Abstract
Prokaryotes memorize invader information by incorporating alien DNA as spacers into CRISPR arrays. Although the spacer size has been suggested to be predefined by the architecture of the acquisition complex, there is usually an unexpected heterogeneity. Here, we explored the causes of this heterogeneity in Haloarcula hispanica I-B CRISPR. High-throughput sequencing following adaptation assays demonstrated significant size variation among 37 957 new spacers, which appeared to be sequence-dependent. Consistently, the third nucleotide at the spacer 3΄-end (PAM-distal end) showed an evident bias for cytosine and mutating this cytosine in the protospacer sequence could change the final spacer size. In addition, slippage of the 5΄-end (PAM-end), which contributed to most of the observed PAM (protospacer adjacent motif) inaccuracy, also tended to change the spacer size. We propose that both ends of the PAM-protospacer sequence should exhibit nucleotide selectivity (with different stringencies), which fine-tunes the structural ruler, to a certain extent, to specify the spacer size.
INTRODUCTION
The clustered regularly interspaced short palindromic repeats (CRISPRs) and the CRISPR-associated (Cas) proteins together provide adaptive immunity to invading elements (e.g. viruses and plasmids) in bacteria and archaea (1–7). CRISPR-Cas systems are highly diversified and have been classified into 2 classes, 6 types and 17 subtypes (8,9). In general, all these systems function through the following three stages: the adaptation stage, which incorporates the invader sequence (i.e. spacers) into the CRISPR array (10); the CRISPR RNA (crRNA) biogenesis stage, which involves CRISPR transcription and Cas-dependent RNA processing, and thereby transmits the spacer information to the mature crRNA (11–14); and the interference stage, which utilizes the crRNA to guide Cas protein(s) for specific cleavage of the invader DNA/RNA (15–17). Although the crRNA biogenesis and interference processes have been extensively investigated, the adaptation mechanisms remain poorly characterized.
CRISPR adaptation was initially reported in the Streptococcus thermophilus type II system (1), but recent studies have mainly focused on type I systems (10). There are two related but different adaptation pathways in type I systems, i.e. naïve adaptation and priming adaptation (18). The former pathway occurs infrequently and involves only the two core Cas proteins: Cas1 and Cas2. The priming pathway is much more efficient and involves additional Cas proteins (Cas3 and the Cascade effector) and a pre-existing spacer that fully or partially matches the invader DNA. On the one hand, the two pathways exhibit several differences during protospacer (from which spacer is derived) selection. For example, naïve adaptation prefers foreign DNA due to its replication dependence (19), while priming adaptation acquires spacers much more discriminatively from the priming crRNA-targeted foreign DNA (20–22). In addition, during priming adaptation, protospacer selection usually shows a more stringent PAM (protospacer adjacent motif) conservation (23) and a strand bias (20,22,24–26). On the other hand, the two pathways are related in that new spacers acquired through naïve adaptation can fuel the priming pathway, which may be more specifically termed ‘interference-driven acquisition’ (26). Notably, although naïve adaptation is theoretically a prerequisite for the priming pathway, recent studies have demonstrated that priming may be the predominant adaptation mode in various type I systems (20,22,27).
Scientists had been puzzled by how the spacer size is determined during acquisition until the recent reports of the substrate-binding structure of the Escherichia coli Cas1–Cas2 complex (28,29). It seems that the structural constraints of this complex provide a molecular ruler that predetermines the spacer length to be 32 bp. Consistently, the vast majority (∼95%) of new spacers are of this size during adaptation assays in E. coli (23). However, spacer size varies greatly in other systems, although Cas1 and Cas2 are the most conserved Cas proteins. For example, the spacer size ranges mainly from 34 to 37 bp in the I-B, I-C, I-D and I-U subtypes and from 35 to 45 bp in the I-A subtype (30). In addition, in a large number of organisms, the CRISPR arrays of these subtypes frequently harbor spacers that are 5–6 bp or even 15 bp longer than the average size (30). In fact, the spacer size can even vary greatly in a single organism. For example, we previously described the significant spacer size variation (33–54 bp) in the six I-B CRISPRs in Haloferax mediterranei (14). An early bioinformatics study by Grissa et al. also highlighted the spacer size variation among different types (subtypes) or organisms (31), and exemplified this variation in a single organism: spacers are 38–53 bp in Pyrobaculum aerophilum and 51–72 bp in Methanopyrus kandleri. Therefore, in addition to the ruler mechanism, there seem to be additional factors or mechanisms that are involved in controlling the spacer size in most, if not all, CRISPR systems.
Here, we investigated the spacer size heterogeneity during priming adaptation mediated by the Haloarcula hispanica I-B CRISPR-Cas. Interestingly, the sizes of 37 957 new spacers generally fit a normal distribution, which is consistent with our analysis of 604 I-B spacers that pre-exist in haloarchaeal genomes. Significantly, the spacer size appeared to be group-specific, i.e. dependent on the protospacer sequence. We revealed that the third nucleotide at the 3΄-end of new or pre-existing spacers tended to be a cytosine, and manipulating the position of this cytosine (relative to the PAM) could alter the final spacer size. Therefore, we propose that, in addition to the PAM, the sequence of the protospacer itself is also sensed by the I-B acquisition machinery, which may be involved in size control during protospacer processing. Taking advantage of our ‘single-guide priming adaptation’ system (see below), we also utilized the high-throughput sequencing data to evaluate the protospacer selectivity of the I-B acquisition machinery, including self/non-target DNA avoidance, PAM conservation and locational preference (on the viral target DNA).
MATERIALS AND METHODS
Strains and culturing conditions
The H. hispanica strains that were utilized in our study are listed in Supplementary Table S1. DF60, a ΔpyrF strain of H. hispanica ATCC 33960 (32) was used as the parental strain. In its derivative DF60P, the wild-type CRISPR was replaced by the priming-CRISPR which constantly produces the priming guide s13-crRNA (33). The pCR-A or pSg-A plasmid (see below) was transformed into DF60P to generate the DF60PA or SgPA strain. DF60 and DF60P were cultured in an AS-168 medium (per liter, 200 g of NaCl, 20 g of MgSO4·7H2O, 2 g of KCl, 3 g of trisodium citrate, 1 g of sodium glutamate, 50 mg of FeSO4·7H2O, 0.36 mg of MnCl2·4H2O, 5 g of Bacto casamino acids and 5 g of yeast extract, pH 7.2) with uracil at a final concentration of 50 mg/l. For DF60PA and SgPA, the yeast extract was subtracted from the medium to provide selection pressure.
Escherichia coli JM109 was used for plasmid engineering, and cultured in the lysogeny broth. Ampicillin was used at a final concentration of 100 mg/l.
Plasmid construction and transformation
Information for the plasmids is provided in Supplementary Table S1. The pCR-A plasmid carries ∼460-bp chromosomal sequence to facilitate its integration into the chromosome and an adaptation-CRISPR, which consists of a CRISPR leader and a single repeat (33). The last 10 bp of its repeat sequence was mutated to generate pSg-A. The pVS plasmid carries an HHPV-2 fragment containing the target sequence of s13-crRNA, and point mutations were introduced into its derivatives, p7908mut and p7981mut.
