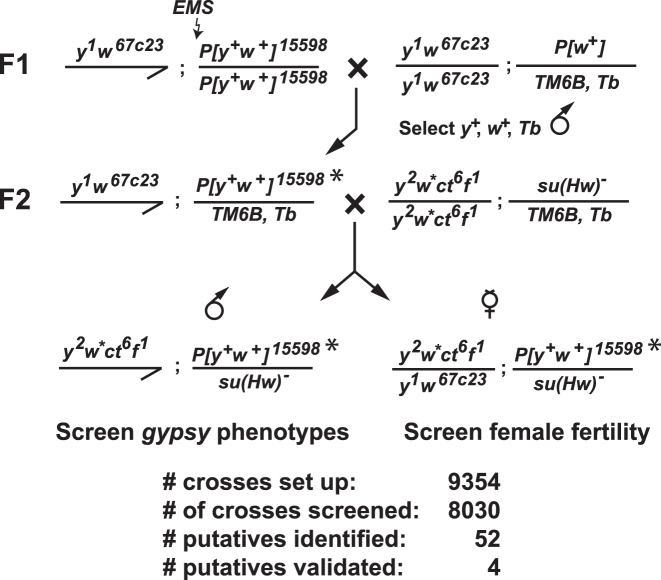
The strategy for isolating new su(Hw) alleles is shown in Figure 1. Two-to-four day old y1w67c23; P{EPgy2}CG6499EY02782 males (Bloomington # 15598) were desiccated for 12 to 24 h and then fed 25 mM ethyl methanesulfonate (EMS) in 10% sucrose (w/v). This parental genotype carries a yellow+, white+ marked third chromosome that allowed us to identify the mutagenized chromosome. After 24 h, mutagenized males were transferred to bottles with standard corn meal/agar medium and mated with y1w67c23; P[w+]/TM6B, Tb virgin females. In total, over 9000 males were mutagenized. Mated females were transferred to a new bottle every 3 to 4 days. Next, 8000+ mutagenized F1 y1w67c23; P[EPgy2]CG6499EY02782*/TM6B, Tb males were crossed to y2w1118ct6f1; su(Hw)2/TM6B, Tb virgin females. The resulting F2 y2w1118ct6f1; P[EPgy2]CG6499EY02782*/su(Hw)2 males were screened for suppression of the gypsy-induced mutations cut6 (ct6) and forked1 (f1), while the resulting F2 y2w1118ct6f1/y1w67c23; P[EPgy2]CG6499EY02782/su(Hw)2 females were mated with wild-type males to test for fertility. Putative su(Hw) mutants were retested by complementation using extant su(Hw) alleles. Stocks of four new su(Hw) alleles were established [named su(Hw)M393, su(Hw)A460, su(Hw)A1933, su(Hw)A2663] by crossing y2w1118ct6f1; P[EPgy2]CG6499EY02782, su(Hw)m/TM6B, Tb males to y2w1118ct6f1; su(Hw)2/TM6B, Tb virgin females. As EMS has the potential to generate multiple mutations, properties of the newly generated su(Hw) EMS mutations were only studied in heteroallelic combination with other su(Hw) mutants.
Figure 1.
Ethyl methanesulfonate (EMS) mutagenesis screen for new su(Hw) mutants. Males homozygous for a third chromosome marked with a yellow (y)+, white+(w)+ P-element (P[y+, w+]15598) were fed EMS and were mated to females carrying the third chromosome balancer TM6B, Tb. F1 males were selected that carried the mutagenized third chromosome (P[y+, w+]15598*) and the TM6B, Tb balancer chromosome. These males were mated to virgin females carrying the multiply marked X-chromosome with three gypsy-induced mutations (y2, ct6, f1) and a su(Hw) null allele carried with the TM6B, Tb balancer. The non-Tb F2 male progeny were screened for reversal of the gypsy-induced mutations and the non-Tb F2 female progeny were screened for infertility. If either outcome was obtained, then P[y+, w+]15598*/ TM6B, Tb males and females were crossed to establish a stock. Of more than 8000 chromosomes screened, 52 putative su(Hw) alleles were identified in the F2 generation. Four stocks were generated with confirmed su(Hw) mutations.
Molecular characterization of su(Hw) alleles
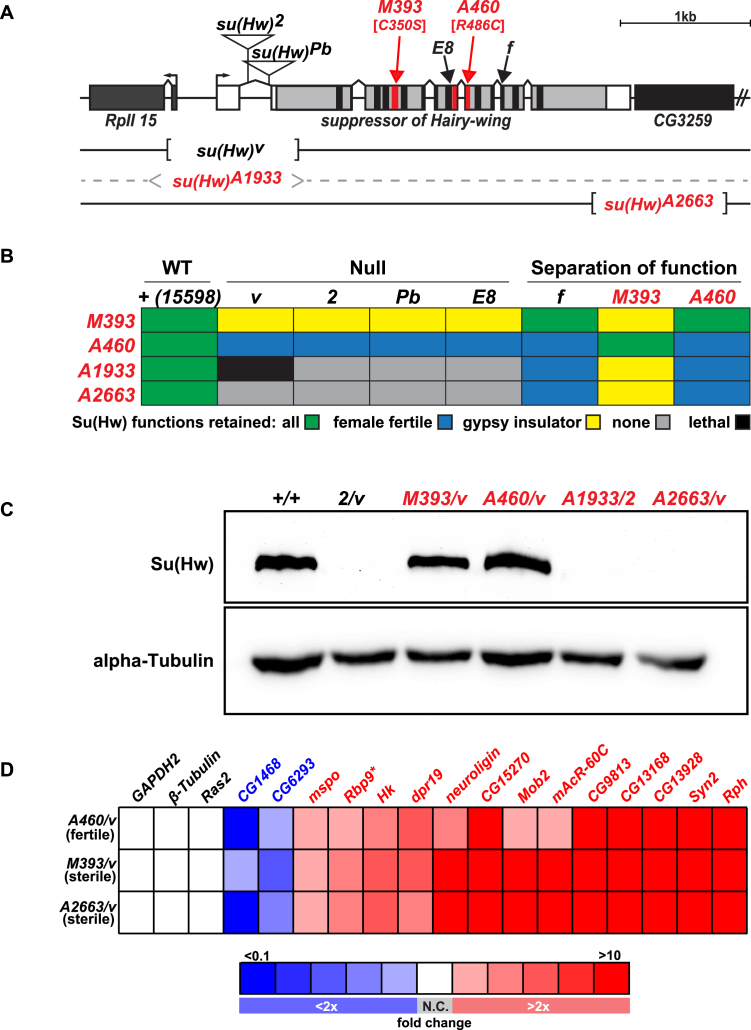
Molecular lesions in the newly identified su(Hw) alleles were defined using polymerase chain reaction (PCR) analysis of DNA isolated from parental and su(Hw)m/ su(Hw)v trans-heterozygous adult flies. PCR primers were located 5΄ and 3΄ of the coding sequence, with the 5΄ primer anchored within the su(Hw)v deletion to restrict PCR amplification to the new su(Hw) allele (Figure 2A). Sequence analysis of PCR products revealed that su(Hw)M393 carries single base substitution that changed a zinc-chelating cysteine to a serine codon in ZF4 (C350S), su(Hw)A460 carries a single base substitution that changed an arginine to a cysteine codon in ZF8 (R486C) and su(Hw)A2663 mutation carries a 1099 bp deletion that removes from +3534 to +4633 base pairs relative to the su(Hw) transcription start site. No PCR product was obtained from the su(Hw)A1933 genomic DNA, suggesting this allele carries a deletion within the su(Hw) locus, a prediction supported by complementation analyses (Figure 2B). Failure of su(Hw)A1933 to complement the viability of su(Hw)v suggests that the su(Hw)A1933 lesion also disrupts the essential RpII15 gene.
Figure 2.
Properties of su(Hw) alleles generated during EMS mutagenic screen. (A) Shown is a diagram of the su(Hw) locus, including su(Hw) and the 5΄ RPII15 and 3΄ CG3259 genes (rectangles). The 5΄ and 3΄ UTRs of the su(Hw) gene are shown in white and the coding region in gray, with the locations of the ZFs shown in black or red. Lesions associated with mutant alleles are depicted, including the insertions in su(Hw)2 and su(Hw)Pb, the ZF point mutations in su(Hw)M393 [C350S], su(Hw)E8, su(Hw)A460 [R486C] and su(Hw)f, and the location of the deletions in su(Hw)v, su(Hw)A1933 and su(Hw)A2663. (B) Complementation data obtained from crosses between extant and new su(Hw) alleles, including both null and separation-of-function mutants. Trans-heterozygotes showed the following phenotypes: (i) had all Su(Hw) functions (green), (ii) had female fertility only (blue), (iii) had gypsy-insulator function only (yellow), (iv) had no Su(Hw) function (gray) or (v) was adult lethal (black). The su(Hw)+ stock was the parental stock used in the EMS screen that carried the marked P[y+, w+]15598 third chromosome. (C) Western blot of protein extracts obtained from su(Hw)+/+ and su(Hw)−/-ovaries probed with antibodies against Su(Hw) and alpha-Tubulin (loading control). (D) Heat map of qRT-PCR analyses of gene expression changes of Su(Hw) target genes in RNA isolated from the su(Hw)A460/v (fertile), su(Hw)M393/v (sterile) and su(Hw)A2663/v (sterile) relative to the su(Hw)+/+ parental line (15 598). Genes studied are listed above the table, with three non-target genes (black) and fifteen target genes analyzed, including two upregulated (blue) and thirteen downregulated (red) genes. Boxes represent less than 2-fold (white, no change: N.C.) or 2–4-fold, 4–6-fold, 6–8-fold, 8–10-fold and greater than 10-fold changes, denoted as darkening shades of red and blue for increased and decreased, respectively. Data represent the average fold change of three biological replicates.