For plasmid engineering, high-fidelity KOD-Plus DNA polymerase (TOYOBO, Osaka, Japan) was used to amplify the DNA inserts, which were validated by DNA sequencing. The primers are listed in Supplementary Table S2. The plasmids and DNA inserts were digested and ligated with New England Biolabs (Beverly, MA, USA) restriction enzymes and T4 DNA ligase, respectively. To introduce point mutations into pVS, bridge polymerase chain reaction (PCR) was conducted. For example, primers 7908M-R and 7908M-F with designed mutations were separately used to amplify the 5΄-half and the 3΄-half of the viral sequence, and then the two halves were connected by bridge PCR. Plasmids were transformed into DF60 or DF60P cells according to the online protocol (http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2009_v7.2mds.pdf).
Spacer acquisition assays
The spacer acquisition assay was conducted with the SgPA strain as previously described (33). Briefly, the culture of this strain was serially sub-inoculated at a ratio of 1:15 (microbial culture: fresh medium) three times. During each inoculation, fresh HHPV-2 dilution was added at a calculated MOI (multiplicity of infection) of ∼10, and the virus-archaeon mixture was cultured for 7 days prior to the next inoculation. The final culture was centrifuged to collect the cells, which were then lysed by distilled water. The lysate was subjected to PCR analysis using primers surrounding the a-CRISPR structure, i.e. Exp-Fa and Exp-Ra in Supplementary Table S2. The PCR products were separated on a 1.2% agarose gel.
Spacer acquisition from the pVS plasmid or its derivates was similarly investigated. These plasmids were transformed into DF60 other than SgPA for pyrF-based selection. Single transformant colonies were picked and cultured in an AS-168 medium with uracil added for 5 days. The cells were collected by centrifugation and lysed by distilled water. The lysate was subjected to PCR analyses using a forward primer specific for the new spacer (Exp-7908-F and Exp-7981-F) and a backward primer specific for the original spacer2 (s2-primer).
High-throughput sequencing and analysis
Six independent SgPA colonies were separately subjected to serial sub-inoculation and virus infection as described above, and their final cultures were pooled prior to PCR analysis. From the agarose gel, PCR bands corresponding to the ‘expanded’ a-CRISPR were purified using the AxyPrep™ DNA Gel Extraction Kit (Corning, NY, USA). The purified DNA samples were subjected to HiSeq2500 sequencing (Biomarker, Beijing, China). After assembly of the pair-end data and filtration of the low-quality data, reads containing multiple (two or three) repeats were selected. For the reads containing two repeats, the intervening sequence was considered as the initially acquired spacer (s-1). For the reads with three repeats, the leader-distal new spacer was initially acquired, while the leader-proximal spacer was the secondly acquired (s-2). Using the BLASTN program against the HHPV-2 or H. hispanica genome, the protospacer sequence was preliminarily identified for each spacer and then manually calibrated in the case of mismatches. The 3 bp 5΄-upstream of each protospacer was considered its PAM. Perl scripts were run to analyze the protospacer sequences and their distribution on the HHPV-2 genome. Nucleotide conservation was analyzed using the WebLogo web server (http://weblogo.berkeley.edu/logo.cgi).
RESULTS
New spacers were almost exclusively derived from the target DNA of the priming guide
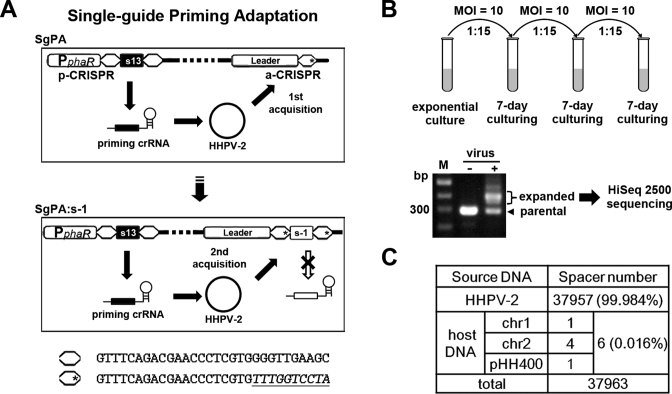
In H. hispanica, we previously established a priming adaptation system with two separate CRISPRs, i.e. the priming-CRISPR (p-CRISPR), which provides a priming guide (s13-crRNA) that targets the HHPV-2 DNA and the adaptation-CRISPR (a-CRISPR), which accepts new spacers from the virus (33). Here, we further modified the a-CRISPR repeat by mutating its leader-distal 10 bp (Figure 1A), which was previously shown not to be required for spacer incorporation (33). However, this 10-bp sequence encodes the Cas6-processing site on the repeat RNA and the 8-nt 5΄-handle on the mature crRNA (14), both of which are essential for CRISPR function (34). Therefore, it could be expected that, although new spacers could incorporate into the mutated a-CRISPR, they would be unable to give rise to additional crRNA-guides (as indicated in Figure 1A). Consistently, we failed to detect the interference effects of the new spacers in this strain (Supplementary Figure S1). Therefore, in this modified system, there would be only one single type of crRNA-guide (s13-crRNA from the p-CRISPR) and this mutant was named ‘Single-guide Priming Adaptation’ (SgPA). We believe that SgPA should be an ideal model to specifically investigate the protospacer selection of the priming adaptation machinery because, no matter the new spacers are invader-derived or self-targeting and no matter they are interference-proficient or not (regardless of the repeat mutation), they would not provide selective advantages or a penalty to the host cell. In addition, the secondary priming or interference-driven acquisition (26) effects of the new spacers would also be avoided in this system.
Figure 1.
The SgPA (single-guide priming adaptation) system acquired new spacers almost exclusively from the viral DNA that was targeted by the priming guide. (A) Diagram showing the design of the SgPA system. As previously described (33), the priming-CRISPR (p-CRISPR) produces the only crRNA-guide (s13-crRNA, which partially matches the HHPV-2 DNA) and new spacers from the viral DNA are incorporated into the adaptation-CRISPR (a-CRISPR). The last 10 bp of the a-CRISPR repeat were mutated (indicated with an asterisk) to prevent the newly acquired spacers (e.g. s-1) from encoding additional crRNA-guides. The repeat sequence with or without the mutation (underlined) is shown at the bottom. (B) Spacer acquisition assay using the SgPA strain. After serial sub-inoculation and repeated virus infection, the genomic DNA was subjected to PCR reaction using primers surrounding a-CRISPR (Exp-Fa and Exp-Ra in Supplementary Table S2). The ‘expanded’ PCR bands were gel-extracted and subjected to HiSeq 2500 sequencing. MOI, multiplicity of infection. Lane M, dsDNA size marker. (C) Nearly all (99.984%) of the new spacers were derived from the viral DNA, while only six spacers were derived from the host DNA sequences on the two chromosomes (chr1 and chr2) or the plasmid (pHH400).
An exponential SgPA culture was serially sub-inoculated three times, and during each inoculation, fresh HHPV-2 dilution was added with an MOI of ∼10 (Figure 1B). As expected, expansion of a-CRISPR was evidently detected by PCR with its surrounding primers. As indicated in Figure 1B, the ‘expanded’ DNA bands were purified and subjected to illumina sequencing. After data processing, a total of 37 963 spacer sequences were extracted from the high-throughput sequencing reads. Significantly, 99.984% of the spacers (37 957 of 37 963) were derived from the HHPV-2 DNA that was targeted by the priming guide and only six were mapped on the host DNA that was not crRNA-targeted (Figure 1C). Interestingly, only one spacer was derived from the 2.9-Mb chromosome (chr1), and one spacer was derived from the 0.4-Mb mega-plasmid (pHH400), but four spacers matched the 0.5-Mb mini-chromosome (chr2). In addition, three of the six self-targeting spacers contained mismatches to the protospacer that was simultaneously accompanied by an incorrect PAM (Supplementary Table S3). As stated above, there were no selection effects against a self-targeting spacer regardless of whether it was functional or not; hence, it could be inferred that, without a corresponding priming guide, spacer acquisition from the host DNA is not only very rare (at a rate of ∼0.016%) but is also error-prone.
PAM specificity was highly stringent with a low error rate
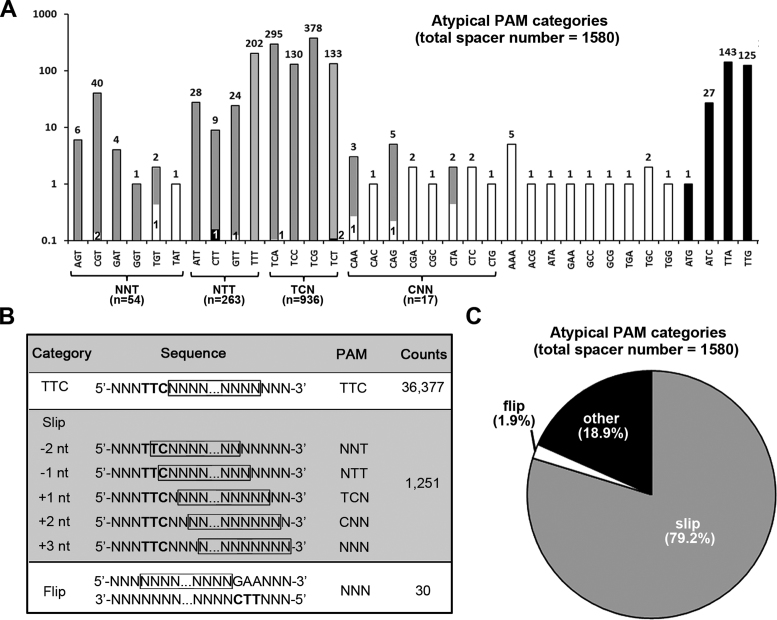
In contrast to the PAM inaccuracy that was frequently observed for the self-targeting spacers, most (95.84%) of the virus-targeting spacers were derived from sequences flanked by the canonical PAM 5΄-TTC-3΄, and only 4.16% (1580) were derived from protospacers that were preceded by a non-TTC trinucleotide. However, among the 63 ( = 43-1) possible non-TTC trinucleotides, only 35 were observed as a PAM and at a very different frequency (Figure 2A), which suggests the presence of underlying error mechanisms. Therefore, we examined the contexts of the 1580 protospacers.
Figure 2.
Atypical PAMs caused by different errors during spacer acquisition. (A) A histogram showing the observed frequency of each of the 35 non-canonical PAMs. Note that the vertical coordinates (frequency) are in a logarithmic scale, but the column is divided proportionally to the frequency. Each column or its divisions representing atypical PAMs probably caused by ‘slipping’ errors are shown in gray, those representing PAMs caused by ‘flipping’ errors are in white, and the others are in black. The frequency of the protospacers with an atypical PAM is labeled above the columns. (B) The typical PAM sequence 5΄-TTC-3΄ (in bold) occurred at a ‘non-canonical’ position for the ‘slip’ and ‘flip’ category spacers. The protospacer sequences are shown in frame. For the ‘slip’ category, 5΄-TTC-3΄ occurred at the protospacer 5΄-end with a − or + slippage, which indicates an upstream (5΄-direction) or downstream (3΄-direction) slipping error of the PAM-end cutting during protospacer selection. For the ‘flip’ category, 5΄-TTC-3΄ occurred at the 3΄-end, but on the complementary strand of the protospacer. (C) A pie chart summarizing the ratio of spacers with an atypical PAM that may have been caused by slipping (gray), flipping (white) or other (black) errors.
On the one hand, a TTC trinucleotide was indeed observed around the 5΄-end of 1251 of these protospacers, but at a ‘non-canonical’ position: the TTC trinucleotide was either straddling the 5΄-end or 1–3 nt away from the protospacer (Figure 2B). We propose that the spacer acquisition machinery may have slipped upstream or downstream from these TTC locations, leading to the observation of an atypical PAM. As illustrated in Figure 2B, a -2 or -1 nt slippage on the TTC sequence would cause an NNT or NTT PAM, while a +1, +2 or +3 nt slippage would lead to a TCN, CNN or NNN PAM. In fact, for 261 of the 263 protospacers with an NTT PAM (i.e. ATT, TTT, CTT or GTT), a C or TC always served as the first nucleotide(s) (Supplementary Table S4), suggesting a -1 or -2 nt slippage event. Instead, 933 of the 936 protospacers with a TCN PAM (i.e. TCA, TCT, TCC or TCG) were preceded by a conserved T, suggesting a +1 nt slippage event. In addition, most protospacers with an NNT PAM (e.g. AGT, CGT, GAT, GGT and TGT) began with a conserved TC, while those with a CNN PAM (e.g. CAA, CAG and CTA) were usually preceded by a TT. In summary, these conserved contexts substantially support that the 1251 protospacers with a non-canonical PAM should have arisen via the ‘slipping’ mechanism. On the other hand, for 30 protospacers with a very rare PAM (e.g. AAA, ACG and ATA), we identified a conserved 5΄-GAA-3΄ immediately downstream of their 3΄-end (see the ‘flip’ category in Figure 2B). It appears that its complement (5΄-TTC-3΄ on the other strand) may have been sensed by the acquisition machinery, but the following sequence was flipped and integrated into the CRISPR cassette in the opposite direction, like the frequently observed ‘flippage’ events in E. coli (35). Finally, there remain 299 protospacers with a non-canonical PAM that could not be explained by the ‘slipping’ or ‘flipping’ errors. Notably, this collection mainly includes 143, 125 and 27 protospacers preceded by TTA, TTG and ATC, respectively (Figure 2A). These trinucleotides highly resemble the canonical PAM TTC and, thus, have probably been misrecognized to initiate spacer acquisition.
To summarize, the ‘slipping’ and ‘flipping’ mechanisms could explain 79.2% and 1.9% of the PAM inaccuracy, respectively, while the remaining 18.9% may be mainly caused by PAM misrecognition (Figure 2C). Therefore, in this view, although there was no selection for a functional PAM in this assay, the PAM specificity of the acquisition machinery on the target DNA appeared to be highly stringent with an error rate of no more than 0.8% (299/37957).