Quantitative PCR (qPCR) analyses of gene expression
Gene expression analyses were completed using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) of total RNA isolated from ∼50 ovary pairs per biological replicate, as described previously (18). Expression levels were determined using the housekeeping gene RpL32 as an internal control.
Analysis of Su(Hw) binding in vitro
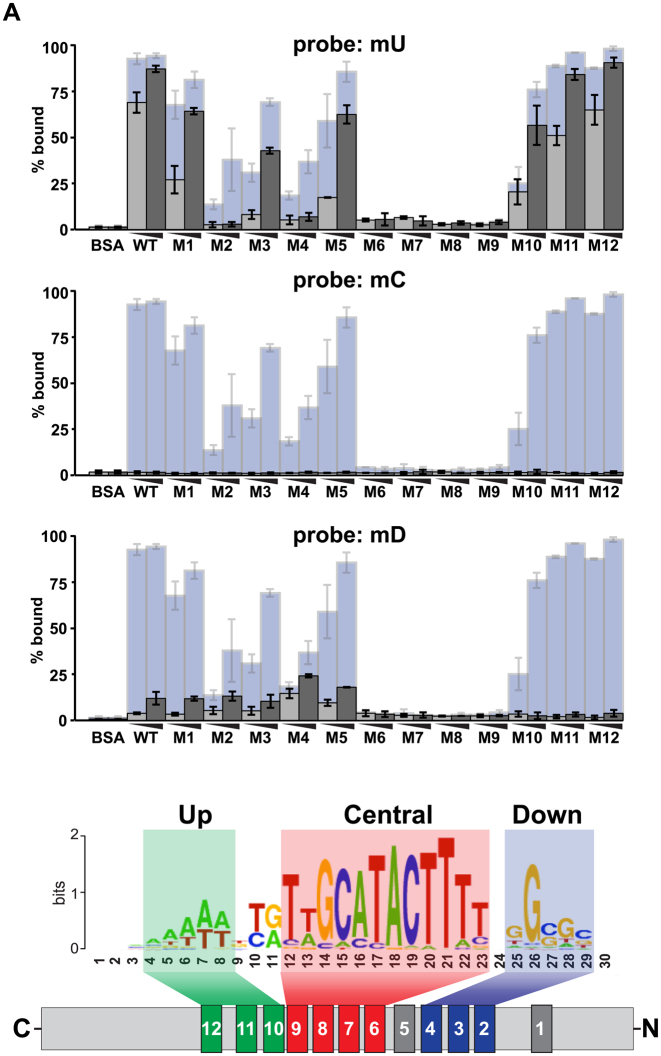
The in vitro DNA binding properties of full-length wild-type and Su(Hw) ZF mutants were studied. For each ZF mutant, the su(Hw) cDNA was mutated using a QuikChange II XL Site-Directed Mutagenesis Kit, such that codons for zinc-chelating amino acids were mutated. DNA sequencing confirmed that only the expected change was introduced. The collection of ZF mutants includes: H238A (ZF1); H308A (ZF2); H337A (ZF3); C350S (ZF4) regenerating Su(Hw)M4M393; C382A, C385A (ZF5); H431A (ZF6); H459Y (ZF7); H487A (ZF8); H515A (ZF9); C525Y (ZF10); H571A (ZF11), H614A(ZF12), see Supplementary Table S1. Su(Hw) wild-type ZF mutant proteins were purified from Escherichia coli DE3 cells, as described previously (28). To determine the amount of each full-length protein, Bradford analyses were performed on purified proteins using bovine serum albumin as a standard, followed by polyacrylamide and western analyses of these proteins to verify amounts (Supplementary Figure S1A). To assess whether disruption of a single ZF significantly affected the overall Su(Hw) structure, we conducted limited trypsin digestion. Overall, digests of Su(Hw)WT and Su(Hw) ZF mutants were indistinguishable (data not shown), suggesting that folding of the mutant proteins remains unchanged. Apparent DNA binding affinities of each purified wild-type and ZF Su(Hw) mutant were determined using Electrophoretic Mobility Shift Assay (EMSAs), with binding reactions containing two proteins amounts (0.3 and 1.0 μg). Conditions for these analyses were described previously (28). The DNA probes in this study included SBSs located within a 212 bp fragment. The perfect match (PM) SBS was derived from an endogenous SBS at cytological location 4C15, by introducing two nucleotide substitutions to establish a perfect consensus. For probes mU, mC, mD (Figure 7) and mUmD, each probe was mutated such that five nucleotides of the designated module(s) were changed in the following pattern, A to C, T to G, C to A and G to T.
Figure 7.
Mapping Su(Hw) ZF recognition of binding modules within the SBS motif. (A) Top: quantification of binding properties of Su(Hw)WT and Su(Hw) ZF mutants to DNA probes carrying mutations in either the upstream core (mU), the central core (mC) or the downstream core (mD) mutant probes. For each of these probes, five nucleotides in the core were mutated away from the consensus sequence. Indicated is the percent bound probe observed using 0.3 μg (light gray) or 1.0 μg (dark gray) of Su(Hw) protein. For comparison, data are shown using the PM probe (light blue, Figure 3). Data shows the average of two experiments. Bottom: shown is a schematic of Su(Hw) binding with a full consensus SBS, showing which ZFs are involved in recognition of specific cores.
Generation of transgenic su(Hw) Drosophila stocks
The in vivo functions of Su(Hw) ZF mutants were determined through analysis of transgenic lines, each expressing a Su(Hw) ZF mutant protein engineered to carry the same amino acid substitutions as described for the bacterially produced proteins [Su(Hw)MZF]. These mutations were inserted into a 6-kb genomic fragment that included 1.3 kb of 5΄ and 0.5 kb of 3΄ DNA of the su(Hw) gene, PCR amplified from bacterial artificial chromosome CH322-158DO6 (Children's Hospital Oakland Research Institute BACPAC Resources Center). Point mutations were introduced into this fragment, as described above. P[su(Hw)MZF] transgenic lines expressed wild-type Su(Hw) or Su(Hw) mutated for ZF1 to ZF3, and ZF5 to ZF12. All expression transgenes were integrated into the attP2 site that is located at cytological location 68A4 on chromosome 3L (29). Transgenic P[su(Hw)MZF] flies were crossed into a su(Hw)v background, generating P[su(Hw)MZF], su(Hw)v recombinant chromosomes; each confirmed by PCR and sequencing.
Western analyses and quantification
Su(Hw) protein levels were analyzed using western blot analyses of ovary extracts. Blots used a 1:500 dilution of guinea pig anti-Su(Hw) antibody (30), detected using a 1:20 000 dilution of secondary antibody, HRP-conjugated donkey anti-guinea pig IgG (Jackson ImmunoResearch). As a loading control, blots were subsequently incubated with the mouse anti-alpha-tubulin IgG primary antibody (Sigma, T5168) and HRP-conjugated rabbit anti-mouse IgG secondary antibody (Sigma, A9044). Imaging of western blots was done using a cooled CCD camera (Ultra-Violet Products BioImaging) and quantification was accomplished using LabWorks software (Ultra-Violet Products BioImaging) and Microsoft Excel.
Polytene chromosome staining
Polytene chromosome analysis was done as described previously (31). Images were processed using ImageJ and Adobe Photoshop. Guinea pig anti-Su(Hw) primary antibody was used at a 1:250 dilution. Goat anti-guinea pig Alexa Fluor 488 (A11073) secondary antibody was used at a 1:1000 dilution.
ChIP-seq, peak detection, validation and motif analysis
Genome-wide association of Su(Hw)M4M393 was determined using ChIP, using ∼200 ovary pairs dissected from su(Hw)M393/v females younger than 6 h old per experiment, as described previously (31). Single-end libraries for Illumina high-throughput sequencing were prepared from ∼100 ng of DNA from each fraction (Cincinnati Children's Hospital Medical Center Genetic Variation and Gene Discovery Core Facility, Cincinnati, OH, USA). Illumina Genome Analyzer IIx fastq files were processed as described previously (31). ChIP-seq datasets were evaluated using Partek v. 6.5. The Su(Hw)M4M393 ChIP-seq1 generated over 32 million mapped reads for anti-Su(Hw) IP and over 34 million mapped reads for the control pre-immune IP. A replicate experiment, Su(Hw)M4M393 ChIP-seq2, generated over 40 million mapped reads for anti-Su(Hw) IP and over 47 million mapped reads for the control pre-immune IP. ChIP-seq1 identified 636 sites using a 1% false discovery rate (FDR) and a 3× fold-enrichment (IP versus IgG) cutoff, and ChIP-seq2 recovered 329 sites using a 1% FDR and a 1.5× fold-enrichment (IP versus IgG) cutoff. Over 90% of SBSs identified in ChIP-seq2 were identified in Su(Hw)M4M393 ChIP-seq1 (Supplementary Figure S2). ChIP-seq data are submitted to the NIH GEO/Sequence Read Archive database, accession number GSE86243.
Validation of ChIP-seq experiments
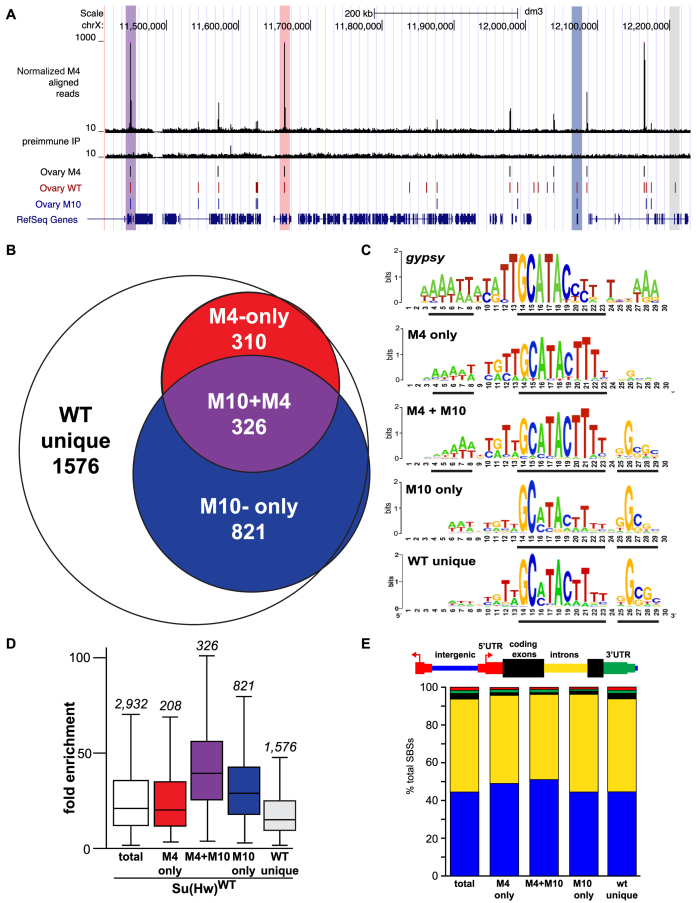
Several strategies were used to validate Su(Hw)M4M393 ChIP-seq and to align with modENCODE ChIP-seq guidelines (32). First, SBSs validation was completed using ChIP-qPCR. The guinea pig anti-Su(Hw) antibody used for all ChIP experiments has been extensively characterized and meets modENCODE criteria for primary and secondary characterization of ChIP-seq antibodies (17,30,31). Biologically independent chromatin samples were isolated from ∼100 ovary pairs per replicate, dissected from females younger than 6 h old and stored in PBS at −80°C. ChIP was completed as described previously (30,31). Subsequent qPCR reactions were completed using IQ SYBR Green supermix (Bio-Rad) with primers designed to amplify 100–200 bp fragments centered on each SBS, using a 7900 HT PCR instrument (Applied Biosystems). Percent input was calculated using the following formula (% input = 2∧[Ct(input)-Ct(IP)] × 1/DF × 100) where DF is the dilution factor between input and IP samples. Second, to define the motif underlying each collection of SBSs, sequences from each class were submitted to MEME v.4.4.0 (33). The sequence submitted for each site ranged in size from 100–1000 bp, based on the endpoints of the called site from Partek v. 6.5. In all cases the top motif generated was related to the established SBS consensus sequence (Figure 6C). Third, visual inspection of ChIP-seq data was performed using the UCSC Genome Browser and in comparison to previously published Su(Hw) ChIP-seq or ChIP-chip datasets (Figure 6A or data not shown; (31,34–37).
Figure 6.
Su(Hw)M4 localizes to a subset of endogenous SBSs. (A) Shown is a UCSC Genome Browser view of a region of chromosome X with the ChIP-seq track obtained for Su(Hw)M4. Tracks include Su(Hw)M4 aligned reads, pre-immune IP control reads, called peaks from the Su(Hw)M4, Su(Hw)WT and Su(Hw)M10 ovary datasets (31) and RefSeq genes. Highlighted are peaks that represent an overlap of Su(Hw)M4, Su(Hw)WT and Su(Hw)M10 (purple); an overlap of Su(Hw)M4 and Su(Hw)WT (red); an overlap of Su(Hw)WT and Su(Hw)M10 (blue); or no overlap with Su(Hw)M4 or Su(Hw)M10 (WT unique, gray). (B) Venn diagram depicting the overlap between Su(Hw)WT, Su(Hw)M10 and Su(Hw)M4 SBSs. The number of SBSs in each category is listed. (C) MEME-generated sequence logo for the gypsy insulator and SBS classes including M4-only, M4+M10, M10-only and WT unique. The black lines beneath each logo identify the most conserved cores in each motif. (D) Shown is a graph of ChIP-seq fold enrichment for SBSs in the following categories: total [white box; (31)], M4-only (red), M4+M10 (purple), M10-only (blue) and WT unique (gray). Each box represents the 25th to 75th percentile interval, with the median enrichment indicated by the line. Whiskers represent the non-outlier range. The total number of SBSs in each class is shown above the box plot. (E) Shown is the genomic distribution of total SBSs and SBS classes relative to gene features including intergenic regions (blue), introns (yellow), coding exons (black), 5΄ UTRs (red) and 3΄ UTRs (green).
Analyses of SBS subclass characteristics
Overlaps of SBS classes with published genome-wide binding datasets for other proteins were determined using features in the UCSC Genome Browser. The proportion of overlap in a given SBS subclass was compared to the proportion of overlap for all SBSs with a large sample Z-test. The Z-test was performed as follows. First, the proportion of overlap for both the total SBSs and a given SBS class of interest was calculated as P = NSBSs/Noverlapping SBSs. Next, common p was calculated as common P = (Noverlapping SBSs group1 + Noverlapping SBSs group2) ÷ (NSBSs group1 + NSBSs group2). Standard Error of the mean (SE) was calculated as SE = SQRT [(common p * (1-common p)* ((1 ÷ NSBSs group1) + (1 ÷ NSBSs group2))]. The z-score was calculated as z = [(pgroup1 -pgroup2) ÷ SE] and converted to a P-value using P-value = NORMSDIST(z).
RESULTS
An F2 genetic screen identifies new su(Hw) separation-of-function (SOF) alleles
To gain a better understanding of Su(Hw) function, we conducted a forward F2 genetic screen (Figure 1). Previously, su(Hw) mutants were identified solely through assessment of gypsy insulator function. In contrast, our screen assayed for loss of gypsy insulator function and female fertility. After screening more than 8000 chromosomes, we identified four new su(Hw) alleles that fell into three complementation classes (Figure 2). Class I includes su(Hw)A1933 and su(Hw)A2663, two alleles that failed to complement both the gypsy insulator and sterility phenotypes when heterozygous with extant su(Hw) null alleles (Figure 2B). Class II includes su(Hw)A460, an allele that failed to complement gypsy insulator function but complemented the sterility of extant su(Hw) null alleles (Figure 2B). This SOF phenotype has been observed previously, represented by the extant allele su(Hw)f. Class III includes su(Hw)M393, an allele that complemented gypsy insulator function but failed to complement the sterility of extant su(Hw) null alleles (Figure 2B). No extant su(Hw) allele shows this complementation pattern. Notably, su(Hw)M393 and su(Hw)A460 trans-heterozygotes complement each other (Figure 2B). These data demonstrate that the insulator and fertility functions of Su(Hw) are genetically separable.
The molecular lesions associated with the new su(Hw) alleles were defined. The two class I null alleles carry deletions within the su(Hw) locus. For su(Hw)A1933, the deletion extended into the essential upstream gene, RpII15, demonstrated by lethality when heterozygous with su(Hw)v, an allele that also carries an RpII15 deletion (Figure 2A and B). For su(Hw)A2663, the deletion extended into the downstream CG3259 gene (Figure 2A). Western analyses showed that no Su(Hw) protein was produced in su(Hw)A1933/2 and su(Hw)A2663/vanimals (Figure 2C). The class II and III SOF alleles carry mutations in ZFs. For su(Hw)A460, the lesion changed amino acid 486 in ZF8 from an arginine to a cysteine. For su(Hw)M393, the lesion changed amino acid 350 in ZF4 from a cysteine to a serine. Western analyses of protein extracts from su(Hw)A460/v and su(Hw)M393/v SOF animals showed that wild-type levels of full-length Su(Hw) protein were produced (Figure 2C). These data suggest that SOF alleles generate Su(Hw) mutated for a single ZF within an otherwise wild-type protein.
Most Su(Hw) ZFs contribute to in vitro DNA binding
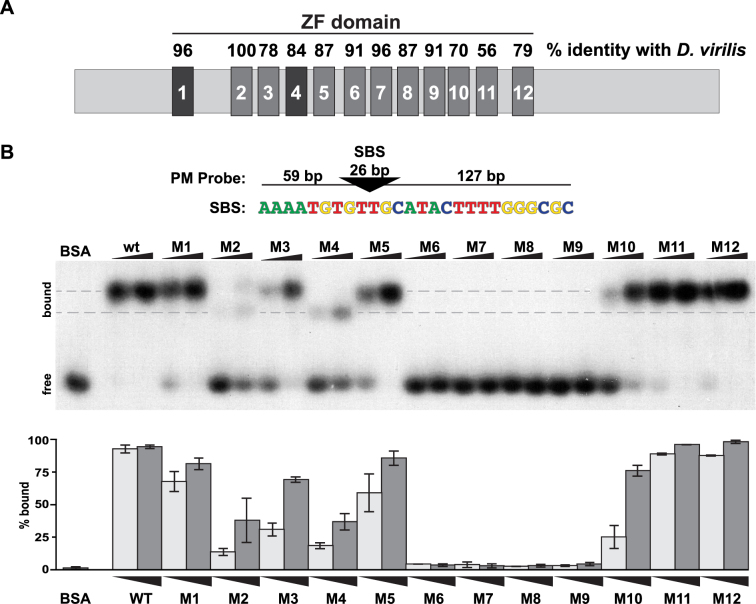
Prompted by our findings that ZF mutations separate Su(Hw) in vivo functions, we hypothesized that loss of different ZFs might change DNA binding properties of Su(Hw) leading to changes in function. To test this prediction, we investigated how individual ZFs contribute to the in vitro DNA binding properties of Su(Hw) using electrophoretic mobility shift assays (EMSAs; Figure 3). For these studies, wild-type and Su(Hw) ZF mutants were bacterially expressed and purified (Supplementary Figure S1A). Each Su(Hw) ZF mutant (designated Su(Hw)MZF#) carried an amino acid substitution of at least one zinc-chelating amino acid within a single ZF (Supplementary Table S1), which we reasoned resulted in a complete loss of ZF function. DNA binding of these mutant proteins was assessed using a double-stranded DNA probe that carried an endogenous SBS (4C15) engineered so that it was a PM to the Su(Hw) binding motif.