Spacer size is heterogeneous and its distribution is group-specific
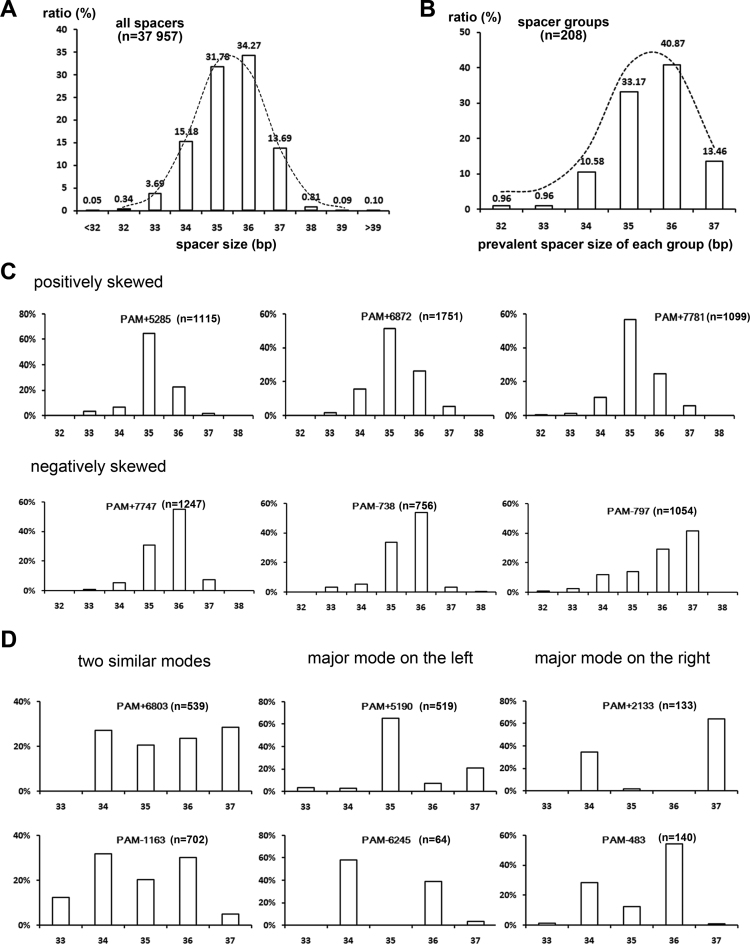
Spacer size is usually heterogeneous (30,31). The 13 spacers in the wild-type H. hispanica I-B CRISPR vary in size from 33 to 37 bp. From the CRISPRdb database (31), we collected 604 I-B spacers in haloarchaea and observed a wide size distribution mainly between 32 and 39 bp, with only 33.1% being of the most prevalent size 36 bp (Supplementary Figure S2). Consistently, in our assay, the size of the newly acquired spacers also varied mainly from 32 to 39 bp, with the most prevalent 36 bp accounting for only 34.3% (Figure 3A). Interestingly, for both the experimentally acquired new spacers and the in silico analyzed ‘old’ spacers, the spacer size generally fit a normal distribution (Figure 3A and Supplementary Figure S2). Hence, we inferred that the size heterogeneity might have derived from some random factors that may influence the size control during protospacer selection, such as cutting errors (e.g. slipping at the PAM end) or the diversified protospacer sequence (see below).
Figure 3.
The size heterogeneity of all spacers or a specific spacer group. (A) A histogram showing the size distribution of all 37 957 virus-targeting spacers. (B) The most prevalent spacer size for each of 208 groups varied from 32 to 37 bp. The distributions in both panels A and B generally fit a (skewed) normal curve (the dotted lines). (C and D) The spacer size variation for different spacer groups. The horizontal and vertical coordinates represent the spacer size (bp) and the spacer ratio of this size, respectively. Examples in panel C fit a unimodal distribution (one mode), while those in panel D fit a bimodal distribution (two modes). For unimodal distribution, a positive or negative skew means more samples are larger or smaller than the modal value (i.e. the most prevalent size). For bimodal distribution, the two modes may be (nearly) equal or unequal. If unequal, the major mode could be on the left or on the right of this distribution, which means that the smaller or larger modal value is the most prevalent size. Note that a spacer group includes spacers with a specific TTC trinucleotide (on the ‘+’ or ‘−’ strand) as their PAM (see main text for details).
As proposed above, although ‘slipping’ or ‘flipping’ errors may lead to an incorrect PAM being observed (Figure 2), the bona fide recognized PAM should be stringently a 5΄-TTC-3΄ sequence on the virus DNA. Hence, we classified spacers with a specific 5΄-TTC-3΄ recognized as their PAM into one group. For example, the plus-strand 5΄-TTC-3΄ at positions 6448–6450 served as the PAM for the 1612 spacers in group ‘PAM+6450’, of which 1424 and 188 spacers fall into the ‘TTC’ and ‘+1 slip’ categories, respectively. We sought to determine whether the spacers in a single group would also fit the overall size distribution observed in Figure 3A. First, we calculated the prevalent spacer size in each group (note that only the ‘TTC’ category spacers were analyzed). Remarkably, the prevalent spacer size in each of 208 groups varied from 32 to 37 bp and fit a normal curve, albeit with a negative skew (Figure 3B). This variation indicates that the spacer size distribution should be group-specific; thus, we then analyzed the distribution in each spacer group (once again, only the ‘TTC’ category spacers were considered). Figure 3C illustrates that the spacer size in some groups fit a unimodal distribution (with a single mode); however, the groups shown in Figure 3D fit a bimodal distribution (with two modes). The unimodal distribution could be positively or negatively skewed, indicating that more spacers in this group were larger or smaller than the modal value (the most prevalent size). Instead, the bimodal distribution could have two (nearly) equal modes or a major mode on the left (the smaller modal value is preferred) or on the right (the larger modal value is preferred). These data elaborated that spacer size distribution varied greatly among different groups.
Slippage of the PAM-end cutting contributed to spacer size heterogeneity
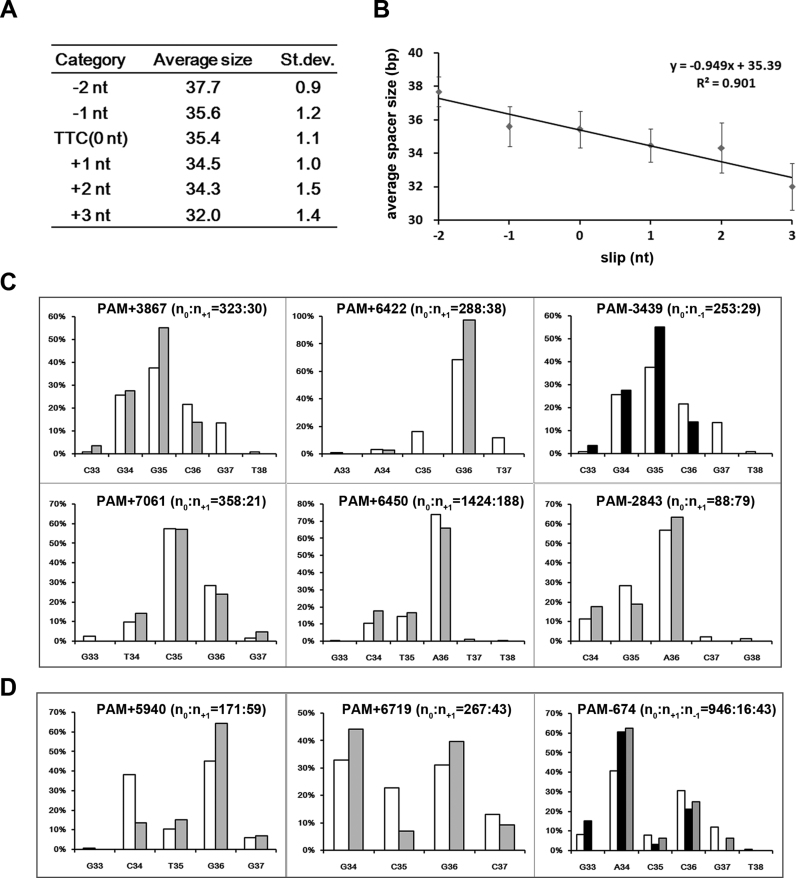
To test the hypothesis that random cutting errors, such as the slipping events at the PAM-end, may disturb spacer size control, we calculated the average spacer size for each ‘slip’ category (Figure 4A). The average spacer size in the ‘TTC’ category was 35.4 bp; however, the ‘-2 nt slip’ and ‘-1 nt slip’ categories have a larger average (37.7 and 35.6 bp, respectively), while the ‘+1 nt slip’, ‘+2 nt slip’ and ‘+3 nt slip’ categories have a smaller average (34.5, 34.3 and 32.0 bp, respectively). When plotted on a scatter diagram (Figure 4B), the average spacer size showed a considerable linear correlation with the ‘slippage value’ (R2 = 0.901). This correlation indicates that ‘slipping’ errors tend to alter the final spacer size.