Figure 3.
Analyses of in vitro binding properties of Su(Hw) ZF mutants. (A) Shown is a diagram of the ∼110 kD Su(Hw) protein with each ZF represented as a large rectangle. The ZFs are tightly clustered, with amino acid spacing ranging from 47 amino acids between ZF1 and ZF2 to 3 amino acids between ZF8 and ZF9. The domain includes two C2HC ZFs (ZF1, ZF4) and 10 C2H2 ZFs. The percent of amino acid identity between the Drosophila melanogaster Su(Hw) ZFs and the corresponding Drosophila virilis ZFs is indicated above each rectangle. (B) Top: the structure of the 212-bp DNA probe (PM) is shown, with the 26-bp SBS (triangle) located 59 bp from the 5΄ and 127 bp from the 3΄ end. The nucleotide sequence of the Su(Hw) binding consensus is indicated below the triangle. Bottom: shown is a representative image of an EMSA, comparing DNA binding of Su(Hw)WT and Su(Hw)M proteins that carry a replacement of zinc-chelating amino acids in the indicated ZF, referred to as Mutant (M)1 to M12. Bovine serum albumin (1.0 μg) serves as a negative control. Binding was assayed using two protein amounts (0.3 and 1.0 μg). The migration of free and bound DNA is indicated on the left. Gray dashes indicate the locations of two Su(Hw)–DNA complexes with different migrations. Quantification of the percent of bound probe for each Su(Hw) protein is graphed (0.3 μg, light gray; 1.0 μg, dark gray). Quantification represents an average of two replicates, with the bars showing the range of values obtained.
EMSA analyses identified three broad categories of DNA binding (Figure 3B). One category includes four ZF mutants that show near wild-type binding, with >80–90% of the probe shifted at the higher protein level of 1 μg per reaction [Su(Hw)M1, Su(Hw)M5, Su(Hw)M11 and Su(Hw)M12]. A second category includes four ZF mutants with intermediate levels of DNA binding, with 5–40% of the probe shifted at the higher protein concentration [Su(Hw)M2, Su(Hw)M3, Su(Hw)M4, Su(Hw)M10]. The third category includes four ZF mutants that showed little to no DNA binding, with <5% of the probe shifted at any protein concentration [Su(Hw)M6, Su(Hw)M7, Su(Hw)M8 and Su(Hw)M9]. These analyses suggest that eight Su(Hw) ZFs are involved in DNA recognition, with ZF6 through ZF9 having an essential role for binding to the PM probe.
Our in vitro EMSA analyses established that the SOF su(Hw)A460 mutation resides in a ZF essential for DNA binding. This paradox suggests that the R486C substitution in Su(Hw)A460 only partially disrupts ZF8 function, either through changes in DNA identification because R486 is in the ZF recognition helix or through altered zinc chelation and ZF8 structure due to the addition of an extra cysteine (3). To test this prediction, we generated and purified the R486C Su(Hw) mutant and tested its in vitro DNA binding properties. Indeed, we found that this mutant protein bound 75% (12/16) of randomly selected endogenous SBSs in vitro (Supplementary Figure S2), reinforcing the idea that this substitution causes a partial loss of ZF8 function. We refer to this mutant protein as Su(Hw)m8A460, using a lower case ‘m’ to denote the predicted partial loss of function. The second SOF mutant su(Hw)M393 carries a mutation of a zinc-chelating cysteine in ZF4, identical to the amino acid substitution in Su(Hw)M4. As such, we predict that this change causes complete loss of ZF4 function. Our in vitro EMSA analyses showed that Su(Hw)M4 binding to the PM probe was reduced, but not lost (Figure 3B), implying that Su(Hw)M4M393 remains capable of recognising some SBSs.
Migration of Su(Hw) ZF mutants was not uniform in EMSAs (Figure 3B). We observed Su(Hw)M2 and Su(Hw)M4 formed protein–DNA complexes with increased mobility relative to Su(Hw)WT and other ZF mutants (Figure 3B, lower dotted line). Increased mobility was also observed with probes carrying different endogenous SBSs (data not shown). These changes occur even though Su(Hw)M2 and Su(Hw)M4 are the same size as other Su(Hw) proteins (Supplementary Figure S1) and the binding conditions were identical. Based on these observations, we predict that some Su(Hw) ZF mutants adopt different DNA-bound conformations from the wild-type Su(Hw) protein.
Fertility of su(Hw)A460/v females correlates with maintained gene repression
Previous studies established that Su(Hw) has a prominent role in repressing neuronal genes in the ovary, with regulation of Rbp9 critical for female fertility (17). Based on these observations, we predicted that expression of Su(Hw) target genes would be altered in sterile su(Hw)M393/v females, but not fertile su(Hw)A460/v females. Expression levels of Su(Hw) target genes were measured in RNA isolated from the su(Hw) SOF alleles and su(Hw)+/+ ovaries. We found that all Su(Hw) target genes (15/15) were mis-regulated in su(Hw)M393/v ovaries (Figure 2D), with observed changes similar to those found in ovaries isolated from su(Hw) null females. Gene expression was also altered in fertile su(Hw)A460/v females. In fact, most Su(Hw) target genes tested (8/15) were mis-regulated in su(Hw)A460/v ovaries. Even so, RNA levels of the critical fertility gene, Rbp9, showed only a ∼2-fold increase, which is below the level associated with female sterility (17). Together, these analyses emphasize that Su(Hw) is required for transcriptional regulation in the ovary, with fertility linked to maintenance of Rbp9 repression.
Chromosome association in vivo correlates with DNA binding in vitro
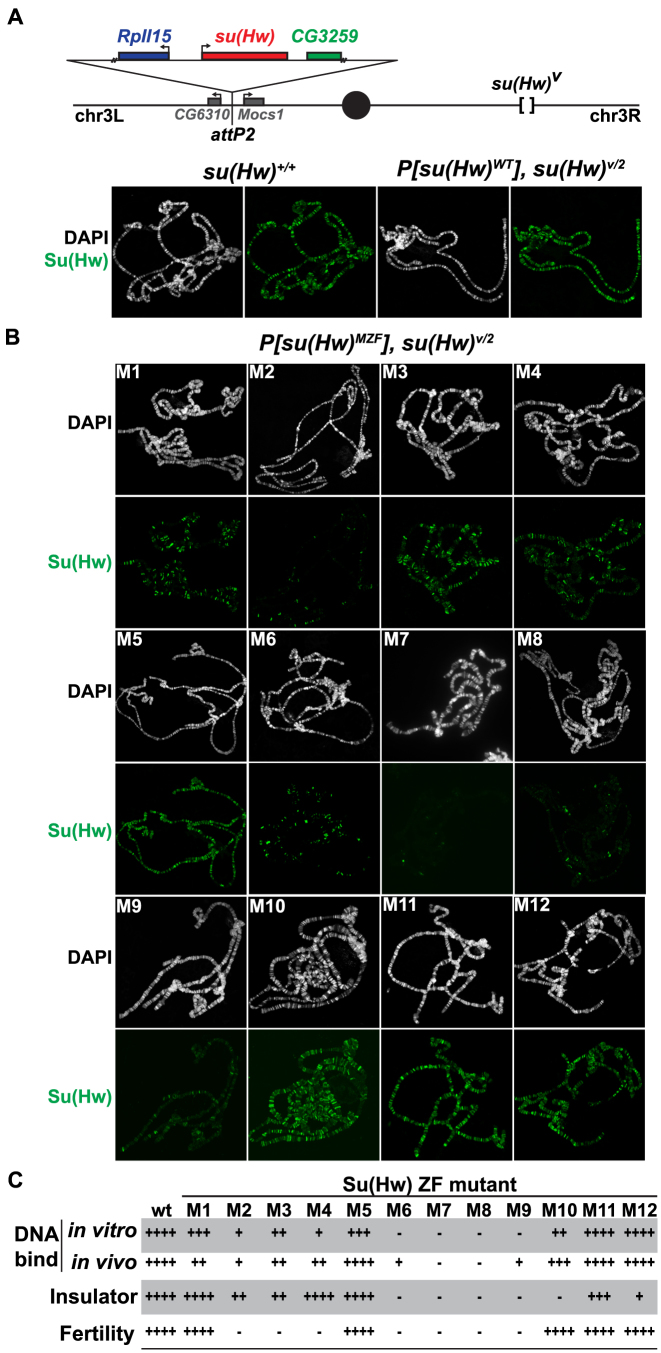
To define how individual ZFs contribute to in vivo Su(Hw) function, we generated transgenic lines expressing Su(Hw) with a defective ZF. Each transgene was integrated at an identical site on chromosome 3L (Figure 4A, attP2). Transgenes were crossed into a su(Hw) mutant background to produce P[su(Hw)*], su(Hw)v/2 flies, so that Su(Hw) protein was produced only from the transgene. To determine whether mutation of any ZF changed the steady-state level of Su(Hw), we completed western analysis. We found wild-type levels of full-length protein in all lines (Supplementary Figure S1B), allowing us to complete functional tests that defined individual contributions of the twelve Su(Hw) ZFs.
Figure 4.