Figure 4.
Slipping events tended to alter the size but not the 3΄-end of a spacer. (A) The average size (bp) was calculated for spacers from the ‘TTC’ category and those from each ‘slip’ category. The standard deviation (St. dev.) is also shown. (B) The linear correlation between the average spacer size and the slippage value (0 nt for the ‘TTC’ category). (C and D) The 3΄-terminal nucleotide distribution for the ‘TTC’ category spacers (columns in white; n = n0), the ‘+1 nt slip’ category spacers (columns in gray; n = n+1), or the ‘−1 nt slip’ category spacers (columns in black; n = n-1) from a specific group. Nine representative groups in which slipping errors occurred at their PAM-end most frequently are shown (see Supplementary Table S5). Regardless of whether the distribution fit a unimodal (panel C) or a bimodal (panel D) pattern, the distribution appeared similar between different categories of the same group. The horizontal and vertical coordinates, respectively, represent the PAM-distal nucleotides (33–38 bp downstream of the PAM) and the ratio of the spacers that terminated at each position.
Subsequently, we investigated nine spacer groups in which slippage (usually +1 nt or −1 nt) frequently occurred (Supplementary Table S5). Compared to the ‘TTC’ category, their ‘−1 nt slip’ or ‘+1 nt slip’ category spacers usually showed a prevalent size increased or decreased by 1 bp. The most significant example may be PAM-674, where 34-bp spacers prevailed in the ‘TTC’ category, while 35 and 33-bp spacers, respectively, prevailed in the ‘−1 nt slip’ and ‘+1 nt slip’ categories. Consistently, when calculating the average spacer size, we noticed that every 1 nt slippage altered this value by 0.919 ± 0.336 bp (Supplementary Table S5). Apparently, these slipping events contributed to the observed size heterogeneity. This contribution reflects that the PAM-distal cutting on a protospacer seemed not to be influenced by the PAM-end errors. Hence, for the nine groups, we analyzed the frequency of each PAM-distal nucleotide serving as the 3΄-terminus. Similar to the spacer size, distribution of the terminal nucleotide within a spacer group also fit a unimodal (Figure 4C) or bimodal (Figure 4D) distribution. However, no matter whether the distribution was unimodal or bimodal, it was highly conserved between the ‘TTC’ and the ‘slip’ categories. Again, taking PAM-674 as an example, the 3΄-terminal nucleotide of its ‘TTC’, ‘+1 nt slip’ and ‘−1 nt slip’ category spacers showed a fairly similar bimodal distribution: the 34th nucleotide (A34) downstream of the 5΄-TTC-3΄ PAM was primarily preferred as the terminal one, and the 36th nucleotide (C36) was the secondarily preferred (Figure 4D). Therefore, our data imply an intrinsic nucleotide preference at the 3΄-end.
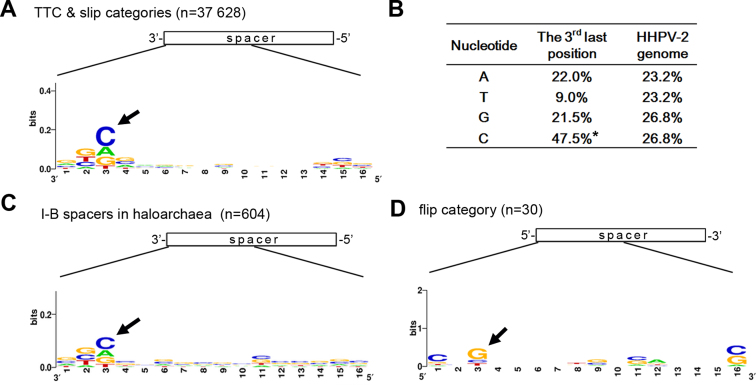
A cytosine was preferred as the third 3΄-end nucleotide
To investigate this potential preference, we collected the 16 nt at the 3΄-end of each spacer from the ‘TTC’ and the ‘slip’ categories (with a total spacer number of 37 628), and generated a sequence logo (Figure 5A). Nucleotide preference was detected for the third 3΄-end base position, which tended to be occupied by a cytosine (C). In fact, 47.5% of the 37 628 spacers have a cytosine at this position, which was significantly higher than the 26.8% cytosine content of the viral DNA (Figure 5B). Similarly, we also constructed a sequence logo for the 604 haloarchaeal spacers that were collected from the CRISPRdb database (31). Significantly, the cytosine preference was observed again at the same position (Figure 5C). In addition, these two sequence logos both showed that thymine (T) seemed to be the most disfavored (like shown in Figure 5B). Consistently, for the example groups in Figures 3 and 4, the third 3΄-end nucleotide of their prevalent spacer sequences was predominantly cytosine and never thymine (Supplementary Table S5 and S6). It is worth mentioning that, for the ‘TTC’ and ‘slip’ category spacers, we have also analyzed the sequence composition downstream of each protospacer, but failed to detect evident base bias (data not shown). Interestingly, by analyzing the 5΄-end nucleotides of the 30 ‘flip’ category spacers (Figure 5D), we found that the third nucleotide preferred to be a guanine (G), which is the complement of C. This coincidence not only supported our hypothesis that these protospacers have been flipped and incorporated into CRISPR in the opposite direction, but also reinforced the nucleotide preference at the protospacer 3΄-end.
Figure 5.
Nucleotide preference at the spacer 3΄-end. (A) Nucleotide conservation at the 3΄-end of the 37 628 spacers from the ‘TTC’ and the ‘slip’ categories. (B) The frequency of A/T/G/C at the third last position of the ‘TTC’ and ‘slip’ category spacers, which significantly differs from the nucleotide composition of the HHPV-2 genome. Note that the cytosine bias (indicated by an asterisk) at the third last position is significant (P < 0.001, χ2 test). (C) Nucleotide preference at the 3΄-end of 602 haloarchaeal spacers that were collected from the CRISPRdb database (Supplementary Figure S2). (D) Nucleotide conservation at the 5΄-end of the 30 ‘flip’ category spacers. Numbers along the X axis indicate positions with respect to the spacer 3΄-end (in panels A and C) or 5΄-end (in panel D). Black arrows indicate the preferred nucleotides.
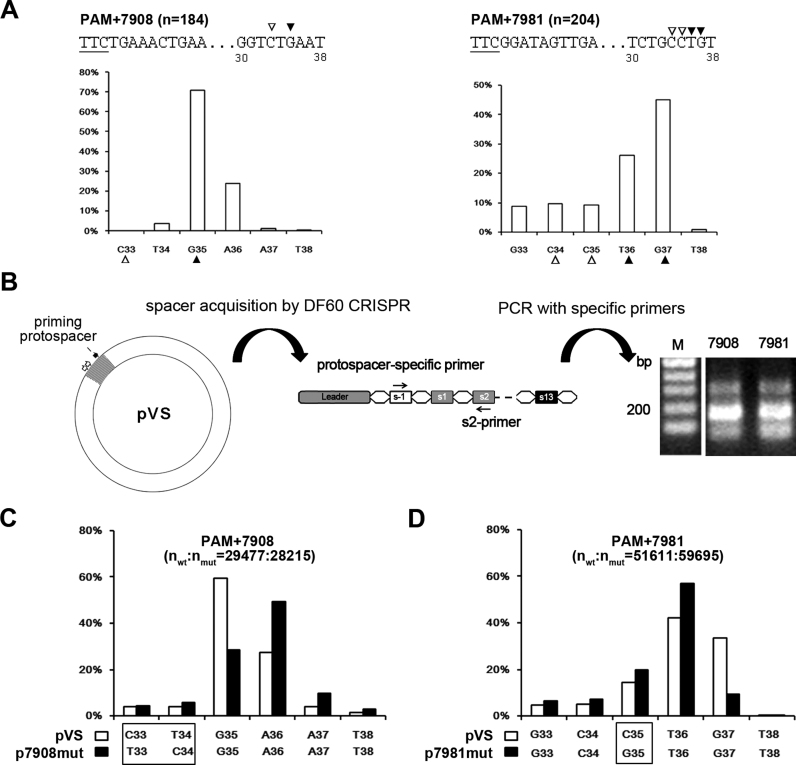
In Figure 6A, we show another two examples. The spacers from the PAM+7908 group predominantly terminated at the 35th nucleotide (G35) following the PAM, with the 33rd nucleotide (C33) being a cytosine. By contrast, for PAM+7981, the 37th nucleotide G (G37) and the 36th nucleotide T (T36) were both preferred as the terminal nucleotide because the 35th and the 34th nucleotides (C35 and C34) are both cytosine. Previously, we described a plasmid-based adaptation assay: when pVS (carrying an HHPV-2 DNA fragment targeted by s13-crRNA) was introduced into the DF60 strain (possessing the wild-type CRISPR), efficient adaptation to this plasmid was primed (20). We wondered whether the nucleotide preference is conserved when the PAM+7908 and PAM+7981 spacers are acquired in this scenario. Therefore, we specifically detected their acquisition using specific primers and the s2-primer (Figure 6B), and subjected these PCR products to illumina sequencing. Similar to the results of the virus assay, the last nucleotide of the PAM+7908 spacers from pVS was predominantly G35 (Figure 6C), while the last nucleotide of the PAM+7981 spacers was usually T36 or G37 (Figure 6D) (note that T36 was more preferred as the last nucleotide, which is slightly different from the virus results). Significantly, when we mutated the 33-CT-34 dinucleotide within the PAM+7908 protospacer into 33-TC-34 (generating p7908mut), A36 instead of G35 was favored as the last nucleotide of new spacers (Figure 6C). Consistently for PAM+7981, when nucleotide C35 was mutated to a guanine (generating p7981mut), G37 was no longer preferred as the last nucleotide (Figure 6D). Therefore, these data substantially support the preference for a cytosine at the third last position of a new spacer, which seemed to be conserved during adaptation to viruses and plasmids.
Figure 6.
Protospacer mutations at the 3΄-end could alter the final spacer size. (A) Variation of the 3΄-end of spacers from groups PAM+7908 and PAM+7981. The horizontal and vertical coordinates, respectively, represent the PAM-distal nucleotides (33–38 bp downstream of the PAM) and the ratio of the spacers terminating at each position. The third nucleotide (indicated by an empty triangle) at the prevalent 3΄-end (indicated by a solid triangle) tended to be a cytosine. The PAM (underlined) and its following sequences are shown for each group. Position numbers shown beneath the sequence are relative to the PAM. (B) An assay to detect the acquisition of the PAM+7908 and PAM+7981 spacers from pVS which carries an HHPV-2 fragment (in gray). The viral fragment includes sequences (two empty arrows) from which PAM+7908 and PAM+7981 spacers were derived and the ‘priming protospacer’ that is targeted by s13-crRNA; thus, adaptation to this plasmid could be efficiently primed in DF60. A new-spacer-specific primer and s2-primer were together used to detect the acquisition of the PAM+7908 and PAM+7981 spacers. Lane M, dsDNA size marker. (C and D) The 3΄-end of the PAM+7908 and PAM+7981 spacers tended to change when their protospacer was mutated. The different nucleotide(s) between the wild-type (wt) pVS and its mutant (mut) p7908mut/p7981mut are framed. These plasmids were separately subjected to the adaptation assay illustrated in panel B, and the PCR products were subjected to high-throughput sequencing to characterize the spacer 3΄-end variation.
Protospacer distribution on the viral genome fit the sliding hypothesis
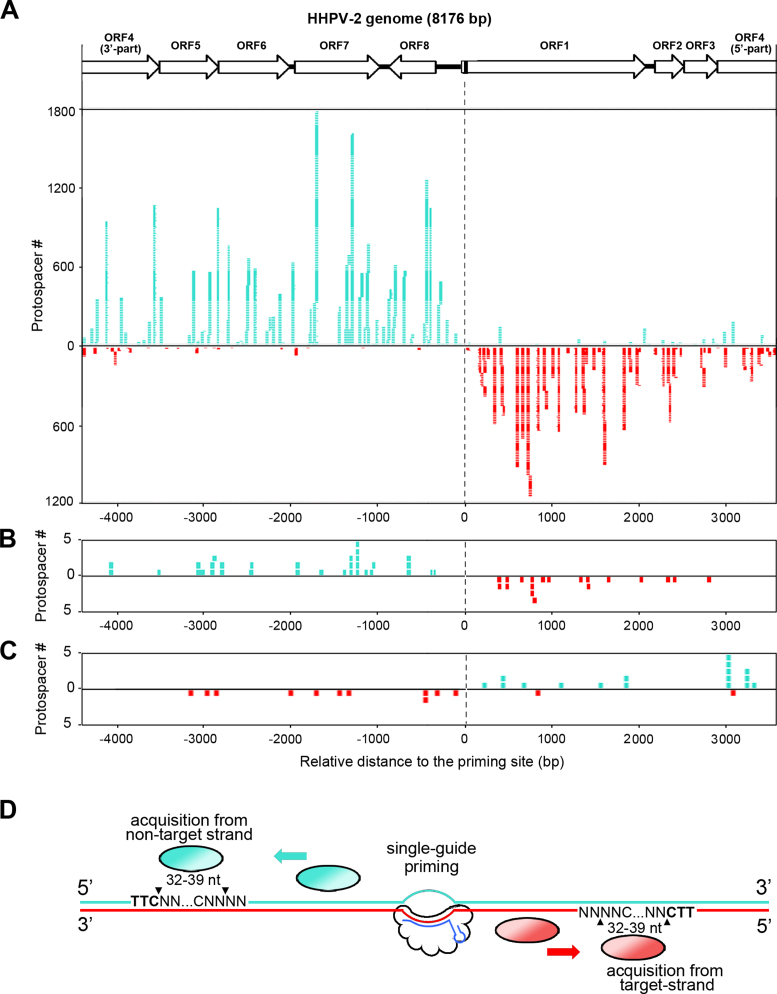
Primed spacer acquisition from both strands of the target DNA was initially reported for the I-B CRISPR in H. hispanica (20) and the I-F in Pectobacterium atrosepticum (22), and was later described for the I-F in Pseudomonas aeruginosa (36) and the I-E in E. coli (37). Here for H. hispanica, we analyzed the distribution of protospacers, from which the 37 957 virus-targeting spacers derived, on the HHPV-2 genome (Figure 7A). The vast majority of the protospacers upstream of the priming protospacer were on the non-target strand (the strand replaced by s13-crRNA during R-loop formation), while those downstream of the priming protospacer were mainly on the target strand (the strand base pairing to s13-crRNA). On both strands, highly acquired protospacers were located near the 5΄-side of the priming site, while the distal protospacers were less frequently acquired, which nearly fit the 3΄-5΄ sliding hypothesis for the acquisition machinery (Figure 7D). Yet notably, this distance seemed not to be the only deciding factor for acquisition efficiency because some neighboring protospacers were acquired very differently. In Figure 7B, we specifically analyzed the protospacer of the secondly acquired spacers (the leader-proximal spacer in a read containing two new spacers) and observed a similar distribution pattern, which suggests that the first new spacer (s-1) should not disturb the subsequent acquisition events. This is consistent with our initial design that new spacers were not allowed to encode functional crRNAs that may elicit the secondarily-primed or interference-driven acquisition (26) (Figure 1A). Interestingly, the protospacer distribution of the 30 ‘flip’ category spacers showed the opposite pattern (Figure 7C): upstream (of the priming protospacer) protospacers were usually located on the target strand, while the downstream protospacers were usually on the non-target strand. This pattern further supports our ‘flipping’ prediction for these spacers.