Analyses of in vivo function of Su(Hw) ZF mutants. (A) Top: schematic of the third chromosome organization in su(Hw) transgenic flies. Transgenes were inserted into the attp2 landing site located between the CG6310 and Mocs1 genes. Each transgene carried a 6-kb genomic fragment, which included 1.2 kb of 5΄ DNA encompassing the complete RpII15 gene (blue rectangle), a wild-type or mutant su(Hw) gene (red rectangle) and 0.5 kb of 3΄ DNA that encompassed a portion of CG3258 (green rectangle). Transgenes were recombined onto a su(Hw)v chromosome. Bottom: shown is the salivary gland polytene chromosome distribution of endogenous Su(Hw) [su(Hw)+/+] and wild-type Su(Hw) expressed from the transgene [P[su(Hw)WT, su(Hw)v/2]. Chromosomes were stained with the DAPI (white) and antibodies against Su(Hw) (green). (B) Shown is the polytene chromosome distribution of the Su(Hw) mutant proteins. Mutant proteins were either expressed from a transgene (M1–M3, M5–M9, M11–M12) or from endogenous su(Hw) mutant alleles (M4, M10). (C) The shown table summarizes the function associated with Su(Hw) ZF mutants. The four categories include in vitro DNA binding based on EMSA assays, in vivo DNA binding based polytene chromosome assays, gypsy insulator activity based on body, wing and bristle (y2, ct6, f1) phenotypes and female fertility based. The scale corresponds to ++++ = 75–100%, +++ = 50–74%, ++ = 25–49%, + = 5–24%, − = <5% of wild-type function.
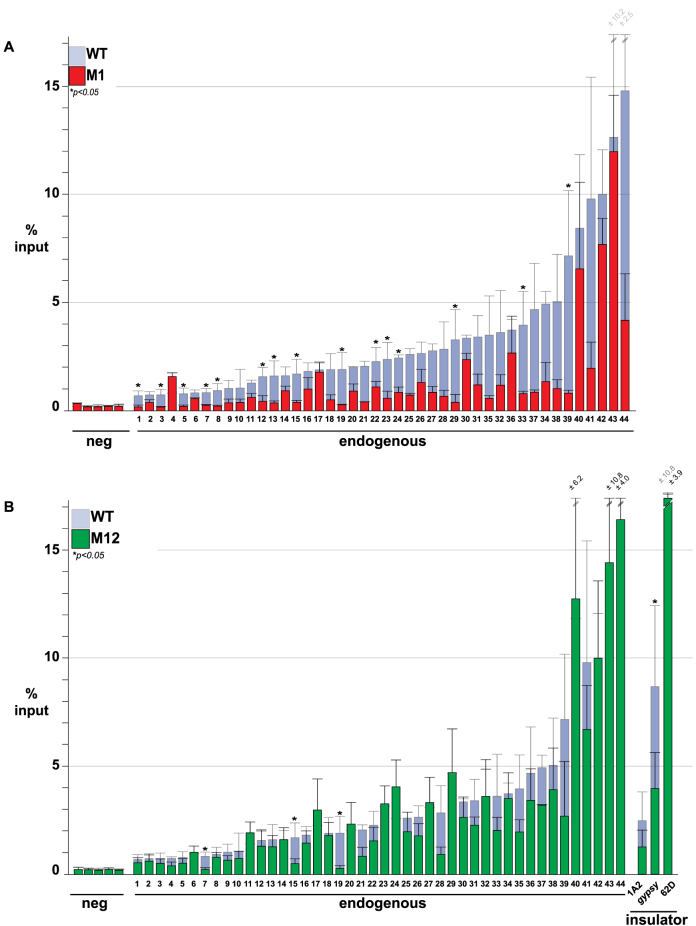
To obtain a measure of genome-wide SBS occupancy of transgenically expressed Su(Hw), we examined Su(Hw) binding to salivary gland polytene chromosomes. As a control, we stained P[su(Hw)WT], su(Hw)v/2 chromosomes, observing that the transgenic wild-type Su(Hw) globally bound chromosomes (Figure 4A). Next, we analyzed P[su(Hw)MZF], su(Hw)v/2 chromosomes (Figure 4B). In all but one case, the level of genome-wide association correlated with the in vitro DNA binding of the Su(Hw) ZF mutant (Figure 4B and C). The one exception was Su(Hw)M1, a ZF mutant that showed near wild-type in vitro binding but reduced in vivo occupancy (Figure 4B). To understand the extent of the in vivo SBS loss, we used chromatin immunoprecipitation coupled with qPCR (ChIP-qPCR). These studies showed decreased occupancy of Su(Hw)M1 at the majority of sites tested (84%, 37/44; Figure 5A). To test whether reduced in vivo occupancy reflected an inability to recognize the endogenous SBS, we assayed in vitro Su(Hw)M1 binding to three SBSs that were unoccupied in vivo. In all cases, Su(Hw)M1 bound these SBSs at levels comparable to Su(Hw)WT (Supplementary Figure S3). These data imply that Su(Hw) ZF1 is not required for DNA recognition in vitro, but is necessary for association with a subset of SBSs in vivo. We hypothesize that ZF1 might be required to facilitate occupancy or stabilize Su(Hw) association in the context of chromatin.
Figure 5.
Analysis of Su(Hw)M1 and Su(Hw)M12 association at endogenous SBSs. (A) Shown are ChIP-qPCR data for Su(Hw)WT (light gray), Su(Hw)M1 (red) and Su(Hw)M12 (green) at genomic regions lacking an SBS (5 negative control sites), carrying an SBS (44 endogenous sites) and known insulators (1A2, 62D, gypsy). Data for Su(Hw)M1 and Su(Hw)M12 are graphed on top of the Su(Hw)WT data for comparison. Data represent average percent input of two-four biological replicates. Error bars indicate standard deviation. P < 0.05 (Student's t-test).
Two clusters of Su(Hw) ZFs are required for gypsy insulator function
Su(Hw) insulator function depends upon recruitment of two cofactors, CP190 and Mod(mdg4)67.2 [Mod67.2] (38,39). Only half of endogenous SBSs overlap with these partner proteins (35,36). While Su(Hw) interacts with Mod67.2 using a region outside of the ZF domain (40), the region of Su(Hw) required for CP190 recruitment is currently unknown. To establish whether any ZF is needed for recruitment or stabilization of these insulator co-factors, salivary polytene chromosomes from the Su(Hw)MZF lines were co-stained with antibodies against Su(Hw) and either Mod67.2 or CP190. These studies showed that Su(Hw) ZF mutants extensively co-localize with Mod67.2 and CP190 (data not shown), implying that disruption of individual Su(Hw) ZFs has little effect on partnership with the gypsy insulator proteins.
We next questioned whether gypsy-insulator function might be altered in the su(Hw) ZF mutant backgrounds. Previous studies have connected insulator function to the formation of topologically associated domains (TADs) that define structural domains of regulatory interaction (41–43). Such observations suggest that a genome-wide reduction in Su(Hw) occupancy might lead to diminished insulator effectiveness, due to changes in TAD structure. To evaluate this hypothesis, we tested whether the Su(Hw) ZF mutants were capable of establishing insulator function at the gypsy-induced ct6 and f1 alleles. In general, our data correlate chromosome association and insulator function, wherein ZFs that are essential for in vitro and in vivo DNA binding are also critical for gypsy insulator function (ZFs 6–9). In addition, we confirmed that ZF10 is required for insulator function as seen previously (28,30,31). In contrast, reduced genome-wide Su(Hw) occupancy does not alter insulator function, demonstrated by the robust enhancer blocking activity of the Su(Hw)M1 and Su(Hw)M4 proteins (Figure 4B and C). One unexpected finding was that Su(Hw)M12 lost gypsy-insulator function. This observation was surprising because our in vitro binding assays demonstrated that Su(Hw)M12 bound the PM probe and the gypsy insulator (Figure 3B, data not shown) and our in vivo studies show strong chromosome association (Figure 4B). To understand the loss of insulator function, we directly examined Su(Hw)M12 in vivo binding using ChIP-qPCR. We measured Su(Hw)M12 occupancy at the gypsy insulator and 46 SBSs (Figure 5B), including two endogenous SBSs with insulator function, 1A-2 and 62D (18,19,28). These studies showed that Su(Hw)M12 bound most SBSs (41/46), including 62D, but had reduced occupancy at gypsy and 1A-2 (Figure 5B). Taken together, our data suggest that insulator function depends directly on gypsy insulator occupancy and not on the global distribution of Su(Hw). Further, we show that two ZF clusters are required for insulator function, ZFs 6–9 and ZFs 10 and 12, with the C-terminal cluster needed for Su(Hw) binding within a chromatin context.
Su(Hw) ZFs required for fertility differ from those needed for insulator function
To identify ZFs required for female fertility, we completed two assays. First, we assessed whether P[su(Hw)MZF], su(Hw)v/2 females produced offspring when mated to su(Hw)+/+ males. We found that sterility occurred upon loss of ZF2 to ZF4, as well as ZF6 to ZF9 (Figure 4C). Second, we examined Su(Hw) repressor function, by measuring levels of Rbp9 transcripts in ovary RNA isolated from su(Hw)M1, su(Hw)M5, su(Hw)M11, su(Hw)M12 females. In all mutant lines, we found that Rbp9 mRNA levels were not significantly different from those in su(Hw)WT (data not shown). Based on these data, we conclude that the Su(Hw) fertility function depends on two ZF clusters, ZF2 to ZF4 and ZF6 to ZF9. These data reveal that ZF requirements differ for Su(Hw) insulator and fertility functions, in line with our genetic screen that identified SOF alleles.