Figure 7.
Distribution of protospacers on the viral genome supported the sliding hypothesis. (A) The protospacer distribution of the 37 957 spacers on the HHPV-2 genome. The circular viral genome is depicted in linear with ORF4 being split into two parts. The black bar in ORF1 indicates the priming protospacer. Each of the turquoise or red stacking strips represents a protospacer mapped on the non-target or target strand. (B) The protospacer distribution of the secondly acquired spacers (the leader-proximal one of the two new spacers in a single illumina read). (C) The protospacer distribution of the 30 ‘flip’ category spacers. (D) A schematic representation of the sliding hypothesis of the Haloarcula hispanica spacer acquisition machinery. The priming guide (s13-crRNA in our assay; shown in blue) base pairs to the target strand (in red) and replaces the non-target strand (in turquoise) of its protospacer. Then, the spacer acquisition machinery is recruited to the priming site, moves along the non-target or target strand in the 3΄-5΄ direction to find a 5΄-TTC-3΄ PAM sequence (in bold) and acquires its downstream 32–39 nt with the third last one being preferentially a cytosine.
DISCUSSION
Grissa et al. have documented that the size of CRISPR spacers is highly heterogeneous not only in arrays of different types (or subtypes), but also in those of the same type (subtype) from different organisms (31). This study also described the great size variation in a single organism, for example, the CRISPRs in P. aerophilum and M. kandleri carry spacers of 38–53 bp and 51–72 bp, respectively. We also previously described the great size variation in the six I-B CRISPRs from H. mediterranei (14). A recent study specifically discussed the remarkable spacer size heterogeneity that is common in type I systems, and it was suggested that the Cascade complexes in these systems may show an altered stoichiometry to accommodate the crRNA encoded by these varying-sized spacers (30). Consistently, recent studies in type I systems demonstrated that a normal-sized spacer could be significantly extended or shortened while maintaining its interference capability (30,38,39). Therefore, these documents elaborated the remarkable heterogeneity in spacer size and the corresponding flexibility of the interfering complex. However, how this heterogeneity is generated during spacer acquisition, and what characteristics or behaviors of the acquisition machinery should be responsible for this heterogeneity, are poorly understood. Here, we investigated the spacer size heterogeneity of I-B CRISPRs. Both ‘old’ spacers (pre-existing in the sequenced haloarchaeal genomes) and new spacers (acquired during our adaptation assay) were analyzed and shown to vary greatly in size (mainly 32–39 bp). Significantly, these two spacer collections showed a fairly similar normal distribution, which centered between 35 and 36 bp (with the two values accounting for more than 60%). Apparently, this center should be determined by a common mechanism, probably the molecular ruler that is provided by the structural constraints of the acquisition complex, as proposed for the E. coli I-E system (28,29). The structure of the E. coli Cas1-Cas2 revealed a molecular ruler that predetermines the spacer length to be 32 bp, and consistently, 95% of newly acquired spacers during an in vivo assay were reported to be of this size (23). However, in the I-B system, the putative ruler should be rather relaxed because the spacer size distribution is much less concentrated and appeared to be group-specific, that is, dependent on the protospacer sequence. We further showed that this sequence dependence, at least partly, derives from the preference of the adaptation machinery to generate the 3΄-terminal (PAM-distal) cut 2 bp downstream of a cytosine. When this cytosine of two spacer groups (PAM+7908 and PAM+7981) was moved or mutated, the prevalent spacer size was substantially changed. In fact, nucleotide preference at the spacer 3΄-end has also been observed in E. coli. Yosef et al. determined an AA motif at the 3΄-end of spacers that were highly acquired during adaptation and, thus, defined it as an ‘acquisition affecting motif’ (40). Therefore, it appeared that the structural constraints of the E. coli Cas1–Cas2 complex are more rigid, thus providing a strict molecular ruler that defines a precise spacer length, and, in this case, its nucleotide selectivity on the very protospacer-3΄-end (30 bp downstream of the PAM) will determine the acquisition efficiency. By contrast, the structural constraints of the H. hispanica acquisition machinery tend to be relaxed, which allows the complex sensing a favorite nucleotide (cytosine) within a wider position range (mainly 30–35 bp downstream of the PAM). Consistently, for the highly (or infrequently) acquired spacer groups, base bias was not evidently observed at any specific positions downstream of the PAM; thus, the acquisition efficiency of a protospacer seemed to be determined by other factors (such as its relative position to the priming site). Therefore, the spacer size variation in H. hispanica seems mainly due to the relaxed structural constraints and the nucleotide preference of the acquisition machinery. In addition, we also showed that cutting errors at the protospacer 5΄-end (termed ‘slipping’) tended to alter the final spacer size but not to change the 3΄-terminus, which further supports the relaxed structural constraints and nucleotide preference.
We also stressed the sequence selectivity on the protospacer 5΄-end, i.e. the well-known PAM specificity, of the acquisition machinery. Though in appearance, 4.16% of the new spacers possessed an incorrect PAM, we propose that 3.30% and 0.08% were respectively due to the ‘slipping’ and ‘flipping’ errors during protospacer cutting or inserting (into the CRISPR), and provided convincing evidences. For all of the ‘slip’ category spacers, a 5΄-TTC-3΄ trinucleotide was observed around the protospacer 5΄-end, but not at the canonical PAM positions. While for all the ‘flip’ category spacers, the complement of the 5΄-GAA-3΄ trinucleotide was observed immediately downstream of the protospacer 3΄-end. In addition, the ‘flip’ category spacers showed a guanine preference at their third base position (corresponding to the cytosine preference at the third last base position of the normal spacers) (Figure 5), and a protospacer distribution that was opposite to that observed for the normal spacers (Figure 7C). Therefore, PAM sensing errors should occur at a very low rate (no more than 0.79%), which underlined the stringent PAM specificity of the acquisition machinery. It should be noted that there was no selection for/against a (non-)functional spacer (or PAM) in our SgPA system; thus, the above-mentioned ratios should reflect the bona fide error rates during adaptation.
In addition, the design of the SgPA system also facilitated revealing some other protospacer selection rules. On the one hand, only 0.016% of the new spacers were derived from the host DNA that was not targeted by the priming crRNA, and these spacers were usually accompanied by an incorrect PAM. This result indicated that naïve adaptation, which does not require the priming step, should be very inefficient and error-prone, if it does exist in this system. On the other hand, because new spacers did not elicit secondary priming (or interference-driven) acquisition or interference-based selection, the protospacer distribution of all the virus-targeting spacers on the HHPV-2 genome should honestly reflect the intrinsic protospacer selectivity of the primed adaptation machinery on the viral DNA. As shown in Figure 7, our data suggest the acquisition complex should start from the priming site, move along the non-target or target strand in the 3΄-5΄ direction sensing the PAM sequence and then acquire its downstream 32–39 nt as a new spacer, of which the third nucleotide at the 3΄-end tends to be a cytosine. We noticed that, in addition to the protospacer location, some other factors may influence acquisition efficiency, like local sequences and possibly the viral DNA replication and gene transcription behaviors.
In summary, we characterized the protospacer selectivity of the I-B adaptation machinery in H. hispanica, and emphasized its sequence dependence including: (i) the PAM specificity, which may be sometimes underestimated due to ‘slipping’ or ‘flipping’ errors, and (ii) the nucleotide preference at the PAM-distal end, which fine-tunes the ruler mechanism and explains the spacer size heterogeneity. It will be interesting to investigate whether the spacer size control mechanism, which relies on both the structural constraints and the nucleotide preference of the acquisition machinery, is common for the vast CRISPR systems with a varying spacer size.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31571283, 31271334]; CAS/SAFEA International Partnership Program for Creative Research Teams. Funding for open access charge: National Natural Science Foundation of China [31571283].
Conflict of interest statement. None declared.
REFERENCES
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 2. Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- 3. Wiedenheft B., Sternberg S.H., Doudna J.A.. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012; 482:331–338. [DOI] [PubMed] [Google Scholar]
- 4. Barrangou R., Marraffini L.A.. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014; 54:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang H., Peng N., Shah S.A., Huang L., She Q.. Archaeal extrachromosomal genetic elements. Microbiol. Mol. Biol. Rev. 2015; 79:117–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garrett R.A., Shah S.A., Erdmann S., Liu G., Mousaei M., León-Sobrino C., Peng W., Gudbergsdottir S., Deng L., Vestergaard G. et al. CRISPR-Cas adaptive immune systems of the sulfolobales: Unravelling their complexity and diversity. Life (Basel). 2015; 5:783–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang J., White M.F.. Hot and crispy: CRISPR-Cas systems in the hyperthermophile Sulfolobus solfataricus. Biochem. Soc. Trans. 2013; 41:1422–1426. [DOI] [PubMed] [Google Scholar]
- 8. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wright A.V., Nuñez J.K., Doudna J.A.. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016; 164:29–44. [DOI] [PubMed] [Google Scholar]
- 10. Sternberg S.H., Richter H., Charpentier E., Qimron U.. Adaptation in CRISPR-Cas systems. Mol. Cell. 2016; 61:797–808. [DOI] [PubMed] [Google Scholar]
- 11. Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J.. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008; 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carte J., Pfister N.T., Compton M.M., Terns R.M., Terns M.P.. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010; 16:2181–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A.. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010; 329:1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li M., Liu H., Han J., Liu J., Wang R., Zhao D., Zhou J., Xiang H.. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J. Bacteriol. 2013; 195:867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marraffini L.A., Sontheimer E.J.. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008; 322:1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K.. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P.. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009; 139:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fineran P.C., Charpentier E.. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012; 434:202–209. [DOI] [PubMed] [Google Scholar]
- 19. Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R.. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015; 520:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M., Wang R., Zhao D., Xiang H.. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014; 42:2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M., Wang R., Xiang H.. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 2014; 42:7226–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richter C., Dy R.L., McKenzie R.E., Watson B.N., Taylor C., Chang J.T., McNeil M.B., Staals R.H., Fineran P.C.. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014; 42:8516–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savitskaya E., Semenova E., Dedkov V., Metlitskaya A., Severinov K.. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013; 10:716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swarts D.C., Mosterd C., van Passel M.W., Brouns S.J.. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012; 7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E.. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012; 3:945. [DOI] [PubMed] [Google Scholar]
- 26. Staals R.H.J., Jackson S.A., Biswas A., Brouns S.J.J., Brown C.M., Fineran P.C.. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat. Commun. 2016; 7:12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xue C., Seetharam A.S., Musharova O., Severinov K., Brouns S.J., Severin A.J., Sashital D.G.. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015; 43:10831–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., Wang Y.. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell. 2015; 163:840–853. [DOI] [PubMed] [Google Scholar]
- 29. Nuñez J.K., Harrington L.B., Kranzusch P.J., Engelman A.N., Doudna J.A.. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature. 2015; 527:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuznedelov K., Mekler V., Lemak S., Tokmina-Lukaszewska M., Datsenko K.A., Jain I., Savitskaya E., Mallon J., Shmakov S., Bothner B. et al. Altered stoichiometry Escherichia coli Cascade complexes with shortened CRISPR RNA spacers are capable of interference and primed adaptation. Nucleic Acids Res. 2016; 44:10849–10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grissa I., Vergnaud G., Pourcel C.. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007; 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu H., Han J., Liu X., Zhou J., Xiang H.. Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J. Genet. Genomics. 2011; 38:261–269. [DOI] [PubMed] [Google Scholar]
- 33. Wang R., Li M., Gong L., Hu S., Xiang H.. DNA motifs determining the accuracy of repeat duplication during CRISPR adaptation in Haloarcula hispanica. Nucleic Acids Res. 2016; 44:4266–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maier L.K., Stachler A.E., Saunders S.J., Backofen R., Marchfelder A.. An active immune defense with a minimal CRISPR (clustered regularly interspaced short palindromic repeats) RNA and without the Cas6 protein. J. Biol. Chem. 2015; 290:4192–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shmakov S., Savitskaya E., Semenova E., Logacheva M.D., Datsenko K.A., Severinov K.. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 2014; 2:5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vorontsova D., Datsenko K.A., Medvedeva S., Bondy-Denomy J., Savitskaya E.E., Pougach K., Logacheva M., Wiedenheft B., Davidson A.R., Severinov K. et al. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res. 2015; 43:10848–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strotskaya A., Savitskaya E., Metlitskaya A., Morozova N., Datsenko K.A., Semenova E., Severinov K.. The action of Escherichia coli CRISPR-Cas system on lytic bacteriophages with different lifestyles and development strategies. Nucleic Acids Res. 2017; 45:1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo M.L., Jackson R.N., Denny S.R., Tokmina-Lukaszewska M., Maksimchuk K.R., Lin W., Bothner B., Wiedenheft B., Beisel C.L.. The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucleic Acids Res. 2016; 44:7385–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gleditzsch D., Müller-Esparza H., Pausch P., Sharma K., Dwarakanath S., Urlaub H., Bange G., Randau L.. Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system. Nucleic Acids Res. 2016; 44:5872–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yosef I., Shitrit D., Goren M.G., Burstein D., Pupko T., Qimron U.. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14396–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.