Genome-wide identification of Su(Hw)M4M393 bound SBSs reveals sequence variation
To gain a molecular understanding of how ZF4 loss affects Su(Hw) function, we mapped Su(Hw)M4M393 binding genome-wide using ChIP-seq. Chromatin was isolated from ovaries dissected from females younger than six hours old. In this way, the Su(Hw)M4M393 SBS dataset could be directly compared with ovarian SBS datasets obtained for Su(Hw)WT and Su(Hw)M10f (31). Based on a 1% FDR and a 3-fold enrichment cutoff, we identified 777 SBSs. Of these, 82% (636) overlap with previously identified ovarian SBSs [Figure 6A, B; (31,36)]. A second α-Su(Hw) ChIP-seq was performed that identified only 329 M4-retained SBSs. Importantly, 92% of these sites overlap with sites identified in ChIP-seq1. We reasoned that the reduced SBS recovery in the ChIP-seq2 dataset might be due to a lower efficiency of ChIP. To test this prediction, we determined whether Su(Hw)M4M393 bound in vivo to 18 SBSs that were recovered in ChIP-seq1 but not ChIP-seq2, as well as 12 sites that were recovered in both experiments. In these studies, ChIP-qPCR was completed using chromatin isolated from ovaries obtained from su(Hw)M393/v and compared with ChIP-qPCR of chromatin isolated from su(Hw)E8/vfemales (Supplementary Figure S4), a mutant encoding a full-length protein unable to bind DNA. We found that Su(Hw)M4M393 bound all SBSs. Additionally, SBSs identified in both ChIP-seq datasets had a higher average Su(Hw)M4M393 occupancy. Taken together, these data support our inference that differences in the ChIP-seq datasets reflect a reduced antibody precipitation in ChIP-seq2. We conclude that the SOF Su(Hw)M4M393 mutant binds ∼20% of SBSs genome-wide.
To understand differences between the SOF ZF mutants, we compared features of SBSs occupied by either Su(Hw)M4M393 [insulator only] or Su(Hw)M10f [fertility only]. We found that SBSs fell into four subclasses (Figure 6B). Subclass I was SBSs bound only by Su(Hw)WT [WT-unique, 1576 sites], subclass II was SBSs bound by Su(Hw)WT and Su(Hw)M10f [M10-only, 821 sites], subclass III was SBSs bound by Su(Hw)WT, Su(Hw)M4M393 and Su(Hw)M10f [M4+M10, 326 sites], and subclass IV was SBSs bound by Su(Hw)WT and Su(Hw)M4M393 [M4-only, 310 sites]. To determine whether DNA sequence differences exist between subclasses, we used the motif search program MEME. These analyses uncovered that the Su(Hw) binding consensus is larger than previously recognized [(36); Figure 6C]. The extended consensus contains three modules, corresponding to a new upstream AT-rich module, a central GCATACTTT module and a downstream GC-rich module. The sequence motif observed for M4-only SBSs contains the upstream and central module, which largely matches the consensus sequence for SBSs in the gypsy insulator (Figure 6C). In contrast, the sequence motif observed for M10-only SBSs contains the central and downstream modules. The WT-unique sites carry the same consensus sequence as M10-only sites. As WT-unique sites represent low occupancy SBSs, we predict that the loss of either ZF4 or ZF10 disrupts Su(Hw) association at these sites (Figure 6D). Taken together, our data reveal that the Drosophila genome includes sequence subclasses of SBSs, with the most frequent subclass (M10-only) carrying the two modules that represent the previously identified Su(Hw) binding motif (36).
We determined how Su(Hw) bound to the newly identified modules in the compound consensus sequence. In these studies, we used EMSA to assess the relative affinities of Su(Hw)WT and Su(Hw) ZF mutant proteins for double stranded DNA probes carrying the PM consensus with mutations in the different modules, upstream (mU), central (mC) or downstream (mD) (Figure 7). Several observations were made. First, all mutant probes displayed weaker Su(Hw) binding than was observed for the PM probe (Figure 7), suggesting that a consensus sequence with three modules represents the highest affinity SBS. This observation is supported by data showing that in vivo Su(Hw) occupancy is highest at the M4+M10 sites (Figure 6D). Second, the central module is necessary but not sufficient for Su(Hw) binding. EMSA studies demonstrate that loss of this module affects binding of all Su(Hw) proteins, but a probe carrying only the central module cannot bind Su(Hw)WT (mUmD, Supplementary Figure S5). Third, binding of Su(Hw)WT and ZF mutants to the mU probe was the strongest relative to other mutant probes. Consistent with this observation, the mU probe sequence corresponds to M10-only sites that represent the second highest occupancy subclass in the genome (Figure 6D). DNA recognition of mU requires ZF2 to ZF4, as loss of these ZFs reduces Su(Hw) binding to levels similar to that observed with loss of ZF6 to ZF9. Fourth, Su(Hw) demonstrated a lower affinity to the mD probe than to the mU probe. These results are consistent with the observation that M4-only sites show lower in vivo occupancy than M10-only sites (Figure 6D). DNA recognition of the mD probe requires ZF10–ZF12, because loss of any of these ZFs abolishes Su(Hw) binding. Taken together, these data reveal that Su(Hw) binding requires a site with at least two modules, each recognized by distinct ZF clusters.
SBS subclasses possess distinct functional characteristics
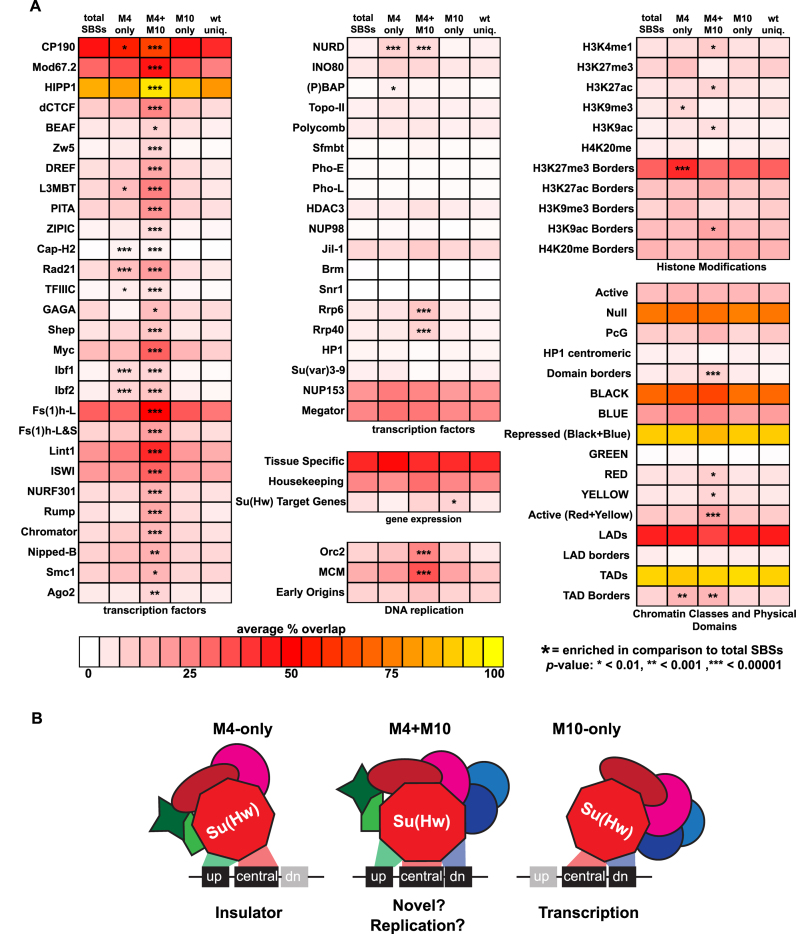
Our genetic studies suggest that sequence composition of an SBS corresponds with Su(Hw) regulatory function. Based on these observations, we predicted that SBS sequence subclasses possess distinct properties. To test this possibility, we analyzed whether SBS subclasses differed in genomic location or chromatin features (Figure 8). These analyses revealed that M4-only SBSs show characteristics predicted for genomic insulators, while M10-only SBSs show characteristics associated with transcriptional repression.
Figure 8.
Properties of SBS classes. (A) Shown is a heat map depicting the SBS subclass overlap with chromatin features including transcription factors, gene expression, DNA replication, histone modifications and physical and epigenetic domains. Scale for the heat map reflects the average percent overlap of SBSs within each class with genome-wide data for each feature. Statistical significance was calculated using a large sample Z-test and is indicated with an asterisk(s). (B) Proposed model of Su(Hw) function at SBS classes. DNA binding data demonstrate that binding different SBSs requires distinct ZFs and that these classes are enriched for different protein cofactors, suggesting that ZFs not engaged in DNA binding might be engaged in cofactor recruitment that might impact site-specific Su(Hw) function.
Most SBSs reside in introns and intergenic regions, consistent with high frequency of these features within the genome (34,35). The distribution of SBS subclasses relative to gene features was examined (Figure 6E). We found that M4-only and M4+M10 sites show a modest enrichment in intergenic regions (Figure 6E), locations expected for sites involved in insulator function. Notably, both subclasses carry the 5΄ AT-rich module, a feature found in the gypsy insulator consensus sequence (Figure 6C). This similarity provides a second connection with insulator function. The M10-only motif is enriched among SBSs in Su(Hw) target genes (Figure 8A) and near exons and transcription start sites (17). As the majority of Su(Hw) target genes are repressed, these observations link the M10-only class with gene silencing.
We investigated the relationship between SBS sequence subclasses and chromatin features. TAD borders are composed of high occupancy sites for multiple architectural proteins (44), with SBSs enriched at boundaries of repressive chromatin domains (45). Consistent with this observation, the highest occupancy subclass, M4+M10, displays enriched overlap with all architectural proteins relative to total SBSs (Figure 8A). Further, this sequence subclass displays relative enrichment of active chromatin marks and DNA replication factors (Orc2, MCMs), suggesting a link to DNA replication. Based on previous studies that connected Su(Hw) recruitment of the histone acetyltransferase SAGA and ORC (46), we predict that SBSs that overlap with ORC correspond to the M4+M10 sequence subclass. Recent observations connect TADs to replication domains (47), indicating that Su(Hw) might contribute to TAD formation through effects on origin usage in DNA replication. The M4-only class also shows significant enrichment of specific architectural proteins, including CP190, the cohesin subunit Rad21 and the DNA binding Insulator Binding Factors, Ibf1 and Ibf2. Our finding that a SBS subclass is enriched for the insulator factors Ibf1 and Ibf2 is supported by two observations in the literature. First, large-scale protein interaction studies recovered Ibf1 as a Su(Hw) co-factor (23). Second, the SBS motif found at Ibf1/2 bound sites corresponds to the M4-only motif (48). Finally, we found that M4-only and M4+M10 SBSs exhibit relative enrichment at H3K27me3 and TAD boundaries. Together, these observations suggest a differential enrichment of chromatin factors with SBS subclasses, linking the M4-only and M4+M10 SBSs with insulator function.
DISCUSSION
Drosophila Su(Hw) is a multivalent transcriptional regulator that confers activation, repression or insulation. Here, we define the role of the ZF domain in Su(Hw) function, prompted by our identification of SOF su(Hw) alleles obtained in an unbiased F2 screen (Figures 1 and 2). Our studies reveal a link between the regulatory versatility of Su(Hw) function and the underlying DNA sequence of an SBS.
Features of the polydactyl Su(Hw) ZF DNA binding domain
Su(Hw) has a conserved twelve ZF domain that binds a compound consensus sequence of ∼26 nucleotides comprised of three modules (Figure 6C). Using a combination of in vitro and in vivo assays, we show that DNA binding by Su(Hw) requires that the binding site contains an intact central module and one of the other two modules (Figure 7). Distinct ZFs clusters bind each module (Figures 3 and 7). The ZF2–ZF4 cluster binds the downstream module, the ZF6–ZF9 cluster binds the central module and the ZF10–ZF12 cluster binds the upstream module. Among these ZF clusters, only the ZF6–ZF9 cluster is essential, as mutation of any one of these fingers abolishes in vitro and in vivo DNA binding (Figures 3, 4 and 7). Notably, this cluster has conserved features found in other tandem DNA binding proteins (3), including a five amino acid linker that carries the TGE(K/R)P sequence corresponding to the hallmark feature of DNA-binding fingers that dock within the major groove (5).
A single DNA-binding ZF interacts with three consecutive nucleotides (3). As such, only eight or nine ZFs are needed to recognize a ∼26 nucleotide motif. Consistent with this prediction, we find that not all of the twelve Su(Hw) ZFs are required for DNA recognition, including four ZFs (ZF1, ZF5, ZF11, ZF12) that when lost had only modest effects in our in vitro assay (Figures 3 and 7). Even so, these non-essential ZFs are strongly conserved in drosophilids (Figure 3A). ZF1 displays the highest amino acid conservation (96% identity) over 40 million years of evolution, suggesting an alternative contribution to Su(Hw) function. Indeed, we find that ZF1 is required for in vivo occupancy (Figures 4 and 5). The mechanism responsible for this ZF1 contribution is currently unknown. One possibility is that ZF1 interacts with RNA to stabilize Su(Hw) association at certain sites, as recently demonstrated for the multifunctional Yin-yang 1 (YY1) TF (49–51). Alternatively, ZF1 might direct protein-protein interactions (12). As Su(Hw)M1 fails to bind a subset of SBSs in vivo that it can bind in vitro, we speculate that ZF1 might direct association with a chromatin remodeling complex that facilitates in vivo occupancy. In support of this idea, SBSs show enrichment of the NURF chromatin remodeling complex (Figure 8A), which has been implicated in insulator function (52–54). Further experiments are needed to distinguish between these possibilities.
Our data are reminiscent of properties described for the multifunctional transcription factor CTCF that carries a contiguous eleven ZF domain. Analyses of genome-wide CTCF binding in human cells revealed a large compound CTCF consensus sequence spanning ∼41 bp (23). Individual CTCF sites were found to carry combinations of four modules, recognized using clusters of different ZFs that bind each module (24). Similar to SBSs, the majority of CTCF sites are composed of a two-module combination (23). Based on these findings, a CTCF code was postulated, predicting that the pleiotropic functions of CTCF are conferred by recognition of diverse sequences through combinatorial use of its ZF domain (24,55). The strong parallel between previous findings for CTCF and our data for Su(Hw) indicate the presence of a ‘Su(Hw) code’ within the Drosophila genome.
Functional diversity of Su(Hw) is linked to SBS sequence
The ‘Su(Hw) code’ hypothesis predicts that the diversity of Su(Hw) function correlates with distinct SBS sequence subclasses. Here, we provide several lines of evidence that support this prediction. We identified two ZF mutants that separate Su(Hw) functions. Loss of ZF4 [su(Hw)M393] within the ZF2 to ZF4 cluster causes female sterility due to altered regulation of Su(Hw) target genes, including de-repression of Rbp9, whereas gypsy insulator function is retained. ChIP-seq analyses of Su(Hw)M4M393 revealed that loss of ZF4 significantly reduces Su(Hw) occupancy to ∼20% of total SBSs. Su(Hw)M4M393 retained sites correspond to SBSs that carry the upstream module (M4-only and M4+M10 sites; Figure 6). In contrast, loss of ZF10 [su(Hw)f] within the ZF10 to ZF12 cluster causes a loss of gypsy insulator function, whereas female fertility and Rbp9 repression is retained (17). ChIP-seq analyses of Su(Hw)M10f revealed that loss of ZF10 reduces Su(Hw) occupancy to ∼40% of total SBSs [Figure 6B; (31)]. Su(Hw)M10f retained sites correspond to SBSs that carry the downstream module (M10-only, M4+M10 sites; Figure 6). Together, these findings link SBS sequence subclasses with the diverse functions of Su(Hw). Specifically, we predict that the M10-only subclass represents SBSs involved in gene repression, whereas the M4-only and M4+M10 subclasses represent sequences with insulator function.
Integrative analyses of modENCODE data support the classification of SBS subclasses into functional groups (Figure 8). The M4-only subclass shows two features, including a relative enrichment of chromatin factors associated with insulator function (Cap-H2, Rad21 and Ibf1/2) and enriched positioning at TAD borders that are associated with H3K27me3. These data reinforce a link between the M4-only subclass and Su(Hw) insulator function. Similarly, the M4+M10 class represents the highest Su(Hw) occupancy class (Figure 6C), a feature of insulator function. Lastly, the majority M10-only sites (∼80% of SBSs) localize within repressive black chromatin (21). Genome-wide studies suggest that most SBSs are associated with repression of nearby gene expression (22) and Su(Hw) target genes show enrichment for M10-only SBSs [Figure 8; (17)]. As shown in our model (Figure 8), the distinct features of each SBS sequence subclass suggest possible categories for regulatory function.
We consider several mechanisms for how SBS sequence might contribute to Su(Hw) regulatory output. First, binding affinity might be critical for aspects of Su(Hw) function, as demonstrated for CTCF insulator function. High affinity CTCF sites are commonly found within borders of TADs (56), and appear to be critical for maintaining TAD structure throughout the cell cycle. Interestingly, the high affinity M4+M10 sites are enriched for replication proteins (Figure 8), indicating that regulation of DNA replication origin usage might contribute to TAD formation. Alternatively, the high affinity CTCF sites have been associated with promoters of unidirectionally transcribed genes, leading to the prediction that these sites form a barrier to RNA polymerase II elongation that prevents antisense transcription (57). Second, SBS sequence subclasses might alter the conformation of Su(Hw) when it binds DNA, thereby imparting specific regulatory function. As demonstrated in our EMSA analyses, Su(Hw) ZF mutants migrate differently in complex with DNA (Figure 3), reinforcing the idea that different ZF usage might alter Su(Hw) or DNA conformation. Allosteric effects of DNA on TF function have been observed previously. For example, studies of the glucocorticoid receptor (GR) have shown that GR binding to sites with a single base pair difference results in distinct GR conformations and regulatory activity (58,59). Third, differences in ZF engagement upon SBS binding might influence co-factor recruitment by promoting availability of some ZFs for protein-protein or protein–RNA interactions. In addition, it has been reported that CTCF regulation of the human p53 gene might employ such as mechanism. These studies found that CTCF regulation involves one ZF cluster for binding DNA and a second for binding the antisense Wrap53 RNA (60). Additionally, the CTCF ZF domain has been shown to recruit proteins, such as YY1 (61). Information concerning the role of Su(Hw) ZFs in co-factor recruitment is limited. We have found that loss of individual ZFs did not alter global colocalization of co-factors CP190 and Mod67.2 (data not shown). However, previous studies demonstrated that the Su(Hw) ZF domain was required for recruitment of ENY2 (62), a third Su(Hw) co-factor whose genome-wide distribution is unknown. Given that Su(Hw) has diverse regulatory outputs, we suggest that multiple mechanisms are likely to contribute to SBS subclass function.
In summary, Su(Hw) carries a large ZF domain that directs association with a modular consensus-binding site. Differential usage of ZFs imparts binding to SBSs with varied combinations of sequence modules, a strategy that might underlie the functional diversity of Su(Hw). Significantly, Su(Hw) represents the major class of metazoan transcription factors. Indeed, nearly half of all human transcription factors are C2H2 ZF proteins (7). Among these, the average number of ZFs is ten (4). Our findings add to growing evidence that a DNA binding code might be common among polydactyl ZF transcription factors (11,23,24), with these proteins using combinations of ZF clusters to bind genomic loci with different binding motif modules. Together, these findings predict that DNA codes might ultimately impact the regulatory output of this class of multifunctional transcription factors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bing for his advice on bioinformatic analyses, Amber Hohl for help with the genetic screen, and Miles Pufall and members of the Geyer Lab for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [GM042539 to P.K.G]. Funding for open access charge: NIH [GM042539].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lelli K.M., Slattery M., Mann R.S.. Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 2012; 46:43–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Emerson R.O., Thomas J.H.. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009; 5:e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Enuameh M.S., Asriyan Y., Richards A., Christensen R.G., Hall V.L., Kazemian M., Zhu C., Pham H., Cheng Q., Blatti C. et al. Global analysis of Drosophila Cys2-His2 zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 2013; 23:928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Najafabadi H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M. et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015; 33:555–562. [DOI] [PubMed] [Google Scholar]
- 5. Wolfe S.A., Nekludova L., Pabo C.O.. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000; 29:183–212. [DOI] [PubMed] [Google Scholar]
- 6. Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010; 79:213–231. [DOI] [PubMed] [Google Scholar]
- 7. Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M.. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009; 10:252–263. [DOI] [PubMed] [Google Scholar]
- 8. Razin S.V., Borunova V.V., Maksimenko O.G., Kantidze O.L.. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochemistry (Mosc). 2012; 77:217–226. [DOI] [PubMed] [Google Scholar]
- 9. Nikolaev L.G., Akopov S.B., Didych D.A., Sverdlov E.D.. Vertebrate protein CTCF and its multiple roles in a Large-Scale regulation of genome activity. Curr. Genomics. 2009; 10:294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ohlsson R., Renkawitz R., Lobanenkov V.. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001; 17:520–527. [DOI] [PubMed] [Google Scholar]
- 11. Han B.Y., Foo C.S., Wu S., Cyster J.G.. The C2H2-ZF transcription factor Zfp335 recognizes two consensus motifs using separate zinc finger arrays. Genes Dev. 2016; 30:1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brayer K.J., Segal D.J.. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 2008; 50:111–131. [DOI] [PubMed] [Google Scholar]
- 13. Brown R.S. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 2005; 15:94–98. [DOI] [PubMed] [Google Scholar]
- 14. Geyer P.K., Corces V.G.. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Gene Dev. 1992; 6:1865–1873. [DOI] [PubMed] [Google Scholar]
- 15. Roseman R.R., Johnson E.A., Rodesch C.K., Bjerke M., Nagoshi R.N., Geyer P.K.. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics. 1995; 141:1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roseman R.R., Pirrotta V., Geyer P.K.. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993; 12:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soshnev A.A., Baxley R.M., Manak J.R., Tan K., Geyer P.K.. The Drosophila Suppressor of Hairy-wing insulator protein has an essential role as a transcriptional repressor in the ovary. Development. 2013; 140:3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soshnev A.A., Li X., Wehling M.D., Geyer P.K.. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 2008; 4:e1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parnell T.J., Viering M.M., Skjesol A., Helou C., Kuhn E.J., Geyer P.K.. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Golovnin A., Birukova I., Romanova O., Silicheva M., Parshikov A., Savitskaya E., Pirrotta V., Georgiev P.. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development. 2003; 130:3249–3258. [DOI] [PubMed] [Google Scholar]
- 21. Filion G.J., van Bemmel J.G., Braunschweig U., Talhout W., Kind J., Ward L.D., Brugman W., de Castro I.J., Kerkhoven R.M., Bussemaker H.J. et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010; 143:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L., Lin M.F. et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rhee H.S., Pugh B.F.. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011; 147:1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakahashi H., Kwon K.R., Resch W., Vian L., Dose M., Stavreva D., Hakim O., Pruett N., Nelson S., Yamane A. et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013; 3:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harrison D.A., Gdula D.A., Coyne R.S., Corces V.G.. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Gene Dev. 1993; 7:1966–1978. [DOI] [PubMed] [Google Scholar]
- 26. Parkhurst S.M., Harrison D.A., Remington M.P., Spana C., Kelley R.L., Coyne R.S., Corces V.G.. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Gene Dev. 1988; 2:1205–1215. [DOI] [PubMed] [Google Scholar]
- 27. Harrison D.A., Mortin M.A., Corces V.G.. The RNA polymerase II 15-kilodalton subunit is essential for viability in Drosophila melanogaster. Mol. Cell. Biol. 1992; 12:928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuhn-Parnell E.J., Helou C., Marion D.J., Gilmore B.L., Parnell T.J., Wold M.S., Geyer P.K.. Investigation of the properties of non-gypsy suppressor of hairy-wing-binding sites. Genetics. 2008; 179:1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Markstein M., Pitsouli C., Villalta C., Celniker S.E., Perrimon N.. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008; 40:476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baxley R.M., Soshnev A.A., Koryakov D.E., Zhimulev I.F., Geyer P.K.. The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis. Dev. Biol. 2011; 356:398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soshnev A.A., He B., Baxley R.M., Jiang N., Hart C.M., Tan K., Geyer P.K.. Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 2012; 40:5413–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 34. Bushey A.M., Ramos E., Corces V.G.. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009; 23:1338–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Negre N., Brown C.D., Shah P.K., Kheradpour P., Morrison C.A., Henikoff J.G., Feng X., Ahmad K., Russell S., White R.A. et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010; 6:e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwartz Y.B., Linder-Basso D., Kharchenko P.V., Tolstorukov M.Y., Kim M., Li H.B., Gorchakov A.A., Minoda A., Shanower G., Alekseyenko A.A. et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012; 22:2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matzat L.H., Dale R.K., Lei E.P.. Messenger RNA is a functional component of a chromatin insulator complex. EMBO Rep. 2013; 14:916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Georgiev P.G., Gerasimova T.I.. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 1989; 220:121–126. [DOI] [PubMed] [Google Scholar]
- 39. Pai C.Y., Lei E.P., Ghosh D., Corces V.G.. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004; 16:737–748. [DOI] [PubMed] [Google Scholar]
- 40. Ghosh D., Gerasimova T.I., Corces V.G.. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001; 20:2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dixon J.R., Gorkin D.U., Ren B.. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016; 62:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ali T., Renkawitz R., Bartkuhn M.. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 2016; 37:17–26. [DOI] [PubMed] [Google Scholar]
- 44. Van Bortle K., Nichols M.H., Li L., Ong C.T., Takenaka N., Qin Z.S., Corces V.G.. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014; 15:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hou C., Li L., Qin Z.S., Corces V.G.. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell. 2012; 48:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vorobyeva N.E., Mazina M.U., Golovnin A.K., Kopytova D.V., Gurskiy D.Y., Nabirochkina E.N., Georgieva S.G., Georgiev P.G., Krasnov A.N.. Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of origin recognition complex-binding sites in the Drosophila genome. Nucleic Acids Res. 2013; 41:5717–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rivera-Mulia J.C., Gilbert D.M.. Replication timing and transcriptional control: beyond cause and effect-part III. Curr. Opin. Cell Biol. 2016; 40:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cuartero S., Fresan U., Reina O., Planet E., Espinas M.L.. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 2014; 33:637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wai D.C., Shihab M., Low J.K., Mackay J.P.. The zinc fingers of YY1 bind single-stranded RNA with low sequence specificity. Nucleic Acids Res. 2016; 44:9153–9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jeon Y., Lee J.T.. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011; 146:119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sigova A.A., Abraham B.J., Ji X., Molinie B., Hannett N.M., Guo Y.E., Jangi M., Giallourakis C.C., Sharp P.A., Young R.A.. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015; 350:978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kwon S.Y., Grisan V., Jang B., Herbert J., Badenhorst P.. Genome-Wide mapping targets of the metazoan chromatin remodeling factor NURF reveals nucleosome remodeling at enhancers, core promoters and gene insulators. PLoS Genet. 2016; 12:e1005969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bohla D., Herold M., Panzer I., Buxa M.K., Ali T., Demmers J., Kruger M., Scharfe M., Jarek M., Bartkuhn M. et al. A functional insulator screen identifies NURF and dREAM Components to be required for Enhancer-Blocking. PLoS One. 2014; 9:e107765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li M., Belozerov V.E., Cai H.N.. Modulation of chromatin boundary activities by nucleosome-remodeling activities in Drosophila melanogaster. Mol. Cell. Biol. 2010; 30:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohlsson R., Lobanenkov V., Klenova E.. Does CTCF mediate between nuclear organization and gene expression?. Bioessays. 2010; 32:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rudan M.V., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., Hadjur S.. Comparative Hi-C reveals that CTCF Underlies evolution of chromosomal Domain Architecture. Cell Rep. 2015; 10:1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ghirlando R., Felsenfeld G.. CTCF: making the right connections. Gene Dev. 2016; 30:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Meijsing S.H., Pufall M.A., So A.Y., Bates D.L., Chen L., Yamamoto K.R.. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009; 324:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Watson M.L., Baehr L.M., Reichardt H.M., Tuckermann J.P., Bodine S.C., Furlow J.D.. A cell-autonomous role for the glucocorticoid receptor in skeletal muscle atrophy induced by systemic glucocorticoid exposure. Am. J. Physiol. Endocrinol. Metab. 2012; 302:E1210–E1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Donohoe M.E., Zhang L.F., Xu N., Shi Y., Lee J.T.. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007; 25:43–56. [DOI] [PubMed] [Google Scholar]
- 62. Kurshakova M., Maksimenko O., Golovnin A., Pulina M., Georgieva S., Georgiev P., Krasnov A.. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol. Cell. 2007; 27:332–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.